Abstract
Background
Pseudomonas aeruginosa, the rhamnolipids-producer, is one of dominant bacteria in oil reservoirs. Although P. aeruginosa strains are facultative bacteria, the anaerobic biosynthesis mechanism of rhamnolipids is unclear. Considering the oxygen scarcity within oil reservoirs, revealing the anaerobic biosynthesis mechanism of rhamnolipids are significant for improving the in-situ production of rhamnolipids in oil reservoirs to enhance oil recovery.
Results
Pseudomonas aeruginosa SG anaerobically produced rhamnolipids using glycerol rather than glucose as carbon sources. Two possible hypotheses on anaerobic biosynthesis of rhamnolipids were proposed, the new anaerobic biosynthetic pathway (hypothesis 1) and the highly anaerobic expression of key genes (hypothesis 2). Knockout strain SGrmlB failed to anaerobically produce rhamnolipids using glycerol. Comparative transcriptomics analysis results revealed that glucose inhibited the anaerobic expression of genes rmlBDAC, fabABG, rhlABRI, rhlC and lasI. Using glycerol as carbon source, the anaerobic expression of key genes in P. aeruginosa SG was significantly up-regulated. The anaerobic biosynthetic pathway of rhamnolipids in P. aeruginosa SG were confirmed, involving the gluconeogenesis from glycerol, the biosynthesis of dTDP-l-rhamnose and -hydroxy fatty acids, and the rhamnosyl transfer process. The engineered strain P. aeruginosa PrhlAB constructed in previous work enhanced 9.67% of oil recovery higher than the wild-type strain P. aeruginosa SG enhancing 8.33% of oil recovery.
Conclusion
The highly anaerobic expression of key genes enables P. aeruginosa SG to anaerobically biosynthesize rhamnolipids. The genes, rmlBDAC, fabABG, rhlABRI, rhlC and lasI, are key genes for anaerobic biosynthesis of rhamnolipid by P. aeruginosa. Improving the anaerobic production of rhamnolipids better enhanced oil recovery in core flooding test. This study fills the gaps in the anaerobic biosynthesis mechanism of rhamnolipids. Results are significant for the metabolic engineering of P. aeruginosa to enhance anaerobic production of rhamnolipids.
Keywords: Pseudomonas aeruginosa, Rhamnolipids, Anaerobic biosynthesis, Glycerol, RmlBDAC, Microbial enhanced oil recovery
Background
At present, the demand for oil in economic development continues to increase, while the recoverable oil reserves in oil reservoirs are decreasing, and it is more difficult to find new oil and gas resources. It is of great strategic significance to enhance the oil recovery of existing oil fields through scientific research and innovation. Microbial oil recovery technology (MEOR) is an economical, effective and environmentally friendly oil recovery technology [1]. It is feasible to enhance oil recovery using microorganisms to in-situ produce biosurfactants in oil reservoirs [2,4]. It is also proved that the stable microbial growth and efficient production of biosurfactants such as rhamnolipids in reservoirs are the keys to the successful implementation of MEOR [1, 3, 4]. Rhamnolipids can effectively reduce the oilwater interfacial tension, emulsify crude oil and change the wettability of reservoir rock, so it is one of the excellent oil displacement agents used to improve oil recovery [1, 4]. The environments in oil reservoirs are anoxic or anaerobic [5]. The injection of air into oil reservoirs is difficult to ensure the effective oxygen supply of aerobic microorganisms [4]. However, studies on anaerobic synthesis of biosurfactants are still weak [6], which limits to enhance oil recovery by anaerobic production of biosurfactants in oil reservoirs.
Although rhamnolipids as a natural biosurfactants is extensively studied and applied in MEOR and other fields [7, 8], studies on anaerobic biosynthesis of rhamnolipids are also scarce [6]. Many rhamnolipid-producing strains have been repoted, such as Pseudomonas sp. and Burkholderia sp. [8]. Among the rhamnolipid-producing strains, Pseudomonas aeruginosa has the highest rhamnolipid yield [8]. Therefore, P. aeruginosa was mainly studied for producing rhamnolipids [8, 9]. P. aeruginosa are facultative bacterial strains that can grow and metabolize at both aerobic and anaerobic conditions [6, 10, 11]. But most of the studies on rhamnolipids production by P. aeruginosa were focused on aerobic biosynthesis.
Studies have discovered that some P. aeruginosa strains can anaerobically produce rhamnolipids when using glycerol as carbon source [12,15]. But the related anaerobic biosynthesis mechanisms of rhamnolipids are unclear. How to further enhance the anaerobic yield of rhamnolipids requires the modification of key genes or the regulation of the anaerobic biosynthetic pathways, which all depend on the anaerobic biosynthesis mechanisms of rhamnolipids.
This study aims to explore the underlying anaerobic biosynthetic mechanism of rhamnolipids when P. aeruginosa SG using glycerol as carbon source. Strain P. aeruginosa SG was used for rhamnolipids production [13, 14]. P. aeruginosa SG was cultured with different carbon sources (glucose and glycerol). Cell growth and rhamnolipids production were comparatively studied. Two possible hypotheses on anaerobic biosynthesis of rhamnolipid using glycerol were proposed. RNA sequencing (RNA-seq) was performed on the anaerobic culture of strain SG using glucose and glycerol as carbon source, respectively. The transcriptomic data were analyzed for the anaerobic biosynthesis pathways of rhamnolipids and related genes regulation. The method of gene knock-out was used to identify the key genes and the anaerobic biosynthetic pathway of rhamnolipids when P. aeruginosa SG using glycerol. Application potential by anaerobic production of rhamnolipids for MEOR was evaluated and discussed. Results will help to regulate P. aeruginosa to produce more rhamnolipids under anaerobic conditions. This study also provides scientific guidance for the development of MEOR technology through in situ production of rhamnolipids.
Materials and methods
Bacterial strains and culture conditions
Strain P. aeruginosa SG was used as a rhamnolipids-producer in this study. P. aeruginosa SG can anaerobically produce rhamnolipids [13, 14]. P. aeruginosa SG was cultured in LB (LuriaBertani) medium to prepare seed culture at 35C and 180rpm for 12h. To avoid interference of rhamnolipids produced in seed culture, the seed culture was centrifuged at 5000g for 2min. The bacterial cells were washed twice using sterile distilled water. The suspension liquid of bacterial cells with sterile distilled water was transferred into the fermentation medium. The fermentation medium except for carbon source contained 4g/l of NaNO3, 3g/l of KH2PO4, 4g/l of K2HPO43H2O, 0.5g/l of KCl, 0.5g/l of NaCl, 0.2g/l of CaCl22H2O, 1.0g/l of MgSO47H2O. Glucose and glycerol were used as carbon sources with the concentrations of 40g/l. The aerobic culture was performed in 250ml-triangular flasks containing 100ml fermentation medium, at 35C and 180rpm for 6days. The anaerobic culture was performed in 100ml-serum bottles containing 80ml anaerobic fermentation medium. The anaerobic medium was prepared as described in previous studies [13, 16]. The anaerobic culture was incubated at 35C and 50rpm for 8days. Three parallels were set for each experiments. The inoculum amount was 3% (v/v). The non-inoculated medium was used as the negative control. Escherichia coli DH5, E. coli S17-1 and recombinant strains were cultured in LB medium at 35C and 180rpm. During gene knock-out process, LB medium containing ampicillin of 100mg/l and kanamycin of 50mg/l were used for recombinant E. coli strains. LB medium containing kanamycin of 350mg/l was used for recombinant P. aeruginosa strains.
Analytical methods
The anaerobic culture was sampled from serum bottles using sterile syringes. Cell growth of strain SG was represented by OD600 values in the anaerobic culture. Then samples were centrifuged at 10,000g for 10min, respectively. The cell free supernatant was collected. The surfactants anaerobically produced by P. aeruginosa SG using glycerol were rhamnolipids which were confirmed by analytical methods of FTIR and HPLC-MS [14]. In this study, rhamnolipids concentration was also represented by the diameter of oil spreading circle formed by cell free supernatant. The diameter of oil spreading circle was measured as described in previous studies [17, 18]. Then residue supernatant surface tension was measured by surface tensiometer (BZY-1, Shanghai Hengping Instrument and Meter Factory, Shanghai, China).
Gene rmlB knock-out
In this study, using the suicidal plasmid pK18mobSacB of P. aeruginosa, gene rmlB was knocked out to block the biosynthetic pathway of dTDP-L-rhamnose controlled by rmlBDAC operon genes. The rmlBDAC operon genes control the biosynthesis of dTDP-rhamnose [19]. The gene fragment rmlBD was obtained by PCR using the genomic DNA of P. aeruginosa SG as the template. The PCR primers were rmBD-f: 5-CGGAAGCTTATGTGGACCGCTCGAT-3 and rmBD-r: 5-GCCGAATTCCTGTTGCAGCTTGCGGT-3. The rmlBD fragment was 1969bp with restriction sites of Hind III and EcoR I at the 5 end and 3 end, respectively. The rmlBD fragment was cloned into pMD19T (simple) vector to construct plasmid pMD19-rmlBD. The rmlBD fragment contains two Eco52 I restriction sites at 831bp and 1206bp. After pMD19T-rmlBD successively digested with Eco52 I enzyme and T4 DNA ligase, plasmid pMD19-rmlBD was constructed and purified by gel extraction kit (Takara, Japan). The rmlBD fragment was cloned into the Hind III and EcoR I sites of plasmid pK18mobSacB to construct recombinant plasmid pK18-rmlBD. The recombinant plasmid pK18-rmlBD was transformed into strain E. coli S17-1. Using conjugation method [20], the plasmids pK18-rmlBD were transferred into the strain P. aeruginosa SG. Mutants were cultured on LB medium containing Ampicillin and kanamycin for the first recombination. The LB medium containing 20% sucrose and Ampicillin was used to pick out the knockout strain P. aeruginosa SGrmlB. The anaerobic cell growth and anaerobic rhamnolipid production of wild-type strain SG and the knockout strain SGrmlB were comparatively determined.
RNA extraction, library construction and RNA sequencing
P. aeriginosa SG cells were sampled from its anaerobic culture with OD600=0.6. P. aeriginosa SG cultured with glucose and glycerol as carbon sources were marked as GluAn and GlyAn, respectively. Three replicates, GluAn-1, GluAn-2, GluAn-3 and GlyAn-1, GlyAn-2, GlyAn-3, were sampled for GluAn and GlyAn. The bacterial cells were collected by centrifugation (4C, 4000g, 5min). The harvested bacterial cells were used for the total RNA extraction. The Takara RNA kit (Takara, Japan) was used for RNA extraction and purification according to the manufacturer provided protocol. Then RNA purity of GluAn samples and GlyAn samples was evaluated by Nanodrop2000 with OD260/280 values and OD260/230 values. RNA fragment length of GluAn samples and GlyAn samples was determined using Agilent 2100 Bioanalyzer. The tested mRNA was enriched by removing rRNA. After enrichment, mRNA is fragmented into short fragments. First strand of cDNA was synthesized by reverse transcription using random hexamers and mRNA as template, and then the buffer, dNTPs and DNA polymerase I were added to synthesize the two-strand cDNA. Then AMPure XP Beads were used to purify double-stranded cDNA. The purified double stranded cDNA was treated with terminal repair, addition of A and joint. The cDNA libraries were constructed using the method of chain-specific library by the Allwegene BioTech in Beijing (China). RNA sequencing was performed using Illumina Hiseq 4000 with PE150 double terminal sequencing strategy by the Allwegene BioTech in Beijing (China).
Transcriptome analysis
The obtained raw reads were filtered to remove the reads with sequencing adapter and the low-quality Reads. The sequencing data were statistically analyzed for raw reads numbers, clean reads numbers, sequence error rate, Q20 and Q30 (proportion of clean data with Phred value greater than 20 and 30) and GC content (%). The high quality data, clean reads, were used for transcriptome analysis [21]. The obtained clean reads were mapped to the reference genome of P. aeruginosa PAO1 using Bowtie2 alignment software. The regional distribution and Reads density distribution were evaluated. The gene annotation, gene structure analysis and the new transcript prediction were performed.
FPKM (Fragments per kilobase of exon model per million mapped reads) values were calculated using HTSeq software [22] for the gene expression level analysis of each sample. The number of genes and the expression level of a single gene at different expression levels were counted respectively, and the FPKM value of one was used as the threshold to judge whether the gene was expressed or not. Using DESeq method [23], gene differential expression analysis was performed according to the readCount data obtained from gene expression level analysis. The differentially expressed genes between GluAn samples and GlyAn samples were identified with log2(FoldChange) value>1 and q-value<0.05. Hierarchical clustering analysis of differentially expressed genes was performed based on their expression levels, log10(FPKM+1). The GO (Gene Ontology, http://www.geneontology.org/) enrichment analysis was carried out using GOseq software [24], and the probability of GO term enriched by differential genes was calculated. The differentially expressed genes were enriched into molecular function, biological process, cellular component. KEGG (Kyoto Encyclopedia of Genes and Genomes, http://www.kegg.jp) enrichment analysis was performed for pathways enrichment of the differentially expressed genes [25].
Pathways description and genes discovery
The metabolic pathways related to fatty acid metabolism, glycometabolism and rhamnosyl transfer process were mined from the transcripts information that has be enriched into KEGG pathways. Referring to the known information of aerobic biosynthetic pathways of rhamnolipid in model strain P. aeruginosa PAO1, the anaerobic biosynthetic pathways of rhamnolipid were attempted to analyze, starting with the metabolism of glycerol. Two precursors of rhamnolipid biosynthesis, dTDP-l-rhamnose and -hydroxy fatty acids, were of particular consideration. In the transcriptome of P. aeruginosa SG using glycerol under anaerobic conditions, the anaerobic biosynthesis of dTDP-l-rhamnose and -hydroxy fatty acids and the transfer process of rhamnosyl were the target biosynthetic pathways. When P. aeruginosa SG using glucose as carbon source under anaerobic conditions, the related biosynthetic pathways were also comparatively analyzed.
The transcriptome data of P. aeruginosa SG using glucose and glycerol as carbon source under anaerobic conditions were compared and analyzed. The related key genes that were significantly up-regulated or activated when using glycerol as carbon source were mined from the differentially expressed genes between GluAn samples and GlyAn samples. Referring to the known key genes of aerobic biosynthetic pathways of rhamnolipid in model strain P. aeruginosa PAO1, the candidate key genes involving in the anaerobic biosynthesis of rhamnolipid using glycerol were picked out from the significantly up-regulated genes. The genes related to the biosynthesis of dTDP-l-rhamnose and -hydroxy fatty acids, and the rhamnosyl transfer process were the target genes.
Core flooding test
Enhanced oil recovery by in-situ anaerobic production of rhamnolipids was evaluated by core flooding test. The experimental protocol of core flooding test was referred to the previous study [13]. The engineered strain P. aeruginosa PrhlAB with higher anaerobic yield of rhamnolipids was used in core flooding tests. The engineered strain P. aeruginosa PrhlAB was constructed by increasing the copy number of rhlAB genes in P. aeruginosa SG [15]. The rock core has length of 293mm, diameter of 38mm, absolute permeability of 0.388m2, pore volume of 69.9ml, respectively. The Xinjiang oilfield production water and crude oil (density of 0.886g/cm3 and viscosity of 6.3mPas) were used. After the first water flooding, 0.5 PV of culture solution [strain PrhlAB and medium (1:20, v/v)] was injected into rock core. The medium contained 40g/l glycerol, 4g/l of NaNO3, 3g/l of KH2PO4, 4g/l of K2HPO43H2O, 0.5g/l of KCl, 0.5g/l of NaCl, 0.2g/l of CaCl22H2O. Then the rock core was incubated at 39C for 8days. During the core flooding process, the volumes of displaced oil (ml) and displaced water (ml) were recorded. Enhanced oil recovery efficiency (%) was calculated.
Results and discussion
Rhamnolipids production and cell growth using glucose and glycerol
Under aerobic conditions, strain P. aeruginosa SG decreased the culture surface tension to 27.0mN/m and 27.3mN/m using both glucose and glycerol as carbon sources, respectively. The oil spreading circles diameters of the aerobic culture were 482mm and 565mm when P. aeruginosa SG using glucose and glycerol as carbon sources, respectively. The surface activity and oil spreading activity indicate that P. aeruginosa SG produced biosurfactants [17, 18]. The produced biosurfactants by P. aeruginosa was rhamnolipids [14, 15]. Under aerobic conditions, P. aeruginosa SG produced rhamnolipids using both glucose and glycerol as carbon sources.
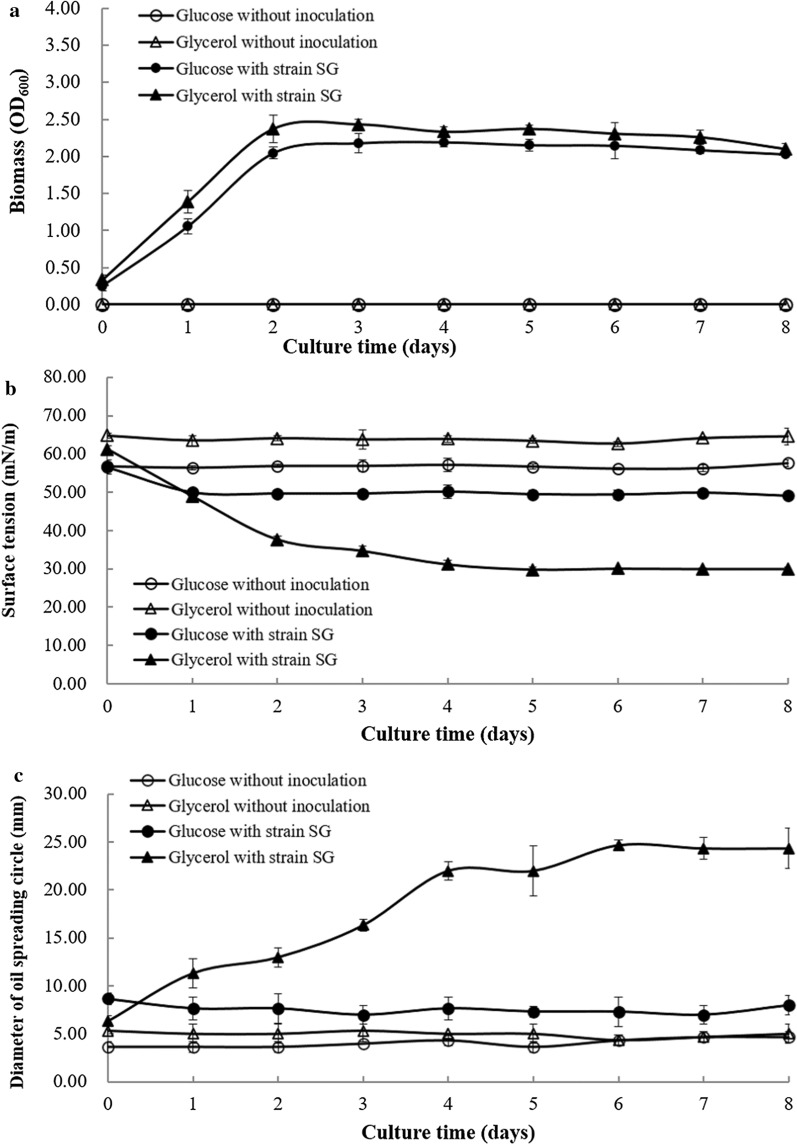
But rhamolipids production by strain SG under anaerobic conditions were quite different. Under anaerobic conditions, the cell growth (OD600), surface tension and oil spreading circles diameters were shown in Fig.1, From the biomass (OD600) point of view, strain P. aeruginosa SG grew well under anaerobic conditions using both glucose and glycerol as carbon sources (Fig.1a). However, the surface tension values of anaerobic culture were decreased from 64 to 30mN/m using glycerol (Fig.1b). While the surface tension values of anaerobic culture were decreased from 57 to 48mN/m using glucose. Results indicated that strain P. aeruginosa SG produced biosurfactants when using glycerol as carbon source. As shown in Fig.1c, the oil spreading circles diameters of anaerobic culture were 24mm and 8mm when strain P. aeruginosa SG using glucose or glycerol as carbon sources, respectively. The oil spreading activity also demonstrates that P. aeruginosa SG produced biosurfactants when using glycerol as carbon source [17, 18]. The anaerobically produced biosurfactants by P. aeruginosa SG was identified as rhamnolipids [13]. Results confirmed that P. aeruginosa SG can anaerobically grow well using both glucose and glycerol, but P. aeruginosa SG anaerobically produces rhamnolipids when using glycerol as carbon sources.
Fig. 1.
Anaerobic growth and production of rhamnolipids by P. aeruginosa SG using glucose and glycerol as carbon sources a biomass (OD600), b surface tension and c oil spreading activity
Research hypotheses for anaerobic biosynthesis of rhamnolipids using glycerol
Under aerobic conditions, dTDP-rhamnose and -hydroxy fatty acids are two required precursors for rhamnolipids biosynthesis [19]. The gluconeogenesis pathway, the EntnerDoudoroff pathway and de novo synthesis of fatty acids are all central metabolic pathways in P. aeruginosa. These metabolic pathways can also support the growth of P. aeruginosa under anaerobic conditions [10, 26]. Therefore, under anaerobic conditions, glucose can be transformed into glucose-6-phosphate and then glucose-1-phosphate that was used for dTDP-l-rhamnose biosynthesis. Through catabolic pathways, both glucose and glycerol can link with the de novo synthesis pathway of fatty acids under anaerobic conditions. And then -hydroxy fatty acids can be synthesized by RhlA enzyme. Theoretically, the two precursors, dTDP-l-rhamnose and -hydroxy fatty acids, can be synthesize using both glucose and glycerol under anaerobic conditions. Therefore, rhamnolipids should be anaerobically synthesized by P. aeruginosa using both glycerol and glucose as carbon sources. In fact, P. aeruginosa anaerobically synthesized rhamnolipids mere using glycerol rather than glucose as carbon source.
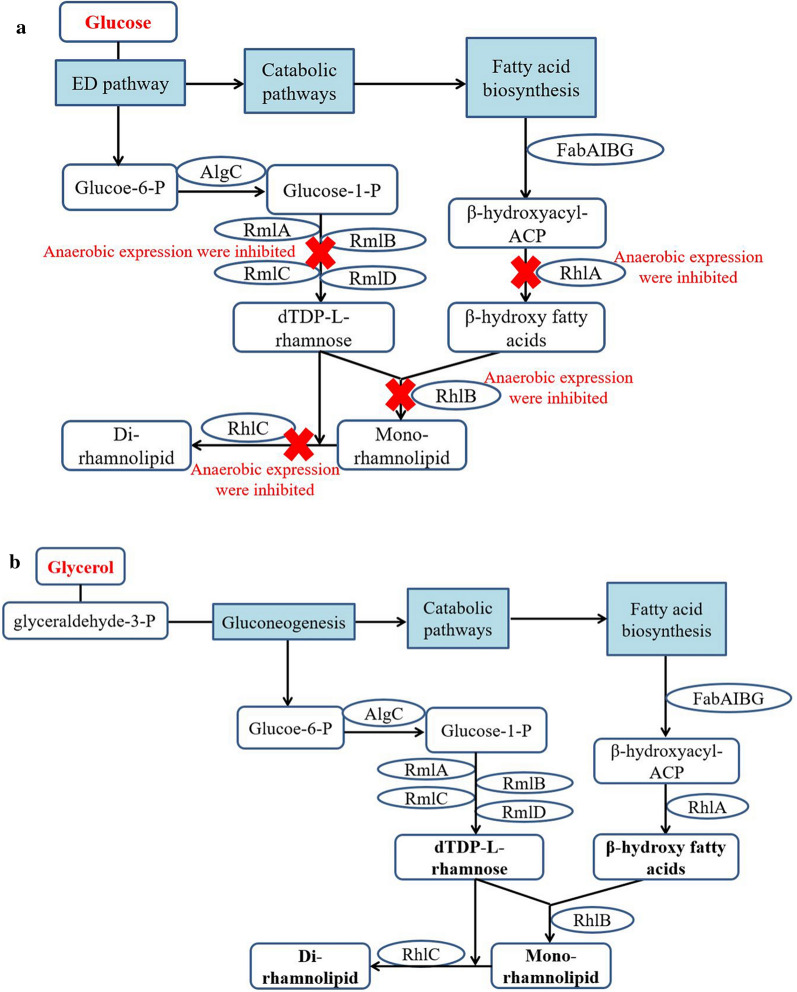
Here, two possible hypotheses on anaerobic biosynthesis of rhamnolipid by P. aeruginosa using glycerol were proposed. Hypothesis 1: in P. aeruginosa, the anaerobic biosynthetic pathway of rhamnolipid is different from the aerobic biosynthetic pathway of rhamnolipids. This hypothesis suggests that anaerobic conditions block the biosynthesis of dTDP-l-rhamnose from glucose-1-phosphate, which results in P. aeruginosa being unable to anaerobically synthesize rhamnolipids from glucose. There may be other pathways to metabolize glycerol to synthesize dTDP-l-rhamnose without the effect of rmlBDAC operon genes, which leads to the anaerobic biosynthesis of rhamnolipids using glycerol. Hauser and Karnovsky (1957) cultured P. aeruginosa using 14C-labeled glycerol--14C and glycerol--14C as carbon sources. They found that the C6 skeleton of rhamnose was composed of two 14C-labeled C3 units that were derived from two molecules glycerol (14C-labeled C3 unit) without carbon chain rearrangement [27]. Hypothesis 2 was shown in Fig.2. In P. aeruginosa, the rhamnolipids biosynthetic pathway is same under both aerobic and anaerobic conditions, but the anaerobic expression of key genes for rhamnolipids synthesis is inhibited using glucose as carbon source, and these key genes can be highly expressed under anaerobic conditions using glycerol as carbon source. Previous studies compared the gene expression differences of P. aeruginosa using glucose as carbon source under aerobic and anaerobic conditions, and the results showed that the expressions of rhlR, rhlI and rhlAB genes related to rhamnolipids synthesis were inhibited under anaerobic conditions using glucose [28,30].
Fig. 2.
The anaerobic biosynthetic pathways of rhamnolipid in P. aeruginosa SG a using glucose as carbon source; b using glycerol as carbon source
From the two hypotheses point of view, the anaerobic synthesis of dTDP-rhamnose is one key metabolic pathway for rhamnolipids biosynthesis under anaerobic conditions. Knocking out the rmlBDAC operon can identify whether the rmlBDAC genes involved in the anaerobic biosynthesis of rhamnolipid using glycerol. If the knockout strain SGrmlB can anaerobically produce rhamnolipids using glycerol as carbon source, there is a new biosynthetic pathway of dTDP-l-rhamnose under anaerobic conditions (hypothesis 1). If not, the rmlBDAC genes control the anaerobic biosynthesis of dTDP-rhamnose, and the expression of rmlBDAC genes and other key genes are inhibited under anaerobic conditions (hypothesis 2). The transcriptomics results can provide the related information.
Anaerobic production of rhamnolipids by strain SG and SGrmlB using glycerol
dTDP-Rhamnose and -hydroxy fatty acids are two required precursors for rhamnolipids biosynthesis. The rmlBDAC operon genes control the biosynthesis of dTDP-rhamnose, and -hydroxy fatty acids are derived from the de novo synthesis of fatty acids [19]. The de novo synthesis of fatty acids is central metabolic pathway in P. aeruginosa. Therefore, anaerobic synthesis of dTDP-rhamnose is one key metabolic pathway for rhamnolipid biosynthesis under anaerobic conditions.
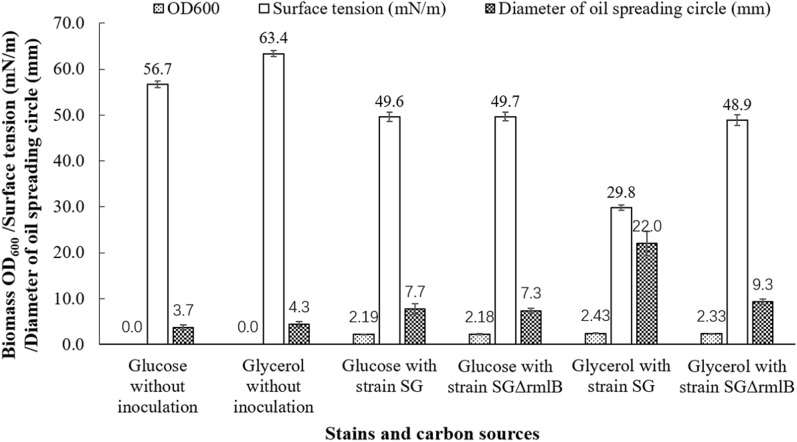
The anaerobic cell growth and anaerobic rhamnolipids production of wild-type strain SG and the knockout strain SGrmlB were comparatively determined. If the knockout strain SGrmlB can anaerobically produce rhamnolipids using glycerol as carbon source, there is a new biosynthetic pathway of dTDP-l-rhamnose for rhamnolipids synthesis using glycerol under anaerobic conditions. If not, the rmlBDAC operon genes control the anaerobic biosynthesis of dTDP-rhamnose then for rhamnolipids synthesis using glycerol under anaerobic conditions. As shown in Fig.3, the knockout strain SGrmlB can anaerobically grow using both glucose and glycerol as carbon sources with OD600=2.18 and 2.33. The wild-type strain SG can anaerobically produce rhamnolipids using glycerol as carbon source, reducing surface tension to 29.8mN/m and forming oil spreading circle with diameter of 22.0mm. Although glycerol used as carbon source, the knockout strain SGrmlB can not anaerobically produce rhamnolipids, reducing surface tension to 48.9mN/m and forming oil spreading circle with diameter of 9.3mm. Results indicated that the rmlBDAC operon genes involved in the anaerobic biosynthesis of rhamnolipids using glycerol.
Fig. 3.
Anaerobic growth and production of rhamnolipids by the wild-type strain P. aeruginosa SG and the knockout strain P. aeruginosa SGrmlB using glucose and glycerol as carbon sources
Basic transcriptome results of strain SG using glycerol or glucose under anaerobic conditions
To understand why glycerol can be used for anaerobically synthesizing rhamnolipids by strain SG while glucose cannot, RNA sequencing (RNA-seq) was performed on strain SG using glucose and glycerol as carbon source under anaerobic conditions, respectively. The RNA-seq data were shown in Table 1. A total of 90270732 Raw reads were obtained. After quality control, 84703416 Clean Reads were obtained. Totally, 5875 genes were mapped to the reference genome of P. aeruginosa PAO1 (NC_002516). The RNA sequencing data have been deposited in NCBIs Sequence Read Archive (SRA) with Accession Number PRJNA670563 (https://dataview.ncbi.nlm.nih.gov/object/PRJNA670563?reviewer=q57rp98cvqi8nvid6dv8urgint). FPKM values of the mapped genes were calculated for analysis of the gene expression level. The FPKM value of one was used as the threshold to judge whether the gene was expressed or not. The correlation of gene expression levels between samples is an important indicator to test the reliability of experiments and the reasonableness of sample selection [31]. The closer the Pearson correlation coefficient squared (R2) is to 1, the higher the similarity of expression patterns between samples is and the less different genes among samples. In this study, Pearson correlation coefficient squared (R2) between samples in GluAn and samples in GlyAn were all greater than 0.92, which indicated the ideal sampling and experimental conditions.
Table 1.
RNA-seq data of strain SG using glycerol and glucose as carbon sources under anaerobic conditions
| Sample name | Raw reads | Clean reads | Clean bases (Gb) | GC content (%) | Total mapped reads | Mapped percentage (%) |
|---|---|---|---|---|---|---|
| GluAn-1 | 13878806 | 12993448 | 1.94 | 63.60 | 12542479 | 96.53 |
| GluAn-2 | 16251408 | 15319168 | 2.30 | 63.86 | 14794181 | 96.57 |
| GluAn-3 | 14265600 | 13323478 | 2.00 | 63.58 | 12748319 | 95.68 |
| GlyAn-1 | 16223066 | 15051700 | 2.26 | 64.14 | 14573577 | 96.82 |
| GlyAn-2 | 14887764 | 13860002 | 2.08 | 63.94 | 13433828 | 96.93 |
| GlyAn-3 | 14764088 | 14155620 | 2.12 | 63.94 | 13733081 | 97.02 |
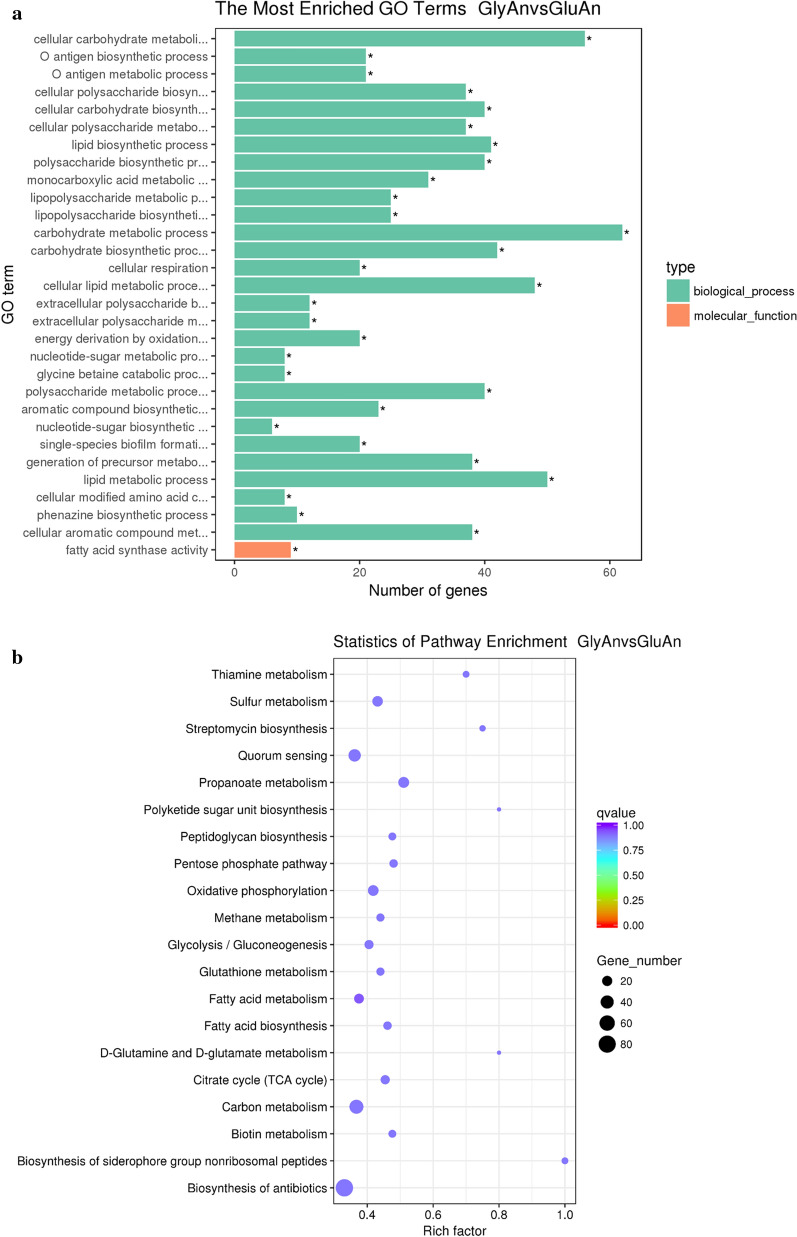
According to the readCount data obtained from gene expression level analysis, a total of 972 significantly differentially expressed genes with log2(FoldChange) value>1 and q-value<0.05 were identified between strain P. aeruginosa SG using glycerol and glucose as carbon sources under anaerobic conditions. There were 549 up regulated genes and 423 down regulated genes in GlyAn samples vs GluAn samples. As shown in Fig.4a, the up regulated genes were significantly enriched into 37 GO terms belonging to two major functional categrories, molecular function and biological process. KEGG pathways enrichment analysis showed that the up regulated genes were enriched into 20 pathways (Fig.4b). Some of the enriched metabolic pathways are closely related to the biosynthesis of rhamnolipids, such as fatty acid biosynthesis and metabolism, quorum sensing, gluconeogenesis.
Fig. 4.
GO annotation and KEGG pathways enrichment of the differentially expressed genes between using GlyAn and GluAn a the most enriched GO Terms, the asterisk indicated that the GO item was significantly enriched (P<0.05); and b the statistics of KEGG pathways enrichment
Differentially expressed genes involved in anaerobic biosynthesis of rhamnolipids
As listed in Table 2, the genes related to the biosynthesis of dTDP-l-rhamnose and -hydroxy fatty acids, and the rhamnosyl transfer process pathways were picked out from the differentially expressed genes between GlyAn samples and GluAn samples. The precursor, dTDP-l-rhamnose, provides the glycosyl part for the biosynthesis of rhamnolipids. It can be initiated by the gluconeogenesis pathway or the EntnerDoudoroff pathway, and the algC gene and rmlBDAC operon genes control the dTDP-l-rhamnose biosynthesis [32, 33]. The another precursor, -hydroxy fatty acids, provides the lipid chain part for the biosynthesis of rhamnolipids. It can be initiated by the de novo synthesis of fatty acids. Fatty acid synthase system II, Ketolipoyl reductase (FabG) and peptide-transferase (RhlA) catalyze the biosynthesis of -hydroxy fatty acids [32, 33]. The two precursors, dTDP-l-rhamnose and -hydroxy fatty acids, were assembled to mono-rhamnolipids and di-rhamnolipids catalyzed by rhamnotransferase I (RhlB) and rhamnotransferase II (RhlC), respectively [34]. Rhamnotransferase I (RhlB) and rhamnotransferase II (RhlC) were coded by genes rhlB and rhlC, respectively. The genes lasRI and rhlRI in quorum-sensing pathways are responsible for activating the expression of genes, rhlAB and rhlC [34, 35]. Under anaerobic conditions, the related structural genes and regulatory genes, rmlBDAC, rhlAB, rhlC, fabA, fabG and rhlRI, lasI, were significantly up-regulated or activated when using glycerol as carbon source. Glycerol metabolism up-regulated the expression of genes in Quorum-sensing system, which promote the expression of genes rmlBDAC, fabA, fabG, rhlAB and rhlC. The anaerobic high expression of key genes is significant for P. aeruginosa producing rhamnolipids under anaerobic conditions.
Table 2.
The differentially expressed genes related to the biosynthesis of dTDP-l-rhamnose and -hydroxy fatty acids, and the rhamnosyl transfer process
| Gene id | Gene name | Readcount-GlyAn | Readcount-GluAn | log2(foldchange) | p-value | Gene description |
|---|---|---|---|---|---|---|
| PA5322 | algC | 3951.9 | 3245.6 | 0.28 | 0.43 | phosphomannomutase AlgC |
| PA5161 | rmlB | 4139.6 | 1464.7 | 1.49 | 9.58E09 | dTDP-d-glucose 4,6-dehydratase |
| PA5162 | rmlD | 1650.9 | 538.10 | 1.62 | 1.02E09 | dTDP-4-dehydrorhamnose reductase |
| PA5163 | rmlA | 2270.2 | 789.6 | 1.52 | 8.69E09 | glucose-1-phosphate thymidylyltransferase |
| PA5164 | rmlC | 2170.5 | 649.9 | 1.74 | 3.76E11 | dTDP-4-dehydrorhamnose 3,5-epimerase |
| PA1609 | fabB | 2195.5 | 1304.6 | 0.75 | 1.65E03 | beta-ketoacyl-ACP synthase I |
| PA1610 | fabA | 945.2 | 472.6 | 1.00 | 3.96E05 | beta-hydroxydecanoyl-ACP dehydrase |
| PA2965 | fabF1 | 1117.3 | 966.8 | 0.21 | 0.40 | beta-ketoacyl-acyl carrier protein synthase II |
| PA1373 | fabF2 | 109.6 | 395.1 | 1.85 | 2.57E12 | 3-oxoacyl-acyl carrier protein synthase II |
| PA2968 | fabD | 702.6 | 439.7 | 0.68 | 6.68E03 | malonyl-CoA-[acyl-carrier-protein] transacylase |
| PA2967 | fabG | 2143.7 | 926.5 | 1.21 | 5.82E07 | 3-oxoacyl-[acyl-carrier-protein] reductase |
| PA3333 | fabH2 | 677.1 | 10.0 | 6.08 | 2.65E70 | 3-oxoacyl-[acyl-carrier-protein] synthase III |
| PA3476 | rhlI | 5429.4 | 92.4 | 5.88 | 1.56E87 | autoinducer synthesis protein RhlI |
| PA3477 | rhlR | 13664.1 | 732.3 | 4.22 | 1.93E56 | transcriptional regulator RhlR |
| PA3478 | rhlB | 26462.6 | 243.7 | 6.76 | 1.28E91 | rhamnosyltransferase chain B |
| PA3479 | rhlA | 42259.3 | 370.3 | 6.83 | 1.27E69 | rhamnosyltransferase chain A |
| PA1130 | rhlC | 1418.9 | 16.1 | 6.46 | 2.61E86 | rhamnosyltransferase 2 |
| PA1432 | lasI | 1260. 4 | 196.1 | 2.68 | 1.95E23 | autoinducer synthesis protein LasI |
Pathways and key genes for anaerobic synthesis of rhamnolipids using glycerol
In this study, the knockout strain SGrmlB could not anaerobically synthesize rhamnolipids even using glycerol as carbon source. Results showed that the pathways to metabolize glycerol to synthesize dTDP-l-rhamnose was also controlled by rmlBDAC operon genes under anaerobic conditions. The gene expression results showed that the related genes for rhamnolipids biosynthesis were highly expressed under anaerobic conditions using glycerol. The genes, rmlBDAC, rhlAB, rhlC, fabA, fabG and rhlRI, lasI, were significantly up-regulated expression using glycerol as carbon source, compared to using glucose under anaerobic conditions.
Based on the above results, Hypothesis 2 is more convincing (Fig.2). The anaerobic biosynthesis mechanism of rhamnolipids in P. aeruginosa was revealed at the gene level using glycerol as carbon source. The anaerobic biosynthetic pathway of rhamnolipids in P. aeruginosa SG were confirmed, involving the gluconeogenesis from glycerol, the biosynthesis of dTDP-l-rhamnose and -hydroxy fatty acids, and the rhamnosyl transfer process, which is consistent with the aerobic biosynthetic pathways in model strain P. aeruginosa PAO1. The key genes involved in anaerobic biosynthesis of rhamnolipids are rmlBDAC, rhlABRI, rhlC, fabA, fabG and lasI. Glycerol metabolism up-regulated the expression of genes in Quorum-sensing system (rhlRI and lasI), which promote the expression of genes rmlBDAC, rhlAB and rhlC, thus enable P. aeruginosa to anaerobically produce rhamnolipids using glycerol.
Enhanced oil recovery by anaerobic production of rhamnolipids
After the first water flooding, 45.21% of the oil was recovered from the core model. At the end of the second water flooding, 54.88% of oil was recovered. In-situ anaerobic production of rhamnolipids by engineered strain P. aeruginosa PrhlAB enhanced 9.67% of oil recovery. In our previous study [13], in-situ anaerobic production of rhamnolipids by wild strain P. aeruginosa SG enhanced 8.33% of oil recovery. Increasing the copy number of rhlAB genes in P. aeruginosa SG, the engineered strain P. aeruginosa PrhlAB can produce more rhamnolipids under anaerobic conditions [15]. Improving the anaerobic production of rhamnolipids better enhanced oil recovery in core flooding test. Future research will be focused on further enhancing the anaerobic production yield based on the anaerobic biosynthetic mechanism of rhamnolipids.
Other prospect of the anaerobic production of rhamnolipids
P. aeruginosa strains are facultative bacteria [10], but the vast majority of studies on the biosynthesis of rhamnolipids in P. aeruginosa were focused on aerobic biosynthesis [19, 34, 35]. Anaerobic biosynthesis of rhamnolipids is promising to the in-situ applications in anoxic environments, such as deep soil, oil reservoirs, sediments [4, 6, 13]. Rhamnolipids products have also been partially commercialized in some fields. Nevertheless, the necessary aeration and defoaming processes in the aerobic fermentation of rhamnolipids lead to high production costs, which affects the large-scale industrial application of rhamnolipids [12, 36]. The foam problem in production of rhamnolipids can be avoided by anaerobic fermentation without aeration. The low anaerobic production yield of rhamnolipids was the bottleneck for in-situ enhanced oil recovery, and the foamless fermentation. Enhancing the anaerobic production yield of rhamnolipids is of great importance. In the future, the revealed biosynthetic pathways and key genes involved in anaerobic biosynthesis of rhamnolipids would be regulated and modified to enhance P. aeruginosa to anaerobically produce more rhamnolipids. Besides, -hydroxy fatty acids are precursor substrates of rhamnolipids synthesis as well. Fatty acid synthesis requires a lot of reducing power (NADPH or NADH2). Compared with anaerobic conditions, aerobic conditions are conducive to cell synthesis of reducing power, which promotes fatty acid synthesis and ultimately enhances rhamnolipids synthesis. Increasing the reducing power level in P. aeruginosa cells will also enhance the production of rhamnolipids under anaerobic conditions. Results of this study would be significant for the metabolic engineering of P. aeruginosa to enhance anaerobic production of rhamnolipids.
Conclusion
P. aeruginosa can anaerobically produce rhamnolipid using glycerol rather than glucose. Two possible hypotheses on anaerobic biosynthesis of rhamnolipid using glycerol were proposed. The rmlBDAC operon genes involved in the anaerobic biosynthesis of rhamnolipid using glycerol. The expression of key genes was inhibited under anaerobic conditions when P. aeruginosa using glucose. The highly anaerobic expression of key genes enables P. aeruginosa to anaerobically biosynthesize rhamnolipids using glycerol. The anaerobic biosynthetic pathway of rhamnolipid using glycerol was confirmed. Improving the anaerobic production of rhamnolipids better enhanced oil recovery. This study fills the gaps in the anaerobic biosynthesis mechanism of rhamnolipids.
Acknowledgements
This work was financially mainly supported by the National Natural Science Foundation of China (31700117). We thank Dr. Qingfeng Cui for his kind help in core flooding tests.
Authors contributions
FZ conceived and designed the study, carried out the experiments and drafted the manuscript. QZW assisted in bacterial cultivation and data analysis. YZ assisted in analyzing the RNA-seq data. LYL participated in gene knockout experiments. All authors read and approved the final manuscript.
Funding
This work was financially supported by the National Natural Science Foundation of China (31700117) and the Introduced Talent Research Start-up Fund of Qufu Normal University, and the Young Talents Invitation Program of Shandong Provincial Colleges and Universities.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Correspondence to Feng Zhao.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Brown LR. Microbial enhanced oil recovery (MEOR) Curr opin Microbiol. 2010;13(3):316–20. doi: 10.1016/j.mib.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 2.Youssef NH, Simpson DR, Duncan KE, et al. In situ biosurfactant production by Bacillus strains injected into a limestone petroleum reservoir. Appl Environ Microbiol. 2007;73(4):1239–47. doi: 10.1128/AEM.02264-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Youssef NH, Simpson DR, McInerney MJ, et al. In-situ lipopeptide biosurfactant production by Bacillus strains correlates with improved oil recovery in two oil wells approaching their economic limit of production. Int Biodeterior Biodegrad. 2013;81:127–32. doi: 10.1016/j.ibiod.2012.05.010. [DOI] [Google Scholar]
- 4.Zhao F, Li P, Guo C, Shi R, Zhang Y. Bioaugmentation of oil reservoir indigenous Pseudomonas aeruginosa to enhance oil recovery through in-situ biosurfactant production without air injection. Bioresour Technol. 2018;251:295–302. doi: 10.1016/j.biortech.2017.12.057. [DOI] [PubMed] [Google Scholar]
- 5.Bian XY, Mbadinga SM, Liu YF, Yang SZ, Liu JF, Ye RQ, Gu JD, Mu BZ. Insights into the anaerobic biodegradation pathway of n-alkanes in oil reservoirs by detection of signature metabolites. Sci Rep. 2015;5(1):1–12. doi: 10.1038/srep09801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Domingues PM, Almeida A, Leal LS, et al. Bacterial production of biosurfactants under microaerobic and anaerobic conditions. Rev Environ Sci Bio. 2017;16(2):239–72. doi: 10.1007/s11157-017-9429-y. [DOI] [Google Scholar]
- 7.Zou H, Du W, Ji M, Zhu R. Enhanced electrokinetic remediation of pyrene-contaminated soil through pH control and rhamnolipids addition. Environ Eng Sci. 2016;33(7):507–13. doi: 10.1089/ees.2016.0019. [DOI] [Google Scholar]
- 8.Chong H, Li Q. Microbial production of rhamnolipids: opportunities, challenges and strategies. Microb Cell Fact. 2017;16(1):137. doi: 10.1186/s12934-017-0753-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mller MM, Kgler JH, Henkel M, Gerlitzki M, Hrmann B, Phnlein M, Syldatk C, Hausmann R. Rhamnolipids-next generation surfactants? J Biotechnol. 2012;162:366–380. doi: 10.1016/j.jbiotec.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 10.Arai H. Regulation and function of versatile aerobic and anoxic respiratory metabolism in Pseudomonas aeruginosa. Front Microbiol. 2011;2:1–13. doi: 10.3389/fmicb.2011.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schobert M, Jahn D. Anaerobic physiology of Pseudomonas aeruginosa in the cystic fibrosis lung. Int J Med Microbiol. 2010;300:549–56. doi: 10.1016/j.ijmm.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 12.Chayabutra C, Wu J, Ju LK. Rhamnolipids production by Pseudomonas aeruginosa under denitrification: effects of limiting nutrients and carbon substrates. Biotechnol Bioeng. 2001;72:25–33. doi: 10.1002/1097-0290(20010105)72:1<25::AID-BIT4>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 13.Zhao F, Zhang J, Shi R, Han S, Ma F, Zhang Y. Production of biosurfactant by a Pseudomonas aeruginosa isolate and its applicability to in situ microbial enhanced oil recovery under anoxic conditions. RSC Adv. 2015;5(45):36044–50. doi: 10.1039/C5RA03559G. [DOI] [Google Scholar]
- 14.Zhao F, Shi R, Ma F, Han S, Zhang Y. Oxygen effects on rhamnolipids production by Pseudomonas aeruginosa. Microb Cell Fact. 2018;17(1):39. doi: 10.1186/s12934-018-0888-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao F, Cui Q, Han S, Dong H, Zhang J, Ma F, Zhang Y. Enhanced rhamnolipid production of Pseudomonas aeruginosa SG by increasing copy number of rhlAB genes with modified promoter. RSC Adv. 2015;5(86):70546–52. doi: 10.1039/C5RA13415C. [DOI] [Google Scholar]
- 16.Javaheri M, Jenneman GE, McInerney MJ, Knapp RM. Anaerobic production of a biosurfactants by Bacillus licheniformis JF-2. Appl Environ Microbiol. 1985;50(3):698–700. doi: 10.1128/AEM.50.3.698-700.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Youssef NH, Duncan KE, Nagle DP, Savage KN, Knapp RM, McInerney MJ. Comparison of methods to detect biosurfactant production by diverse microorganisms. J Microbiol Methods. 2004;56:339–47. doi: 10.1016/j.mimet.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 18.Zhao F, Liang X, Ban Y, Han S, Zhang J, Zhang Y, Ma F. Comparison of methods to quantify rhamnolipids and optimization of oil spreading method. Tenside Surfact Det. 2016;53(3):243–8. doi: 10.3139/113.110429. [DOI] [Google Scholar]
- 19.Sobern-Chvez G, Lepine F, Dziel E. Production of rhamnolipids by Pseudomonas aeruginosa. Appl Microbiol Biotechnol. 2005;68:718–25. doi: 10.1007/s00253-005-0150-3. [DOI] [PubMed] [Google Scholar]
- 20.Schfer A, Tauch A, Jger W, Kalinowski J, Thierbach G, Phler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 21.Nagalakshmi U, Waern K, Snyder M. RNA-Seq: a method for comprehensive transcriptome analysis. Curr Prot Mole Biol. 2010;89(1):4–11. doi: 10.1002/0471142727.mb0411s89. [DOI] [PubMed] [Google Scholar]
- 22.Anders S, Pyl PT, Huber W. HTSeq-a python framework to work with high-throughput sequencing data. Bioinformatics. 2014;31(2):166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Young MD, Wakefield MJ, Smyth GK, et al. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11(2):R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanehisa M, Araki M, Goto S, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36(suppl 1):480–4. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hunt JC, Phibbs PV. Regulation of alternate peripheral pathways of glucose catabolism during aerobic and anaerobic growth of Pseudomonas aeruginosa. J Bacteriol. 1983;154(2):793–802. doi: 10.1128/JB.154.2.793-802.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hauser G, Karnovsky ML. Rhamnose and rhamnolipide biosynthesis by Pseudomonas aeruginosa. J Biol Chem. 1957;224(1):91–105. doi: 10.1016/S0021-9258(18)65013-6. [DOI] [PubMed] [Google Scholar]
- 28.Filiatrault MJ, Wagner VE, Bushnell D, Haidaris CG, Iglewski BH, Passador L. Effect of anaerobiosis and nitrate on gene expression in Pseudomonas aeruginosa. Infect Immun. 2005;73(6):3764–72. doi: 10.1128/IAI.73.6.3764-3772.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wagner VE, Bushnell D, Passador L, et al. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J Bacteriol. 2003;185(7):2080–95. doi: 10.1128/JB.185.7.2080-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alvarez-Ortega C, Harwood CS. Responses of Pseudomonas aeruginosa to low oxygen indicate that growth in the cystic fibrosis lung is by aerobic respiration. Mol Microbiol. 2007;65(1):153–65. doi: 10.1111/j.1365-2958.2007.05772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hansen KD, Wu Z, Irizarry RA, Leek JT. Sequencing technology does not eliminate biological variability. Nat Biotechnol. 2011;29:572–3. doi: 10.1038/nbt.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olvera C, Goldberg JB, Snchez R, Sobern-Chvez G. The Pseudomonas aeruginosa algC gene product participates in rhamnolipid biosynthesis. FEMS Microbiol Lett. 1999;179(1):85–90. doi: 10.1111/j.1574-6968.1999.tb08712.x. [DOI] [PubMed] [Google Scholar]
- 33.Rahim R, Burrows LL, Monteiro MA, Perry MB, Lam JS. Involvement of the rml locus in core oligosaccharide and O polysaccharide assembly in Pseudomonas aeruginosa. Microbiology. 2000;146(11):2803–14. doi: 10.1099/00221287-146-11-2803. [DOI] [PubMed] [Google Scholar]
- 34.Reis RS, Pereira AG, Neves BC, Freire DMG. Gene regulation of rhamnolipid production in Pseudomonas aeruginosaa review. Bioresour Technol. 2011;102:6377–84. doi: 10.1016/j.biortech.2011.03.074. [DOI] [PubMed] [Google Scholar]
- 35.Dobler L, Vilela LF, Almeida RV, et al. Rhamnolipids in perspective: gene regulatory pathways, metabolic engineering, production and technological forecasting. New Biotechnol. 2016;33(1):123–35. doi: 10.1016/j.nbt.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 36.Long X, Shen C, He N, et al. Enhanced rhamnolipids production via efficient foam-control using stop valve as a foam breaker. Bioresour Technol. 2017;224:536–43. doi: 10.1016/j.biortech.2016.10.072. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article.