Highlights
-
l
Performed DNA-based ddPCR to accurately monitor the MRD of AE (+) AML.
-
l
Identified quantification of the fusion gene correlates strongly with clinical prognosis.
Keywords: Droplet digital PCR, Acute myeloid leukemia, AML1-ETO fusion gene
Abstract
Relapse of childhood AML1-ETO (AE) acute myeloid leukemia is the most common cause of treatment failure. Optimized minimal residual disease monitoring methods is required to prevent relapse. In this study, we used next-generation sequencing to identify the breakpoints in the fusion gene and the DNA-based droplet digital PCR (ddPCR) method was used for dynamic monitoring of AE-DNA. The ddPCR technique provides more sensitive and precise quantitation of the AE gene during disease progression and relapse. Quantification of the AE fusion gene by ddPCR further contributes to improved prognosis. Our study provides valuable methods for dynamic surveillance of AE fusion DNA and assistance in determining the prognosis.
1. Introduction
The overall survival (OS) of childhood acute myeloid leukemia (AML) has been improved through dynamic monitoring of minimal residual disease (MRD) and optimization of treatment [1]. Treatment failure resulting from disease relapse in AML, including AML1-ETO (AE) (also known as RUNX1-RUNX1T1) (+) AML, remains a challenge [2]. Developing more sensitive MRD detection methods might improve current treatment strategies and prevent relapse.
For AML with fusion genes, the main methods of MRD detection include multiparameter flow cytometry (MFC) and real-time quantitative polymerase chain reaction (RT-qPCR) [3]. Although a high MRD level detected by MFC after induction therapy is associated with inferior prognosis [4], this technique has not been fully standardized due to the limited sensitivity and antigenic changes following treatment [5]. Compared to MFC, RT-qPCR analysis provides better sensitivity and AE transcript levels can be used to predict the risk of relapse [6,7]. However, RT-qPCR is a relatively quantitative method and relies on the gene expression level. Besides, AE transcripts can still be detected by RT-qPCR in some AE(+) AML patients with long-term remission, RT-qPCR may thus fail to predict relapse [8]. These factors limit the value of the RT-qPCR method for accurate detection of the fusion genes. Droplet digital PCR (ddPCR) is a recently developed technology that can achieve highly accurate absolute quantification. It has shown great advantages in detecting mutations [9,10]. However, the use of the DNA-based ddPCR method for monitoring MRD with fusion genes in childhood AML has not yet been reported.
Therefore, the purpose of this study was to explore whether DNA-based ddPCR could monitor MRD sensitively and predict prognosis in childhood AE(+) AML.
2. Materials and methods
2.1. Patients
Forty-nine AML patients were identified to carry AE fusion gene and fifteen AML patients were identified to carry CBFβ-MYH11 fusion gene. The pre-treatment DNA samples from 64 patients were used for targeted sequencing (TS), and 6 samples from AE(+) AML patients were simultaneously analyzed by whole-genome sequencing (WGS). DNA samples from 20 AE(+) AML patients collected at multiple time points during treatment were used for ddPCR. Detailed information is showed in Supplementary Table S1. Clinical data were collected including age, sex, white blood cells, genetic mutation results, MFC results, and RT-qPCR results of AE transcripts. Chemotherapy treatment referred to the AML-99 regimen [11]. The OS period is defined as the time from diagnosis to the date of death or the last follow-up. The progression-free survival (PFS) period is defined as the time from diagnosis to disease progression, death, or the date of the last follow-up. The median follow-up time was 52 months (range: 8 to 91 months). Samples were not excluded from any analysis in any experiments.
2.2. Cell lines
The AML cell lines were purchased from ATCC. Kasumi-1 cells were cultured in RPMI-1640 (Gibco, C11875500BT) supplemented with 20% fetal bovine serum (Fetal Bovine Serum, FBS, Biological Industries, 04-001-1ACS). THP-1 was cultured in RPMI-1640 supplemented with 10% FBS at 37 °C, 5% CO2.
2.3. DNA extraction
DNA samples from cell lines and patient derived bone marrow cells were extracted using Universal Genomic DNA KIT (CWBIO, CW2298M) according to the manufacturer's instructions. The purity of DNA was measured by spectrophotometer and quantified by Qubit fluorescence method (Qubit dsDNA BR Assay Kit, Invitrogen Q32850).
2.4. Next-generation sequencing
Next-generation sequencing, including targeted sequencing and whole-genome sequencing, were used to predict the gene fusion sites of patients. The sequencing library was constructed according to the manufacturer's instructions and sequenced by Hiseq 4000. The raw sequencing data were mapped to the human reference genome (UCSC hg19) and the Clipping reveal Structure (CREST) software was used to predict the gene fusion sites based on split reads analysis.
2.5. Fusion gene verification
Primers for fusion sites verification of 49 AE(+) patients and 15 CBFβ-MYH11(+) patients are listed in Supplementary Table S2 and Table S3. TransStart® Taq DNA Polymerase (TransGen Biotech, AP141-03) was used for PCR amplification of the fragments containing the fusion sites. The PCR reaction was performed with an initial denaturation of 94 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 60 °C for 30 s, and extension at 72 °C for 1 min, with an extension at 72 °C for 10 min. The PCR products were verified by Sanger sequencing through ABI 3730xl DNA Analyzer (Applied Biosystems, USA). Sequence was mapped to the reference human genome sequence (UCSC, hg19).
2.6. DNA-based ddPCR
BioRad QX200 DropletDigital PCR (BioRad, USA) instrument was used for ddPCR. The ddPCR was performed with different total amounts of DNA (0.5–100 ng) from Kasumi-1 cell line about correlation between the proportion of AE fusion gene copies and the total amount of DNA. Experiments of serially diluted cell suspensions were performed from DNA samples of Kasumi-1 and THP-1 cell lines with 20 ng DNA per sample. The amount of DNA samples used for ddPCR from each patient was 20 ng. The primers used for the Kasumi-1 cell line and 20 patients are shown in Supplementary Table S4. The ddPCR reaction was performed with an initial denaturation of 95 °C for 5 min, followed by 40 cycles of denaturation at 95 °C for 30 s, annealing at 60 °C for 1 min, and extension at 60 °C for 1 min, with a post cycling step of 4 °C for 5 min and 90 °C for 5 min. The numbers of droplets of each sample were over 12,000 and ddPCR for each sample was repeated three times.
2.7. Statistical analysis
Pearson chi-squared test was used for comparison between groups of categorical data. Spearman correlation analysis was used to analyze the correlation between variables. The survival rates of the patients were evaluated by the Kaplan-Meier method, the grouping thresholds were determined by the Receiver Operating Characteristic curve to calculate the area under the curve, and the comparisons between groups were determined by the Log-Rank test. A two-tailed P value less than 0.05 was considered statistically significant. The statistical data was analyzed by SPSS 22.0 software, and GraphPad Prism 8.0 software were used for graphing.
3. Results
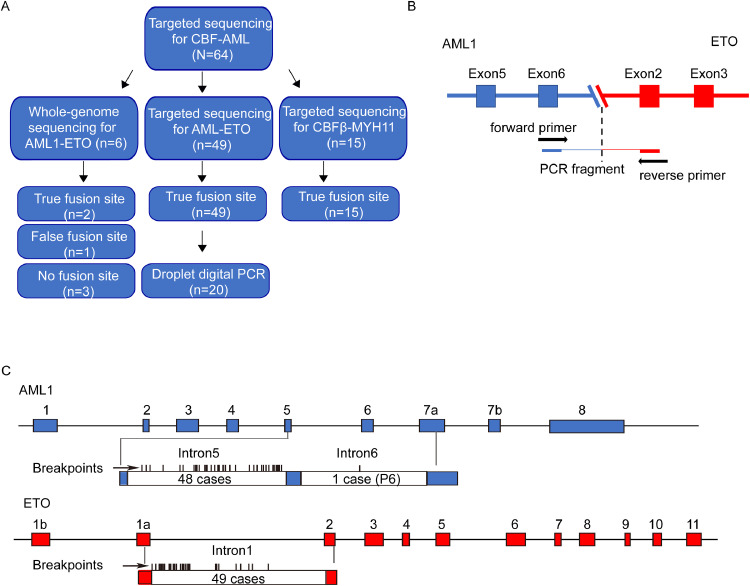
3.1. Targeted sequencing allows precise identification of personalized fusion gene breakpoints
To identify the specific breakpoints of the AE fusion gene, we performed TS of AML1 and ETO in 49 de novo AE(+) AML patient samples. Six samples were further explored by WGS (Fig. 1A, Supplementary Table S1). The results of AE fusion gene identification by TS and their validation by Sanger sequencing (Fig. 1B) are shown in Supplementary Table S2. The fusion sites were successfully identified in 49 AE(+) AML patients through TS, and TS was shown to be more accurate and less biased than WGS for fusion sites identification (Fig. 1A). The AML1 breakpoints of 48 patients were enriched in intron 5 with ETO breakpoints in intron 1 (Fig. 1C). These findings were consistent with the previously reported intron regions containing AE breakpoints [12]. The only exception was Patient 6 (P6), with the breakpoint in AML1 located in intron 6. TS was also used to predict the fusion sites of CBFβ and MYH11 in an additional 15 CBFβ-MYH11(+) AML samples; the results are shown in Supplementary Table S3.
Fig. 1.
Next-generation sequencing assists in accurate identification of the AML fusion gene site. A. Schematic diagram showing the fusion site detection by targeted sequencing and whole-genome sequencing. B. Schematic diagram showing the method for verification of the AE fusion site by Sanger sequencing. C. Results of breakpoint tracing in AML1 intron 5 and intron 6 and in ETO intron 1 for 49 patients.
3.2. DNA-based ddPCR can be used for accurate monitoring of MRD in AE(+) AML patients
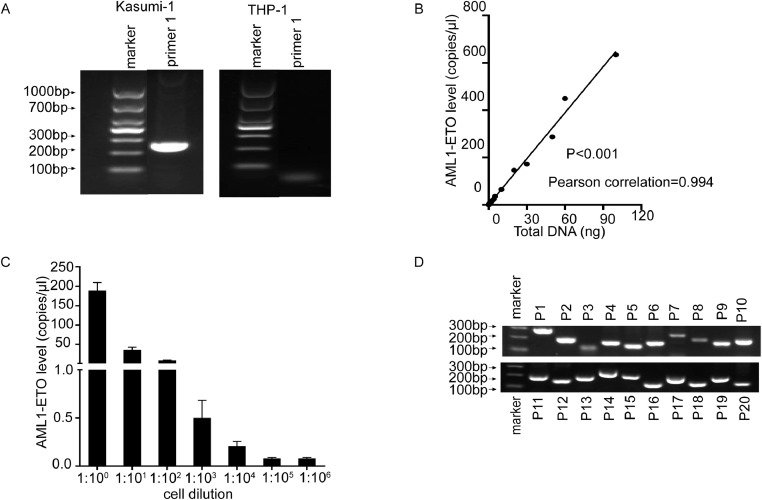
Next, we used cell lines to verify the accurate detection of fusion DNA by ddPCR (Fig. 2A, Supplementary Table S4). The results showed that the proportion of AE fusion gene copies was significantly positively correlated with the total amount of DNA (P < 0.001) (Fig. 2B). Using ddPCR, we also detected the AE fusion gene in all DNA samples from serially diluted cell suspensions (Fig. 2C), which demonstrate the applicability of DNA-based ddPCR for surveillance and quantification of AE-DNA.
Fig. 2.
Results for the preliminary exploration of method by ddPCR. A. The primer specificity was verified by conventional PCR in Kasumi-1 and THP-1 cell lines (primers 1 for ddPCR). The expected amplicon sizes for positive results were 269 bp. B. Correlation analysis of the ddPCR method using different amounts of DNA from Kasumi-1 and positive AE fusion gene counts. C. ddPCR analysis of THP-1 and Kasumi-1 cell suspensions serially diluted from 1:100 to 1:106. D. Agarose gel electrophoresis results of conventional PCR testing using personalized primers for ddPCR of 20 patients.
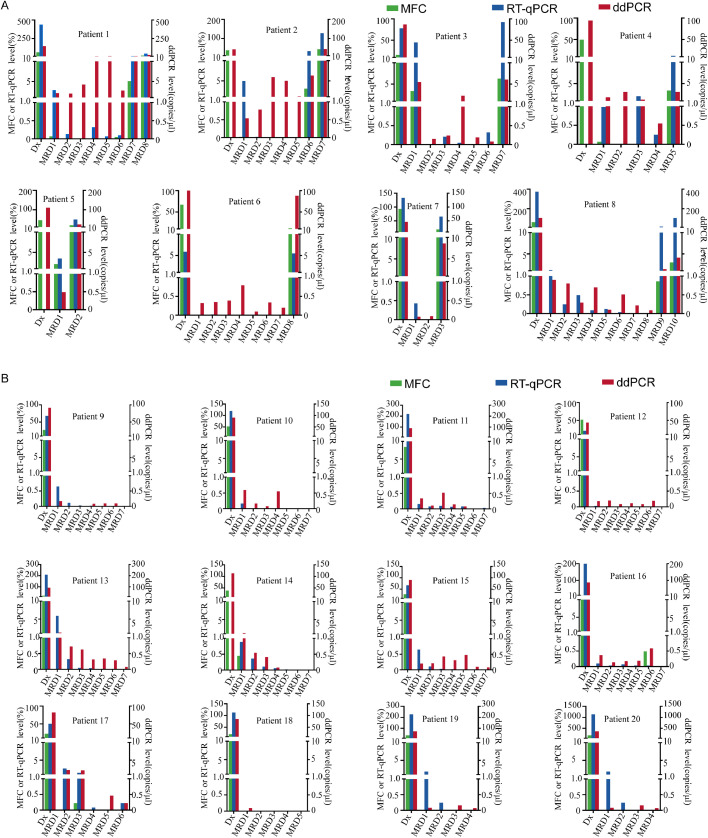
To achieve dynamic MRD monitoring of AE fusion genes at the DNA level, we performed ddPCR (Fig. 2D, Supplementary Table S4) and compared the results with MFC and RT-qPCR. The MRD detection results from three methods (Fig. 3) showed that, when the MFC-MRD of different patients at different time points were negative, the fusion gene detected at the RNA and DNA levels was positive (21.0%, 30/143). Furthermore, multiple MRD results showed negative results for RT-qPCR but positive for ddPCR at the DNA level (73.8%, 31/42)
Fig. 3.
Comparison between the multiparameter flow cytometry, real-time-qPCR, and droplet digital PCR methods for detection of the AE fusion gene in 143 samples obtained from 20 patients before and during treatment. A. Results for eight patients with disease progression after treatment. B. Results for 12 patients with sustained complete remission defined by multiparameter flow cytometry after treatment.
For eight patients (P1 to P8) with disease progression, the AE fusion gene was detected by ddPCR even when the RT-qPCR results were negative at multiple time-points. Once significant disease progression was indicated by MFC analysis, we also detected increased AE fusion gene expression using the ddPCR method (Fig. 3A). Through dynamic monitoring of 12 patients (P9 to P20) with long-term follow-up results after treatment showing continuous remission by MFC and RT-qPCR, these patients tested positive by DNA-based ddPCR at different time-points (Fig. 3B). These results indicated that DNA-based ddPCR detection of AE fusion genes is a sensitive and accurate technique for MRD monitoring of childhood AML.
3.3. DNA quantification of the fusion gene correlates with clinical prognosis
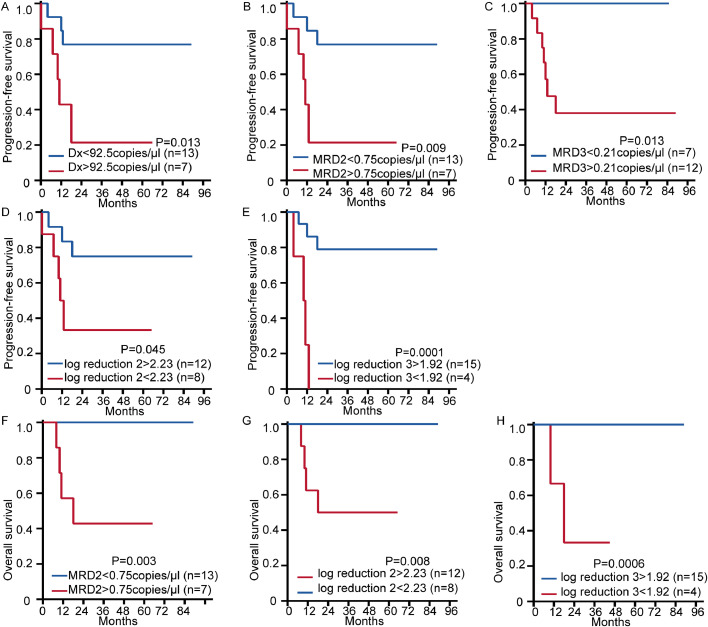
Correlation analysis showed that in addition to the KIT mutation, sex, age, and white blood cell counts were not related to the DNA quantification of AE at the time of diagnosis (P > 0.05). And we found that the groups with higher AE-DNA levels at the time of diagnosis, MRD2, and MRD3 showed poor PFS. Inferior PFS was also observed in the groups that showed less reduction in the AE-DNA levels at MRD2 or MRD3 phase (P < 0.05). Patients with higher levels of AE-DNA at MRD2 phase and less reduction at MRD2 or MRD3 also had poorer OS (P < 0.05, Table 1, and Fig. 4). These results demonstrated that quantification of the fusion gene at the DNA level by ddPCR contributed to the prognosis prediction and risk stratification, which may further improve the current therapeutic strategies for patients with the AE fusion.
Table 1.
Clinical characteristics and outcomes of 20 AML1-ETO(+) AML patients.
| Median (range) | N (%) | PFS (%) | OS (%) | |
|---|---|---|---|---|
| Age (years) | 9 (2–15) | P = 0.215 | P = 0.656 | |
| >10 | 11 (55) | 71.6±14.0 | 81.8±11.6 | |
| <10 | 9 (45) | 44.4±16.6 | 76.2±14.8 | |
| Sex | P = 0.572 | P = 0.486 | ||
| Male | 11 (55) | 55.6±16.6 | 88.9±10.5 | |
| Female | 9 (45) | 61.4±15.3 | 72.7±13.4 | |
| WBC count ( × 109/L) | 33.6 (1.8-186.8) | P = 0.024 | P = 0.198 | |
| >14.1 | 10 (50) | 37.5±16.1 | 67.5±15.5 | |
| <14.1 | 10 (50) | 80.0±12.6 | 90.0±9.5 | |
| Gene mutation | P = 0.081 | P = 0.194 | ||
| KIT mutation | 6 (30) | 33.3±19.2 | 100.0±0.0 | |
| Without KIT mutation | 14 (70) | 70.1±12.6 | 71.4±12.1 | |
| ddPCR-Dx (copies/μl) | 88.4 (43.3-146.0) | P = 0.013 | P = 0.054 | |
| >92.5 | 7 (35) | 21.4±17.8 | 57.1±18.7 | |
| <92.5 | 13 (65) | 76.9±11.7 | 91.7±8.0 | |
| ddPCR-MRD1 (copies/μl) | 1.1 (0.08-5.5) | P = 0.116 | P = 0.061 | |
| >0.43 | 11 (55) | 42.4±15.6 | 63.6±14.5 | |
| <0.43 | 9 (45) | 77.8±13.9 | 100.0±0.0 | |
| Log reduction1 | P = 0.116 | P = 0.061 | ||
| >2.4 | 9(45) | 77.8±13.9 | 100.0±0.0 | |
| <2.4 | 11(55) | 42.4±15.6 | 63.6±14.5 | |
| ddPCR-MRD2 (copies/μl) | 1.9 (0.0-5.5) | P = 0.009 | P = 0.003 | |
| >0.75 | 7(35) | 21.4±17.8 | 42.9±18.7 | |
| <0.75 | 13(65) | 76.9±11.7 | 100.0±0.0 | |
| Log reduction2 | P = 0.045 | P = 0.008 | ||
| >2.2 | 12(60) | 75.0±12.5 | 100.0±0.0 | |
| <2.2 | 8(40) | 33.3±18.0 | 50.0±17.7 | |
| ddPCR-MRD3 (coies/μl) | 1.3 (0.0-8.6) | P = 0.013 | P = 0.146 | |
| >0.21 | 12(63) | 38.1±14.7 | 72.7±13.4 | |
| <0.21 | 7(37) | 100.00±0.00 | 100.0±0.0 | |
| Log reduction3 | P < 0.001 | P = 0.007 | ||
| >1.9 | 15(79) | 79.0±10.8 | 93.3±6.4 | |
| <1.9 | 4(21) | 0.00±0.00 | 33.3±27.2 |
Fig. 4.
Kaplan–Meier curve analysis showing the correlation of survival with the amount of fusion gene DNA at different stages detected by ddPCR. Groups were divided based on the area under the curve as different cut-off values for univariate survival analysis; the cut-off values for each pair of groups are shown in Table 1. (A–C) PFS for groups with different amounts of DNA at diagnosis: (A), after the first consolidation therapy (MRD2) (B), and after the second consolidation therapy (MRD3) (C). (D–E) PFS for groups with different quantitative DNA decline speeds at MRD2 stage: (D), and MRD3 stage (E) compared with the time of diagnosis. (F–H) Overall survival for groups with different amounts of DNA: (F) after the first consolidation therapy (MRD2) (G) different quantitative DNA decline speeds at MRD2 stage (H) different quantitative DNA decline speeds at MRD3 stage. All P < 0.05 determined by log-rank test. Dx:diagnosed; Log reduction: Log10(MRD/Dx).
4. Discussion
MRD monitoring in childhood AML is essential to measure the treatment effectiveness and identify patients at high risk of relapse [2]. In the current study, we used DNA-based ddPCR to detect fusion genes for MRD monitoring in children with AE(+) AML. This technique was shown to be more sensitive and accurate than the most widely used MRD monitoring methods, such as MFC and RT-qPCR. In addition, we found that MRD status and quantification of DNA by ddPCR was a powerful predictor of prognosis or relapse risk in patients with AE(+) AML.
The techniques currently for MRD monitoring of variations in mutant genes are MFC, PCR-based methods, and NGS with limitations [13,14]. To overcome these drawbacks of these methods, we employed ddPCR to achieve accurate and sensitive detection of the AE fusion gene in childhood AML. This technology allows for the absolute quantification from only a small amount of DNA and exhibits excellent performance on detecting MRD in other diseases [15,16,17,18]. The DNA-based ddPCR monitoring method evaluated in this study was designed to target the AE fusion gene, which is not affected by the clonal evolution of gene mutations and AML progression. These findings indicated that ddPCR monitoring of the quantitative variations in fusion genes at the DNA level could assist in predicting prognosis.
Study showed that low level of AML1-ETO expression did not necessarily predict relapse [8]. When RT-qPCR showed low-positive result, it could not represent the absolute level of MRD due to the gene expression dependence of this method. The result acquired by RT-PCR thus do not reflect the proportion of AE(+) AML cells existing in the bone marrow or the proportion of cells expressing AE(+) transcript in AE(+) population, which will affect the low-level RT-qPCR results for prognosis prediction. In this study, we noticed that some DNA-based ddPCR results also showed low-level of fusion gene. Although the clinical significance of this result is not clear for now, DNA based fusion gene detection method is independent of gene expression and directly reflects the absolute number of cells from MRD because each single cell has one fusion gene template for detection, which is the major difference from RNA-based method. Besides, combined with ddPCR, this method can achieve absolute quantification of fusion gene positive cells at the molecular level. Therefore, we proposed that low-level of fusion gene DNA reflects MRD information that differ from RT-qPCR and the former is more clinically meaningful. However, studies with larger sample size are needed for further verification. For patients with positive MRD, Gemtuzumab ozogamicin or hematopoietic stem cell transplantation can be useful for treatment in addition to traditional chemotherapy, which can reduce MRD and prevent relapse [19,20].
This study has demonstrated the utility of ddPCR for dynamic monitoring of AE fusion genes at the DNA level in childhood AE(+) AML and revealed that DNA quantification of AE fusion genes correlates with the prognosis of AML patients. We provided a new perspective on MRD monitoring although this method is an essentially exploratory work due to the limitation of sample size and selection bias. This new assay needs to be properly validated in a larger prospective clinical trial
Conflict of Interest disclosure
The authors declare no potential conflicts of interest.
Acknowledgments
Acknowledgements
The authors would like to thank the patients who consented to participate in this study and the cooperation of the participating centers and their staff. This work was supported by grants from the Ministry of Science and Technology of China (2019YFA0110803), the CAMS Innovation Fund for Medical Sciences (2016-I2M-1-002 and 2019-12M-1-006), the National Nature Science Foundation of China (81770175, 81890992 and 8190011827), and the Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (2018RC310019).
Ethics approval and consent to participate
This study was conducted according to the principals of the Declaration of Helsinki and was approved by the Ethics Committee of Institute of Hematology & Blood Diseases Hospital, Chinese Academy of Medical Sciences & Peking Union Medical College. All participants were provided with written informed consent.
Author contributions
All authors had full access to all of the data in the study and are fully responsible for the content and editorial decisions for this manuscript. Xiaoyan Chen: Methodology, Investigation, Original draft preparation. Suyu Zong: Investigation, Software, Validation. Meihui Yi: Data curation, Investigation. Chao Liu: Visualization, Investigation. Bingrui Wang: Investigation. Yongjuan Duan: Investigation. Xuelian Cheng: Investigation. Min Ruan: Investigation. Li Zhang: Investigation. Yao Zou: Investigation. Yumei Chen: Investigation. Wenyu Yang: Software, Validation. Ye Guo: Supervision. Xiaojuan Chen: Supervision. Tianyuan Hu: Software, Validation, Supervision. Tao Cheng: Supervision. Xiaofan Zhu: Conceptualization, Methodology, Writing- Reviewing and Editing. Yingchi Zhang: Conceptualization, Methodology, Writing- Reviewing and Editing.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101119.
Contributor Information
Xiaofan Zhu, Email: xfzhu@ihcams.ac.cn.
Yingchi Zhang, Email: zhangyingchi@ihcams.ac.cn.
Appendix. Supplementary materials
References
- 1.Rubnitz J.E. Current management of childhood acute myeloid leukemia. Paediatr. Drugs. 2017;19:1–10. doi: 10.1007/s40272-016-0200-6. [DOI] [PubMed] [Google Scholar]
- 2.Zwaan C.M., Kolb E.A., Reinhardt D., Abrahamsson J., Adachi S., Aplenc R. Collaborative efforts driving progress in pediatric acute myeloid leukemia. J. Clin. Oncol. 2015;33:2949–2962. doi: 10.1200/JCO.2015.62.8289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Selim A.G., Moore A.S. Molecular minimal residual disease monitoring in acute myeloid leukemia: challenges and future directions. J. Mol. Diagn. 2018;20:389–397. doi: 10.1016/j.jmoldx.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 4.Loken M.R., Alonzo T.A., Pardo L., Gerbing R.B., Raimondi S.C., Hirsch B.A. Residual disease detected by multidimensional flow cytometry signifies high relapse risk in patients with de novo acute myeloid leukemia: a report from Children's Oncology Group. Blood. 2012;120:1581–1588. doi: 10.1182/blood-2012-02-408336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buldini B., Maurer-Granofszky M., Varotto E., Dworzak M.N. Flow-cytometric monitoring of minimal residual disease in pediatric patients with acute myeloid leukemia: recent advances and future strategies. Front. Pediatr. 2019;7:412. doi: 10.3389/fped.2019.00412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voso M.T., Ottone T., Lavorgna S., Venditti A., Maurillo L., Lo-Coco F. MRD in AML: the role of new techniques. Front. Oncol. 2019;9:655. doi: 10.3389/fonc.2019.00655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pigazzi M., Manara E., Buldini B., Beqiri V., Bisio V., Tregnago C. Minimal residual disease monitored after induction therapy by RQ-PCR can contribute to tailor treatment of patients with t(8;21) RUNX1-RUNX1T1 rearrangement. Haematologica. 2015;100:e99–101. doi: 10.3324/haematol.2014.114579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inaba H, Coustan-Smith E, Cao X, Pounds SB, Shurtleff SA, Wang KY. Comparative analysis of different approaches to measure treatment response in acute myeloid leukemia. J. Clin. Oncol. 2012;30(29):3625–3632. doi: 10.1200/JCO.2011.41.5323. [Comparative Study; Evaluation Study; Journal Article; Multicenter Study; Research Support, N.I.H., Extramural]2012-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flach J., Shumilov E., Joncourt R., Porret N., Novak U., Pabst T. Current concepts and future directions for hemato-oncologic diagnostics. Crit. Rev. Oncol. Hematol. 2020;151 doi: 10.1016/j.critrevonc.2020.102977. [DOI] [PubMed] [Google Scholar]
- 10.Cilloni D., Petiti J., Rosso V., Andreani G., Dragani M., Fava C. Digital PCR in myeloid malignancies: ready to replace quantitative PCR? Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20092249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsukimoto I., Tawa A., Horibe K., Tabuchi K., Kigasawa H., Tsuchida M. Risk-stratified therapy and the intensive use of cytarabine improves the outcome in childhood acute myeloid leukemia: the AML99 trial from the Japanese Childhood AML cooperative study group. J. Clin. Oncol. 2009;27:4007–4013. doi: 10.1200/JCO.2008.18.7948. [DOI] [PubMed] [Google Scholar]
- 12.Lin S., Mulloy J.C., Goyama S. RUNX1-ETO leukemia. Adv Exp Med Biol. 2017;962:151–173. doi: 10.1007/978-981-10-3233-2_11. [DOI] [PubMed] [Google Scholar]
- 13.Dix C., Lo T.H., Clark G., Abadir E. Measurable residual disease in acute myeloid leukemia using flow cytometry: a review of where we are and where we are going. J. Clin. Med. 2020;9 doi: 10.3390/jcm9061714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin J.A., O'Brien M.A., Hills R.K., Daly S.B., Wheatley K., Burnett A.K. Minimal residual disease monitoring by quantitative RT-PCR in core binding factor AML allows risk stratification and predicts relapse: results of the United Kingdom MRC AML-15 trial. Blood. 2012;120:2826–2835. doi: 10.1182/blood-2012-06-435669. [DOI] [PubMed] [Google Scholar]
- 15.Della S.I., De Novi L.A., Santoro A., Salemi D., Tam W., Cavalli M. Digital droplet PCR and next-generation sequencing refine minimal residual disease monitoring in acute lymphoblastic leukemia. Leuk Lymphoma. 2019;60:2838–2840. doi: 10.1080/10428194.2019.1607325. [DOI] [PubMed] [Google Scholar]
- 16.Wakamatsu M., Okuno Y., Murakami N., Miwata S., Kitazawa H., Narita K. Detection of subclonal SETBP1 and JAK3 mutations in juvenile myelomonocytic leukemia using droplet digital PCR. Leukemia. 2020 doi: 10.1038/s41375-020-0817-x. [DOI] [PubMed] [Google Scholar]
- 17.Machova P.K., Zizkova H., Zuna J., Motlova E., Hovorkova L., Gottschalk A. Analysis of chronic myeloid leukaemia during deep molecular response by genomic PCR: a traffic light stratification model with impact on treatment-free remission. Leukemia. 2020;34:2113–2124. doi: 10.1038/s41375-020-0882-1. [DOI] [PubMed] [Google Scholar]
- 18.Drandi D., Kubiczkova-Besse L., Ferrero S., Dani N., Passera R., Mantoan B. Minimal residual disease detection by droplet digital PCR in multiple myeloma, mantle cell lymphoma, and follicular lymphoma: a comparison with real-time PCR. J. Mol. Diagn. 2015;17:652–660. doi: 10.1016/j.jmoldx.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 19.Hu G.H., Cheng Y.F., Lu A.D., Wang Y., Zuo Y.X., Yan C.H. Allogeneic hematopoietic stem cell transplantation can improve the prognosis of high-risk pediatric t(8;21) acute myeloid leukemia in first remission based on MRD-guided treatment. BMC Cancer. 2020;20:553. doi: 10.1186/s12885-020-07043-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Hear C., Inaba H., Pounds S., Shi L., Dahl G., Bowman W.P. Gemtuzumab ozogamicin can reduce minimal residual disease in patients with childhood acute myeloid leukemia. Cancer Am. Cancer Soc. 2013;119:4036–4043. doi: 10.1002/cncr.28334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.