Abstract
Background
The purpose of this study was to report the RNA sequencing profile according to the presence or absence of sarcopenia in elderly patients with osteoporotic hip fracture. Therefore, an important genetic factor candidate for sarcopenia causing hip fracture in elderly with osteoporosis has been identified.
Methods
The patient group involved subjects over 65 years who had undergone hip fracture surgery. Among 323 hip fracture (HF) patients identified from May 2017 to December 2019, 162 HF patients (90 non-sarcopenia and 72 sarcopenia groups), excluding subjects with high energy trauma and non-osteoporosis, were finally included in the analysis. For RNA sequencing, each patient with hand grip strength (HGS) values in the top 10% were enrolled in the control group and with the bottom 10% in the patient group. After excluding patients with poor tissue quality, 6 patients and 5 patients were selected for sarcopenia and non-sarcopenia groups, respectively. For qPCR validation, each patient with HGS values in the top 20% and bottom 20% was enrolled in the control and patient groups, respectively. After excluding patients with poor tissue quality, 12 patients and 12 patients were enrolled in the sarcopenia and non-sarcopenia groups, respectively. Sarcopenia was defined according to the Asia Working Group for Sarcopenia (AWGS) criteria for low muscle strength (hand grip strength below 18 kg in women and 28 kg in men) and low muscle mass (SMI below 5.4 kg/m2 in women and 7.0 kg/m2 in men). The libraries were prepared for 100 bp paired-end sequencing using TruSeq Stranded mRNA Sample Preparation Kit (Illumina, CA, USA). The gene expression counts were supplied to Deseq2 to extract possible gene sets as differentially expressed genes (DEG) that discriminate between sarcopenia and non-sarcopenia groups that were carefully assigned by clinical observation. For the classification of the candidate genes from DEG analysis, we used the public databases; gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG). Quantitative real-time PCR was performed for validation.
Results
Samples collected were subjected to RNAseq using the Illumina platform. A total of 11 samples from both sarcopenia and non-sarcopenia groups were sequenced. Fifteen genes (RUNX 1, NGFR, CH3L1, BCL3, PLA2G2A, MYBPH, TEP1, SEMA6B, CSPG4, ACSL5, SLC25A3, NDUFB5, CYC1, ACAT1, and TCAP) were identified as differentially expressed genes (DEG) in both the groups.
In the qPCR results, the expression levels of SLC25A3 and TCAP gene in the OS group were significantly lower than in the non-OS groups whereas an increase in RUNX1 mRNA level was observed in the OS samples (p < 0.05).
Conclusions
In summary, this study detected gene expression difference according to the presence or absence of sarcopenia in elderly osteoporosis female patients with hip fracture. We have also identified 15 important genes (RUNX 1, NGFR, CH3L1, BCL3, PLA2G2A, MYBPH, TEP1, SEMA6B, CSPG4, ACSL5, SLC25A3, NDUFB5, CYC1, ACAT1, TCAP), a few GO categories and biological pathways that may be associated with the osteosarcopenia. Our study may provide effective means for the prevention, diagnosis and treatment sarcopenia in elderly osteoporosis female patients.
The Translational potential of this article
These findings provide a novel insight into the effects of aging on the response in women with postmenopausal osteoporosis. Further studies are underway to identify the specific signalling pathways involved. These results reveal potential therapeutic targets that could aid the regenerative capacity of aging skeletal muscle.
Keywords: Differential gene expression, RNA sequencing, Hip fracture, Osteoporosis, Sarcopenia
1. Introduction
The world's population is expected to age rapidly in most of the regions and people are living a longer life. This situation is similar in Korea and the country became an aging society (elderly population ≥7% of the total population) in 2000 [1]. In 2018, Korea became an aged society (defined as an elderly population ≥14% of the total population), and by 2026 will be a super-aged society (elderly population ≥20% of the total population) [2].
In elderly populations, there exists a high prevalence of many chronic diseases including osteoporosis and sarcopenia [3]. Although osteoporosis-related studies involving the elderly population have reported clinical outcomes and established treatment guidelines in the last decades; in recent years, increased attention has been given to sarcopenia and related studies [4,5]. In addition, sarcopenia has been the International Classification of Disease, Tenth Revision, Clinical Modification (ICD-10-CM) as M62.84 since October 1, 2016 [6].
Sarcopenia has been mostly manifested in metabolic diseases, such as diabetes, obesity, cachexia, and some specific diseases, including chronic renal failure, congestive heart failure, and chronic obstructive pulmonary disease. However, sarcopenia is significantly associated with osteoporosis in the elderly population. Several studies have confirmed that sarcopenia and osteoporosis (osteosarcopenia) share common risk factors and biological pathways, and osteosarcopenia is associated with a significant physical disability, representing a significant threat to loss of independence in later life [2,7]. The combination of these two diseases exacerbates negative health outcomes and has been described as a “hazardous duet” adding the propensity of falling from sarcopenia to vulnerability of bones in those with osteoporosis. The combined effect of sarcopenia and osteoporosis represents a serious problem in the elderly [8].
Previously, a biomarker study on sarcopenia in patients with hip fractures and a few genomic studies involving sarcopenia according to aging were carried out.
However, the purpose of this study was to report the RNA sequencing profile based on the presence or absence of sarcopenia in elderly osteoporotic female patients with hip fractures. Consequently, an important genetic factor candidate for sarcopenia causing a hip fracture in elderly women with osteoporosis has been identified.
2. Methods
2.1. Study subject
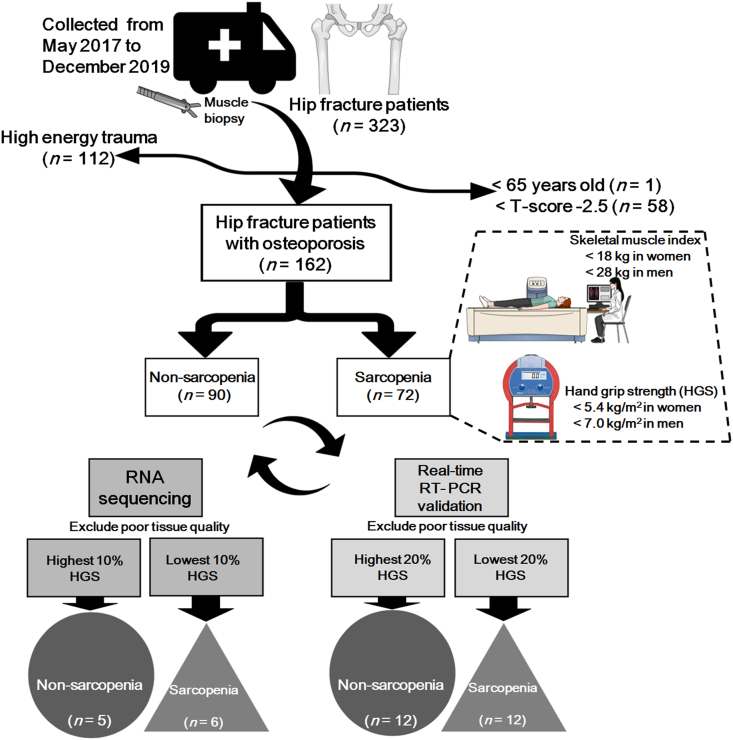
The patient group involved subjects over 65 years who had undergone hip fracture surgery. Among 323 hip fracture (HF) patients identified from May 2017 to December 2019, 162 HF patients (90 non-sarcopenia and 72 sarcopenia groups), excluding subjects with high energy trauma and non-osteoporosis, were finally included in the analysis. For RNA sequencing, each patient with hand grip strength (HGS) values in the top 10% were enrolled in the control group and with the bottom 10% in the patient group. After excluding patients with poor tissue quality, 6 patients and 5 patients were selected for sarcopenia and non-sarcopenia groups, respectively. For qPCR validation, each patient with HGS values in the top 20% and bottom 20% was enrolled in the control and patient groups, respectively. After excluding patients with poor tissue quality, 12 patients and 12 patients were enrolled in the sarcopenia and non-sarcopenia groups, respectively (Fig. 1).
Figure 1.
Flow-chart of patient selection.
The study protocol was approved by the Institutional Review Board (IRB) of Gyeongsang National University Hospital (IRB number: 2019-02-013).
2.2. Muscle samples preparation
Muscle samples were collected by 5 × 5 × 3 mm gluteus maximus obtained from the surgical opening. Tissues were stored at −80 °C in Tissue-Tek (Miles, Elkhart, IN, USA).
2.3. Diagnosis for osteosarcopenia
Body composition was measured by whole-body DXA (DPX-NT; GE Medical Systems Lunar, Madison, WI, USA). Bone mineral content, fat mass, and lean soft tissue mass were measured separately for each part of the body, including the arms and legs. The lean soft tissue masses of the arms and legs were almost equal to the skeletal muscle mass. As absolute muscle mass correlates with height, the skeletal muscle mass index (SMI) was calculated by the following formula: lean mass [kg]/height2 [m2], which is directly analogous to body mass index (BMI: weight [kg]/height2 [m2]). Arm SMI was defined as (arm lean mass [kg]/height2 [m2]). Leg SMI was defined as (leg lean mass [kg]/height2 [m2]). Appendicular SMI was defined as the sum of the arm and leg SMIs.
Muscle strength was assessed by handgrip strength using an analogue dynamometer (TK 5001 Grip-A, Takei, Tokyo, Japan). In the sitting position, the elbow was flexed at 90° with the shoulder attached to the torso, with the wrist maintaining a neutral posture (0°) and the grip with maximal strength was measured.
Sarcopenia was defined according to the Asia Working Group for Sarcopenia (AWGS) criteria for low muscle strength (hand grip strength below 18 kg in women and 28 kg in men) and low muscle mass (SMI below 5.4 kg/m2 in women and 7.0 kg/m2 in men) (16).
Bone mineral content (BMC) and bone mineral density (BMD) at the total femur, femoral neck, and lumbar spine (L1-L4) sites were measured by trained technicians using dual-energy X-ray absorptiometry (Lunar Prodigy, GE Healthcare, Madison, WI, USA). Osteoporosis was diagnosed with supplying by the DXA manufacturer (from Japanese population) using T-score criteria of the World Health Organization: Normal, T-score of the total femur, femoral neck, and lumbar spine at either site ≥ −1; Osteopenia, −2.5 < the lowest T-score < −1; Osteoporosis, the lowest T-score ≤ −2.5.
Osteoporosis was defined as a BMD 2.5 standard deviation (SD) below the peak bone mass of a young, healthy, gender-, and race-matched reference population according to the World Health Organization (WHO) diagnostic classification. The relation between BMD (T-score) and SMI was used for the classification of osteosarcopenia (T-score ≤ −2.5 and low SMI) and normal (high T-score and high SMI). Nutritional status was assessed before surgery with the complete Mini nutritional assessment (MNA). The screening part consists of questions about food intake, weight loss, mobility, stress, neuropsychological problems and body mass index (BMI). The assessment part consists of more detailed questions about food intake, the self-view of nutritional and health status and measurements of mid-arm and calf circumference. Malnourished patients were defined as having a MNA score less than 17 points.
2.4. Experimental method
The libraries were prepared for 100 bp paired-end sequencing using TruSeq Stranded mRNA Sample Preparation Kit (Illumina, CA, USA). Namely, mRNA molecules were purified and fragmented from 1 μg of total RNA using oligo (dT) magnetic beads. The fragmented mRNAs were synthesized as single-stranded cDNAs through random hexamer priming. By applying this as a template for second-strand synthesis, double-stranded cDNA was prepared. After the sequential process of end repair, A-tailing and adapter ligation, cDNA libraries were amplified with PCR (Polymerase Chain Reaction). The quality of the cDNA libraries was evaluated with the Agilent 2100 BioAnalyzer (Agilent, CA, USA). The libraries were quantified with the KAPA library quantification kit (Kapa Biosystems, MA, USA) according to the manufacturer's library quantification protocol. Following cluster amplification of denatured templates, sequencing was progressed as paired-end (2 × 100bp) using Illumina HiSeq platform sequencer (Illumina, CA, USA).
2.5. Transcriptome data analysis
Low-quality reads were identified according to the following criteria; reads include more than 10 percent of missed bases, reads include more than 40 percent of bases whose quality scores are less than 20 and reads from which average quality scores of each read are less than 20. The entire filtering process was done using the in-house scripts. The filtered short reads were mapped to the human reference coding sequence sequences (CDS), version GRCh38, using software Kallisto [9]. The gene expression counts were supplied to Deseq2 to extract possible gene sets as differentially expressed genes (DEG) that discriminate between sarcopenia and non-sarcopenia groups that were carefully assigned by clinical observation [10]. Based on the fold-changes and adjusted p-values, the candidate genes were listed.
2.6. Gene classification analysis
For the classification of the candidate genes from DEG analysis, we used the public databases; gene ontology (GO) [11] and Kyoto Encyclopedia of Genes and Genomes (KEGG) [12]. The GO annotations were downloaded from the Ensembl databases; Human genes (GRCh38. p13) of Ensembl Genes 100 [13]. GO enrichment analysis was implemented by fisher's exact test and the resulting p-values were corrected by fdr-bh of python module state models [14].
2.7. Quantitative real-time RT-PCR (qPCR)
As a template for PCR amplification, 1.3 μL of previously synthesized cDNA was used for RNA-sequencing. The cDNA was mixed with forward and reverse primers for RUNX1, SLC25A3, BCL3, CHI3L1, MYBPH, ACSL5, NDUFB5, NGFR, PLAG2A, CSPG4, SEMA6B, TEP1, AC068506.1, THBS1, ACAT1, CYC1, TCAP, and HPN in a 384-well plate (MicroAmp® 384-well Reaction Plate with Barcode, Applied Biosystems, Foster City, CA, USA) containing Power SYBR Green PCR Master Mix (Applied Biosystems, Warrington, UK). Each sample was measured in triplicate. To normalize the mRNA levels among the samples, GAPDH was amplified by real-time RT-PCR (qPCR) as a housekeeping gene. The analysis was performed using the ViiA™ 7 real-time PCR system (Applied Biosystems, USA). The sequences of the primers used in the experiments are shown in Table 1. The cycle threshold values measured after the experiment were analyzed by the relative quantification 2−ΔΔC(T) method.
Table 1.
Primer sequences of target genes for quantitative real-time RT- PCR analysis.
| Gene | Forward primer (5′ → 3′) | Reverse primer (5′ → 3′) |
|---|---|---|
| GAPDH | AGCCACATCGCTCAGACAC | GCCCAATACGACCAAATCC |
| RUNX1 | TCTTCACAAACCCACCGCAA | CTGCCGATGTCTTCGAGGTTC |
| SLC25A3 | TATCTCTGGCGCACATCACTA | CTTAGCAGCTTCCATAGGAGC |
| BCL3 | AACCTGCCTACACCCCTATAC | CACCACAGCAATATGGAGAGG |
| CHI3L1 | GAAGACTCTCTTGTCTGTCGGA | AATGGCGGTACTGACTTGATG |
| MYBPH | CCAGGGGAAGCCTAAGCCT | CGAGCGAATGAAGAGGATGGA |
| ACSL5 | TGGCTATCTTACAAACAGGTGTC | TCCACTCTGGCCTATTCTGAG |
| NDUFB5 | AGCTGGAAGTGCGAAAATTGA | ATACCAGGGTCCATCTCCTCT |
| NGFR | CCTACGGCTACTACCAGGATG | CACACGGTGTTCTGCTTGT |
| PLA2G2A | CTGTTGCTACAAACGTCTGGA | GCTCCCCGAGTTGCTAAACTT |
| CSPG4 | GCCACGTTGTCAGTCGATG | CCCATAGGGGACCTCTAGGG |
| SEMA6B | AAACGTGTGTCGGATGAAGGG | GGGTTGAAGGCGTTGGAAC |
| TEP1 | TGGGTCGTTGGTTTGATTCAG | GCTTCACGGCCAGATCCTC |
| AC068506.1 | CACGCACAGATGGAAGAAAT | CACCCAATCTTCCATTGCAT |
| THBS1 | TGCTATCACAACGGAGTTCAGT | GCAGGACACCTTTTTGCAGATG |
| ACAT1 | ATGCCAGTACACTGAATGATGG | GATGCAGCATATACAGGAGCAA |
| CYC1 | CTTCGCGGGGTAGTGTTGG | GGCCAGACTTCGACGACAA |
| TCAP | GTCGGAGGAGAACTGTGAGC | ATGCCCATCCGCATCATCAG |
| HPN | GGGCCATTGTGGCTGTTCT | CGTCCCTTCCGTCTTGTCAAA |
3. Results
3.1. Comparison of parameters based on the presence of osteosarcopenia
Age (P = 0.638), grip strength (P = 0.168), vitamin D (P = 0.635), parathyroid hormone (P = 0.754), and femur neck BMD (P = 0.626) were not significantly different between the two groups. The demographic data of the patients are shown in Table 2.
Table 2.
Comparison of parameters according to presence of sarcopenia.
| Parameters | Non-sarcopenia (n = 5) | Sarcopenia (n = 6) | p-value |
|---|---|---|---|
| Age (years) | 79.89 ± 7.16 | 79.82 ± 9.30 | 0.638 |
| BMI (kg/m2) | 24.89 ± 7.16 | 21.83 ± 9.30 | 0.063 |
| Grip strength (kg) | 12.03 ± 5.00 | 9.58 ± 3.62 | 0.168 |
| SMI (kg) | 6.40 ± 0.65 | 4.83 ± 0.35 | <0.001 |
| Vitamin D (ng/ml) | 11.30 ± 9.23 | 22.29 ± 9.10 | 0.635 |
| PTH (ng/ml) | 71.91 ± 38.92 | 84.24 ± 164.31 | 0.754 |
| Smoking status | 0 | 0 | |
| ∗Physical activity (≥moderate) | 1 | 1 | 0.887 |
| MNA score <17 | 3 | 4 | 0.819 |
BMI; body mass index, SMI; skeletal muscle mass, PTH; Parathyroid hormone, MNA; mini nutritional assessment, ∗ Based on the IPAQ Scoring Protocol: moderate intensity activity ≥ 5 days per week
3.2. Gene associated with the diagnosis of osteosarcopenia
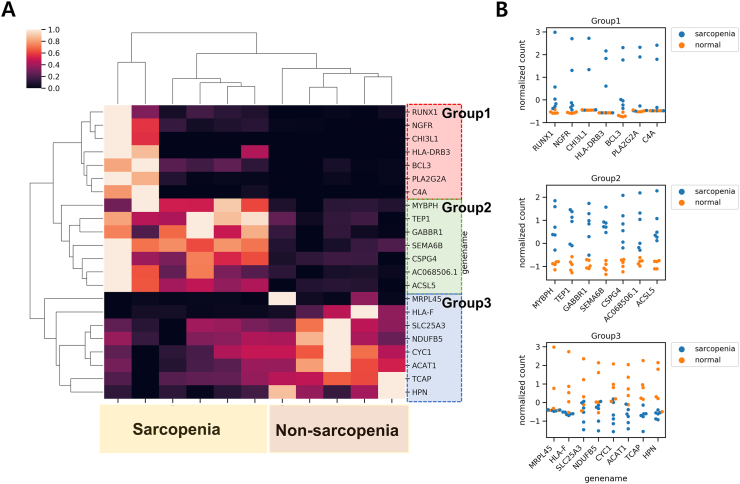
Samples collected were subjected to RNAseq using the Illumina platform. A total of 11 samples from both sarcopenia and non-sarcopenia groups were sequenced (Table S1). Based on the number of produced reads and the mapping rate, we determined the RNAseq results were reliable for the downstream analysis (Fig. S2). The average amount of each sample reads was 6.4 Gbases. More than 97 percent of the sequences were over Q30 base quality scores. The sequences were quantified into gene expressions using the Kallisto software [9]. Fifteen genes (RUNX 1, NGFR, CH3L1, BCL3, PLA2G2A, MYBPH, TEP1, SEMA6B, CSPG4, ACSL5, SLC25A3, NDUFB5, CYC1, ACAT1, and TCAP) were identified as differentially expressed genes (DEG) in both the groups with statistical significance (adjusted p-value < 0.01) by software Deseq2 (Fig. 2A).
Figure 2.
Genes associated to sarcopenia diagnosis (A) DEG heatmap clearly separating sarcopenia from normal patients (B) Swarm plot of known genes in each expression group.
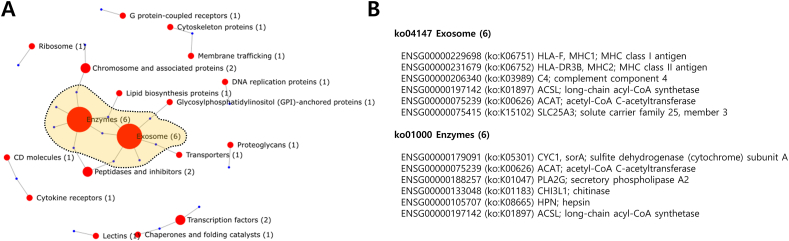
Notably, the expression patterns of the DEGs were classified into three groups. The genes in Group 1 showed elevated expression in the sarcopenia patients, especially the first two cases while the non-sarcopenia patients exhibited low expression levels. The genes in Group 2 showed elevated expression in all the sarcopenia patients compared to the low expression in non-sarcopenia patients. On the other hand, the genes in Group 3 showed elevated expression in non-sarcopenia patients. For the functional annotation, the DEGs were classified by the Kegg Brite database (Fig. 3A) [12].
Figure 3.
Kegg Brite classification of DEGs (A) Network-based Kegg Brite classification; red circles represent Kegg Brite classes, the size of red circles shows the number of corresponding genes, and blue circles represent genes corresponding the classes (B) The gene members in “Exosome” and “Enzyme” classes that are mostly found from the DEGs. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
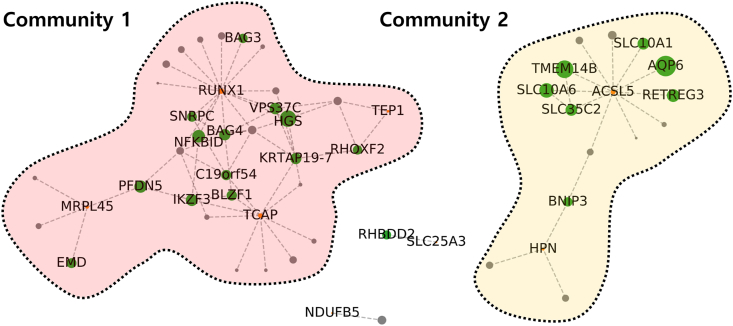
Sarcopenia and osteoporosis have commonly reported pathophysiological characteristics like fat penetration into the muscle and bone [15]. Supporting this knowledge, typically representing classes were “Exosome” and “Enzyme” and the genes commonly classified in both the classes were ACSL; long-chain acyl-CoA synthetase and ACAT; acetyl-CoA C-acetyltransferase. ACSL5 showed clear upregulation while ACAT1 showed downregulation in the sarcopenia patients (Fig. 2B). Especially, ACSL5 has hub genes as interacting partners based on human reference interactome (HuRI) (Fig. 4 and Table S3) [16]. The hub interactors of ACSL5 provide insights into the link between the perturbations in ACSL5 and sarcopenia. The interacting hub genes; solute carrier (SLC) families, transmembrane protein 14 B (TMEM14B), and aquaporin 6 (AQP6) implies that the transport-related protein activities can be related to sarcopenia. Notably, another SLC family SLC25A3 showed significant down-regulation in sarcopenia patients (Fig. 2B).
Figure 4.
One step neighbors of DEGs based on HuRI network. The red nodes represent the DEGs, the green nodes are the known network hubs of one step neighbors of DEGs, and the gray nodes are non-hub interactors of DEGs. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Meanwhile, previous researches showed changes in the muscle and bone structure in response to high levels of fat and suggested an increase in the levels of apoptosis and autophagy [17]. Consistent with the results, HLA-F and HLA-DR3B showed differential expression and were mapped to the pathway, “Phagosome” (ko04145) in Kegg Pathway (Fig. 2B). Moreover, the DEGs; HPN, RUNX1, MRPL45, and TCAP revealed classification of the hub interactors under the GO term “macroautophagy” (GO:0016,236), “regulation of macroautophagy” (GO:0016,241), and “chaperone-mediated autophagy” (GO:0061,684) (Table S3) thereby suggesting that the patients with sarcopenia may have different autophagy regulations.
3.3. qPCR validation results
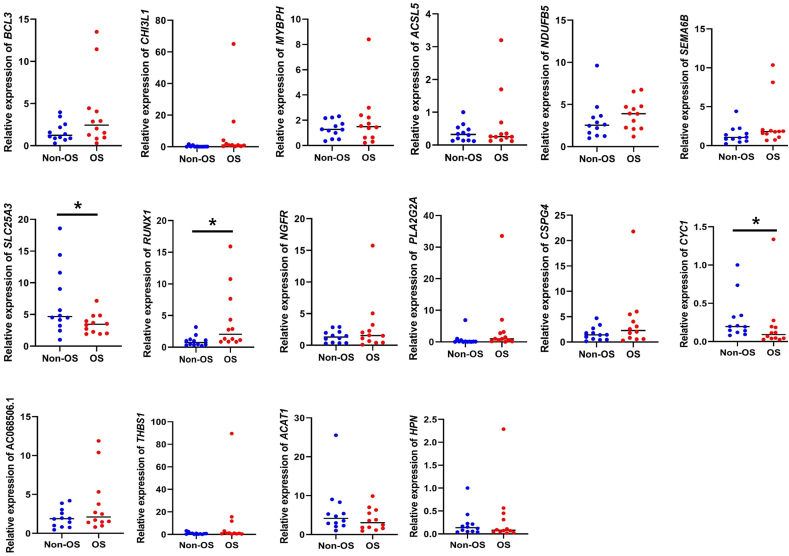
In the qPCR results, no significant differences were observed in BCL3, CHI3L1, MYBPH, ACSL5, NDUFB5, NGFR, PLA2G2A, CSPG4, SEMA6B, TEP1, AC068506.1, THBS1, ACAT1, CYC1, and HPN genes expression level between the non-OS and OS groups (Fig. 5). The expression levels of SLC25A3 and TCAP gene in the OS group were significantly lower than in the non-OS groups whereas an increase in RUNX1 mRNA level was observed in the OS samples (p < 0.05).
Figure 5.
qPCR validation results.
4. Discussion
The main finding of this study was the identification of 15 genes (RUNX 1, NGFR, CH3L1, BCL3, PLA2G2A, MYBPH, TEP1, SEMA6B, CSPG4, ACSL5, SLC25A3, NDUFB5, CYC1, ACAT1, and TCAP) with large differences in mRNA expression in skeletal muscle samples from patients with osteosarcopenia. In addition, phagosome and endocytosis-related pathways showed significant findings in the enrichment analysis. Previous studies related to muscles and Top 15 genes that demonstrated a large difference in expression between the two groups are mentioned below:
4.1. Muscle differentiation capacity and inflammation-related gene
With regards to ChIP-seq, ATAC-seq, and histone H3K4me1/H3K27ac modification analyses related to RUNX 1, the association between Runx1-dependent gene expression and markers related to muscle differentiation such as MyoD and c-Jun was reported [18].
In RNA sequencing studies associated with the NGFR (Nerve Growth Factor Receptor) gene, it was demonstrated that the cell surface receptors ERBB3 and NGFR demarcate myogenic populations, including PAX7 progenitors in human fetal development and hPSC-SMPCs [19].
In addition, studies have shown that the CH3L1 gene acts as a myokine that stimulates human myocyte proliferation during exercise. The reports suggested that CHI3L1 is induced by acute exercise and that CHI3L1/PAR-2 signaling activates myocyte proliferation, which is important for the restructuring of skeletal muscle in the response to exercise training [20].
The BLC3 gene, which has been well-studied in many inflammatory pathways, was also one of the genes demonstrating large differences in expression. In the BCL-3 KO study, inhibition of fiber atrophy and abolition of NF-kappaB reporter activity was observed, and it was proven that it was possible to suppress muscle progression and sarcopenia progression due to chronic inflammatory inflammation [21].
Finally, the difference in the expression level of PLA2G2A was also identified as a distinctive feature. PLA2G2A gene has many known links in endocrine diseases such as obesity or thyroid. Studies on the association between PLA2G2A and T2DM have been conducted, one of the most common causes of sarcopenia, and an increase in polymorphism has been reported in T2DM patients [22]. However, further research on this mechanism is necessary.
4.2. Skeletal muscle fiber, axon, and adipose tissue-related gene
Myosin Binding Protein and Myosin heavy chain genes play an important role in the formation of skeletal muscle. In human muscle, MyHC isoforms such as types I, IIa, and IIx/d are expressed, and mRNA expression has been reported to change with the progression in the aging process [23]. However, the role of MYHC protein in sarcopenia has yet not been clarified.
In an experimental study evaluating the myosin heavy chain plasticity of aging skeletal muscle after aerobic exercise, a decrease in the MHC IIa and IIx at mRNA and protein levels and an increase in MHC I protein levels after training were reported followed by an increase in myosin heavy chain plasticity. The study suggested that a shift towards an oxidative MHC phenotype may be beneficial for metabolic and functional health in older individuals [24]. Telomeres protect chromosome ends and shorten with age in most of the tissues.
Although none of the studies have directly analyzed the TEP1 gene, many studies related to aging of telomere have been reported. Among these studies, one study demonstrated that the C2C12 root canal pretreated with the p38 inhibitor (SB-202190) prevented A23187-induced reduction in Trf1 mRNA expression, indicating a link between Trf1 gene expression and p38 MAPK activation [25].
Semaphorin 6 B is a member of the semaphorin family of signal proteins, which have been implicated in various biological processes such as axon guidance and muscle regeneration. In one study, the authors investigated the effect of fenofibrate as a PPARα activator on the expression of the Sema6B gene in rat skeletal muscle. They conclude that the expression of Sema6B is down-regulated by fenofibrate in rat skeletal muscle [26].
Adipose and fibrous tissues in the muscle are observed in some skeletal muscle pathologies such as Duchenne muscular dystrophy and sarcopenia affects muscle strength and muscle development. In one study based on RNA interference analysis, it was found that CSPG4 is involved in the pro-adipogenic effect of bFGF, TGF-β-induced alpha-smooth muscle actin expression, and stress fiber formation. It was suggested that this finding would facilitate the development of effective therapies for skeletal muscle pathology, by establishing additional markers for MPC detection and characterizing its role in fibrotic/adipose production differentiation [27].
A study on overexpression of Long-Chain Acyl-CoA Synthetase 5 (ACSL-5) revealed an increase in fatty acid oxidation and free radical formation along with attenuation of insulin signaling in primary human skeletal myotubes. The study concluded that ACSL-5 increases mitochondrial fatty acid oxidation in human skeletal muscle; however, it is hypothesized that it may be associated with increased free radical production and reduced insulin signaling [28].
4.3. Mitochondrial energy metabolism-related gene
A study was conducted on the loss of mitochondrial phosphate transporter SLC25A3 in skeletal muscle and its association with mitochondria. The observations suggest that the loss of PiC encoded by acute SLC25A3 can be compensated by an alternative route of Pi transport from the skeletal muscle to the mitochondria. However, a decrease in ATP synthesis and matrix Ca2 + buffering capacity are likely to contribute to initial fatigue. Therefore, further investigations on the SLC25A3 gene involved in mitochondrial energy metabolism are necessitated [29].
The maximum oxygen intake and the amount of Type 1 fiber are interrelated, but the underlying molecular mechanisms are not well understood. To elucidate the mechanisms, the gene expression profile in skeletal muscle biopsy samples was evaluated. Eleven genes (NDUFB4, COX5A, UQCRB, ATP5C1, ATP5G3, ETHE1, FABP3, ISCA1, MYST4, C9orf3, and PKIA) were found to be positively correlated with both Vo(2max) and the percentage of type 1 fibers. Vo(2max) closely reflects the expression of OXPHOS genes, particularly the NDUFB5 and ATP5C1, in skeletal muscle, thereby suggesting good muscle fitness [30]. Another study reported the association between the CYC1 gene and apoptosis in skeletal muscle [31].
In addition, Acyl-CoA metabolism in skeletal muscle regulates systemic glucose homeostasis and is very important in maintaining skeletal muscle strength. The impaired capacity of skeletal muscle to switch between the oxidation of fatty acid (FA) and glucose is linked to disordered metabolic homeostasis. To understand how muscle FA oxidation affects systemic glucose, a study has been conducted on mice with skeletal muscle-specific long-chain acyl-CoA synthase (ACSL). It was reported that acyl -CoA synthesized by other ACSL isoforms was not available for β-oxidation, as the content of muscle acyl -CoA increased significantly due to exhaustion after exercise. Further, it was suggested that compartmentalization of acyl-CoA would seriously impair excessive glucose demand and systemic glucose homeostasis [32].
Although there is no direct association with mitochondrial metabolism, TCAP belonged to Group 2, and a study reported that TCAP knockdown caused muscle atrophy [33].
4.4. Elucidation of the biologic process by enrichment analysis
In the enrichment analysis, biological processes like phagocytosis and endosome were demonstrated to be highly related to osteosarcopenia in elderly patients.
Although an imbalance between proteolysis and synthesis is known as an important mechanism of sarcopenia, it is emphasized that the mechanism of degradation is more important for the elderly. Research on the ubiquitin pathway, calcium calpain pathway, and autophagy has been actively conducted to understand the mechanism of proteolysis. In particular, the muscles of the hip fracture patients who had already undergone osteoporosis were analyzed in the samples. It was found that autophagy-related phagocytosis and endocytosis were significantly involved in RNA sequencing thereby leading to sarcopenia. There have been numerous studies on phagocytosis in skeletal muscle and the mechanism involving impairment of IL15 signaling followed by inhibition of phagocytosis and promotion of skeletal muscle breakdown is most often explained [[34], [35], [36]]. In addition, studies on the relationship between endosome and Ecm29 have been reported [37,38].
4.5. Biologic process of phagosome related gene
KEGG pathway analysis revealed that F-actin [[39], [40], [41]], TUBB (beta-tubulin) [42], MHCI, MHCII [43], and TSP (Thrombospondin) [44] were affected in phagosomes (Fig. 3).
There are several limitations to this study. First, the number of samples included in RNA sequencing was small. For the classification of patients and controls, comparisons between the two groups were performed through clear phenotype differences and accurate labeling. In the future, further studies will be carried out by involving a large sample size. Second, all parameters related to bone and muscle interaction may not have been adjusted. Third, it is difficult to rule out the possibility that the results of the study are not due to the disease itself but due to individual differences. However, to exclude individual differences, demographic factors such as age, sex, and test results were matched, and PCR verification was conducted.
So far, many genomic studies related to sarcopenia have been conducted. Nevertheless, the strength of this study relies on classification based on the presence or absence of sarcopenia in elderly patients with osteoporosis. In other words, an analysis of the mechanism for the development of sarcopenia was performed in elderly patients who had already undergone osteoporosis. In addition, when the two groups were divided, the upper and lower 10% extremes were selected and labeling was performed to avoid the patients in the gray zone. It can be considered that the candidate gene for identifying the risk factor or genetic factor that progresses to sarcopenia in the elderly with osteoporotic hip fracture has already progressed. In the future, the results of this study are expected to be used to develop treatments to prevent re-fracture in osteoporotic hip fracture patients and prevent fractures caused by falls in patients with osteosarcopenia.
5. Conclusions
In summary, this study detected gene expression differences according to the presence or absence of sarcopenia in elderly osteoporosis female patients with hip fractures. We have also identified 15 important genes (RUNX 1, NGFR, CH3L1, BCL3, PLA2G2A, MYBPH, TEP1, SEMA6B, CSPG4, ACSL5, SLC25A3, NDUFB5, CYC1, ACAT1, and TCAP), a few GO categories, and biological pathways that may be associated with osteosarcopenia. It is hypothesized that the results of our study may provide effective means for the prevention, diagnosis, and treatment of sarcopenia in elderly osteoporotic female patients.
Declaration of competing interest
None declared by all authors.
Acknowledgements
This work was funded by the by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MEST) (No. NRF-2019R1F1A1059208). The National Research Foundation of Korea had no role in design or analysis of the project, or the writing of this article. And the authors thank all patients and staff who made this study possible.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jot.2021.04.009.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.United Nations, Department of Economic and Social Affairs, Population Division . 2019. World population ageing; p. 2020. highlights. [Google Scholar]
- 2.Yoo J.-I., Ha Y.-C. Review of epidemiology, diagnosis, and treatment of osteosarcopenia in Korea. J Bone Metab. 2018;25:1–7. doi: 10.11005/jbm.2018.25.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greco E.A., Pietschmann P., Migliaccio S. Osteoporosis and sarcopenia increase frailty syndrome in the elderly. Front Endocrinol. 2019;10 doi: 10.3389/fendo.2019.00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lo J.H., U K.P., Yiu T., Ong M.T., Lee W.Y. Sarcopenia: current treatments and new regenerative therapeutic approaches. J Orthop Transl. 2020;23:38–52. doi: 10.1016/j.jot.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang J., Leung K.-S., Chow S.K.-H., Cheung W.-H. Inflammation and age-associated skeletal muscle deterioration (sarcopaenia) J Orthop Transl. 2017;10:94–101. doi: 10.1016/j.jot.2017.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao L., Morley J.E. Sarcopenia is recognized as an independent condition by an international classification of disease, Tenth revision, clinical modification (ICD-10-CM) code. J Am Med Dir Assoc. 2016;17:675–677. doi: 10.1016/j.jamda.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 7.Kirk B., Miller S., Zanker J., Duque G. A clinical guide to the pathophysiology, diagnosis and treatment of osteosarcopenia. Maturitas. 2020;140:27–33. doi: 10.1016/j.maturitas.2020.05.012. [DOI] [PubMed] [Google Scholar]
- 8.Crepaldi G., Maggi S. Sarcopenia and osteoporosis: a hazardous duet. J Endocrinol Invest. 2005;28:66–68. [PubMed] [Google Scholar]
- 9.Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 10.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hubbard T., Barker D., Birney E., Cameron G., Chen Y., Clark L. The Ensembl genome database project. Nucleic Acids Res. 2002;30:38–41. doi: 10.1093/nar/30.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seabold S., Perktold J. 2010. Statsmodels: econometric and statistical modeling with Python. Austin, Texas. 92–6. [DOI] [Google Scholar]
- 15.Duque G., editor. Osteosarcopenia: bone, muscle and fat interactions. Springer International Publishing; 2019. [DOI] [Google Scholar]
- 16.Luck K., Kim D.K., Lambourne L., Spirohn K., Begg B.E., Bian W. A reference map of the human binary protein interactome. Nature. 2020;580:402–408. doi: 10.1038/s41586-020-2188-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.L S., S T., D M., G D. Good, bad, or ugly: the biological roles of bone marrow fat. Curr Osteoporos Rep. 2018;16:130–137. doi: 10.1007/s11914-018-0427-y. [DOI] [PubMed] [Google Scholar]
- 18.Umansky K.B., Gruenbaum-Cohen Y., Tsoory M., Feldmesser E., Goldenberg D., Brenner O. Runx1 transcription factor is required for myoblasts proliferation during muscle regeneration. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hicks M.R., Hiserodt J., Paras K., Fujiwara W., Eskin A., Jan M. ERBB3 and NGFR mark a distinct skeletal muscle progenitor cell in human development and hPSCs. Nat Cell Biol. 2018;20:46–57. doi: 10.1038/s41556-017-0010-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Görgens S.W., Hjorth M., Eckardt K., Wichert S., Norheim F., Holen T. The exercise-regulated myokine chitinase-3-like protein 1 stimulates human myocyte proliferation. Acta Physiol. 2016;216:330–345. doi: 10.1111/apha.12579. [DOI] [PubMed] [Google Scholar]
- 21.Hunter R.B., Kandarian S.C. Disruption of either the Nfkb1 or the Bcl3 gene inhibits skeletal muscle atrophy. J Clin Invest. 2004;114:1504–1511. doi: 10.1172/JCI21696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khajeniazi S., Marjani A., Shakeri R., Hakimi S. Polymorphism of secretary PLA2G2A gene associated with its serum level in Type2 diabetes mellitus patients in Northern Iran. Endocr Metab Immune Disord - Drug Targets. 2019;19:1192–1197. doi: 10.2174/1871530319666190528111225. [DOI] [PubMed] [Google Scholar]
- 23.Marx J.O., Kraemer W.J., Nindl B.C., Larsson L. Effects of aging on human skeletal muscle myosin heavy-chain mRNA content and protein isoform expression. J Gerontol Ser A. 2002;57:B232–B238. doi: 10.1093/gerona/57.6.B232. [DOI] [PubMed] [Google Scholar]
- 24.Konopka A.R., Trappe T.A., Jemiolo B., Trappe S.W., Harber M.P. Myosin heavy chain plasticity in aging skeletal muscle with aerobic exercise training. J Gerontol A Biol Sci Med Sci. 2011;66:835–841. doi: 10.1093/gerona/glr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ludlow A.T., Lima L.C.J., Wang J., Hanson E.D., Guth L.M., Spangenburg E.E. Exercise alters mRNA expression of telomere-repeat binding factor 1 in skeletal muscle via p38 MAPK. J Appl Physiol Bethesda Md. 1985;113:1737–1746. doi: 10.1152/japplphysiol.00200.2012. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murad H., Kasies F., Azroony R., Alya G., Madania A. Effects of fenofibrate on Semaphorin 6B gene expression in rat skeletal muscle. Mol Med Rep. 2011;4:575–580. doi: 10.3892/mmr.2011.463. [DOI] [PubMed] [Google Scholar]
- 27.Takeuchi S., Nakano S.-I., Nakamura K., Ozoe A., Chien P., Yoshihara H. Roles of chondroitin sulfate proteoglycan 4 in fibrogenic/adipogenic differentiation in skeletal muscle tissues. Exp Cell Res. 2016;347:367–377. doi: 10.1016/j.yexcr.2016.08.023. [DOI] [PubMed] [Google Scholar]
- 28.Kwak H.-B., Woodlief T.L., Green T.D., Cox J.H., Hickner R.C., Neufer P.D. Overexpression of long-chain acyl-CoA synthetase 5 increases fatty acid oxidation and free radical formation while attenuating insulin signaling in primary human skeletal myotubes. Int J Environ Res Publ Health. 2019;16 doi: 10.3390/ijerph16071157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seifert E.L., Anderson-Pullinger L., Debattisti V., Sharpadskaya Y., Hajnoczky G. Loss of mitochondrial phosphate carrier SLC25A3 in skeletal muscle provides evidence for compensatory but lower capacity phosphate uptake into mitochondria. Biochim Biophys Acta BBA - Bioenerg. 2018;1859:e98. doi: 10.1016/j.bbabio.2018.09.290. [DOI] [Google Scholar]
- 30.Parikh H., Nilsson E., Ling C., Poulsen P., Almgren P., Nittby H. Molecular correlates for maximal oxygen uptake and type 1 fibers. Am J Physiol Endocrinol Metab. 2008;294:E1152–E1159. doi: 10.1152/ajpendo.90255.2008. [DOI] [PubMed] [Google Scholar]
- 31.Gesing A., Masternak M.M., Wang F., Lewinski A., Karbownik-Lewinska M., Bartke A. Decreased expression level of apoptosis-related genes and/or proteins in skeletal muscles, but not in hearts, of growth hormone receptor knockout mice. Exp Biol Med Maywood NJ. 2011;236:156–168. doi: 10.1258/ebm.2010.010202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li L.O., Grevengoed T.J., Paul D.S., Ilkayeva O., Koves T.R., Pascual F. Compartmentalized acyl-CoA metabolism in skeletal muscle regulates systemic glucose homeostasis. Diabetes. 2015;64:23–35. doi: 10.2337/db13-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Markert CD, Ning J, Staley JT, Heinzke L, Childers CK, Ferreira JA, et al. TCAP knockdown by RNA interference inhibits myoblast differentiation in cultured skeletal muscle cells. Neuromuscul Disord n.d.;18:413–422. [DOI] [PubMed]
- 34.Duggal N.A., Pollock R.D., Lazarus N.R., Harridge S., Lord J.M. Major features of immunesenescence, including reduced thymic output, are ameliorated by high levels of physical activity in adulthood. Aging Cell. 2018;17 doi: 10.1111/acel.12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quinn L.S., Anderson B.G., Strait-Bodey L., Wolden-Hanson T. Serum and muscle interleukin-15 levels decrease in aging mice: correlation with declines in soluble interleukin-15 receptor alpha expression. Exp Gerontol. 2010;45:106–112. doi: 10.1016/j.exger.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Girard D., Paquet M.E., Paquin R., Beaulieu A.D. Differential effects of interleukin-15 (IL-15) and IL-2 on human neutrophils: modulation of phagocytosis, cytoskeleton rearrangement, gene expression, and apoptosis by IL-15. Blood. 1996;88:3176–3184. [PubMed] [Google Scholar]
- 37.Gorbea C., Goellner G.M., Teter K., Holmes R.K., Rechsteiner M. Characterization of mammalian Ecm29, a 26 S proteasome-associated protein that localizes to the nucleus and membrane vesicles. J Biol Chem. 2004;279:54849–54861. doi: 10.1074/jbc.M410444200. [DOI] [PubMed] [Google Scholar]
- 38.Gorbea C., Pratt G., Ustrell V., Bell R., Sahasrabudhe S., Hughes R.E. A protein interaction network for Ecm29 links the 26 S proteasome to molecular motors and endosomal components. J Biol Chem. 2010;285:31616–31633. doi: 10.1074/jbc.M110.154120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walker S.M., Schrodt G.R. I segment lengths and thin filament periods in skeletal muscle fibers of the Rhesus monkey and the human. Anat Rec. 1974;178:63–81. doi: 10.1002/ar.1091780107. [DOI] [PubMed] [Google Scholar]
- 40.Gunning P., Ponte P., Blau H., Kedes L. alpha-skeletal and alpha-cardiac actin genes are coexpressed in adult human skeletal muscle and heart. Mol Cell Biol. 1983;3 doi: 10.1128/mcb.3.11.1985. 1985–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kojima H., Ishijima A., Yanagida T. Direct measurement of stiffness of single actin filaments with and without tropomyosin by in vitro nanomanipulation. Proc Natl Acad Sci U S A. 1994;91:12962–12966. doi: 10.1073/pnas.91.26.12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laughlin M.H., Padilla J., Jenkins N.T., Thorne P.K., Martin J.S., Rector R.S. Exercise-induced differential changes in gene expression among arterioles of skeletal muscles of obese rats. J Appl Physiol Bethesda Md. 1985;119:583–603. doi: 10.1152/japplphysiol.00316.2015. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moarbes V., Mayaki D., Huck L., Leblanc P., Vassilakopoulos T., Petrof B.J. Differential regulation of myofibrillar proteins in skeletal muscles of septic mice. Phys Rep. 2019;7:e14248. doi: 10.14814/phy2.14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olfert M., Carpenter J., Stricker J., Audet G., Olenich S. Thrombospondin-1 protects heart and skeletal muscle mass from cigarette smoke induced modifications. Faseb J. 2015;29:825–828. doi: 10.1096/fasebj.29.1_supplement.825.8. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.