Abstract
Effective therapeutics to combat emerging viral infections are an unmet need. Historically, treatments for chronic viral infections with single drugs have not been successful, as exemplified by human immunodeficiency virus (HIV) and hepatitis C virus (HCV) infections. Combination therapy for these diseases has led to improved clinical outcomes with dramatic reductions in viral load, morbidity, and mortality. Drug combinations can enhance therapeutic efficacy through additive, and ideally synergistic, effects for emerging and re-emerging viruses, such as influenza, severe acute respiratory syndrome-coronavirus (SARS-CoV), Middle East respiratory syndrome (MERS)-CoV, Ebola, Zika, and SARS-coronavirus 2 (CoV-2). Although novel drug development through traditional pipelines remains a priority, in the interim, effective synergistic drug candidates could be rapidly identified by drug-repurposing screens, facilitating accelerated paths to clinical testing and potential emergency use authorizations.
Keywords: Emerging viral diseases, SARS-CoV-2, COVID-19, Drug combination therapy
Introduction
Emerging viral diseases are most often caused by novel viruses of zoonotic nature, by mutated strains of existing viruses, and by re-emergence of previously existing viruses. Viral infections that are epidemic or pandemic in nature can lead to immense health and economic burdens on society. By far the worst pandemic in recent history was caused by the H1N1 influenza strain in 1918, which led to an estimated 50 million deaths worldwide.1 Over the past few decades, several epidemic-causing viruses, such as Ebola, SARS-CoV, MERS-CoV, and Zika virus (ZIKV), have emerged. The SARS outbreak in 2003 led to some 8098 infections worldwide with an ∼ 10% mortality rate.2 MERS, which first appeared in 2012, has a mortality rate of 35%3 with outbreaks still prevalent in some parts of the world. Ebola infections were first reported in 1976 and have since re-emerged more than 20 times.4 Recent outbreaks in Africa were caused by the Zaire ebolavirus species (Ebola virus, EBOV), which has high mortality rates of 70–90% without treatment.5 ZIKV infections were first reported in 1947 as a mildly infectious disease; however, it was not until 2015–2016 that ZIKV was recognized as an epidemic leading to birth defects and severe congenital central nervous system malformations.6 Most recently, SARS-CoV-2, which appeared in December 2019, has led to a large-scale pandemic resulting in over 130 million infections worldwide as of April 14, 2021.7
Vaccines are an important and effective tool to combat infectious viral diseases. To date, vaccines or effective therapeutics for SARS, MERS, ZIKV, or all Ebola species are not available. The vaccine development and distribution processes are usually lengthy, which can be exacerbated further by viral genome mutations leading to antigenic drift. Until the availability of safe and effective vaccines for emerging viral diseases, the pursuit of suitable antiviral therapeutics is vital for treatments of these diseases.
Drug repurposing redirects existing or previously approved drugs as new therapeutics for new clinical indications in a time and cost-effective manner. Drug repurposing screenings can be performed quickly by using approved drug compound libraries.8 Thus, drug repurposing can be useful in expeditiously discovering potential therapeutics for treatments of emerging viral infections because these drugs can be directly applied to clinical trials or potentially be used as emergency therapy. The most common challenge presented by drug-repurposing screens is the failure to find potent compounds that can be applied clinically because many weak hits are usually identified. These weak compounds are clinically not useful because their IC50 or IC90 values are higher than their Cmax values, which is the highest achievable plasma concentration of a drug. Alternatively, the rate of success of drug repurposing can be improved by identifying suitable drug combinations of two or more drugs that have synergistic effects, as individual drug concentrations can be reduced when used in combination.
Treatment of a disease with combinations of two or more drugs is commonly known as combination therapy. Drug combinations can result in different outcomes, including functional antagonism, increased drug toxicity, and synergistic/additive effects.9 Whereas drug combinations have the potential to cause adverse effects because of drug–drug interactions, prudent use can confer several advantages. Combination therapy can facilitate targeting multiple pathways to promote drug synergy, such that the beneficial effects far outweigh the simple additive effects. In addition to treatment efficacy, synergistic drug combinations enable the reduction of individual drug doses, thereby increasing patient tolerability and decreasing drug toxicities. Combination drug therapy has been used to treat heterogeneous and multifaceted diseases that are difficult to treat, such as cancer,10 hypertension, and severe bacterial/fungal infections. Severe bacterial infections, where the causative agents are unknown, always require more than two drugs for treatment to broaden the antibacterial spectrum and reduce the occurrence of drug resistance. However, these drug combinations were not developed preclinically but were determined empirically in the clinic, primarily because of the unreliability of effective in vivo models and differences between human and animal model drug metabolism profiles. For the treatment of hypertension, at least two (sometimes three) drugs in combination are often used, because combination therapy often outperforms monotherapy. To improve patient compliancy, several drugs have been successfully incorporated as polypills, where multiple drugs are combined into a single pill. Combination therapy can also suppress or delay the onset of drug resistance, which is often inevitable in many diseases. Chronic viral diseases caused by HIV and HCV are classic examples in which combination therapy has become the standard-of-care treatment.
Although combination therapy does not always culminate in disease cures, it can significantly improve the quality of life and increase lifespan because of increased therapeutic efficacy and prevention of drug resistance. Historical experience from severe and chronic viral disease treatments suggests that effective treatment for emerging infectious diseases, such as COVID-19, will also likely entail two to three drug combinations. In this review, we discuss the use of combination drug therapy from a historical perspective, as well as future options for emerging infectious viral diseases.
Historical lessons and clinical implications of drug combination therapy in viral infections
HIV
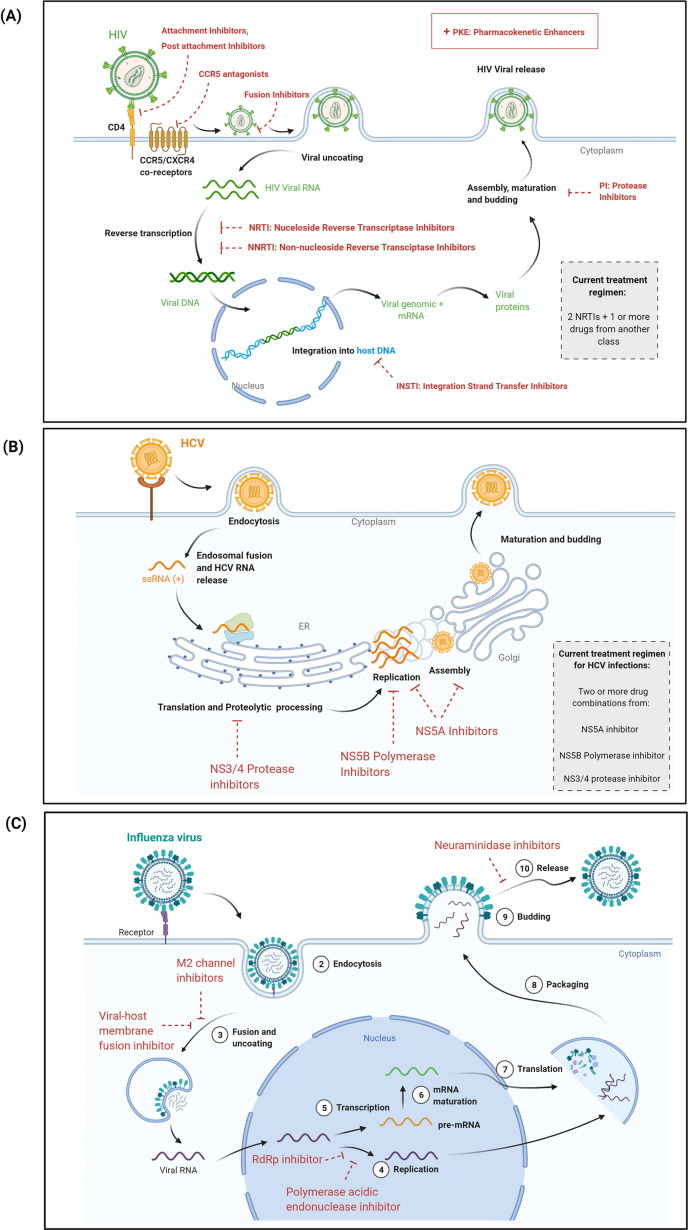
In the absence of treatment intervention, chronic HIV infections lead to Acquired immunodeficiency syndrome (AIDS), a chronic disease in which the immune system is compromised. AIDS cases were first reported in 1981, and HIV was recognized as the causative agent in 1983.11 During the early days of the pandemic, HIV infections were progressive, with extremely poor prognosis and survival rates for patients with advanced-stage HIV infections or AIDS.12, 13 HIV infections are life-long, with no effective cures or approved vaccines. However, tremendous therapeutic advancements have enabled effective suppression of HIV load in patients and a significant reduction in HIV and AIDS-related mortality. The first antiretroviral for HIV treatment, azidothymidine, was approved in 1987. Azidothymidine, a nucleoside reverse transcriptase inhibitor (NRTI), decreased rates of mortality as well as of infections in patients with AIDS.14 However, loss of efficacy to monotherapy arose rapidly with the emergence of drug resistance due to high HIV mutation rates, which frequently occurs in RNA viruses due to viral genome mutations and lack of proofreading.12 Between 1987 and 1995, it was standard practice to treat HIV with mono or dual NRTI therapy; however, this treatment approach was often either less effective or ineffective.12, 15 The standard-of-care treatment for HIV changed dramatically with the introduction of protease inhibitors (PI) to the treatment regimen. Combination antiretroviral therapy (cART) comprising two NRTIs and a PI resulted in significant reductions in HIV infection, progression to AIDS, other secondary infections, and mortality rates, and has been established as standard of treatment for drug-naïve HIV infections.12, 15, 16 Two-drug regimens (for example, Dovato and Juluca) (Table 1 ) can also be used for the initial treatment of HIV infections.17 Based on clinical parameters and contraindications, drug combinations can now be selected from different US Food and Drug Administration (FDA)-approved antivirals with different mechanisms of action (Fig. 1 and Table 1). Single pills comprising combinations of drugs (Table 1) have enabled dramatic improvements in patient compliance and treatment adherence rates, leading to improved clinical outcomes.18, 19 With cART, HIV has become a manageable disease with life-expectancies approaching that of the general population.20
Table 1.
Approved HIV, HCV, and influenza drugs.
| Mode of action | Drug name(s) |
|---|---|
| HIV | |
| NRTIs, nucleoside reverse transcriptase inhibitors | abacavir, emtricitabine, lamivudine, stavudine, tenofovir disoproxil fumarate, zidovudine |
| NNRTIs, non nucleoside reverse transcriptase inhibitors | delavirdine, didanosine, doravirine, efavirenz, etravirine, nevirapine, rilpivirine |
| INSTIs, integration strand transfer inhibitors | bictegravir, cabotegravir, dolutegravir, raltegravir |
| PIs, protease inhibitors | atazanavir, darunavir, fosamprenavir, indinavir, lopinavir (+ritonavir), nelfinavir, saquinavir, tipranavir |
| Fusion inhibitors | enfuvirtide |
| Attachment inhibitors | fostemsavir |
| Postattachment inhibitors | ibalizumab-uiyk |
| CCR5 antagonists | maraviroc |
| PKEs | cobicistat, ritonavir (a PI used as a PKE) |
| HIV drug combinations | |
| 2 NRTI + NNRTI | Atripla: emtricitabine + tenofovir disoproxil fumarate + efavirenz Complera: emtricitabine + tenofovir disoproxil fumarate + rilpivirine Delstrigo: lamivudine + tenofovir disoproxil fumarate + doravirine Odefsey: emtricitabine + tenofovir alafenamide fumarate + rilpivirine Symfi: lamivudine + tenofovir disoproxil fumarate + efavirenz |
| 2 NRTI + INSTI | Biktarvy: emtricitabine + tenofovir alafenamide fumarate + bictegravir Triumeq: abacavir sulfate + lamivudine + dolutegravir sodium |
| 2 NRTI + INSTI + PKE | Genvoya: emtricitabine + tenofovir alafenamide fumarate + elvitegravir + cobicistat Stribild: emtricitabine + tenofovir disoproxil fumarate + elvitegravir + cobicistat |
| NRTI + INSTI | Dovato: lamivudine + dolutegravir |
| INSTI + NNRTI | Juluca: dolutegravir + rilpivirine |
| 2 INSTI + PI + PKE | Symtuza: emtricitabine + tenofovir alafenamide fumarate + darunavir ethanolate + cobicistat |
| HCV | |
| NS5A inhibitor | daclatasvir (discontinued), elbasvir, edipasvir, ombitasvir, pibrentasvir, velpatasvir |
| NS5B polymerase inhibitor | dasabuvir, ribavirin (used in combination only), sofosbuvir |
| NS3/4 PI | boceprevir (discontinued), faldoprevir, glecaprevir, grazoprevir, paritaprevir, ritonavir, simeprevir, telaprevir (discontinued), voxilaprevir |
| HCV drug combinations | |
| NS5A + 2 NS3/4 PIs (+NS5B polymerase inhibitor for Viekira Pak) | Technivie/Viekira Pak (discontinued in the USA): ombitasvir + paritaprevir + ritonavir (+dasabuvir for Viekira Pak) |
| NS5A + NS5B polymerase inhibitors | Harvoni: ledipasvir + sofosbuvir |
| NS3/4 protease + NS5A inhibitors | Maviret: glecaprevir + pibrentasvir |
| NS5B polymerase + NS5A inhibitors | Epclusa: sofosbuvir + velpatasvir |
| NS5A + NS3/4 PIs | Zepatier: elbasvir + grazoprevir |
| NS5B polymerase + NS5A + NS3/4 PIS | Vosevi: sofosbuvir + velpatasvir + voxilaprevir |
| Influenza | |
| Adamantane M2 ion channel inhibitors: inhibit viral uncoating; effective only against influenza A viruses | amantadine (FDA approved in 1966; discontinued 2010) rimantadine (FDA approved in 1993) Not recommended for use in the USA because of antiviral resistancea |
| Viral entry blocker: inhibits viral–host membrane fusion | umifenovir (approved in Russia in 1993 and in China in 2006) |
| Neuraminidase inhibitors: prevent viral release and spread to healthy cells; approved for both influenza A and B viral infections | zanamivir (FDA approved in 1999) laninamivir (approved in Japan in 2010) peramivir (FDA approved in 2014) oseltamivir (FDA approved in 2016) |
| RNA-dependent RNA polymerase inhibitor: inhibits viral synthesis | favipiravir (approved in Japan in 2014 and in China in 2016) |
| Polymerase acidic endonuclease inhibitor: inhibits viral replication | baloxavir marboxil (FDA approved in 2018 for uncomplicated flu) |
Data from CDC.gov; drugs.NCATS.io; FDA.gov; hcvguidelines.org; hivinfo.nih.gov.
Figure 1.
Therapeutic targets for HIV, HCV and influenza. (a) US Food and Drug Administration (FDA)-approved therapeutic interventions for HIV infection. Attachment inhibitors, postattachment inhibitors, CCR5 inhibitors, and fusion inhibitors serve as viral entry inhibitors. Other antivirals include antiretrovirals nucleoside/non-nucleoside reverse transcriptase inhibitors (NRTIs/NNRTIs), integrase strand transfer inhibitors (INSTIs), and protease inhibitors (PIs). Pharmacokinetic enhancers (PKEs) can also be used in combination with other drugs. Combination antiretroviral therapy (cART) generally comprises two NRTI drugs plus one or more drugs from other categories (see also Table 1 in the main text). (b) Therapeutic targets for the treatment of hepatitis C virus (HCV) infections. Treatment of HCV usually comprises combinations of two or more drugs from the following categories: NS3/4 PIs, NS5A inhibitors, and NS5B polymerase inhibitors (see also Table 1 in the main text). Treatment regimens vary depending on different clinical parameters. As an example, initial treatment for a simple, drug-naïve HCV infection can include a combination of sofobuvir + velpatasvir (NS5B polymerase inhibitor + NS5A inhibitor) for 12 weeks or glecaprevir + pibrentasvir (NS3/4 PI + NS5A inhibitor) for 8 weeks.26(c) Current approved drug targets against influenza (A and B) virus. Combination drugs are being tested in clinical trials but have not yet been approved (see also Table 1 in the main text).
HCV
Unlike HIV, HCV replicates in the cytoplasm without viral genome integration into the host cell and thus, HCV infections have the potential to be cured with antiviral drugs.21 Acute HCV infections can be spontaneously cleared by the immune system, which is estimated to occur in ∼20–30% of infected adults.22 The remaining 70–80% of infected individuals develop chronic HCV, which increases the risk for developing cirrhosis, liver cancer, and other liver diseases.22 In addition, because HCV is associated with chronic inflammation, it also increases the risk of developing vascular diseases, insulin resistance/diabetes, and extrahepatic cancers.21
Currently, there are no effective vaccines against HCV; however, antiviral therapeutics have enabled a cure rate of >95% of patients with HCV infections, significantly reducing the risk of death because of HCV-related complications.23 Clinically, the primary outcome for HCV treatment is a sustained virological response (SVR), which is defined as the amount of HCV in the blood lower than detectable limits after 12 weeks or more of therapy completion.24 Of patients who achieve SVR, 99% are cured of HCV, whereas relapse with reappearance of the HCV virus occurs in only < 1% of people in the absence of re-exposure.24
Over the past 20 years, HCV was treated with non-PEGylated and later PEGylated interferon (IFN) either with or without the PI ribavirin (RBV).21 In addition to adverse reactions, treatment with IFN/RBV had poor clinical outcomes, with a cure rate of no more than 50% depending on HCV genotypes (genetic variants of HCV) and other comorbidities.21, 25 The treatment approach changed to triple-combination therapy in 2011 after the approval of NS3/4A PIs, such as telaprevir or boceprevir, which increased the cure rates to 75%.26 With the advent of newer direct-acting antivirals (DAAs), such as sofosbuvir, a uridine prodrug that inhibits HCV NS5B polymerase, and daclatasvir, a NS5A inhibitor, cure rates improved dramatically to 95% without increases in significant adverse effects.21 Current treatment options for HCV with DAAs (Table 1) vary depending on age, HCV genotype, stage of liver disease, or other underlying conditions.27 A simplified initial treatment of drug-naïve HCV may include a combination of sofobuvir + velpatasvir for 12 weeks or glecaprevir + pibrentasvir for 8 weeks27 (Fig. 1 and Table 1). A recent clinical trial showed that using two different three-drug combinations were successful in reducing treatment timeline from 12 to 6 weeks in patients with drug-naïve HCV.28 Thus, combination therapy in HCV has demonstrated the additional benefit of reduced treatment time with good efficacy and minimal adverse effects. In addition, combination therapy can be a valuable tool in combating drug resistance and HCV infections with other comorbidities. For example, in drug-experienced HCV patients who had relapsed after DAA treatment, treatment with new combinations of drugs achieved high success rates.29, 30, 31, 32 Moreover, combinations of drugs for HIV/HCV co-infections tested in clinical trials demonstrated that HIV suppression and high SVR for HCV could be achieved in the absence of major adverse effects.33
Influenza
A century after the 1918 pandemic, influenza infections are still prevalent, at ∼ 1 billion cases worldwide and up to 650 000 deaths yearly.34 Although vaccines against influenza are available, they need to be modified and administered on a yearly basis because of frequent viral mutations. Over the past decade, the effectiveness of influenza vaccines has varied between 20% and 60%, although four strains of the virus are targeted.35
Several antiviral therapeutics for the treatment of influenza infections have been developed (Fig. 1 and Table 1). Influenza viruses mutate at a very high rate, facilitating drug resistance against antivirals and presenting treatment challenges. Antiviral resistance against the adamantanes amantadine and rimantadine was widespread during the 1980s and early 2000s, ultimately leading to global resistance in 45% of all influenza type A cases.36 Although most influenza strains are still susceptible to many neuraminidase inhibitors,34, 37 resistance to the neuraminidase inhibitor oseltamivir was observed in 98–100% of circulating strains during the 2008–2009 influenza season.37 To overcome this challenge, drug combinations can be used, because the development of resistance to multiple drugs simultaneously is less likely.38 A recent study tested the combination of favipiravir + oseltamivir as a treatment for severe influenza in a small cohort of patients.39 This study found that combination therapy might hasten recovery compared with monotherapy with oseltamivir alone. Additionally, several monoclonal antibodies (mAbs) in Phase II trials (Table 2 ) have been found to be safe and can decrease duration of illness in cases of uncomplicated influenza.40, 41, 42, 43 Although treatment with the mAb MEDI8852 in combination with oseltamivir did not show improvement in clinical outcomes compared with monotherapy with each individual drug in uncomplicated influenza,41 it might be beneficial in the treatment of severe illness or future resistant influenza strains. The safety and efficacy of the mAb VIS410 is currently being tested as monotherapy or in combination with oseltamivir in patients severely ill with influenza A (ClinicalTrials.gov ID: NCT03040141).
Table 2.
List of monoclonal antibodies currently in clinical trials for use against viruses.
| Virus | Monoclonal antibody | Clinical status | Refs/Clinicaltrials.gov ID |
|---|---|---|---|
| Influenza | VIS410 | Phase II | 42, 43; NCT02989194, NCT03040141 |
| MEDI8852 | Phase II | 40, 41; NCT02603952 | |
| Ebola | MAb114 | Phase I | NCT03478891 |
| Triple cocktail ZMapp | Phase I/II | NCT02363322 | |
| Triple antibody cocktail atoltivimab+odesivimab+maftivimab | FDA approved in October 2020 for EBOV | FDA.gov | |
| SARS-CoV-2 | Casirivimab+imdevimab | FDA EUA approval in November 2020 | FDA.gov |
| Bamlanivimab | FDA EUA approval in November 2020 | FDA.gov |
Drug combination therapy for emerging viral diseases
For many new and emerging infectious diseases, rapid therapeutic development options are imperative because the traditional development process for novel drugs still requires a decade or more from initial studies to FDA approval. Moreover, even if effective antiviral monotherapies are identified, drug resistance can soon develop. Therefore, as for the viral diseases caused by influenza, HIV, and HCV, successful treatments for severe viral diseases, such as EBOV, ZIKV, and the emergent coronavirus COVID-19, will likely require vaccines as well as drug combination therapies of specific antiviral agents.
The most desired goal for combination therapy is to enhance therapeutic efficacy through synergistic and/or additive responses by targeting multiple viral targets as well as host cell pathways. Drugs that target host cell pathways, such as host cell viral receptors and proteases for viral protein priming, in combination with antiviral agents, can reduce the development of drug resistance because mutations of host cell proteins after viral infections generally do not occur.
Using animal models to identify appropriate drug treatments for viral diseases can be challenging and problematic. Human viral disease symptoms do not always manifest in laboratory animals commonly used as in vivo disease models and drug treatments and/or doses can vary considerably between different models. In addition, the drug metabolism enzymes in animals are very different from those in humans. A large proportion of drugs that test with in vivo efficacy in preclinical models fail to succeed in human clinical trials because of inefficacy and/or unexpected toxicities. For example, nonhuman primates treated with remdesivir for EBOV showed remarkable efficacy with 100% cure rates.44 However, this did not translate to humans, because remdesivir treatment did not improve clinical outcomes in patients but instead increased undesired host toxicities.45 Moreover, the translational value of testing drug–drug interactions in animal models is low, because drug–drug interactions in humans cannot be accurately assessed in animal models.
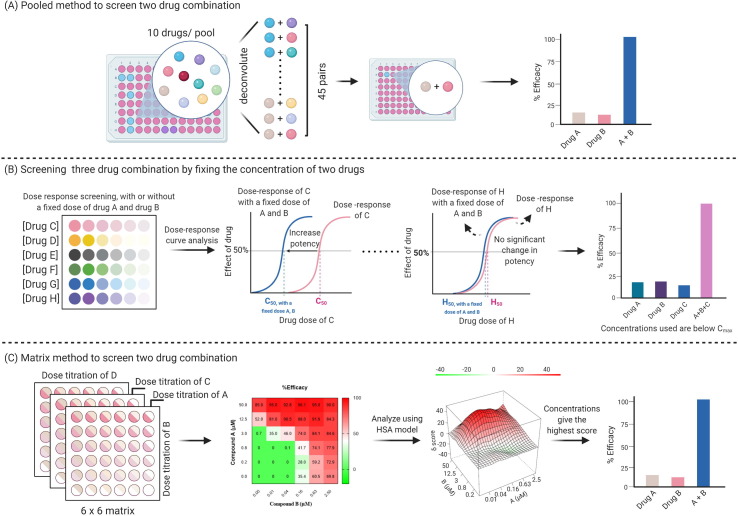
With synergistic drug combinations, lower individual drug doses can be used to treat diseases effectively without increasing adverse effects. Although drug combinations can be tested directly in the clinic because of medical urgency, in vitro screening results can help identify efficacious combinations. Methods are available to determine the pharmacological effects of drug combinations and to quantify synergism in vitro, and are reviewed in detail elsewhere.46 Several approaches have been explored to reduce the testing pairs to a manageable range. These include pooled screens (Fig. 2 a), prioritized compound collections, and predetermined concentrations when dealing with large compound libraries or for a higher order combination, such as three- or four-drug combinations (Fig. 2b,c). As an example, a pooled screening approach was designed to search for effective two-drug combinations for HIV infection from 1000 compounds.47 A pool of ten compounds was examined in a single well where each pool was deconvoluted into 45 two-drug combinations. In the initial screen, 116 pairs reduced viral infection rate by at least 50%.47 Among them, 41 pairs produced synergistic effects according to both Bliss independence48 and Highest Single Agent49 models.
Figure 2.
Schematic workflows of drug combination screening. (a) Pooled method to screen two-drug combinations. Responses of multiple ten-drug pools were examined in the primary test. Each effective pool was deconvoluted and tested as 45 two-drug combinations to reveal effective drug pairs. (b) Screening three-drug combinations by fixing the concentration of two drugs. In this method, the dose response of a third drug was investigated with or without a fixed dose of two drugs, such as drug A and B. The presence of drug A and B might result in the dose–response curve of the third drug shifting to the left, increase potency (as shown for drug C in the figure), or lead to no significant change (as shown for drug H in the figure). Combinations with increased potency can be further tested. (c) Matrix method to screen two-drug combinations. Effects of a given drug pair can be examined in a 6 × 6 matrix format. The matrix comprises a series of dilution concentrations of one drug (example drug A) in each column and a series of dilution concentrations of another drug (example drug B) in each row. Results can then be analyzed based on different models. A Highest Single Agent (HSA) model was used as a representation here. The matrix method can reveal effective drug pairs, as well as the concentration of each drug that gives the greatest synergistic effect.
Matrix drug combination studies are a common method used for identification of drug combination pairs with synergistic effects.9, 50 Incorporated with large-scale automation platforms, multidose response matrices of two-drug combinations can be screened in a high-throughput manner. A recent study examined 73 two-drug combinations using an in vitro SARS-CoV-2 cytopathic assay in a 6 × 6 dose matrix format, which identified 16 synergistic and eight antagonistic combinations based on the Highest Single Agent model.9 Compound selection and compound pair prioritization were facilitated by an in silico approach. This study demonstrates the potential for increasing use of computational methods to curate combination pairs in the future. In general, synergistic and additive combinations can be determined by utilizing different reference models. Each model owns unique assumptions and, therefore, there are no consensus agreements on, or preferences for, any one model because conclusions can vary with different models used and with input parameters.51 The theories and applications of these models are reviewed in detail elsewhere.52, 53, 54, 55, 56
A left shift in the compound concentration–response curve in the presence of a single concentration of a second drug or two other drugs has also been used to identify drug synergy. For example, in a study that examined EBOV entry inhibitors using three-drug combinations, compounds with activity were first categorized based on the information including in vitro and in vivo efficacy, and pharmacokinetic and toxicity profiles.57 A dose–response curve shift by the third drug was used to monitor the synergistic effect of combinations when the concentrations of two drugs were fixed below their Cmax values for clinical relevance. Three sets of the three-drug combinations could inhibit > 90% of EBOV entry. More importantly, the concentrations of each individual drug used in the combinations were below their Cmax values, suggesting good clinical translation potential.
Current treatment options for emerging viral diseases
EBOV, MERS, and ZIKV infections are still endemic in certain parts of the world, but fortunately have not developed into diseases on a pandemic scale. Drug-repurposing screens have helped identify potential treatment options for MERS,58 ZIKV,59, 60 and EBOV57; however, none are currently clinically approved as a therapeutic for those diseases. Well-controlled randomized trials for EBOV have been challenging for feasibility and ethical reasons.61 Moreover, combination drug trials for EBOV treatment have not been performed, because patient recruitment has been limited. Clinical trials conducted on therapeutics for EBOV thus far have been on single drugs, which have largely been unsuccessful, likely because of limited activities and efficacies of single drugs in viral infections, as well as limited plasma/tissue drug concentration availability. In October 2020, the FDA approved an Ebola glycoprotein-directed mAb cocktail for the treatment of EBOV (Table 2). However, these antibody treatments require intravenous administration and are more effective in decreasing mortality rates in patients who are not severely ill.62 Therefore, better therapeutics for the treatment of EBOV disease are still needed.
Since the emergence of COVID-19 in late 2019, several hundred vaccines have progressed to preclinical development stages, with several others in Phase III clinical trials. At the time of writing, three COVID-19 vaccines, including two mRNA vaccines (Moderna’s mRNA-1273 and Pfizer’s BNT162b2)63, 64, 65 and the single-dose Janssen vaccine, have been granted Emergency Use Authorization (EUA) by the FDA and other regulatory agencies. Although vaccines will be key in providing much-needed relief from the COVID-19 pandemic, it is unknown how long the protection from vaccines will last. Moreover, it remains unknown whether SARS-CoV-2 will appear seasonally each year, and whether mutations will render the vaccines less effective. New SARS-CoV-2 variants with mutations in the spike protein have already emerged from the UK, South Africa, and Brazil and are purported to be more transmissible.66 In addition, the effects of selective pressures on SARS-CoV-2 after vaccination have not yet been encountered, which could result in new mutations with resistance to COVID-19 vaccines. Thus, the development of therapeutics against COVID-19 remains urgently needed.
Drug repurposing and repositioning have been extensively used for the treatment of patients with COVID-19. At the time of writing (April 2021), there were 2800 registered clinical trials on ClinicalTrials.gov based on a search of ‘COVID-19′ with ‘drug’, and over 1200 publications for the search term of ‘drug repurposing for COVID-19′ in PubMed. However, most repurposed drugs as single-drug treatments have not shown effective clinical efficacy against COVID-19. Thus far, remdesivir is the only small molecule approved by the FDA and other regulatory agencies for the treatment of patients hospitalized with COVID-19. Although remdesivir effectively blocks SARS-CoV2 infections in vitro,67 it has failed to show clinical efficacy or decrease mortality rates consistently.68, 69 A double-blind, randomized, placebo-controlled trial showed that remdesivir could shorten the recovery time in adults with COVID-19 from 15 to 10 days70 (ClinicalTrials.gov ID: NCT04280705). However, in a study conducted in 11 330 adults in 30 countries, remdesivir showed no improvement in overall mortality, initiation of ventilation, and duration of hospital stay68 (ClinicalTrials.gov ID: NCT04315948). EUA has also been granted to mAbs for the treatment of mild–moderate COVID-19 (Table 2). Preliminary reports showed that convalescent plasma has benefits in cases of severe COVID-19 illness but not in patients critically ill with the virus.71
The success rate of many currently used treatments depends on disease severity. In most cases, COVID-19 leads to mild or asymptomatic disease.72 In severe cases, the host immune response contributes to COVID-19 disease pathophysiology by precipitating a cytokine storm, which can lead to acute respiratory distress, hypoxemia, and organ failure.73 Thus, in addition to antivirals, immune modulators can also help manage adverse COVID-19 disease outcomes. Glucocorticoids, such as dexamethasone, have been shown to decrease mortality rates in patients with COVID-19 who received invasive mechanical ventilation/supplemental oxygen for treatment72 or in patients with acute respiratory distress.74 A clinical trial that tested the efficacy and safety of triple-combined therapy with IFNβ1b + lopinavir-ritonavir + RBV in a small cohort of patients showed better patient outcomes than treatment with lopinavir-ritonavir alone.75 Another study found that, in patients hospitalized with COVID-19, a combination of remdesivir and baricitinib compared with remdesivir alone not only reduced recovery time, but also led to fewer adverse events.76 We foresee that more clinical results of drug combination therapy for COVID-19 will be reported.
Concluding remarks and perspectives
To date, effective therapeutics are still an unmet medical need for the treatment of SARS, MERS, ZIKV, EBOV, and COVID-19. Treatments of these emerging infectious viral diseases might benefit from combination therapy, as has been achieved for chronic viral diseases, such as HIV and HCV. Similarly, multiple drugs in the same mechanistic category together with those against different targets will likely be needed for effective treatment of COVID-19. Although studies have found synergistic drug combinations to be effective against EBOV, MERS, ZIKV, and SARS-CoV-2 viral infections in vitro,9, 57, 59, 77, 78 to our knowledge, these combinations have not yet been clinically tested. As an example, in the screening study by Bobrowski et al., significant synergistic combinations against SARS-CoV-2 were found with the antivirals remdesivir, nitazoxanide, and umifenovir.9 Of these, and as noted earlier, remdesivir has been the only drug thus far to receive FDA approval for the treatment of COVID-19. However, the WHO ultimately recommended against the use of remdesivir in treating COVID-19, because it failed to support positive patient-important outcomes.68 Remdesivir is approved for patients hospitalized with COVID-19 and requires intravenous administration, which severely limits its use. Additionally, the efficacy of remdesivir is not high, potentially because of metabolic differences in different tissues.79 Remdesivir was originally developed against the RNA-dependent RNA polymerase (RdRp) of EBOV. Thus, SARS-CoV-2 specific RdRp inhibitors need to be designed and developed that, importantly, can be taken orally to treat earlier-stage disease in an outpatient setting. In this latter regard, treatment with the oral RdRp inhibitor molnupiravir, first developed for influenza, has been shown to reduce SARS-CoV-2 infection in animal models and is currently in Phase II/III clinical trials for patients both hospitalized and nonhospitalized with COVID-19.80, 81 Treatment with combinations of mAbs with small-molecule antiviral drugs has been pursued for cancer therapy.82 Such combinations might also improve efficacy for COVID-19 and could be directly applied to patients with COVID-19 in the clinic if both the mAbs and small-molecule antivirals are approved.
At present, COVID-19 continues to present as a major global crisis, with infections and deaths steadily on the rise. Although three COVID-19 vaccines have been authorized for use, with more likely on the way, vaccinations might not be feasible in some individuals because of underlying conditions or personal concerns and might not be available to certain underserved populations. In addition, vaccine efficacy might diminish over time because of the emergence of viral mutations. The infectivity of SARS-CoV-2 remains much higher than that of SARS-CoV, and asymptomatic and presymptomatic transmissions continue to contribute to widespread SARS-CoV-2 infections.83 Thus, COVID-19 appears to be unlikely to spontaneously disappear in the near future. If COVID-19 were to appear seasonally, the disease might need to be managed in the same way as influenza infections, where drugs are given to patients with severe disease to decrease complications and prevent deaths. The treatment goal for COVID-19 is to minimize hospital visits and rates of admission, reduce the average number of days of infection/disease course, as well as significantly decrease mortality rates. Therefore, effective and multiple therapeutics options are necessary.
The discovery of new drug combinations and improvement of therapeutic efficiency of existing drugs through drug repurposing can be greatly enhanced through the aid of artificial intelligence, which includes computational and mathematical modeling. In silico models, such as quantitative structure–activity relationship (QSAR) studies, can predict the biological activities of molecules on a given target based on their chemical structures as well as predict drug synergy.84 Such strategies have been used for drug discovery against SARS-CoV-2.85, 86 and are reviewed in.87, 88 Furthermore, artificial intelligence could be used in concert with personalized genomic analysis as well as personalized drug screening89 to develop specific drug combinations for the treatment of emerging infectious diseases to meet the therapeutic needs of different patient populations.
In addition to artificial intelligence, ‘humanized’ animal models will add to our knowledge armament against SARS-CoV-2. For example, the symptoms of COVID-19 were successfully recapitulated in mice expressing the human angiotensin converting enzyme 2 (hACE2) transgene,90, 91, 92 which is required for SARS-CoV-2 entry into host cells. By expressing hACE2 and human immune cells in immunodeficient mice, antivirals, immune modulators, and their combinations can be better tested for therapeutic efficacy.93
Although drug development is a time-intensive process with associated high costs, efforts to develop different classes of anti-SARS-CoV-2 drug are also warranted. Mirroring the drugs developed for HIV infections, several targets of SARS-CoV-2 have been considered for drug development, including (i) viral entry inhibition including spike protein-ACE2 binding; (ii) main protease (also called 3CL protease); (iii) papain-like protease (PLpro); (iv) RNA-dependent RNA polymerase (RdRp); and (5) replicase.94 More effective drug combinations for improving COVID-19 clinical outcomes might be discovered once these specific SARS-CoV-2 drugs become available in the future.
Declarations of interest
The authors declare that they have no competing interests.
Acknowledgments
Figures were created in BioRender.com. This work was supported by the Intramural Research Programs of the National Center for Advancing Translational Sciences, National Institutes of Health.
References
- 1.1918 Pandemic. Centers for Disease Control and Prevention Website. www.cdc.gov/flu/pandemic-resources/1918-pandemic-h1n1.html. Updated March 20 2019 [Accessed May 14, 2021]
- 2.Severe Acute Respiratory Syndrome. Centers for Disease Control and Prevention Website. www.cdc.gov/sars/about/fs-sars.html/. Updated December 6, 2017. [Accessed May 14, 2021]
- 3.Middle East Respiratory Syndrome Virus. WHO Website. www.who.int/news-room/fact-sheets/detail/middle-east-respiratory-syndrome-coronavirus-(mers-cov). Updated March 11, 2019. [Accessed May 14, 2021]
- 4.Ebola Virus Disease. WHO Website. www.who.int/en/news-room/fact-sheets/detail/ebola-virus-disease. Updated February 23, 2021. [Accessed May 14, 2021]
- 5.Ebola Virus Disease. Centers for Disease Control and Prevention Website. www.cdc.gov/vhf/ebola/clinicians/vaccine/index.html. Updated February 25, 2021. [Accessed May 14, 2021]
- 6.Gubler D.J., Vasilakis N., Musso D. History and emergence of Zika virus. J Infect Dis. 2017;216(suppl_10):S860–S867. doi: 10.1093/infdis/jix451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coronavirus Resource Center. Johns Hopkins University Website. https://coronavirus.jhu.edu/map.html. Updated April 14, 2021. [Accessed May 14, 2021]
- 8.Skuta C., Popr M., Muller T., Jindrich J., Kahle M., Sedlak D., et al. Probes & Drugs portal: an interactive, open data resource for chemical biology. Nat Methods. 2017;14:759–760. doi: 10.1038/nmeth.4365. [DOI] [PubMed] [Google Scholar]
- 9.Bobrowski T., Chen L., Eastman R.T., Itkin Z., Shinn P., Chen C.Z., et al. Synergistic and antagonistic drug combinations against SARS-CoV-2. Mol Ther. 2021;29:873–885. doi: 10.1016/j.ymthe.2020.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cancer Trends Progress Report. National Cancer Institute. https://progressreport.cancer.gov/ [Accessed May 14, 2021]
- 11.Gallo R.C., Montagnier L. The discovery of HIV as the cause of AIDS. N Engl J Med. 2003;349:2283–2285. doi: 10.1056/NEJMp038194. [DOI] [PubMed] [Google Scholar]
- 12.Peters B.S., Conway K. Therapy for HIV: past, present, and future. Adv Dent Res. 2011;23:23–27. doi: 10.1177/0022034511399082. [DOI] [PubMed] [Google Scholar]
- 13.Bloch M., John M., Smith D., Rasmussen T.A., Wright E. Managing HIV-associated inflammation and ageing in the era of modern ART. HIV Med. 2020;21(Suppl. 3):2–16. doi: 10.1111/hiv.12952. [DOI] [PubMed] [Google Scholar]
- 14.Fischl M.A., Richman D.D., Grieco M.H., Gottlieb M.S., Volberding P.A., Laskin O.L., et al. The efficacy of azidothymidine (AZT) in the treatment of patients with AIDS and AIDS-related complex. A double-blind, placebo-controlled trial. N Engl J Med. 1987;317:185–191. doi: 10.1056/NEJM198707233170401. [DOI] [PubMed] [Google Scholar]
- 15.Akanbi M.O., Scarsi K.K., Taiwo B., Murphy R.L. Combination nucleoside/nucleotide reverse transcriptase inhibitors for treatment of HIV infection. Expert Opin Pharmacother. 2012;13:65–79. doi: 10.1517/14656566.2012.642865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cihlar T., Fordyce M. Current status and prospects of HIV treatment. Curr Opin Virol. 2016;18:50–56. doi: 10.1016/j.coviro.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 17.Guidelines for the Use of Antiretroviral Agents in Adults and Adolescents Living with HIV Website. https://clinicalinfo.hiv.gov/en/guidelines/adult-and-adolescent-arv/what-start-initial-combination-regimens-antiretroviral-naive. Updated December 18, 2019. [Accessed May 14, 2021]
- 18.Scott Sutton S., Magagnoli J., Hardin J.W. Impact of pill burden on adherence, risk of hospitalization, and viral suppression in patients with HIV Infection and AIDS receiving antiretroviral therapy. Pharmacotherapy. 2016;36:385–401. doi: 10.1002/phar.1728. [DOI] [PubMed] [Google Scholar]
- 19.Cohen J., Beaubrun A., Bashyal R., Huang A., Li J., Baser O. Real-world adherence and persistence for newly-prescribed HIV treatment: single versus multiple tablet regimen comparison among US Medicaid beneficiaries. AIDS Res Ther. 2020;17:12. doi: 10.1186/s12981-020-00268-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Samji H., Cescon A., Hogg R.S., Modur S.P., Althoff K.N., Buchacz K., et al. Closing the gap: increases in life expectancy among treated HIV-positive individuals in the United States and Canada. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0081355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parlati L., Hollande C., Pol S. Treatment of hepatitis C virus infection. Clin Res Hepatol Gastroenterol. Published online November 30. 2020 doi: 10.1016/j.clinre.2020.11.008. [DOI] [PubMed] [Google Scholar]
- 22.Stuart J.D., Salinas E., Grakoui A. Immune system control of hepatitis C virus infection. Curr Opin Virol. 2020;46:36–44. doi: 10.1016/j.coviro.2020.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hepatitis C. WHO Website. www.who.int/news-room/fact-sheets/detail/hepatitis-c. Updated July 27, 2020. [Accessed May 14, 2021]
- 24.Viral Hepatitis and Liver Disease. US Department of Veterans Affairs Website. www.hepatitis.va.gov. Updated January 7, 2021. [Accessed May 14, 2021]
- 25.Kish T., Aziz A., Sorio M. Hepatitis C in a new era: a review of current therapies. P T. 2017;42:316–329. [PMC free article] [PubMed] [Google Scholar]
- 26.Pawlotsky J.M. Hepatitis C virus: standard-of-care treatment. Adv Pharmacol. 2013;67:169–215. doi: 10.1016/B978-0-12-405880-4.00005-6. [DOI] [PubMed] [Google Scholar]
- 27.Recommendations for testing managing and treating hepatitis C. AASLD/IDSA HCV guidance panel website. http://hcvguidelines.org. Updated August 27, 2020. [Accessed May 14, 2021]
- 28.Kohli A., Osinusi A., Sims Z., Nelson A., Meissner E.G., Barrett L.L., et al. Virological response after 6 week triple-drug regimens for hepatitis C: a proof-of-concept phase 2A cohort study. Lancet. 2015;385:1107–1113. doi: 10.1016/S0140-6736(14)61228-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bourlière M., Gordon S.C., Flamm S.L., Cooper C.L., Ramji A., Tong M., et al. Sofosbuvir, velpatasvir, and voxilaprevir for previously treated HCV infection. N Engl J Med. 2017;376:2134–2146. doi: 10.1056/NEJMoa1613512. [DOI] [PubMed] [Google Scholar]
- 30.Sarrazin C., Cooper C.L., Manns M.P., Reddy K.R., Kowdley K.V., Roberts S.K., et al. No impact of resistance-associated substitutions on the efficacy of sofosbuvir, velpatasvir, and voxilaprevir for 12 weeks in HCV DAA-experienced patients. J Hepatol. 2018;69:1221–1230. doi: 10.1016/j.jhep.2018.07.023. [DOI] [PubMed] [Google Scholar]
- 31.Belperio P.S., Shahoumian T.A., Loomis T.P., Backus L.I. Real-world effectiveness of sofosbuvir/velpatasvir/voxilaprevir in 573 direct-acting antiviral experienced hepatitis C patients. J Viral Hepat. 2019;26:980–990. doi: 10.1111/jvh.13115. [DOI] [PubMed] [Google Scholar]
- 32.Said E.M., Abdulaziz B.A., El Kassas M., El Attar I.H., Emadeldeen M., Abd-Elsalam S.M. High success rates for the use of sofosbuvir/ombitasvir/paritaprevir/ritonavir + ribavirin and sofosbuvir/simeprevir/daclatasvir + ribavirin in retreatment of chronic hepatitis C infection after unsuccessful sofosbuvir/daclatasvir therapy: a real-life experience. Arch Virol. 2020;165:1633–1639. doi: 10.1007/s00705-020-04639-x. [DOI] [PubMed] [Google Scholar]
- 33.Huhn G.D., Ramgopal M., Jain M.K., Hinestrosa F., Asmuth D.M., Slim J., et al. HIV/HCV therapy with ledipasvir/sofosbuvir after randomized switch to emtricitabine-tenofovir alafenamide-based single-tablet regimens. PLoS ONE. 2020;15 doi: 10.1371/journal.pone.0224875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hussain M., Galvin H.D., Haw T.Y., Nutsford A.N., Husain M. Drug resistance in influenza A virus: the epidemiology and management. Infect Drug Resist. 2017;10:121–134. doi: 10.2147/IDR.S105473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Influenza. Centers for Disease Control and Prevention website. www.cdc.gov/flu/vaccines-work/effectiveness-studies.htm. Updated December 11, 2020. [Accessed May 14, 2021]
- 36.Nguyen J.T., Hoopes J.D., Le M.H., Smee D.F., Patick A.K., Faix D.J., et al. Triple combination of amantadine, ribavirin, and oseltamivir is highly active and synergistic against drug resistant influenza virus strains in vitro. PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0009332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Melville K., Rodriguez T., Dobrovolny H.M. Investigating different mechanisms of action in combination therapy for influenza. Front Pharmacol. 2018;9:1207. doi: 10.3389/fphar.2018.01207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perelson A.S., Rong L., Hayden F.G. Combination antiviral therapy for influenza: predictions from modeling of human infections. J Infect Dis. 2012;205:1642–1645. doi: 10.1093/infdis/jis265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y., Fan G., Salam A., et al. Comparative effectiveness of combined favipiravir and oseltamivir therapy versus oseltamivir monotherapy in critically ill patients with influenza virus infection. J Infect Dis. 2020;221:1688–1698. doi: 10.1093/infdis/jiz656. [DOI] [PubMed] [Google Scholar]
- 40.Mallory R.M., Ali S.O., Takas T., Kankam M., Dubovsky F., Tseng L. A phase 1 study to evaluate the safety and pharmacokinetics of MEDI8852, an anti-influenza A monoclonal antibody, in healthy adult volunteers. Biologicals. 2017;50:81–86. doi: 10.1016/j.biologicals.2017.08.007. [DOI] [PubMed] [Google Scholar]
- 41.Ali S.O., Takas T., Nyborg A., Shoemaker K., Kallewaard N.L., Chiong R., et al. Evaluation of MEDI8852, an anti-influenza A monoclonal antibody, in treating acute uncomplicated influenza. Antimicrob Agents Chemother. 2018;62:e00694–18. doi: 10.1128/AAC.00694-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hershberger E., Sloan S., Narayan K., Hay C.A., Smith P., Engler F., et al. Safety and efficacy of monoclonal antibody VIS410 in adults with uncomplicated influenza A infection: results from a randomized, double-blind, phase-2, placebo-controlled study. EBioMedicine. 2019;40:574–582. doi: 10.1016/j.ebiom.2018.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sloan S.E., Szretter K.J., Sundaresh B., Narayan K.M., Smith P.F., Skurnik D., et al. Clinical and virological responses to a broad-spectrum human monoclonal antibody in an influenza virus challenge study. Antiviral Res. 2020;184 doi: 10.1016/j.antiviral.2020.104763. [DOI] [PubMed] [Google Scholar]
- 44.Warren T.K., Jordan R., Lo M.K., Ray A.S., Mackman R.L., Soloveva V., et al. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mulangu S., Dodd L.E., Davey R.T., Jr, Tshiani Mbaya O., Proschan M., Mukadi D., et al. A randomized, controlled trial of Ebola virus disease therapeutics. N Engl J Med. 2019;381:2293–2303. doi: 10.1056/NEJMoa1910993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shyr ZA, Cheng Y, Zheng W. Drug combinations. In: Kenakin T, ed. Comprehensive Pharmacology. Oxford: Elsevier (in press).
- 47.Tan X., Hu L., Luquette L.J., 3rd, et al. Systematic identification of synergistic drug pairs targeting HIV. Nat Biotechnol. 2012;30:1125–1130. doi: 10.1038/nbt.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bliss C.I. The toxicity of poisons applied jointly 1. Ann Appl Biol. 1939;26(3):585–615. [Google Scholar]
- 49.Berenbaum M.C. What is synergy? Pharmacol Rev. 1989;41:93–141. [PubMed] [Google Scholar]
- 50.Heske C.M., Davis M.I., Baumgart J.T., Wilson K., Gormally M.V., Chen L., et al. Matrix screen identifies synergistic combination of PARP inhibitors and nicotinamide phosphoribosyltransferase (NAMPT) inhibitors in Ewing sarcoma. Clin Cancer Res. 2017;23:7301–7311. doi: 10.1158/1078-0432.CCR-17-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Greco W., Unkelbach H.-D., Pöch G.S.J., Kundi M.W.B. Consensus on concepts and terminology for combined action assessment: the Saariselkä agreement. Arch Complex Environ Stud. 1992;4:65–69. [Google Scholar]
- 52.Tang J., Wennerberg K., Aittokallio T. What is synergy? The Saariselka agreement revisited. Front Pharmacol. 2015;6:181. doi: 10.3389/fphar.2015.00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Foucquier J., Guedj M. Analysis of drug combinations: current methodological landscape. Pharmacol Res Perspect. 2015;3 doi: 10.1002/prp2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Geary N. Understanding synergy. Am J Physiol Endocrinol Metab. 2013;304:E237–E253. doi: 10.1152/ajpendo.00308.2012. [DOI] [PubMed] [Google Scholar]
- 55.Meyer C.T., Wooten D.J., Lopez C.F., Quaranta V. Charting the fragmented landscape of drug synergy. Trends Pharmacol Sci. 2020;41:266–280. doi: 10.1016/j.tips.2020.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vlot A.H.C., Aniceto N., Menden M.P., Ulrich-Merzenich G., Bender A. Applying synergy metrics to combination screening data: agreements, disagreements and pitfalls. Drug Discov Today. 2019;24:2286–2298. doi: 10.1016/j.drudis.2019.09.002. [DOI] [PubMed] [Google Scholar]
- 57.Sun W., He S., Martínez-Romero C., Kouznetsova J., Tawa G., Xu M., et al. Synergistic drug combination effectively blocks Ebola virus infection. Antiviral Res. 2017;137:165–172. doi: 10.1016/j.antiviral.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McFee R.B. Middle East Respiratory Syndrome (MERS) Coronavirus. Dis Mon. 2020;66 doi: 10.1016/j.disamonth.2020.101053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu M., Lee E.M., Wen Z., Cheng Y., Huang W.K., Qian X., et al. Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat Med. 2016;22:1101–1107. doi: 10.1038/nm.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang S., Xu M., Lee E.M., Gorshkov K., Shiryaev S.A., He S., et al. Emetine inhibits Zika and Ebola virus infections through two molecular mechanisms: inhibiting viral replication and decreasing viral entry. Cell Discov. 2018;4:31. doi: 10.1038/s41421-018-0034-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sissoko D., Laouenan C., Folkesson E., M'Lebing A.B., Beavogui A.H., Baize S., et al. Experimental treatment with favipiravir for Ebola virus disease (the JIKI Trial): a historically controlled, single-arm proof-of-concept trial in Guinea. PLoS Med. 2016;13 doi: 10.1371/journal.pmed.1001967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Levine M.M. Monoclonal antibody therapy for Ebola virus disease. N Engl J Med. 2019;381:2365–2366. doi: 10.1056/NEJMe1915350. [DOI] [PubMed] [Google Scholar]
- 63.Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., et al. An mRNA vaccine against SARS-CoV-2 - preliminary report. N Engl J Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Anderson E.J., Rouphael N.G., Widge A.T., Jackson L.A., Roberts P.C., Makhene M., et al. Safety and immunogenicity of SARS-CoV-2 mRNA-1273 vaccine in older adults. N Engl J Med. 2020;383:2427–2438. doi: 10.1056/NEJMoa2028436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Emerging SARS-CoV-2 Variants. Centers for Disease Control and Prevention Website. https://www.cdc.gov/coronavirus/2019-ncov/more/science-and-research/scientific-brief-emerging–variants.html. Updated January 28, 2021. [Accessed May 14, 2021]
- 67.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.WHO Solidarity Trial Consortium, Pan H., Peto R., Henao-Restrepo A.M., Preziosi M.P., Sathiyamoorthy V., et al. Repurposed antiviral drugs for Covid-19 - interim WHO Solidarity Trial results. N Engl J Med. 2021;384:497–511. doi: 10.1056/NEJMoa2023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Y., Zhou F., Zhang D., Zhao J., Du R., Hu Y., et al. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395:1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., et al. Remdesivir for the treatment of Covid-19 - final report. N Engl J Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wooding D.J., Bach H. Treatment of COVID-19 with convalescent plasma: lessons from past coronavirus outbreaks. Clin Microbiol Infect. 2020;26:1436–1446. doi: 10.1016/j.cmi.2020.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.RECOVERY Collaborative Group Dexamethasone in hospitalized patients with Covid-19 – preliminary report. N Engl J Med. 2020;58:133. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fajgenbaum D.C., June C.H. Cytokine storm. N Engl J Med. 2020;383:2255–2273. doi: 10.1056/NEJMra2026131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tomazini B.M., Maia I.S., Cavalcanti A.B., Berwanger O., Rosa R.G., Veiga V.C., et al. Effect of dexamethasone on days alive and ventilator-free in patients with moderate or severe acute respiratory distress syndrome and COVID-19: the CoDEX randomized clinical trial. JAMA. 2020;324:1307–1316. doi: 10.1001/jama.2020.17021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hung I.F., Lung K.C., Tso E.Y., Liu R., Chung T.W., Chu M.Y., et al. Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet. 2020;395:1695–1704. doi: 10.1016/S0140-6736(20)31042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kalil A.C., Patterson T.F., Mehta A.K., Tomashek K.M., Wolfe C.R., Ghazaryan V., et al. Baricitinib plus remdesivir for hospitalized adults with Covid-19. N Engl J Med. 2021;384:795–807. doi: 10.1056/NEJMoa2031994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dyall J., Nelson E.A., DeWald L.E., Guha R., Hart B.J., Zhou H., et al. Identification of combinations of approved drugs with synergistic activity against Ebola virus in cell cultures. J Infect Dis. 2018;218(suppl_5):S672–S678. doi: 10.1093/infdis/jiy304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shin J.S., Jung E., Kim M., Baric R.S., Go Y.Y. Saracatinib inhibits Middle East respiratory syndrome-coronavirus replication in vitro. Viruses. 2018;10:283. doi: 10.3390/v10060283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yan V.C., Muller F.L. Advantages of the parent nucleoside GS-441524 over remdesivir for Covid-19 treatment. ACS Med Chem Lett. 2020;11:1361–1366. doi: 10.1021/acsmedchemlett.0c00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wahl A., Gralinski L.E., Johnson C.E., Yao W., Kovarova M., Dinnon K.H., 3rd, et al. SARS-CoV-2 infection is effectively treated and prevented by EIDD-2801. Nature. 2021;591:451–457. doi: 10.1038/s41586-021-03312-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cox R.M., Wolf J.D., Plemper R.K. Therapeutically administered ribonucleoside analogue MK-4482/EIDD-2801 blocks SARS-CoV-2 transmission in ferrets. Nat Microbiol. 2021;6:11–18. doi: 10.1038/s41564-020-00835-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.van der Zanden S.Y., Luimstra J.J., Neefjes J., Borst J., Ovaa H. Opportunities for small molecules in cancer immunotherapy. Trends Immunol. 2020;41:493–511. doi: 10.1016/j.it.2020.04.004. [DOI] [PubMed] [Google Scholar]
- 83.COVID-19. Centers for Disease Control and Prevention Website. https://www.cdc.gov/coronavirus/2019-ncov/hcp/clinical-guidance-management-patients.html. Updated February 16, 2021. [Accessed May 14, 2021]
- 84.Sidorov P., Naulaerts S., Ariey-Bonnet J., Pasquier E., Ballester P.J. Predicting synergism of cancer drug combinations using NCI-ALMANAC data. Front Chem. 2019;7:509. doi: 10.3389/fchem.2019.00509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ton A.T., Gentile F., Hsing M., Ban F., Cherkasov A. Rapid identification of potential inhibitors of SARS-CoV–2 main protease by deep docking of 1.3 billion compounds. Mol. Inform. 2020;39 doi: 10.1002/minf.202000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6:14. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mohamed K., Yazdanpanah N., Saghazadeh A., Rezaei N. Computational drug discovery and repurposing for the treatment of COVID-19: a systematic review. Bioorg Chem. 2021;106 doi: 10.1016/j.bioorg.2020.104490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhou Y., Wang F., Tang J., Nussinov R., Cheng F. Artificial intelligence in COVID-19 drug repurposing. Lancet Digit Health. 2020;2:e667–e676. doi: 10.1016/S2589-7500(20)30192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gorshkov K., Chen C.Z., Marshall R.E., Mihatov N., Choi Y., Nguyen D.T., et al. Advancing precision medicine with personalized drug screening. Drug Discov Today. 2019;24:272–278. doi: 10.1016/j.drudis.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bao L., Deng W., Huang B., Gao H., Liu J., Ren L., et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583:830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- 91.Winkler E.S., Bailey A.L., Kafai N.M., Nair S., McCune B.T., Yu J., et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat Immunol. 2020;21:1327–1335. doi: 10.1038/s41590-020-0778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jiang R.D., Liu M.Q., Chen Y., Shan C., Zhou Y.W., Shen X.R., et al. Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell. 2020;182:50–58. doi: 10.1016/j.cell.2020.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Muñoz-Fontela C., Dowling W.E., Funnell S.G.P., Gsell P.S., Riveros-Balta A.X., Albrecht R.A., et al. Animal models for COVID-19. Nature. 2020;586:509–515. doi: 10.1038/s41586-020-2787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shyr Z.A., Gorshkov K., Chen C.Z., Zheng W. Drug discovery strategies for SARS-CoV-2. J Pharmacol Exp Ther. 2020;375:127–138. doi: 10.1124/jpet.120.000123. [DOI] [PMC free article] [PubMed] [Google Scholar]