SUMMARY
The tumor immune microenvironment plays a critical role in cancer progression and response to immunotherapy in clear cell renal cell carcinoma (ccRCC), yet the composition and phenotypic states of immune cells in this tumor are incompletely characterized. We performed single-cell RNA and T cell receptor sequencing (scRNA-seq/scTCR-seq) on 164,722 individual cells from tumor and adjacent non-tumor tissue in patients with ccRCC across disease stages – early, locally advanced, and advanced/metastatic. Terminally exhausted CD8+ T cells were enriched in metastatic disease and were restricted in TCR diversity. Within the myeloid compartment, pro-inflammatory macrophages were decreased, and suppressive M2-like macrophages were increased in advanced disease. Terminally exhausted CD8+ T cells and M2-like macrophages co-occurred in advanced disease and expressed ligands and receptors that support T cell dysfunction and M2-like polarization. This immune dysfunction circuit is associated with a worse prognosis in external cohorts and identifies potentially targetable immune inhibitory pathways in ccRCC.
INTRODUCTION
The tumor immune microenvironment (TME) is a complex ecosystem that plays a critical role in cancer progression and response to immunotherapies (Binnewies et al., 2018). For clear cell renal cell carcinoma (ccRCC), large-scale genomic studies have provided important insights into the somatic alterations that impact tumor progression (Turajlic et al., 2018a; Turajlic et al., 2018b) and response to immune checkpoint blockade (Braun et al., 2020), but the immune cell changes that occur with advancing disease stage are incompletely characterized. Across solid tumors, infiltration by CD8+ T cells is associated with an improved prognosis, but paradoxically in ccRCC, such infiltration has been associated with a worse prognosis (Fridman et al., 2017). Characterization of the composition and cellular states of tumor-infiltrating immune cells has the potential to illuminate how the immune system might permit or even contribute to cancer progression or response to immunotherapy.
Prior efforts to characterize the ccRCC immune microenvironment using bulk transcriptomic sequencing (Clark et al., 2019; Rooney et al., 2015; Senbabaoglu et al., 2016) or large selected panels of protein markers (Chevrier et al., 2017; Giraldo et al., 2017) have advanced our understanding of how immune composition may impact clinical outcome, but were limited in their ability to completely characterize heterogeneous immune cell populations and their phenotypic states within the TME. By contrast, single-cell RNA sequencing (scRNA-seq) enables comprehensive parallel characterizations of cellular composition and transcriptional states, and has already yielded important insights into the cell-of-origin in ccRCC (Young et al., 2018). Single-cell transcriptomic studies in other tumor types have revealed not only the important role of CD8+ T cells within the TME (Zhang et al., 2018; Zheng et al., 2017), but also that of myeloid subsets, including tumor-associated macrophages (TAMs) (Zhang et al., 2020). Further, paired single-cell T cell receptor sequencing (scTCR-seq) has facilitated the tracking of individual T cell clonotypes in different tissue compartments, yielding new insights into the role of clonotype expansion in response to immune therapies (Wu et al., 2020).
Herein, we performed single-cell transcriptomic profiling of immune cells in ccRCC and adjacent non-tumor tissue from 13 patients across clinical stages to define the changes in infiltrating immune cells with advancing disease. We identified a higher proportion of exhausted CD8+ T cell and immunosuppressive M2-like macrophages in advanced disease. We detected numerous inhibitory interactions between these populations that are enriched in metastatic disease and are associated with poorer survival across several external cohorts. Overall, these data support a model of progressive immune dysfunction with advancing disease stage in ccRCC and identify potentially targetable inhibitory interactions between the infiltrating T cell and myeloid compartments.
RESULTS
Single-cell profiling of the immune microenvironment in ccRCC
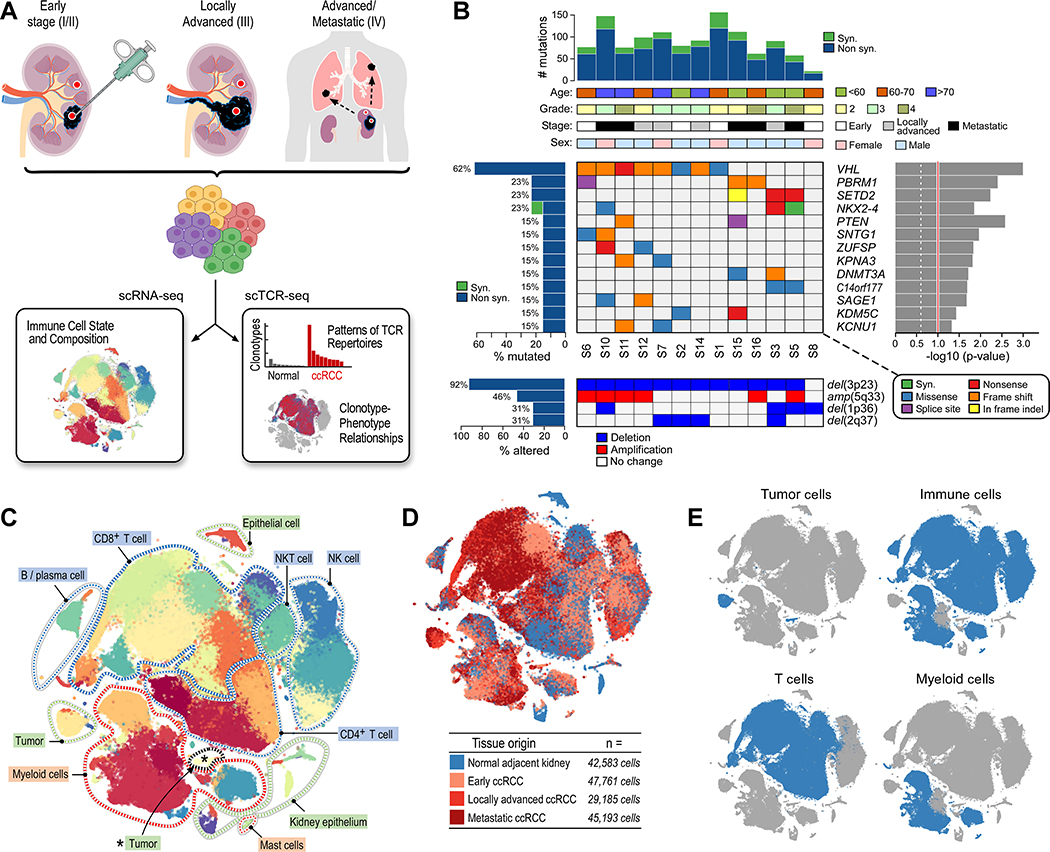
We prospectively collected fresh tumor specimens and adjacent non-tumor tissue from 13 patients with pathologically-confirmed ccRCC at early (stage I/II, n = 4), locally advanced (stage III, n = 4), and advanced/metastatic (stage IV, n = 5) stages (Table S1). ScRNA-seq and scTCR-seq was performed using the droplet-based 10x Genomics platform (Figure 1A). Whole-exome sequencing (WES) data generated from tumor and adjacent normal tissue DNA revealed the detection of genomic alterations typical for ccRCC (Cancer Genome Atlas Research, 2013), including >90% of tumors with loss of regions of chromosome 3p and >60% with mutations in VHL (Figure 1B). After initial quality control and filtering, transcriptomic data were available for 164,722 cells (Data S1–2, STAR Methods).
Figure 1.
Single-cell profiling of clear cell renal cell carcinoma. (A) Single-cell transcriptomic and T cell receptor profiling of clear cell renal cell carcinoma and adjacent normal tissue across disease stages. (B) Clinico-genomic description of the patient cohort. Top histogram, mutation rate per sample; top tracks, clinical and pathologic characteristics; Right histogram, MutSig2CV significance for recurrently mutated genes (genes with mutations in ≥2 samples displayed); Left histograms, frequency of somatic alterations; Upper heatmap, distribution of mutation events; bottom heatmap, distribution of copy number alterations. (C) scRNA-seq analysis reveals substantial transcriptional heterogeneity within known immune populations (cell populations labeled). (D) All disease stages and normal adjacent tissue are well represented in the dataset. tSNE colored by tissue origin. (E) Predominantly immune cells were analyzed by scRNA-seq. See also Figure S1, Tables S1–S2, and Data S1–2.
Graph-based clustering analysis resulted in 39 cell clusters (Figures 1C, S1A; Table S2). Using known lineage markers, we identified numerous CD4+ and CD8+ T cell populations (expressing CD3D, CD3E, and CD3G), NK populations (expressing NCAM1/CD56, NCR1/NKp46), small populations of B cells (expressing CD19, MS4A1/CD20), plasma cells (expressing CD38, SDC1/CD138, TNFRSF17/BCMA), and multiple myeloid subsets including monocytes/macrophages (expressing ITGAM/CD11b), conventional dendritic cells (expressing THBD/CD141, CLEC9A), plasmacytoid dendritic cells (expressing CLEC4C), and mast cells (expressing TPSAB1/Tryptase, KIT) (Figure S1B). Among non-immune clusters, which comprised only 5% of the cells (as a consequence of the dissociation protocol), we identified tumor cells (expressing CA9) and normal kidney epithelial cells, including proximal (expressing ALDOB) and distal (expressing UMOD) convoluted tubule cells.
Overall, the matched tumor and adjacent normal tissues originating from each disease stage were well represented in the dataset (Figure 1D). Twenty-nine of 39 clusters (74.4%) contained at least 100 cells from multiple individual samples (median of 5 individual samples per cluster), typically across the different disease stages (Figure S1C). Although whole tumor (or adjacent normal tissue) was used as input for scRNA-seq, our dissociation protocol was optimized to maximize the yield of immune cells (including T and myeloid cells) without cell sorting. Both T cells and myeloid cells were well represented in the dataset (Figure 1E).
Tumor-infiltrating T cells exhibit substantial transcriptional heterogeneity
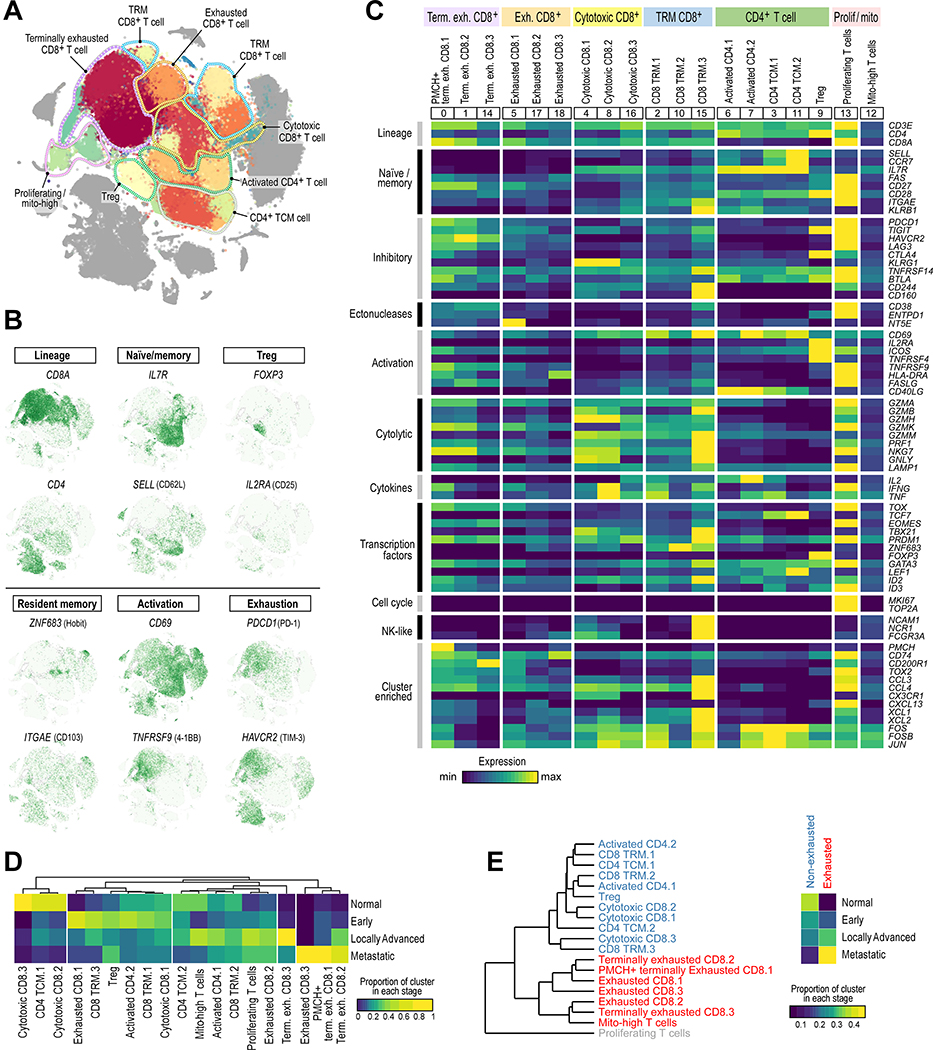
To better understand the transcriptional heterogeneity within tumor-infiltrating T cells, we first identified immune cell populations (expressing PTPRC/CD45), and among those cells, identified cell clusters expressing known T cell markers (CD3D, CD3E, CD3G). With graph-based clustering on this subset of 98,049 T cells, 19 individual T cell populations were identified, falling into 8 broad categories (Figures 2A, S2A). These included T-regulatory (Treg) cells, CD4+ T cells with an activated or central memory phenotype, CD8+ tissue-resident memory T cells, and a large population of exhausted CD8+ T cells (Figure 2B). With the exception of one small CD8+ cluster (‘Exhausted CD8.3’, comprised of only 47 cells), all other T cell clusters contained cells from multiple patients (Figure S2B).
Figure 2.
The T cell landscape of ccRCC reveals transcriptionally heterogeneous cell populations. (A) Sub-clustering of T cells demonstrates substantial transcriptional heterogeneity within both CD8+ and CD4+ T cells. Major groups of T cell populations labeled. (B) tSNE Feature plot representation of marker gene expression within individually identified T cell populations and phenotypic states. (C) Heatmap of T cell lineage and functional markers provides phenotypic information for individual T cell populations. Expression values are scaled between minimum and maximum expression for each gene across all clusters. (D) Different T cell populations are enriched in different disease stages (or in normal adjacent tissue). Heatmap representation of the proportion of each cluster from each disease stage. (E) Hierarchical clustering of T cell clusters demonstrates a close relationship between the exhausted T cell populations (red) compared to the non-exhausted population (blue). T cells that are actively proliferating cluster separately (grey). Exhausted T cell populations are enriched in more advanced disease stages, whereas non-exhausted T cells are enriched in earlier stage ccRCC and normal adjacent tissue. See also Figure S2, Tables S2–S3, and Data S1–2.
We explored the identity of each T cell population through differential gene expression analysis of each cluster compared to all other T cells (Table S3) and by assessing the expression of an extended panel of genes associated with T cell lineage and function (Figures 2C, S2C). Three CD8+ T cell clusters expressed PDCD1/PD-1, the transcriptional regulator of exhaustion TOX (Khan et al., 2019), and high levels of HAVCR2/TIM-3 and other inhibitory checkpoints, consistent with a terminally exhausted phenotype (Miller et al., 2019). This group was transcriptionally heterogeneous, with one cell cluster expressing higher levels of the prohormone PMCH (‘PMCH+ terminally exhausted CD8.1’) and another expressing higher levels of the inhibitory checkpoint CD200R1 (‘Terminally exhausted CD8.3’). A second group of three CD8+ T cell clusters also expressed PDCD1, TOX, and numerous other inhibitory checkpoints, though at a lower level, and were designated as “Exhausted CD8” cells.
A third large group of three CD8+ T cell clusters expressed numerous cytotoxic molecules (PRF1 and numerous granzymes) and higher levels of KLRG1, PRDM1/BLIMP-1, TBX21/T-bet, and ID2, consistent with a cytotoxic / effector population (Kaech and Cui, 2012). The fourth and final group consisted of three CD8+ T cell clusters that expressed ZNF683/Hobit, PRDM1, and ITGAE/CD103, consistent with a tissue-resident memory (TRM) phenotype (Mackay et al., 2016). This group displayed substantial transcriptional heterogeneity, with one cluster (‘CD8 TRM.3’) expressing high levels of cytotoxic genes (GZMB, PRF1), and markers of both CD8+ T cells (CD3E, CD8A) and NK cells (KLRB1, NCAM1/CD56, NCR1/NKp46, FCGR3A/CD16), potentially indicating some mixing of NK cells within this cluster.
Among CD4+ T cells, three broad groups were uncovered. The first group (two clusters) expressed CD69 and CD40LG/CD40L, consistent with an activated phenotype. The second group (two clusters) expressed those activation markers, but also expressed higher levels of IL7R, SELL/CD62L, and CCR7, consistent with a central memory phenotype. The final CD4+ T cell cluster expressed high levels of FOXP3 and IL2RA/CD25, consistent with a Treg population.
Finally, two additional T cell clusters were identified – one expressing high levels of cell cycle genes including MKI67/KI-67, consistent with a proliferating population, and the other with relatively low expression of most described markers and upregulation of mitochondrial genes, most consistent with a dying cell population.
After normalizing for the total number of T cells per sample, we assessed the composition of each cluster with respect to disease stage (Figure 2D). Cytotoxic CD8+ T cell clusters were enriched in normal tissue and early stage ccRCC (p < 2.2 × 10−16, two-sided Fisher’s exact), while terminally exhausted CD8+ T cells were enriched in advanced/metastatic disease (p < 2.2 × 10−16, two-sided Fisher’s exact test). CD4+ T cells were overall enriched in normal tissue and early stage ccRCC (p < 2.2 × 10−16, two-sided Fisher’s exact), though some subpopulations (i.e. ‘Activated CD4.1’ and ‘Treg’) were well represented in more advanced stages. Hierarchical clustering based on the average log-normalized gene expression profile of each cluster revealed two large groupings of cells – CD8+ T cells with an exhaustion phenotype and most other “non-exhausted” T cell populations (a proliferating T cell population, dominated by expression of cell cycle genes and a mix of all T cell clustering separately). We found that non-exhausted T cells were predominantly found in normal kidney and early stage ccRCC, while exhausted CD8+ T cells were strikingly enriched in later stage (i.e. locally advanced and metastatic) disease (p < 2.2 × 10−16, two-sided Fisher’s exact test; Figure 2E).
CD8+ T cells are progressively dysfunctional with advancing disease stage
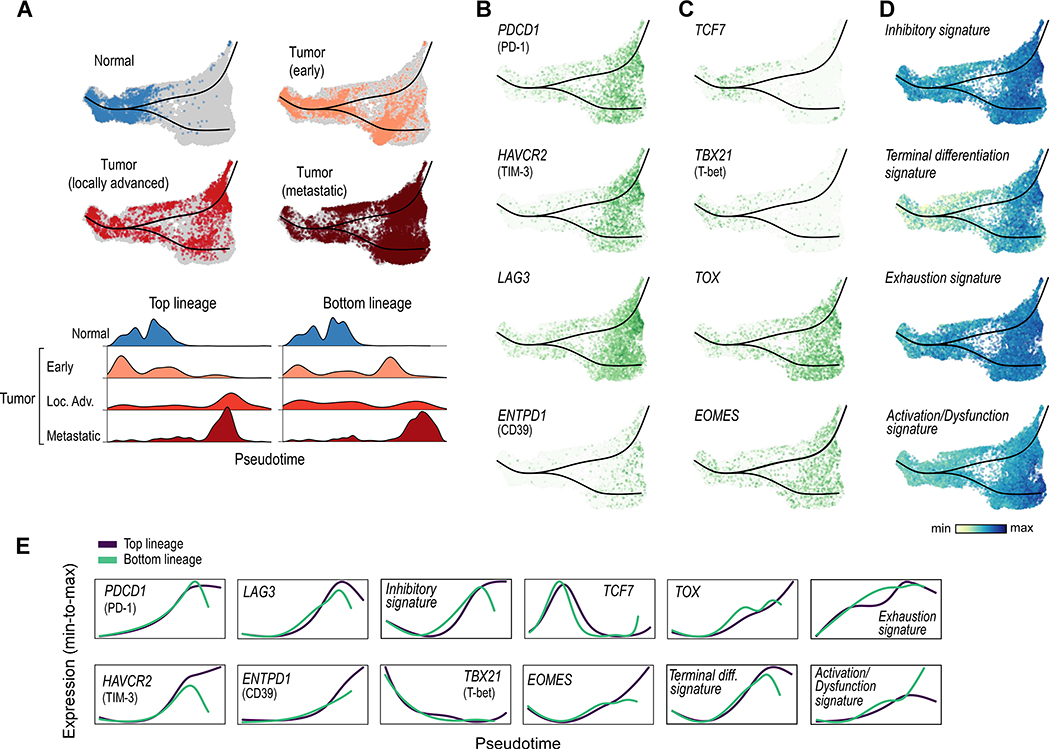
To better understand the changes in CD8+ T cells across disease stages, we used Slingshot (Street et al., 2018) to perform trajectory inference on a 3-dimensional UMAP representation of T cells expressing PTPRC/CD45, CD3D, CD3E, CD3G, CD8A, and CD8B (and not expressing CD4) and tradeSeq (Van den Berge et al., 2020) to visualize the expression of individual genes along a dynamic biologic timeline. This analysis produced a predominantly linear (left-to-right, Figure 3A) trajectory, with one branch point leading to partially overlapping top and bottom lineages.
Figure 3.
CD8+ T cell trajectory analysis reveals increased terminal exhaustion with advancing disease stage. (A) Slingshot trajectory analysis of CD8+ T cells reveals a predominantly linear trajectory (with two highly similar lineages), with CD8+ T cells from normal tissue and early tumors predominantly early in pseudotime (left), and CD8+ T cells from locally advanced and metastatic disease predominantly later in pseudotime (right). (B) Markers of T cell exhaustion are increased at the end of the trajectory. (C) Transcription factors associated with progenitor/self-renewing state (TCF7, TBX21/T-bet) are increased early in pseudotime and decreased later in pseudotime; by contrast, those associated with a terminally exhausted state (TOX, EOMES) are increased late in pseudotime. (D) Signatures of T cell inhibition, dysfunction (exhaustion), and terminal differentiation are increased late in pseudotime. (E) TradeSeq analysis demonstrates the average expression pattern of each gene and signature across pseudotime (scaled from minimum to maximum average expression for each gene or signature). See also Figure S3, Tables S2 and S4, and Data S1–2.
We first observed that the position of individual cells along the trajectory (i.e. ‘in pseudotime’) varied largely according to their disease stage. CD8+ T cells from normal adjacent tissue localized almost entirely in the early portion of the trajectory, cells from early stage tumor were found largely in the early and middle portions of the trajectory, those from locally advanced tumors were spread throughout, and cells from advanced/metastatic tumors were heavily skewed toward the ends of the trajectory (Figure 3A). Through differential gene expression analysis, numerous inhibitory checkpoints had increased expression in CD8+ T cells from metastatic tumors compared to earlier stage tumors (Figure S3A, Table S3). Their expression, including PDCD1/PD-1, HAVCR2/TIM-3, and LAG3, was substantially increased late in pseudotime (Figures 3B, 3E, S3B). The expression of ENTPD1/CD39, which has been associated with tumor-specific CD8+ T cells (Simoni et al., 2018) and with terminal exhaustion in the context of viral infection (Gupta et al., 2015), was also increased later in pseudotime.
We next examined the expression of transcription factors TCF7/TCF1 and TBX21/T-bet, which have been associated with a more progenitor or stem-like state (Miller et al., 2019; Paley et al., 2012) and better outcomes in multiple cancer types (Jansen et al., 2019; Sade-Feldman et al., 2018). Their expression levels were both increased early in the trajectory, but decreased later in pseudotime (Figures 3C, 3E, S3C). By contrast, expression of TOX and EOMES, which are associated with a more terminally exhausted phenotype, progressively increased throughout pseudotime.
To further explore the hypothesis that CD8+ T cells later in pseudotime have a more terminally exhausted phenotype, we investigated the expression of manually curated and previously described gene signatures of exhaustion and terminal differentiation along the trajectory (Figures 3D, 3E, S3D; Table S4). Concordant with the single gene results, the expression of gene signatures of CD8+ T cell inhibition, terminal differentiation (Azizi et al., 2018), exhaustion (Sade-Feldman et al., 2018), and activation/dysfunction (Singer et al., 2016) all progressively increased throughout pseudotime.
The partially overlapping top and bottom CD8+ T cell lineages differed in expression of cell stress-associated genes (e.g. JUN and FOS) and a cell stress signature (Li et al., 2019) (Figures S3E, S3F), a potential technical artifact related to tissue dissociation (O’Flanagan et al., 2019; van den Brink et al., 2017). Gene set enrichment analysis revealed that differential pathways between the top and bottom lineages were mostly associated with general cell stress and heat stress (Figures S3G, S3H), suggesting that the slight differences between these two partially overlapping branches is likely due to technical artifact rather than true biological differences. In sum, these data support a predominantly linear trajectory associated with progressive T cell exhaustion, with CD8+ T cells from more advanced disease found later in pseudotime.
Single-cell TCR analysis reveals lower clonotypic diversity in terminally exhausted T cells
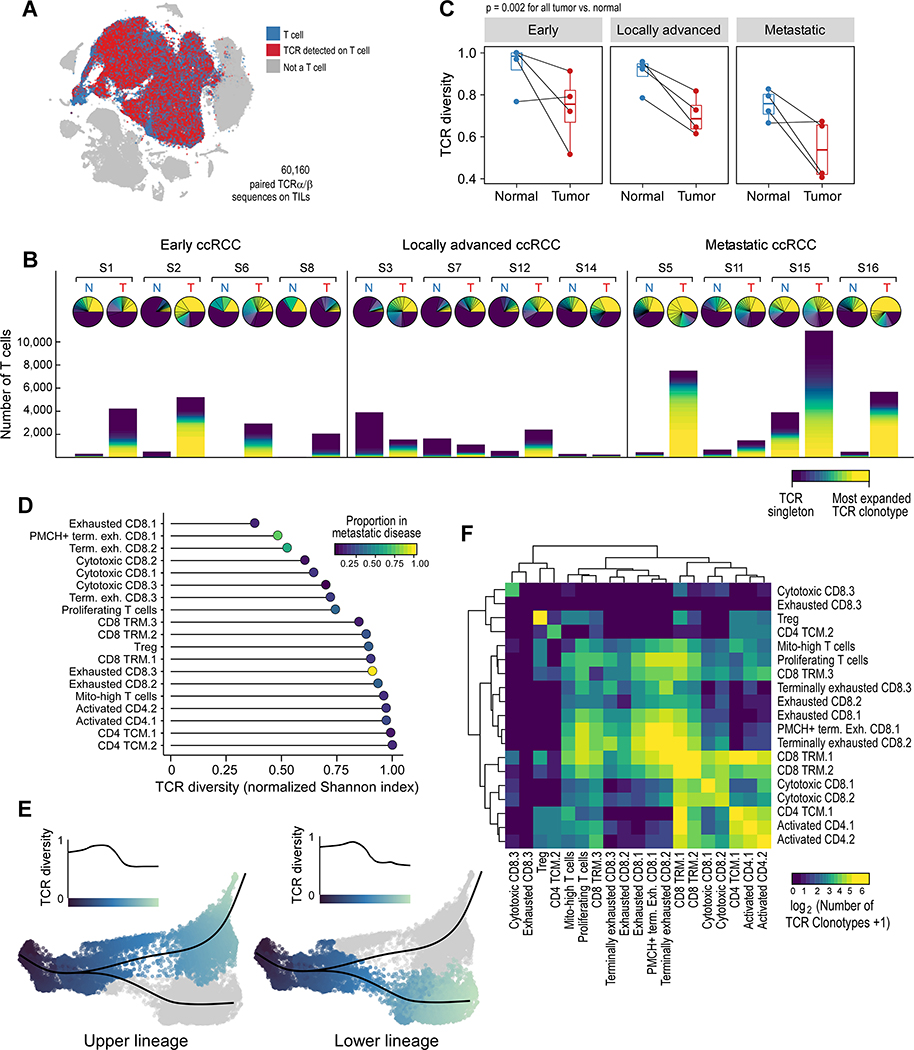
We investigated the TCR repertoire of T cells in tumor and adjacent normal tissue by performing scTCR-seq on the same cells for which we had transcriptome data, enabling a linkage between clonotype and phenotype (Figure 4A, Table S5). The TCR repertoire on a sample-by-sample basis for all tumor and normal pairs revealed striking clonotype enrichment across the 60,160 T cells thus evaluated in numerous samples, with the top clonotype alone accounting for more than 25% of infiltrating T cells in multiple samples (Samples 2, 14, 5 and 16; Figure 4B). Few clonotypes were dually expanded in tumor and adjacent normal tissue, and when it did occur, they were rarely present in similar proportions, arguing against an organized expansion process in both tissue compartments (Wu et al., 2020) (Figure S4A). Overall, TCR diversity was lower in tumors compared to adjacent normal samples (p = 0.002; Figure 4C).
Figure 4.
TCR analysis reveals lower diversity in terminally exhausted T cells. (A) Paired α-β TCRs were detected in T cells. (B) Overall representation of the TCR clonotype repertoire for each patient, in tumor and adjacent normal tissue, organized by disease stage. Upper pie chart, the relative proportion of each clonotype, with the most abundant clonotype in yellow, and “singlets” (i.e. clonotypes found in only 1 T cell) in dark purple. Lower stacked bar chart, the absolute number of each clonotype from each sample, colored by clonotype abundance. (C) Paired analysis of tumor-normal TCR diversity reveals a lower TCR diversity (more oligoclonal) in tumors compared to adjacent normal kidney (boxplot hinges, 25th to 75th percentiles; central lines, medians, whiskers, highest and lowest values no greater than 1.5x interquartile range, dots, outliers; two-sided paired Wilcoxon rank-sum test). (D) Terminally exhausted CD8+ T cells, enriched in metastatic disease, have lower TCR diversity. (E) CD8+ T cells later in pseudotime, corresponding to terminally exhausted CD8+ T cells, have lower TCR diversity. (F) Heatmap displaying the relative numbers of phenotypically “homogeneous” clonotypes (i.e. clonotypes found predominantly in 1 cluster) and phenotypically “heterogeneous” clonotypes (i.e. clonotypes predominantly distributed across multiple clusters). See also Figure S4, Tables S2 and S5, and Data S1–2.
We next explored whether distinct T cell populations exhibit differences in TCR diversity (Figure 4D). Non-Treg CD4+ T cells had the highest TCR diversity (i.e. most polyclonal), while CD8+ terminally exhausted T cells appeared to have the lowest TCR diversity. Using a local diversity method (STAR Methods), we calculated the TCR diversity along the CD8+ T cell trajectory (Figure 4E). Concordant with the cluster-based analysis, TCR diversity decreased at the trajectory termini, corresponding to the localization of the terminally exhausted CD8+ T cells in pseudotime. When we randomly permuted cells across the entire trajectory (Figure S4B), we observed that changes in TCR diversity over pseudotime remained substantially different from the null distributions generated by such permutations. Overall, these data support a lower clonotypic diversity in terminally exhausted CD8+ T cells, and are consistent with prior antigen experience.
To assess whether the specificity of infiltrating T cells would be strongly linked with the phenotypic state, we investigated whether individual T cell clonotypes were mostly found within one phenotypic cluster or distributed across multiple clusters (Figure 4F). Examples of both clonotype patterns were observed. Clonotypes in Tregs were generally homogeneous and infrequently shared across other clusters. By contrast, clonotypes were commonly shared among numerous CD4+ populations (‘CD4 TCM.1’, ‘Activated CD4.1’, and ‘Activated CD4.2’) and among exhausted CD8+ clusters (‘Exhausted CD8.1’, ‘PMCH+ terminally exhausted CD8.1’, and ‘Terminally exhausted CD8.2’). With the exception of cluster ‘CD8 TRM.1’, most heterogeneous clonotypes were shared between clusters that were closely phenotypically related, suggesting that antigen specificity is generally linked to T cell phenotype.
Not all tumor-infiltrating T cells are necessarily tumor-specific, and multiple groups have reported a large population of non-tumor-reactive “bystander” T cells within the TME, some with specificity for common viral antigens (Scheper et al., 2019; Simoni et al., 2018). Using VDJdb, a curated database of known TCR-antigen specificities (Bagaev et al., 2020), we investigated which clonotypes in our dataset were predicted to be bystander T cells with a known viral specificity. Overall, few T cells were predicted to have a viral specificity (Figure S4C, Table S5). Among exhausted and terminally exhausted CD8+ T cells, only 0–0.16% of T cells were predicted to have a viral specificity. Only one cluster that was enriched in normal tissue, ‘Cytotoxic CD8.3’, had a substantial proportion of predicted viral-specific T cells. Overall though, the proportion of viral-specific T cells was not significantly different between tumor and adjacent normal samples (Figure S4D).
Tumor-associated macrophages have decreased inflammatory cytokine production and increased anti-inflammatory M2-like features in advanced disease
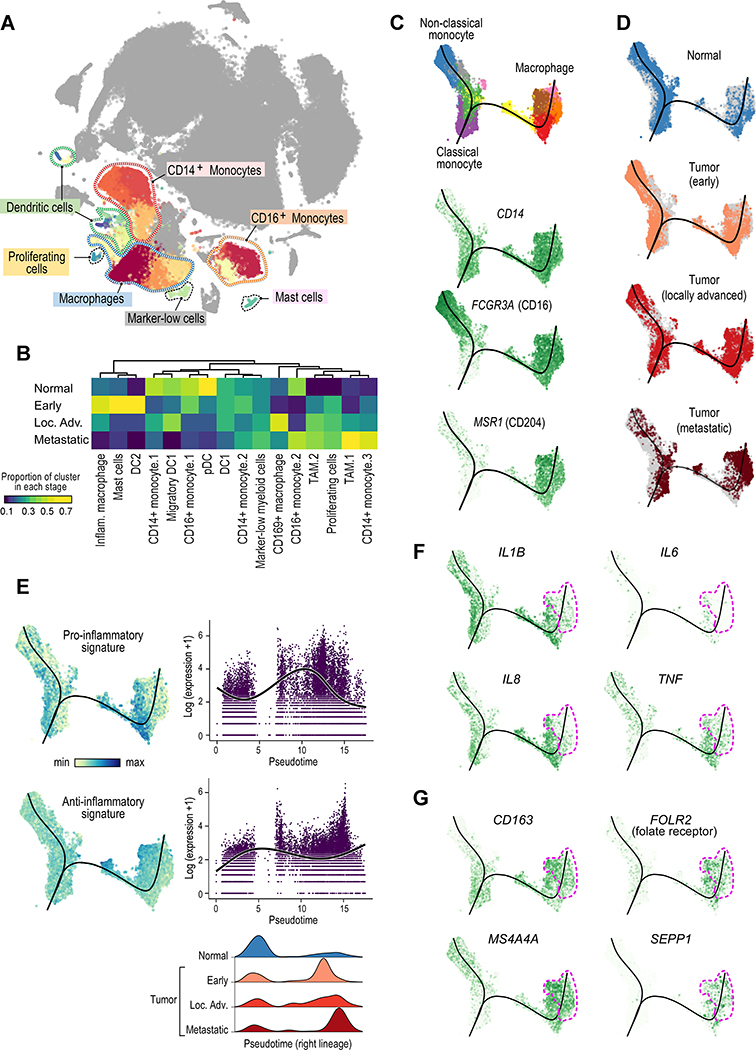
Myeloid cells play an important role in the tumor microenvironment and can contribute to tumorigenesis and treatment resistance (DeNardo and Ruffell, 2019; McDermott et al., 2018). To systematically evaluate myeloid subsets in tumor and adjacent normal tissue, we first identified immune cell populations (those expressing PTPRC/CD45), and among those cells, identified all non-lymphoid cell clusters (i.e. not expressing markers of T, B, NK, or plasma cells). We then performed graph-based clustering on this subset of 27,439 myeloid cells, which revealed 16 individual myeloid cell populations, falling into 6 broader categories (Figures 5A, S5A). Cell types were inferred by differential gene expression (Table S3) and examination of known lineage markers (Figure S5B). Classical monocytes expressed S100A8, S100A9, and CD14. Non-classical monocytes (NCMs) lacked CD14 expression but expressed FCGR3A/CD16. Macrophages expressed high levels of MHC class II molecules. One macrophage cluster (‘Inflammatory macrophage’) expressed high levels of pro-inflammatory cytokines, and a separate macrophage cluster (‘CD169+ macrophage’) expressed lower levels of most markers, but higher levels of SIGLEC1/CD169. Dendritic cells (DCs) expressed FLT3, with DC1 cells also expressing THBD/CD141 and CLEC9A, and a ‘Migratory DC1’ cluster additionally expressing CCR7. One mast cell (MC) cluster expressed TPSAB1/tryptase and KIT. The final group contained a cluster of proliferating cells of mixed lineage (high MKI67 and TOP2A expression) and a cluster with low expression of most marker genes. All myeloid clusters were composed of cells from multiple patients (Figure S5C). Three clusters (‘TAM.1’, ‘TAM.2’, and ‘CD14+ monocyte.3’) were enriched in metastatic disease (p < 2.2 × 10−16, two-sided Fisher’s exact test; Figure 5B).
Figure 5.
Analysis of the myeloid landscape reveals decreased pro-inflammatory macrophages and increased anti-inflammatory M2-like TAMs with advanced disease stage. (A) Sub-clustering of myeloid cells identifies monocytes, macrophages, dendritic cells, and mast cells. (B) Different myeloid cell populations are enriched in different disease stages (or in normal adjacent tissue). Heatmap representation of the proportion of each cluster from each disease stage. (C) Slingshot trajectory analysis of monocyte/macrophage cells recapitulates known lineage relationships, with classical monocytes (CD14+) branching into either non-classical monocytes (CD16+) or into macrophages. (D) Trajectory analysis across disease stages reveals a loss of non-classical monocytes and an increase in macrophages in advancing disease stages. Macrophages from metastatic tumors are predominantly located later in pseudotime (right lineage). (E) Macrophages from earlier stage tumors display a predominantly inflammatory phenotype; by contrast, macrophages from metastatic tumors have a predominantly anti-inflammatory phenotype. (F-G) Trajectory feature plots, with the location of macrophages from metastatic samples marker outlined in pink. F, inflammatory cytokine expression is decreased, while G, expression of M2-like macrophage markers is increased in macrophages from metastatic tumors. See also Figure S5, Tables S2 and S4, and Data S1–2.
Myeloid cells, and TAMs in particular, can have more subtle differences in cell state, existing along a continuum rather than in firmly defined discrete phenotypic states (Chevrier et al., 2017). We therefore performed trajectory inference on monocyte and macrophage populations from tumor and adjacent normal samples (Figure 5C and STAR Methods). The resulting trajectory begins with classical monocytes (expressing CD14), and then bifurcates to a non-classical monocyte population on one side (expressing FCGR3A/CD16) and a macrophage population on the other side (expressing, for example, the macrophage scavenger receptor MSR1/CD204).
There were notable differences between monocytes and macrophages from normal tissue and those from different disease stages (Figure 5D). Cells from normal tissue consisted mostly of classical and non-classical monocytes, with few macrophages. By contrast, cells from tumors were largely skewed towards the macrophage lineage (right branch of the trajectory). In advanced/metastatic disease, there were very few non-classical monocytes. Within the macrophage lineage, macrophages from earlier stage tumors were ordered earlier in pseudotime, and those from advanced tumors were skewed toward late pseudotime values.
We compared macrophages (cells on the right branch of the trajectory, after bifurcation) from advanced/metastatic tumors to those from earlier stages, and found significantly higher expression of pro-inflammatory cytokines and chemokines in earlier stage disease (Figure S5D). Macrophages from metastatic disease had higher expression of HLA class I and II genes (e.g., HLA-A, HLA-DRB1), interferon-responsive genes (e.g. IFI27), cysteine cathepsins (e.g. CTSL, CTSS), complement-related genes (e.g. C1QA, C1QB, SERPING1), and lipid transport-related genes (e.g. APOE, PLTP). We examined the expression of inflammation-related signatures within this trajectory (Azizi et al., 2018) (Table S4) and found that macrophages earlier in pseudotime (corresponding to macrophages from earlier stage tumors) had higher pro-inflammatory signature expression, and that macrophages later in pseudotime (corresponding to those from metastatic/advanced stage tumors) had higher anti-inflammatory signature expression (Figure 5E).
Regarding the expression patterns of individual genes along the trajectory, expression of pro-inflammatory cytokines IL1B, IL6, IL8, and TNF was significantly decreased in metastatic macrophages later in pseudotime (Figures 5F, S5E), concordant with our signature analysis. Conversely, the expression of numerous genes associated with anti-inflammatory M2-like macrophages were increased later in pseudotime (corresponding to macrophages from metastatic tumors), including CD163 (Jayasingam et al., 2019), FOLR2/folate receptor beta (Puig-Kroger et al., 2009), MS4A4A (Mattiola et al., 2019; Sanyal et al., 2017), and SEPP1 (Barrett et al., 2015; Solinas et al., 2010) (Figures 5G and S5F). Overall, these data suggest a decrease in pro-inflammatory macrophages and an increase in M2-like anti-inflammatory macrophages with advancing disease stage.
M2-like macrophages and terminally exhausted CD8+ T cells form inhibitory interactions in more advanced ccRCC
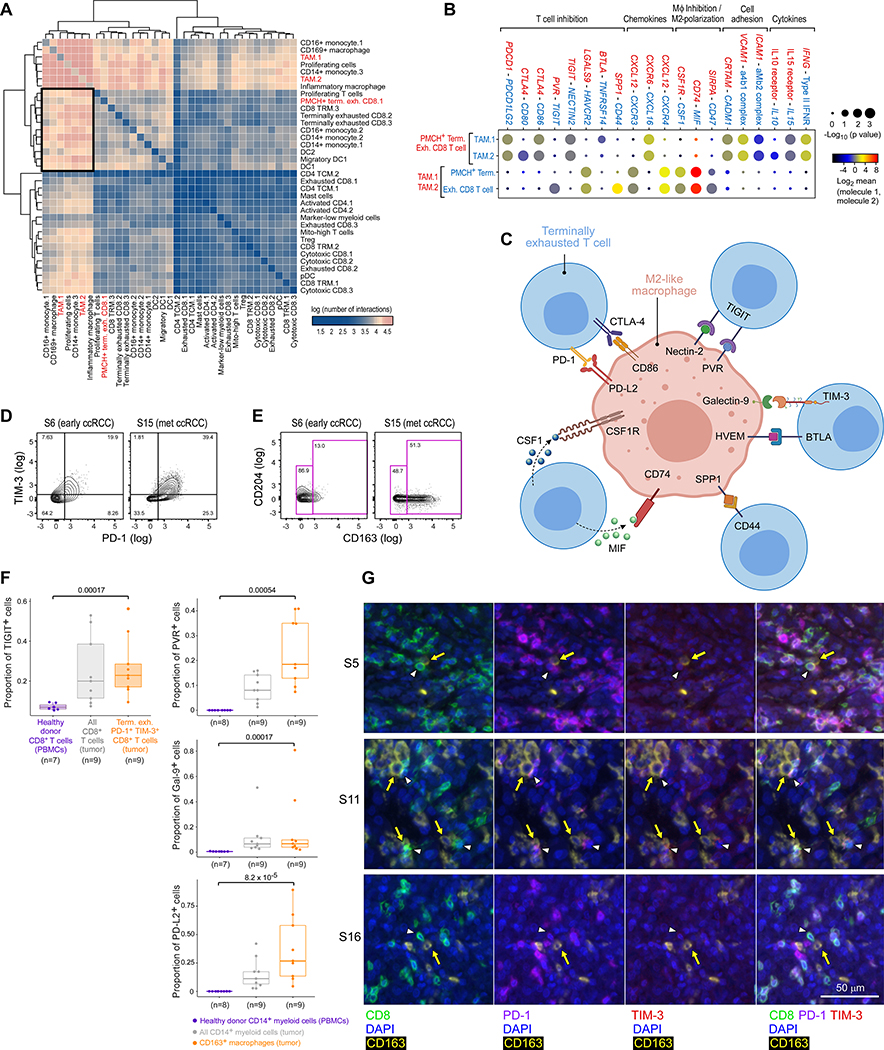
We used a public repository of ligand-receptor interactions, CellPhoneDB v2.0 (Efremova et al., 2020), to infer interactions between all T and myeloid cell populations (Figure 6A,Table S6). Numerous myeloid populations were predicted to have a large number of cell-cell interactions (p < 0.05 by CellPhoneDB permutation testing; Figure 6A, upper left), and conversely most T cell populations had relatively few predicted interactions (Figure 6A, lower right). Terminally exhausted CD8+ T cells, however, were predicted to have a large number of interactions with monocyte and macrophage populations (Figure 6A, black box). The ‘PMCH+ terminally exhausted CD8.1 cluster’, which is the most highly enriched cluster in metastatic disease, was predicted to have a high number of significant ligand-receptor interactions with the ‘TAM.1’, ‘TAM.2’, and ‘CD14+ monocyte.3’ populations, which were the most highly enriched myeloid populations in metastatic disease. We confirmed that the proportion of these populations among total T cells and total myeloid cells, respectively, was highly correlated across all samples (Figure S6A). We identified 77 significant ligand-receptor interactions between the terminally exhausted CD8+ T cell population and these three myeloid populations (Figure S6B and Table S6).
Figure 6.
M2-like macrophages form bi-directional inhibitory interactions with terminally exhausted CD8+ T cells. (A) Heatmap of the number of significant ligand-receptor interactions between each T and myeloid cell population, demonstrating a substantial number of interactions between TAMs and CD8+ terminally exhausted T cell populations (black box). (B) Highlighted significant interactions between PMCH+ terminally exhausted CD8+ T cell population (most enriched in metastatic disease) and TAM populations, showing multiple interactions that inhibit T cells and promote M2-like polarization. (C) Schema of inhibitory and M2-promoting interactions of interest. (D-E) Example flow cytometry gating for (D) PD1–1+ TIM-3+ terminally exhausted CD8+ T cells (previously gated on CD45+ CD14− CD3+ CD8+ live cells), and (E) CD163+ M2-like macrophages (previously gated on CD45+ CD3− CD14+ myeloid cells), from early (S6) and metastatic (S15) ccRCC tumors. (F) Flow cytometry analysis showing the proportion of each CD8+ T cell population that expresses TIGIT, or of each CD14+ myeloid population that expresses PVR, Galectin-9, or PD-L2, demonstrating that PD1–1+ TIM-3+ terminally exhausted CD8+ T cells (shaded orange) and CD163+ M2-like macrophages (orange) from tumors have protein-level surface expression of these receptors and ligands, compared to control cells from healthy donor PBMCs (boxplot hinges, 25th to 75th percentiles; central lines, medians, whiskers, highest and lowest values no greater than 1.5x interquartile range, dots, outliers; two-sided Wilcoxon rank-sum test for pair-wise comparison between the dysfunctional immune population of interest, orange, and the negative controls from healthy donor PBMCs, purple). (G) Multiplex immunofluorescence images of three advanced stage tumors demonstrating in situ interactions between PD-1+ TIM-3+ CD8+ terminally exhausted T cells (white arrowheads) and CD163+ M2-like macrophages (yellow arrows). PBMCs: peripheral blood mononuclear cells. See also Figure S6, Tables S2, S4, and S6, and Data S1–2.
We identified numerous biologically important interactions between that terminally exhausted CD8+ T cell population and the M2-like ‘TAM.1’ and ‘TAM.2’ populations (Figure 6B), including cytokine and chemokine signaling, and cell adhesion. Numerous predicted interactions were related to T cell inhibitory signaling or macrophage inhibition/M2-like polarization (Figure 6C, Table S6). M2-like TAMs expressed ligands for numerous T cell immune checkpoints, including PDCD1LG2/PD-L1 (binds to PDCD1/PD-1), CD80 and CD86 (binds to CTLA4), NECTIN2/CD112 and PVR/CD155 (bind to TIGIT), LGALS9/Galectin-9 (binds to HAVCR2/TIM-3), TNFRSF14/HVEM (binds to BTLA), and SPP1 (binds to CD44) (Klement et al., 2018; Mahoney et al., 2015; Qin et al., 2019). Terminally exhausted T cells also expressed genes for ligands that support M2-like polarization, including MIF (binds to CD74) and CSF1 (binds to CSF1R) (Figueiredo et al., 2018; Neubert et al., 2018). By using high-dimensional flow cytometry analysis of available tumor and normal samples (n = 15), we confirmed the protein-level expression of inferred receptors and ligands on terminally exhausted CD8+ T cells (expressing PD-1 and TIM-3; Figure 6D) and M2-like macrophages (expressing CD163; Figure 6E), respectively. Terminally exhausted CD8+ T cells expressed the inhibitory receptors TIGIT (Figure 6F) and, to a lesser extent, CTLA-4 (Figure S6C), compared to control CD8+ T cells from healthy donor peripheral blood mononuclear cells (PBMCs). M2-macrophages displayed higher expression of the cognate ligands PVR (binds TIGIT), Galectin-9 (binds TIM-3), PD-L2 (binds PD-1), and PD-L1 (binds PD-1; Figures 6F and S6C), than control CD14+ myeloid cells from healthy adult volunteers. For confirmed receptor-ligand pairs involving the receptors PD-1, TIM-3, and TIGIT, locally advanced and metastatic ccRCC had a higher proportion of M2-like macrophages expressing ligands and terminally exhausted CD8+ T cells expressing cognate receptors, compared to normal kidney and early ccRCC (Figure S6D).
To further examine spatial interactions between M2-like macrophages (expressing CD163) and terminally exhausted CD8+ T cells (expressing PD-1 and TIM-3) within the tumor microenvironment, we performed multiplex immunofluorescence (IF) of available, high-quality tumor and normal samples from the original discovery set (n = 22). The average density of M2-like macrophages and terminally exhausted T cells was higher in locally advanced and metastatic ccRCC compared to normal kidney and early ccRCC (p = 0.019; Figure S6E). Within three metastatic ccRCC tumors (for which adjacent IF images for CD163 and for CD8, PD-1, and TIM-3 could be merged), we observed numerous instances of co-localization in situ between M2-like macrophages and terminally exhausted CD8+ T cells (Figure 6G), further supporting the inferred interactions between these two dysfunctional immune populations in advanced disease.
Inhibitory interactions between M2-like macrophages and terminally exhausted CD8+ T cells in advanced disease are associated with worse overall survival
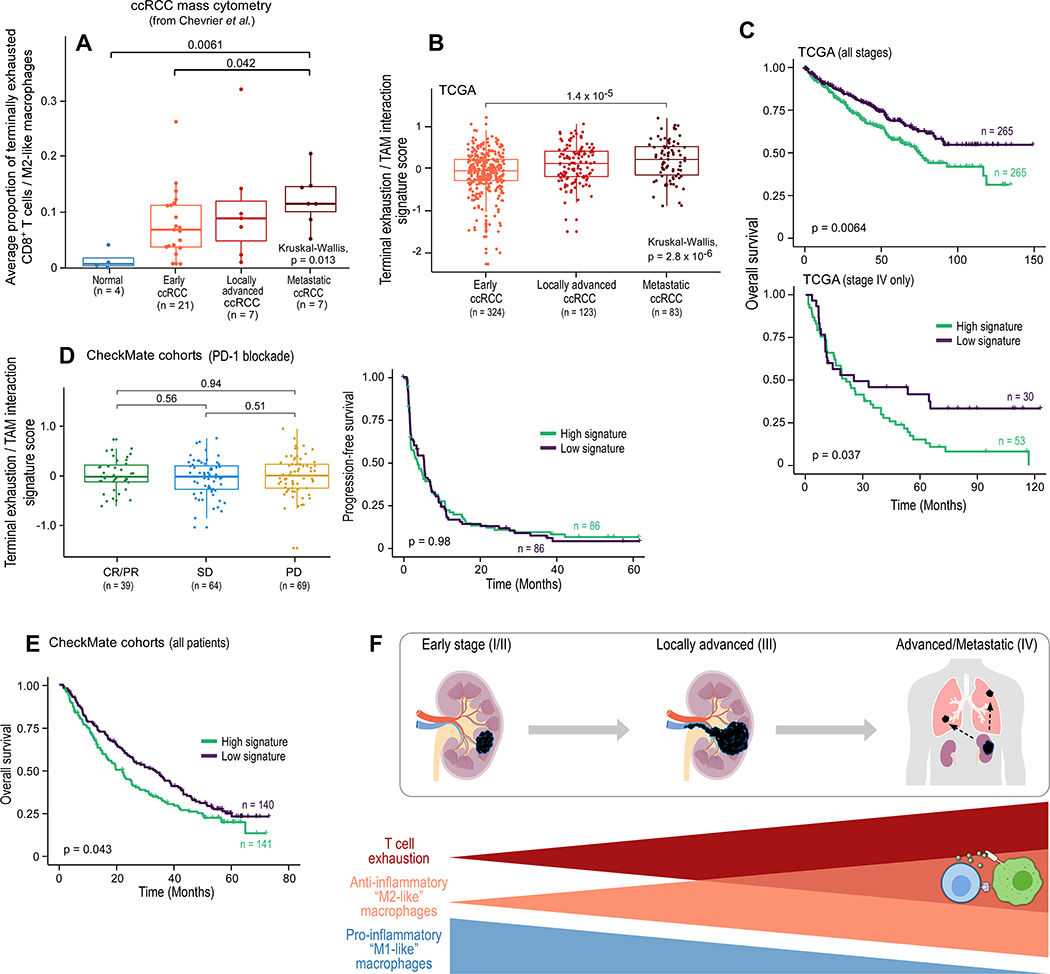
To explore these findings in a larger, independent cohort of ccRCC patients, we re-analyzed publicly available mass cytometry data from ccRCC tumor and normal kidneys (Chevrier et al., 2017). We confirmed the presence of terminally exhausted CD8+ T cells (expressing CD8, CD137/4–1BB, PD-1, and TIM-3, and lacking expression of CD4 or KI-67, matching the phenotype from our scRNA-seq analysis; Figure S7A) and M2-like macrophages (expressing high levels of CD163; Figure S7B). Moreover, concordant with our initial findings, the average proportion of terminally exhausted CD8+ T cells and M2-like macrophages increased with advancing disease stage (p = 0.013; Figure 7A).
Figure 7.
Progressive immune dysfunction with advancing disease stage in ccRCC. (A) Average proportion of terminally exhausted CD8+ T cells (as a proportion of all CD8+ T cells) and CD163+ M2-like macrophages (as a proportion of all myeloid cells) for the indicated disease stage in the external ccRCC mass cytometry cohort from Chevrier et al., 2017 (two-sided Wilcoxon rank-sum test for pair-wise comparison; Kruskal-Wallis test for global p value). (B) A gene expression signature of CD8+ T cell terminal exhaustion and interactions with M2-like TAMs for the indicated disease stage in the external TCGA KIRC cohort (two-sided Wilcoxon rank-sum test for pair-wise comparison; Kruskal-Wallis test for global p value). (C) Overall survival for the overall and advanced (stage IV) TCGA ccRCC cohorts based on high terminal exhaustion/TAM interaction signature (≥ median) versus low signature expression. (D) Signature analysis of tumors from the CheckMate cohorts of advanced ccRCC examining the response (two-sided Wilcoxon rank-sum test) or progression free survival following treatment with PD-1 blockade based on high versus low signature expression. (E) Overall survival for the entire CheckMate cohort, based on high terminal exhaustion/TAM interaction signature (≥ median) versus low signature expression. (F) Schematic representation of the results of the scRNA-seq analysis, demonstrating an increase in T cell exhaustion, a decrease in inflammatory macrophages, and an increase in M2-like macrophages with advancing disease stage. Terminally exhausted CD8+ T cells and M2-like macrophages form an immune dysfunction circuit in advanced ccRCC. For boxplots, hinges are 25th to 75th percentiles; central lines are medians, whiskers are highest and lowest values no greater than 1.5x interquartile range, and dots are outliers. For survival analyses, two-sided log-rank test. See also Figure S7.
We hypothesized that the inhibitory interactions between terminally exhausted CD8+ T cells and M2-like macrophages may constitute an immune dysfunction circuit, and by suppressing anti-tumor immune activity in advanced disease, could contribute to a worse prognosis. To investigate this hypothesis, we devised a composite expression signature (terminal exhaustion / TAM interaction signature; STAR Methods), and explored this signature in multiple external ccRCC datasets. In the ccRCC cohort (KIRC) of TCGA (Liu et al., 2018), expression of the terminal exhaustion / TAM interaction signature was increased in more advanced disease stages (p = 2.8 × 10−6; Figure 7B), consistent with the findings in our discovery cohort and the independent mass cytometry cohort (Figure 7A). High expression of this gene signature was associated with a worse overall survival across all disease stages (p = 0.0064). Importantly, this association was not observed in earlier stages of ccRCC (Figures S7C and S7D), but specifically within patients with stage IV (advanced/metastatic) disease (p = 0.037; Figure 7C). To investigate whether this dysfunctional immune interaction may affect response to therapy, we examined this signature in 281 patients with advanced/metastatic ccRCC treated with either an immune checkpoint inhibitor (PD-1 blockade) or an mTOR inhibitor (Braun et al., 2020). The terminal exhaustion / TAM interaction signature was not associated with response or progression-free survival (PFS) with either PD-1 blockade (p = 0.98 for PFS; Figure 7D) or mTOR inhibition (p = 0.3; Figure S7E), and therefore was not predictive of response to therapy. However, in agreement with the findings in the TCGA KIRC cohort, patients with baseline high expression of this signature had a worse overall survival (p = 0.043; Figure 7E), further supporting an association between this inhibitory circuit and a worse overall prognosis.
DISCUSSION
Building on prior single-cell transcriptomic efforts to understand the cell-of-origin (Young et al., 2018) and tumor microenvironment signatures (Wang et al., 2018), we comprehensively analyzed the immune compartment in varying clinical stages of disease. We observed that dysfunctional CD8+ T cells, consistent with a terminally exhausted phenotype previously observed in ccRCC (Chevrier et al., 2017; Giraldo et al., 2017; Jansen et al., 2019; Pignon et al., 2019), were more abundant in advanced disease. These cells also expressed higher levels of the transcription factor EOMES, which has been previously linked to a terminal exhaustion phenotype (Li et al., 2018; Sade-Feldman et al., 2018), but also in some cases to a more progenitor-exhausted phenotype (typically in the context of murine models of chronic viral infection) (Chen et al., 2019; Hudson et al., 2019; Im et al., 2016). Terminally exhausted CD8+ T cells were highly clonal, with some clonotypes accounting for more than 25% of total T cells in an individual sample. Such a high degree of clonotype enrichment raises important questions regarding the antigenic targets of these tumor-infiltrating T cells. These highly clonally enriched T cells were not predicted to be viral-specific, and expressed higher levels of PDCD1/PD-1 and ENTPD1/CD39, arguing that they are unlikely to be non-specific bystanders within the TME. Further work is needed to better understand the antigenic targets in ccRCC.
Through trajectory analysis of the myeloid compartment, we observed a decrease in pro-inflammatory macrophages and an increase in M2-like TAMs in advanced ccRCC, which have been associated with a worse prognosis (Hakimi et al., 2019; Xu et al., 2014). In additional to classical M2 markers (such as CD163 and MSR1/CD204), these M2-like TAMs expressed high levels of HLA class II, complement-related genes, and multiple cysteine cathepsins, which have previously been associated with tumor cell proliferation, infiltration, and resistance to cytotoxic chemotherapy (Jakos et al., 2019). Of note, macrophage phenotypes often exist along a continuum and may not fit clearly into a simple M1 vs. M2 polarization model (Azizi et al., 2018; Chevrier et al., 2017). We have therefore chosen to adopt the “M2-like” terminology (Sica and Mantovani, 2012), to indicate macrophages that, while expressing many typical features for M2 polarization, also express atypical features (such as high expression of HLA molecules and certain chemokines, such as CXCL9).
M2-like TAMs co-occurred with terminally exhausted CD8+ T cells across samples, and we therefore computationally inferred cell-cell communication between these T cell and myeloid populations. This analysis identified multiple significant cell-cell interactions forming a bi-directional circuit, whereby M2-like TAMs express ligands for multiple T cell inhibitory receptors (PD-1, CTLA-4, TIGIT, TIM-3, among others), and terminally exhausted CD8+ T cells produce factors that encourage M2-like polarization (CSF1 and MIF). High-dimensional flow cytometry and multiplex IF experimentally supported the inferred interactions between M2-like macrophages and terminally exhausted CD8+ T cells, and the increase in these dysfunctional immune populations in advanced disease stage was confirmed in multiple independent cohorts. Further, this bi-directional inhibitory circuit was also detected in a scRNA-seq analysis of metastatic ccRCC (Bi et al., submitted). Overall, these data support a model of progressive immune dysfunction with advancing disease stage (Figure 7F).
This study does have several notable limitations. Multi-region sampling would be needed to fully capture the intra-tumoral heterogeneity of geographically distinct tumor regions. Additionally, we sampled “normal” tissue near the tumor, which may have pre-neoplastic changes (or rare nests of tumor cells), and may therefore represent a more intermediate state rather than true healthy, normal tissue (Aran et al., 2017). Further, although our work focused on the immune compartment in ccRCC, tumor-immune cell interactions may also play an important role in disease progression and response to therapy. Our experimental protocol involved a cryopreservation step that can diminish more sensitive cell populations (including tumor cell populations) (Denisenko et al., 2020; Slyper et al., 2020), limiting the ability to perform such an analysis in the current study. Moreover, droplet-based scRNA-seq analysis results in a loss of spatial orientation in the TME, which is particularly important for understanding the immune microenvironment. Fully understanding these important cell-cell interactions will ultimately require spatially mapping cell populations at high resolution, now enabled by spatial transcriptomic technologies (Eng et al., 2019; Rodriques et al., 2019) and high-dimensional protein-detection methods (Goltsev et al., 2018; Lin et al., 2018). Finally, additional experimental and clinical efforts are needed to establish causality between terminally exhausted CD8+ T cells and M2-like macrophages, and to determine whether inhibiting this immune circuit would effectively reinvigorate an anti-tumor immune response.
Moving forward, the current study provides a roadmap for understanding and potentially altering immune dysfunction in ccRCC. Existing immunotherapies in advanced kidney cancer focus on improving T cell function through inhibition of the PD-1 or CTLA-4 immune checkpoints (Choueiri and Motzer, 2017; Motzer et al., 2018), but the majority of patients still do not have durable responses to these agents. This work identifies multiple other T cell inhibitory pathways and argues that the myeloid compartment plays a substantial role in the immune dysfunction seen in more advanced disease. Ultimately, the inhibitory interactions we identify between the T and myeloid cells represent potential therapeutic targets, with the goal of further improving anti-tumor immunity in advanced ccRCC.
STAR METHODS
Resource Availablity
Lead Contact:
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Catherine J. Wu (cwu@partners.org).
Materials Availability:
This study did not generate new unique reagents.
Data and Code Availability:
All raw DNA and RNA sequencing files are deposited in dbGaP under accession number phs002252.v1.p1 at https://www.ncbi.nlm.nih.gov/gap/. Processed data is provided in supplemental tables S2 and S3. All computer code used in this study is publicly available, as listed in the Key Resource Table.
Key Resources Table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Human CD3 BUV661 (clone UCHT1, 1:33 dilution) | BD Biosciences | Cat # 612964 |
| Anti-Human CD8 BUV563 (clone RPA-T8, 1:100 dilution) | BD Biosciences | Cat # 612914 |
| Rat Anti-CD11b BUV395 (clone M1/70, 1:50 dilution) | BD Biosciences | Cat # 565976 |
| Anti-Human CD14 BV750 (clone 63D3, 1:50 dilution) | BioLegend | Cat # 367136 |
| Anti-Human CD16 BUV496 (clone 3G8, 1:20 dilution) | BD Biosciences | Cat # 612944 |
| Anti-Human CD45 BUV805 (clone HI30, 1:33 dilution) | BD Biosciences | Cat # 612892 |
| Anti-Human CD68 BV711 (clone Y1/82A, 1:20 dilution) | BD Biosciences | Cat # 565594 |
| Anti-Human CD163 PE/Dazzle (clone GHI/61, 1:20 dilution) | BioLegend | Cat # 333624 |
| Anti-Human CD204 APC (clone 7C9C20, 1:20 dilution) | BioLegend | Cat # 371906 |
| Anti-Human PVR BV510 (clone SK11.4, 1:20 dilution) | BioLegend | Cat # 337634 |
| Anti-Human TIGIT BV605 (clone A15153G, 1:20 dilution) | BioLegend | Cat # 372712 |
| Anti-Human PDL1 PercpCy5.5 (clone 29E.2A3, 1:20 dilution) | BioLegend | Cat # 329738 |
| Anti-Human PDL2 BV421 (clone 24F.10C12, 1:20 dilution) | BioLegend | Cat # 329616 |
| Anti-Human Galectin-9 PE-Cy7 (clone 9M1–3, 1:20 dilution) | BioLegend | Cat # 348916 |
| Anti-Human PD1 BV650 (clone EH12.2H7, 1:20 dilution) | BioLegend | Cat # 329950 |
| Anti-Human TIM-3 BD BB515 (clone 7D3, 1:50 dilution) | Biosciences | Cat # 565568 |
| Anti-Human HLA-DR BV785 (clone L243, 1:20 dilution) | BioLegend | Cat # 307642 |
| Anti-Human CD86 BUV737 (clone FUN-1, 1:20 dilution) | BD Biosciences | Cat # 612784 |
| Anti-Human CTLA-4 PE-Cy5 (clone BNI3, 1:10 dilution) | BD Biosciences | Cat # 555854 |
| Anti-Human TCF1 PE (clone 7F11A10, 1:20 dilution) | BioLegend | Cat # 655208 |
| Anti-Human CAIX AF700 (clone 303123, 1:20 dilution) | R&D Systems | Cat # FAB2188N-100UG |
| Mouse monoclonal anti-CD8 | Agilent | C8/144B |
| Goat monoclonal anti-TIM3 | R&D biosystems | AF2365 |
| Mouse monoclonal anti-LAG3 | LifeSpan Biosciences | 17B4 |
| Mouse monoclonal anti-PD1 | Gordon Freeman laboratory, Dana Farber Cancer Institute | EH33 |
| Rabbit Monoclonal anti-CD163 | Abcam | EPR19518 |
| Biological Samples | ||
| Primary human ccRCC samples | Dana-Farber/Brigham and Women’s Hospital, Brigham and Women’s Faulkner Hospital, Beth Israel Deaconess Medical Center | N/A |
| Human adjacent normal tissue samples from ccRCC patients | Dana-Farber/Brigham and Women’s Hospital, Brigham and Women’s Faulkner Hospital, Beth Israel Deaconess Medical Center | N/A |
| Human buffy coats | Research Blood Components, LLC. | #002 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Collagenase D | Roche | Cat#11088858001 |
| Dispase (5 Units/mL) | STEMCELL | Cat#07913 |
| DNase I (2000 Units/mL) | New England Biolabs | Cat#M0303L |
| ACK lysing buffer | Gibco | Cat#A1049201 |
| Dynabeads™ MyOne™ SILANE | Life Technologies | Cat#2000048 |
| Critical Commercial Assays | ||
| MACS Dead Cell Removal Kit | Miltenyi | Cat#130–090-101 |
| Chromium Single Cell 5’ Library & Gel Bead Kit | 10x Genomics | Cat#1000006 |
| Chromium Single Cell 5’ Library Construction Kit | 10x Genomics | Cat#1000020 |
| Chromium Single Cell V(D)J Enrichment Kit, Human T cell | 10x Genomics | Cat#1000005 |
| Chromium i7 Multiplex Kit | 10x Genomics | Cat#120262 |
| Chromium Chip A Single Cell Kit | 10x Genomics | Cat#120236 |
| eBioscience Foxp3 Transcription Factor Staining Buffer Set | ThermoFisher | Cat # 00-5523-00 |
| Zombie NIR Fixable Viability Dye | BioLegend | Cat # 423106 |
| Deposited Data | ||
| Data files for scRNA-seq | This paper | dbGaP: phs002252.v1.p1 |
| Bulk RNA-seq and clinical data for TCGA KIRC cohort | TCGA | https://gdc.cancer.gov/about-data/publications/pancanatlas |
| Bulk RNA-seq and clinical data for CheckMate cohorts | Braun et al., 2020 | https://static-content.springer.com/esm/art%3A10.1038%2Fs41591-020-0839-y/MediaObjects/41591_2020_839_MOESM2_ESM.xlsx |
| VDJdb | Bagaev et al., 2020 | https://vdjdb.cdr3.net |
| Software and Algorithms | ||
| Cell Ranger v2.1.0 | 10x Genomics | http://10xgenomics.com/ |
| GATK4 v2.7.0 | McKenna et al., 2010 | https://github.com/gatk-workflows/gatk4-somatic-snvs-indels/ |
| ABSOLUTE v10 | Carter et al., 2012 | https://software.broadinstitute.org/cancer/cga/absolute |
| ISAR-GISTIC v1 | Zack et al., 2013 | https://hub.docker.com/r/arielyh/erv_pipeline |
| GISTIC2 v1 | Mermel et al., 2011 | https://hub.docker.com/r/broadgdac/tool_gistic2 |
| Seurat v3.1.4 | Butler et al., 2018 | https://satijalab.org/seurat/ |
| scran v1.12.1 | Lun et al., 2016 | https://bioconductor.org/packages/release/bioc/html/scran.html |
| scater v1.12.2 | McCarthy et al., 2017 | https://bioconductor.org/packages/release/bioc/html/scater.html |
| batchelor v1.0.1 | Haghverdi et al., 2018 | https://bioconductor.org/packages/release/bioc/html/batchelor.html |
| clustree v0.4.3 | Zappia and Oshlack, 2018 | https://cran.r-project.org/package=clustree |
| edgeR v3.26.8 | Robinson et al., 2010 | https://www.bioconductor.org/packages/release/bioc/html/edgeR.html |
| uwot v0.1.8 | Mellville et al., 2020 | https://cran.r-project.org/package=uwot |
| Slingshot v1.7.0 | Street et al. 2018 | https://bioconductor.org/packages/release/bioc/html/slingshot.html |
| tradeSeq v1.2.01 | Van den Berge et al., 2020 | https://www.bioconductor.org/packages/release/bioc/html/tradeSeq.html |
| CellPhoneDB v2.0 | Efremova et al., 2020 | https://github.com/Teichlab/cellphonedb |
| ReactomePA v1.26.0 | Yu and He, 2016 | https://bioconductor.org/packages/release/bioc/html/ReactomePA.html |
| survival v3.1–12 | Comprehensive R Archive Network (CRAN) | https://cran.r-project.org/package=survival |
| survminer v0.4.3 | Comprehensive R Archive Network (CRAN) | https://cran.r-project.org/package=survminer |
| FlowJo v10.7 | FlowJo LLC. | https://www.flowjo.com/ |
| HALO platform v3.0 | Indica Lab | https://indicalab.com/halo/ |
| FlowSOM v1.21.0 | Van Gassen et al. 2015 | http://bioconductor.org/packages/release/bioc/html/FlowSOM.html |
Experimental Model and Subject Details
Human specimens:
Institutional Review Board approval (Dana-Farber Cancer Institute) and informed consent was obtained prior to tissue acquisition and analysis. Thirteen patients (4 females and 9 males) with pathologically confirmed clear cell renal carcinoma were enrolled in this study. The patient ages ranged from 50 to 83 years, with a median age of 64.5 years. No patient had received prior systemic therapy for their cancer. Classified by AJCC version 8 (Amin et al., 2017), four patients had early stage (stage I or II), four patients had locally advanced (stage III), and five patients had advanced/metastatic disease (stage IV). Primary tumor and adjacent normal kidney samples were obtained for all patients with the exception of one patient with advanced disease (S5), for which an adrenal-based tumor and adjacent normal adrenal tissue was collected. For one patient with locally advanced disease (S12), an additional tumor specimen from the venous tumor thrombus was obtained. For one additional patient with advanced disease (S11), an additional metastatic soft tissue nodule was collected (though it was highly necrotic). Clinical characteristics are summarized in Table S1.
Method Details
Sample processing:
Fresh tumor and adjacent normal tissue from surgical resections were rinsed in HBSS (Gibco) until there was no visible blood. A section of each tissue was removed and frozen on dry ice (for downstream WES). The remaining tissue was minced by the cross-blade technique in digest media, consisting of collagenase D (0.11 Units/mL, Roche), dispase (0.56 Units/mL, STEMCELL), DNase I (50 Units/mL, New England Biolabs), calcium chloride (5mM), and HBSS. Minced tissue was incubated in digest media 37°C for 5 minutes with occasional stirring. The resulting digest was diluted with MEM media (Corning) supplemented with 10% FBS (Gibco) and penicillin/streptomycin (Gibco), and was poured over a 100μM cell strainer. Remaining tissue was pressed through the cell strainer using the plunger end of a syringe. Cells were washed with supplemented MEM media, and cells were resuspending in ACK lysis buffer (Gibco) and incubated at room temperature for 3 minutes for red blood cell lysis. Cells were washed, and then cryopreserved in a sterile-filtered solution of 10% dimethyl sulfoxide (Corning) and 90% FBS using a CoolCell freezing container (Corning), and stored in liquid nitrogen. Prior to single-cell sequencing, cells were rapidly thawed in warm DMEM (Gibco) supplemented with 10% FBS and pelleted. Dead cells were immunomagnetically depleted using the Dead Cell Removal Kit (Miltenyi), according to the manufacturer’s instructions. Cells were rinsed and resuspended in sterile-filtered PBS (Corning) with 0.04% BSA (Sigma Aldrich) on wet ice. Cell suspensions immediately proceed to single-cell library preparation.
Whole exome sequencing (WES):
DNA extraction and sequencing was performed for tumor and adjacent normal tissue through a standardized workflow at the Broad Institute Genomics Platform (Cambridge, MA), as previously described (Miao et al., 2018). Briefly, DNA extraction from fresh frozen tissue was performed using the AllPrep DNA/RNA Mini (Qiagen). WES library preparation was performed using the KAPA Library Prep kit. Hybridization and capture were performed using the Rapid Capture Enrichment Kit (Illumina). Library construction, hybridization, and captures steps were performed in an automated fashion using the Agilent Bravo liquid handling system. Libraries were quantified, and equimolar amounts of libraries were pooled and sequenced using Illumina HiSeq2500 with 2 × 76 paired-end sequencing.
Single-cell library preparation and sequencing:
Viable cells were resuspended in PBS with 0.04% BSA at a cell concentration of 1000 cells/μL. 17,000 cells were loaded onto a 10x Genomics Chromium™ instrument (10x Genomics) according to the manufacturer’s instructions. The scRNA-seq libraries were processed using Chromium™ single cell 5’ library & gel bead kit and coupled scTCR-seq libraries were obtained using Chromium™ single cell V(D)J enrichment kit (human T cell) (10x Genomics). Quality control for amplified cDNA libraries and final sequencing libraries were performed using Bioanalyzer High Sensitivity DNA Kit (Agilent). Both scRNA-seq and scTCR-seq libraries were normalized to 4nM concentration and pooled in a volume ratio of 4:1. The pooled libraries were sequenced on Illumina NovaSeq S4 platform. The sequencing parameters were: Read 1 of 150bp, Read 2 of 150bp and Index 1 of 8bp. The sequencing data were processed using the Cell Ranger pipeline (version 2.1.0 using hg19 reference, 10x Genomics), including sequencing data demultiplexing, data alignment and TCR clonotype assembly.
Quantification and Statistical Analysis
Mutation and copy number analysis:
For mutation analysis, we performed a comparative analysis of whole-exome sequencing data of tumor and matched germline samples using the Mutect2 tool in GATK4 (v2.7.0) to identify somatic mutations, insertions and deletions in patients (McKenna et al., 2010). Potential artifacts in the calls arising from orientation bias and alignment errors were removed through a set of filters (https://github.com/gatk-workflows/gatk4-somatic-snvs-indels/). The final list of somatic alterations for each patient was annotated using Funcotator. Significantly recurrent mutations in the cohort were identified using the MutSig2CV (Lawrence et al., 2014) tool (v1.3) using a nominal p-value threshold of 0.05.
We called and filtered copy number events by processing exome data for each patient using GATK4 ModelSegments (McKenna et al., 2010). We then used ABSOLUTE (Carter et al., 2012) to estimate purity, ploidy and cancer cell fractions of somatic alterations. We used ISAR-GISTIC (Zack et al., 2013) to estimate purity and ploidy-corrected segment values. Finally, the GISTIC2 (Mermel et al., 2011) tool was used to identify recurrent amplifications and deletions in the cohort.
Data alignment, pre-processing, filtering, batch correction, normalization, clustering, and doublet detection:
The raw counts data containing all sequenced barcodes was imported into R, where cells were called using Seurat v3.1.4 (Butler et al., 2018). Barcodes with fewer than 1,000 total UMIs, fewer than 500 genes expressed, or more than 15% of all UMIs originating from mitochondrial genes were filtered out, resulting in 164,722 barcodes believed to represent real cells. Genes were filtered for high expression or potential marker status by retaining those with at least 5 UMIs in at least 30 cells (6,260 genes).
Using the Bioconductor package scran v1.12.1 (Lun et al., 2016), cells were separated by a coarse, initial clustering and within-group sum factors were calculated for normalization with the scater v1.12.2 package (McCarthy et al., 2017). Multi-batch principal components analysis (PCA) was performed on the normalized data and the top 200 PCs were corrected for batch effects by the mutual nearest neighbors (MNN) method of Haghverdi, et al. (Haghverdi et al., 2018), using the batchelor v1.0.1 package. For visualization purposes, this batch-corrected embedding was summarized in two dimensions by t-distributed stochastic neighbor embedding (t-SNE) (Maaten and Hinton, 2008).
Clustering was performed on the batch-corrected PCs by the Louvain community detection method, using Seurat. This method was run multiple times over a range of possible resolution parameters (which determine the number of clusters), then the clustree v0.4.3 package (Zappia and Oshlack, 2018) was used to compare results and select an appropriate resolution based on cluster stability.
Each cell was assigned a doublet score by the “doubletCells” method of scran (Lun et al., 2016). This method simulates doublets by combining random pairs of observed cells, then embeds them in a low-dimensional space generated by running PCA on the log-normalized expression values of the original cells. Observed cells are assigned a doublet score determined by the ratio of the local density of simulated doublets to the local squared density of original cells. Four cell clusters had a higher average doublet score (data not shown), corresponding to the same four cell clusters that expressed markers from multiple cell lineages (Figure S1B). These doublet cell clusters (3.4% of the total cell population) were excluded from downstream analysis.
Differential gene expression analysis:
All differential expression testing was performed using a negative binomial generalized linear model (GLM) in the edgeR v3.26.8 package (Robinson et al., 2010). This model used the raw UMI counts as the outcome, with an offset to adjust for each cell’s total library size and an additional categorical covariate to adjust for batch effects. A likelihood ratio test was used to test binary predictors, such as one cluster versus all other clusters or one sample type versus another sample type. Results were ranked by FDR-adjusted p-values and, for one-versus-all analysis of clusters, only the top 1,000 genes were retained. Results from differential gene expression analysis are provided in Table S3.
Analysis of cell clusters across ccRCC disease stages and adjacent normal tissue:
To account for differences in cell numbers between samples, within-cluster counts were normalized by sample. For each sample, all T and myeloid cell clusters were first normalized by the total number of T or myeloid cells in that sample, respectively, to obtain the proportion of each T and myeloid cell cluster within each sample. Next, the average (arithmetic mean) value of each cluster was calculated for each ccRCC disease stage and for normal adjacent tumor, to identify enriched clusters in individual disease stages. The values for each individual cluster were then scaled between 0 and 1, providing the relative proportion of each cluster that comes from each disease stage. Heatmaps were generated using the R package pheatmap (v1.0.12, with column clustering; Figures 2D and 5B).
Marker gene heatmaps and hierarchical clustering of T cell clusters:
For the marker heatmaps (Figures 2C, S1B, and S5B), the average expression of each gene was calculated for each cluster and log-transformed. The expression value of each gene was then normalized by the maximum average gene expression in any cluster (minimum-to-maximum scale).
For hierarchical clustering of T cell populations (Figure 2E), each T cell cluster was represented by the vector of gene-wise average log(counts+1). Hierarchical clustering was performed by using the Euclidean distances between these vectors.
Trajectory analysis:
In order to be classified as a CD8+ T cell for the purposes of trajectory inference, a cell from the T cell population had to have at least one UMI for each of the genes CD3D, CD3E, CD3G, CD8A, and CD8B, and zero UMIs for CD4. UMAP dimensionality reduction (Maaten and Hinton, 2008) was performed on the MNN-corrected PCs for this reduced set of “classic” CD8+ T cells (using the uwot v0.1.8 package (Melville et al., 2020)) and a small, outlying cluster was removed. For the myeloid trajectory, clusters were manually selected from the myeloid population, representing all monocyte and macrophages populations (excluding dendritic cells, mast cells, proliferating cells, and the “marker-low” cells). UMAP was again used for dimensionality reduction and a small, outlying population was removed.
Following dimensionality reduction for both trajectories, hierarchical clustering was performed on the UMAP coordinates. Slingshot v1.7.0 (Street et al., 2018) was used to fit a minimum spanning tree (MST) to these clusters and determine the approximate trajectory structure. This piecewise linear trajectory was smoothed using simultaneous principal curves to arrive at the final trajectories and pseudotime values. For the CD8 clustering, the number of clusters was set to 7, but we found that our results were quite robust to this choice. Using 2 to 20 clusters and either hierarchical and k-means clustering (38 total cases), we re-ran this analysis and found that the resulting pseudotime values were always highly correlated with the original results (r > 0.9 in all cases, not shown). We performed a similar analysis for the monocyte/macrophage clustering.
Following trajectory inference, we tested for associations between pseudotime and individual gene expression levels by fitting negative binomial general additive models (NB-GAM) via the tradeSeq v1.2.01 package (Van den Berge et al., 2020). These models use smoothing splines to express a gene’s average expression level as a function of pseudotime. This allows us to identify genes with non-constant patterns of expression along each trajectory and, furthermore, test for differences in these patterns between lineages.
Gene set enrichment analysis:
To more systematically identify differences between the top and bottom CD8+ T cell trajectories, we performed differential gene expression analysis (using edgeR, as described earlier) between the ‘PMCH+ terminally exhausted CD8.1’ cluster (which accounted for a substantial proportion of the bottom lineage) and the ‘Terminally exhausted CD8.2’ cluster (which accounted for a substantial proportion of the top lineage). Out of the top 1000 differentially expressed genes, we utilized the genes that were relatively upregulated in the ‘PMCH+ terminally exhausted CD8.1’ cluster for downstream analysis. Using the Bioconductor package ReactomePA v1.26.0 (Yu and He, 2016), we identified enriched Reactome pathways using the “gsePathway” function (with parameters nPerm=10000; pvalueCutoff=0.2; and pAdjustMethod=”BH”). Example GSEA plots were generated using the “gseaplot” function.
TCR analysis:
Clonotype information was derived from the Cell Ranger V(D)J Annotation pipeline v2.1.0. All assembled contigs were filtered to retain only those that were assigned a raw clonotype ID and categorized as being both full length and productive. Cell barcodes were then filtered to retain only those with two contigs, categorized as one TCR alpha chain and one TCR beta chain. Each clonotype was assigned a unique identifier, consisting of the predicted amino acid sequences of the CDR3 regions of these two chains, which was used to match clonotypes across samples.
Within-sample and within-cluster diversity scores were calculated using entropy with a natural logarithm. For the estimation of diversity along trajectories, a local diversity method was developed and implemented in R. This method is similar to loess regression, in that it uses a fixed percentage of the data and a tricube weighting function centered on a given point to estimate the value of a function at that point. Instead of regression, because clonotype labels represent a categorical outcome variable, the function we calculate at each point is normalized entropy, normalized by the total number of unique clonotypes present along the lineage.
Prediction of viral-specific TCRs:
T cell receptors (TCRs) contain complementarity-determining regions (CDRs) that determine antigen specificities. We used a database of known TCR specificities established by published T cell specificity assays, VDJdb (https://vdjdb.cdr3.net) (Bagaev et al., 2020; Shugay et al., 2018), to probe the specificity of each TCR in our dataset. TCR sequences in VDJdb were matched to single-cell TCR sequences from ccRCC tumor and normal tissue samples. The VDJdb dataset was filtered for Human species clonotypes. Sample clonotypes were considered a match to a known viral-specific clonotype if the complementarity-determining regions (CDRs) matched to the same viral epitope for both the α- and β-chains of a TCR clonotype. An exact match was required for the variable (V), diversity (D), and the joining (J) regions of the β-chain, and the V-J regions of the α-chain (i.e. exact match for CDR1 and CDR2). For CDR3 we allowed up to a one amino acid substitution between the sequenced and database TCRs. We then calculated the number of viral-specific clonotypes in each T cell cluster and in each sample.
Receptor-ligand interaction analysis:
For all T cell and myeloid clusters, we identified all significant pair-wise interactions using CellPhoneDB v2.0 (Efremova et al., 2020). Using the 19 identified T cell and 16 myeloid clusters, we generated the required metadata and count files (using log-normalized counts). We performed our analysis using the Python package (https://github.com/Teichlab/cellphonedb), utilizing the “statistical method” with default parameters. The full output (means and p values) are provided in Table S6.
Isolation of human PBMCs.
Human buffy coats were purchased from Research Blood Components, LLC. PBMCs were isolated with LSM Lymphocyte Separation Medium (MP Biomedicals, 085049) and density centrifugation at room temperature at 650xg. The isolated PBMCs were cryopreserved and stored in liquid nitrogen until the time of analysis.
Flow cytometry staining.
Cryopreserved single-cell suspensions from human renal and tumor tissue (and PBMCs for use as controls) were thawed. Samples were stained for viability with Zombie NIR Fixable Viability Dye Kit (1:500 dilution, BioLegend, 423106) at 25° C for 20 minutes in the dark. Samples were then washed in PBS+2% FBS, prior to staining with a master mix of surface antibodies in PBS + 2% fetal bovine serum (FBS) at 25°C for 60 minutes in the dark. Appropriate antibody concentrations were determined previously by titration (see Key Resource Table). Samples were fixed per manufacturers’ directions for 60 min (eBioscience™, 00–5523-00) and stained for intracellular targets in 1X permeabilization buffer (eBioscience, 00–5523-00), at 25°C for 60 minutes in the dark. Samples were washed with PBS + 2% FBS and resuspended in 1X stabilizing fixative for use in flow cytometry (BD™, 338036).
Flow cytometry instrumentation and data analysis.
Sample data was acquired using a 28 Color BD FACSymphony provided by the Harvard Medical School Immunology Department’s flow cytometry facility. Flow data was analyzed using FlowJo™ v10.7 (FlowJo LLC.). Gating was determined via comparison with Fluorescence Minus One (FMO) controls (using human PBMCs), in addition to comparison with human PBMCs as a reference
Multiplex immunofluorescence assay and image analysis.
CD8, PD-1, TIM-3, and LAG-3 multiplex immunofluorescence (IF) was performed on a Bond RX Autostainer (Leica Biosystems) using the Perkin Elmer Opal tyramide signal system. The anti-CD8 antibody (1:5,000, C8/144B, mouse monoclonal antibody, Agilent) was detected using the Opal 520 fluorophore (1:150, FITC); the anti-TIM3 antibody (1:1,000, AF2365 goat monoclonal antibody, R&D Systems) was detected using the Opal 540 fluorophore (1:50, Cy3); the anti-LAG3 antibody (1/10,000, 17B4 mouse monoclonal antibody, LifeSpan Biosciences) was detected using the Opal 560 fluorophore (1:150, Texas Red); the validated anti-PD-1 antibody (1:5,000, EH33 mouse monoclonal antibody, Dr. Freeman laboratory, Dana-Farber Cancer Institute, Boston, MA) was detected using the Opal 690 fluorophore (1:50, Cy5). CD163 single IF was performed on a sequential slide using a Bond III autostainer (Leica Biosystems) using the Alexa tyramide signal system. The anti-CD163 (1:7,500, EPR19518 rabbit monoclonal antibody, Abcam) was detected using the Alexa 555 fluorophore (1:100, FITC).
The slides were imaged using the Vectra 3 automated quantitative pathology imaging system (PerkinElmer), and whole slide multispectral images were acquired at 100x magnification. At least 5 stamps of 931×698 um were selected per slide, in addition to a 2793 × 2094 um one, in areas of high immune infiltration (hotspots) using Perkin Elmer Phenochart v 1.0 software. The large stamps were synchronized between the sequential slides whenever possible. Each stamp was then acquired at 200x magnification using the Vectra 3. Inform 2.2 software was then used in order to deconvolute the multispectral images, as previously described (Pignon et al., 2019).
Digital whole slide multispectral images in .qptiff file format and hotspot deconvoluted images in .tiff format were uploaded into Indica Lab HALO platform version 3.0. For each whole slide image, the tumor area was manually annotated by a pathologist (TD). Empty spaces, necrosis, erythrocytes, and fibrous septa were excluded using the HALO platform tissue classification module. CD8 cells were phenotyped using the Indica Lab High-Plex FL v2.0 module, using DAPI-based nuclear segmentation and detection of FITC positive cells by adapting a dye cytoplasm positive threshold for each slide.
The immune cells were similarly phenotyped in the hotspots images using the 4 channels dedicated to the 4 antibodies of interest (FITC, Cy3, Texas Red, Cy5). CD163 cells were phenotyped using the Indica Lab High-Plex FL 3.1.0 using DAPI-based nuclear segmentation and detection of FITC positive cells by adapting a dye cytoplasm positive threshold for each slide. Paired large hotspot images, when available, were registered using the Indica Lab registration module and fused using the Indica Lab Serial Registration module. A unique algorithm was created for each whole slide, and each group of hotspots and its accuracy was validated through visual inspection by two pathologists (TD, SS).
Analysis of mass cytometry data.
FCS files (n = 78) for each of the two panels in the study by Chevrier et al. (2017) were downloaded from Cytobank (https://cytobank.org/). These files were already pre-processed as described in the original reference. Samples from patients without annotation of cancer stage (n = 26), patients who were not treatment-naïve (i.e. received neoadjuvant therapy; samples 2, 7, 38, 82), from a non-primary, local recurrence (sample 68), from patients who later developed metastatic recurrence (samples 34, 41, 45, 78), or from patients who died from a different malignancy (acute myeloid leukemia, sample 53) were excluded from analysis. Additionally, samples excluded in the original study (17 and 25) and duplicate samples (26b) were excluded from our analysis. Normal samples were given the stage ‘Normal’, any metastatic grade samples were annotated as stage IV, leaving a total of n = 39 samples to be processed. Stages I-II were considered ‘Early’, and stages III and IV were considered ‘Locally Advanced’ and ‘Metastatic/Advanced’, respectively.
FlowSOM v1.21.0 (Van Gassen et al., 2015) was applied in a two-step process for both panels’ data separately. For both panels, the first step was clustering using all available markers to 20 meta-clusters. For the T cell panel, CD3+ cells were selected (clusters 7, 8, 10, 11, 14, 15, 16, 18) and subclustered using FlowSOM on all markers except CD15, CD86, CD20, CD7, CD68, and CD206, which were also excluded in the original analysis of the dataset by Chevrier et al. For the TAM panel, the group of myeloid cell clusters was selected based on expression of myeloid markers such as CD14, CD68, and CD86, and lacking expression of B and T cell markers CD20 and CD3, respectively (clusters 6, 7, 9, 10, 11, 12, 17, 18), these cells were subclustered using FlowSOM on all available markers. In both cases, a resolution of 25 meta-clusters was selected, and marker distributions for each cluster were inspected to determine the populations of interest (for terminally exhausted CD8+ T cells, expression of CD8, PD-1, TIM-3, CD137 and without CD4 or Ki67 expression; for M2-like macrophages, high expression of CD163). The analysis was carried out using R v4.0.2.
Statistical testing was carried out using Kruskal-Wallis and Wilcoxon tests for the per sample average cluster proportions of the two cell types of interest within the CD8+ T cell and myeloid subsets, respectively. Visualizations were created using uwot v0.1.8, ggplot2 v3.3.2, ggridges v0.5.2, pheatmap v1.0.12, and using functions derived from the work of Nowicka et al. (Nowicka et al., 2017).
Terminally exhausted/TAM interaction signature analysis in TCGA and CheckMate cohorts:
A terminally exhausted/TAM interaction signature was generated by combining genes that were significantly upregulated in the ‘PMCH+ terminally exhausted CD8.1’ cluster (at least 2-fold higher expression than other T cell clusters) with the genes that comprise the components of the 77 significant ligand-receptor interactions identified between this same T cell cluster, and the ‘TAM.1’, ‘TAM.2’, and ‘CD14+ monocyte.3’ clusters (also enriched in metastatic disease). The genes comprising this signature are reported in Table S4. Bulk RNA-seq and clinical data were obtained for the TCGA ccRCC cohort (KIRC, downloaded from: https://gdc.cancer.gov/about-data/publications/pancanatlas) (Cancer Genome Atlas Research, 2013; Liu et al., 2018), and for the CheckMate cohorts of patients with advanced ccRCC treated with either PD-1 blockade or mTOR inhibition as part of prospective clinical trials (Braun et al., 2020). For each cohort, the expression values for each gene was converted to a z-score (centered at 0), to ensure that highly expressed genes did not overly influence the signature score compared to more lowly expressed genes. For each sample, a signature score was calculated by taking an average (arithmetic mean) of the z-score expression values for all genes in the Terminally exhausted/TAM interaction signature.
For the TCGA KIRC cohort, differences in expression of the signature score between clinical stages were assessed using the Kruskal-Wallis test to compare multiple groups, and a two-sided Wilcoxon rank-sum test to perform pairwise comparisons (R package stats v3.5.2). For the CheckMate cohorts, pair-wise differences in signature score expression between clinical response groups (complete response/partial response [CR/PR], stable disease [SD], and progressive disease [PD]) were assessed using a two-sided Wilcoxon rank-sum test. All comparisons used an alpha level of 0.05.
The association between signature score (high, or greater than or equal to the median signature score for that cohort, vs. low, or less than the median signature score for the cohort) and progression-free (PFS) and overall survival (OS) time was assessed by Kaplan-Meier analysis. We used the two-sided log-rank test (at significance level of 0.05) to detect significant differences in PFS and OS between the signature-high and signature-low groups. All survival analyses were performed using the R packages survival v3.1–12 and survminer v0.4.3.
Supplementary Material
Table S1. Patient-level clinical and somatic alteration data table, related to Figure 1.
Table S2. Cell metadata, including sample origin, mapping of each cell to its designated cluster, and annotation of whether it was included in the CD8+ T cell or monocyte/macrophage trajectory analyses, related to Figures 1–6, and S1–6.
Table S3. Differential gene expression analysis, including top 1000 differentially expressed genes for each cluster (compared to all other clusters) for graph-based clustering of all cells, T cells only, and myeloid cells only, related to Figures 2C, S1B, S3A, S5B, and S5D.
Table S4. Gene signatures used in this study, related to Figures 3D–F, 5E, 6D–G, S3D–E, and S6C.
Table S5. TCR clonotype information for each T cell, including the cell barcode (enabling mapping back to gene expression data in Table S2), TCR clonotype, and annotation of whether it was predicted to be viral-specific, related to Figures 4 and S4.
Table S6. Ligand-receptor means and p-values for all T and myeloid cell interactions (from CellPhoneDB v2.0), related to Figures 6A–C, and S6B.
Data S1. ScRNA-seq raw count matrix (part 1 of 2), after quality control filtering, with genes as rows and cell barcodes as columns, related to Figure 1–6, S1–3, and S5.
Data S2. ScRNA-seq raw count matrix (part 2 of 2), after quality control filtering, with genes as rows and cell barcodes as columns, related to Figure 1–6, S1–3, and S5.
Acknowledgements:
We are grateful to P. Bachireddy, D. Neuberg, B. Iorgulescu, G. Oliveira, E. Parry, M. Ficial, R. Bhatt., N. Hacohen, and M. Atkins for discussions and input. We also appreciate the patients who generously provided their samples for this research. This work is supported in part by the Dana-Farber / Harvard Cancer Center Kidney Cancer SPORE (P50-CA101942-12). D.A.B. acknowledges support by the DF/HCC Kidney Cancer SPORE Career Enhancement Program (P50CA101942-15), DOD CDMRP (KC170216, KC190130), and the DOD Academy of Kidney Cancer Investigators (KC190128). K.S. acknowledges support from the NIH/NIGMS (5R35GM131802, 5R01GM083084) and the ENCODE Data Analysis Center (NIH/NHGRI 5R01HG009446). C.B.P. and L.R.O. acknowledge funding support from the Independent Research Fund Denmark (8048-00078B). S.H.G. is supported by a Kay Kendall Leukaemia Fund Fellowship. L.H. is supported by the Fondation de France during her post-doctoral research fellowship at DFCI. Z.B. acknowledges research support from Bristol-Myers Squibb and Genentech. S.L. is supported by the NCI Research Specialist Award (R50CA251956). D.B.K. is supported in part by the NIH/NCI (R21 CA216772-01A1 and NCI-SPORE-2P50CA101942-11A1). W.X. is supported in part by the DF/HCC Kidney Cancer SPORE Career Enhancement Program (P50CA101942) and the ASCO Conquer Cancer Foundation Young Investigator Award. S.A.S acknowledges support by the NCI (R50RCA211482). R.A.I. acknowledges support from the NIH/NIGMS (R35GM131802). T.K.C. is supported in part by the Dana-Farber/Harvard Cancer Center Kidney SPORE (P50CA101942) and Cancer Center Support Grant (P30CA006516), the Kohlberg Chair at Harvard Medical School, the Trust Family, Michael Brigham, and Loker Pinard Funds for Kidney Cancer Research at DFCI, and various National Cancer Institute (NCI), Department of Defense (DOD), Foundations and industry grants. C.J.W. acknowledges support from NIH: NCI-1RO1CA155010 and NIH/NCI U24 CA224331. This work was supported in part by The G. Harold and Leila Y. Mathers Foundation. C.J.W. is a scholar of the Leukemia and Lymphoma Society, and is supported in part by the Parker Institute for Cancer Immunotherapy.
Declaration of Interests:
D.A.B. reported nonfinancial support from Bristol-Myers Squibb, honoraria from LM Education/Exchange Services, and personal fees from Octane Global, Defined Health, Dedham Group, Adept Field Solutions, Slingshot Insights, Blueprint Partnerships, Charles River Associates, Trinity Group, and Insight Strategy, outside of the submitted work. S.H.G. has patents licensed to Novalgen Inc, outside of the submitted work. Z.B. reported nonfinancial support from Bristol-Myers Squibb and Genentech/imCore. D.B.K. has acted as an advisor of and has received consulting fees from Neon Therapeutics and owns equity in Agenus, Armata pharmaceuticals, Breakbio, Biomarin Pharmaceutical, Bristol-Myers Squibb, Celldex Therapeutics, Chinook Therapeutics, Editas Medicine, Exelixis, Gilead Sciences, IMV, Lexicon Pharmaceuticals, Moderna, and Regeneron Pharmaceuticals. K.M.M. reports research support from Bristol-Myers Squibb. B.A.M. reports consulting fees from Bayer, Astellas, Astra Zeneca, Seattle Genetics, Dendreon, Calithera, Exelixis, Nektar, Pfizer, Janssen, Genentech, Eisai and EMD Serono, and research support to DFCI from Bristol Myers Squibb, Calithera, Exelixis, Seattle Genetics. D.F.M. has received consulting fees from Bristol-Myers Squibb, Pfizer, Merck, Alkermes, EMD Serono, Eli Lilly, Iovance, Eisai, and research support from Bristol-Myers Squibb, Merck, Genentech, Pfizer, Exelixis, X4 Pharma, and Alkermes. S.A.S. reported nonfinancial support from Bristol-Myers Squibb outside the submitted work. S.A.S. previously advised and has received consulting fees from Neon Therapeutics. S.A.S. reported nonfinancial support from Bristol-Myers Squibb, and equity in Agenus Inc., Agios Pharmaceuticals, Breakbio Corp., Bristol-Myers Squibb, and Lumos Pharma, outside the submitted work. S.S. reported personal fees from Merck, AstraZeneca, Bristol-Myers Squibb, CRISPR Therapeutics AG, AACR, and NCI, grants from Bristol-Myers Squibb, AstraZeneca, Novartis, and Exelixis, and royalties from Biogenex. A.H.S. has patents/pending royalties on the PD-1 pathway from Roche and Novartis. A.H.S. is on advisory boards for Bicara, Janssen Immunology, Surface Oncology, Elstar, SQZ Biotechnologies, Elpiscience, Selecta, and Monopteros, and consults for Novartis. A.H.S. has received research funding from AbbVie, Novartis, Roche, UCB, Ipsen, Quark and Merck. D.F.M. reported personal fees from Bristol-Myers Squibb, Pfizer, Merck, Novartis, Exelixis, Array BioPharm, Genentech, Alkermes, Jounce Therapeutics, X4 Pharma, Peloton, EMD Serono, and Eli Lilly; research support from Bristol-Myers Squibb, Prometheus Laboratories, Merck, Genentech, Pfizer, Exelixis, Novartis, X4 Pharma, Alkermes, and Peloton. T.K.C. reported grants and personal fees from Astra Zeneca, personal fees from Bayer, grants and personal fees from Bristol-Myers Squibb, personal fees from Cerulean, grants and personal fees from Eisai, personal fees from Foundation Medicine Inc, grants and personal fees from Exelixis, grants and personal fees from Genentech, personal fees from Roche, grants and personal fees from GlaxoSmithKline, grants and personal fees from Merck, from Novartis, Peloton, and Pfizer, personal fees from Prometheus Labs, grants and personal fees from Corvus, personal fees from Ipsen, grants from Tracon, grants from Astellas outside the submitted work. The other authors declare no potential conflicts of interest. C.J.W. is an equity holder of BioNTech.
REFERENCES
- Amin MB, American Joint Committee on Cancer., and American Cancer Society. (2017). AJCC cancer staging manual, Eight edition / editor-in-chief, Amin Mahul B.; editors, Edge Stephen B. and 16 others; Gress Donna M. - Technical editor; Meyer Laura R. - Managing editor. edn (Chicago IL: American Joint Committee on Cancer, Springer; ). [Google Scholar]
- Aran D, Camarda R, Odegaard J, Paik H, Oskotsky B, Krings G, Goga A, Sirota M, and Butte AJ (2017). Comprehensive analysis of normal adjacent to tumor transcriptomes. Nat Commun 8, 1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azizi E, Carr AJ, Plitas G, Cornish AE, Konopacki C, Prabhakaran S, Nainys J, Wu K, Kiseliovas V, Setty M, et al. (2018). Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment. Cell 174, 1293–1308 e1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagaev DV, Vroomans RMA, Samir J, Stervbo U, Rius C, Dolton G, Greenshields-Watson A, Attaf M, Egorov ES, Zvyagin IV, et al. (2020). VDJdb in 2019: database extension, new analysis infrastructure and a T-cell receptor motif compendium. Nucleic Acids Res 48, D1057–D1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett CW, Reddy VK, Short SP, Motley AK, Lintel MK, Bradley AM, Freeman T, Vallance J, Ning W, Parang B, et al. (2015). Selenoprotein P influences colitis-induced tumorigenesis by mediating stemness and oxidative damage. J Clin Invest 125, 2646–2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, Coussens LM, Gabrilovich DI, Ostrand-Rosenberg S, Hedrick CC, et al. (2018). Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med 24, 541–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun DA, Hou Y, Bakouny Z, Ficial M, Sant’ Angelo M, Forman J, Ross-Macdonald P, Berger AC, Jegede OA, Elagina L, et al. (2020). Interplay of somatic alterations and immune infiltration modulates response to PD-1 blockade in advanced clear cell renal cell carcinoma. Nat Med. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nature Biotechnology 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research, N. (2013). Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 499, 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, Laird PW, Onofrio RC, Winckler W, Weir BA, et al. (2012). Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol 30, 413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Ji Z, Ngiow SF, Manne S, Cai Z, Huang AC, Johnson J, Staupe RP, Bengsch B, Xu C, et al. (2019). TCF-1-Centered Transcriptional Network Drives an Effector versus Exhausted CD8 T Cell-Fate Decision. Immunity 51, 840–855 e845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevrier S, Levine JH, Zanotelli VRT, Silina K, Schulz D, Bacac M, Ries CH, Ailles L, Jewett MAS, Moch H, et al. (2017). An Immune Atlas of Clear Cell Renal Cell Carcinoma. Cell 169, 736–749 e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choueiri TK, and Motzer RJ (2017). Systemic Therapy for Metastatic Renal-Cell Carcinoma. N Engl J Med 376, 354–366. [DOI] [PubMed] [Google Scholar]
- Clark DJ, Dhanasekaran SM, Petralia F, Pan J, Song X, Hu Y, da Veiga Leprevost F, Reva B, Lih TM, Chang HY, et al. (2019). Integrated Proteogenomic Characterization of Clear Cell Renal Cell Carcinoma. Cell 179, 964–983 e931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNardo DG, and Ruffell B (2019). Macrophages as regulators of tumour immunity and immunotherapy. Nat Rev Immunol 19, 369–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denisenko E, Guo BB, Jones M, Hou R, de Kock L, Lassmann T, Poppe D, Clement O, Simmons RK, Lister R, et al. (2020). Systematic assessment of tissue dissociation and storage biases in single-cell and single-nucleus RNA-seq workflows. Genome Biol 21, 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremova M, Vento-Tormo M, Teichmann SA, and Vento-Tormo R (2020). CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc 15, 1484–1506. [DOI] [PubMed] [Google Scholar]
- Eng CL, Lawson M, Zhu Q, Dries R, Koulena N, Takei Y, Yun J, Cronin C, Karp C, Yuan GC, et al. (2019). Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature 568, 235–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo CR, Azevedo RA, Mousdell S, Resende-Lara PT, Ireland L, Santos A, Girola N, Cunha R, Schmid MC, Polonelli L, et al. (2018). Blockade of MIF-CD74 Signalling on Macrophages and Dendritic Cells Restores the Antitumour Immune Response Against Metastatic Melanoma. Front Immunol 9, 1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman WH, Zitvogel L, Sautes-Fridman C, and Kroemer G (2017). The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol 14, 717–734. [DOI] [PubMed] [Google Scholar]
- Giraldo NA, Becht E, Vano Y, Petitprez F, Lacroix L, Validire P, Sanchez-Salas R, Ingels A, Oudard S, Moatti A, et al. (2017). Tumor-Infiltrating and Peripheral Blood T-cell Immunophenotypes Predict Early Relapse in Localized Clear Cell Renal Cell Carcinoma. Clin Cancer Res 23, 4416–4428. [DOI] [PubMed] [Google Scholar]
- Goltsev Y, Samusik N, Kennedy-Darling J, Bhate S, Hale M, Vazquez G, Black S, and Nolan GP (2018). Deep Profiling of Mouse Splenic Architecture with CODEX Multiplexed Imaging. Cell 174, 968–981 e915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta PK, Godec J, Wolski D, Adland E, Yates K, Pauken KE, Cosgrove C, Ledderose C, Junger WG, Robson SC, et al. (2015). CD39 Expression Identifies Terminally Exhausted CD8+ T Cells. PLoS Pathog 11, e1005177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghverdi L, Lun ATL, Morgan MD, and Marioni JC (2018). Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nature Biotechnology 36, 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakimi AA, Voss MH, Kuo F, Sanchez A, Liu M, Nixon BG, Vuong L, Ostrovnaya I, Chen YB, Reuter V, et al. (2019). Transcriptomic Profiling of the Tumor Microenvironment Reveals Distinct Subgroups of Clear Cell Renal Cell Cancer: Data from a Randomized Phase III Trial. Cancer Discov 9, 510–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson WH, Gensheimer J, Hashimoto M, Wieland A, Valanparambil RM, Li P, Lin JX, Konieczny BT, Im SJ, Freeman GJ, et al. (2019). Proliferating Transitory T Cells with an Effector-like Transcriptional Signature Emerge from PD-1(+) Stem-like CD8(+) T Cells during Chronic Infection. Immunity 51, 1043–1058 e1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im SJ, Hashimoto M, Gerner MY, Lee J, Kissick HT, Burger MC, Shan Q, Hale JS, Lee J, Nasti TH, et al. (2016). Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 537, 417–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakos T, Pislar A, Jewett A, and Kos J (2019). Cysteine Cathepsins in Tumor-Associated Immune Cells. Front Immunol 10, 2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen CS, Prokhnevska N, Master VA, Sanda MG, Carlisle JW, Bilen MA, Cardenas M, Wilkinson S, Lake R, Sowalsky AG, et al. (2019). An intra-tumoral niche maintains and differentiates stem-like CD8 T cells. Nature 576, 465–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasingam SD, Citartan M, Thang TH, Mat Zin AA, Ang KC, and Ch’ng ES (2019). Evaluating the Polarization of Tumor-Associated Macrophages Into M1 and M2 Phenotypes in Human Cancer Tissue: Technicalities and Challenges in Routine Clinical Practice. Front Oncol 9, 1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech SM, and Cui W (2012). Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol 12, 749–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan O, Giles JR, McDonald S, Manne S, Ngiow SF, Patel KP, Werner MT, Huang AC, Alexander KA, Wu JE, et al. (2019). TOX transcriptionally and epigenetically programs CD8(+) T cell exhaustion. Nature 571, 211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klement JD, Paschall AV, Redd PS, Ibrahim ML, Lu C, Yang D, Celis E, Abrams SI, Ozato K, and Liu K (2018). An osteopontin/CD44 immune checkpoint controls CD8+ T cell activation and tumor immune evasion. J Clin Invest 128, 5549–5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES, and Getz G (2014). Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, van der Leun AM, Yofe I, Lubling Y, Gelbard-Solodkin D, van Akkooi ACJ, van den Braber M, Rozeman EA, Haanen J, Blank CU, et al. (2019). Dysfunctional CD8 T Cells Form a Proliferative, Dynamically Regulated Compartment within Human Melanoma. Cell 176, 775–789 e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, He Y, Hao J, Ni L, and Dong C (2018). High Levels of Eomes Promote Exhaustion of Anti-tumor CD8(+) T Cells. Front Immunol 9, 2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JR, Izar B, Wang S, Yapp C, Mei S, Shah PM, Santagata S, and Sorger PK (2018). Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee AV, et al. (2018). An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 173, 400–416 e411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun ATL, Bach K, and Marioni JC (2016). Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biology 17, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maaten L.v.d., and Hinton G (2008). Visualizing Data using t-SNE. Journal of Machine Learning Research 9, 2579–2605. [Google Scholar]
- Mackay LK, Minnich M, Kragten NA, Liao Y, Nota B, Seillet C, Zaid A, Man K, Preston S, Freestone D, et al. (2016). Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science 352, 459–463. [DOI] [PubMed] [Google Scholar]
- Mahoney KM, Rennert PD, and Freeman GJ (2015). Combination cancer immunotherapy and new immunomodulatory targets. Nat Rev Drug Discov 14, 561–584. [DOI] [PubMed] [Google Scholar]
- Mattiola I, Tomay F, De Pizzol M, Silva-Gomes R, Savino B, Gulic T, Doni A, Lonardi S, Astrid Boutet M, Nerviani A, et al. (2019). The macrophage tetraspan MS4A4A enhances dectin-1-dependent NK cell-mediated resistance to metastasis. Nat Immunol 20, 1012–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy DJ, Campbell KR, Lun ATL, and Wills QF (2017). Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 33, 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott DF, Huseni MA, Atkins MB, Motzer RJ, Rini BI, Escudier B, Fong L, Joseph RW, Pal SK, Reeves JA, et al. (2018). Clinical activity and molecular correlates of response to atezolizumab alone or in combination with bevacizumab versus sunitinib in renal cell carcinoma. Nat Med 24, 749–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. (2010). The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20, 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melville J, Lun A, and Djekidel MN (2020). uwot: The Uniform Manifold Approximation and Projection (UMAP) Method for Dimensionality Reduction.
- Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, and Getz G (2011). GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 12, R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao D, Margolis CA, Gao W, Voss MH, Li W, Martini DJ, Norton C, Bosse D, Wankowicz SM, Cullen D, et al. (2018). Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science 359, 801–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BC, Sen DR, Al Abosy R, Bi K, Virkud YV, LaFleur MW, Yates KB, Lako A, Felt K, Naik GS, et al. (2019). Subsets of exhausted CD8(+) T cells differentially mediate tumor control and respond to checkpoint blockade. Nat Immunol 20, 326–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motzer RJ, Tannir NM, McDermott DF, Aren Frontera O, Melichar B, Choueiri TK, Plimack ER, Barthelemy P, Porta C, George S, et al. (2018). Nivolumab plus Ipilimumab versus Sunitinib in Advanced Renal-Cell Carcinoma. N Engl J Med 378, 1277–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubert NJ, Schmittnaegel M, Bordry N, Nassiri S, Wald N, Martignier C, Tille L, Homicsko K, Damsky W, Maby-El Hajjami H, et al. (2018). T cell-induced CSF1 promotes melanoma resistance to PD1 blockade. Sci Transl Med 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowicka M, Krieg C, Crowell HL, Weber LM, Hartmann FJ, Guglietta S, Becher B, Levesque MP, and Robinson MD (2017). CyTOF workflow: differential discovery in high-throughput high-dimensional cytometry datasets. F1000Res 6, 748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Flanagan CH, Campbell KR, Zhang AW, Kabeer F, Lim JLP, Biele J, Eirew P, Lai D, McPherson A, Kong E, et al. (2019). Dissociation of solid tumor tissues with cold active protease for single-cell RNA-seq minimizes conserved collagenase-associated stress responses. Genome Biol 20, 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paley MA, Kroy DC, Odorizzi PM, Johnnidis JB, Dolfi DV, Barnett BE, Bikoff EK, Robertson EJ, Lauer GM, Reiner SL, et al. (2012). Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science 338, 1220–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignon JC, Jegede O, Shukla SA, Braun DA, Horak CE, Wind-Rotolo M, Ishii Y, Catalano PJ, Grosha J, Flaifel A, et al. (2019). irRECIST for the Evaluation of Candidate Biomarkers of Response to Nivolumab in Metastatic Clear Cell Renal Cell Carcinoma: Analysis of a Phase II Prospective Clinical Trial. Clin Cancer Res 25, 2174–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig-Kroger A, Sierra-Filardi E, Dominguez-Soto A, Samaniego R, Corcuera MT, Gomez-Aguado F, Ratnam M, Sanchez-Mateos P, and Corbi AL (2009). Folate receptor beta is expressed by tumor-associated macrophages and constitutes a marker for M2 anti-inflammatory/regulatory macrophages. Cancer Res 69, 9395–9403. [DOI] [PubMed] [Google Scholar]
- Qin S, Xu L, Yi M, Yu S, Wu K, and Luo S (2019). Novel immune checkpoint targets: moving beyond PD-1 and CTLA-4. Mol Cancer 18, 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriques SG, Stickels RR, Goeva A, Martin CA, Murray E, Vanderburg CR, Welch J, Chen LM, Chen F, and Macosko EZ (2019). Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution. Science 363, 1463–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney MS, Shukla SA, Wu CJ, Getz G, and Hacohen N (2015). Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160, 48–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sade-Feldman M, Yizhak K, Bjorgaard SL, Ray JP, de Boer CG, Jenkins RW, Lieb DJ, Chen JH, Frederick DT, Barzily-Rokni M, et al. (2018). Defining T Cell States Associated with Response to Checkpoint Immunotherapy in Melanoma. Cell 175, 998–1013 e1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal R, Polyak MJ, Zuccolo J, Puri M, Deng L, Roberts L, Zuba A, Storek J, Luider JM, Sundberg EM, et al. (2017). MS4A4A: a novel cell surface marker for M2 macrophages and plasma cells. Immunol Cell Biol 95, 611–619. [DOI] [PubMed] [Google Scholar]
- Scheper W, Kelderman S, Fanchi LF, Linnemann C, Bendle G, de Rooij MAJ, Hirt C, Mezzadra R, Slagter M, Dijkstra K, et al. (2019). Low and variable tumor reactivity of the intratumoral TCR repertoire in human cancers. Nat Med 25, 89–94. [DOI] [PubMed] [Google Scholar]
- Senbabaoglu Y, Gejman RS, Winer AG, Liu M, Van Allen EM, de Velasco G, Miao D, Ostrovnaya I, Drill E, Luna A, et al. (2016). Tumor immune microenvironment characterization in clear cell renal cell carcinoma identifies prognostic and immunotherapeutically relevant messenger RNA signatures. Genome Biol 17, 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shugay M, Bagaev DV, Zvyagin IV, Vroomans RM, Crawford JC, Dolton G, Komech EA, Sycheva AL, Koneva AE, Egorov ES, et al. (2018). VDJdb: a curated database of T-cell receptor sequences with known antigen specificity. Nucleic Acids Res 46, D419–D427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sica A, and Mantovani A (2012). Macrophage plasticity and polarization: in vivo veritas. J Clin Invest 122, 787–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoni Y, Becht E, Fehlings M, Loh CY, Koo SL, Teng KWW, Yeong JPS, Nahar R, Zhang T, Kared H, et al. (2018). Bystander CD8(+) T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature 557, 575–579. [DOI] [PubMed] [Google Scholar]
- Singer M, Wang C, Cong L, Marjanovic ND, Kowalczyk MS, Zhang H, Nyman J, Sakuishi K, Kurtulus S, Gennert D, et al. (2016). A Distinct Gene Module for Dysfunction Uncoupled from Activation in Tumor-Infiltrating T Cells. Cell 166, 1500–1511 e1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slyper M, Porter CBM, Ashenberg O, Waldman J, Drokhlyansky E, Wakiro I, Smillie C, Smith-Rosario G, Wu J, Dionne D, et al. (2020). A single-cell and single-nucleus RNA-Seq toolbox for fresh and frozen human tumors. Nat Med 26, 792–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CC, Beckermann KE, Bortone DS, De Cubas AA, Bixby LM, Lee SJ, Panda A, Ganesan S, Bhanot G, Wallen EM, et al. (2018). Endogenous retroviral signatures predict immunotherapy response in clear cell renal cell carcinoma. J Clin Invest 128, 4804–4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinas G, Schiarea S, Liguori M, Fabbri M, Pesce S, Zammataro L, Pasqualini F, Nebuloni M, Chiabrando C, Mantovani A, et al. (2010). Tumor-conditioned macrophages secrete migration-stimulating factor: a new marker for M2-polarization, influencing tumor cell motility. J Immunol 185, 642–652. [DOI] [PubMed] [Google Scholar]
- Street K, Risso D, Fletcher RB, Das D, Ngai J, Yosef N, Purdom E, and Dudoit S (2018). Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19, 477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Harashima N, Kajigaya S, Yokoyama H, Cherkasova E, McCoy JP, Hanada K, Mena O, Kurlander R, Tawab A, et al. (2008). Regression of human kidney cancer following allogeneic stem cell transplantation is associated with recognition of an HERV-E antigen by T cells. J Clin Invest 118, 1099–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turajlic S, Xu H, Litchfield K, Rowan A, Chambers T, Lopez JI, Nicol D, O’Brien T, Larkin J, Horswell S, et al. (2018a). Tracking Cancer Evolution Reveals Constrained Routes to Metastases: TRACERx Renal. Cell 173, 581–594 e512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turajlic S, Xu H, Litchfield K, Rowan A, Horswell S, Chambers T, O’Brien T, Lopez JI, Watkins TBK, Nicol D, et al. (2018b). Deterministic Evolutionary Trajectories Influence Primary Tumor Growth: TRACERx Renal. Cell 173, 595–610 e511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Berge K, Roux de Bezieux H, Street K, Saelens W, Cannoodt R, Saeys Y, Dudoit S, and Clement L (2020). Trajectory-based differential expression analysis for single-cell sequencing data. Nat Commun 11, 1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Brink SC, Sage F, Vertesy A, Spanjaard B, Peterson-Maduro J, Baron CS, Robin C, and van Oudenaarden A (2017). Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat Methods 14, 935–936. [DOI] [PubMed] [Google Scholar]
- Van Gassen S, Callebaut B, Van Helden MJ, Lambrecht BN, Demeester P, Dhaene T, and Saeys Y (2015). FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A 87, 636–645. [DOI] [PubMed] [Google Scholar]
- Wang T, Lu R, Kapur P, Jaiswal BS, Hannan R, Zhang Z, Pedrosa I, Luke JJ, Zhang H, Goldstein LD, et al. (2018). An Empirical Approach Leveraging Tumorgrafts to Dissect the Tumor Microenvironment in Renal Cell Carcinoma Identifies Missing Link to Prognostic Inflammatory Factors. Cancer Discov 8, 1142–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Madireddi S, de Almeida PE, Banchereau R, Chen YJ, Chitre AS, Chiang EY, Iftikhar H, O’Gorman WE, Au-Yeung A, et al. (2020). Peripheral T cell expansion predicts tumour infiltration and clinical response. Nature 579, 274–278. [DOI] [PubMed] [Google Scholar]
- Xu L, Zhu Y, Chen L, An H, Zhang W, Wang G, Lin Z, and Xu J (2014). Prognostic value of diametrically polarized tumor-associated macrophages in renal cell carcinoma. Ann Surg Oncol 21, 3142–3150. [DOI] [PubMed] [Google Scholar]
- Young MD, Mitchell TJ, Vieira Braga FA, Tran MGB, Stewart BJ, Ferdinand JR, Collord G, Botting RA, Popescu DM, Loudon KW, et al. (2018). Single-cell transcriptomes from human kidneys reveal the cellular identity of renal tumors. Science 361, 594–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, and He QY (2016). ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol Biosyst 12, 477–479. [DOI] [PubMed] [Google Scholar]
- Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, Lawrence MS, Zhsng CZ, Wala J, Mermel CH, et al. (2013). Pan-cancer patterns of somatic copy number alteration. Nat Genet 45, 1134–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappia L, and Oshlack A (2018). Clustering trees: a visualization for evaluating clusterings at multiple resolutions. GigaScience 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Li Z, Skrzypczynska KM, Fang Q, Zhang W, O’Brien SA, He Y, Wang L, Zhang Q, Kim A, et al. (2020). Single-Cell Analyses Inform Mechanisms of Myeloid-Targeted Therapies in Colon Cancer. Cell 181, 442–459 e429. [DOI] [PubMed] [Google Scholar]
- Zhang L, Yu X, Zheng L, Zhang Y, Li Y, Fang Q, Gao R, Kang B, Zhang Q, Huang JY, et al. (2018). Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature 564, 268–272. [DOI] [PubMed] [Google Scholar]
- Zheng C, Zheng L, Yoo JK, Guo H, Zhang Y, Guo X, Kang B, Hu R, Huang JY, Zhang Q, et al. (2017). Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell 169, 1342–1356 e1316. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Patient-level clinical and somatic alteration data table, related to Figure 1.
Table S2. Cell metadata, including sample origin, mapping of each cell to its designated cluster, and annotation of whether it was included in the CD8+ T cell or monocyte/macrophage trajectory analyses, related to Figures 1–6, and S1–6.
Table S3. Differential gene expression analysis, including top 1000 differentially expressed genes for each cluster (compared to all other clusters) for graph-based clustering of all cells, T cells only, and myeloid cells only, related to Figures 2C, S1B, S3A, S5B, and S5D.
Table S4. Gene signatures used in this study, related to Figures 3D–F, 5E, 6D–G, S3D–E, and S6C.
Table S5. TCR clonotype information for each T cell, including the cell barcode (enabling mapping back to gene expression data in Table S2), TCR clonotype, and annotation of whether it was predicted to be viral-specific, related to Figures 4 and S4.
Table S6. Ligand-receptor means and p-values for all T and myeloid cell interactions (from CellPhoneDB v2.0), related to Figures 6A–C, and S6B.
Data S1. ScRNA-seq raw count matrix (part 1 of 2), after quality control filtering, with genes as rows and cell barcodes as columns, related to Figure 1–6, S1–3, and S5.
Data S2. ScRNA-seq raw count matrix (part 2 of 2), after quality control filtering, with genes as rows and cell barcodes as columns, related to Figure 1–6, S1–3, and S5.
Data Availability Statement
All raw DNA and RNA sequencing files are deposited in dbGaP under accession number phs002252.v1.p1 at https://www.ncbi.nlm.nih.gov/gap/. Processed data is provided in supplemental tables S2 and S3. All computer code used in this study is publicly available, as listed in the Key Resource Table.
Key Resources Table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Human CD3 BUV661 (clone UCHT1, 1:33 dilution) | BD Biosciences | Cat # 612964 |
| Anti-Human CD8 BUV563 (clone RPA-T8, 1:100 dilution) | BD Biosciences | Cat # 612914 |
| Rat Anti-CD11b BUV395 (clone M1/70, 1:50 dilution) | BD Biosciences | Cat # 565976 |
| Anti-Human CD14 BV750 (clone 63D3, 1:50 dilution) | BioLegend | Cat # 367136 |
| Anti-Human CD16 BUV496 (clone 3G8, 1:20 dilution) | BD Biosciences | Cat # 612944 |
| Anti-Human CD45 BUV805 (clone HI30, 1:33 dilution) | BD Biosciences | Cat # 612892 |
| Anti-Human CD68 BV711 (clone Y1/82A, 1:20 dilution) | BD Biosciences | Cat # 565594 |
| Anti-Human CD163 PE/Dazzle (clone GHI/61, 1:20 dilution) | BioLegend | Cat # 333624 |
| Anti-Human CD204 APC (clone 7C9C20, 1:20 dilution) | BioLegend | Cat # 371906 |
| Anti-Human PVR BV510 (clone SK11.4, 1:20 dilution) | BioLegend | Cat # 337634 |
| Anti-Human TIGIT BV605 (clone A15153G, 1:20 dilution) | BioLegend | Cat # 372712 |
| Anti-Human PDL1 PercpCy5.5 (clone 29E.2A3, 1:20 dilution) | BioLegend | Cat # 329738 |
| Anti-Human PDL2 BV421 (clone 24F.10C12, 1:20 dilution) | BioLegend | Cat # 329616 |
| Anti-Human Galectin-9 PE-Cy7 (clone 9M1–3, 1:20 dilution) | BioLegend | Cat # 348916 |
| Anti-Human PD1 BV650 (clone EH12.2H7, 1:20 dilution) | BioLegend | Cat # 329950 |
| Anti-Human TIM-3 BD BB515 (clone 7D3, 1:50 dilution) | Biosciences | Cat # 565568 |
| Anti-Human HLA-DR BV785 (clone L243, 1:20 dilution) | BioLegend | Cat # 307642 |
| Anti-Human CD86 BUV737 (clone FUN-1, 1:20 dilution) | BD Biosciences | Cat # 612784 |
| Anti-Human CTLA-4 PE-Cy5 (clone BNI3, 1:10 dilution) | BD Biosciences | Cat # 555854 |
| Anti-Human TCF1 PE (clone 7F11A10, 1:20 dilution) | BioLegend | Cat # 655208 |
| Anti-Human CAIX AF700 (clone 303123, 1:20 dilution) | R&D Systems | Cat # FAB2188N-100UG |
| Mouse monoclonal anti-CD8 | Agilent | C8/144B |
| Goat monoclonal anti-TIM3 | R&D biosystems | AF2365 |
| Mouse monoclonal anti-LAG3 | LifeSpan Biosciences | 17B4 |
| Mouse monoclonal anti-PD1 | Gordon Freeman laboratory, Dana Farber Cancer Institute | EH33 |
| Rabbit Monoclonal anti-CD163 | Abcam | EPR19518 |
| Biological Samples | ||
| Primary human ccRCC samples | Dana-Farber/Brigham and Women’s Hospital, Brigham and Women’s Faulkner Hospital, Beth Israel Deaconess Medical Center | N/A |
| Human adjacent normal tissue samples from ccRCC patients | Dana-Farber/Brigham and Women’s Hospital, Brigham and Women’s Faulkner Hospital, Beth Israel Deaconess Medical Center | N/A |
| Human buffy coats | Research Blood Components, LLC. | #002 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Collagenase D | Roche | Cat#11088858001 |
| Dispase (5 Units/mL) | STEMCELL | Cat#07913 |
| DNase I (2000 Units/mL) | New England Biolabs | Cat#M0303L |
| ACK lysing buffer | Gibco | Cat#A1049201 |
| Dynabeads™ MyOne™ SILANE | Life Technologies | Cat#2000048 |
| Critical Commercial Assays | ||
| MACS Dead Cell Removal Kit | Miltenyi | Cat#130–090-101 |
| Chromium Single Cell 5’ Library & Gel Bead Kit | 10x Genomics | Cat#1000006 |
| Chromium Single Cell 5’ Library Construction Kit | 10x Genomics | Cat#1000020 |
| Chromium Single Cell V(D)J Enrichment Kit, Human T cell | 10x Genomics | Cat#1000005 |
| Chromium i7 Multiplex Kit | 10x Genomics | Cat#120262 |
| Chromium Chip A Single Cell Kit | 10x Genomics | Cat#120236 |
| eBioscience Foxp3 Transcription Factor Staining Buffer Set | ThermoFisher | Cat # 00-5523-00 |
| Zombie NIR Fixable Viability Dye | BioLegend | Cat # 423106 |
| Deposited Data | ||
| Data files for scRNA-seq | This paper | dbGaP: phs002252.v1.p1 |
| Bulk RNA-seq and clinical data for TCGA KIRC cohort | TCGA | https://gdc.cancer.gov/about-data/publications/pancanatlas |
| Bulk RNA-seq and clinical data for CheckMate cohorts | Braun et al., 2020 | https://static-content.springer.com/esm/art%3A10.1038%2Fs41591-020-0839-y/MediaObjects/41591_2020_839_MOESM2_ESM.xlsx |
| VDJdb | Bagaev et al., 2020 | https://vdjdb.cdr3.net |
| Software and Algorithms | ||
| Cell Ranger v2.1.0 | 10x Genomics | http://10xgenomics.com/ |
| GATK4 v2.7.0 | McKenna et al., 2010 | https://github.com/gatk-workflows/gatk4-somatic-snvs-indels/ |
| ABSOLUTE v10 | Carter et al., 2012 | https://software.broadinstitute.org/cancer/cga/absolute |
| ISAR-GISTIC v1 | Zack et al., 2013 | https://hub.docker.com/r/arielyh/erv_pipeline |
| GISTIC2 v1 | Mermel et al., 2011 | https://hub.docker.com/r/broadgdac/tool_gistic2 |
| Seurat v3.1.4 | Butler et al., 2018 | https://satijalab.org/seurat/ |
| scran v1.12.1 | Lun et al., 2016 | https://bioconductor.org/packages/release/bioc/html/scran.html |
| scater v1.12.2 | McCarthy et al., 2017 | https://bioconductor.org/packages/release/bioc/html/scater.html |
| batchelor v1.0.1 | Haghverdi et al., 2018 | https://bioconductor.org/packages/release/bioc/html/batchelor.html |
| clustree v0.4.3 | Zappia and Oshlack, 2018 | https://cran.r-project.org/package=clustree |
| edgeR v3.26.8 | Robinson et al., 2010 | https://www.bioconductor.org/packages/release/bioc/html/edgeR.html |
| uwot v0.1.8 | Mellville et al., 2020 | https://cran.r-project.org/package=uwot |
| Slingshot v1.7.0 | Street et al. 2018 | https://bioconductor.org/packages/release/bioc/html/slingshot.html |
| tradeSeq v1.2.01 | Van den Berge et al., 2020 | https://www.bioconductor.org/packages/release/bioc/html/tradeSeq.html |
| CellPhoneDB v2.0 | Efremova et al., 2020 | https://github.com/Teichlab/cellphonedb |
| ReactomePA v1.26.0 | Yu and He, 2016 | https://bioconductor.org/packages/release/bioc/html/ReactomePA.html |
| survival v3.1–12 | Comprehensive R Archive Network (CRAN) | https://cran.r-project.org/package=survival |
| survminer v0.4.3 | Comprehensive R Archive Network (CRAN) | https://cran.r-project.org/package=survminer |
| FlowJo v10.7 | FlowJo LLC. | https://www.flowjo.com/ |
| HALO platform v3.0 | Indica Lab | https://indicalab.com/halo/ |
| FlowSOM v1.21.0 | Van Gassen et al. 2015 | http://bioconductor.org/packages/release/bioc/html/FlowSOM.html |