Abstract
Background
Sarcocystis species are obligatorily heteroxenous parasites, of which some are zoonotic, representing a public health and economic impact. This study investigated the occurrence of Sarcocystis spp. in cattle sampled from a Belgian slaughterhouse.
Methods
A total of 200 carcasses were included in the study, sampled during 10 sampling days. The sedimentation method was applied to isolate the sarcocysts from both heart and diaphragm muscles collected from each carcass. Multiplex PCR, PCR–RFLP as well as cox1 gene sequencing techniques were applied serially on collected sarcocysts for species identification.
Results
Sarcocystis spp. were detected in 64% (128/200; 95% CI 57–71%) of the sampled carcasses. Female dairy cattle presented the highest Sarcocystis occurrence rate (91%) as well as the highest Sarcocystis species diversity compared to female beef and male beef. Sarcocystis spp. were detected more often in the heart muscles than in the diaphragm among female beef (p < 0.001) and dairy carcasses (p = 0.001), while in male carcasses no significant difference was observed (p = 0.763). The effect of age was not significant in male carcasses (p = 0.872), while the odds of finding sarcocysts significantly increased with age (p = 0.003) within both types of female carcasses. S. cruzi was the most prevalent species and was found in 56.5% (113/200) of the carcasses, followed by S. hominis (21.0%, 42/200), S. bovifelis (12.5%, 25/200), S. bovini (2.0%, 4/200), S. hirsuta (1.5%, 3/200) and S. heydorni (0.5%, 1/200). Six different species were detected in the diaphragm, while only two species were recovered from the heart. S. cruzi was the most prevalent species in heart, while in the diaphragm, this was S. hominis.
Conclusions
The detection of S. hominis in 21% of the sampled carcasses presents a potential food safety issue, and further research is warranted into controlling this infection.
Graphic Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s13071-021-04788-1.
Keywords: Sarcocystis spp., Cattle, Carcass, Belgium, Sarcocystis hominis, Cox1, Observational study
Background
Sarcocystis species (Apicomplexa: Sarcocystidae) are intracellular protozoan parasites, of which some species are zoonotic. Since their first discovery in striated muscles of a domestic mouse in 1843, over 200 Sarcocystis species have been identified. The complete life cycles are known for only 26 species [1]. The complete life cycle of Sarcocystis requires two hosts, generally a herbivore or carnivore intermediate host and a carnivore or omnivore definitive host. Cattle are the intermediate host of several Sarcocystis species (of which some species names are still debated), including S. cruzi with canids as definitive host, S. bovifelis, S. bovini and S. hirsuta with felids as definitive host, and S. hominis and S. heydorni with humans as definitive host [2–4]. Sarcocystis rommeli, which in fact is considered to be S. bovifelis by Gjerde [4], has felids as definitive host [5]. In the case of S. hominis and S. heydorni, intestinal sarcocystiosis may develop after consuming raw or undercooked beef containing mature sarcocysts, with symptoms such as abdominal pain, distension, watery diarrhea and eosinophilia [6]. Oocysts (mature oocyst containing two sporocysts) are formed in the small intestine and are excreted with the stool into the environment either as oocysts or when the oocyst rupture, as sporocysts [7, 8].
Bovine eosinophilic myositis (BEM) is a specific inflammatory myopathy that might be associated with S. cruzi and S. hominis infections [9–12]. Although usually no clinical symptoms are observed in animals, the gross lesions of BEM (green, focal stripes or patches) are detected in the cattle carcass after slaughter and cause carcass condemnation and economic losses [9, 13, 14]. In the study of Vangeel et al. [13], S. hominis, S. cruzi and S. hirsuta were detected in lesions of BEM, indicating that BEM was not only associated with one specific Sarcocystis species. On the contrary, not all the Sarcocystis infections present BEM.
Sarcocystis infection in cattle is reported worldwide, with an occurrence rate between 36.2% and 100% [15–20]. Sarcocystis cruzi and S. hominis are the most commonly reported species and are found in different muscles in cattle, e.g. heart, diaphragm, tongue and esophagus [21–24]. However, the results from those studies are difficult to compare as different sample types from different locations were collected, and different techniques were used to define infection status and species identification. Specifically, for Belgium, Vangeel et al. [23] found complete sarcocysts or fragments of sarcocysts in 94% of raw minced beef samples from retail stores. Molecular results revealed that 97.4% of the collected thick-walled sarcocysts were S. hominis. Based on the results, it was assumed that most carcasses are infected with this zoonotic species in Belgium.
Although high occurrences of Sarcocystis spp. in different cattle muscles, minced meat or beef burger samples have been reported, the species identification methods were not consistent and not adapted to the newly defined (though debated) S. bovifelis, S. bovini and S. heydorni species. In Belgium, there are no recent studies determining the occurrence of Sarcocystis species in slaughtered cattle. Therefore, heart and diaphragm samples were collected from carcasses in a Belgian slaughterhouse, followed by sarcocyst isolation and identification based on sedimentation and updated molecular methods. Besides, the associations between the presence, intensity and diversity of Sarcocystis spp. and different animal characteristics (age, type and sex) were explored.
Methods
Sample collection
A Belgian cattle slaughterhouse, located in Flanders—the northern part of Belgium—was visited 10 times from July to October 2019. During each visit, around 20 consecutive carcasses were sampled. Heart and diaphragm muscles (100–150 g of each) were collected from each of the 201 selected carcasses and stored in separate sealed bags. All samples were transported to the laboratory and stored at 4 °C until analysis. For all sampled carcasses, the age, category (A–E, according to Council Regulation (EC) No. 1234/2007 [25]), classification [conformation (S, E, U, R, O, P) and fat cover (1–5) class] were obtained from the slaughterhouse.
Microscopic examination following sedimentation
A modified sedimentation method according to Vangeel et al. [23] was applied in this study. For each sample (heart and diaphragm), fat tissue and fasciae were removed, after which 100 g was minced with 30 ml of saline solution for approximately 5 s at high speed in a blender. A glass funnel ending with a rubber tube on a tripod was closed with a clip near the end of the rubber tube, with a sieve in combination with a single layer of gauze on top of the funnel. The minced meat was put on the gauze, and preheated (37 °C) saline solution was added until the minced meat was fully covered. The whole apparatus was incubated at 37 °C for 4 h. Every hour, the minced meat in the sieve was gently stirred with a spoon.
At the end of the incubation, 10 ml of the solution from the bottom of the apparatus was collected into a glass flask by opening the spring clip on the rubber tube. This sediment was examined for the presence of sarcocysts in a graded petri dish with a stereo microscope (×40). During microscopic examination, the morphology of the sarcocysts was recorded, e.g. thin-walled or thick-walled, with pointed or rounded edge [23, 26]. The microscopic examination was stopped when 15 sarcocysts were identified. Identified sarcocysts were stored separately in 1.5-ml tubes with 20 µl of saline solution. In addition, the number of identified sarcocysts was recorded. When 15 or more sarcocysts were identified, the number of sarcocysts was recorded as 15. All the sarcocysts were stored at −20 °C. From each sample, at least one sarcocyst per morphologic group (see above, e.g. thick-walled and rounded edge) was selected for molecular identification to species level.
Molecular identification of Sarcocystis species
The DNA of a single sarcocyst was extracted using the Tissue and Hair Extraction Kit (Promega, USA) according to the manufacturer’s recommendations. Three techniques were applied to identify the Sarcocystis species. All selected sarcocysts were subjected to multiplex polymerase chain reaction (mPCR) [27], after which the unidentifiable sarcocysts were subjected to PCR-restriction fragment length polymorphism (PCR–RFLP); primers 18S9L: 5’-GGATAAACCGTGGTAATTCTATG-3’ and 18S1H: 5’-GGCAAATGCTTTCGCAGTAG-3’ were from Hamidinejat et al. [28]. Sarcocysts that could also not be identified with the latter technique were sent for sequencing. All the primers in this study were from Sigma-Aldrich (USA).
Multiplex PCR
The PCR was conducted according to Rubiola et al. [27], except that 1 mM instead of 0.5 mM of Sarco Rev primer was used in the PCR mixture. The PCR results were separated in 2% agarose (0.5× TAE buffer, 70 min at 120 V) and visualized using UV light. Sarcocysts with only a band at 200–250 base pairs (bp), the size of the Sarcocystis spp. fragment, were considered unidentified and were tested using PCR–RFLP. For sarcocysts without the 200–250-bp band nor any of the species-specific bands, the sarcocyst was recorded as Sarcocystis-negative.
PCR–RFLP
After amplification of the 18S rDNA (using primers 18S9L and 18S1H according to Hamidinejat et al. [28]), the fragments (approx. 900 bp) were digested with the FokI [29] and BfaI [30] enzymes. The PCR was modified from Jehle et al. [29]. The 50-µL PCR reaction mix contained 25 µL of GoTaq G2 Hot Start Master Mix (Promega), 0.5 µL of each primer (working solution: 50 pmol/µl), 19 µL of nuclease-free water and 5 µL of sample DNA. The PCR was performed with the following protocol: pre-denaturation at 94 °C for 2 min, 35 cycles of denaturation at 94 °C for 40 s, annealing at 61 °C for 60 s and elongation at 72 °C for 80 s, and a final elongation at 72 °C for 5 min. The PCR products were separated in 2% agarose (0.5× TAE -buffer, 60 min at 110 V) and visualized using UV light. Ten microliters of obtained PCR product was digested using 30 µL digestion mix containing 5U of the enzymes (FokI, BfaI (FastDigest, Thermo Scientific)) at 37 °C for 2 h, respectively. The results were visualized in 2.5% agarose (0.5× TAE buffer, 90 min at 120 V). The size (in bp) of the expected 18S rDNA digestion fragments for each species distinguished by agarose gel electrophoresis PCR–RFLP is shown in Additional file 1.
Sequencing
If the species was not identified by either the mPCR or the PCR–RFLP, then the sarcocyst was sent to Eurofins Genomics (Luxembourg) for sequencing based on the mitochondrial cytochrome c oxidase subunit I gene (cox1). Sarc cox1 SF and Sarc cox1 SR 9 primers were used according to Gjerde [4], and the PCR protocol was applied according to Hoeve-Bakker et al. [24]. The sequences obtained were compared to available sequences from the GenBank database using BLAST. The nucleotide sequences of these unidentified Sarcocystis spp. were deposited in the GenBank database under the following accession numbers: UNS1: MW756133, UNS2: MW756134, and UNS3: MW756135.
Statistical analysis
A prevalence of 90% was assumed to calculate the sample size, which was based on the study of Vangeel et al. [23]. At least 138 samples would be needed to ensure precision of 5% of the 95% confidence interval (CI) around the prevalence estimate. For a prevalence of 85%, 196 samples would be required to obtain the same precision. To account for a lower prevalence than initially anticipated and potential losses, a total of 201 carcasses were sampled. One carcass was classified as category Z (i.e. from an animal above 8 but below 12 months old) and was excluded from the analysis set, resulting in a final sample of 200 carcasses. All analyses were performed using R version 4.0.2 [31]. The final dataset that was used for the analyses is deposited open access in Zenodo [32].
Based on the conformation, carcasses were categorized into two types: the S/E/U conformation categories were assumed to be beef cattle, and R/O/P were categorized as dairy cattle. When including the sex of the animal, three distinct groups were created: male beef, female beef and female dairy cattle. Binomial and categorical variables were summarized using tables, continuous variables (age) were inspected visually using histograms, and count data (intensity and number of different species) using bar plots. An organ was considered positive when at least one sarcocyst was molecularly confirmed as Sarcocystis spp. If sarcocysts were recovered by microscopy, but results were not confirmed by molecular identification, then the organ was considered negative. A carcass was considered positive if the diaphragm and/or heart was positive. The Sarcocystis infection results were summarized for the different groups of cattle (male beef, female beef and female dairy cattle). The 95% Clopper–Pearson confidence intervals around binomial proportions (number of positives/total number tested) were calculated using the DescTools package [33]. The proportion of Sarcocystis-positive organs (heart versus diaphragm) were compared using the McNemar chi-square test for paired proportions. Differences in Sarcocystis positivity between the three groups were tested using logistic regressions, with the Sarcocystis result (positive/negative) as outcome and the group as exploratory variable. The relation between Sarcocystis positivity and age was assessed within males and females separately, because almost all males were younger than females. Age (in years) was included as a continuous predictor variable. For females, the type (beef/dairy) was included as main effect, and the interaction between type and age was also evaluated. The different models were compared using the Akaike information criterion (AIC), and the model with the lowest AIC (which was the model without type–age interaction) was used. To compare species diversity (expressed as the total number of unique Sarcocystis species within a carcass) between the different groups, nonparametric pairwise Wilcoxon Rank Sum tests were performed, adjusting for multiple testing using the Holm correction.
A phylogenetic tree was constructed using MEGA 7.0 [34] via the neighbor-joining method.
Results
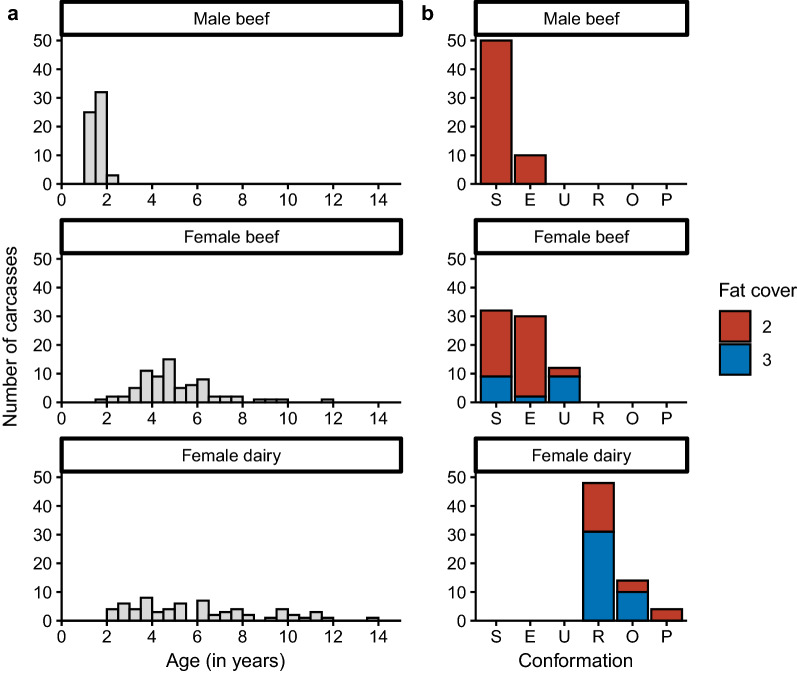
In total, the results of 200 carcasses were analyzed. Based on sex and type, carcasses originated from three distinct groups of animals: 60 male beef, 74 female beef and 66 female dairy cattle. The distribution of age, conformation and fat content within each of these groups is visualized in Fig. 1. The mean age of male beef, female beef and female dairy cattle was 1.6 years (range from 1.1 to 2.2 years), 5.1 years (1.7–11.8 years) and 6.1 years (2.1–13.6 years), respectively.
Fig. 1.
a Age distribution and b classification (conformation and fat cover) of bovine carcasses. Data are presented for each of the three different groups of carcasses: male beef (n = 60), female beef (n = 74) and female dairy (n = 66)
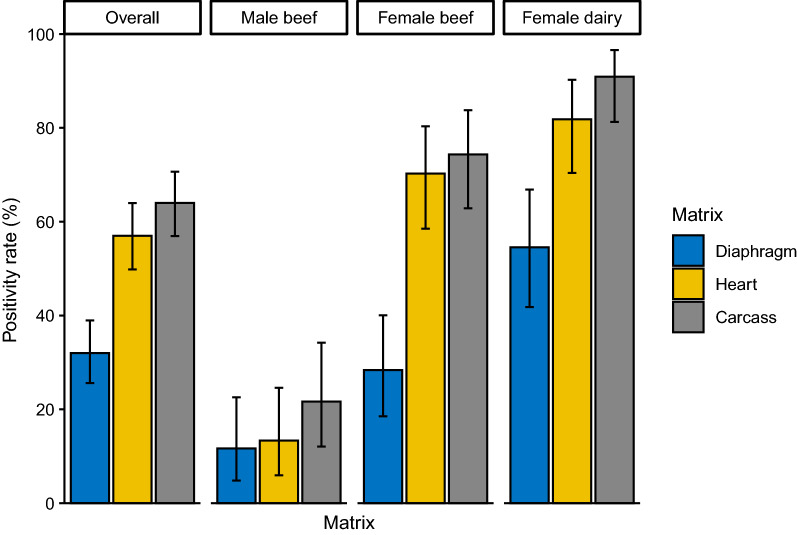
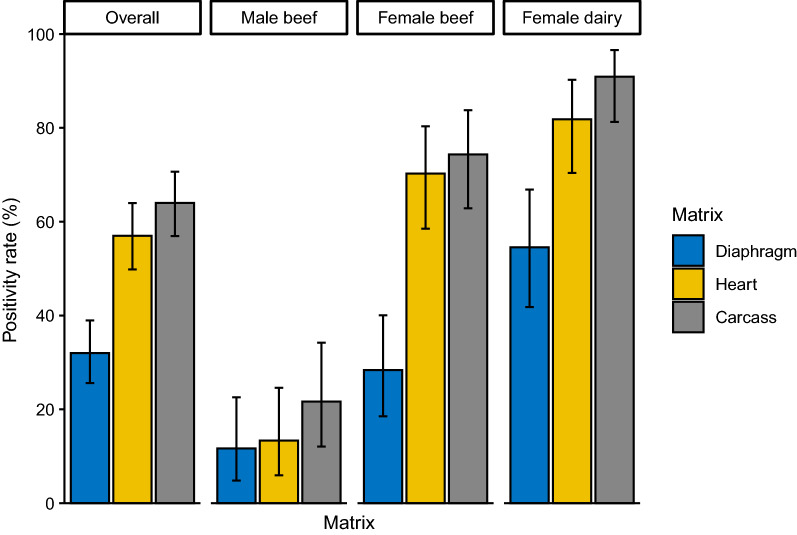
Overall, Sarcocystis spp. were detected in 64% (128/200; 95% CI 57–71%) of all the sampled carcasses. Sarcocystis spp. were found in the heart of 114 carcasses (57%, 95% CI 50–64%) and the diaphragm of 64 carcasses (32%, 95% CI 26–39%). In 50 carcasses (25%), both the heart and diaphragm were Sarcocystis-positive, while only the heart and diaphragm were positive in 64 and 14 carcasses, respectively. The proportion of positive heart samples was significantly higher than the diaphragm samples (McNemar’s statistic = 32, df = 1, p < 0.001).
Sarcocystis spp. occurrence in female beef, female dairy and male beef carcasses
Carcasses from female dairy carcasses had the highest Sarcocystis occurrence rate (91%; 95% CI 81–97%) among the three different groups (Fig. 2). In the beef type, males had a lower Sarcocystis positivity rate (22%; 95% CI 12–34%) compared to females (74%; 95% CI 63–84%). The estimated odds of Sarcocystis spp. infection in the male beef group was 0.10 (95% CI 0.04–0.21; p < 0.001) times smaller compared to female beef cattle, and the odds in female dairy carcasses was 3.5 (95% CI 1.4–10; p = 0.014) times larger than in female beef carcasses.
Fig. 2.
The occurrence of Sarcocystis spp. in cattle carcasses during slaughter. Data are presented overall (n = 200) and per group: male beef (n = 60), female beef (n = 74) and female dairy carcasses (n = 66). The error bars indicate the 95% confidence interval
When considering the different muscles, Sarcocystis spp. were detected more often in the heart muscle than in the diaphragm among female beef carcasses (70% vs. 28%, McNemar test statistic = 26; df = 1, p < 0.001) and dairy carcasses (82% vs. 55%, n = 66; McNemar test statistic = 11, df = 1, p = 0.001; Fig. 2). Within male beef carcasses, no significant difference was observed (McNemar = 0.1, df = 1, p = 0.763).
Sarcocystis spp. occurrence for different ages
Since carcasses from males (range from 1.1 to 2.2 years) were generally much younger than females (range from 1.7 to 13.6 years) (Fig. 1), the effect of age was evaluated separately for carcasses from male and female cattle. For female carcasses, the odds of finding sarcocysts significantly increased with age (ORadjusted = 1.66 per year, 95% CI 1.22–2.41, p = 0.003) within both types (Fig. 3). The age-adjusted odds ratio for female dairy carcasses was 3.3 times higher (95% CI 1.24–10.3; p = 0.024) than for female beef carcasses. In female carcasses below 7 years old, Sarcocystis was observed in 71% (47/66) of the beef and 89% (39/44) of the dairy type, whereas all the female beef (8/8, 100%) and all but one dairy carcass (21/22, 95%) above 7 years old had a Sarcocystis infection (Fig. 3). Within the carcasses of male cattle, the effect of age was not significant (OR = 1.2 [0.1–15.5]; p = 0.872). The effect of age was also explored for presence of infection in diaphragm and heart samples separately and is visualized in Additional file 2.
Fig. 3.
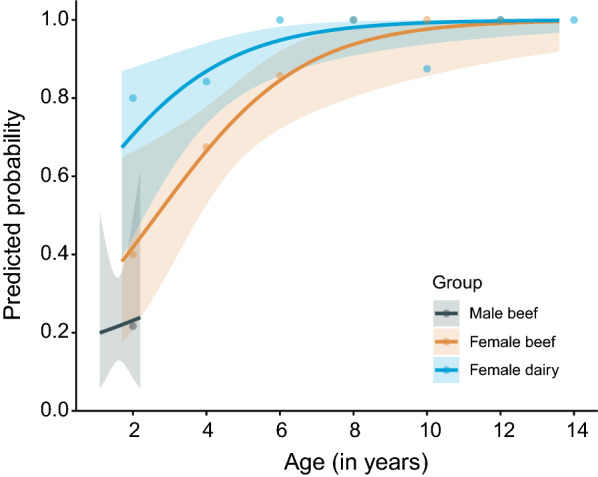
Sarcocystis positivity with age. The lines represent the predicted probabilities with the 95% confidence interval, in carcasses from male beef (n = 60), female beef (74) and female dairy cattle (n = 66). The points represent the observed proportions of Sarcocystis-positive carcasses, binned per 2 years and plotted at the midpoint of the age category (e.g. the points at 2 years represent the positivity rate of carcasses between 1 and 3 years within each of the groups)
Intensity of sarcocysts
The number of sarcocysts in the 10 ml of sediment of the aliquot of the sample was recorded when conducting the microscopic examinations, censored at 15 sarcocysts. In Sarcocystis-positive heart samples for which the intensity was recorded (n = 104), the median intensity was the upper limit (i.e. 15 or above). In contrast, the median number of sarcocysts in positive diaphragm samples (n = 64) was three. The intensity of sarcocysts was similar in the different groups of cattle (data not shown).
Species distribution
After microscopic examination of the sediment, 140 morphologically identified sarcocysts from 115 heart samples were further molecularly identified (one sarcocyst from 94 hearts, two sarcocysts from 18 hearts, three sarcocysts from two hearts and four sarcocysts from one heart). For diaphragm samples, 115 morphologically identified sarcocysts were further molecularly identified (one sarcocyst from 31 diaphragms, two sarcocysts from 25 diaphragms, three sarcocysts from 10 diaphragms and four sarcocysts from one diaphragm). Firstly, based on the mPCR of the morphologically identified sarcocysts, five sarcocysts from hearts and seven sarcocysts from diaphragms were Sarcocystis spp.-negative. In addition, a total of 28 sarcocysts could not be identified by mPCR at species level, thus they were subjected to PCR–RFLP for species identification. The species of 24 of these sarcocysts could not be identified by PCR–RFLP, and 22 of them were sent for cox1 sequencing. For two sarcocysts the cox1 PCR was negative, so these sarcocysts were considered Sarcocystis-positive, but the species was considered unidentified. After sequencing, the species of three sarcocysts could not be identified (Additional file 3), resulting in a total of five sarcocysts for which the species remained unidentified (two from the heart and three from the diaphragm).
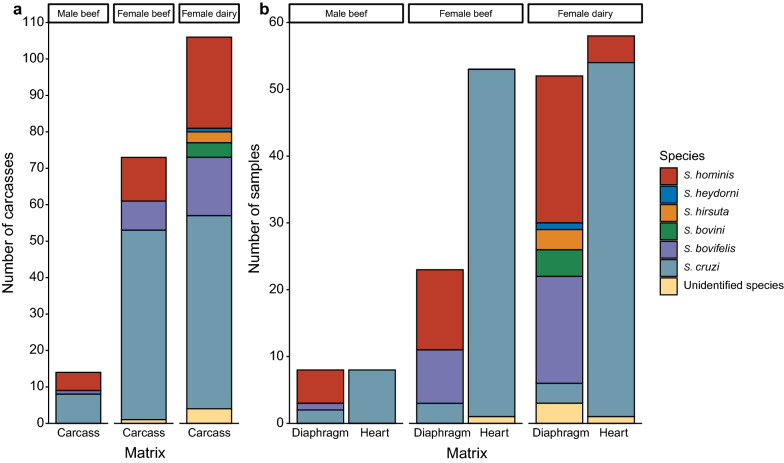
Overall, six different species were identified from the 200 carcasses (both organs combined; Fig. 4a). Sarcocystis cruzi was the most prevalent species and was found in 56.5% (113/200) of the carcasses, followed by S. hominis (21.0%, 42/200), S. bovifelis (12.5%, 25/200), S. bovini (2.0%, 4/200), S. hirsuta (1.5%, 3/200) and S. heydorni (0.5%, 1/200). In five carcasses (2.5%), unidentified Sarcocystis species were detected. Within each of the three cattle groups, S. cruzi was the most prevalent species, followed by S. hominis and S. bovifelis (Fig. 4a). Sarcocystis heydorni, S. bovini and S. hirsuta were found in female dairy cattle only.
Fig. 4.
Sarcocystis species distribution (a) in carcasses and (b) in diaphragm/heart samples. Results are from Sarcocystis-positive carcasses of male beef (n = 13), female beef (n = 55) and female dairy cattle (n = 60). For each species, the height of the bar represents the number of positive carcasses/samples. The overall total number of positive samples does not correspond to the total number of Sarcocystis-positive carcasses/samples due to the presence of mixed infections (multiple species in one carcass/organ)
In heart muscles, only two species, S. cruzi (56.5%, 113/200) and S. hominis (2.0%, 4/200), were identified, and for two sarcocysts the exact species could not be identified (Fig. 4b). Six Sarcocystis species were observed from diaphragm muscles. Sarcocystis hominis (19.5%, 39/200) had the highest infection rate, followed by S. bovifelis, S. cruzi, S. bovini, S. hirsuta and S. heydorni. For three diaphragm samples, the species could not be identified (Fig. 4b).
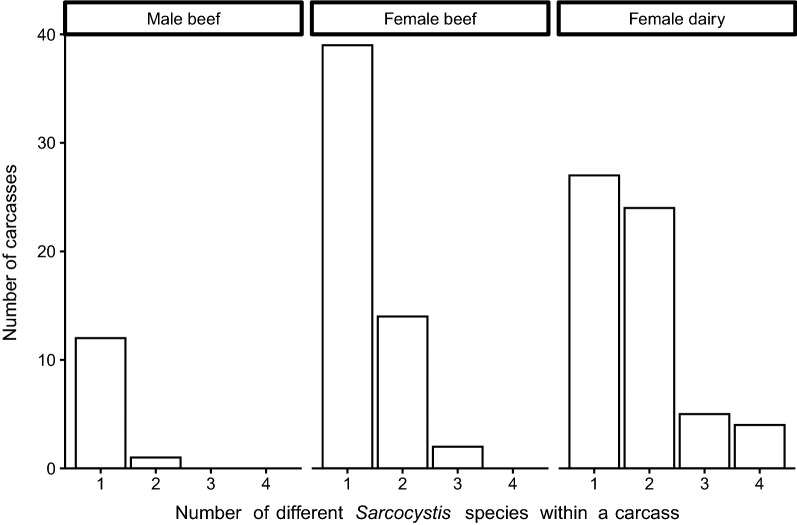
In 30% of the 128 Sarcocystis-infected carcasses (39/128), two different species were observed, and in 8.6% (11/128) of the carcasses, 3–4 different species were observed (Fig. 5). In the majority of the infected heart and diaphragm muscles, only one Sarcocystis species was observed (109 out of 114 positive heart samples, and 51 out of 64 positive diaphragm samples). In five heart muscles, two different species were observed, while in seven and six diaphragm muscles, two and three species were observed, respectively. Only in female dairy carcasses, up to four different species were detected within one carcass (n = 4 out of 60 Sarcocystis-positive carcasses; Fig. 5). The number of different species in female dairy carcasses was significantly higher than in male beef carcasses (W = 579, p = 0.007) and female beef cattle (W = 2120, p = 0.007). The difference in the number of different species between male beef and female beef carcasses was not statistically significant (W = 280, p = 0.111).
Fig. 5.
Number of different Sarcocystis species within a carcass (based on heart and diaphragm). Results are presented for Sarcocystis spp.-positive male beef (n = 13), female beef (n = 55) and female dairy carcasses (n = 60)
Discussion
The final results in our study showed an overall prevalence of 64% in 200 cattle carcasses. Vangeel et al. [23] reported a 94% occurrence rate of Sarcocystis spp. in raw minced beef originating from retail stores in Belgium. This difference could be explained by the fact that minced meat is a mixture of meat from several carcasses as well as different muscles. The chances of finding Sarcocystis spp. in these ‘mixed’ samples is higher than in a single/separate sample from one carcass.
Muscles from the heart, tongue, esophagus and diaphragm are the target-colonizing areas of Sarcocystis spp. [1]. In the present study, a significantly higher occurrence rate was observed in heart (57%) than in diaphragm (32%) samples. Similarly, Yang et al. [18] also reported more Sarcocystis spp. in heart muscles (49%) than in diaphragms (14%). In the study of Latif et al. [16], the occurrence rate in the heart (8%) was lower than in the diaphragm (27%) in cattle, while in water buffaloes, the heart was the most often infected organ (67%), with no Sarcocystis spp. detected in the diaphragm. High occurrence rates, ranging between 58% and 99.5% in hearts and 58%–90.9% in diaphragms in cattle, have been reported from different countries, e.g. Iran [19, 35], Italy [22], the Netherlands [24], Brazil [17] and Argentina [36]. This shows that although heart and diaphragm are target-colonizing areas of Sarcocystis spp., the occurrence rate can vary between countries/studies. Besides the differences between organs/muscles mentioned above, also differences in detection and identification techniques used (e.g. morphological identification versus molecular identification) and differences in study populations (e.g. age, sex, breed), render comparison between the studies complicated.
Differences in breed as well as sex and age of the host, climatic factors and management practices [37] may all result in differences in the prevalence estimates of Sarcocystis spp. in cattle. This was also highlighted in our study, where the Sarcocystis spp. occurrence rate differed markedly between the different groups of animals, with the highest rate observed in female dairy, followed by female beef, and the lowest in male beef cattle. In contrast to our results, Savini et al. [37] found the highest prevalence of Sarcocystis spp. infections in esophagus samples of entire males (92%) (n = 51) compared to either castrated males (60%) (n = 261) or females (51%) (n = 170). On the other hand, Hornok et al. [21] found no significant difference between infection rates of individually-collected heart and esophagus samples from bulls (67%) and cows (64%). However, the mean age of PCR-positive cattle (6.2 ± 4.4 years) was significantly higher than that of PCR-negative ones (4.7 ± 4 years). Similarly, we also found a significant increase in Sarcocystis infections with increasing age for the carcasses of beef and dairy cows. This increase was partially similar to the study of Savini et al. [37], in which an increasing prevalence of Sarcocystis spp. infection was found in cattle up to the age of 4 years, after which the prevalence significantly decreased. Savini et al. [37] hypothesized that this might be explained by the reduction of sarcocysts over time because of the host response or a reflection of (natural or managerial) selection, where the generally healthier animals are kept for longer periods. In the present study, we did not observe such a decrease, as the vast majority of animals above 7 years were infected with Sarcocystis species. Savini et al. [37] attributed the increased prevalence with age (until 4 years) to the greater opportunity for older animals to be exposed to oocysts/sporocysts. In Belgium, dairy cattle are often kept outdoors, permitting contact with potentially contaminated water and soil/pasture. Additionally, dairy cattle are generally kept longer than beef cattle, allowing more exposure time, and leading to an older age when arriving at the slaughterhouse. Indeed, the higher occurrence of Sarcocystis spp. in carcasses of dairy cows than of beef cows was partially explained by the higher slaughter age in the former group. Since the bulls in the present study were slaughtered at a considerably younger age than the cows, we did not want to extrapolate the age effect from cows to bulls. Nevertheless, we believe that the lower slaughter age of male beef cattle may also (partially) explain the lower occurrence rate in carcasses of bulls in our study. Still, it is obvious that other factors besides age may have contributed to the differences between groups, since carcasses of dairy cows still had a significantly higher odds of being infected than beef cows after correcting for the age effect. We did not collect/evaluate other variables, such as the breed, feed, outdoor access and geographical information during this study as this was not our main aim, but future research studying such factors should be encouraged, as an increased epidemiological understanding of these parasites may contribute to the control of Sarcocystis infections in cattle.
Six Sarcocystis species were identified through molecular tools, i.e. S. cruzi, S. hominis, S. bovifelis, S. hirsuta, S. heydorni and S. bovini. Sarcocystis cruzi followed by S. hominis had the highest prevalence among cattle, which was similar with the study of Hoeve-Bakker et al. [24] and Hornok et al. [21]. Significantly more different species were observed in our female dairy group. This might be explained by longer exposure times (outdoor access, age) of this specific cattle type. Interestingly, when looking into different muscles, the diaphragm had a higher species diversity (S. cruzi, S. hominis, S. bovifelis, S. hirsuta, S. heydorni and S. bovini and unidentified species) than the heart (S. cruzi, S. hominis and unidentified species). Sarcocystis cruzi was the most prevalent species in the heart, while in the diaphragm this was S. hominis. Sarcocystis hominis and S. heydorni are zoonotic Sarcocystis species and can cause intestinal sarcocystiosis in humans [7]. When S. hominis- and/or S. heydorni-infected diaphragms enter the food chain, they might pose a high risk for public health, depending on the further processing. In the Belgian study of Vangeel et al. [23], 63 of 67 minced meat samples were found infected, with thick-walled sarcocysts detected in 61 samples. Molecular analyses (including PCR and sequencing) of the latter samples could reliably identify 39 PCR products, of which 38 were confirmed as S. hominis. This seems much higher than the occurrences observed in the current study. Yet again, results are difficult to compare as minced meat is composed of several organs of several carcasses.
Conclusions
In this study, Sarcocystis spp. were detected in 64% of sampled carcasses. Nevertheless, extrapolation of these results to the Belgian cattle population at slaughter point should be done carefully, as the sampled population does not fully represent the total population. Indeed, the conformation of R in the dairy type was slightly oversampled compared to the total population of slaughtered cattle in Flanders. Moreover, beef cattle in Flanders are rather specific, with its typical Belgian White Blue breed that is characterized by muscular hypertrophy (and related management practices), which may limit generalizability of our data to other cattle populations. Significantly more infections were detected in female dairy cattle (91%), and the odds of infection increased with age in female animals. More infections were detected in the heart muscles, where S. cruzi was the most prevalent species, while in the diaphragm, this was S. hominis. The detection of S. hominis in 21% of the sampled carcasses presents a potential food safety issue, and further research is warranted into controlling this infection.
Supplementary Information
Additional file 1: Expected fragment sizes (in base pairs) from digestion of 18S rDNA PCR products of bovine Sarcocystis species with FokI and BfaI.
Additional file 2: Sarcocystis positivity rate in diaphragm and heart samples among different groups.
Additional file 3: Phylogenetic tree of unidentified Sarcocystis species (cox1 sequencing).
Acknowledgements
We thank Sandra Vangeenberghe for the technical support and laboratory analyses. The slaughterhouse personnel are acknowledged for their kind cooperation.
Abbreviations
- AIC
Akaike information criterion
- BEM
Bovine eosinophilic myositis
- bp
Base pairs
- CI
Confidence interval
- cox1
Cytochrome c oxidase subunit I gene
- mPCR
Multiplex polymerase chain reaction
- OR
Odds ratio
- RFLP
Restriction fragment length polymorphism
Authors' contributions
HZ: methodology, investigation, writing—original draft, visualization. IVD: methodology, formal analysis, data curation, writing—original draft, writing—review & editing, visualization. TWK: investigation; BS: methodology, investigation, writing—review & editing. SG: conceptualization, methodology, resources, writing—original draft, writing—review & editing, supervision, project administration. All authors read and approved the final manuscript.
Funding
Hang Zeng received funding from China Scholarship Council, Grant Number 201706910091.
Availability of data and materials
The dataset supporting the conclusions of this article is available in the Zenodo repository, https://doi.org/10.5281/zenodo.4611489. The nucleotide sequences of the unidentified Sarcocystis spp. UNS1, UNS2, UNS3 are in GenBank under the following accession numbers: UNS1: MW756133, UNS2: MW756134, UNS3: MW756135.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Dubey JP, Calero-Bernal R, Rosenthal BM, Speer CA, Fayer R. Sarcocystosis of animals and humans. 2. Boca Raton: CRC Press; 2015. [Google Scholar]
- 2.Dubey JP, van Wilpe E, Calero-Bernal R, Verma SK, Fayer R. Sarcocystis heydorni, n. sp. (Apicomplexa: Sarcocystidae) with cattle (Bos taurus) and human (Homo sapiens) cycle. Parasitol Res. 2015;114:4143–7. doi: 10.1007/s00436-015-4645-2. [DOI] [PubMed] [Google Scholar]
- 3.Gjerde B. Molecular characterisation of Sarcocystis bovifelis, Sarcocystis bovini n. sp., Sarcocystis hirsuta and Sarcocystis cruzi from cattle (Bos taurus) and Sarcocystis sinensis from water buffaloes (Bubalus bubalis) Parasitol Res. 2016;115:1473–92. doi: 10.1007/s00436-015-4881-5. [DOI] [PubMed] [Google Scholar]
- 4.Gjerde B. The resurrection of a species: Sarcocystis bovifelis Heydorn et al. 1975 is distinct from the current Sarcocystis hirsuta in cattle and morphologically indistinguishable from Sarcocystis sinensis in water buffaloes. Parasitol Res. 2016;2016(115):1–21. doi: 10.1007/s00436-015-4785-4. [DOI] [PubMed] [Google Scholar]
- 5.Lindsay DS, Dubey JP. Neosporosis, toxoplasmosis, and sarcocystosis in ruminants: an update. Vet Clin North Am-Food Anim Pract. 2020;36:205–222. doi: 10.1016/j.cvfa.2019.11.004. [DOI] [PubMed] [Google Scholar]
- 6.Chen X, Zuo Y, Zuo W. Observation on the clinical symptoms and sporocyst excretion in human volunteers experimentally infected with Sarcocystis hominis. Zhongguo ji sheng chong xue yu ji sheng chong bing za zhi (Chinese J Parasitol Parasit Dis) 1999;17:25–27. [PubMed] [Google Scholar]
- 7.Dubey JP. Foodborne and waterborne zoonotic sarcocystosis. Food Waterborne Parasitol. 2015;1:2–11. doi: 10.1016/j.fawpar.2015.09.001. [DOI] [Google Scholar]
- 8.Fayer R. Sarcocystis spp. in human infections. Clin Microbiol Rev. 2004;17:894–902. doi: 10.1128/CMR.17.4.894-902.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wouda W, Snoep JJ, Dubey JP. Eosinophilic Myositis due to Sarcocystis hominis in a beef cow. J Comp Pathol. 2006;135:249–253. doi: 10.1016/j.jcpa.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 10.Bundza A, Feltmate TE. Eosinophilic myositis/lymphadenitis in slaughter cattle. Can Vet J La Rev Vet Can. 1989;30:514–6. [PMC free article] [PubMed] [Google Scholar]
- 11.Gajadhar AA, Marquardt WC. Ultrastructural and transmission evidence of Sarcocystis cruzi associated with eosinophilic myositis in cattle. Can J Vet Res. 1992;56:41–46. [PMC free article] [PubMed] [Google Scholar]
- 12.Gajadhar AA, Yates WD, Allen JR. Association of eosinophilic myositis with an unusual species of Sarcocystis in a beef cow. Can J Vet Res. 1987;51:373–378. [PMC free article] [PubMed] [Google Scholar]
- 13.Vangeel L, Houf K, Geldhof P, De Preter K, Vercruysse J, Ducatelle R, et al. Different Sarcocystis spp. are present in bovine eosinophilic myositis. Vet Parasitol. 2013;197:543–8. doi: 10.1016/j.vetpar.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 14.Rimaila-Pärnänen E, Nikander S. Generalized eosinophilic myositis with sarcosporidiosis in a Finnish cow. Nord Vet Med. 1980;32:96–99. [PubMed] [Google Scholar]
- 15.Marques SM, Barbosa J, Quadros R, Pilati C. Occurrence of Sarcocystis cruzi in cattle in the state of Santa Catarina, Brazil. Int J Infect Dis. 2018;73:303. doi: 10.1016/j.ijid.2018.04.4105. [DOI] [Google Scholar]
- 16.Latif B, Vellayan S, Heo CC, Kannan Kutty M, Omar E, Abdullah S, et al. High prevalence of muscular sarcocystosis in cattle and water buffaloes from Selangor. Malaysia Trop Biomed. 2013;30:699–705. [PubMed] [Google Scholar]
- 17.Ferreira MST, Vogel FSF, Sangioni LA, Cezar AS, Braunig P, de Avilla BS, et al. Sarcocystis species identification in cattle hearts destined to human consumption in southern Brazil. Vet Parasitol Reg Stud Reports. 2018;14:94–98. doi: 10.1016/j.vprsr.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 18.Yang YR, Dong H, Su RJ, Wang YH, Wang RH, Jiang YB, et al. High prevalence of Sarcocystis spp. infections in cattle (Bos taurus) from central China. Parasitol Int. 2018;67:800–4. doi: 10.1016/j.parint.2018.08.006. [DOI] [PubMed] [Google Scholar]
- 19.Ayazian Mavi S, Teimouri A, Mohebali M, Sharifi Yazdi MK, Shojaee S, Rezaian M, et al. Sarcocystis infection in beef and industrial raw beef burgers from butcheries and retail stores: a molecular microscopic study. Heliyon. 2020;6:e04171. doi: 10.1016/j.heliyon.2020.e04171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Böttner A, Charleston WAG, Pomroy WE, Rommel M. The prevalence and identity of Sarcocystis in beef cattle in New Zealand. Vet Parasitol. 1987;24:157–168. doi: 10.1016/0304-4017(87)90036-7. [DOI] [PubMed] [Google Scholar]
- 21.Hornok S, Mester A, Takács N, Baska F, Majoros G, Fok É, et al. Sarcocystis-infection of cattle in Hungary. Parasit Vectors. 2015;8:4–9. doi: 10.1186/s13071-014-0617-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bucca M, Brianti E, Giuffrida A, Ziino G, Cicciari S, Panebianco A. Prevalence and distribution of Sarcocystis spp. cysts in several muscles of cattle slaughtered in Sicily, Southern Italy. Food Control. 2011;22:105–8. doi: 10.1016/j.foodcont.2010.05.015. [DOI] [Google Scholar]
- 23.Vangeel L, Houf K, Chiers K, Vercruysse J, D’Herde K, Ducatelle R. Molecular-based identification of Sarcocystis hominis in Belgian minced beef. J Food Prot. 2007;70:1523–1526. doi: 10.4315/0362-028X-70.6.1523. [DOI] [PubMed] [Google Scholar]
- 24.Hoeve-Bakker BJA, van der Giessen JWB, Franssen FFJ. Molecular identification targeting cox1 and 18S genes confirms the high prevalence of Sarcocystis spp. in cattle in the Netherlands. Int J Parasitol. 2019;49:859–66. doi: 10.1016/j.ijpara.2019.05.008. [DOI] [PubMed] [Google Scholar]
- 25.European Union. Council Regulation (EC) No 1234/2007 of 22 October 2007 establishing a common organisation of agricultural markets and on specific provisions for certain agricultural products (Single CMO Regulation). 2007.
- 26.Gjerde B. Morphological and molecular characteristics of four Sarcocystis spp. in Canadian moose (Alces alces), including Sarcocystis taeniata n. sp. Parasitol Res. 2014;113:1591–604. doi: 10.1007/s00436-014-3806-z. [DOI] [PubMed] [Google Scholar]
- 27.Rubiola S, Civera T, Ferroglio E, Zanet S, Zaccaria T, Brossa S, et al. Molecular differentiation of cattle Sarcocystis spp. by multiplex PCR targeting 18S and COI genes following identification of Sarcocystis hominis in human stool samples. Food Waterborne Parasitol. 2020;18:00074. doi: 10.1016/j.fawpar.2020.e00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamidinejat H, Jalali MHR, Gharibi D, Molayan PH. Detection of Sarcocystis spp. in cattle (Bos taurus) and water buffaloes (Bubalus bubalis) in Iran by PCR–RFLP. J Parasit Dis. 2015;39:658–62. doi: 10.1007/s12639-014-0426-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jehle C, Dinkel A, Sander A, Morent M, Romig T, Luc PV, et al. Diagnosis of Sarcocystis spp. in cattle (Bos taurus) and water buffalo (Bubalus bubalis) in Northern Vietnam. Vet Parasitol. 2009;166:314–20. doi: 10.1016/j.vetpar.2009.08.024. [DOI] [PubMed] [Google Scholar]
- 30.Hajimohammadi B, Dehghani A, Ahmadi MM, Eslami G, Oryan A, Khamesipour A. Prevalence and species identification of Sarcocystis in raw hamburgers distributed in Yazd, Iran using PCR-RFLP. J Food Qual Hazards Control. 2014;1:15–20. [Google Scholar]
- 31.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria; 2020.
- 32.Zeng H, Gabriël S, Soba B, Van Damme I. Dataset of the publication “Sarcocystis species in bovine carcasses from a Belgian abattoir: a cross-sectional study.” Zenodo; 2021. 10.5281/zenodo.4611489. [DOI] [PMC free article] [PubMed]
- 33.Signorell A. DescTools: Tools for descriptive statistics. R package version 0.99.38. 2020. http://cran.r-project.org/package=DescTools.
- 34.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nourani H, Matin S, Nouri A, Azizi H. Prevalence of thin-walled Sarcocystis cruzi and thick-walled Sarcocystis hirsuta or Sarcocystis hominis from cattle in Iran. Trop Anim Health Prod. 2010;42:1225–1227. doi: 10.1007/s11250-010-9552-z. [DOI] [PubMed] [Google Scholar]
- 36.Moré G, Abrahamovich P, Jurado S, Bacigalupe D, Marin JC, Rambeaud M, et al. Prevalence of Sarcocystis spp. in Argentinean cattle. Vet Parasitol. 2011;177:162–5. doi: 10.1016/j.vetpar.2010.11.036. [DOI] [PubMed] [Google Scholar]
- 37.Savini G, Dunsmore JD, Robertson ID, Seneviratna P. The epidemiology of Sarcocystis spp. in cattle of Western Australia. Epidemiol Infect. 1992;108:107–13. doi: 10.1017/S0950268800049554. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Expected fragment sizes (in base pairs) from digestion of 18S rDNA PCR products of bovine Sarcocystis species with FokI and BfaI.
Additional file 2: Sarcocystis positivity rate in diaphragm and heart samples among different groups.
Additional file 3: Phylogenetic tree of unidentified Sarcocystis species (cox1 sequencing).
Data Availability Statement
The dataset supporting the conclusions of this article is available in the Zenodo repository, https://doi.org/10.5281/zenodo.4611489. The nucleotide sequences of the unidentified Sarcocystis spp. UNS1, UNS2, UNS3 are in GenBank under the following accession numbers: UNS1: MW756133, UNS2: MW756134, UNS3: MW756135.