Abstract
The extracellular matrix (ECM) is an intricate megastructure made by bacterial cells to form architecturally complex biostructures called biofilms. Protection of cells, modulation of cell-to-cell signalling, cell differentiation and environmental sensing are functions of the ECM that reflect its diverse chemical composition. Proteins, polysaccharides and eDNA have specific functionalities while cooperatively interacting to sustain the architecture and biological relevance of the ECM. The accumulated evidence on the chemical heterogeneity and specific functionalities of ECM components has attracted attention because of their potential biotechnological applications, from agriculture to the water and food industries. This review compiles information on the most relevant bacterial ECM components, the biophysical and chemical features responsible for their biological roles, and their potential to be further translated into biotechnological applications.
Keywords: Bacterial biofilms, Extracellular matrix, Amyloid proteins, Exopolysaccharides, Biotechnological applications
1. Introduction
The extracellular matrix (ECM), both in eukaryotes and prokaryotes, is a mixture of high-molecular-weight polymers that are secreted to the external medium and are produced by nearly all types of cell (Dragos and Kovacs, 2017). By definition, the eukaryotic ECM can be understood as the non-cellular three-dimensional macromolecular network composed of a mixture of components such as collagens, proteoglycans/glycosaminoglycans (PGs), elastin, fibronectin, laminins, and several other glycoproteins. This structure can be found in tissues and organs providing support to the cellular components and providing biochemical and biomechanical cues for tissue morphogenesis, differentiation and homeostasis [1], [2].
Fibril-forming collagen type I and II are the major constituents of the extracellular matrix of eukaryotic tissues, which can be found associated to other ECM proteins, collagens and PGs thus constructing large fibrilar structures [3]. These structures, in combination with other ECM molecules, define the 3D matrix network [2], [4]. It is thus conceivable that the composition and structural organization of the ECM influences relevant biological processes such as adhesion, migration, proliferation and differentiation of eukaryotic cells [5]. Indeed, the ECM has been described as a reservoir for the localization and concentration of growth factors and signalling molecules, which form gradients critical for the establishment of developmental patterning during morphogenesis [6], [7].
In contrast to eukaryotes, in which cells are intrinsically grouped forming tissues and organs, bacterial cells live as independent individuals or forming multicellular communities, known as biofilms, growing over surfaces and providing several benefits as the better adaptation to different environmental conditions, improved attachment to hosts and to the access to nutrients [8], [9], [10], [11]. Analogous to eukaryotic tissues, bacterial cells within biofilms are embedded in a secreted and multifunctional ECM that provides i) structural support to the community, ii) improved cellular adhesion, iii) regulation of the flux of signals and nutrients to ensure cell differentiation [12], [13], and iv) a formidable physicochemical barrier against external assaults [14], [15]. The microbial ECM is heterogeneously composed of proteins, exopolysaccharides, nucleic acids, lipids and secondary metabolites, each of which preserves similar functionality but is chemically variable across bacterial taxa. In this mini-review, we introduce the main components of the prokaryotic ECM, their functions in the maturation of the biofilm structure and bacterial interactions with the environment, and we highlight the biophysical peculiarities that allow their biotechnological exploitation.
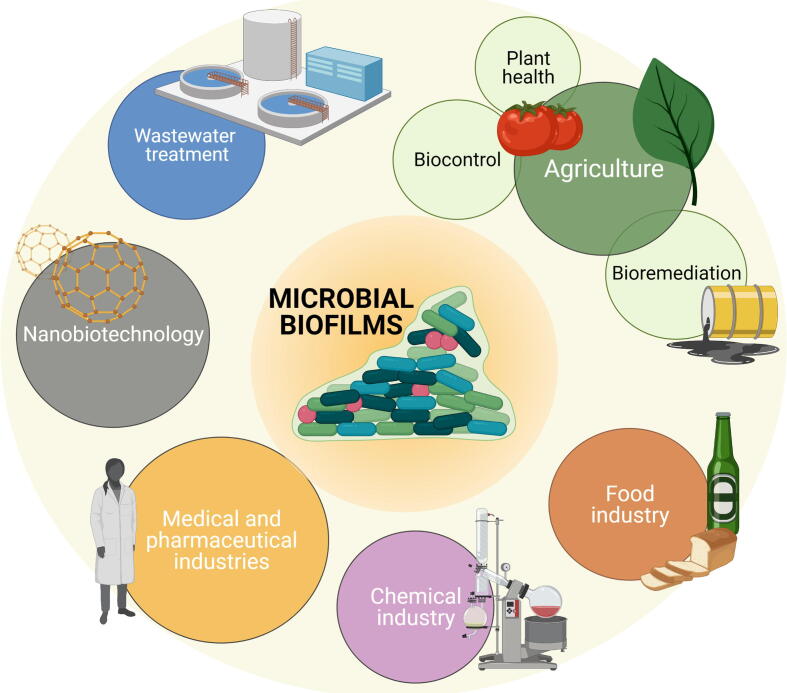
2. Biotechnological applications of biofilms
Bacterial biofilms are widely distributed in nature and largely contribute to the modification of the environment in a variety of ways. However, the fact that bacterial biofilms have been extensively studied with human bacterial pathogens has led to biased negative perceptions associated with contamination and pathogenicity [16]. Water, food and agricultural industries, sustainable agriculture, and the production of recombinant proteins and chemicals are examples of biotechnology research fields that benefit from the unique properties of bacterial biofilms (Fig. 1) [17], [18], [19]. In addition, the possibility of combining diverse strains in multispecies biofilms expands their biotechnological applications, diversifying the variety of products that would be impossible to obtain with single strain cultures.
Fig. 1.
Applicability of microbial biofilms to different branches of the biotechnological industry. Microbial biofilms are currently used in different agricultural techniques involved in biocontrol, bioremediation and plant health. In addition, they can be found in wastewater treatment plants, in the nanobiotechnology industry involved in the production of nanotubes and nanowires, in the food and chemical industries and also in areas of the pharmaceutical and medical industries. Created with BioRender.com.
Bacterial biofilms can be immobilized in bioreactors using different strategies such as adsorption, entrapment, or covalent bond. Adsorption based on cell fixing is the most commonly used method in biofilm reactors (fluidized bed reactors, continuous stirred tank reactors, airlift reactors, and packed bed reactors), and it is probably the most natural method because it leverages the inherent ability of bacterial cells to adhere to any given support [18], [20], [21]. Immobilization of cells on alginate beads is also interesting for industrial bioprocesses and has been successfully used in the preservation of cell viability, the degradation and biotransformation of pollutants, and the production of enzymes, probiotics and other valuable products [22], [23]. The water industry was the first to implement biofilm reactors, including the use of biofilters or moving bed biofilm reactors for wastewater treatment [24]. Recently, the development and use of biofilm-based systems has increased to produce a variety of valuable chemicals, although many steps need to be completely understood to optimize the production and reach the highest yield [17], [25].
Bacterial biofilms have also shown great potential in the food industry and the implementation of sustainable agricultural practices. In food industry, for instance, biofilms formed by probiotics in the gut epithelial mucosal surface have shown advantages and health benefits. Biofilms formed by probiotic bacteria such as Lactotoccus reuteri lead to the accumulation of bacteriocins which provides protection against foodborne pathogens [26], [27]. In the agriculture field, biofilm-forming microbes have been shown to contribute to crop yield in multifaceted ways, including the promotion of plant growth and protection from abiotic disorders (dissection or high salinity) and microbial pathogens [28], [29], [30]. Root-colonizing bacteria, such as Azotobacter spp., Azospirillum spp., Bacillus spp., Beijerinckia spp., Pseudomonas spp., Rhizobium, and Bradyrhizobium spp. are known to enhance the growth of plants by improving the availability of phosphorous, potassium and zinc, fixing atmospheric di-nitrogen, or triggering the production of hormones such as auxins, gibberellins, and cytokinins [31]. Effective colonization and the establishment of biofilms on diverse plant organs create protective microbial barriers that reduce the growth of pathogens by limiting the availability of essential nutrients and micronutrients for growth and pathogenicity or by producing a variety of antimicrobials (i.e., 2, 4-diacetylphloroglucinol, cyclic lipopeptides, tropolone, pyrrolnitrin, pyoluteorin, phenazine, zwittermicin A, xanthobaccin, oligomycin A, or kanosamine) to effectively eradicate or reduce the population density of pathogenic competitors [32], [33], [34], [35], [36], [37], [38]. Novel strategies in biocontrol are using nanoparticle-entrapped biofilms to fight against bacterial and fungal pathogens [39]. This methodology has shown promising results as those observed by the combination of ZnO nanoparticles and P. chlororaphis O6 inhibiting Fusarium growth [40], and the combination of nano-silica and Pseudomonas sp. enhancing the biocontrol activity against maize pathogens [41].
A benefit of bacterial biofilms to agriculture is their use in soil bioremediation [42]. It has been demonstrated that cyanobacteria are able to accumulate high concentrations of toxic compounds such as insecticides or heavy metals that persist in crop soils after their application. This ability can be used for the immobilization of cyanobacteria forming biofilms on alginate and silica gel, thus increasing their resistance to toxic compounds and a series of procedural advantages such as a less water requirement for cultivation and easy harvesting [43]. Pseudomonas putida and other bacterial species are very interesting from the environmental and industrial points of view due to their remarkable ability to tolerate high concentration of toxic compounds and to degrade pollutants as xenobiotics, an effect that can be increased when bacterial cells are forming biofilms [44], [45], [46], [47]. Accumulation of toxic compounds supposes a substantial threat to public health and the environment and the above-mentioned species are attracting attention as very promising microorganisms to be implemented in the bioremediation of contaminated soils and waters [48]. Finally, complex multispecies biofilms are of great interest for biotechnological applications as many possible combinations of diverse microbes can improve bioprocesses through cooperative methods, including metabolic cross-talk and sharing of resources. Examples are the degradation of crude oil [49] and the desulphurization of dibenzothiophene (DBT) to form sulphur-free 2-hydroxybiphenyl [50].
During biofilm lifestyle, bacterial cells also produce a wide array of secondary metabolites (lipopeptides, bacteriocins, antibiotics, amino acids, toxins, etc) which play critical roles in the ecology of bacterial communities, either as inhibitors of competitors growth or acting as signal molecules that modulate microbial interspecies or interkingdom communication and behaviour [51], [52], [53]. The diversity of secondary metabolites and their functionalities are reasons for their enormous biotechnological interest in a variety of fields of industry (medical, food industry or agricultural). Deeper review of the utilization of secondary metabolites as antimicrobials in medicine can be found in these works [54], [55], [56]. Different than antimicrobial therapies, secondary metabolites produced by microbial species (daunomycin, mitomycin C, adriamycin, etc.) are also applied in the treatment of different types of cancer [57], as anti-malarial compounds (gliotoxin) [58] or antiplasmodials (trichodermol) [59]. In addition, it should be specially mentioned how diverse bacterial secondary metabolites are contributing to plant health and crop yield by targeting microbial pathogens: i) Lipopeptides not only inhibit fungal and bacterial pathogen growth, but also trigger the plant immunity system or promote plant growth [55], [60]; ii) small molecules as bacillaene, difficidin and macrolactin, produced by B. amyloliquefaciens, inhibit Gram-positive and Gram-negative pathogens such as Erwinia amylovora [61]. Food and flavour industries are also prominent areas where secondary metabolites are gaining interest. Examples of that are 3-octanone, 1-octen-3-ol and 3-octanol produced by Trichoderma spp. in mushroom flavour and aroma [62], and the use of probiotics for the production of antioxidant compounds [63], [64].
3. Extracellular matrix components in bacterial biofilms
Biofilms are chemically complex and diverse, which may define their extensive impact in the environment. Thus, the knowledge on the individual structural components of this bacterial megastructures are essential to potentiate the aforementioned benefits of biofilms and to discover unprecedented biotechnological uses for each component.
3.1. Exopolysaccharides (EPSs).
The extracellular matrix of bacterial biofilms is commonly composed of proteins, exopolysaccharides, nucleic acids, lipids and other minor biomolecules such as secondary metabolites. EPSs are probably the most abundant component of the ECM and are considered important elements related to the virulence of bacterial pathogens or for bacterial protection [12], [65]. The formation of biofilms is the result of a highly coordinated developmental programme; thus, different structural and regulatory elements are spatially and temporally expressed. Studies with diverse bacterial species have shown the importance of EPS in different stages of biofilm development, from the initial cellular adhesion to surfaces to the formation of complex structures and the final dispersion of the biofilm. Thus, it is not surprising that EPSs are important contributors to the architecture of biofilms composed of largely diverse bacterial species, as seen in E. coli, S. mutans, Vibrio, B. subtilis and Pseudomonas, among others [65]. In addition to this well-known structural function, the physicochemical properties of EPSs provide biofilms with a very effective impenetrable barrier that prevents or delays the entrance of antimicrobials into the biofilm, which gives ample time to initiate the expression of resistance genes by individual cells [66], [67].
The staphylococcal polysaccharide intercellular adhesion (PIA) and PIA-related polymers in Staphylococcus and other Gram-negative species, cellulose in Pseudomonas, Vibrio and Salmonella, or alginate, Psl, Pel in Pseudomonas are common bacterial EPS that can be found in different bacteria [68], [69]. The production of several EPSs with different compositions by the same strain, as exemplified by P. aeruginosa, allows for the adaptation to environmental changes, for the colonization of diverse niches, or for the fight against other microorganisms. P. aeruginosa isolated from cystic fibrosis patients overproduces alginate, leading to mucoid colonies; however, this EPS is not essential for biofilm formation and mainly relies on the production of Pel and Psl in laboratory strains [70], [71]. The EPS produced by S. mutants is one of the main contributors to the virulence of these strains and the formation of dental biofilms. The EPSs produced by glucosyltransferases encoded by gtfB, gtfC and gtfD and fructosyltransferases (Ftfs) leads to the synthesis of a mixture of different types of soluble and insoluble glucans and fructans thus promoting the local colonization of microorganisms on the teeth while forming a protective extracellular matrix [72], [73], [74]. The EPS produced by B. subtilis is encoded by the epsO-A operon which is mainly regulated by sinI-sinR, and is mainly composed of glucose, galactose, and N-acetylgalactosamine. At the genomic level, EpsA and EpsB constitute a sensor tyrosine kinase system, and they participate in the activation of other protein targets important for EPS synthesis, such as the glycosyltransferase EpsE [75]. EpsE, located in the cell membrane, seems to be involved in the inhibition of flagellar rotation through a clutch-like mechanism [76]. EpsC, EpsM and EpsN have been implicated in the synthesis of N,N’-diacetylbacillosamine, while EpsHIJK are involved in the synthesis of poly-N-acetylglucosamine [77]. Together, the EPSs of B. subtilis are directly involved in the formation of the wrinkles typical of in vitro biofilms formed by this strain [78] and are also involved in population motility [79]. In closely related B. cereus, different eps regions are found, and these regions appear to have complementary or specific roles. In the B. cereus type strain ATCC14567, EPS1 is orthologous to the EPS of B. subtilis and is more relevant in terms of social bacterial motility; however, EPS2 is involved in bacterial adhesion to surfaces, cell-to-cell interaction, cellular aggregation and biofilm formation [80]. The diversity of EPSs is interpreted as bacterial ecological adaptations to the different niches in which each strain is found during the different stages of the bacterial life cycle. Some of the most studied EPSs in Bacillus strains are levan type I and II, which consist of β-2,6-linked D-fructose units and a fructose polymer with a glucose residue linked to the terminal fructose by a α-glycoside bond, respectively, and they can be synthesized outside the cell following the extrusion of the extracellular enzyme levansucrase [81], [82]. For example, these levan-based EPSs participate in the assembly of a levan capsule in Pseudomonas that protects cells against attack from bacteriophages or avoids desiccation of the cells and also as a polysaccharide that helps the aggregation of Paenibacillus on root-adhering soil on wheat plants [83].
In recent decades, the physicochemical characteristics and variability in the structure of EPSs have received special attention for their industrial and medical applications (Table 1). Industrial use of EPSs can be found in many different areas such as the food industry, agriculture, and cosmetics. Examples of the most relevant EPSs routinely considered in the food industry are chitosan, xanthan gum, kefiran and inulin. These polymers are used as viscosity-increasing agents, gelling agents, stabilizers in multiphase solutions, additionally to their use as lubricants, flocculants, or flavour enhancers contributing to increase food quality [84], [85], [86]. Gelrite and xanthan gum are used in agriculture as sprays and pesticides, in the biodegradation of gasoline and in the transportation of gel-encapsulated bacteria for bioaugmentation of contaminated aquifers [86], [87]. In addition, EPSs are also interesting in the agricultural field given that they can potentiate beneficial effects such as the increase in root water retention, the improvement of salt tolerance or the avoidance of the toxicity provoked by flocculants derived from aluminium, polyacrylamide derivatives and polyethyleneimine used in water treatment plants [88]. In the cosmetics industry, EPSs such as xanthan gum, pullulan and kelcogels are also used as rheological stabilizers and fragrance carriers.
Table 1.
Properties and applications of exopolysaccharides.
| Properties | Applications | Refs. | |
|---|---|---|---|
| Exopolysaccharides | |||
| Xanthan | Coating Emulsifying properties Thickening agent High viscosity at low shear rates Freeze-thaw stability Odorless |
Foods Petroleum industry Pharmaceuticals Cosmetics and personal care products Agriculture |
[167], [168] |
| Gellan | Hydrocolloid Stability over wide pH range Gelling capacity Thermoreversible gels Adhesive Versatile texture High clarity Dispersibility Biocompatibility |
Foods Pet food Pharmaceuticals Research: agar substitute and gel electrophoresis |
[168], [169] |
| Alginate | Hydrocolloid Gelling capacity Film-forming Stabilizer Thickening agent Foam stabilizer |
Foods Medicine
|
[169], [170] |
| Cellulose | High crystallinityInsolubility in most solvents High tensile strength Moldability |
Foods Biomedical
|
[171], [172] |
| Dextran | Non-ionic Good stability Newtonian fluid behavior Improve moisture retention and viscosity Low solubility in water Antigenic properties |
Foods Pharmaceutical industry Chromatographic media |
[173] |
| Levan | Low viscosity High water solubility Anti-tumor activity Anti-inflammatory Adhesive strength Film-forming capacity Biocompatibility |
Food (prebiotic) Feed Medicines Cosmetics Industry |
[174], [175] |
| Proteins | |||
| Amyloids | Polymerization Adhesive Bioflocculants |
Building blocks for nanostructures, nanosensors and nanotubes. Amyloid-based gels in cell adhesion and wound healing Drug delivery systems |
[131], [132], [135] |
| BslA | Hydrophobicity | Food and cosmetic industries as stabilizers | [124] |
| Extracellular DNA | |||
| eDNA | Easy digestion | Biomedicine | [157], [166], [159] |
| Negative charge | Forensic (eukaryotic eDNA) | ||
In addition to the above-mentioned fields, EPSs are also exploited for medical and pharmaceutical applications. The physiochemical properties of EPSs as xanthan, pullulan, dextran, hyaluronan, levan, alginate, cellulose and gellan (Table 1), in addition to chemical modifications such as acetylation or oxidation, have contributed to extend their utility [86]. For instance, alginate is used in the formation of nanoparticles for controlled drug release and it is also used as adjuvant for vaccines; pullulan, dextran, or bacterial cellulose are employed for the development of new micelle systems that can improve drug solubility and stability; and bacterial cellulose is also applied in the field of wound healing due to its permeability [86], [89], [90].
3.2. Proteins
In addition to the EPS, proteins are also important components of the ECM with very interesting roles and prospective biotechnological applications. Proteins in biofilms can be classified roughly into two main groups based on functionality: adhesins and functional amyloids. Adhesins are proteins that can be found in gram-negative and gram-positive species [91], [92], [93]. These proteins are cell-surface exposed proteins that promote cell-to-cell contacts within a biofilm or adhesion of bacterial cells to biotic or abiotic surfaces. Examples of adhesins are the biofilm-associated proteins Bap and SasG and fibronectin binding proteins EnBPA and EnBPB of S. aureus [94], [95]; adhesin p1 of Streptococcus mutans [96], and members of the antigen-43 family of autotransporter adhesins in E. coli, such as TibA, and the autotransporter AIDA-I, are involved in the adherence of E. coli to human cells [97], [98], [99]. The adhesion proteins LapA and LapF and the recently discovered MapA in Pseudomonas strains have also been described as key elements in the colonization of surfaces, seed adhesion and biofilm development and maturation [91], [100], [101].
The other main group of proteinaceous components of biofilms is functional amyloids. These proteins, synthesized as monomers, progress timely into aggregates to finally render insoluble fibers with a common quaternary structure characterized by a cross-β pattern, in which hydrogen-bonded β-strands run perpendicularly to the axis of the fibril [102]. Amyloids were initially associated to diverse human disorders (Alzheimer, Parkinson and Huntington, among others) [103], however, amyloid fibres were later found in bacteria associated with the ECM of both gram-positive and gram-negative bacterial species. Functions of amyloids in microbes include the involvement in adhesion and biofilm formation, spore coating and protection, or in the dissemination of virulence factors and evasion of the host immune system, among others [104], [105], [106]. Examples of these proteins are the curli amyloid fibres of E. coli and Salmonella spp. [107], the Fap amyloid fibrils found in Pseudomonas spp. Biofilms [108], TasA of B. subtilis and B. cereus [109], [110] and the harpins found in gram-negative pathogenic strains of Erwinia amylovora and P. syringae, among others [111], [112], [113]. Along with the curli fibres of E. coli, one of the best characterized functional amyloids is TasA produced by B. subtilis [114], [115], [116], [117]. TasA is encoded by the tapA-sipW-tasA operon and needs the activity of TapA and SipW for correct fibre assembly in vivo. In fact, SipW is a bifunctional signal peptidase in charge of processing and translocating TasA and TapA to the exterior of the cell, and in addition, SipW seems to act as a regulatory element in the expression of the tapA and eps operons [118]. In addition, TapA is required for biofilm formation and polymerization of TasA fibres in vivo [115]. However, as exemplified in other amyloids, TasA has the intrinsic ability to form amyloid fibres in vitro in the absence of TapA, which also preserves the structural peculiarities of amyloid proteins but fails to form structurally defined fibres [110].
Although not included in the two main groups of proteins described above, BslA is another protein component of the ECM of B. subtilis with the outstanding ability to self-polymerize into a structural polymer. This protein is a cell surface-associated amphiphilic protein that forms a protective hydrophobic coat on the surface of the biofilm to prevent the penetration of hydrophobic fluids, and its deletion alters the microstructure of colonies [119], [120]. BslA functions in cooperation with other ECM components as TasA/TapA and EPS to ease the maturation of the biofilm [99], [120]. Hydrophobin proteins are typically found in fungi, where they assemble spontaneously into amphipathic monolayers at hydrophobic–hydrophilic interfaces. The surfactant and amphipathic nature of the hydrophobin layers helps in the formation of essential aerial structures of filamentous fungi, such as hyphae, fruiting bodies, and spores [121], [122], in interactions with the environment and in protection against the host defence system [123]. Hydrophobins, such as the previously mentioned BslA, might also have applications in the food or cosmetic industries as stabilizers. The use of BslA in the production process of ice creams is a good example because the combination of the protein with air, fat and water yields a stable mixture permitting the ice cream to stay frozen for longer periods of time and retarding the growth of ice crystals [124].
In addition to the critical contribution of amyloid proteins to the progress towards the different stages of the biofilm life cycle, complementary roles in bacterial growth and survival, detoxification of toxic compounds, resistance to antibiotics and even electron transport have been discovered [125], [126]. Furthermore, the interesting biophysical properties of amyloid proteins, exemplified by their outstanding resistance to chemical and thermal denaturants or pH changes [127], [128], [129], are promoting studies on their use and implementation in a variety of biotechnological processes. In this sense, bacterial amyloids are attracting greater interest as potential natural building blocks for the design of new nanostructures and nanomaterials: nanowires and nanotubes for electronics, nanosensors, amyloid-based gels for cell adhesion and wound healing, or as drug delivery systems [130], [131], [132]. Specifically, studies performed with E. coli have permitted the development of a system to fabricate multiscale patterning fibres as versatile scaffolds able to synthetize fluorescent quantum dots, gold nanowires and nanoparticles [133]. A biofilm-integrated nanofiber display (BIND) system has served as the base for the exploitation of curli fibres to create a biocatalytic biofilm in which functional nanofibers immobilized the industrially relevant enzyme α-amylase [134]. Amyloids can also be used as bioflocculants in microalgae cultivation and are becoming popular as a preferred biomass for biofuel production, promoting the use of cost- and energy-efficient technologies [135]. Because of their variability, functional amyloid proteins with their different biophysical properties are promising sources for the development of potential biotechnological tools and use in technological applications (Table 1).
3.3. Extracellular DNA.
The last main component of the bacterial extracellular matrix is extracellular genomic DNA (eDNA). Initially, it was assumed that eDNA was derived from lysed cells and that mere remnants of eDNA were present and had no relevance on biofilm structure [136], [137]. However, this concept was rebutted by many studies showing species-specific amounts of eDNA in different single and multispecies biofilms and showing organized patterns forming grid-like structures of filamentous networks [138], [139], [140], [141], [142]. eDNA can be released from lysed cells by mechanisms analogous to holin-antiholin systems, as in S. aureus [143], but the possibility of secretion by different eDNA secretion mechanisms has also been described, such as secretion by outer membrane vesicles in P. aeruginosa, S. aureus, B. subtilis, Klebsiella pneumoniae, S. epidermidis, and V. cholerae. The addition of DNase to growing or mature biofilms resulted in the inhibition of biofilm formation or disruption of established biofilms, leading to the establishment of a direct relation between the age of the biofilm and its disruption (young biofilms were more sensitive to DNase than older biofilms) [144], [145]. Currently, it is accepted that eDNA is a structural component of the ECM that provides structural stability to bacterial biofilms by interacting with other ECM components, such as exopolysaccharides and proteins, modulating cell surface properties and promoting cell-to-cell and cell-to-surface adhesion [146], [147], [148]. For example, eDNA in P. aeruginosa has been described to physically interact with the exopolysaccharide Psl, forming fibres facilitating bacterial adhesion and growth [149], while in B. subtilis, eDNA appears to interact with EPS in the early phases of biofilm development [150]. Several studies have also demonstrated the ability of eDNA to act as a chelator of cationic antimicrobials and explained their role in increasing resistance against antibiotics [139], [151], [152]. In addition to eDNA-polysaccharide interactions, eDNA has also been shown to attract and bind amyloid proteins, causing the polymerisation of the matrix and stimulating autoimmunity [153], [154].
eDNA is rarely used as a biotechnological resource (Table 1); however, there are many applications where this component of the ECM has attracted attention. eDNA is mostly fragmented, which suggests an interesting way to fine tune the attachment of bacterial cells to surfaces, as experimentally reported for Listeria monocytogenes [155]. Therefore, eDNA seems to work as a unique element for the control of biofilm attachment and structural stability, and modification of the release of this molecule may be used to alter the mechanical properties of biofilms [140]. Another application is related to the increased resistance of biofilms to antibiotics. The negative charge of eDNA contributes to the creation of a shield that protects biofilm-associated cells from aminoglycosides and cationic antimicrobial peptides that are positively charged by its binding, thus avoiding the penetration of these peptides inside the bacterial cells [156].
Considering its relevance for biofilm formation, eDNA can also be used as a target in the search for antibiofilm agents [157], [158], [159]. Primarily, the use of DNase to digest eDNA leads to biofilm disruption, but the use of antibodies against DNA-binding proteins located at the intersection of crossed eDNA strands has also been investigated [160]. Continuing in this therapeutic direction, nanomaterial-cleaving eDNA in S. aureus biofilms has been proposed as a promising treatment against biofilm-related infections [161].
Very interestingly, although not bacterial, eukaryotic eDNA seems to be useful for medical diagnostics, given the correlation of eDNA concentration with different pathologies, including cancer and autoimmune disorders [162], [163], [164]. In fact, eukaryotic eDNA is used during pregnancy, as eDNA from foetal cells circulates in the maternal blood allowing the detection of foetal genetic disorders [165]. Finally, the application of eDNA in forensics is also interesting, as it can be located and quantified on human epithelial cells or other surfaces, providing a new tool in forensic analysis of touch samples [159], [166].
4. Future perspectives and conclusions
The ECM is a complex structure that is chemically and functionally diverse. Mechanistic studies are providing increased knowledge on the chemical structure and biophysical peculiarities of the molecules that compose this structure. Additional studies on the features defining the stages of biofilm development are necessary to generate a solid body of knowledge that will enable further manipulation or even specific design of polymers for current or new biotechnological applications. The application of the bacterial biofilms or the different components of the extracellular matrix in industrial and agricultural processes is arising as a very promising strategy, both from economic and ecologic point of views, reducing production costs while increasing productivity, fighting against pollutants and serving as an ecological tool against agricultural plagues. Major challenges rely on the ability to scale the production of such biotechnological products at industrial level and thus, their commercial applications due to the high cost of the industrial processes and the low yield that is currently obtained from their production. Studies on the fine genetic regulation of the production of ECM components, the isolation of new producer strains and the utilization of the most adequate substrates should serve to select the more suitable natural microorganisms, especially when genetic manipulation is not permitted or affordable. The unspecific interaction of certain ECM with themselves or medium components impose limitations to further downstream processes. Thus, specific studies on the physicochemical singularities of each ECM component are a matter of interest to define the best ways to improve extraction and purification methods and how to proceed in the bioreactor, improving not only yield but also the quality and utility of the bioproduct.
CRediT authorship contribution statement
Carlos Molina-Santiago: Writing - original draft. Antonio Vicente: Writing - original draft. Diego Romero: Conceptualization, Writing - original draft, Writing - review & editing, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by an ERC Starting Grant (BacBio 637971) and by a grant from the Plan Nacional de I + D + I of Ministerio de Ciencia e Innovación (PID2019-107724 GB-I00)). C.M.S is funded by the program Juan de la Cierva Incorporación (IJC2018-036923-I).
References
- 1.Frantz C., Stewart K.M., Weaver V.M. The extracellular matrix at a glance. J Cell Sci. 2010;123(Pt 24):4195–4200. doi: 10.1242/jcs.023820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Theocharis A.D., Skandalis S.S., Gialeli C., Karamanos N.K. Extracellular matrix structure. Adv Drug Deliv Rev. 2016;97:4–27. doi: 10.1016/j.addr.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Mouw J.K., Ou G., Weaver V.M. Extracellular matrix assembly: a multiscale deconstruction. Nat Rev Mol Cell Biol. 2014;15(12):771–785. doi: 10.1038/nrm3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chagnot C., Listrat A., Astruc T., Desvaux M. Bacterial adhesion to animal tissues: protein determinants for recognition of extracellular matrix components. Cell Microbiol. 2012;14(11):1687–1696. doi: 10.1111/cmi.12002. [DOI] [PubMed] [Google Scholar]
- 5.Patti J.M., Hook M. Microbial adhesins recognizing extracellular matrix macromolecules. Curr Opin Cell Biol. 1994;6(5):752–758. doi: 10.1016/0955-0674(94)90104-x. [DOI] [PubMed] [Google Scholar]
- 6.Kim S.H., Turnbull J., Guimond S. Extracellular matrix and cell signalling: the dynamic cooperation of integrin, proteoglycan and growth factor receptor. J Endocrinol. 2011;209(2):139–151. doi: 10.1530/JOE-10-0377. [DOI] [PubMed] [Google Scholar]
- 7.J.K. Kular S. Basu R.I. Sharma The extracellular matrix: Structure, composition, age-related differences, tools for analysis and applications for tissue engineering J Tissue Eng. 5 2014 2041731414557112 [DOI] [PMC free article] [PubMed]
- 8.Kolter R., Greenberg E.P. Microbial sciences: the superficial life of microbes. Nature. 2006;441(7091):300–302. doi: 10.1038/441300a. [DOI] [PubMed] [Google Scholar]
- 9.Oliveira N.M., Martinez-Garcia E., Xavier J., Durham W.M., Kolter R., Kim W. Biofilm Formation As a Response to Ecological Competition. PLoS Biol. 2015;13(7) doi: 10.1371/journal.pbio.1002191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morikawa M., Kagihiro S., Haruki M., Takano K., Branda S., Kolter R. Biofilm formation by a Bacillus subtilis strain that produces gamma-polyglutamate. Microbiology (Reading). 2006;152(Pt 9):2801–2807. doi: 10.1099/mic.0.29060-0. [DOI] [PubMed] [Google Scholar]
- 11.Romero D. Bacterial determinants of the social behavior of Bacillus subtilis. Res Microbiol. 2013;164(7):788–798. doi: 10.1016/j.resmic.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 12.Steinberg N., Kolodkin-Gal I. The Matrix Reloaded: Probing the Extracellular Matrix Synchronizes Bacterial Communities. J Bacteriol. 2015;197(13):2092–2103. doi: 10.1128/JB.02516-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steinberg N., Keren-Paz A., Hou Q., Doron S., Yanuka-Golub K., Olender T. The extracellular matrix protein TasA is a developmental cue that maintains a motile subpopulation within Bacillus subtilis biofilms. Sci Signal. 2020;13(632) doi: 10.1126/scisignal.aaw8905. [DOI] [PubMed] [Google Scholar]
- 14.Molina-Santiago C., Pearson J.R., Navarro Y., Berlanga-Clavero M.V., Caraballo-Rodriguez A.M., Petras D. The extracellular matrix protects Bacillus subtilis colonies from Pseudomonas invasion and modulates plant co-colonization. Nat Commun. 2019;10(1):1919. doi: 10.1038/s41467-019-09944-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vlamakis H., Aguilar C., Losick R., Kolter R. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev. 2008;22(7):945–953. doi: 10.1101/gad.1645008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Percival S.L., Suleman L., Vuotto C., Donelli G. Healthcare-associated infections, medical devices and biofilms: risk, tolerance and control. J Med Microbiol. 2015;64(Pt 4):323–334. doi: 10.1099/jmm.0.000032. [DOI] [PubMed] [Google Scholar]
- 17.Edel M., Horn H., Gescher J. Biofilm systems as tools in biotechnological production. Appl Microbiol Biotechnol. 2019;103(13):5095–5103. doi: 10.1007/s00253-019-09869-x. [DOI] [PubMed] [Google Scholar]
- 18.Berlanga M., Guerrero R. Living together in biofilms: the microbial cell factory and its biotechnological implications. Microb Cell Fact. 2016;15(1):165. doi: 10.1186/s12934-016-0569-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soares A., Azevedo A., Gomes L.C., Mergulhão F.J. Recombinant protein expression in biofilms. AIMS Microbiol. 2019;5(3):232–250. doi: 10.3934/microbiol.2019.3.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halan B., Buehler K., Schmid A. Biofilms as living catalysts in continuous chemical syntheses. Trends Biotechnol. 2012;30(9):453–465. doi: 10.1016/j.tibtech.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 21.Costa A.M., Mergulhao F.J., Briandet R., Azevedo N.F. It is all about location: how to pinpoint microorganisms and their functions in multispecies biofilms. Future Microbiol. 2017;12:987–999. doi: 10.2217/fmb-2017-0053. [DOI] [PubMed] [Google Scholar]
- 22.Huang H.Y., Tang Y.J., King V.A., Chou J.W., Tsen J.H. Properties of Lactobacillus reuteri chitosan-calcium-alginate encapsulation under simulated gastrointestinal conditions. Int Microbiol. 2015;18(1):61–69. doi: 10.2436/20.1501.01.235. [DOI] [PubMed] [Google Scholar]
- 23.Bayat Z., Hassanshahian M., Cappello S. Immobilization of Microbes for Bioremediation of Crude Oil Polluted Environments: A Mini Review. Open Microbiol J. 2015;9:48–54. doi: 10.2174/1874285801509010048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bouwer E.J., Crowe P.B. Biological Processes in Drinking. Water Treatment. 1988;80(9):82–93. [Google Scholar]
- 25.Gross R., Buehler K., Schmid A. Engineered catalytic biofilms for continuous large scale production of n-octanol and (S)-styrene oxide. Biotechnol Bioeng. 2013;110(2):424–436. doi: 10.1002/bit.24629. [DOI] [PubMed] [Google Scholar]
- 26.Ünal Turhan E., Erginkaya Z., Korukluoğlu M., Konuray G. Beneficial Biofilm Applications in Food and Agricultural Industry. In: Malik A., Erginkaya Z., Erten H., editors. Health and Safety Aspects of Food Processing Technologies. Springer International Publishing; Cham: 2019. pp. 445–469. [Google Scholar]
- 27.Jones S.E., Versalovic J. Probiotic Lactobacillus reuteri biofilms produce antimicrobial and anti-inflammatory factors. BMC Microbiol. 2009;9:35. doi: 10.1186/1471-2180-9-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meena K.K., Sorty A.M., Bitla U.M., Choudhary K., Gupta P., Pareek A. Abiotic Stress Responses and Microbe-Mediated Mitigation in Plants: The Omics Strategies. Front Plant Sci. 2017;8:172. doi: 10.3389/fpls.2017.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosier A., Medeiros F.H.V., Bais H.P. Defining plant growth promoting rhizobacteria molecular and biochemical networks in beneficial plant-microbe interactions. Plant Soil. 2018;428(1):35–55. [Google Scholar]
- 30.Berlanga-Clavero M.V., Molina-Santiago C., de Vicente A., Romero D. More than words: the chemistry behind the interactions in the plant holobiont. Environ Microbiol. 2020;22(11):4532–4544. doi: 10.1111/1462-2920.15197. [DOI] [PubMed] [Google Scholar]
- 31.Rana S., Upadhyay L.S.B. Microbial exopolysaccharides: Synthesis pathways, types and their commercial applications. Int J Biol Macromol. 2020;157:577–583. doi: 10.1016/j.ijbiomac.2020.04.084. [DOI] [PubMed] [Google Scholar]
- 32.Altaf M.M., Ahmad I. CABI; Wallingford: 2016. Biofilm formation on plant surfaces by rhizobacteria: impact on plant growth and ecological significance; pp. 81–95. [Google Scholar]
- 33.Cazorla F.M., Duckett S.B., Bergstrom E.T., Noreen S., Odijk R., Lugtenberg B.J. Biocontrol of avocado dematophora root rot by antagonistic Pseudomonas fluorescens PCL1606 correlates with the production of 2-hexyl 5-propyl resorcinol. Mol Plant Microbe Interact. 2006;19(4):418–428. doi: 10.1094/MPMI-19-0418. [DOI] [PubMed] [Google Scholar]
- 34.Gupta O.P., Karkute S.G., Banerjee S., Meena N.L., Dahuja A. Contemporary Understanding of miRNA-Based Regulation of Secondary Metabolites Biosynthesis in Plants. Front Plant Sci. 2017;8:374. doi: 10.3389/fpls.2017.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Penha R.O., Vandenberghe L.P.S., Faulds C., Soccol V.T., Soccol C.R. Bacillus lipopeptides as powerful pest control agents for a more sustainable and healthy agriculture: recent studies and innovations. Planta. 2020;251(3):70. doi: 10.1007/s00425-020-03357-7. [DOI] [PubMed] [Google Scholar]
- 36.Aleti G., Lehner S., Bacher M., Compant S., Nikolic B., Plesko M. Surfactin variants mediate species-specific biofilm formation and root colonization in Bacillus. Environ Microbiol. 2016;18(8):2634–2645. doi: 10.1111/1462-2920.13405. [DOI] [PubMed] [Google Scholar]
- 37.Kiesewalter H.T., Lozano-Andrade C.N., Strube M.L., Kovacs A.T. Secondary metabolites of Bacillus subtilis impact the assembly of soil-derived semisynthetic bacterial communities. Beilstein J Org Chem. 2020;16:2983–2998. doi: 10.3762/bjoc.16.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rayanoothala P., Divya M., Mahapatra S., Das S. Microbial Biofilm: Formation, Quorum Sensing, and Its Applications in Plant Disease Management. In: Singh K.P., Jahagirdar S., Sarma B.K., editors. Emerging Trends in Plant Pathology. Springer Singapore; Singapore: 2021. pp. 385–397. [Google Scholar]
- 39.Bhatia R., Gulati D., Sethi G. Biofilms and nanoparticles: applications in agriculture. Folia Microbiol. 2021;66(2):159–170. doi: 10.1007/s12223-021-00851-7. [DOI] [PubMed] [Google Scholar]
- 40.Dimkpa C.O., McLean J.E., Britt D.W., Anderson A.J. Antifungal activity of ZnO nanoparticles and their interactive effect with a biocontrol bacterium on growth antagonism of the plant pathogen Fusarium graminearum. Biometals. 2013;26(6):913–924. doi: 10.1007/s10534-013-9667-6. [DOI] [PubMed] [Google Scholar]
- 41.Rangaraj S., Gopalu K., Muthusamy P., Rathinam Y., Venkatachalam R., Narayanasamy K. Augmented biocontrol action of silica nanoparticles and Pseudomonas fluorescens bioformulant in maize (Zea mays L.). RSC. Advances. 2014;4(17):8461–8465. [Google Scholar]
- 42.Karn S.K., Duan J., Jenkinson I.R. Book Review: Role of Biofilms in Bioremediation. 2017;5:22. [Google Scholar]
- 43.Rangsayatorn N., Pokethitiyook P., Upatham E.S., Lanza G.R. Cadmium biosorption by cells of Spirulina platensis TISTR 8217 immobilized in alginate and silica gel. Environ Int. 2004;30(1):57–63. doi: 10.1016/S0160-4120(03)00146-6. [DOI] [PubMed] [Google Scholar]
- 44.Fernandez M., Duque E., Pizarro-Tobias P., Van Dillewijn P., Wittich R.M., Ramos J.L. Microbial responses to xenobiotic compounds. Identification of genes that allow Pseudomonas putida KT2440 to cope with 2,4,6-trinitrotoluene. Microb. Biotechnol. 2009;2(2):287–294. doi: 10.1111/j.1751-7915.2009.00085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramos J.L., Duque E., Gallegos M.T., Godoy P., Ramos-Gonzalez M.I., Rojas A. Mechanisms of solvent tolerance in gram-negative bacteria. Annu Rev Microbiol. 2002;56:743–768. doi: 10.1146/annurev.micro.56.012302.161038. [DOI] [PubMed] [Google Scholar]
- 46.Molina-Santiago C., Udaondo Z., Gomez-Lozano M., Molin S., Ramos J.L. Global transcriptional response of solvent-sensitive and solvent-tolerant Pseudomonas putida strains exposed to toluene. Environ Microbiol. 2017;19(2):645–658. doi: 10.1111/1462-2920.13585. [DOI] [PubMed] [Google Scholar]
- 47.Benedetti I., de Lorenzo V., Nikel P.I. Genetic programming of catalytic Pseudomonas putida biofilms for boosting biodegradation of haloalkanes. Metab Eng. 2016;33:109–118. doi: 10.1016/j.ymben.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 48.Kour D, Rana KL, Kaur T, Yadav N, Yadav AN, Rastegari AA, et al. Chapter 18 - Microbial biofilms: Functional annotation and potential applications in agriculture and allied sectors. In: Yadav MK, Singh BP, editors. New and Future Developments in Microbial Biotechnology and Bioengineering: Microbial Biofilms: Elsevier; 2020. p. 283-301.
- 49.McGenity T.J., Folwell B.D., McKew B.A., Sanni G.O. Marine crude-oil biodegradation: a central role for interspecies interactions. Aquat Biosyst. 2012;8(1):10. doi: 10.1186/2046-9063-8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martinez I., Mohamed M.E., Rozas D., Garcia J.L., Diaz E. Engineering synthetic bacterial consortia for enhanced desulfurization and revalorization of oil sulfur compounds. Metab Eng. 2016;35:46–54. doi: 10.1016/j.ymben.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 51.Cámara-Almirón J., Molina-Santiago C., Pérez-García A., de Vicente A., Cazorla F.M., Romero D. Understanding Bacterial Physiology for Improving Full Fitness. In: De Cal A., Melgarejo P., Magan N., editors. How Research Can Stimulate the Development of Commercial Biological Control Against Plant Diseases. Springer International Publishing; Cham: 2020. pp. 47–60. [Google Scholar]
- 52.C. Molina-Santiago D. Vela-Corcía D. Petras L. Díaz-Martínez A.I. Pérez-Lorente S. Sopeña-Torres et al. Chemical interplay and complementary adaptative strategies toggle bacterial antagonism and co-existence. 2021 01 2021 pp. 11.426172 [DOI] [PMC free article] [PubMed]
- 53.Sansinenea E., Ortiz A. Secondary metabolites of soil Bacillus spp. Biotechnol Lett. 2011;33(8):1523–1538. doi: 10.1007/s10529-011-0617-5. [DOI] [PubMed] [Google Scholar]
- 54.Vaishnav P., Demain A.L. Unexpected applications of secondary metabolites. Biotechnol Adv. 2011;29(2):223–229. doi: 10.1016/j.biotechadv.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 55.Keswani C., Mishra S., Sarma B.K., Singh S.P., Singh H.B. Unraveling the efficient applications of secondary metabolites of various Trichoderma spp. Appl Microbiol Biotechnol. 2014;98(2):533–544. doi: 10.1007/s00253-013-5344-5. [DOI] [PubMed] [Google Scholar]
- 56.Quinn G.A., Banat A.M., Abdelhameed A.M., Banat I.M. Streptomyces from traditional medicine: sources of new innovations in antibiotic discovery. J Med Microbiol. 2020;69(8):1040–1048. doi: 10.1099/jmm.0.001232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Demain A.L., Vaishnav P. Natural products for cancer chemotherapy. Microb Biotechnol. 2011;4(6):687–699. doi: 10.1111/j.1751-7915.2010.00221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tanaka Y., Shiomi K., Kamei K., Sugoh-Hagino M., Enomoto Y., Fang F. Antimalarial activity of radicicol, heptelidic acid and other fungal metabolites. J Antibiot (Tokyo). 1998;51(2):153–160. doi: 10.7164/antibiotics.51.153. [DOI] [PubMed] [Google Scholar]
- 59.Isaka M., Punya J., Lertwerawat Y., Tanticharoen M., Thebtaranonth Y. Antimalarial Activity of Macrocyclic Trichothecenes Isolated from the Fungus Myrothecium verrucaria. J Nat Prod. 1999;62(2):329–331. doi: 10.1021/np980323x. [DOI] [PubMed] [Google Scholar]
- 60.Scarselletti R., Faull J.L. In vitro activity of 6-pentyl-α-pyrone, a metabolite of Trichoderma harzianum, in the inhibition of Rhizoctonia solani and Fusarium oxysporum f. sp. lycopersici. Mycol Res. 1994;98(10):1207–1209. [Google Scholar]
- 61.Magno-Perez-Bryan M.C., Martinez-Garcia P.M., Hierrezuelo J., Rodriguez-Palenzuela P., Arrebola E., Ramos C. Comparative Genomics Within the Bacillus Genus Reveal the Singularities of Two Robust Bacillus amyloliquefaciens Biocontrol Strains. Mol Plant Microbe Interact. 2015;28(10):1102–1116. doi: 10.1094/MPMI-02-15-0023-R. [DOI] [PubMed] [Google Scholar]
- 62.Combet E., Eastwood D.C., Burton K.S., Combet E., Henderson J., Henderson J. Eight-carbon volatiles in mushrooms and fungi: properties, analysis, and biosynthesis. Mycoscience. 2006;47(6):317–326. [Google Scholar]
- 63.Kim J.Y., Lee M.Y., Ji G.E., Lee Y.S., Hwang K.T. Production of γ-aminobutyric acid in black raspberry juice during fermentation by Lactobacillus brevis GABA100. Int J Food Microbiol. 2009;130(1):12–16. doi: 10.1016/j.ijfoodmicro.2008.12.028. [DOI] [PubMed] [Google Scholar]
- 64.Tallapragada P., Chapter D.R. Microbial Production of Secondary Metabolites as Food Ingredients. In: Holban A.M., Grumezescu A.M., editors. Vol. 11. Academic Press; 2017. pp. 317–345. (Microbial Production of Food Ingredients and Additives). [Google Scholar]
- 65.Limoli D.H., Jones C.J., Wozniak D.J. Bacterial Extracellular Polysaccharides in Biofilm Formation and Function. Microbiol Spectr. 2015;3(3) doi: 10.1128/microbiolspec.MB-0011-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Walters M.C., 3rd, Roe F., Bugnicourt A., Franklin M.J., Stewart P.S. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob Agents Chemother. 2003;47(1):317–323. doi: 10.1128/AAC.47.1.317-323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jefferson K.K., Goldmann D.A., Pier G.B. Use of confocal microscopy to analyze the rate of vancomycin penetration through Staphylococcus aureus biofilms. Antimicrob Agents Chemother. 2005;49(6):2467–2473. doi: 10.1128/AAC.49.6.2467-2473.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arciola C.R., Campoccia D., Ravaioli S., Montanaro L. Polysaccharide intercellular adhesin in biofilm: structural and regulatory aspects. Front Cell Infect Microbiol. 2015;5:7. doi: 10.3389/fcimb.2015.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Karygianni L., Ren Z., Koo H., Thurnheer T. Biofilm Matrixome: Extracellular Components in Structured Microbial Communities. Trends Microbiol. 2020;28(8):668–681. doi: 10.1016/j.tim.2020.03.016. [DOI] [PubMed] [Google Scholar]
- 70.Colvin K.M., Irie Y., Tart C.S., Urbano R., Whitney J.C., Ryder C. The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ Microbiol. 2012;14(8):1913–1928. doi: 10.1111/j.1462-2920.2011.02657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moradali M.F., Ghods S., Rehm B.H.A. Pseudomonas aeruginosa Lifestyle: A Paradigm for Adaptation. Survival, and Persistence. Front Cell Infect Microbiol. 2017;7:39-. doi: 10.3389/fcimb.2017.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Koo H., Xiao J., Klein M.I., Jeon J.G. Exopolysaccharides produced by Streptococcus mutans glucosyltransferases modulate the establishment of microcolonies within multispecies biofilms. J Bacteriol. 2010;192(12):3024–3032. doi: 10.1128/JB.01649-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bowen W.H., Koo H. Biology of Streptococcus mutans-derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms. Caries Res. 2011;45(1):69–86. doi: 10.1159/000324598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Klein M.I., Hwang G., Santos P.H., Campanella O.H., Koo H. Streptococcus mutans-derived extracellular matrix in cariogenic oral biofilms. Front Cell Infect Microbiol. 2015;5:10. doi: 10.3389/fcimb.2015.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guttenplan S.B., Blair K.M., Kearns D.B. The EpsE flagellar clutch is bifunctional and synergizes with EPS biosynthesis to promote Bacillus subtilis biofilm formation. PLoS Genet. 2010;6(12) doi: 10.1371/journal.pgen.1001243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Blair K.M., Turner L., Winkelman J.T., Berg H.C., Kearns D.B. A molecular clutch disables flagella in the Bacillus subtilis biofilm. Science. 2008;320(5883):1636–1638. doi: 10.1126/science.1157877. [DOI] [PubMed] [Google Scholar]
- 77.Roux D., Cywes-Bentley C., Zhang Y.F., Pons S., Konkol M., Kearns D.B. Identification of Poly-N-acetylglucosamine as a Major Polysaccharide Component of the Bacillus subtilis Biofilm Matrix. J Biol Chem. 2015;290(31):19261–19272. doi: 10.1074/jbc.M115.648709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marvasi M., Visscher P.T., Casillas M.L. Exopolymeric substances (EPS) from Bacillus subtilis: polymers and genes encoding their synthesis. FEMS Microbiol Lett. 2010;313(1):1–9. doi: 10.1111/j.1574-6968.2010.02085.x. [DOI] [PubMed] [Google Scholar]
- 79.Nagorska K., Ostrowski A., Hinc K., Holland I.B., Obuchowski M. Importance of eps genes from Bacillus subtilis in biofilm formation and swarming. J Appl Genet. 2010;51(3):369–381. doi: 10.1007/BF03208867. [DOI] [PubMed] [Google Scholar]
- 80.Caro-Astorga J., Alvarez-Mena A., Hierrezuelo J., Guadix J.A., Heredia-Ponce Z., Arboleda-Estudillo Y. Two genomic regions encoding exopolysaccharide production systems have complementary functions in B. cereus multicellularity and host interaction. Sci Rep. 2020;10(1):1000. doi: 10.1038/s41598-020-57970-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Abdel-Fattah W.R., Chen Y., Eldakak A., Hulett F.M. Bacillus subtilis phosphorylated PhoP: direct activation of the E(sigma)A- and repression of the E(sigma)E-responsive phoB-PS+V promoters during pho response. J Bacteriol. 2005;187(15):5166–5178. doi: 10.1128/JB.187.15.5166-5178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.El-Refai H.A., Abdel-Fattah A.F., Mostafa F.A. Enzymic synthesis of levan and fructo-oligosaccharides by Bacillus circulans and improvement of levansucrase stability by carbohydrate coupling. World J Microbiol Biotechnol. 2009;25(5):821–827. [Google Scholar]
- 83.Bezzate S., Aymerich S., Chambert R., Czarnes S., Berge O., Heulin T. Disruption of the Paenibacillus polymyxa levansucrase gene impairs its ability to aggregate soil in the wheat rhizosphere. 2000;2(3):333–342. doi: 10.1046/j.1462-2920.2000.00114.x. [DOI] [PubMed] [Google Scholar]
- 84.Barcelos M.C.S., Vespermann K.A.C., Pelissari F.M., Molina G. Current status of biotechnological production and applications of microbial exopolysaccharides. Crit Rev Food Sci Nutr. 2020;60(9):1475–1495. doi: 10.1080/10408398.2019.1575791. [DOI] [PubMed] [Google Scholar]
- 85.Ferreira A.R., Alves V.D., Coelhoso I.M. Polysaccharide-Based Membranes in Food Packaging Applications. Membranes (Basel) 2016;6(2) doi: 10.3390/membranes6020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kambourova M., Oner E.T., Poli A. Chapter 15 - Exopolysaccharides from Prokaryotic Microorganisms—Promising Sources for White Biotechnology Processes. In: Pandey A., Höfer R., Taherzadeh M., Nampoothiri K.M., Larroche C., editors. Industrial Biorefineries & White Biotechnology. Elsevier; Amsterdam: 2015. pp. 523–554. [Google Scholar]
- 87.Moslemy P., Neufeld R.J., Millette D., Guiot S.R. Transport of gellan gum microbeads through sand: an experimental evaluation for encapsulated cell bioaugmentation. J Environ Manage. 2003;69(3):249–259. doi: 10.1016/j.jenvman.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 88.Ruden C. Acrylamide and cancer risk–expert risk assessments and the public debate. Food Chem Toxicol. 2004;42(3):335–349. doi: 10.1016/j.fct.2003.10.017. [DOI] [PubMed] [Google Scholar]
- 89.Moscovici M. Present and future medical applications of microbial exopolysaccharides. Front Microbiol. 2015;6:1012. doi: 10.3389/fmicb.2015.01012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Torres F.G., Commeaux S., Troncoso O.P. Biocompatibility of bacterial cellulose based biomaterials. J Funct Biomater. 2012;3(4):864–878. doi: 10.3390/jfb3040864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Martinez-Gil M., Ramos-Gonzalez M.I., Espinosa-Urgel M. Roles of cyclic Di-GMP and the Gac system in transcriptional control of the genes coding for the Pseudomonas putida adhesins LapA and LapF. J Bacteriol. 2014;196(8):1484–1495. doi: 10.1128/JB.01287-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Clarke S.R., Foster S.J. Surface adhesins of Staphylococcus aureus. Adv Microb Physiol. 2006;51:187–224. doi: 10.1016/S0065-2911(06)51004-5. [DOI] [PubMed] [Google Scholar]
- 93.Wang X., Preston J.F., 3rd, Romeo T. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J Bacteriol. 2004;186(9):2724–2734. doi: 10.1128/JB.186.9.2724-2734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Roche F.M., Meehan M., Foster T.J. The Staphylococcus aureus surface protein SasG and its homologues promote bacterial adherence to human desquamated nasal epithelial cells. Microbiology (Reading). 2003;149(Pt 10):2759–2767. doi: 10.1099/mic.0.26412-0. [DOI] [PubMed] [Google Scholar]
- 95.O'Neill E., Pozzi C., Houston P., Humphreys H., Robinson D.A., Loughman A. A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins. FnBPA and FnBPB. J Bacteriol. 2008;190(11):3835–3850. doi: 10.1128/JB.00167-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sullan R.M., Li J.K., Crowley P.J., Brady L.J., Dufrene Y.F. Binding forces of Streptococcus mutans P1 adhesin. ACS Nano. 2015;9(2):1448–1460. doi: 10.1021/nn5058886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Paxman J.J., Lo A.W., Sullivan M.J., Panjikar S., Kuiper M., Whitten A.E. Unique structural features of a bacterial autotransporter adhesin suggest mechanisms for interaction with host macromolecules. Nat Commun. 2019;10(1):1967. doi: 10.1038/s41467-019-09814-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sherlock O., Schembri M.A., Reisner A., Klemm P. Novel roles for the AIDA adhesin from diarrheagenic Escherichia coli: cell aggregation and biofilm formation. J Bacteriol. 2004;186(23):8058–8065. doi: 10.1128/JB.186.23.8058-8065.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hobley L., Harkins C., MacPhee C.E., Stanley-Wall N.R. Giving structure to the biofilm matrix: an overview of individual strategies and emerging common themes. FEMS Microbiol Rev. 2015;39(5):649–669. doi: 10.1093/femsre/fuv015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Boyd C.D., Smith T.J., El-Kirat-Chatel S., Newell P.D., Dufrene Y.F., O'Toole G.A. Structural features of the Pseudomonas fluorescens biofilm adhesin LapA required for LapG-dependent cleavage, biofilm formation, and cell surface localization. J Bacteriol. 2014;196(15):2775–2788. doi: 10.1128/JB.01629-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Collins A.J., Pastora A.B., Smith T.J., MapA O.GA. a Second Large RTX Adhesin Conserved across the Pseudomonads, Contributes to Biofilm Formation by Pseudomonas fluorescens. J Bacteriol. 2020;202(18) doi: 10.1128/JB.00277-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Knowles T.P., Vendruscolo M., Dobson C.M. The amyloid state and its association with protein misfolding diseases. Nat Rev Mol Cell Biol. 2014;15(6):384–396. doi: 10.1038/nrm3810. [DOI] [PubMed] [Google Scholar]
- 103.Rambaran R.N., Serpell L.C. Amyloid fibrils: abnormal protein assembly. Prion. 2008;2(3):112–117. doi: 10.4161/pri.2.3.7488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shanmugam N., Baker M., Ball S.R., Steain M., Pham C.L.L., Sunde M. Microbial functional amyloids serve diverse purposes for structure, adhesion and defence. Biophys Rev. 2019;11(3):287–302. doi: 10.1007/s12551-019-00526-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pham C.L., Shanmugam N., Strange M., O'Carroll A., Brown J.W., Sierecki E. Viral M45 and necroptosis-associated proteins form heteromeric amyloid assemblies. EMBO Rep. 2019;20(2) doi: 10.15252/embr.201846518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Van Gerven N., Van der Verren S.E., Reiter D.M., Remaut H. The Role of Functional Amyloids in Bacterial Virulence. J Mol Biol. 2018;430(20):3657–3684. doi: 10.1016/j.jmb.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chapman M.R., Robinson L.S., Pinkner J.S., Roth R., Heuser J., Hammar M. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science. 2002;295(5556):851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zeng G., Vad B.S., Dueholm M.S., Christiansen G., Nilsson M., Tolker-Nielsen T. Functional bacterial amyloid increases Pseudomonas biofilm hydrophobicity and stiffness. Front Microbiol. 2015;6:1099. doi: 10.3389/fmicb.2015.01099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Caro-Astorga J., Perez-Garcia A., de Vicente A., Romero D. A genomic region involved in the formation of adhesin fibers in Bacillus cereus biofilms. Front Microbiol. 2014;5:745. doi: 10.3389/fmicb.2014.00745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.El Mammeri N., Hierrezuelo J., Tolchard J., Camara-Almiron J., Caro-Astorga J., Alvarez-Mena A. Molecular architecture of bacterial amyloids in Bacillus biofilms. FASEB J. 2019;33(11):12146–12163. doi: 10.1096/fj.201900831R. [DOI] [PubMed] [Google Scholar]
- 111.Barny M.-A. Erwinia amylovora hrpN mutants, blocked in harpin synthesis, express a reduced virulence on host plants and elicit variable hypersensitive reactions on tobacco. Eur J Plant Pathol. 1995;101(3):333–340. [Google Scholar]
- 112.Lee J., Klessig D.F., Nurnberger T. A harpin binding site in tobacco plasma membranes mediates activation of the pathogenesis-related gene HIN1 independent of extracellular calcium but dependent on mitogen-activated protein kinase activity. Plant Cell. 2001;13(5):1079–1093. doi: 10.1105/tpc.13.5.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Camara-Almiron J., Caro-Astorga J., de Vicente A., Romero D. Beyond the expected: the structural and functional diversity of bacterial amyloids. Crit Rev Microbiol. 2018;44(6):653–666. doi: 10.1080/1040841X.2018.1491527. [DOI] [PubMed] [Google Scholar]
- 114.Romero D., Aguilar C., Losick R., Kolter R. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc Natl Acad Sci U S A. 2010;107(5):2230–2234. doi: 10.1073/pnas.0910560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Romero D., Vlamakis H., Losick R., Kolter R. An accessory protein required for anchoring and assembly of amyloid fibres in B. subtilis biofilms. Mol Microbiol. 2011;80(5):1155–1168. doi: 10.1111/j.1365-2958.2011.07653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Camara-Almiron J., Navarro Y., Diaz-Martinez L., Magno-Perez-Bryan M.C., Molina-Santiago C., Pearson J.R. Dual functionality of the amyloid protein TasA in Bacillus physiology and fitness on the phylloplane. Nat Commun. 2020;11(1):1859. doi: 10.1038/s41467-020-15758-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Diehl A., Roske Y., Ball L., Chowdhury A., Hiller M., Moliere N. Structural changes of TasA in biofilm formation of Bacillus subtilis. PNAS. 2018;115(13):3237–3242. doi: 10.1073/pnas.1718102115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Terra R., Stanley-Wall N.R., Cao G., Lazazzera B.A. Identification of Bacillus subtilis SipW as a bifunctional signal peptidase that controls surface-adhered biofilm formation. J Bacteriol. 2012;194(11):2781–2790. doi: 10.1128/JB.06780-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kobayashi K., Iwano M. BslA(YuaB) forms a hydrophobic layer on the surface of Bacillus subtilis biofilms. Mol Microbiol. 2012;85(1):51–66. doi: 10.1111/j.1365-2958.2012.08094.x. [DOI] [PubMed] [Google Scholar]
- 120.Hobley L., Ostrowski A., Rao F.V., Bromley K.M., Porter M., Prescott A.R. BslA is a self-assembling bacterial hydrophobin that coats the Bacillus subtilis biofilm. PNAS. 2013;110(33):13600–13605. doi: 10.1073/pnas.1306390110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wessels J.G. Hydrophobins: proteins that change the nature of the fungal surface. Adv Microb Physiol. 1997;38:1–45. doi: 10.1016/s0065-2911(08)60154-x. [DOI] [PubMed] [Google Scholar]
- 122.Valsecchi I., Dupres V., Stephen-Victor E., Guijarro J.I., Gibbons J., Beau R. Role of Hydrophobins in Aspergillus fumigatus. J Fungi (Basel) 2017;4(1) doi: 10.3390/jof4010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Quarantin A., Hadeler B., Kroger C., Schafer W., Favaron F., Sella L. Different Hydrophobins of Fusarium graminearum Are Involved in Hyphal Growth, Attachment, Water-Air Interface Penetration and Plant Infection. Front Microbiol. 2019;10:751. doi: 10.3389/fmicb.2019.00751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Stanley-Wall N.R., MacPhee C.E. Connecting the dots between bacterial biofilms and ice cream. Phys Biol. 2015;12(6) doi: 10.1088/1478-3975/12/6/063001. [DOI] [PubMed] [Google Scholar]
- 125.DeBenedictis E.P., Hamed E., Keten S. Mechanical Reinforcement of Proteins with Polymer Conjugation. ACS Nano. 2016;10(2):2259–2267. doi: 10.1021/acsnano.5b06917. [DOI] [PubMed] [Google Scholar]
- 126.Yates M.D., Strycharz-Glaven S.M., Golden J.P., Roy J., Tsoi S., Erickson J.S. Measuring conductivity of living Geobacter sulfurreducens biofilms. Nat Nanotechnol. 2016;11(11):910–913. doi: 10.1038/nnano.2016.186. [DOI] [PubMed] [Google Scholar]
- 127.Larsen P., Nielsen J.L., Dueholm M.S., Wetzel R., Otzen D., Nielsen P.H. Amyloid adhesins are abundant in natural biofilms. Environ Microbiol. 2007;9(12):3077–3090. doi: 10.1111/j.1462-2920.2007.01418.x. [DOI] [PubMed] [Google Scholar]
- 128.Erskine E., MacPhee C.E., Stanley-Wall N.R. Functional Amyloid and Other Protein Fibers in the Biofilm Matrix. J Mol Biol. 2018;430(20):3642–3656. doi: 10.1016/j.jmb.2018.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Erskine E., Morris R.J., Schor M., Earl C., Gillespie R.M.C., Bromley K.M. Formation of functional, non-amyloidogenic fibres by recombinant Bacillus subtilis TasA. Mol Microbiol. 2018;110(6):897–913. doi: 10.1111/mmi.13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Taglialegna A., Navarro S., Ventura S., Garnett J.A., Matthews S., Penades J.R. Staphylococcal Bap Proteins Build Amyloid Scaffold Biofilm Matrices in Response to Environmental Signals. PLoS Pathog. 2016;12(6) doi: 10.1371/journal.ppat.1005711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Taglialegna A., Lasa I., Valle J. Amyloid Structures as Biofilm Matrix Scaffolds. J Bacteriol. 2016;198(19):2579–2588. doi: 10.1128/JB.00122-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hauser C.A., Maurer-Stroh S., Martins I.C. Amyloid-based nanosensors and nanodevices. Chem Soc Rev. 2014;43(15):5326–5345. doi: 10.1039/c4cs00082j. [DOI] [PubMed] [Google Scholar]
- 133.Chen A.Y., Zhong C., Lu T.K. Engineering living functional materials. ACS Synth Biol. 2015;4(1):8–11. doi: 10.1021/sb500113b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Botyanszki Z., Tay P.K., Nguyen P.Q., Nussbaumer M.G., Joshi N.S. Engineered catalytic biofilms: Site-specific enzyme immobilization onto E. coli curli nanofibers. Biotechnol Bioeng. 2015;112(10):2016–2024. doi: 10.1002/bit.25638. [DOI] [PubMed] [Google Scholar]
- 135.Sarang M.C., Nerurkar A.S. Amyloid protein produced by B. cereus CR4 possesses bioflocculant activity and has potential application in microalgae harvest. Biotechnol Lett. 2020;42(1):79–91. doi: 10.1007/s10529-019-02758-3. [DOI] [PubMed] [Google Scholar]
- 136.Sutherland I.W. The biofilm matrix–an immobilized but dynamic microbial environment. Trends Microbiol. 2001;9(5):222–227. doi: 10.1016/s0966-842x(01)02012-1. [DOI] [PubMed] [Google Scholar]
- 137.Sutherland I. Biofilm exopolysaccharides: a strong and sticky framework. Microbiology (Reading). 2001;147(Pt 1):3–9. doi: 10.1099/00221287-147-1-3. [DOI] [PubMed] [Google Scholar]
- 138.Whitchurch C.B., Tolker-Nielsen T., Ragas P.C., Mattick J.S. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295(5559):1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 139.Mulcahy H., Charron-Mazenod L., Lewenza S. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog. 2008;4(11) doi: 10.1371/journal.ppat.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kassinger S.J., van Hoek M.L. Biofilm architecture: An emerging synthetic biology target. Synth Syst Biotechnol. 2020;5(1):1–10. doi: 10.1016/j.synbio.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Steinberger R.E., Holden P.A. Extracellular DNA in single- and multiple-species unsaturated biofilms. Appl Environ Microbiol. 2005;71(9):5404–5410. doi: 10.1128/AEM.71.9.5404-5410.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Allesen-Holm M., Barken K.B., Yang L., Klausen M., Webb J.S., Kjelleberg S. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol Microbiol. 2006;59(4):1114–1128. doi: 10.1111/j.1365-2958.2005.05008.x. [DOI] [PubMed] [Google Scholar]
- 143.Rice K.C., Mann E.E., Endres J.L., Weiss E.C., Cassat J.E., Smeltzer M.S. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. PNAS. 2007;104(19):8113–8118. doi: 10.1073/pnas.0610226104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Seper A., Fengler V.H., Roier S., Wolinski H., Kohlwein S.D., Bishop A.L. Extracellular nucleases and extracellular DNA play important roles in Vibrio cholerae biofilm formation. Mol Microbiol. 2011;82(4):1015–1037. doi: 10.1111/j.1365-2958.2011.07867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mann E.E., Rice K.C., Boles B.R., Endres J.L., Ranjit D., Chandramohan L. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS ONE. 2009;4(6) doi: 10.1371/journal.pone.0005822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Das T., Kutty S.K., Kumar N., Manefield M. Pyocyanin facilitates extracellular DNA binding to Pseudomonas aeruginosa influencing cell surface properties and aggregation. PLoS ONE. 2013;8(3) doi: 10.1371/journal.pone.0058299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kawarai T., Narisawa N., Yoneda S., Tsutsumi Y., Ishikawa J., Hoshino Y. Inhibition of Streptococcus mutans biofilm formation using extracts from Assam tea compared to green tea. Arch Oral Biol. 2016;68:73–82. doi: 10.1016/j.archoralbio.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 148.Novotny L.A., Amer A.O., Brockson M.E., Goodman S.D., Bakaletz L.O. Structural stability of Burkholderia cenocepacia biofilms is reliant on eDNA structure and presence of a bacterial nucleic acid binding protein. PLoS ONE. 2013;8(6) doi: 10.1371/journal.pone.0067629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang S., Liu X., Liu H., Zhang L., Guo Y., Yu S. The exopolysaccharide Psl-eDNA interaction enables the formation of a biofilm skeleton in Pseudomonas aeruginosa. Environ Microbiol Rep. 2015;7(2):330–340. doi: 10.1111/1758-2229.12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Peng N., Cai P., Mortimer M., Wu Y., Gao C., Huang Q. The exopolysaccharide-eDNA interaction modulates 3D architecture of Bacillus subtilis biofilm. BMC Microbiol. 2020;20(1):115. doi: 10.1186/s12866-020-01789-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hathroubi S., Hancock M.A., Bosse J.T., Langford P.R., Tremblay Y.D., Labrie J. Surface Polysaccharide Mutants Reveal that Absence of O Antigen Reduces Biofilm Formation of Actinobacillus pleuropneumoniae. Infect Immun. 2016;84(1):127–137. doi: 10.1128/IAI.00912-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Schilcher K., Andreoni F., Dengler Haunreiter V., Seidl K., Hasse B., Zinkernagel A.S. Modulation of Staphylococcus aureus Biofilm Matrix by Subinhibitory Concentrations of Clindamycin. Antimicrob Agents Chemother. 2016;60(10):5957–5967. doi: 10.1128/AAC.00463-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Randrianjatovo-Gbalou I., Rouquette P., Lefebvre D., Girbal-Neuhauser E., Marcato-Romain C.E. In situ analysis of Bacillus licheniformis biofilms: amyloid-like polymers and eDNA are involved in the adherence and aggregation of the extracellular matrix. J Appl Microbiol. 2017;122(5):1262–1274. doi: 10.1111/jam.13423. [DOI] [PubMed] [Google Scholar]
- 154.Schwartz K., Ganesan M., Payne D.E., Solomon M.J., Boles B.R. Extracellular DNA facilitates the formation of functional amyloids in Staphylococcus aureus biofilms. Mol Microbiol. 2016;99(1):123–134. doi: 10.1111/mmi.13219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Harmsen M., Lappann M., Knochel S., Molin S. Role of extracellular DNA during biofilm formation by Listeria monocytogenes. Appl Environ Microbiol. 2010;76(7):2271–2279. doi: 10.1128/AEM.02361-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Chiang W.C., Nilsson M., Jensen P.O., Hoiby N., Nielsen T.E., Givskov M. Extracellular DNA shields against aminoglycosides in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother. 2013;57(5):2352–2361. doi: 10.1128/AAC.00001-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Penesyan A., Gillings M., Paulsen I.T. Antibiotic discovery: combatting bacterial resistance in cells and in biofilm communities. Molecules (Basel, Switzerland). 2015;20(4):5286–5298. doi: 10.3390/molecules20045286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wnorowska U., Niemirowicz K., Myint M., Diamond S.L., Wroblewska M., Savage P.B. Bactericidal activities of cathelicidin LL-37 and select cationic lipids against the hypervirulent Pseudomonas aeruginosa strain LESB58. Antimicrob Agents Chemother. 2015;59(7):3808–3815. doi: 10.1128/AAC.00421-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Nagler M., Insam H., Pietramellara G., Ascher-Jenull J. Extracellular DNA in natural environments: features, relevance and applications. Appl Microbiol Biotechnol. 2018;102(15):6343–6356. doi: 10.1007/s00253-018-9120-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Rocco C.J., Davey M.E., Bakaletz L.O., Goodman S.D. Natural antigenic differences in the functionally equivalent extracellular DNABII proteins of bacterial biofilms provide a means for targeted biofilm therapeutics. Mol Oral Microbiol. 2017;32(2):118–130. doi: 10.1111/omi.12157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Thiyagarajan D., Das G., Ramesh A. Extracellular-DNA-Targeting Nanomaterial for Effective Elimination of Biofilm. 2016;2(9):879–887. [Google Scholar]
- 162.Laktionov P.P., Tamkovich S.N., Rykova E.Y., Bryzgunova O.E., Starikov A.V., Kuznetsova N.P. Cell-surface-bound nucleic acids: Free and cell-surface-bound nucleic acids in blood of healthy donors and breast cancer patients. Ann N Y Acad Sci. 2004;1022:221–227. doi: 10.1196/annals.1318.034. [DOI] [PubMed] [Google Scholar]
- 163.Laktionov P., Rykova E., Toni M., Spisni E., Griffoni C., Bryksin A. Knock down of cytosolic phospholipase A2: an antisense oligonucleotide having a nuclear localization binds a C-terminal motif of glyceraldehyde-3-phosphate dehydrogenase. Biochim Biophys Acta. 2004;1636(2–3):129–135. doi: 10.1016/j.bbalip.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 164.Raptis L., Menard H.A. Quantitation and characterization of plasma DNA in normals and patients with systemic lupus erythematosus. J Clin Invest. 1980;66(6):1391–1399. doi: 10.1172/JCI109992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Fan H.C., Gu W., Wang J., Blumenfeld Y.J., El-Sayed Y.Y., Quake S.R. Non-invasive prenatal measurement of the fetal genome. Nature. 2012;487(7407):320–324. doi: 10.1038/nature11251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wang C., Stanciu C.E., Ehrhardt C.J., Yadavalli V.K. Nanoscale characterization of forensically relevant epithelial cells and surface associated extracellular DNA. Forensic Sci Int. 2017;277:252–258. doi: 10.1016/j.forsciint.2017.06.019. [DOI] [PubMed] [Google Scholar]
- 167.Sworn G. Thickeners and Gelling Agents; Food Stabilisers: 2009. Xanthan Gum; pp. 325–342. [Google Scholar]
- 168.Ling C.-C. Bacterial Polysaccharides: Current Innovations and Future Trends. Edited by Matthias Ullrich. 2009;10(15):2539–2540. [Google Scholar]
- 169.Fialho A.M., Moreira L.M., Granja A.T., Popescu A.O., Hoffmann K., Sá-Correia I. Occurrence, production, and applications of gellan: current state and perspectives. Appl Microbiol Biotechnol. 2008;79(6):889. doi: 10.1007/s00253-008-1496-0. [DOI] [PubMed] [Google Scholar]
- 170.Rehm BHA. Alginates from Bacteria. Biopolymers Online.
- 171.Cheng K.C., Catchmark J.M., Demirci A. Enhanced production of bacterial cellulose by using a biofilm reactor and its material property analysis. Journal of biological engineering. 2009;3:12. doi: 10.1186/1754-1611-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Bae S.O., Sugano Y., Ohi K., Shoda M. Features of bacterial cellulose synthesis in a mutant generated by disruption of the diguanylate cyclase 1 gene of Acetobacter xylinum BPR 2001. Appl Microbiol Biotechnol. 2004;65(3):315–322. doi: 10.1007/s00253-004-1593-7. [DOI] [PubMed] [Google Scholar]
- 173.Rehm B.H.A. Bacterial polymers: biosynthesis, modifications and applications. Nat Rev Microbiol. 2010;8(8):578–592. doi: 10.1038/nrmicro2354. [DOI] [PubMed] [Google Scholar]
- 174.de Oliveira M.R., da Silva R.S.S.F., Buzato J.B., Celligoi M.A.P.C. Study of levan production by Zymomonas mobilis using regional low-cost carbohydrate sources. Biochem Eng J. 2007;37(2):177–183. [Google Scholar]
- 175.Silbir S., Dagbagli S., Yegin S., Baysal T., Goksungur Y. Levan production by Zymomonas mobilis in batch and continuous fermentation systems. Carbohydr Polym. 2014;99:454–461. doi: 10.1016/j.carbpol.2013.08.031. [DOI] [PubMed] [Google Scholar]