Abstract
New SARS-CoV-2 variants emerged in the United Kingdom and South Africa in December 2020 in concomitant with the Brazillian variant in February 2021 (B.1.1.248 lineage) and currently sparking worldwide during the last few months. The new strain 501.V2 in South Africa bears three mutations in the spike receptor-binding domain (RBD); K417 N, E484K, and N501Y, while the Brazilian B.1.1.248 lineage has 12 mutations. In the current study, we simulate the complex ACE2-SARS-CoV-2 spike RBD system in which the RBD is in the wild-type and mutated isoforms. Additionally, the cell-surface Glucose Regulated Protein 78 (CS-GRP78) associated with the ACE2-SARS-CoV-2 spike RBD complex (ACE2-S RBD) is modeled at the presence of these mutant variants of the viral spike. The results showed that E484K and N501Y are critical in viral spike recognition through either ACE2 or CS-GRP78. The mutated variants (the UK, South African, and Brazilian) of the spike RBD tightly bind to GRP78 more than in the case of the wild-type RBD. These results point to the potent role of GRP78 with ACE2 in the attachment of the new variants, which could be a key for the design of inhibitors to block SARS-CoV-2 attachment and entry to the host cell.
Keywords: SARS-CoV-2, 501.V2, B.1.1.248 lineage, Structural bioinformatics, GRP78, Spike RBD
Graphical abstract
1. Introduction
New variants of SARS-CoV-2 have been spread worldwide in the last few months, drawing attention and raising doubts [1]. At the same time of vaccine availability by pharmaceutical companies, doubts were raised regarding the efficacy of vaccines against the emerging mutations in the receptor-binding domain (RBD) of the spike protein [1,2]. Some potential mutations found in the new variants of SARS-CoV-2 could be problematic owing to its viral-host cell recognition engagement, for example, the N501Y mutation, which is shared in the three variants, the South African (501.V2), Brazilian (B.1.1.248 lineage) and the UK (VOC-202012/01) [3]. This mutation could affect the host-cell receptor ACE2 (angiotensin-converting enzyme 2) recognition as it lies in its binding surface residue [4].
We previously reported the Glucose Regulated Protein 78 (GRP78), also called Heat Shock Protein A5 (HSPA5) or Bip, as a possible recognition element for SARS-CoV-2 attachment and entry [5,6]. The substrate-binding domain β (SBDβ) of the GRP78 is the docking domain for the C480–C488 region in the SARS-CoV-2 spike RBD. CS-GRP78 is predicted to bind to the spike alongside the putative host-cell receptor, ACE2 [7,8]. The generated hypotheses are approved by a new experimental study where Lee et al. have located GRP78 in association with both ACE2 and the spike proteins upon the infection of cells by SARS-CoV-2. They propose that the substrate binding domain is crucial for the association [9].
Computational methods are used as an integral part of the experiments for accelerating the availability of approved drugs and vaccines against many viral pathogens, including SARS-CoV-2 [[10], [11], [12]]. Molecular docking and dynamics simulations predict the fate of small molecule binding to biomolecules and predict protein behavior in silico at the physiological conditions [[13], [14], [15], [16]]. In this study, protein-protein docking combined with molecular dynamics simulations is used to study the potential mutations reported in the new variants of SARS-CoV-2 spike RBD and its contribution to host-cell recognition binding affinity on both ACE2 and GRP78 receptors.
Despite its incorporation in the ACE2 binding surface, the N501Y mutation showed a remarkable increase in binding of the ACE2-spike RBD complex to the host-cell surface Glucose Regulated Protein 78 (CS-GRP78) [17]. On the other hand, the E484K is located at the spike RBD's binding motif (C480–C488 region) that we previously reported to be recognized by cell-surface GRP78 [5]. In essence, the new mutations are considerable binding targets for CS-GRP78.
2. Materials and methods
2.1. Molecular docking
HADDOCK webserver was used to predict the binding of GRP78 to the spike/ACE2 complex [18]. The PyMOL V2.2.2 software was utilized to prepare the corresponding RBD mutations (K417 N, E484K, and N501Y) found in SARS-CoV-2 variants 501.V2 and B.1.1.248 [19]. We docked GRP78 (PDB ID: 5E84) with both wild-type spike RBD-ACE2 (PDB ID: 6M17) complex (WT ACE2-RBD) and the mutated RBD-ACE2 complex (Mut ACE2-RBD). Unnecessary chains and molecules were removed from the PDB files using PyMOL V2.2.2, while hydrogens were added using Chimera software [20,21]. GRP78 and SARS-CoV-2 Spike RBD's active site residues were set as T428, V429, V432, T434, F451, S452, V457 & I489, and C480–C488, respectively, while other options of HADDOCK were kept default [22,23]. Any attached carbohydrate moieties (NAG) were kept in the structures due to their crucial role in viral spike recognition.
2.2. Molecular dynamics simulation
Molecular Dynamic Simulation (MDS) on the spike RBD WT and mutated (K417 N, E484K, and N501Y) is performed using Nanoscale molecular dynamics software (NAMD) version 2.13 [24]. The necessary files for MDS were generated using the CHARMM-GUI webserver [[25], [26], [27], [28]]. The system's temperature was set to the physiological 310 K, while the salt concentration was set to 154 mM NaCl. Minimization in 20000 steps was performed on the RBDs in the NVT ensemble (constant number of atoms, constant volume, and constant temperature) using a conjugate gradient algorithm. Equilibration is then performed in an NPT ensemble (constant number of atoms, constant pressure, and constant temperature) for 1 ns period. Nose-Hoover Langevin piston controlled the pressure at the atmospheric value (1.01325 bar). In contrast, Langevin dynamics managed the temperature at the physiological 310 K. Finally, a production run for 25 ns was done in the NVT ensemble. The CHARMM36 force field is used, while the TIP3P water model is utilized in the simulation using NAMD 2.13 software [29,30]. Different in-house written scripts and the visualizing molecular dynamics (VMD) software tools are being used in data analysis [31,32].
3. Results and discussion
The values of the HADDOCK scores for the docking of GRP78 to the WT ACE2-RBD and the Mut ACE2-RBD (mutated) are −74.3 ± 0.9 and −91.5 ± 3.5, respectively. This indicates better binding for the GRP78 against the mutated variant of SARS-CoV-2 compared to the wild-type. Table 1 lists the scores and interactions established upon docking GRP78 into the two complexes (WT ACE2-RBD and Mut ACE2-RBD). GRP78 is tightly bound to the mutated RBD with six H-bonds and five hydrophobic contacts instead of two H-bonds and three hydrophobic contacts in the case of WT ACE2-RBD. The essential E484K mutant in the SARS-CoV-2 spike RBD is responsible for three H-bonds to GRP78 residue, T458. The two representative conformations of the 25 ns molecular dynamics simulation trajectories (state 1 and state 2 in Table 1) for the mutated RBD show comparable results regarding the scores and the interacting residues. K484 is responsible for at least two interactions (H-bonds or hydrophobic interactions) in the mutated RBD.
Table 1.
The interaction patterns after docking GRP78 (5E84) into the WT ACE2-RBD and Mut ACE2-RBD complexes (6M17). Bold residues are the interacting residues found in both WT and Mut ACE2-RBD complexes, while the red-colored residues are the crucial mutations in the RBD. State 1 and state 2 are the two representative conformations for the RBD Mut complex after clustering the MDS trajectories.
| H-bonding |
Hydrophobic interaction |
||||||
|---|---|---|---|---|---|---|---|
| complex | HADDOCK score | No. | Amino acids involved from RBD | Amino acids involved from GRP78 | No. | Amino acids involved from RBD | Amino acids involved from GRP78 |
| WT ACE2-RBD-GRP78 | −74.3 ± 0.9 | 2 | N481 and F486 | E427 and G454 | 3 | T478, P479, and V483 | V453 (2) and V457 |
| Mut ACE2-RBD-GRP78 | −91.5 ± 3.5 | 6 | T478, K484(3), and F486(2) | S452, G454(2), and T458(3) | 5 | E471, I472, N481, and V483(2) | T428, V429(2), V453, and I459 |
| Mut ACE2-RBD-GRP78 (state 1) | −93.3 ± 3.0 | 5 | P479, N481, K484(2), and F486 | E427, G430, S452, G454, and T456 | 6 | V483(4), F486, and F490 | I426, T428, V429, F451, T456, and I459 |
| Mut ACE2-RBD-GRP78 (state 2) | −82.9 ± 1.4 | 5 | C480, G482(2), V483, and K484 | V429, I450, and S452(3) | 4 | P479, V483, K484, and F486 | T428, V429, F451, and V490 |
GRP78 does not only interacts with spike RBD, but also it interacts with ACE2 RBD. Three H-bonds (T456 and R488(2)) and two hydrophobic contacts (A486 and P491) are responsible for binding the host-cell chaperone (GRP78) to the mutant spike RBD. Instead, the interactions in the WT isoform of RBD were through three H-bonds (A486, R488, G489) and a salt bridge (R488).
In the mutant RBD isoform, the N501Y may be responsible for better binding of the RBD to the host cell receptors, as we reported previously in the UK variant of SARS-CoV-2 (VOC-202012/01) [17]. Y501 in the mutated RBD variants forms H-bond to K353 of ACE2. The established H-bond may be responsible for a more stable complex in the new variants of SARS-CoV-2 (UK, South African, and Brazilian).
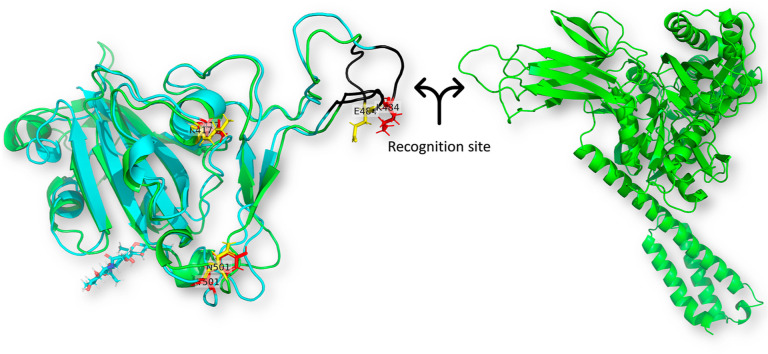
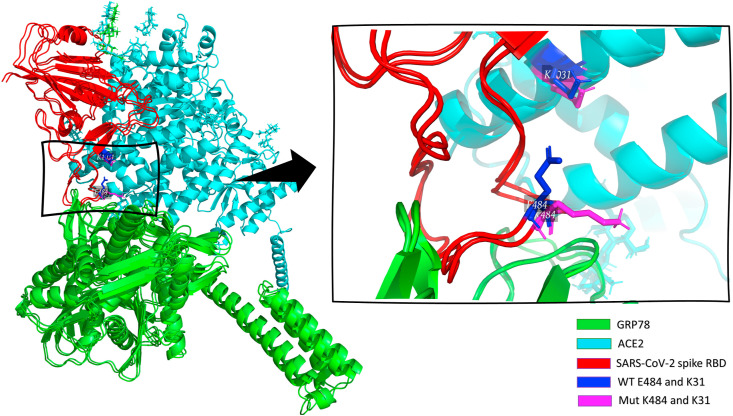
To check the effect of spike RBD mutation E484K alone on the ACE2 binding, we perform another mutated spike RBD isoform with only E484K mutant. Fig. 1 shows the superposition of the WT ACE2-RBD-GRP78 and the Mut ACE2-RBD-GRP, where ACE2, spike RBD, and GRP78 are depicted in cyan, red, and green cartoon, respectively. The enlarged panel shows the orientation of E484 (WT in blue sticks) versus K484 (Mut in magenta sticks) of the spike RBD relative to K31 of the ACE2.
Fig. 1.
The superposition of the docked complexes of GRP78 (green) to ACE2 (cyan)-SARS-CoV-2 Spike RBD (red) complex WT and Mutated (E484K). The enlarged panel on the right side shows the interacting residues in colored sticks in which WT RBD is blue, while the E484K mutant variant in magenta. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Surprisingly, the WT has a salt bridge between E484 (RBD) and K31 (ACE2); this bridge is broken in the E484K isoform. As compensation, more H-bonds are formed in the mutated isoform compared to the WT (19 versus 12 for the Mut and WT, respectively). There is a net 5% increase in the HADDOCK score in the E484K isoform (−120.0 ± 4.1) compared to the WT (−126.1 ± 3.3). This increase in the score is also reported in the UK, N501Y RBD, variant (HADDOCK score −120.8 ± 1.7) and the triple mutant (K417 N, E484K, and N501Y) RBD variant (HADDOCK score −121.0 ± 3.2).
Table 2 shows the docking scores and the interactions established upon docking of the ACE2 into SARS-CoV-2 spike RBD in WT and the 501.V2 variant (K417 N, E484K, and N501Y) without GRP78. We performed this docking experiment to check the ACE2-spike RBD binding in the WT and a mutated variant without the other receptor GRP78 in charge. As shown in the table, the salt bridge formed in the WT ACE2-spike RBD complex is not present in the mutated variant due to the E484K mutation. Additionally, K417 N mutation is responsible for the loss of one H-bond in the Mut ACE2-spike RBD variant. These two broken bonds may be accountable for reduced binding (increased HADDOCK score) in the mutated variant. On the other hand, the N501Y mutation is responsible for forming one H-bond and one hydrophobic contact in the mutated variants of SARS-CoV-2. Hence, it contradicts the reduction in the binding from the other two modifications. The net HADDOCK score increase (decreased binding affinity) in the mutated variant is only 4% from the WT RBD isoform for the ACE2-RBD system.
Table 2.
The interaction patterns after docking ACE2 to WT and Mut spike RBD. Bold residues are the interacting residues found in both WT and Mut ACE2-spike RBD complexes, while the red-colored residues are the crucial mutations in the spike RBD.
| H-bonding |
Hydrophobic interaction |
Salt Bridge |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| complex | HADDOCK score | No. | Amino acids involved from RBD | Amino acids involved from ACE2 | No. | Amino acids involved from RBD | Amino acids involved from ACE2 | No. | Amino acids involved from RBD | Amino acids involved from ACE2 |
| WT ACE2-RBD | −126.1 ± 3.3 | 12 | K417, Y449, Y473, N487, Y489, Q493, S494, T500(3), G502, and V503 | E23, D30, H34(2), D38, Y41, Y83(2), T324, K353, D355, and R357 | 9 | F456(2), Y473, A475, F486(2), Y489, and T500(2) | Q24, T27(3), D30, Y41, M82, Y83, and D355 | 1 | E484 | K31 |
| Mut ACE2-RBD | −121.0 ± 3.2 | 14 | G446, Y449, Y453, Q474, N487(3), Y489(2), Q493(2), Q498, Y501, and G502 | Q24(3), T27 K31(2), H34, D38(2), Q42, Y83(2), and K353(2) | 10 | F456(2), Y473, A475, F486(2), Q493, Q498, Y501, and Y505 | Q24, T27(2), D30, H34, Y41, M82, Y83, and K353(2) | |||
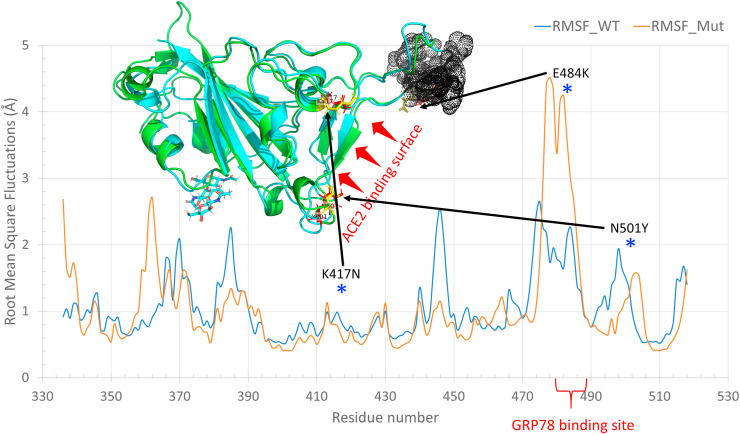
Fig. 2 represents the superposition of the per-residue Root Mean Square Fluctuations (RMSF) in Å (calculated during 25 ns MDS) in the case of WT spike RBD (blue line) and the mutated spike RBD (orange line). The three mutations (K417 N, E484K, and N501Y) are labeled with asterisks on the RMSF graph and colored sticks in the structures. The structures of the spike RBDs are represented in colored cartoons, WT spike RBD is shown in green, while Mut spike RBD is depicted in cyan. The GRP78 binding region (black dots) is the most deviated part for the mutated RBD compared to the WT in the RMSF graph. The RMSF values for this region that bear E484K of the Mut RBD is twice that of WT RBD, which is in good agreement with the docking study. Additionally, the RMSF value for the N501, in the mutated spike RBD, is about 1.5 Å that of Y501 residue in the WT spike RBD.
Fig. 2.
The per -residue RMSF calculated for the 25 ns period MDS on the SARS-CoV-2 spike RBD both WT (blue line) and mutated (orange line). Black dots represent the GRP78 binding region (C480–C488) of the spike RBD. The three mutants are shown in the RMSF graph (blue asterisk) and the structures (yellow and red sticks for WT and Mut, respectively). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
In the mutated isoform of the spike RBD, the residues E484 and less extent N501 show more dynamics than the WT spike RBD residues K484 and Y501. These two mutations lie in the ACE2 binding interface and can affect both ACE2 and GRP78 recognition, which yet to be determined experimentally. This recognition can be prohibited by anti-GRP78 peptides, antibodies, and phytochemicals that we are currently working on [33].
4. Conclusion
The two mutations E484K and N501Y in the spike RBD of SARS-CoV-2 of the new variants (UK, South African, and Brazilian) imbalance the weight in viral spike recognition through either ACE2 or CS-GRP78. The mutated variants exhibit a tighter binding of the spike RBD to CS-GRP78 than ACE2. These findings point to CS-GRP78 as an effective collaborator in the introduction of WT and Mut. SARS-CoV-2, accordingly, consider targeting cell-surface GRP78 in the upcoming vaccine development and drug candidates.
Author contribution
A.E. own the research idea, wrote the manuscript, and designed Fig. I.I. performed the calculations and constructed tables. A.E. revise the manuscript. All authors revise and approve the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Funding
Cairo University funded the research through the COVID-19 fund.
Declaration of competing interest
All the authors declare no competing interest in this work.
Acknowledgment
Prof. Dr. Amy S. Lee is appreciated for her valuable discussions and support. Bibliotheca Alexandrina supercomputing facility, Alexandria, Egypt, is used in the MDS calculations. Cairo University is support the research through the COVID-19 grant.
References
- 1.Wise J. Covid-19: new coronavirus variant is identified in UK. BMJ. 2020;371 doi: 10.1136/bmj.m4857. m4857. [DOI] [PubMed] [Google Scholar]
- 2.Rahimi F., Talebi Bezmin Abadi A. Archives of medical research; 2021. Implications of the Emergence of a New Variant of SARS-CoV-2, VUI-202012/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leung K., Shum M.H., Leung G.M., Lam T.T., Wu J.T. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Euro Surveill. 2021;26 doi: 10.2807/1560-7917.ES.2020.26.1.2002106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Santos J.C., Passos G.A. 2021. The High Infectivity of SARS-CoV-2 B.1.1.7 Is Associated with Increased Interaction Force between Spike-ACE2 Caused by the Viral N501Y Mutation, bioRxiv. 2020.2012.2029.424708. [Google Scholar]
- 5.Ibrahim I.M., Abdelmalek D.H., Elshahat M.E., Elfiky A.A. COVID-19 spike-host cell receptor GRP78 binding site prediction. J. Infect. 2020;80:554–562. doi: 10.1016/j.jinf.2020.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai Y., Zhang J., Xiao T., Peng H., Sterling S.M., Walsh R.M., Jr., Rawson S., Rits-Volloch S., Chen B. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020;369:1586–1592. doi: 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elfiky A.A., Ibrahim I.M., Ismail A.M., Elshemey W.M., A possible role for GRP78 in cross vaccination against COVID-19, J. Infect. 82 (2) 282–327. [DOI] [PMC free article] [PubMed]
- 8.Elfiky A.A. SARS-CoV-2 spike-heat shock protein A5 (GRP78) recognition may be related to the immersed human coronaviruses. Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.577467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlos A.J., Ha D.P., Yeh D.-W., Van Krieken R., Gill P., Machida K., Lee A.S. 2021. GRP78 Binds SARS-CoV-2 Spike Protein and ACE2 and GRP78 Depleting Antibody Blocks Viral Entry and Infection in Vitro, bioRxiv. 2021.2001.2020.427368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahdian S., Ebrahim-Habibi A., Zarrabi M. Drug repurposing using computational methods to identify therapeutic options for COVID-19. J. Diabetes Metab. Disord. 2020:1–9. doi: 10.1007/s40200-020-00546-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elfiky A.A., Azzam E.B., Shafaa M.W. The anti-HCV, Sofosbuvir, versus the anti-EBOV Remdesivir against SARS-CoV-2 RNA dependent RNA polymerase in silico. Mol. Divers. 2021 doi: 10.1007/s11030-020-10178-z. Article in Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ogunyemi O.M., Gyebi G.A., Elfiky A.A., Afolabi S.O., Ogunro O.B., Adegunloye A.P., Ibrahim I.M. Alkaloids and flavonoids from African phytochemicals as potential inhibitors of SARS-Cov-2 RNA-dependent RNA polymerase: an in silico perspective. Antivir. Chem. Chemother. 2020;28 doi: 10.1177/2040206620984076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masoudi-Sobhanzadeh Y. Computational-based drug repurposing methods in COVID-19. Bioimpacts. 2020;10:205–206. doi: 10.34172/bi.2020.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elfiky A.A., Ibrahim I.M., Ismail A.M., Elshemey W.M. A possible role for GRP78 in cross vaccination against COVID-19. J. Infect. 2021;82:282–327. doi: 10.1016/j.jinf.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elfiky A.A., Ibrahim I.M. Zika virus envelope - heat shock protein A5 (GRP78) binding site prediction. J. Biomol. Struct. Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1784794. [DOI] [PubMed] [Google Scholar]
- 16.Basal W., Elfiky A., Eid J. Chaga medicinal mushroom inonotus obliquus (agaricomycetes) terpenoids may interfere with SARS-CoV-2 spike protein recognition of the host cell: a molecular docking study. Int. J. Med. Mushrooms. 2021;23(3):1–10. doi: 10.1615/IntJMedMushrooms.2021037942. [DOI] [PubMed] [Google Scholar]
- 17.Elfiky A.A., Ibrahim I.M. Host-cell recognition through GRP78 is enhanced in the new UK variant of SARS-CoV-2, in silico. J. Infect. 2021;82(5):186–230. doi: 10.1016/j.jinf.2021.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Dijk A.D., Bonvin A.M. Solvated docking: introducing water into the modelling of biomolecular complexes. Bioinformatics. 2006;22:2340–2347. doi: 10.1093/bioinformatics/btl395. [DOI] [PubMed] [Google Scholar]
- 19.V. 2.4.1, the PyMOL Molecular Graphics System, Version 2.4.1 Schrödinger, (LLC).
- 20.DeLano W.L. PyMOL. 2002 [Google Scholar]
- 21.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 22.Yang J., Nune M., Zong Y., Zhou L., Liu Q. Close and allosteric opening of the polypeptide-binding site in a human Hsp70 chaperone BiP. Structure. 2015;23:2191–2203. doi: 10.1016/j.str.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ibrahim I.M., Abdelmalek D.H., Elshahat M.E., Elfiky A.A. COVID-19 spike-host cell receptor GRP78 binding site prediction. J. Infect. 2020;80:554–562. doi: 10.1016/j.jinf.2020.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R.D., Kale L., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brooks B.R., Brooks C.L., 3rd Mackerell A.D., Jr Nilsson L., Petrella R.J., Roux B., Won Y., Archontis G., Bartels C., Boresch S., Caflisch A., Caves L., Cui Q., Dinner A.R., Feig M., Fischer S., Gao J., Hodoscek M., Im W., Kuczera K., Karplus M. CHARMM: the biomolecular simulation program. J. Comput. Chem. 2009;30(10):1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee J., Cheng X., Swails J.M., Yeom M.S., Eastman P.K., Lemkul J.A., Wei S., Buckner J., Jeong J.C., Qi Y., Jo S., Pande V.S., Case D.A., Brooks C.L., MacKerell A.D., Klauda J.B., Im W. CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. J. Chem. Theory Comput. 2016;12:405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jo S., Kim T., Iyer V.G., Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29(11):1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 28.Park S.-J., Lee J., Patel D.S., Ma H., Lee H.S., Jo S., Im W.J.B. Glycan Reader is improved to recognize most sugar types and chemical modifications in the Protein Data. Bank. 2017;33:3051–3057. doi: 10.1093/bioinformatics/btx358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mark P., Nilsson L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. 2001;105:9954–9960. [Google Scholar]
- 30.Huang J., MacKerell A.D., Jr. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 2013;34:2135–2145. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Humphrey W., Dalke A., Schulten K. vol. 14. 1996. VMD: Visual Molecular Dynamics, Journal of Molecular Graphics; pp. 27–38. 33-38. [DOI] [PubMed] [Google Scholar]
- 32.Elfiky A.A., Ismail A.M., Elshemey W.M. Recognition of gluconeogenic enzymes; Icl1, Fbp1, and Mdh2 by Gid4 ligase: a molecular docking study. J. Mol. Recogn. 2020;33 doi: 10.1002/jmr.2831. [DOI] [PubMed] [Google Scholar]
- 33.Elfiky A.A., Baghdady A.M., Ali S.A., Ahmed M.I. GRP78 targeting: hitting two birds with a stone. Life Sci. 2020;260 doi: 10.1016/j.lfs.2020.118317. [DOI] [PMC free article] [PubMed] [Google Scholar]