Abstract
Machine learning methods have been widely applied to big data analysis in genomics and epigenomics research. Although accuracy and efficiency are common goals in many modeling tasks, model interpretability is especially important to these studies towards understanding the underlying molecular and cellular mechanisms. Deep neural networks (DNNs) have recently gained popularity in various types of genomic and epigenomic studies due to their capabilities in utilizing large-scale high-throughput bioinformatics data and achieving high accuracy in predictions and classifications. However, DNNs are often challenged by their potential to explain the predictions due to their black-box nature. In this review, we present current development in the model interpretation of DNNs, focusing on their applications in genomics and epigenomics. We first describe state-of-the-art DNN interpretation methods in representative machine learning fields. We then summarize the DNN interpretation methods in recent studies on genomics and epigenomics, focusing on current data- and computing-intensive topics such as sequence motif identification, genetic variations, gene expression, chromatin interactions and non-coding RNAs. We also present the biological discoveries that resulted from these interpretation methods. We finally discuss the advantages and limitations of current interpretation approaches in the context of genomic and epigenomic studies. Contact:xiaoman@mail.ucf.edu, haihu@cs.ucf.edu
Keywords: deep neural network, feature interpretation, model interpretation, genomics, epigenomics
Introduction
The recent development in deep neural networks (DNNs) has been applied to various tasks and achieved state-of-the-art performance [1–5]. In comparison with early shallow neural networks, DNNs have more complex architectures, including classical feedforward neural networks such as fully connected neural networks and convolutional neural networks (CNNs), recurrent neural networks (RNNs) and their improved versions [3]. DNNs have essentially revolutionized the problem-solving approaches in computer vision and natural language processing (NLP) fields [4, 5]. For example, CNNs have dominated image recognition [6], object detection [7–9] and activity recognition [10]. RNNs have been widely applied to text mining and machine translation [11, 12].
With the rapid accumulation of large-scale high-throughput omics data in the past decades, such as genomic and epigenomic data, DNNs have also received increased attention in bioinformatics [13, 14]. They have demonstrated superior performance to traditional machine learning approaches in many bioinformatics problems such as sequence motif finding [14–16], chromatin interaction prediction [17–19] and genetic variant detection [20].
However, with their black-box nature, DNN-based approaches often encounter difficulties in explaining the relationships between inputs and predicted outputs, also known as the explainability and interpretability issue. There are various perspectives on the difference between interpretability and explainability. For example, Doshi-Velez et al. recently described interpretability as ‘the ability to explain or to present in understandable terms to a human’, while Gilpin et al. defined interpretability as ‘the science of comprehending what a model did’ [21, 22]. These studies referred to interpretability as the first step of explainability, and it alone is necessary but not sufficient for reasoning and understanding. However, in Montavon et al., interpretation was defined as ‘the mapping of an abstract concept into a domain that the human can make sense of’, whereas explanation as ‘the collection of features of the interpretable domain that have contributed for a given example to produce a decision’ [23]. In this survey, we also assume that interpretability is the first step toward explainability. The challenge in interpretability and explainability faced by current DNNs is especially true for many bioinformatics studies. This is because bioinformatics tasks often are not completed at the prediction stage. Bioinformatics data modeling, in general, requires an in-depth understanding of why the input variables/features lead to the output classification to gain insights into phenotypes and their underlying biological mechanisms [24].
The necessity to understand how DNNs perform in each of the prediction tasks motivated the recent surge of research on model interpretations [23, 25–30] with applications in various bioinformatics domains, including genomics and epigenomics research. In this study, we will first review current methods in DNN interpretations. We then survey most data-intensive genomics and epigenomics problems to illustrate the recent application and development of DNN interpretation techniques.
Current methods for DNN interpretation
In the past decade, there has been an increasing interest in finding ways to interpret the features discovered by neural networks, particularly in computer vision and NLP [4, 5]. The beginning of the method development can be traced back to the visualization and interpretation for CNNs in image analysis [25, 29]. Various DNN interpretation categorizations have been proposed since then.
However, there has not yet been a unified terminology to classify the developed methods. For example, Grun et al. classified the existing methods into three categories: input modification, deconvolutional and input reconstruction, among which the deconvolutional methods can be further divided into three subcategories: deconvnet, backpropagation and guided backpropagation approaches [26] (Figure 1). Singh et al. used different terms for their categorization: deconvolution, saliency maps, class optimization and others [31] (Figure 1). Zhang et al. categorized CNN visualization methods into gradient-based methods and up-convolutional net techniques [32]. The gradient-based methods [25, 29, 33, 34] take advantage of gradients to visualize patterns of convolutional layers, whereas the up-convolutional net methods [35] take a different interpretation direction by converting feature maps to images. A more recent survey summarized the interpretation methods into two primary categories: input perturbation and backpropagation [24]. Backpropagation techniques were further categorized into saliency maps and input reconstruction approaches. Although the terminologies are not unified, they have similar concepts underneath. For instance, saliency maps used by Singh et al. is similar to a backpropagation-based deconvolutional method used by Grun et al. Similarly, the class optimization approach by Singh et al. is similar to the input reconstruction approach by Grun et al. In addition to these categories with CNN models, the attention mechanism is a popular interpretation strategy used with RNN models specially in NLP tasks, where the attention to different parts of the sequence input on the output can be used as features [36] (Figure 1). In the following, we present the most popular strategies based on Grun et al.’s and Singh et al.’s classifications on CNNs and the attention mechanism on RNNs.
Figure 1.
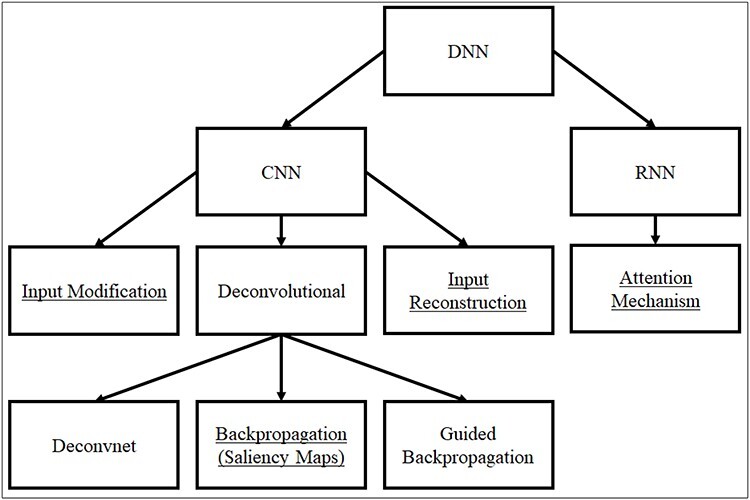
A classification of common DNN interpretation approaches. The underlined ones are used in genomic and epigenomic studies.
Input modification methods
Input modification methods determine the input feature importance by estimating an altered input’s impact on its immediate layers or the DNN output. For example, an input modification technique called occlusion was developed to estimate the importance of different image parts to the image classification [25]. The occlusion method works by systematically covering up a portion of the input image with a gray square and then calculating the filter activation difference in the convolutional layer. By moving the square left to right and top to bottom of an image, the importance of different parts of an image was measured. A heatmap can be created from the resulting changes in activation that represent the activation map of an image. The objects in the image that affect the probability of its predicted classes can also be identified [25]. Although this approach can help create an interesting and clear visualization map for the whole input image, the huge number of training passes needed for every input image to create the visualization map is a significant overhead.
An input modification approach was later developed to identify the receptive field of each unit in each convolutional layer [9]. This approach, in general, followed the above image occlusion method with the exception that, a square patch with randomized pixel values was defined in place of a square patch of fixed color for image occlusion, since the latter could introduce a bias towards a fixed feature. Similar to the above occlusion method, a discrepancy map could also be constructed to show how much each pixel affects the activation of the unit in question. By repeating this process over a large number of images, the discrepancy maps could be aggregated into a combined discrepancy map that was invariant across images, showing which pixels comprised the receptive field of a unit and allowing for semantic interpretations of what each unit was learning to recognize. The significant overhead of generating a huge number of extra data set for each input image also remained as a disadvantage in this study.
Another input modification technique was used by Zintgraf et al. that measured the effect of removing an input dimension on the classification decision and used this piece of information to assign a relevance score to that input dimension [37]. Because relatively few classifiers were permitted to replace the value of an input dimension with an unknown value, and because retraining a neural network to exclude the dimension in question would be prohibitively expensive, removing the dimension was simulated by marginalizing it. Given every feature except the one in question, the conditional probability of predicting a particular class was then approximated with a formula based on the probability of predicting that class given all features and the empirical distribution of the feature whose absence was to be simulated. The accuracy of this approximation was improved by replacing the empirical distribution of the feature to be marginalized with either a conditional probability of that feature’s value given the value of its neighbors or multivariate analysis in which a sliding window of features were removed simultaneously. This formulation was adopted as a deconvolutional technique, in which instead of considering the impact of input modification on the final class prediction, the impact of an unknown hidden unit on the intermediate output was calculated for the corresponding layer to measure the relevance of that unit to that layer’s output. This technique was then propagated, layer by layer, similar to Montavon et al. [38].
Deconvolutional methods
Deconvolutional methods attempt to use the network structure to make sense of the predictions. Iteratively computing how much each unit of the lower layer contributes to the activation of interest and tracing the work of convolutional layers backward until reaching the input layers, deconvolutional methods are able to identify the contribution of input units to the output predictions.
Deconvnet. Deconvnet, a deconvolutional approach developed by Zeiler and Fergus, attached a deconvolutional layer to each convolutional layer in the network to probe the activity of a CNN trained for image classification [25]. The output of each convolutional layer was fed to the associated deconvolutional layer, which reconstructed the activity in the previous layer that led to those activations. Attaching a deconvolutional layer to each convolutional layer allowed this process to be extended from the prediction output all the way down to the input layer, to determine which pixels and structures of an input image were most responsible for the activation of the predicted class. One disadvantage to this method is that it suffers from significant overhead, requiring the training of a second neural network to analyze the features of the first and requiring a separate deconvolutional neural network for each layer of the original convolutional network that one wants to analyze. In addition to the extra space required to store all these networks, they must also be trained, taking additional time as well.
Similarly, deconvolutional layers were used to reverse the work of convolutional layers in image classification [35]. Instead of attaching a deconvolutional layer to each individual convolutional layer, a separate deconvolution network was trained using the feature vector generated confidently by a set of images as input. The deconvolution network was used to predict a pre-image that had been created from the weighted average of those images. This facilitated a way to visualize the feature interpretation by every layer of a CNN. This was similar to the decoding portion of an autoencoder. This approach shared the drawback of the above Zeiler and Fergus’ method [25], as it required storing and training a second network.
Backpropagation, guided backpropagation and saliency map. A backpropagation-based deconvolutional method called saliency map was introduced by Simonyan et al. [29]. A saliency map was constructed for each image, reflecting the contribution of each pixel to the classification decision. Instead of using deconvolutional layers to reverse the work of a trained CNN, a single pass of backpropagation through a trained model was used to determine the relative importance of pixels in an input image. The deconvolutional technique employed here improved deconvnet [25] by eliminating the need to train and store multiple deconvolution networks, achieving a similar goal using only the existing model weights and backpropagation. A deconvolutional approach called guided backpropagation was further developed by combining the deconvnet and backpropagation-based methods [34]. Note that deconvnet masks out the negative contribution of a neuron using the rectified linear unit (ReLU) only during the backward pass, while backpropagation clips out the negative activation of a neuron using the ReLU in the forward pass and then updates the weights during the backward pass, and the guided backpropagation clips out both the negative activation in forward pass and negative contribution in the backward pass, using ReLU units both during the forward and backward propagation steps [26, 34]. In contrast to the earlier deconvolutional approaches, guided backpropagation methods can be applied to networks that do not contain max-pooling layers.
Saliency map technique was also considered by Montavon et al. that used deep Taylor decomposition to map the output activations of a CNN layer to the relevance of that layer’s inputs [38]. Deep Taylor expansion works by taking the first-order Taylor expansion of the function approximated by a unit in one layer in terms of the units in the previous layer. These approximations are then summed across a layer. A heatmap can be constructed, showing how relevant each unit in the previous layer was to the output of the current layer [38]. Propagating this backward through the network also allows the construction of a heatmap showing the relevance of pixels in the input image to the prediction made by the output layer [38]. This approach improves over the previous technique in that it also generates saliency maps showing the features recognized by the input layer as well as each of the intermediate layers, but it is able to do so with much less overhead, due to using only Taylor expansions.
A deconvolutional technique for processing images or videos by CNN layers was created by Yosinski et al., where the activation of each unit in each layer in real-time in response to changing images or video frames could be visualized [30]. While straightforward visualization of layer activations was not new, showing the impact of changes in input on the activations in real-time was an improvement over earlier deconvolutional techniques and allowed the authors to gain insight into how CNNs learn intermediate object detectors (such as shoulders and faces) to help make higher-level classification decisions, even when not trained on those classes explicitly.
Another deconvolutional technique called ‘population encoding’ was developed in Wang et al. to visualize the features learned by each layer of a CNN by clustering the layer’s responses [8]. At each intermediate layer of the network, responses were sampled at a spatial grid of that layer. Every response in a specific grid position of that layer corresponded to a patch of the input image. K-means++ clustering was performed to cluster similar responses. Clusters were ranked based on their goodness according to a weighted sum of the Davies–Bouldin index and the normalized count of each cluster, and similar clusters were merged, with higher-ranked clusters absorbing lower-ranked clusters that had at least 50% similar contents. The consolidated clusters corresponded to visual features (patches) of the original input image that were important in identifying the class under consideration. This technique incurs greater overhead than previous methods but is able to provide more detailed information about the features recognized by each layer of the network, rather than providing a saliency map of only the input layer.
A backpropagation-based deconvolutional method called class activation map (CAM) was created in Zhou et al. [39]. A CAM is an activation map associated with a given class, representing the discriminative power of regions in an image. CAM works by first performing a global average pooling on the last convolutional layer in CNNs, then projecting back the weights of the output layer to the last convolutional layer and finally carrying out a weighted sum of the feature maps in the last convolutional layer. A CAM is essentially the local explanation for a particular classification result. In the case of image classification, a CAM indicates a region that is responsible for the image classification results.
Input reconstruction methods
Input reconstruction methods attempt to construct a synthetic input that either maximizes the activation of a specific class or matches the output of certain standards to understand better what features are learned along the way [26]. For example, an input reconstruction technique was used in Simonyan et al. to generate an image for each class that maximized the prediction score of that class, with the intent of using that image as an archetypical example of the network’s model of that class [29].
An input reconstruction method was developed to help answer the question of whether CNNs were able to learn the correspondence between different parts of an object [40]. The output vector of each layer was used to construct an image out of a library of patches of pixels obtained from the input corpus using K-means clustering. The resemblance of the reconstructed image with the input image was obtained by comparison. Like the previous methods, this approach also incurs a significant overhead by creating and storing a library of image patches.
Another input reconstruction technique used regularized optimization in image space to visualize features at each layer of a neural network as well [30]. The regularizations applied to images in this technique included the L2 norm, Gaussian blurring, clipping pixels with a small norm and clipping pixels with a small contribution. These regularizations corrected several confounding factors found in images. Using all four was found to result in more natural and interpretable images, potentially making the features discovered using other neural network interpretation techniques more meaningful.
An input reconstruction method called ‘activation maximization’ was applied to visualize multifaceted feature representations [27]. It was observed that one class, such as bell peppers or convertibles, often has many facets: bell peppers come in multiple colors and can be whole or cut open, and convertibles can come in multiple colors and look different when viewed from different sides. The neurons in the output layer responsible for recognizing one of these classes must be able to recognize multiple distinct facets of the class in order to do so. An algorithm was developed to visualize multiple facets that the neuron responsible for recognizing a particular class must recognize. This algorithm extracted the encoding of the penultimate layer of the network for each image in an input set corresponding to a particular class, used the principal component analysis to reduce the dimensionality of those encodings, used an algorithm called t-SNE to produce 2D embeddings from those reduced encodings, performed K-means clustering on the embeddings, calculated a mean image for each cluster by averaging the 15 nearest images to each cluster’s centroid and used each mean image to initialize activation maximization. Each cluster represented one facet of the class in question, and the mean image calculated was an exemplar of that facet. Activation maximization was used to refine that exemplar into one more natural looking and more easily interpretable by a human. This technique not only allows humans to observe what features are important for identifying each class but also allow a more detailed examination of how the network uses those disparate features to make its final decision and incurs relatively little overhead while doing so.
Attention mechanism
The above three categories of DNN interpretation methods were initially created for CNN-based DNNs. In the RNN-based DNNs, attention mechanisms are frequently used to selectively emphasize parts of the input during prediction, in addition to other interpretation methods [41]. For example, for the document classification problem in NLP, a hierarchical attention mechanism on a model with gated recurrent units was developed to focus on the important words in the important sentences [42]. Separate attention weights can be used for word and sentence encoder modules, and the important words in the important sentences can be visualized after normalizing the word weights by sentence weights. Another example, attention mechanism has been used in the analysis of clinical data as well [43]. In this study, a deep learning model with stacked long short-term memory (LSTM) was used which paid attention to the topics generated from the clinical notes of patients. Using the attention mechanism, this study found lists the most important topics from the early clinical notes of the patient that were useful for the model to predict the mortality of patients with high efficiency.
Deep learning model interpretation in bioinformatics
In this section, we survey DNN interpretation methods adopted in genomics and epigenomics research and their resulted biological understanding. We majorly focus on popular bioinformatics problems including DNA/RNA binding sequence motif identification [14–16, 20, 44, 45], gene expression prediction [13, 20, 44–46], epigenetic problems such as chromatin accessibility, interaction and DNA methylation predictions [17, 19, 47–50], as well as various directions in non-coding RNA (ncRNA) studies [31, 51, 52].
The adopted interpretation methods in these bioinformatics studies can be categorized into five major classes: input modification, input reconstruction, saliency maps, convolution kernel analysis and attention mechanisms (Table 1, Figure 2). Studies that we failed to categorize with the above are listed in others (Table 1). Note that the convolution kernel analysis, a direct analysis of the convolutional filters/kernels, has not been described in Section 2, as it is a bioinformatics-specific approach that is popular for sequential motif finding problems in bioinformatics [24, 53, 54]. In this analysis, a motif-representing position weight matrix (PWM) is often generated either by input sequence alignments [14, 45] or directly calculating a frequency of input subsequences [44, 48] that activates specific patches of a filter in the first convolutional layer.
Table 1.
Deep learning feature interpretation techniques used by studies in genomics and epigenomics
| Input modification | Input reconstruction | Saliency maps | Convolution kernel analysis | Attention mechanisms (RNN) | Others | |
|---|---|---|---|---|---|---|
| Motif finding | Alipanahi et al., 2015 Lanchantin et al., 2017 | Lanchantin et al., 2016 Lanchantin et al., 2017 | Lanchantin et al., 2017 | Alipanahi et al., 2015 | Lee and Yoon, 2015 | |
| Epigenomics | Zhou et al., 2015 Kelley et al., 2016 | Kelley et al., 2016 Quang et al., 2016 Angermueller et al., 2017 Yin et al., 2019 | ||||
| Chromatin interaction prediction | Singh et al., 2019 | Farre et al., 2018 | Kelley et al., 2018 | Singh et al., 2019 Li et al., 2019 | Zeng et al., 2018 | |
| Gene expression prediction | Singh et al., 2016 | Zeng et al., 2019 | Singh et al., 2017 Sekhon et al., 2018 | Denas and Taylor, 2013 | ||
| ncRNA identification and regulation | Hill et al., 2018 | Park et al, 2017 | Manzanarez-Ozuna et al., 2018 |
Figure 2.
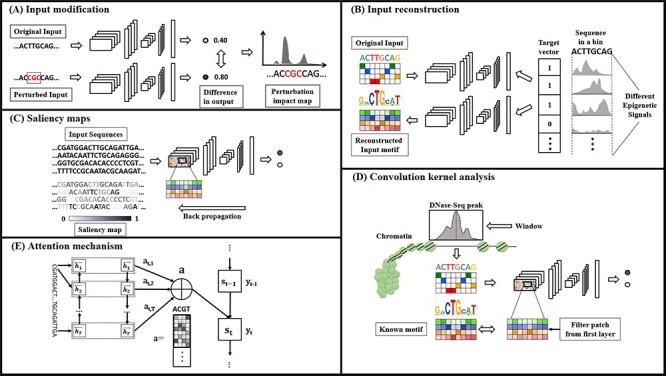
DNN feature interpretation approaches used in genomics and epigenomics. (A) In input modification approach, the effect of changes in the input sequence on the classification decision is measured and visualized as perturbation map. (B) Input reconstruction approach rebuilds an exemplar input that best represents the training weights of the model, which is later compared with the original input. (C) The contribution of the input matrix segments is measured and visualized as saliency maps using backpropagation. (D) In motif finding problems, the first convolution filter of the CNN model is visualized and compared with known motifs. (E) The attention weight mechanism focuses on specific part of the input of an RNN and generates an attention map for that specific input segment on generation of certain output segment.
For the remaining four classes mentioned in Section 2, input modification in bioinformatics is often known by the term in silico mutagenesis. In this method, parts of the input are perturbed with a controlled amount of noise. The subsequent difference in classification performance is used to calculate the importance of that perturbed parts in the model training. Finding the most significant area by exhaustive perturbation of the input segments involves high computation cost. Hence, most of the time, the sequence segment to be perturbed is determined beforehand by either random selection or by selecting a reasonable window centering on a known single nucleotide polymorphism or motif along the input sequence. Bioinformatics studies using input reconstruction strategies follow a more straightforward technique of reconstructing the input representation for a fixed output label or class and comparing it with an input that had been evident as a standard representative of that class in previous studies. Saliency map-based approaches are backpropagation strategies. They track the activation of the filters in the different convolutional layers to find the input contributions. These approaches were proved better than input perturbation in terms of computational cost, since often, they only required a single backward pass to obtain the complete interpretation. Bioinformatic studies using RNN models tended to apply attention mechanisms to find the input contribution on a hidden state of the LSTM by using backpropagation strategies.
DNA/RNA sequence alterations and their impacts on protein binding
DNA- and RNA-binding proteins show specific binding site preferences when they bind to DNA/RNA sequences [53, 55, 56]. Their binding site preference patterns are usually called binding motifs and can often be represented by PWMs [57–59]. Binding motif identification is essential to understanding gene regulation [60–62] and is thus regarded as a classical bioinformatics problem that has been attempted by many algorithms [53, 63–66]. However, due to the intrinsic properties of motifs such as randomness and degeneracy, the problem remains challenging [54, 60, 67–69]. Recent DNN-based methods have demonstrated their superior power in motif identification (Table 1). Their interpretation of motifs also represents early efforts towards DNN interpretation in the bioinformatics domain.
DeepBind. DeepBind was among the first studies to perform a large-scale genome-wide identification of sequence binding specificity of proteins using DNN models [14]. Trained with experimental data, these CNN-based models showed their scalability and ability to characterize DNA- and RNA-binding protein specificity. In order to visualize a discovered transcription factor (TF) binding pattern, a convolution kernel analysis approach was applied: all test sequences were fed through the convolution and rectification stages of a DeepBind model, sequences that contributed to activating a convolution kernel in at least one position by a particular threshold were aligned. From this alignment, subsequences of the average length of a TF binding motif centered around the maximum activating position were extracted to generate a PWM [70]. Since genetic variants in TF binding sites (TFBSs) are likely to perturb gene regulation [71], TF binding affinities were further utilized to identify the functional effects of non-coding genetic variants. For this task, an input modification technique called mutation map was applied. The mutation map was calculated by first obtaining the network’s prediction for a given input sequence, and then, for each position, substituting the original nucleotide found there with one of the alternative nucleotides. The network’s prediction for the mutated sequence was compared with its prediction for the original sequence to obtain the sequence’s sensitivity to the mutation at a specific position. The overall sensitivity score was obtained for substitution at every position of the input sequence with all possible alternative nucleotides. Thus, the mutation map showed the importance of every position of the input sequence to the network’s classification decision. Using these two interpretation strategies, DeepBind generated several examples that were supported with experimentally validated phenomena. Single nucleotide variations from Human Gene Mutation Database [72] were used to show the experimental support. For instance, a TFBS of SP1 was detected in the low density lipoprotein receptor (LDLR) gene promoter region using the convolution kernel analysis approach. Then the mutation map created by the input modification strategy showed how a single nucleotide mutation in that region disrupted the SP1 binding site, which was experimentally shown to lead to familial hypercholesterolemia [73].
DeMo. A DNN model named DeMo was proposed to predict TFBSs as well [15]. The architecture of DeMo features multiple convolutional layers and a highway multilayer perceptron. DeMo models were trained with 108 ChIP-Seq TF data sets on leukemia cell line K562 as DeepBind [14], where 101 bp sequences centered around ChIP-Seq peaks were converted into one-hot encoding serving as positive training samples. The negative training samples were generated by shuffling the sequence nucleotides in the positive samples. An input reconstruction technique inspired by Simonyan et al. [29] was adopted for visualization and interpretation. The approach involved backpropagation through an already trained model with fixed weights to find input sequences that maximally activated the output for a chosen class. The sequences were then converted into a PWM using Laplace smoothing. The motif PWMs found from the reconstructed input sequences were compared with known motif PWMs in the JASPAR database for 57 of the 108 TF test data sets by the tool TOMTOM [69, 74]. The 36 out of the 57 JASPAR motifs could be identified by their method with a significant match (q-value<0.5). The affinities of their motifs on the TFBS test sequences were also compared against JASPAR motifs, showing that the motifs outscored the JASPAR motifs for 29 out of the 57 TFs.
DeMo Dashboard. Later, DeMo was extended into a DNN-based toolkit called Deep Motif Dashboard (DeMo Dashboard) to predict TFBS and visualize TFBS motifs [16]. DeMo Dashboard toolkit includes three different models based on CNN, RNN and CNN-RNN hybrid, respectively. Toward TFBS motif visualization, DeMo Dashboard implemented three techniques: saliency map, temporal output scores and class-specific visualization. A saliency map was generated for a test sequence by calculating the derivative of the DNN score function with respect to the input vector. Pointwise multiplication between the saliency map and the one-hot encoding of the test sequence indicated the importance of each nucleotide position to the classification decision. The temporal output score method was much akin to the input modification technique. In this strategy, a test sequence was fed to the RNN and the CNN-RNN models sequentially, one nucleotide at a time. The prediction scores of the RNN models were observed to find out the exact position in the input sequence, where the model’s prediction was changed. Since the CNN model processed the whole input sequence at once, this strategy was applied only on the RNN and CNN-RNN models. Unlike the above two techniques for which the visualization was specific to an input test sequence, the class-specific visualization method aimed to generalize the visualization to represent the class, which was essentially an input reconstruction method similar to that in previous work [15], where stochastic gradient descent through an already-trained network was used to construct an optimal sequence that maximized the prediction probability of a class, i.e. a TFBS. Using these three techniques, DeMo Dashboard reported a good number of TFBS motifs that significantly matched the JASPAR motifs. The saliency map approach showed that in order to make a decision about a TFBS, the CNN focused on small subsequences of the input sequence, while RNN concentrated on the entire input sequence. The temporal output scores showed that the positions that made the model change their decision to positive were often in the JASPAR motif area. Considering these two strategies, CNN-RNN worked the best, since it was able to achieve the temporal information about the TFBS while concentrating on the motif area. In the case of the input reconstruction strategy, the CNN model did the best at generating concise TF motif representations from a known TF class label.
A deep belief network-based approach. Besides TFBS identification, alternative splice site identification also reveals sequence alterations can have critical effects on gene transcriptional regulation. A deep belief network (DBN) was trained to recognize non-canonical splice sites of gene exons [75]. The non-canonical splice sites could be any combination of nucleotides of varied lengths apart from the canonical splice sites such as AG (acceptor) and GT (donor). The DNA sequence containing canonical splice sites was used to train the model, which later was tested on sequences containing non-canonical splice sites. The DBN contained two weight matrices: 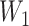 , which mapped the input features to the hidden layer and
, which mapped the input features to the hidden layer and 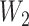 , which mapped the hidden layer to the output layer. The columns of
, which mapped the hidden layer to the output layer. The columns of 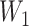 were regarded as feature vectors and the corresponding rows of
were regarded as feature vectors and the corresponding rows of 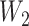 as labels. Each row of
as labels. Each row of 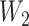 had one element per output class, measuring the confidence of the model’s prediction of that class. Rows in which all elements had similar values represented feature vectors that could not confidently discriminate between classes. In contrast, rows with a larger difference in values specified features that could more effectively discriminate between classes. The feature vectors generated for different sequence inputs were ranked based on their variance, which was indicative of their discriminability of class labels. The five vectors with the highest discrimination were extracted and compared to determine which motifs contributed to effective class discrimination. Along with the canonical splice sites, some non-canonical splice sites were identified such as GCA or NAA in intron boundaries or contiguous A’s in exon boundaries. This technique had the same advantage of relatively low overhead that was seen in several of the deconvolutional approaches of the previous section.
had one element per output class, measuring the confidence of the model’s prediction of that class. Rows in which all elements had similar values represented feature vectors that could not confidently discriminate between classes. In contrast, rows with a larger difference in values specified features that could more effectively discriminate between classes. The feature vectors generated for different sequence inputs were ranked based on their variance, which was indicative of their discriminability of class labels. The five vectors with the highest discrimination were extracted and compared to determine which motifs contributed to effective class discrimination. Along with the canonical splice sites, some non-canonical splice sites were identified such as GCA or NAA in intron boundaries or contiguous A’s in exon boundaries. This technique had the same advantage of relatively low overhead that was seen in several of the deconvolutional approaches of the previous section.
It can be observed from these instances above, as was the case with the broader field of machine learning, both deconvolutional techniques extracting motifs from convolutional filters, and input reconstruction techniques creating an exemplar sequence for a given class are popular in motif finding (Table 1). Deconvolutional techniques are a natural fit for motif finding, as the local receptive field of each filter of a convolutional layer will naturally specialize in detecting short, local patterns, such as motifs and sequence features. This makes the information learned by the filters of convolutional layers a natural source from which to extract motifs that the network learned to recognize as significant. Input reconstruction techniques, on the other hand, are a natural fit for associating motifs with classes of biological interest, since it associates a class with its exemplary input, as learned by the model, and motifs associated with that class can be extracted from that exemplary input. Unlike other fields in machine learning, however, input reconstruction techniques appear to be comparatively more popular (Table 1).
Epigenetic effects of DNA sequence alternations
Epigenomics is a field to study the locations and functions of chemical modifications of genetic materials such as DNAs and histone proteins in a cell. Deep learning has found success in recent years in analyzing large-scale high-throughput epigenomic data. Studies in this direction mostly focused on employing epigenomic and DNA sequence data to predict epigenetic effects of DNA sequence alterations such as chromatin accessibility, DNA methylation and histone modifications (Table 1).
DeepSea. A CNN model named DeepSEA was built to simultaneously predict multiple chromatin effects of DNA sequence alterations, including TF binding, chromatin accessibility and histone marker activities [20]. DeepSEA consists of three convolutional layers, one fully connected layer and a sigmoid output layer. To generate input data, DeepSea split the genome into 200 bp bins. Only the bins with at least one chromatin feature in consideration were kept for further analysis, which covered a total of 17% of the whole genome. A 1 kilobase (kb) long sequence centered around each of the 200 bp bins was converted to one-hot coding, forming the input matrix. The output was a binary vector corresponding to the predictions for all 919 chromatin features. Trained with ENCODE and Roadmap Epigenomics data, DeepSEA was able to predict chromatin features such as TF binding profiles, DNase I hypersensitive site (DHS) profiles and histone mark profiles. Since specific DNA sequence features are associated with protein activities that regulate chromatin accessibility [66, 76], in order to measure the importance of the DNA sequence features, in silico saturated mutagenesis, an input perturbation approach was implemented. Similar to the mutation map [14], one bp was mutated at a time resulting in 3000 extra sequences for every 1 kb long sequence. Then the log-likelihood of every mutated sequence with respect to the original sequence was computed to get the most important positions of that sequence. In this way, FOXA1, FOXA2 and GATA1 affinity were identified by C-to-T, T-to-C and A-to-G alterations, respectively, in different cell types.
Basset. Another CNN-based DNN model named Basset was created, only focusing on the prediction of chromatin accessibility [44]. The full architecture of Basset contains three convolutional layers and two fully connected layers. The input of Basset was a one-hot code represented 600 bp long sequence centering on a DHS. The output was the predicted probability of DNase I hypersensitivity. Basset was trained with DHS data from 164 human cell types. In order to identify sequence features, convolution kernel analysis was performed [44]. In detail, for each filter in the initial convolutional layer, sequences that activated this filter were grouped into a subset, the nucleotide frequencies corresponding to each position in sequences belong to this subset were computed and were converted to a PWM. The motif database CIS-BP [77] was then queried for binding proteins that matched a given PWM using a threshold for a significant similarity (false discovery rate ¡0.1). As a result, 45% of the filter-corresponding PWMs were aligned significantly to protein binding motifs in CIS-BP. Many identified PWMs were found supported by previous experimental evidence, such as known enrichment of higher GC content in TF-bound DHSs, CpG islands and poly-AT stretches and nonconsecutive C’s and G’s. To further pinpoint exact nucleotides that impact chromatin accessibility, Basset also implemented the input modification approach, in silico saturation mutagenesis [44], which generated a heatmap showing influence each possible mutation at each nucleotide position had on the predicted accessibility. Two scores, loss score and gain score, were defined for each position corresponding to the largest possible decrease and increase of accessibility, respectively. If any position in a sequence motif was associated with a high loss or gain score in case of mutation, then the nucleotide at this position was potentially driving accessibility. For example, performing in silico saturation mutagenesis in the AP-1 motif-containing accessible region in embryonic stem cells showed decreased accessibility.
DanQ. A hybrid (CNN+LSTM) DNN model named DanQ was created to predict the chromatin effects of non-coding DNA sequence alterations [45]. The convolutional layer could identify local sequence features such as DNA motifs. Also, the bidirectional LSTM layers were able to learn dependencies between DNA motifs. DanQ utilized the same data set of DeepSEA for training and testing [20]. Similar to the approach used by DeepBind [14] and Basset [44], DanQ also used the convolution kernel analysis technique to identify PWM-represented sequence motifs. Applying a comparison significance cutoff E-value ¡ 0.01 with 320 known motifs, DanQ identified 166 of them in this process. The 166 motifs were also clustered into 118 different varieties to show their model’s ability to learn discriminative functionality of the input genome sequences.
DeepCpG. A DNN model called DeepCpG was designed to predict single-cell methylation states [78]. The model was a combination of a CNN and a gated recurrent unit (GRU) network [26]. The convolution kernel analysis approach was implemented for sequence feature visualization, in which the filters learned by the first convolutional layer in the CNN component were visualized as sequence logos to capture the learned motifs. The importance of the 128 discovered motifs was assessed by measuring their co-occurrence in sequence windows and their association with increased or decreased methylation states predicted by the network. Principal component analysis and hierarchal clustering were used to analyze co-occurrence, and motifs with similar nucleotide compositions were found to co-occur more frequently. In addition, motifs rich in CG nucleotides were found to correlate positively with demethylation and negatively with increased methylation. The 128 motifs were also compared with known motifs, with 20 of the 128 motifs matching known motifs, 17 of which were motifs of TFs with known interactions in the methylation process, and 13 of which were known to interact with two enzymes prominently associated with methylation. These results were encouraging in showing that the network independently learned to recognize features with known biological relevance to the matter of interest helped establish a framework for evaluating visualization criteria and the features learned by a network.
DeepHistone. A DNN model called DeepHistone was designed to accurately predict histone modification sites based on sequences and DNase-Seq data [49]. The architecture of DeepHistone contained three modules corresponding to a DNA sequence, chromosome accessibility and a joint module, respectively. Sequence and chromosome accessibility modules are built on densely connected CNNs. The joint module then combines the previous modules into a feedforward network that produced the final prediction. Similar to DeepCpG [78], DeepHistone implemented the convolution kernel analysis technique to visualize the learned sequence motifs. The sequence motifs were represented as PWMs generated from the first convolutional layer of each CNN and compared with the JASPAR motifs [69]. DeepHistone has shown its ability to focus on biologically significant motifs and motifs that were functional in the cell lines of interest. In different cancer cell lines, several of the motifs recovered corresponded to motifs known to be associated with those forms of cancer. For example, in a lung cancer cell line, DeepHistone retrieved E2F3, a TF that was known to be overexpressed in lung cancer tissue. DeepHistone also retrieved PROX1 and NR2F6 in a cervical cancer cell line, both prominently associated with the progression and spread of cervical cancer. In addition to sequence feature identification, DeepHistone also implemented the input modification technique to investigate the contribution from DNA sequence and chromosome accessibility modules. By modifying the original two-module input into one-module input only architecture, DeepHistone compared the prediction from the alternative DeepHistone (DNA-only) and DeepHistone (DNase-only) models and revealed more contributions from sequence than chromatin accessibility information.
Chromatin interactions
The interaction between different types of gene regulatory regions such as enhancers and promoters is critical to fully understanding gene transcriptional regulation [17–19, 71, 79–86]. Lately, although numerous enhancer regions in different human cell lines have been predicted and experimentally validated [79, 83, 87–91], the driving factors behind the interaction between an enhancer and a promoter are still debatable [79, 83, 87–92]. Researchers working in this area have already started introducing deep learning models to solve this problem (Table 1). Some of the studies also worked on finding important motif features learned by their models. Studies using convolutional layers tend to analyze the convolution kernels/filters after training converges. Some designed their kernels according to a matrix representation of the known TF motifs to see if the input sequence activates the neuron using that filter. Others converted the kernels to a similar matrix representation of the known TF motifs and then compared them to see what important motifs they models were able to learn. Another common trend is the traditional input perturbation technique. Here the sequence inputs are scanned for a known TF motif, and a certain portion of the motif occurring region is mutated with random noise to observe the change in prediction scores.
SPEID. SPEID is among the first DNN models for enhancer–promoter interaction (EPI) prediction using only known enhancer and promoter sequences [17]. The SPEID model included mainly a pair of modules corresponding to enhancers and promoters, each consisting of convolution, activation, max-pool layers, an LSTM and a fully connected layer. The input modification method was implemented to study how changes in input sequences affect the predictions for those sequences. In this procedure, annotated human TF binding motifs were used to scan each enhancer and promoter sequence. The identified occurrences of a TF binding motif in the sequences were then replaced with random noises and the resulted prediction changes were calculated. Averaged prediction accuracy changes were then defined as the importance scores of the corresponding TFs. Correlations of the importance scores of the TFs were shown across six different cell lines. Some of the identified important TFs were also reported by previous studies or supported by experimental evidence to be involved in modulating chromatin loop formation and gene regulation processes. In addition to the input modification approach, convolution kernel analysis was also implemented in SPEID, in which the filters of the convolutional layer were converted into PWMs and then compared with the annotated TF motifs. This procedure was performed for the enhancer and promoter modules separately. TFs that matched with at least three filters with sufficient statistical significance were extracted and compared with the TFs found by TargetFinder [18].
DeepTACT. A DNN model named DeepTACT was created to predict 3D chromatin contacts [48]. DeepTACT was trained by chromatin accessibility data measured with DNase-Seq along with enhancer and promoter sequence data. Two separate DeepTACT models were designed with the same architecture but two different inputs: promoter–promoter and promoter–enhancer interaction data. DeepTACT consisted of three modules: a sequence module that takes promoter and/or enhancer sequences as inputs and contains two separate convolutional layers; an openness module that takes the DNase-Seq data corresponding to the input promoter and promoter/enhancer sequences and also contains two separate convolutional layers; finally, an integration module that merges the output of the two modules with an RNN containing a bidirectional LSTM and an attention layer. Similar to SPEID [17], DeepTACT applied the convolution kernel technique to explain the discovered sequence motifs. The filters of the four convolutional layers in the sequence and openness modules of their model were first converted into PWMs. The PWMs were then compared with annotated TF motif PWMs. A list of closely related TFs was captured by the two ensemble models that were built on promoter–promoter and promoter–enhancer interaction data.
A predictor of chromatin state representative sequences. A DNN model was built with one convolution filter (forward model) to predict a 1D chromatin state sequence representation of the chromatin structure in Drosophila [50]. This model consisted of one convolution layer and a fully connected neural network and was trained to predict a 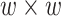 Hi-C contact matrix using a genomic sequence of length
Hi-C contact matrix using a genomic sequence of length 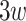 and its overlapping peaks with
and its overlapping peaks with 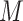 chromatin factors. So, the input was a
chromatin factors. So, the input was a 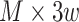 matrix calculated for every chromatin factor and site pair, ranging from 0 to 1, based on the fraction of the site that contained the corresponding factor peak. To create the
matrix calculated for every chromatin factor and site pair, ranging from 0 to 1, based on the fraction of the site that contained the corresponding factor peak. To create the 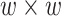 output matrix, Hi-C data of 10 kb resolution in Drosophila was used. The
output matrix, Hi-C data of 10 kb resolution in Drosophila was used. The 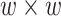 represented mapping between
represented mapping between 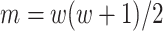 unique sites in its upper diagonal. The only convolution layer with filter width 1 helped the model generate a 1D chromatin state sequence representation from the 2D chromatin factor representation matrix of size
unique sites in its upper diagonal. The only convolution layer with filter width 1 helped the model generate a 1D chromatin state sequence representation from the 2D chromatin factor representation matrix of size 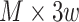 during the training of the forward model. After training the model, an input reconstruction strategy was used to find the 1D chromatin state sequence that was responsible for the chromatin contact of every pair of sites among the
during the training of the forward model. After training the model, an input reconstruction strategy was used to find the 1D chromatin state sequence that was responsible for the chromatin contact of every pair of sites among the 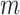 site mappings. When focusing on a neuron representing the contact between a pair of sites among the
site mappings. When focusing on a neuron representing the contact between a pair of sites among the 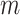 site pairs, the gradient of the chromatin contact values of those site pairs was calculated with respect to the weights learned by the model after training. After a full pass of backpropagation, a 1D chromatin state sequence of length
site pairs, the gradient of the chromatin contact values of those site pairs was calculated with respect to the weights learned by the model after training. After a full pass of backpropagation, a 1D chromatin state sequence of length 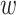 was reconstructed. This state sequence would be highly similar to the 1D chromatin state sequence generated during the training of the forward model, suggesting that given only the chromatin structure, it was possible to generate a likely chromatin state representation that produced it. The reconstructed 1D sequence was a fairly efficient representation of the 2D contact map which provided insights on the difference between the structural and sequential aspects of a chromatin model.
was reconstructed. This state sequence would be highly similar to the 1D chromatin state sequence generated during the training of the forward model, suggesting that given only the chromatin structure, it was possible to generate a likely chromatin state representation that produced it. The reconstructed 1D sequence was a fairly efficient representation of the 2D contact map which provided insights on the difference between the structural and sequential aspects of a chromatin model.
Basenji. A CNN model named Basenji was trained to predict cell-specific epigenetic and transcriptional profiles such as chromatin accessibility [47]. The model consisted of a CNN with max-pooling layers, dilated convolution layers and a fully connected neural network. Dilated convolution layers were multiple layers of convolution filters with gaps where the gap size increased by a factor of two in each layer. The model could be trained with any input DNA sequence up to 131 kb to predict a target vector. Each entry of the target vector represented different epigenetic signals such as CAGE, DNase-Seq, ChIP-Seq and so on, across every 128 bp window of the input sequence. In order to interpret the contribution of the input region on the prediction, a saliency map was calculated with the dot product of the 128 bp bin representation found after the max-pooling step and the gradient of the summed predicted values across the input sequence with respect to the bin representations. For every saliency map, a P-value was calculated by generating saliency maps with shuffled input sequences. The positive saliency score in a region represented enhancing influence of the distal regulatory regions, while the negative score represented silencing influence. The relative saliency scores on putative enhancer, promoter and CCCTC-binding factor (CTCF) binding regions against shuffled backgrounds revealed a significant difference.
EP2vec. Apart from using feedforward networks or RNNs directly to learn the sequence features, NLP techniques were applied in several works to extract features from the input sequences to train a machine learning model. A model named EP2vec was developed to predict EPIs [19]. EP2vec used paragraph vector [93] that originated from word2vec [94] to extract feature vectors from the enhancer and promoter sequences. Here paragraph vector was used to generate a d-dimensional sentence vector for each sequence. EP2vec was trained with the same enhancer and promoter data set as SPEID. Once the training was converged, a sentence vector and word vector embeddings were obtained for each sequence. The learned sequence vectors for each enhancer and promoter sequence pair were further concatenated to train a gradient boosted regression trees model to predict the interaction between that enhancer and promoter. Using the extracted sentence vector and word vectors, EP2vec also determined the importance of each k-mer word within its sequence, based on the similarity of the corresponding sentence vector and word vectors. A similarity score was calculated for every k-mer in a sequence based on the multiplication between the k-mer-associated word vector and the sequence-associated sentence vector. The importance score of each k-mer was then determined by normalization based on the similarity scores of all the k-mers generated from that sequence. The two sets of most important word vectors (for enhancers and promoters) were used to compare with the known motifs. In this way, known motifs that were highly represented in a sequence were identified. EP2vec discovered cell-specific TF motifs involved in a diverse developmental role in the human body.
Despite being able to find a large number of experimentally supported regulatory TF motifs in different cell lines, the feature interpretation part of the above-mentioned studies is confined to the motif finding in enhancer and promoter sequences, respectively. Therefore, the interpretation is not directly related to the physical interactions between the enhancers and promoters. Certain pairs of interacting TFs are shown to bind enhancers and promoters, respectively, which contribute to the interaction of enhancers and promoters [95–97]. Therefore, Identifications of features that are directly relevant to EPIs will benefit further modeling EPIs.
Gene expression prediction
Gene expression in multicellular organisms is normally measured by the amount of messenger RNA (mRNA) transcripts generated in the cell at a given time. The underlying mechanism of gene expression regulation is quite complex, involving various inter-connected transcriptional networks formed by TFs, RNA polymerase II, cis- and trans-regulatory regions, different histone modification events and so on [68, 83, 98–100]. Large amount of data such as TF ChIP-Seq, histone modification ChIP-Seq, Hi-C and RNA-seq have enabled exploiting DNN models to study the regulatory mechanisms underlying gene expression. The DNN feature interpretation methods in these studies, as detailed below, are diverse, covering the convolution kernel analysis, attention mechanisms, input reconstruction and others (Table 1).
A predictor of gene functional elements. One of the very early attempts to utilize DNNs was to understand gene expression [51]. In this study, a CNN model was created to predict a gene’s transcriptional expression level labeled as either an ‘induced’ or ‘repressed’ state. The input data was formatted as an 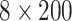 input matrix corresponding to the TF ChIP-Seq binding profiles of four TFs over a 20 kb long DNA sequence surrounding gene transcription start sites (TSSs) in two different types of cell lines. The model architecture contained three convolutional layers, a fully connected layer and a softmax layer. Two deconvolutional methods, deconvnet and backpropagation, were implemented for model interpretation [25, 29]. To illustrate the biological discovery, the authors averaged the input representations for the best 10 examples. The input representations suggested an interplay between TFs in the relevant cells. For example, the induced genes showed a joint enrichment for a pair of TFs, GATA1 and TAL1, in one type of cells while a joint enrichment for another pair, GATA2 and TAL1, in other types of cells. Also, the alignment of GATA1 and GATA2 signals in one type of cells at the proximity of the TSS was observed in their interpretation, suggesting supportive action of both TFs in gene inducement.
input matrix corresponding to the TF ChIP-Seq binding profiles of four TFs over a 20 kb long DNA sequence surrounding gene transcription start sites (TSSs) in two different types of cell lines. The model architecture contained three convolutional layers, a fully connected layer and a softmax layer. Two deconvolutional methods, deconvnet and backpropagation, were implemented for model interpretation [25, 29]. To illustrate the biological discovery, the authors averaged the input representations for the best 10 examples. The input representations suggested an interplay between TFs in the relevant cells. For example, the induced genes showed a joint enrichment for a pair of TFs, GATA1 and TAL1, in one type of cells while a joint enrichment for another pair, GATA2 and TAL1, in other types of cells. Also, the alignment of GATA1 and GATA2 signals in one type of cells at the proximity of the TSS was observed in their interpretation, suggesting supportive action of both TFs in gene inducement.
DeepChrome. Another DNN model called DeepChrome was also created to predict gene expression [46]. DeepChrome focused on using histone modification data to predict gene expression in different human cell types. DeepChrome was trained with gene expression levels and five different histone modification signals in 56 different cell types [101]. To visualize the combinatorial histone interactions and their effect on the prediction, the authors implemented a four-step input reconstruction technique to construct an optimal input corresponding to a classification label. First, a random input was initialized. Then the input vector was updated by the gradient of the loss function with respect to the current input so that the loss function was minimized for the desired label. Each iteration performed stochastic gradient descent on the gradient calculated using the same learning rate used to train the model. After the optimal input and the label of interest were trained, the third step was to clamp the value of each bin with 0 and then normalize it to keep within an interval [0, 1]. Finally, a value ¿ 0.25 was used to identify active bins. The frequency of active bins for each histone mark was counted, and those histone marks with significantly above average counts were considered to have a significant impact on achieving the gene expression level indicated by the output label of interest. The result was a heatmap that showed which histone modification signals were important to achieving a particular gene expression level, independent of the specific gene in question. This input reconstruction method enabled multiple discoveries supported by various studies before. For instance, the correlation between H3K4me3 and H3K36me3 histone marks and H3K4me3 and H3K4me1 histone marks were observed in their generated heatmaps. Also, previously reported coexistence of H3K9me3 and H3K27me3 repressor marks was observed around repressed genes.
AttentiveChrom. Using the same training data set as DeepChrome [46], another model called AttentiveChrom was developed [31]. AttentiveChrom contained a hierarchy of LSTMs to model and analyze the complex dependencies among chromatin factors that regulated gene expression. In this study, the H3K27me3, H3K9me3 markers were considered as the repressed gene markers, H3K4me1 was defined as the enhancer marker, H3K4me3 was defined as the promoter marker, and H3K36me3 was the gene body structure marker. Inspired by Yang et al. [42], AttentiveChrom jointly trained two levels of attention, one attending to the important chromatin markers and the other attending to the important positions within chromatin markers. The attention weights trained by these attention layers served as a form of visualization giving insight into specific important positions considered by the model when making classification decisions. A high correlation between the attention weights was reported for a new histone signal data and the preprocessed separate histone mark profiles for five different histone signals used in the model. For the ‘ON’ genes, high attention weights were found around the promoter, enhancer and gene structure markers, while they were low or average around the repress markers. The opposite was observed for the ‘OFF’ genes. Similar results were also shown for different cell types separately using cell-specific data.
DeepDiff. A model called DeepDiff was built to predict differential gene expression in different cell lines from histone modification signals [102]. Similar to the attention mechanism technique [31], DeepDiff model was built on a hierarchy of LSTMs with two jointly trained levels of attention weights and trained on the same data sets. For a specific gene in a specific cell line, a 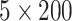 matrix was generated representing the five histone marker signals across the 20 kb long region around the gene TSS. Two such matrices created in this way corresponding to two different cell types served as the input of DeepDiff. Alternative inputs were defined as difference and concatenated histone marker signals in the given two cell types. Like DeepChrome [46], the attention weights learned by the network were used as a means of visualizing the features learned by the algorithm. HM level and bin level attention mechanisms were added to represent the contribution of every histone modification pattern and every bin of every histone modification pattern in prediction results, respectively. In order to interpret the differential gene expression using the attention weights, one of the best performing samples was selected from the test set. The learned attention weights were then recorded for the five histone modification markers corresponding to the predicted top five upregulated/downregulated genes in the cancer cells. Among the five histone markers, H3K4me1 and H3K4me3 received relatively high weights in the upregulated genes and relatively low weights in the downregulated genes. In contrast, H3K27me3 received relatively low weight in the upregulated genes and relatively high weight in the downregulated genes, which were experimentally demonstrated in particular cell lines.
matrix was generated representing the five histone marker signals across the 20 kb long region around the gene TSS. Two such matrices created in this way corresponding to two different cell types served as the input of DeepDiff. Alternative inputs were defined as difference and concatenated histone marker signals in the given two cell types. Like DeepChrome [46], the attention weights learned by the network were used as a means of visualizing the features learned by the algorithm. HM level and bin level attention mechanisms were added to represent the contribution of every histone modification pattern and every bin of every histone modification pattern in prediction results, respectively. In order to interpret the differential gene expression using the attention weights, one of the best performing samples was selected from the test set. The learned attention weights were then recorded for the five histone modification markers corresponding to the predicted top five upregulated/downregulated genes in the cancer cells. Among the five histone markers, H3K4me1 and H3K4me3 received relatively high weights in the upregulated genes and relatively low weights in the downregulated genes. In contrast, H3K27me3 received relatively low weight in the upregulated genes and relatively high weight in the downregulated genes, which were experimentally demonstrated in particular cell lines.
DeepExpression. DeepExpression [13] is another DNN model to predict gene expression using the promoter sequence features and distal EPI features. The model contains two separate modules: one for extracting features from DNA sequences in promoter regions and the other for extracting features from EPI signals. To interpret the underlying feature patterns, DeepExpression used a convolution kernel analysis similar to the interpretation process adopted by SPEID [17] and DeepTACT [48]. The filters of the first convolutional layer in the proximal promoter module that activated a neuron were converted to motifs, and these motifs were then compared with known motifs with a low E-value cutoff. Identified TFs are shown involved in embryonic development, cell cycle regulation, tissue-specific gene expression, cell signaling, apoptosis, tumorigenesis and so on.
ncRNA identification and regulation
ncRNAs are RNA transcripts that are not translated into proteins. The majority of transcripts produced by the human genome are ncRNAs in contrast to only 12~% transcripts as coding RNAs. Despite the abundance of ncRNAs in the human genome, their localization, function and regulation are largely unknown. For instance, for one of the most widely studied types of ncRNAs, microRNAs (miRNAs), we do not yet have a complete picture of their biogenesis, interactions and regulatory mechanisms [103]. Recently, DNN models have been applied to computational studies of ncRNAs. Most of these studies focused on model prediction accuracy. A few of them attempted model interpretation by investigating the importance of extracted features (Table 1).
DeepMiRGene. An RNN model was developed to identify precursor miRNAs (pre-miRNAs) [103]. Both nucleotide and secondary structure information of RNA sequences were integrated as the input of the model. To determine which part of the sequential input was important for the classification decision, the authors implemented an RNN-based CAM approach. An attention mechanism was applied in the LSTM part of their model to learn long-term dependencies of the primary and secondary structure of an RNA molecule. An activation map was formed by performing global average pooling on the attention weighted output to obtain a weight vector. The activation map for a given input sequence was then generated based on the pointwise multiplication of the attention weighted output and the weight vector. Although all pre-miRNA motifs or sequence patterns learned by the model were not reported in this study, the presented CAM technique can be useful for visualizing the most significant sequence features in different studies.
A predictor of miRNA regulation. Using the expression values of 179 miRNAs and mRNA-Smad7 in 1074 samples of patients with breast cancer, a DNN model was designed to predict mRNA-Smad7 expression regulation by miRNAs [52]. A genetic algorithm was used to find the structure of the DNN model with the best predictive ability, along with the best input data set for the model. Forty-four miRNAs were selected to train their best DNN model. The relative importance of each of these miRNAs on the expression of mRNA-Smad7 was evaluated using the Olden algorithm [28, 104]. The Olden algorithm is a classical feature interpretation technique that focuses on the connection weights and direction of the neural network to evaluate feature importance. A greater weight with a positive direction represents greater relative importance. Twenty-three miRNAs were identified as the most important in the predictive decisions of the DNN, among which five have experimentally validated roles in breast cancer. Six miRNAs were shown to be involved in other cancer types, which contributed 23% of the relative importance of their model.
mRNN. A DNN model called mRNN was developed to understand the coding potential of RNAs. mRNN was a GRU-based RNN [105]. Each RNA sequence was converted into one-hot encoding serving as the input matrix. The model output was generated as a score representing the coding potential of the input sequence. Full-length human transcript sequences for mRNA and long ncRNAs (lncRNAs) from GENCODE were used to train mRNN [106]. Four input modification techniques were implemented to interpret the model. First, a region from the 5’ UTR, CDS and 3’ UTR regions of an mRNA was randomly selected and shuffled, the analysis of which suggested higher coding potential of the CDS than the UTR regions. Second, a single nucleotide was mutated (point mutation analysis) at every position of an mRNA transcript. Only 0.227% of the point mutations were able to change the prediction for mRNAs from coding to noncoding. Among these classification-flipping mutations, 67.1% fell within coding regions, and 42.6% of these coding region mutations created an early stop codon. The third input modification technique involved mutating a pair of nucleotides. In order to remove the effect of the point mutation from the pair-wise mutation, a synergy score was defined to exclude the change in the prediction score resulted from a single nucleotide mutation. Finally, the sharpest changes in prediction score (spikes) were identified when truncating an input sequence in different places. In a test set with 500 mRNAs and 500 lncRNAs, the most significant spikes were identified in 82% of the mRNAs and only 9% of lncRNAs. The distribution of the significant spike positions for mRNAs peaked within the CDS, shortly after the start codon. Within a 50 bp window around the center of the spikes, 11 significantly enriched codons were identified. Mutating these codons decreased the spike heights 97.4% of the time.
Although the recent bioinformatic studies regarding ncRNAs tend to apply deep learning techniques with an increasing rate, only a handful of them considered probing their DNNs to explore interpretation possibilities. There is currently a lack of DNN studies on ncRNA-disease association or ncRNA classification that attempted to identify feature interpretations. Two studies we summarized above adopted the old technique involving neuron weights and the computation-intensive input perturbation techniques. One of the future directions in the field of ncRNA research is thus to exploit state-of-the-art feature interpretation techniques.
Discussion
During the past several years, following the triumph of DNNs in various research areas of computer vision and NLP dealing with big data, numerous studies have applied them to solve problems in genomics and epigenomics where a large number of experimental data sets are publicly available. Although the majority of these studies focus only on the prediction performance of DNNs, a number of recent studies attempted model interpretations by identifying the underlying features that were vital to the predictions (Table 2). Since bioinformatics covers a wide spectrum of research topics, we confined this study to five recent topics where DNNs were most adopted and illustrated the recent development and application of DNN interpretation techniques in genomics and epigenomics.
Table 2.
Examples of the features identified by the DNN studies on genomics and epigenomics
| Area | Studies | Feature interpretation results |
|---|---|---|
| Motif finding | Lee and Yoon, 2015 | Along with the canonical splice sites (GT and AG), some non-canonical splice sites were identified such as, GCA or NAA in intron boundaries or contiguous A’s in exon boundaries. |
| Alipanahi et al., 2015 | Several examples were generated that were supported by experiments. For instance, a single nucleotide mutation that disrupted an SP1 binding site in the LDLR promoter leads to familial hypercholesterolemia. | |
| Lanchantin et al., 2016 | The 36 out of 57 JASPAR motifs were identified with a significant match (q-value ¡ 0.5), 29 of which are better than the corresponding JASPAR motifs. | |
| Lanchantin et al., 2017 | A good number of motifs were reported that significantly matched the JASPAR motifs. | |
| Epigenomics | Zhou et al., 2015 | FOXA1, FOXA2 and GATA1 affinity were identified by C-to-T, T-to-C and A-to-G alterations, respectively, in different cell types. |
| Kelley et al., 2016 | The 45% of the filters aligned significantly to protein binding motifs in CIS-BP. Some findings were supported by previous experimental evidence, such as known enrichment of higher GC content in TF-bound DHSs, CpG islands and poly-AT stretches and nonconsecutive C’s and G’s. | |
| Quang et al., 2016 | The 166 of the 320 known motifs could be identified by the interpretation method. The model was shown to learn discriminative functionality of the input genome sequences. | |
| Angermueller et al., 2017 | Motifs with similar nucleotide compositions were found to co-occur more frequently. In addition, motifs rich in CG nucleotides were found to correlate positively with demethylation and negatively with increased methylation. The 20 of the 128 motifs matched known motifs, 17 of which were transcription factors with known interactions in the methylation process, and 13 of which were known to interact with two enzymes prominently associated with methylation. | |
| Yin et al., 2019 | E2F3 was retrieved in a lung cancer cell line, and PROX1 and NR2F6 were retrieved in a cervical cancer cell line. | |
| Chromatin interaction prediction | Singh et al., 2019 | The importance scores of the TFs were fairly correlated across six different cell lines. Some of these important TFs (CTCF, SRF, JUND, SPR1, SP1, EBF1, JUN, BCL11A, ZIC4, E2F3, FOXK1, etc.) were known to be involved in modulating chromatin loop formation and gene regulation processes. |
| Li et al., 2019 | A list of closely related TFs was reported. | |
| Farre et al., 2018 | The reconstructed input sequences were fairly correlated (0.73) with the original input sequences in their test data set. | |
| Kelley et al., 2018 | Promoters were found to have extreme scores at both high and low ends, which were far lesser for enhancers showing lower repressive contribution of enhancers that promoters on gene expression. | |
| Zeng et al., 2018 | Known motifs that are highly represented in a sequence could be identified. Cell-specific TF motifs could also be retrieved that are reported to be involved in diverse developmental roles in the human body. | |
| Gene expression prediction | Denas et al., 2013 | The interpretation method suggested an interplay between GATA1 and GATA2 in the G1E cells. The induced genes showed a joint enrichment for both GATA1 and TAL1 in the ER4 cells and analogous signal alignment between GATA2 and TAL1 in the G1E cells. Also, the alignment of GATA1 and GATA2 signals in ER4 cells at the proximity of the TSS observed in their interpretation suggesting supportive action of both GATA proteins in gene inducement in ER4 cells. |
| Singh et al., 2016 | Correlation between H3K4me3 and H3K36me3 histone marks and H3K4me3 and H3K4me1 histone marks were observed in their generated heatmaps. Also, previously reported coexistence of H3K9me3 and H3K27me3 repressor marks were observed around repressed genes. | |
| Singh et al., 2017 | A high correlation between the attention weights for a new histone signal data and the preprocessed separate histone mark profiles were reported for the five different histone signals used in their model. | |
| Sekhon et al., 2018 | H3K4me1 and H3K4me3 received relatively high weights in upregulated genes and relatively low weights in downregulated genes, while H3K27me3 received relatively low weight in upregulated genes and relatively high weight in downregulated genes, results which had been experimentally demonstrated for these particular cell lines. | |
| Zeng et al., 2019 | NANOG, FOXQ1, ETS1, MYC, etc. were reported as the top TFs, which tend to involve in embryonic development, cell cycle regulation, tissue-specific gene expression, cell signaling, apoptosis, tumorigenesis, etc. | |
| ncRNA identification and regulation | Park et al, 2017 | Visualization of the attention weights heatmap along the positive and negative human pre-miRNA sequence data showed a clear difference in signal along the 10–50% sequence positions where a mature miRNA is located within the pre-miRNA sequences. |
| Manzanarez-Ozuna et al., 2018 | The 23 miRNAs were generated which played the most important roles in the predictive decisions of the DNN. Five of these miRNAs play roles in breast cancer, which together accumulated 40% of the relative importance assigned by their predictive model. Among the rest, six of miRNAs were involved in other cancer types which contributed 23% of the relative importance of their model. | |
| Hill et al., 2018 | Only 0.227% of the point mutations were able to change the prediction for true mRNAs from coding to noncoding. Among these classification-flipping mutations, 67.1% fell within coding regions and 42.6% of these coding region mutations created an early stop codon (UGA/UAG/UAA). The most significant pair of mutations occurred at a CAC codon, and together created a premature UAA stop codon, which the classification from coding to non-coding. For the other transcript, pairs of positions, not in the same codon, changed the synergy score more when mutated together than when mutated separately. |
The surveyed DNN interpretation methods mostly focused on evaluating the importance of individual features rather than interacted feature groups (Table 2). For example, to evaluate a sequence variant’s impact on function, studies often aim to mutate one or two nucleotides and observe their influence on the predictions. However, most bioinformatics problems involve interacting entities such as genes in the same pathways, proteins involved in the same complex and different types of RNA species regulating each other. Incorporating feature interactions into the model interpretation is necessary for realistic problem understanding. Recent DNN development toward this direction starts to consider the relationship among different features, including both linear and nonlinear relationships [107–109]. In the meantime, although many methods have been developed, there is not a unified way to evaluate the methods. When it comes to the omics domain, methods that achieve good explanation and interpretability would require the evaluations specific to particular problems.
If we follow our definitions of explanation and interpretation, i.e. interpretation is the first step toward explanation, then the learned features from the surveyed DNN models thus have limited capability of explaining the model’s decision-making. Nevertheless, to a certain degree, a lot of current DNN models reviewed here focusing on feature identification are able to interpret how and why the prediction is made using these features in a given scenario. Even so, the interpretation of DNN models towards understanding the biological mechanism is still far from reach. Most existing DNN-based methods heavily rely on traditional databases and literature searches for biological interpretation. For example, DeepSEA was able to utilize trained DNNs to predict sequence alterations’ effects on protein binding such as TF binding, histone marker activities and chromatin accessibility. However, when these effects were further considered to predict their functional consequence, resources outside the models such as mutation and genome-wide association databases would be required for additional prediction and interpretation. The predicted functional consequences of a specific nucleotide would need to be subsequently validated based on a literature search. Therefore, the challenge remains for advanced DNN development such that they are capable of incorporating existing knowledge in various types of formats, streamlining their prediction procedure and directly providing biological interpretations. Such development will provide essential insight into bioinformatics research.
Key Points
DNNs have recently gained popularity in various types of genomics and epigenomics studies and achieved high accuracy in predictions and classifications.
DNN model interpretation is important in bioinformatics to gain insights into phenotypes and their underlying biological mechanisms.
DNN model interpretation in bioinformatics studies can be classified into five major classes.
The interpretation of DNN models towards understanding the biological mechanism is still far from reach.
Amlan Talukder is a graduate student from the Department of Computer Science, University of Central Florida. He mainly works on miRNAs and epigenomics.
Clayton Barham is a graduate student from the Department of Computer Science, University of Central Florida. He mainly works on TSS-seq and CAGE data analysis.
Xiaoman Li is an associate professor from Burnett School of Biomedical Science, University of Central Florida. He works on chromatin interactions and metagenomics.
Haiyan Hu is an associate professor from the Department of Computer Science, University of Central Florida. She works on miRNAs, epigenomics and gene transcriptional regulation.
Contributor Information
Amlan Talukder, Computer Science, University of Central Florida, Orlando, FL 32816, USA.
Clayton Barham, Computer Science, University of Central Florida, Orlando, FL 32816, USA.
Xiaoman Li, Burnett School of Biomedical Science, University of Central Florida, Orlando, FL 32816, USA.
Haiyan Hu, Computer Science, University of Central Florida, Orlando, FL 32816, USA.
Authors’ contributions
X.L. and H.H. conceived the idea. A.T., C.B., X.L. and H.H. analyzed the studies and wrote this manuscript. All authors read and approved the final manuscript.
Conflict of Interest
There is no conflict of interest declared.
Funding
This work has been supported by the National Science Foundation [grants 2015838 and 1661414] and the National Institute of Health [grant R15HGM123407].
Reference
- 1. Garcia-Garcia A, Orts-Escolano S, Oprea S, et al. A review on deep learning techniques applied to semantic segmentation. arXiv preprint, arXiv:1704.06857, 2017.
- 2. Zhu Xiao Xiang, Tuia Devis, Mou Lichao, et al. Deep learning in remote sensing: a comprehensive review and list of resources. IEEE Geosci Remote Sens Mag 2017; 5(4): 8–36. [Google Scholar]
- 3. LeCun Yann, Bengio Yoshua, Hinton Geoffrey. Deep learning. Nature 2015; 521(7553):436–44. [DOI] [PubMed] [Google Scholar]
- 4. Voulodimos A, Doulamis N, Doulamis A, et al. Deep learning for computer vision: a brief review. Comput Intell Neurosci 2018;2018:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Young T, Hazarika D, Poria S, et al. Recent trends in deep learning based natural language processing. IEEE Comput Intell Mag 2018;13(3):55–75. [Google Scholar]
- 6. Krizhevsky Alex, Sutskever Ilya, Hinton Geoffrey E.. ImageNet classification with deep convolutional neural networks. Commun ACM 2017;60(6):84–90. [Google Scholar]
- 7. Li K, Ma W, Sajid U, et al. Object detection with convolutional neural networks. CoRR,abs/1912.01844, 2019. [Google Scholar]
- 8. Wang J, Zhang Z, Xie C, et al. Unsupervised learning of object semantic parts from internal states of CNNs by population encoding. arXiv preprint, arXiv:1511.06855, 2015.
- 9. Zhou B, Khosla A, Lapedriza A, et al. Object detectors emerge in deep scene cnns. In: International Conference on Learning Representations, San Diego, CA, USA, 2015.
- 10. Yao Guangle, Lei Tao, Zhong Jiandan. A review of convolutional-neural-network-based action recognition. Pattern Recognit Lett 2019;118:14–22. [Google Scholar]
- 11. Mikolov Tomas, Zweig Geoffrey. Context dependent recurrent neural network language model. In: 2012 IEEE Spoken Language Technology Workshop (SLT), Miami, FL, USA, IEEE, 2012. [Google Scholar]
- 12. Zhang X, Zhao J, LeCun Y. Character-level convolutional networks for text classification. In: Advances in Neural Information Processing Systems, 2015;649–57.
- 13. Zeng Wanwen, Wang Yong, Jiang Rui. Integrating distal and proximal information to predict gene expression via a densely connected convolutional neural network. Bioinformatics 2019;36:496–503. [DOI] [PubMed] [Google Scholar]
- 14. Alipanahi Babak, Delong Andrew, Weirauch Matthew T, Frey Brendan J. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat Biotechnol 2015;33(8):831–8. [DOI] [PubMed] [Google Scholar]
- 15. Lanchantin J, Singh R, Lin Z, et al. Deep motif: visualizing genomic sequence classifications. arXiv preprint, arXiv:1605.01133, 2016.
- 16. Lanchantin J, Singh R, Wang B, et al. Deep motif dashboard: visualizing and understanding genomic sequences using deep neural networks. In: Pacific Symposium on Biocomputing, Kohala Coast, Hawaii, USA. World Scientific, 2017, 254–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Singh Shashank, Yang Yang, Póczos Barnabás, Ma Jian. Predicting enhancer–promoter interaction from genomic sequence with deep neural networks. Quant Biol 2019;7(2):122–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Whalen Sean, Truty Rebecca M, Pollard Katherine S. Enhancer–promoter interactions are encoded by complex genomic signatures on looping chromatin. Nat Genet 2016;48(5):488–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zeng Wanwen, Wu Mengmeng, Jiang Rui. Prediction of enhancer–promoter interactions via natural language processing. BMC Genomics 2018;19(S2):84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhou Jian, Troyanskaya Olga G. Predicting effects of noncoding variants with deep learning-based sequence model. Nat Methods 2015;12(10):931–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Doshi-Velez F, Kim B. Towards a rigorous science of interpretable machine learning. arXiv preprint, arXiv:1702.08608, 2017.
- 22. Leilani H, Gilpin DB, Yuan BZ, et al. Explaining explanations: an overview of interpretability of machine learning. In: 2018 IEEE 5th International Conference on Data Science and Advanced Analytics (DSAA), Turin, Italy, IEEE, 2018, 80–9. [Google Scholar]
- 23. Montavon Grégoire, Samek Wojciech, Müller Klaus-Robert. Methods for interpreting and understanding deep neural networks. Digit Signal Process 2018;73:1–15. [Google Scholar]
- 24. Eraslan Gökcen, Avsec žiga, Gagneur Julien, Theis Fabian J.. Deep learning: new computational modelling techniques for genomics. Nat Rev Genet 2019;20(7):389–403. [DOI] [PubMed] [Google Scholar]
- 25. Zeiler MD, Fergus R. Visualizing and understanding convolutional networks. In: European Conference on Computer Vision, Zurich, Switzerland, Springer, 2014, 818–33. [Google Scholar]
- 26. Grün F, Rupprecht C, Navab N, et al. A taxonomy and library for visualizing learned features in convolutional neural networks. arXiv preprint arXiv:1606.0775, 2016. [Google Scholar]
- 27. Nguyen A, Yosinski J, Clune J. Multifaceted feature visualization: uncovering the different types of features learned by each neuron in deep neural networks. arXiv preprint, arXiv:1602.03616, 2016.
- 28. Olden Julian D, Jackson Donald A. Illuminating the ‘black box’: a randomization approach for understanding variable contributions in artificial neural networks. Ecol Model 2002;154(1-2):135–50. [Google Scholar]
- 29. Simonyan K, Vedaldi A, Zisserman A. Deep inside convolutional networks: visualising image classification models and saliency maps. The 2nd International Conference on Learning Representations, Banff, AB, Canada, 2014.
- 30. Yosinski J, Clune J, Nguyen A, et al. Understanding neural networks through deep visualization. In: Deep Learning Workshop, International Conference on Machine Learning (ICML), Lille Grande Palais, France 2015.
- 31. Singh R, Lanchantin J, Sekhon A, et al. Attend and predict: understanding gene regulation by selective attention on chromatin. In: Advances in Neural Information Processing Systems (NIPS), Long Beach, CA, 2017;6785–95. [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang Q-s, Zhu S-C. Visual interpretability for deep learning: a survey. Front Inf Technol Electron Eng 2018;19(1):27–39. [Google Scholar]
- 33. Mahendran Aravindh, Vedaldi Andrea. Visualizing deep convolutional neural networks using natural pre-images. Int J Comput Vis 2016;120(3):233–55. [Google Scholar]
- 34. Springenberg JT, Dosovitskiy A, Brox T, et al. Striving for simplicity: the all convolutional net. In: International Conference on Learning Representations (workshop track), San Diego, CA, USA, 2015.
- 35. Dosovitskiy A, Brox T. Inverting visual representations with convolutional networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA. IEEE Computer Society, 2016, 4829–37.
- 36. Mnih Volodymyr, Heess Nicolas, Graves Alex, et al. Recurrent models of visual attention. In: Advances in Neural Information Processing Systems, Montreal, Quebec,Canada, 2014, 2204–2212.
- 37. Zintgraf LM, Cohen TS, Adel T, et al. Visualizing deep neural network decisions: prediction difference analysis. In: International Conference on Learning Representations, Toulon, France, 2017.
- 38. Montavon Grégoire, Lapuschkin Sebastian, Binder Alexander, Samek Wojciech, Müller Klaus-Robert. Explaining nonlinear classification decisions with deep Taylor decomposition. Pattern Recognit 2017;65:211–22. [Google Scholar]
- 39. Zhou B, Khosla A, Lapedriza A, et al. Learning deep features for discriminative localization. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 2016, 2921–9.
- 40. Long JL, Zhang N, Darrell T. Do convnets learn correspondence? In: Advances in Neural Information Processing Systems, Montreal, Quebec, Canada, 2014, 1601–9.
- 41. Li J, Chen X, Hovy E, et al. Visualizing and understanding neural models in nlp. In North American Chapter of the Association for Computational Linguistics: Human Language Technologies, San Diego, CA, USA, 2016, 681–91. [Google Scholar]
- 42. Yang Z, Yang D, Dyer C, et al. Hierarchical attention networks for document classification. In: Proceedings of the 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies. Association for Computational Linguistics, San Diego California, USA, 2016, 1480–9. [Google Scholar]
- 43. Wang L, Sha L, Lakin JR, et al. Development and validation of a deep learning algorithm for mortality prediction in selecting patients with dementia for earlier palliative care interventions. JAMA Netw Open 2019;2(7): e196972–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. David R. Kelley, Jasper Snoek, Rinn John L.. Basset: learning the regulatory code of the accessible genome with deep convolutional neural networks. Genome Res 2016;26(7):990–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Quang Daniel, Xie Xiaohui. DanQ: a hybrid convolutional and recurrent deep neural network for quantifying the function of DNA sequences. Nucleic Acids Res 2016;44(11):e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Singh Ritambhara, Lanchantin Jack, Robins Gabriel, Qi Yanjun. DeepChrome: deep-learning for predicting gene expression from histone modifications. Bioinformatics 2016;32(17):i639–48. [DOI] [PubMed] [Google Scholar]
- 47. Kelley David R., Reshef Yakir A., Bileschi Maxwell, et al. Sequential regulatory activity prediction across chromosomes with convolutional neural networks. Genome Res 2018;28(5):739–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li Wenran, Wong Wing Hung, Jiang Rui. DeepTACT: predicting 3d chromatin contacts via bootstrapping deep learning. Nucleic Acids Res 2019;47(10):e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yin Qijin, Wu Mengmeng, Liu Qiao, Lv Hairong, Jiang Rui. DeepHistone: a deep learning approach to predicting histone modifications. BMC Genomics 2019;20(S2):11–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Farré Pau, Heurteau Alexandre, Cuvier Olivier, Emberly Eldon. Dense neural networks for predicting chromatin conformation. BMC Bioinform 2018;19(1):372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Denas O, Taylor J. Deep modeling of gene expression regulation in an erythropoiesis model. In: Representation Learning, ICML Workshop. New York, USA: ACM, 2013. [Google Scholar]
- 52. Manzanarez-Ozuna Edgar, Flores Dora-Luz, Gutiérrez-López Everardo, Cervantes David, Juárez Patricia. Model based on GA and DNN for prediction of mRNA-smad7 expression regulated by miRNAs in breast cancer. Theor Biol Med Model 2018;15(1):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Das MK, Dai H-K. A survey of DNA motif finding algorithms. BMC Bioinform 2007;8:S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang Ying, Goodison Steve, Li Xiaoman, Haiyan Hu.. Prognostic cancer gene signatures share common regulatory motifs. Sci Rep 2017;7(1):4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Achar Avinash, Sætrom Pål. RNA motif discovery: a computational overview. Biol Direct 2015;10(1):61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cai Xiaohui, Lin Hou, Naifang Su, et al. Systematic identification of conserved motif modules in the human genome. BMC Genomics 2010;11(1):567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cook K. B., Kazan H., Zuberi K., Morris Q., Hughes T. R.. RBPDB: a database of RNA-binding specificities. Nucleic Acids Res 2010;39(Database):D301–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ding Jun, Dhillon Vikram, Li Xiaoman, Haiyan Hu.. Systematic discovery of cofactor motifs from ChIP-seq data by SIOMICS. Methods 2015;79-80:47–51. [DOI] [PubMed] [Google Scholar]
- 59. Lambert Samuel A., Jolma Arttu, Campitelli Laura F., et al. The human transcription factors. Cell 2018;172(4):650–65. [DOI] [PubMed] [Google Scholar]
- 60. Pique-Regi R, Degner JF, Pai AA, et al. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res 2010;21(3):447–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sharon Eilon, Kalma Yael, Sharp Ayala, et al. Inferring gene regulatory logic from high-throughput measurements of thousands of systematically designed promoters. Nat Biotechnol 2012;30(6):521–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wang Yan, Liu Jing, Huang Bo, et al. Mechanism of alternative splicing and its regulation. Biomed Rep 2014;3(2):152–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ding Jun, Li Xiaoman, Haiyan Hu.. Systematic prediction of cis-regulatory elements in the chlamydomonas reinhardtii genome using comparative genomics. Plant Physiol 2012;160(2):613–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Li X, Kazan H, Lipshitz HD, et al. Finding the target sites of RNA-binding proteins. Wiley Interdiscip Rev RNA 2013;5(1):111–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Jing S, Sarah A. Teichmann, Thomas A. Down. Assessing computational methods of cis-regulatory module prediction. PLoS Comput Biol 2010;6(12):e1001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zheng Yiyu, Li Xiaoman, Haiyan Hu.. Comprehensive discovery of DNA motifs in 349 human cells and tissues reveals new features of motifs. Nucleic Acids Res 2014;43(1):74–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Timothy L. Bailey DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics 2011;27(12):1653–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Meredith L. Howard and Eric H. Davidson. Cis-regulatory control circuits in development. Dev Biol 2004;271(1):109–18. [DOI] [PubMed] [Google Scholar]
- 69. Khan A, Fornes O, Stigliani A, et al. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res 2017;46(D1):D260–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Crooks G. E.. WebLogo: a sequence logo generator. Genome Res 2004;14(6):1188–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Corradin O, Saiakhova A, Akhtar-Zaidi B, et al. Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res 2013;24(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Stenson Peter D., Mort Matthew, Ball Edward V., et al. The human gene mutation database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet 2013;133(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. De Castro-Orós I, Pampín S, Bolado-Carrancio A, et al. Functional analysis of LDLR promoter and 5’ UTR mutations in subjects with clinical diagnosis of familial hypercholesterolemia. Hum Mutat 2011;32(8):868–72. [DOI] [PubMed] [Google Scholar]
- 74. Gupta S, Stamatoyannopoulos JA, Bailey TL, et al. Quantifying similarity between motifs. Genome Biol 2007;8(2):R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lee T, Yoon S. Boosted categorical restricted Boltzmann machine for computational prediction of splice junctions. In: International Conference on Machine Learning, Lille, France, 2015, 2483–92. [Google Scholar]
- 76. Voss Ty C., Hager Gordon L.. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat Rev Genet 2013;15(2):69–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Weirauch Matthew T., Yang Ally, Albu Mihai, et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell 2014;158(6):1431–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Angermueller Christof, Lee Heather J., Reik Wolf, Stegle Oliver. DeepCpG: accurate prediction of single-cell DNA methylation states using deep learning. Genome Biol 2017;18(1):67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Andersson Robin, Gebhard Claudia, et al. An atlas of active enhancers across human cell types and tissues. Nature 2014;507(7493):455–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Corradin Olivia, Scacheri Peter C. Enhancer variants: evaluating functions in common disease. Genome Med 2014;6(10):85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. He B., Chen C., Teng L., Tan K.. Global view of enhancer-promoter interactome in human cells. Proc Natl Acad Sci 2014, 111(21):E2191–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Li Xin, Zheng Yiyu, Hu Haiyan, Li Xiaoman. Integrative analyses shed new light on human ribosomal protein gene regulation. Sci Rep 2016;6(1):28619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Pennacchio Len A., Bickmore Wendy, Dean Ann, Nobrega Marcelo A., Bejerano Gill. Enhancers: five essential questions. Nat Rev Genet 2013;14(4):288–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zhao Changyong, Li Xiaoman, Haiyan Hu.. PETModule: a motif module based approach for enhancer target gene prediction. Sci Rep 2016;6(1):30043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Talukder Amlan, Saadat Samaneh, Li Xiaoman, Haiyan Hu.. EPIP: a novel approach for condition-specific enhancer–promoter interaction prediction. Bioinformatics 2019;35(20):3877–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Wang Saidi, Hu Haiyan, Li Xiaoman. Shared distal regulatory regions may contribute to the coordinated expression of human ribosomal protein genes. Genomics 2020;112(4):2886–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Shlyueva Daria, Stampfel Gerald, Stark Alexander. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet 2014;15(4):272–86. [DOI] [PubMed] [Google Scholar]
- 88. Ernst Jason, Kellis Manolis. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods 2012;9(3):215–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Gao Tianshun, He Bing, Liu Sheng, et al. EnhancerAtlas: a resource for enhancer annotation and analysis in 105 human cell/tissue types. Bioinformatics 2016;32(23):3543–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Hoffman Michael M, Buske Orion J, Wang Jie, et al. Unsupervised pattern discovery in human chromatin structure through genomic segmentation. Nat Methods 2012;9(5):473–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Wang Jing, Dai Xizhen, Berry Lynne D, et al. HACER: an atlas of human active enhancers to interpret regulatory variants. Nucleic Acids Res 2018;47(D1):D106–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Halfon Marc S.. Studying transcriptional enhancers: the founder fallacy, validation creep, and other biases. Trends Genet 2019;35(2):93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Le Q, Mikolov T. Distributed representations of sentences and documents. In: International Conference on Machine Learning, Beijing, China, 2014,1188–96.
- 94. Mikolov T, Chen K, Corrado G, et al. Efficient estimation of word representations in vector space. 1st International Conference on Learning Representations, Scottsdale, AZ, USA, 2013.
- 95. Ren Gang, Jin Wenfei, Cui Kairong, et al. CTCF-mediated enhancer–promoter interaction is a critical regulator of cell-to-cell variation of gene expression. Mol Cell 2017;67(6):1049–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Weintraub Abraham S., Li Charles H., Zamudio Alicia V., et al. YY1 is a structural regulator of enhancer-promoter loops. Cell 2017;171(7):1573–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Zhang Kai, Li Nan, Ainsworth Richard I., Wang Wei. Systematic identification of protein combinations mediating chromatin looping. Nat Commun 2016;7(1):12249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Wang Ying, Ding Jun, Daniell Henry, Hu Haiyan, Li Xiaoman. Motif analysis unveils the possible co-regulation of chloroplast genes and nuclear genes encoding chloroplast proteins. Plant Mol Biol 2012;80(2):177–87. [DOI] [PubMed] [Google Scholar]
- 99. Wang Ying, Li Xiaoman, Haiyan Hu.. H3k4me2 reliably defines transcription factor binding regions in different cells. Genomics 2014;103(2-3):222–8. [DOI] [PubMed] [Google Scholar]
- 100. Zheng Yiyu, Li Xiaoman, Haiyan Hu.. Discover the semantic structure of human reference epigenome by differential latent dirichlet allocation. In: 2017 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Kansas City, MO, USA, IEEE, 2017. [Google Scholar]
- 101. Kundaje Anshul, Meuleman Wouter, et al. Integrative analysis of 111 reference human epigenomes. Nature 2015;518(7539):317–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Sekhon Arshdeep, Singh Ritambhara, Qi Yanjun. DeepDiff: DEEP-learning for predicting DIFFerential gene expression from histone modifications. Bioinformatics 2018;34(17):i891–i900. [DOI] [PubMed] [Google Scholar]
- 103. Park S, Min S, Choi H-S, et al. Deep recurrent neural network-based identification of precursor microRNAs. In: Advances in Neural Information Processing Systems, Long Beach, CA, USA, 2017, 2891–900.
- 104. Olden JD, Joy MK, Death RG. An accurate comparison of methods for quantifying variable importance in artificial neural networks using simulated data. Ecol Model 2004;178(3-4):389–97. [Google Scholar]
- 105. Hill Steven T, Kuintzle Rachael, Teegarden Amy, et al. A deep recurrent neural network discovers complex biological rules to decipher RNA protein-coding potential. Nucleic Acids Res 2018;46(16):8105–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Harrow J., Frankish A., Gonzalez J. M., et al. GENCODE: the reference human genome annotation for the ENCODE project. Genome Res 2012;22(9):1760–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Mairal J. End-to-end kernel learning with supervised convolutional kernel networks. In: Advances in Neural Information Processing Systems, Barcelona, Spain, 2016, 1399–407.
- 108. Wang C, Yang J, Xie L, et al. Kervolutional neural networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 2019, 31–40.
- 109. Zhang L, Edraki M, Qi G-J. Cappronet: deep feature learning via orthogonal projections onto capsule subspaces. In: Advances in Neural Information Processing Systems, Montréal, Canada, 2018, 5814–23.


