Abstract
Over the past year, numerous studies in the peer reviewed and preprint literature have reported on the virological, epidemiological and clinical characteristics of the coronavirus, SARS-CoV-2. To date, 25 studies have investigated and identified SARS-CoV-2-derived T cell epitopes in humans. Here, we review these recent studies, how they were performed, and their findings. We review how epitopes identified throughout the SARS-CoV2 proteome reveal significant correlation between number of epitopes defined and size of the antigen provenance. We also report additional analysis of SARS-CoV-2 human CD4 and CD8 T cell epitope data compiled from these studies, identifying 1,400 different reported SARS-CoV-2 epitopes and revealing discrete immunodominant regions of the virus and epitopes that are more prevalently recognized. This remarkable breadth of epitope repertoire has implications for vaccine design, cross-reactivity, and immune escape by SARS-CoV-2 variants.
Keywords: SARS-CoV-2, CD4, CD8, T cells, epitopes
In this review, Grifoni et al. present a comparative and unified discussion of the latest research investigating SARS-CoV-2-derived human CD4 and CD8 T cell epitopes and host immune response. It further provides information that can impact vaccine design, cross-reactivity, and immune escape monitoring for SARS-CoV-2 variants.
Introduction
Over the past year, a considerable amount of information has been produced by the scientific community on the SARS-CoV-2 virus and its associated disease, COVID-19, with studies in the peer-reviewed and pre-print literature investigating its different virological, epidemiological, and clinical characteristics. In particular, numerous studies have analyzed the immune response to the virus, its role in protection and disease, and its importance in the context of vaccine development and evaluation. Several excellent reviews—some in this special issue—cover these topics (DiPiazza et al., 2020; Jordan, 2021; Karlsson et al., 2020; Sette and Crotty, 2020b, 2021; Swadling and Maini, 2020).
Here, we focus on a specific topic: our current knowledge concerning the definition and recognition of SARS-CoV-2-derived T cell epitopes in humans. While the data related to this topic was initially sparse, 25 different studies have now been published as of March 15, 2021 (Chen et al., 2021; Ferretti et al., 2020; Gangaev et al., 2020; Habel et al., 2020; Joag et al., 2021; Kared et al., 2021; Keller et al., 2020; Le Bert et al., 2021, Le Bert et al., 2020; Lee et al., 2020; Mahajan et al., 2020; Mateus et al., 2020; Nelde et al., 2021; Nielsen et al., 2020; Peng et al., 2020; Poran et al., 2020b, 2020a; Prakash et al., 2020; Rha et al., 2021; Sahin et al., 2020; Saini et al., 2020, 2021, Schulien et al., 2021; Sekine et al., 2020; Shomuradova et al., 2020; Snyder et al., 2020; Tarke et al., 2021a), which collectively report data from 1,197 human subjects (870 COVID-19 and 327 unexposed controls), leading to the identification of over 1,400 different CD4 (n = 382) and CD8 (n = 1052) T cell epitopes. These studies are listed in Table 1 , which also captures whether these studies defined class I/CD8 epitopes and/or class II/CD4 epitopes.
Table 1.
Summary of results of epitope identification studies
| Reference | Restriction | Screening strategy | Readout type | Assay Readout | # of epitopes | Antigens screened | # of donors |
# of different restricting HLA molecules |
||
|---|---|---|---|---|---|---|---|---|---|---|
| COVID-19 | unexposed | MHC class I | MHC class II | |||||||
| Chen, J Cell Mol Med, 2021 (Chen et al., 2021) | class I/CD8 | predicted | in vitro expansion (ex vivo verification) | Proliferation, ICS | 1 | S | 3 | 1 | NI | |
| Ferretti, Immunity, 2020 (Ferretti et al., 2020) | class I/CD8 | predicted | ex vivo | ELISA, cytotoxicity, multimer staining | 28 | entire proteome | 78 | 6 | NI | |
| Gangaev, Research Square, 2021 (Gangaev et al., 2020) | class I/CD8 | predicted | ex vivo | multimer staining | 9 | entire proteome | 18 | 4 | 4 | NI |
| Habel, PNAS, 2020∗ (Habel et al., 2020) | class I/CD8 | overlapping | in vitro expansion, ex vivo | ICS, multimer staining | 14 | S, N, M, ORF1ab | 18 | 12 | 1 | NI |
| Joag, J Immunol, 2021 (Joag et al., 2021) | class I/CD8 | predicted | in vitro expansion | ICS | 1 | N | 10 | 1 | NI | |
| Kared, J Clin Invest, 2021 (Kared et al., 2021) | class I/CD8 | overlapping | ex vivo | multimer staining | 45 | entire proteome | 30 | 6 | NI | |
| Keller, Blood, 2020 (Keller et al., 2020) | both | predicted | in vitro expansion | ELISpot | 12 | S, M, N, E | 11 | NI | 12 | |
| Le Bert, bioRXiv, 2020 (Le Bert et al., 2021) | both | predicted | in vitro expansion | ICS | 3 | M, N, S | 3 | NI | NI | |
| Le Bert, Nature, 2020 (Le Bert et al., 2020) | both | overlapping | in vitro expansion | ICS | 9 | N, nsp7, nsp13 | 36 | 37 | NI | NI |
| Lee, J Virol, 2020 (Lee et al., 2020) | class I/CD8 | predicted | in vitro expansion | degranulation, ICS | 2 | N | 2 | 1 | NI | |
| Mahajan, bioRXiv, 2020 (Mahajan et al., 2020) | both | predicted | in vitro expansion | ICS, AIM | 10 | S | 17 | NI | NI | |
| Mateus, Science, 2020 (Mateus et al., 2020) | both | overlapping and predicted | in vitro expansion | ELISpot | 138 | entire proteome | 40 | NI | 30 | |
| Nelde, Nat. Immunol, 2021 (Nelde et al., 2021) | both | predicted | in vitro expansion | ELISpot, ICS | 49 | entire proteome | 116 | 104 | 9 | NI |
| Nielsen, bioRXiv, 2020 (Nielsen et al., 2020) | class I/CD8 | predicted | ex vivo | multimer staining | 9 | M, N, S | 106 | 1 | NI | |
| Peng, Nat. Immunol, 2020 (Peng et al., 2020) | both | overlapping | ex vivo | ELISpot, multimer staining | 16 | S, N, M, E, ORF3a, ORF6, ORF7a, ORF8 | 42 | 16 | 6 | NI |
| Poran, bioRXiv, 2020 (Poran et al., 2020b, Poran et al., 2020a) | class I/CD8 | predicted | in vitro expansion | multimer staining | 11 | S, N, M, E, ORF1ab | 3 | 1 | NI | |
| Prakash, bioRXiv, 2020 (Prakash et al., 2020) | both | predicted | ex vivo | ELISpot | 27 | entire proteome | 63 | 10 | 1 | NI |
| Rha, Immunity, 2021 (Rha et al., 2021) | class I/CD8 | predicted | ex vivo | proliferation, ICS, multimer staining | 2 | S, M, N | 116 | 1 | NI | |
| Sahin, medRXiv, 2020 (Sahin et al., 2020) | class I/CD8 | predicted | ex vivo | multimer staining | 8 | S | 3 | 3 | NI | |
| Saini, bioRXiv, 2020 (Saini et al., 2020, Saini et al., 2021) | class I/CD8 | predicted | ex vivo | multimer staining | 409 | entire proteome | 18 | 38 | 10 | NI |
| Schulien, Nat. Med, 2021 (Schulien et al., 2021) | class I/CD8 | predicted | in vitro expansion (ex vivo verification) | degranulation, ICS, multimer staining | 40 | entire proteome | 26 | 8 | 9 | NI |
| Sekine, Cell, 2020 (Sekine et al., 2020) | class I/CD8 | predicted | ex vivo | multimer staining | 2 | Orf3a, ORF6, M, N, E, S | 11 | 18 | 2 | NI |
| Shomuradova, Immunity, 2020 (Shomuradova et al., 2020) | class I/CD8 | predicted | ex vivo | multimer staining | 12 | S | 17 | 17 | 1 | NI |
| Snyder, medRXiv, 2020 (Snyder et al., 2020) | class I/CD8 | predicted | ex vivo | AIM | 235 | entire proteome | 47 | NI | NI | |
| Tarke, Cell Rep Med, 2021 (Tarke et al., 2021a) | both | overlapping (CD4), predicted (CD8) | ex vivo | AIM | 734 | entire proteome | 99 | 26 | 35 | |
The 25 different studies to date (2/28/2021) that have identified SARS-CoV-2 derived CD4 and CD8 epitopes are listed; in cases where the pre-print version analyzed has subsequently been published in the peer-reviewed literature, we have indicated both citations. Studies that, to date, are only available on pre-print servers are highlighted by italicized font. Respective columns summarize the scope and approach of each study, whether CD4 and/or CD8 epitopes were assayed, if predicted and/or overlapping peptide sets were used, and the types of T cell assay approaches utilized. Also tabulated are the number of unique epitopes identified and the specific antigens that were targeted for study. Additional columns show the number of COVID-19 positive and/or unexposed donors screened, and the number of unique HLA class I and class II restricting alleles identified. In vitro expansion refers to any assay that involved a period of in vitro culture before harvesting and assaying for T cell activity. An asterisk (∗) highlights a study that also measured epitope specific responses in tissues. NI indicates not investigated.
The list of papers we have reviewed is, to the best of our knowledge, exhaustive as of March 15th, 2021. Relevant papers were selected based on the objective curation process implemented over 20 years ago by the Immune Epitope Database (IEDB; www.iedb.org) based on the combined use of general broad PubMed queries combined with automated text classifiers and manual curation, as described in more detail elsewhere (Fleri et al., 2017; Salimi et al., 2012). In addition, the results of the IEDB curation were manually inspected by the coauthors to guard against papers missed by the IEDB curation workflow, but no additional papers were identified. This review focuses on SARS-CoV-2 epitopes recognized by human T cells, and thus does not discuss related topics, such as studies identifying T cell epitopes recognized in murine systems (Hassert et al., 2020; Takagi and Matsui, 2021), studies characterizing SARS-CoV-2 peptides eluted from HLA molecules (Knierman et al., 2020; Parker et al., 2020, Weingarten-Gabbay et al., 2020), or characterized by HLA binding in the absence of T cell recognition data (Prachar et al., 2020).
Here, we focus on cataloging and describing SARS-CoV-2 epitopes recognized by human T cells, from data collected from the 25 studies cited above. In this review, we have organized the data into a number of sections, initially describing epitope definitions, screening methodologies, and assay readouts. Subsequent sections describe the number of epitopes identified in the various studies, the antigens recognized, and the distribution of epitopes within them, which lead to the definition of immunodominant regions and immunodominant epitopes. Additional sections are devoted to discussion of epitope identification in different populations and cohorts and the related topics of HLA coverage and immunodominant HLA alleles. We also highlight how the breadth of the T cell repertoire informs discussions of pre-existing reactivity and cross-reactivity with common cold corona and other viruses, as well as cross-reactivity with MERS and SARS-CoV-1, and potential implications for immune escape by SARS-CoV-2 variants. This review is therefore relevant to the molecular definition of the targets of adaptive human T cell responses to SARS-CoV-2.
Epitope definitions
A detailed review of the available epitope data requires a clear definition of the concepts and terminology that have been used, to permit different studies that have used different methodologies to be combined and integrated in a coherent fashion. According to classical textbook definitions, “A T-cell epitope is a short peptide derived from a protein antigen. It binds to an MHC molecule and is recognized by a particular T cell” (Murphey et al., 2012). And, similarly, “The parts of complex antigens that are specifically recognized by lymphocytes are called determinants or epitopes” (Abbas et al., 2007).
T cell epitopes are usually peptides composed of 20 naturally occurring amino acids, although the recognition of haptens, sugars, and post-translationally modified peptides has also been described (Petersen et al., 2009; Sun et al., 2016). (For more information on post-translationally modified epitopes, we refer readers to Petersen et al., 2009.) While many post-translationally modified epitopes have been described in cancer and in autoimmunity, few have been described in the case of viral antigens. However, one question of particular interest, also in the context of SARS-CoV-2, is whether glycosylated sites are differentially recognized and, in the context of N > D modifications, which are associated with the removal of the polysaccharide moiety in the course of cellular processing. However, thus far, in the case of SARS-CoV-2 no reports have appeared of post-translationally modified or glycosylated peptides being recognized by T cell responses.
T cells recognize a bimolecular complex of an epitope bound to a specific class I or class II MHC molecule (HLA in humans), which is called its restriction element. HLA class I restricted epitopes are generally 9–10 residues in size, with several also being 8 or 11 residues depending on HLA restriction, while class II restricted epitopes are typically 13–17 residues, although shorter and longer peptides have also been described (Peters et al., 2020; Gfeller et al., 2018; Trolle et al., 2016; Wang et al., 2008a; O’Brien et al., 2008) By the late 1980s, it was recognized that a given peptide can bind multiple HLA allelic variants, especially if those variants are structurally or genetically related (McMichael et al., 1988; O’Sullivan et al., 1991). The HLA variants or types associated with overlapping peptide-binding repertoires are classified into so-called HLA supertypes (Greenbaum et al., 2011; Sidney et al., 2008). Epitopes that bind multiple HLAs are referred to as promiscuous (Kilgus et al., 1991; Panina-Bordignon et al., 1989). In general, any given HLA-peptide complex can be recognized by a multitude of different T cell receptors, which often share a discernible pattern of sequence similarity (Dash et al., 2017; Glanville et al., 2017).
Viral genomes and proteomes are composed of multiple protein antigens. Each of these antigens is recognized in a human population to varying degrees (Sidney et al., 2020; Yewdell and Bennink, 1999). The concept of immunodominance usually refers to how strongly a given antigen is recognized, either in a given assay, individual, or population, while immunoprevalence refers to how often the antigen is recognized in a given population (Oseroff et al., 2008; Tan et al., 2014; Wang et al., 2008b), although in practice the two terms are frequently used interchangeably.
The immunodominance of a given antigen within a genome or proteome is influenced by variables such as levels of transcription and expression, stability, and patterns of expression in different cell types or anatomical sites. In the context of SARS-CoV-2, Poran et al. point out the potential of leveraging proteomic data to infer relative viral protein abundance (Poran et al., 2020b, 2020a). Several other studies have eluted SARS-CoV-2-derived peptides bound to HLA (Knierman et al., 2020; Parker et al., 2020; Weingarten-Gabbay et al., 2020) but have not shown that the epitopes are actually recognized by T cell responses. Future studies will examine the correspondence between eluted ligands and T cell recognition.
The fact that HLA binding is a necessary but not sufficient requisite for T cell recognition is well established (Assarsson et al., 2007; Kotturi et al., 2007; Yewdell and Bennink, 1999; Yewdell, 2006), as it does not guarantee that a peptide will be generated by antigen processing or ensure the availability of a repertoire of T cells capable of recognizing the corresponding epitope-HLA complex (Hataye et al., 2006; Kotturi et al., 2008). In the case of eluted ligands (Croft et al., 2019; Paul et al., 2020), factors to be considered are whether the assay used to detect eluted ligands has sensitivity comparable to T cell activation (a few epitope copies have been shown to be sufficient to activate T cells; Demotz et al., 1990; Sykulev et al., 1996) and the availability of TCR repertoire, which is also modulated by previous infection history, as discussed in more detail below.
Immunodominance and immunoprevalence within a given antigen indicates how frequently and vigorously a particular epitope is recognized given all possible peptide epitopes contained in the antigen (Sidney et al., 2020; Yewdell and Bennink, 1999). Immunodominance/prevalence hierarchies within an antigen are influenced by variables such as HLA binding capacity, antigen processing, and the repertoire of T Cell Receptor (TCR) recognizing a given HLA-epitope combination. Finally, the term breadth of responses is defined on the basis of how many antigens or epitopes are recognized, either at the level of a given individual or in a population as a whole (Sidney et al., 2020; Yewdell and Bennink, 1999).
A variety of screening methodologies
The process of epitope identification entails testing collections of candidate peptides in an assay of choice. The peptide collections utilized can span the entire genome or proteome or focus on selected antigens of interest. Peptide collections can also correspond to sets of overlapping peptides (a popular choice is 15-mers overlapping by 10 residues) that span a sequence or peptides predicted to bind to one or more different HLA types, as indicated in the third column of Table 1. In general, and in the case of SARS-CoV-2 in particular, overlapping peptides are more often used to define class II restricted epitopes (4 of 9 studies; 44%), partly due to the lower predictive efficacy of HLA class II predictions (Peters et al., 2020) relative to class I epitopes (6/25 studies; 24%), for which predicted binders are often used to probe responses (21 of 25 studies; 85%). While the length of HLA class II restricted epitopes varies, the use of 15-mers overlapping by 10 residues ensures that any possible 10-mer is represented in the peptide set, with the addition of flanking residues at either or both ends. Given that the critical core of class II epitopes is 9 residues in size, this ensures that most, if not all, epitopes are identified without having to rely on bioinformatic predictions.
Another issue of relevance is whether responses are measured directly ex vivo or if an in vitro culture restimulation step is introduced. A restimulation step is often used to expand low frequency T cell specificity that would otherwise be difficult to detect. A number of different methodologies are used to detect or expand T cells, ranging from stimulation with whole antigens or antigen fragments, to the use of peptide pools or isolated individual peptides. However, in vitro restimulation is known to substantially alter the phenotypes and/or relative frequency of responding T cells. The expansion of naive T cells can also occur. In the case of SARS-CoV-2, studies have shown that when peripheral blood mononuclear cell (PBMCs) are expanded for 10–14 days before the assessment of SARS-CoV-2 responses, CD4 + T cells expand to a much greater extent than do CD8+ T cells (Habel et al., 2020; Mateus et al., 2020).
To overcome these caveats, it is preferable to assay T cells ex vivo whenever possible. In the case of SARS-CoV-2 T cell epitopes, 14 studies have used direct ex vivo assays (fourth column of Table 1) and 12 have utilized in vitro culture (one study utilized both in vitro and ex vivo approaches). Alternatively, once the epitopes are identified, they can be used to conduct secondary epitope validation experiments with direct ex vivo modalities, as shown by 2 studies (Chen et al., 2021; Schulien et al., 2021). Of particular note, Keller et al. showed that SARS-CoV-2 T cells can be expanded in controlled conditions and raised the possibility that epitope-expanded T cells could be used for adoptive therapy (Keller et al., 2020). The principle and conditions for adoptive therapy have been described and reviewed elsewhere (Riddell and Greenberg, 1995).
Assay readouts
Regardless of whether T cell responses are detected ex vivo or after in vitro expansion, a variety of different assay methodologies are available to investigate specific T cell responses. In selecting an approach, several considerations apply, including ease of implementation, throughput, and comprehensiveness and functionality. Certain assays, such as enzyme-linked immunospot (ELISpot), supernatant determination, and whole blood assays are easier to employ and more amenable to high-throughput testing. However, they are associated with less granular information. For example, the CD4 versus CD8 phenotype (and the expression of other cell markers) of the responding cells is not readily established by these approaches compared to other methods, such as intracellular cytokine staining (ICS) or activation-induced marker (AIM) assays. The methodologies utilized by the various studies reviewed here are listed in Table 1 and include AIM, degranulation, proliferation, ELISA, ELISpot, ICS, cytotoxicity, and multimer-based assays (for 3, 2, 2, 1, 5, 10, 1m and 13 studies, respectively).
Several studies (Kared et al., 2021; Nielsen et al., 2020; Poran et al., 2020b, 2020a; Prakash et al., 2020; Rha et al., 2021; Sekine et al., 2020; Shomuradova et al., 2020; Ferretti et al., 2020; Gangaev et al., 2020; Habel et al., 2020; Sahin et al., 2020; Schulien et al., 2021; Saini et al., 2020, 2021) performed high-resolution analysis of SARS-CoV-2-specific CD8+ T cells using HLA multimers. However, none of the studies reported similar multimer analyses for CD4 + T cells, despite the fact that, in general, HLA class II restricted SARS-CoV-2-specific T cell responses are more pronounced than HLA class I restricted T cell responses (Grifoni et al., 2020; Nelde et al., 2021). This reflects the relatively higher availability of HLA class I multimeric reagents as compared to their HLA class II counterparts. Some studies analyzed epitope-specific responses not only in blood but also in tissues such as tonsil and lung tissue from uninfected donors (Habel et al., 2020). The analysis of tissue-derived T cells can provide insight into disease—for example, by defining the characteristics of tissue resident memory T cells, which may differ from those circulating in the peripheral blood (Masopust and Soerens, 2019).
An issue encountered with ELISpot, ICS, and related assays is that while they, by definition, identify T cells capable of a functional response, they only (also by definition) detect T cells producing a cytokine of choice; therefore, they are “blind” to T cells that produce different cytokines or that do not produce cytokines in large amounts within the window of time of the assay (e.g., T follicular helper [Tfh] CD4 T cells generally produce very low amounts of cytokines). Both AIM (Dan et al., 2016; Locci et al., 2013; Reiss et al., 2017) and HLA tetramer/multimer assays are “agnostic” in this respect, as they detect all cells activated by the epitope (AIM), or all cells expressing a TCR capable of binding a given epitope-HLA complex (tetramer/multimer). Accordingly, it is often observed that AIM and tetramer assays have higher sensitivity because they detect larger numbers of T cells than ELISpot assays. Sahin et al. note that a comparison of data from MHC multimers with bulk IFNγ+ CD8+ T cell responses indicated that a functional T cell assay might underestimate the total cellular immune response (Sahin et al., 2020). Conversely, T cells captured by tetramers might not be functional or exhausted, and therefore might overestimate the cellular response that is relevant for immunity and infection control. However, for SARS-CoV-2, it has been observed that CD8 T cells identified by HLA multimers in COVID-19 subjects are functional and not exhausted (Rha et al., 2021). In conclusion, a variety of epitope screening and assay strategies have been utilized, each with its own features and potential advantages and disadvantages.
Number of epitopes identified in the different studies
The sixth column of Table 1 lists the total number of characterized canonical CD4 and CD8 epitopes identified in each study, which ranged from 1 to 734 (median of 12). It is not possible to estimate the total number of unique identified epitopes by simply adding these numbers together, because the same epitope might have been identified independently in multiple studies (as addressed below in the immunodominance section). This is especially the case for CD4 epitope studies that have utilized overlapping peptides; essentially, the same epitope might have been identified by two largely overlapping peptides.
As such, to assess CD4 epitope redundancy, we refined the data further by taking advantage of the clustering tool provided by the IEDB (Dhanda et al., 2018a), which automatically removes duplications and largely overlapping entries; we also performed additional manual curation. This clustering tool is an algorithm that generates clusters from a set of input epitopes based on representative or consensus sequences. It allows users to cluster peptide sequences on the basis of a specified level of identity by selecting among three different method options. For our purposes, we utilized the default “cluster-break” settings, which generate clusters in which all component epitopes share at minimum a specified level of homology (70% in our case) and no epitope is present in more than one cluster. Because of the closed ends of the class I MHC binding groove, and hence the incapacity of class I binding peptides to assume alternate frames, overlapping CD8 epitopes are considered unique epitopes by default.
For our analyses, we only considered epitopes of 8–14 residues for HLA class I and epitopes of 12–25 residues for class II. We used these parameters as they reflect the canonical sizes for class I and class II ligands and because of reports that overly short or long ligands can often represent “false positives” rather than being derived from peptides truly bound to MHC (Paul et al., 2018). We have not considered instances where the CD4/CD8 (class II/class I restriction) phenotype of responding T cells was not resolved or could not be reasonably inferred. These selection criteria did not lead to the exclusion of any studies, but rather to a few ambiguous epitopes being identified, accounting for a total of 81 unique sequences omitted from this analysis. Accordingly, we determined that the studies listed in Table 1 encompass 1,434 unique epitopes, which include 1,052 different class I and 382 different class II non-redundant epitopes (versus 416 when redundant epitopes were included).
Regarding limitations of the approach, in our review we have not considered data regarding HLA peptide binding (Prachar et al., 2020) or ligands eluted from HLA (Knierman et al., 2020; Parker et al., 2020; Weingarten-Gabbay et al., 2020) in absence of T cell recognition data. As more of this type of data is generated and reaches a critical mass, it will undoubtedly be of interest to correlate these data with T cell epitope recognition data. Our analyses have also not included epitopes defined in animal models. To date, few studies have described murine epitopes, and no data is available regarding the epitopes recognized by other species used in model systems such as Syrian hamsters or non-human primates (NHPs), even though some data has been reported suggesting that CD4 epitopes recognized in humans can be cross-recognized in NHPs (Shaan Lakshmanappa et al., 2021). Further experiments are required to enable the study of epitope-specific responses in SARS-CoV-2 animal studies. Finally, some of the information contained in this review is derived from preprint manuscripts that had not been formally peer reviewed at the time of analysis. The potential for variation in content between preprint and final versions of various studies is recognized by the curation process instituted by the IEDB team (of which B.P., A.S., and R.V. are part), in which each study originally curated at the preprint stage is re-curated when the study appears in the final published version.
Antigenic targets and epitope distribution
Ten of the 25 epitope identification studies (Ferretti et al., 2020; Gangaev et al., 2020; Kared et al., 2021;Mateus et al., 2020; Nelde et al., 2021; Saini et al., 2020, 2021; Schulien et al., 2021; Snyder et al., 2020; Tarke et al., 2021a;, Prakash et al., 2020) screened peptides derived from the entire SARS-CoV-2 proteome (seventh column of Table 1). The main antigenic targets of CD4 and CD8 SARS-CoV-2 T cell responses have been defined by several studies by utilizing overlapping peptides, rather than by resolving the actual epitopes (Grifoni et al., 2020; Tarke et al., 2021a), and are reviewed elsewhere (Altmann and Boyton, 2020; DiPiazza et al., 2020). These studies determined that structural proteins (S, M and N) are dominant targets of T cell responses, with ORF3, ORF8, and nsp3, 4, 6, 7, 12, and 13 (ORF1ab) also being frequently targeted. Other studies focused on specific subsets of SARS-CoV-2 antigens, as detailed in the seventh column of Table 1.
The various studies differ widely in the depth of screening, number of antigens tested, HLA alleles targeted, and number of peptides screened. For example, Peng et al. (2020) screened the whole proteome, with the exception of ORF1ab, using 423 peptides assayed in 42 infected and 16 non-exposed subjects, and they reported broad CD4 and CD8 responses. Conversely, Schulien et al. (2021) tested only 5 peptides predicted to bind to each of ten different HLAs. Tarke et al. (2021a) used PBMC from 99 donors and probed for CD4 responses using 1,925 peptides that spanned the entire SARS-CoV-2 proteome. To probe for CD8 responses, they tested an additional 5,600 peptides predicted to bind to one or more of 28 prominent HLA class I alleles. Snyder et al. (2020) screened 545 peptides distributed over the SARS-CoV-2 proteome for 26 class I alleles, testing about 20 peptides per allele. Nelde et al. (2021) screened a large number of donors (220 in total) with peptides spanning the breadth of antigens (i.e., the whole SARS-CoV-2 proteome) predicted to bind six HLA class I alleles or various HLA-DR class II. Le Bert et al. (2020) focused on peptides derived from N, nsp7, and nsp13, while Ferretti et al. (2020) screened predicted peptides from the entire proteome for 6 HLA alleles in 5 to 9 donors per HLA.
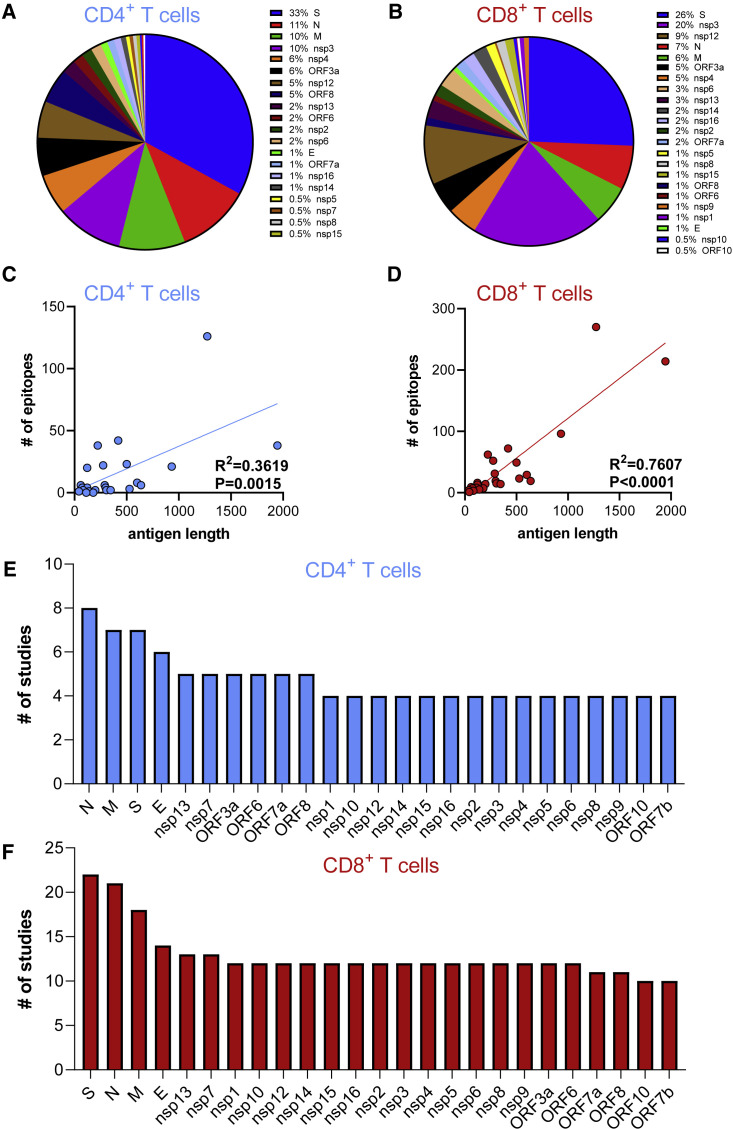
The epitope distribution along the SARS-CoV-2 proteome is analyzed in more detail in Figures 1A and 1B, in which the number of epitopes identified in each antigen is shown for CD4 and CD8 epitopes, respectively. Figures 1C and 1D show the correlation between the number of epitopes and the total number of residues (size) of each antigen. A significant correlation exists between antigen size and the number of epitopes identified for both CD4 (p = 0.0015 and rˆ2 = 0.36) and CD8 epitopes (p < 0.0001 and rˆ2 = 0.76). Certain antigens (N, M, S, and E) were studied in more detail (more studies focused on those antigen targets instead of considering the entire SARS-CoV-2 proteome) (Figures 1E and 1F). This is a significant factor, in addition to antigen length, in influencing the number of epitopes identified. Additionally, we recognized early on that the immunodominance pattern of the CD4 and CD8 T cell response to SARS-CoV-2 largely tracks with the expression level of each of the 25 viral proteins (Grifoni et al., 2020). S, M, and N sgRNAs are highly expressed by SARS-CoV-2 infected cells, and those three proteins are the most immunodominant targets of human CD4 and CD8 T cell responses to SARS-CoV-2 (Grifoni et al., 2020).
Figure 1.
Distribution of CD4 and CD8 epitopes by SARS-CoV-2 antigen
The fraction of known CD4 and CD8 epitopes derived from recognized SARS-CoV-2 antigens is shown in (A) and (B), respectively. The number of epitopes derived from each antigen as a function of antigen size is plotted in (C) and (D) for CD4+ (light blue) and CD8+(red) T cells, respectively; p values were calculated using a simple linear regression. (E) and (F) show the number of studies that probed responses to each antigen. All the source data used in these analyses were derived from the papers cited within Tables 1 and S1.
In conclusion, T cell responses are multi-antigenic, with the structural antigens being broadly recognized but with other proteins, such as nsp3, nsp4, nsp12 and ORF3a, also being vigorously recognized. This difference is not unexpected, since structural proteins are present in high concentrations in the virus and are accessible to the exogenous processing pathway and to HLA class II molecules. Non-structural proteins, which are produced in infected cells, also have access to the endogenous processing pathway and to HLA class I molecules.
Immunome browser analysis identifies patterns of immunodominance
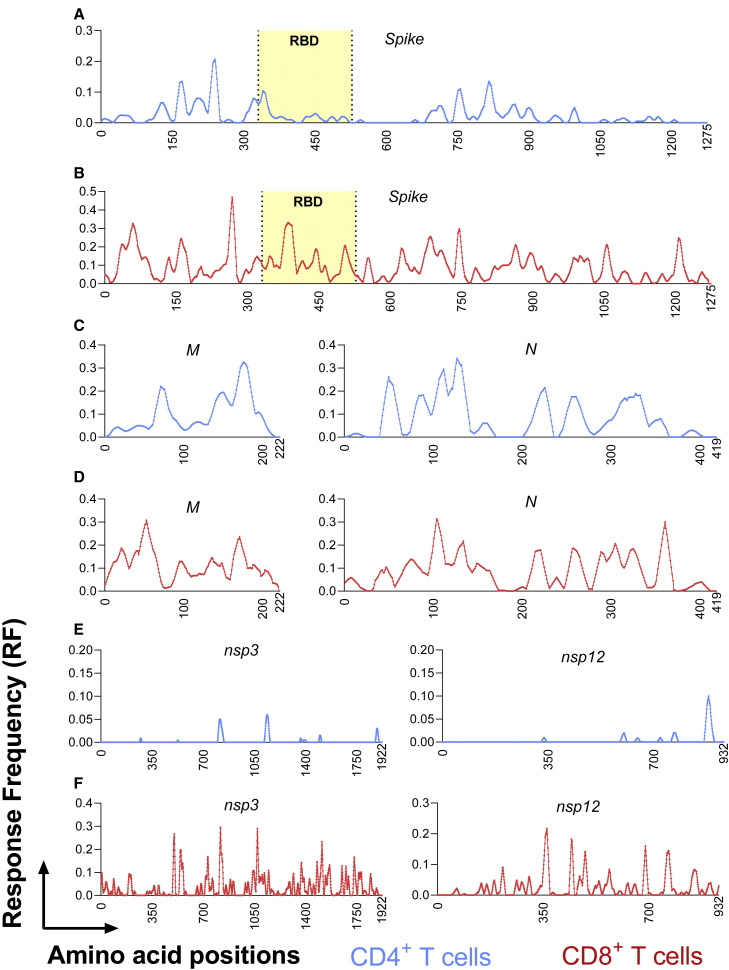
We also assessed whether discrete immunodominant regions would become apparent when we took a global view of the reported epitope data. We utilized the Immunome Browser tool (Vita et al., 2019; Dhanda et al., 2018b), developed and hosted by the IEDB (www.iedb.org). This tool allows patterns of immunodominance to be visualized across the entire SARS-CoV-2 proteome by plotting the 95% confidence interval (CI) of the Response Frequency (RF) for each residue, which is defined as the number of individuals and assays reporting positive responses to a peptide encompassing the particular residue. The lower bound RF values, using an average across a sliding 10-residue window, are plotted for human CD4 and CD8 epitopes in Figure 2 for the antigens S, M, N, nsp3, and nsp12. These antigens were chosen as their epitopes were described in sufficient number to allow us to delineate discrete immunodominant regions.
Figure 2.
Identification of immunodominant antigenic regions
The IEDB’s Immunome Browser tool was utilized to identify potential antigenic regions across the entire SARS-CoV-2 proteome. After searching for SARS-CoV-2-derived CD4+ (light blue) and CD8+(red) T cell epitopes, individual antigens were selected for further evaluation. From the antigen-specific Immunome Browser link, data was downloaded as an Excel file to obtain position-specific lower bound response frequency scores (RF), defined as the number of individuals and assays reporting positive responses to a peptide including that particular residue. For visualization, RF scores for each residue were recalculated to represent a sliding 10-residue window. Position-specific RF values for CD4 (light blue) and CD8 (red) epitopes are shown for the most dominant viral antigens: spike (A and B); M and N (C and D); nsp3 and nsp12 (E and F). The receptor binding domain region of the spike protein, is indicated in yellow in A and B because it is critically recognized by neutralizing antibodies and implicated in viral cell entry.
In the case of the spike protein, several immunodominant regions were observed for CD4 (residues 154–254, 296–370 and 682–925; Figure 2A), compared to a more homogeneous distribution for CD8 (Figure 2B). For the other structural proteins, namely the membrane and nucleocapsid, similar immunodominant regions for CD4 (Figure 2C) and CD8 (Figure 2D) were noted, with the 7–101 and 131–213 residue ranges being more prominent for the membrane protein and the 31–173 and 201–371 ranges for the nucleocapsid. More marked differences in CD4 and CD8 immunodominant regions, and in overall response frequency, were observed in the cases of nsp3 (Figure 2E) and nsp12 (Figure 2F). For both these proteins, defined immunodominant regions for CD4 (789–843, 1118–1158, and 1873–1903 for nsp3 and 863–903 for nsp12) were evident, versus more homogeneous patterns of CD8 recognition similar to that noted for the spike protein (Figure 2B). In conclusion, CD4 + T cells, in general, recognize more defined immunodominant regions than do their corresponding CD8+ counterparts.
Epitope identification in different populations and cohorts
As a whole, the different studies considered here have reported epitope identification results from a total of 1,197 donors (median = 34, range 2 to 220; see the eighth and ninth columns of Table 1). Of those, 870 donors were SARS-CoV-2 infected and 327 were unexposed.
It should be noted that these figures reflect the maximum number of donors utilized in each epitope identification and characterization study, as some assays and some epitopes have been tested in a different number of donors. For example, in some cases 20 donors were tested in ELISpot, but only 10 were evaluated using MHC multimers. Similarly, in several instances, because of the need to match peptide candidates to specific predicted HLA alleles (e.g., HLA-A∗02:01 candidate epitopes may only have been tested in HLA-A∗02:01-positive donors), the actual number of donors in which each peptide was tested might be significantly lower in comparison to other peptides.
Several studies have analyzed differences between the infected and unexposed cohorts and also in the context of potential cross-reactivity of SARS-CoV-2 epitopes with homologous sequences from common cold coronaviruses or other viruses, as discussed in more detail below. Also, as noted elsewhere (Sette and Crotty, 2021), considerable heterogeneity exists in SARS-CoV-2 infection and immune responses as a function of different variables such as age, gender, disease severity, ethnicity, co-morbidities, and time since symptom onset. As yet, the epitope identification studies do not answer the question as to whether differences in the types of epitopes recognized exist as a function of these variables. However, the epitopes defined in these studies, together with data generated from peptide pools, will undoubtedly be key to probing these variables and their role in the differences observed in terms of SARS-CoV-2-specific immune responses by evaluating the overall pattern of reactivity instead of focusing on few antigens or epitope candidates.
One issue to consider in future studies, and touched on further below, is to ensure that different ethnicities are adequately represented in SARS-CoV-2 studies. Thus far, most studies have been performed in donor cohorts that mostly consist of Caucasians and in which other ethnic groups are relatively under-represented.
HLA coverage and epitope identification results
It is well appreciated that HLA molecules are associated with an outstanding degree of diversity. Class I molecules are encoded by 3 main HLA loci (A, B, and C), and class II molecules are encoded by four main loci (DRB1, DRB3/4/5, DP, and DQ). Each locus is highly polymorphic, and because of heterozygosity, each individual might express close to 14 different HLA molecules and a minimum of 7 (if homozygous at all loci). Not only are the various HLA loci highly polymorphic, but the frequencies of their respective alleles vary, sometimes dramatically, across different ethnicities (Gonzalez-Galarza et al., 2020; Robinson et al., 2020). Establishing the extent to which epitope identification studies adequately cover the worldwide population is both a key and non-trivial issue (Greenbaum et al., 2011; McKinney et al., 2013; Sette and Sidney, 1999).
To meaningfully discuss population coverage of HLA allelic variants in the context of epitope identification efforts, we need to define what is meant by population coverage. The total phenotypic coverage provided by a set of HLA alleles represents the fraction of individuals that express at least one of a given set of alleles, while genotypic coverage corresponds to the fraction of genes at a specific locus the set of allelic variants covers. By way of example, an analysis targeting the HLA-A∗01:01, B∗07:02 and DRB1∗01:01 molecules will give a phenotypic coverage (probability that an individual in the average worldwide population will express at least one of these alleles) of approximately 35%. However, these three allelic variants represent only about 5%–10% of the gene variants at each of these three different HLA loci. This is important because in an individual that is “covered,” in the sense of expressing one HLA, the bulk of the T cell response will likely be directed to the other (up to thirteen) class I and class II alleles, leading to a gross misrepresentation of the total response magnitude and target specificity.
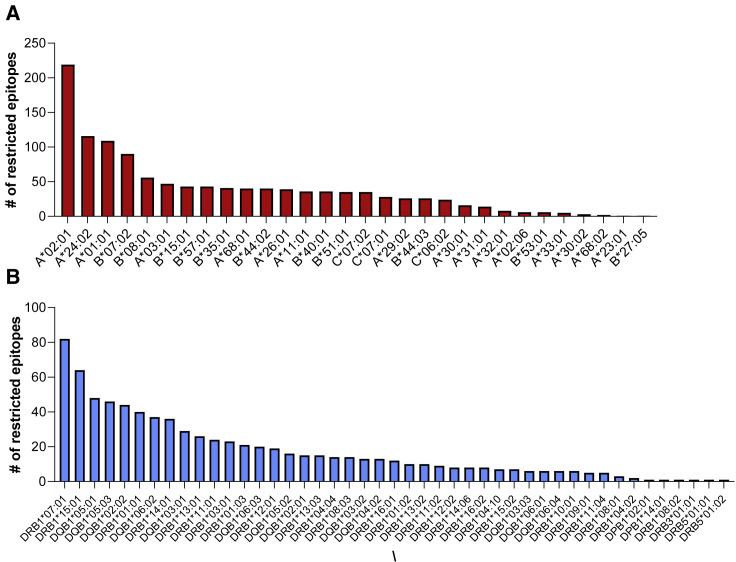
In previous studies, we have devoted significant efforts to analyzing the number of different HLA alleles associated with good genotypic and phenotypic coverage, and found that ∼25 different HLA class II and ∼25 different HLA class I alleles are required to cover 90% or more individuals in an idealized population (43, 61, 62). In the case of SARS-CoV-2 epitope identification studies, HLA restricted epitopes have been identified for 30 HLA class I and for 45 HLA class II alleles (Figures 3A and 3B), including, in both cases, the vast majority of the most common specificities in the general worldwide population (Gonzalez-Galarza et al., 2020; Weiskopf et al., 2013; Greenbaum et al., 2011).
Figure 3.
Defined HLA class I and class II restrictions
HLA-restricted epitopes have been identified for 30 class I (red, A) and 45 class II (light blue, B) molecules. The charts shows the number of restricted epitopes associated with each allele (alleles shown on the horizontal axis).
The median number of epitopes per allele is 35 (range 1 to 219) for class I, and 12 for class II (range 1 to 82). In the case of class I, as might be expected, most restrictions have been identified in the contexts of A∗02:01, A∗24:02, A∗01:01, and B∗07:02, as these are the most common class I alleles worldwide. Similarly, most class II restrictions are for DRB1∗07:01 and DRB1∗15:01, the most common DRB1 specificities worldwide. In both cases, the number of restrictions generally corresponds to overall allele frequency in the respective cohorts. This data exemplifies how the number of epitopes associated with a particular allelic specificity may not necessarily reflect immunodominance, but rather bias due to the availability of corresponding donor samples. Thus, the limited number of epitopes identified for several alleles is because they are rarer and therefore reflective of investigational bias. Additional studies are required to provide fully unbiased investigations of SARS-CoV-2 on a global scale. The number of allelic restrictions identified by the different studies is summarized in the tenth and eleventh columns of Table 1.
Overall, the 25 different studies mapped or inferred 1,191 class I restrictions, including 1,019 unique epitope-allele combinations (Table S1), with individual studies defining between 1 and 523 (median 8). For class II, 783 restrictions were mapped or inferred, with 760 representing unique epitope-allele combinations (Table S1). Only 9 studies investigated CD4 responses, with just 3 identifying class II restrictions (see Table 1). Thus, the number of experimentally defined HLA restrictions are fewer for class II relative to class I, which is consistent with the fact that class I restrictions are more easily inferred or determined and that multimers/tetramers (which implicitly assign restriction) are more broadly available for HLA class I than for HLA class II.
Immunodominance at the level of specific epitopes and alleles
Different studies report numerous peptides as being immunodominant, although each study also used different subjective definitions of immunodominance. While some peptides are repeatedly and independently identified, differences among these studies all contribute to the differences in their outcomes. These include differences in screening procedures, in HLA alleles considered, in the antigens targeted, the sampling of small numbers of individuals, and in how “immunodominance” was defined. For example, Peng et al., (2020) report several immunodominant peptides that they defined as being recognized by 6 or more of the up to 16 subjects screened. Tarke et al., (2021a) also highlight some epitopes as being more dominant, with 49 class II epitopes being recognized in 3 or more donors from an average of 10 donors tested and 41 class I epitopes recognized in 50% or more of the HLA matched donors tested. The same study also found that the response is broad and multi-specific, with ∼8–9 different antigens required to cover about 80% of the total CD4 and CD8 response (Tarke et al., 2021a). Nielsen et al. also concluded that the response is broad, since the top three immunogenic epitopes derived from separate SARS CoV-2 proteins (Nielsen et al., 2020). Keller et al. reported immunodominant epitopes defined as epitopes being recognized in multiple donors from M, N, and S (Keller et al., 2020).
Some specific epitopes are highlighted as being immunodominant in multiple studies. For example, in the context of the HLA-A∗02:01 class I molecule, which is the most studied for CD8 SARS-CoV-2 responses, the S 269–277 epitope (sequence YLQPRTFLL) is detected in 81% of HLA-A2+ individuals in the Nielsen study (Nielsen et al., 2020). The same A2 dominant epitope is also reported by Shomuradova et al., who tested 13 A2 peptides in total, and also identified a less strongly recognized epitope (Shomuradova et al., 2020). In the Habel et al. study, of the 14 peptides screened, S 269–277 generated the strongest IFN-γ+ response, with S 976–984 and ORF1ab 3183–3191 less prominently recognized (Habel et al., 2020). Ferretti et al. identified 3 epitopes recognized in 3 or more subjects (67% of the subjects tested), including S 269–277 (Ferretti et al., 2020). The study by Sahin et al. reports S 269–277 as the most dominant epitope and also identifies epitopes strongly recognized in the context of HLA-A∗24:02 and HLA-B∗35:01 (Sahin et al., 2020). Rha et al. detected S 269–277 responses in 37 of 112 (33%) patients, while S 1220–1228 was detected in only 2 of 40 (5%) patients (Rha et al., 2021), although other studies have observed higher response rates for this latter epitope. Overall, the S 269–277 epitope was found to be positive in 11 independent studies.
Another example of an immunodominant epitope is provided by the HLA-A∗01:01-restricted nsp3 819–828 epitope (sequence TTDPSFLGRY). This epitope was reported by Nelde et al. as being positive in 83% of the donors tested (Nelde et al., 2021). This study also identified a large number of additional dominant CD4 and CD8 restricted epitopes. The same A1-restricted epitope was also reported by Saini et al., who tested over 3,000 peptides for 10 alleles (Saini et al., 2020; Saini et al., 2021) and found 214 peptides that were recognized in 16 out of the 18 samples analyzed. Two additional HLA-A∗01:01 epitopes that overlap with TTDPSFLGRY (nsp3 818–828, sequence HTTDPSFLGRY, and nsp3 819–829, sequence TTDPSFLGRYM) were also identified as particularly dominant. The study by Gangaev et al. screened 50 epitopes for 10 alleles using tetramers (500 total) in 18 donors and identified nine epitopes in total, including the immunodominant nsp3 epitope restricted by HLA-A∗01:01 (Gangaev et al., 2020).
Global analysis of immunodominant epitopes
We further assessed published epitope data to determine whether particular HLA alleles and epitopes are dominantly recognized. In the case of HLA class II, because of the technical issues discussed above, dominant alleles are less readily assigned as restriction elements. In the case of HLA class I, certain alleles, such as HLA-A∗01:01, B∗07:02, B∗08:01 and B∗44:01 were associated with dominant responses (Tarke et al., 2021a). Other alleles, such as HLA A∗02:01, were associated with numerous epitopes but with responses of lower magnitude on average, and alleles such as A∗30:01 and A∗32:01 were associated with weak and infrequent responses. This HLA-allele-specific variation in response frequency/magnitude has been observed previously in the contexts of HIV and Dengue virus, where responses mediated by particular HLA allelic variants were associated with protection or susceptibility to disease (Goulder and Walker, 2012; Weiskopf et al., 2013). Whether HLA types play a role in influencing disease severity in the context of SARS-CoV-2 will have to be established as larger datasets become available.
For present purposes, we have defined the most dominant CD4 and CD8 epitopes as those recognized in 3 or more donors/studies, consistent with the definitions utilized by Mateus et al. and Tarke et al. (Mateus et al., 2020; Tarke et al., 2021a). We utilized this threshold based on previous experience in this matter. By selecting epitopes that have been recognized in multiple different experiments in separate donors allow, we can narrow the number of epitopes and focus on more dominant or prevalent responses while still preserving the goal of representing epitopes presented by a wide variety of HLA alleles. That is because less common HLAs are found, by definition, in fewer individuals, and the studies considered involved a median of 34 donors. Therefore, raising the “bar” further would restrict “immunodominant epitopes” to just those restricted by alleles that are very common in Caucasians.
The immunodominant epitopes identified in this way are highlighted in Table S1 and total 399 epitopes (110 CD4 epitopes and 289 CD8 epitopes). It is important to note that no epitope was not recognized in 100% of the cases/donors it was tested in, as has been observed in other viral systems (e.g., HBV, HIV, Poxviruses, Flu). This is relevant because it argues against the use of single-epitope tetramers to measure responses because of the likelihood of false negative results. Instead, the results argue for the use of peptide pools or multiplexing strategies (Kared et al., 2021; Nelde et al., 2021; Sekine et al., 2020; Shomuradova et al., 2020) to ensure the broad coverage of responses.
Another important consideration, as noted above, is the influence of investigational bias. It is apparent that epitopes from the spike protein, and those restricted by the most common HLA alleles, are overrepresented, which is likely a reflection that the spike antigen and those particular HLA alleles are more frequently studied (Figures 1E and 1F).
Breadth of the T cell repertoire
As summarized above in Figure 1, a total of 1,434 unique, non-redundant CD4 and CD8 epitopes have been defined, with the top 10 antigens accounting for 86% of the total. In these 10 most dominant antigens, a median of 87 epitopes (range of 33 to 396) is recognized. The data presented above demonstrates that T cell responses are multi-antigenic, with structural antigens being broadly recognized, but with other proteins such as nsp3, nsp12, ORF3a, and ORF8 also being vigorously recognized. Furthermore, data from Tarke et al. show that each individual is conservatively estimated to recognize, on average, 19 different CD4 and 17 different CD8 epitopes (Tarke et al., 2021a). Although individuals in our experience target multiple epitopes, the efficacy of the responses and number of epitopes targeted may vary substantially, dependent on HLA, the severity of disease, and other factors.
This breadth of response is apparently at variance with other reports describing only a limited number of epitopes for SARS-COV-2 (Chen et al., 2021; Le Bert et al., 2020; Lee et al., 2020; Nielsen et al., 2020; Rha et al., 2021; Sekine et al., 2020; Kared et al., 2021; Sahin et al., 2020). In some cases, in vitro expansion with artificial antigens has been utilized, and/or a limited number of subjects, cells, and/or epitope candidates were screened. Furthermore, several of the reported narrow repertoire epitopes differ among the different studies, consistent with a stochastic selection effect. Overall, the data curated in the IEDB as of March 15, 2021, reveals that over 1,400 different SARS-CoV-2-derived peptide sequences are reported as being recognized by human T cell responses, and which consist of 382 CD4 and 1,052 CD8 epitopes based on the meta-analysis performed in the current review.
Pre-existing reactivity and cross-reactivity with common cold corona and other viruses
Several studies have detected responses to SARS-CoV-2 sequences in unexposed controls (Sette and Crotty, 2020b, 2021). In some cases, these responses might correspond to infections associated with a lack of antibodies or to a transient antibody response (Sekine et al., 2020; Nelde et al., 2021). However, in other cases, these responses appear to be linked to pre-existing memory responses, which, in some instances, have been mapped to the cross-reactive recognition of the SARS-CoV-2 sequences by T cells induced by endemic “common cold” coronaviruses (17) and potentially other viral species (Bacher et al., 2020; Le Bert et al., 2020). This phenomenon has received considerable attention because of its potential to influence disease severity and vaccination outcomes and because of its potential implications for herd immunity (Bacher et al., 2020; Sette and Crotty, 2020b, 2021; Lipsitch et al., 2020; Sagar et al., 2021).
Epitopes recognized in non-exposed individuals have been defined in 12 studies. In some cases, these SARS-CoV-2 epitopes had significant homology to common cold coronavirus sequences, with cross-reactivity demonstrated at the molecular level in several instances (Mateus et al., 2020). Other studies, as discussed in more detail below, have examined whether SARS-CoV-2 specific T cells might cross-react with other more closely related viruses, such as SARS-CoV-1 and the Middle East Respiratory Syndrome virus (MERS) (see also below). This issue is of relevance in the context of developing vaccines that can elicit T cell responses that broadly recognize coronaviruses of pandemic potential.
The topic of pre-existing immune responses and cross-reactivity with common cold coronaviruses was addressed by several studies that reported a range of findings. Schulien et al. detected cross-reactive T cells in longitudinal samples pre-and-post SARS-CoV-2 infection and reported that these cells were expanded after in vitro restimulation (Schulien et al., 2021). Sekine et al. also detected widespread reactivity in non-exposed individuals using peptide pools (Sekine et al., 2020). Shomuradova et al. detected pre-existing T cell reactivity in unexposed donors using HLA-A2 tetramers but at much lower levels compared to those seen in exposed individuals (Shomuradova et al., 2020). Nelde et al. tested the reactivity of non-exposed donors to epitopes identified in exposed individuals and detected reactivity, albeit at lower levels, for several epitopes (Nelde et al., 2021). Keller et al. detected T cells with minimal cross reactivity with two homologous nucleocapsid peptides from NL63 and OC43 (Keller et al., 2020). Ferretti detected reactivity to OC43 and HKU1 sequences for 2 of 29 dominant epitopes and no reactivity for NL63 and 229E (Ferretti et al., 2020). Rha et al. reported that the SARS-CoV-2 S 269-277 and S 1220-1228 epitopes had low homology to OC43, HKU1, 229E, and NL63 and that MHC class I multimer+ cells were not detected in unexposed subjects (Rha et al., 2021). Prakash identified 24 epitopes, and of those, 11 recalled memory CD8+ T cells from unexposed healthy individuals (Prakash et al., 2020).
A potential explanation for the differences observed in the degree of cross-reactivity of epitope repertoires detected in infected and unexposed subjects is provided by the studies of Mateus et al. (Mateus et al., 2020) and Tarke et al. (Tarke et al., 2021a). These studies demonstrated that, overall, 50% of the epitopes defined in unexposed donors were also recognized in SARS-CoV-2-infected subjects (Mateus et al., 2020; Tarke et al., 2021a), but also that the viral infection created a new repertoire of epitopes recognized only in infected subjects. Conversely, over 80% of the epitopes defined in SARS-CoV-2-infected subjects were not recognized in unexposed donors. This suggests that a pre-existing repertoire of cross-reactive T cells is present in unexposed donors, but that the SARS-CoV-2 infection generates a largely novel repertoire of T cells in addition to the pre-existing one. Consistent with this view, the antigens dominantly recognized in exposed donors tend to only partially overlap with those dominant in non-exposed donors (Le Bert et al., 2020).
The issue of how preexisting memory reactivity might influence immunity has been debated, and a firm conclusion has not been reached as yet (Lipsitch et al., 2020; Sette and Crotty, 2020a, 2020b). While it is not expected that preexisting T cell reactivity might protect against infection, it is possible that preexisting SARS-CoV-2 cross-reactive T cells might modulate disease severity, as reported by a recent study (Sagar et al., 2021), or might even modulate vaccine responsiveness, allowing for a faster or more vigorous response.
The study of protective versus detrimental T cell responses is important for determining the optimal T cell engagement strategies for vaccines. In addition to understanding the relationship between pre-existing immunity to human coronaviruses and host defense against SARS-CoV-2, it is relevant to also consider the contribution of COVID-19-vaccine-boosted cross-reactive immune responses to vaccine-induced protective immunity.
Cross-reactivity with MERS and SARS-CoV-1
As mentioned above, several studies have addressed whether SARS-CoV-2 T cells might cross-react with more closely related viruses such as SARS-CoV-1 and MERS, an issue that is important for the development of vaccines that can elicit T cell responses to coronaviruses of pandemic potential.
As might be expected on the basis of the higher degree of sequence homology, cross-reactivity between SARS-CoV-2 responses and SARS-CoV-1 and MERS was more frequently detected than cross-reactivity between SARS-CoV-2 responses and common cold coronaviruses. More specifically, Le Bert et al. analyzed a cohort of 23 patients who recovered from SARS-1 and found long lasting memory T cells 17 years after the SARS-1 outbreak of 2003 (Le Bert et al., 2020). Habel et al. reported that T cells recognizing selected A2/SARS-CoV-2 CD8+ T cell epitopes can cross-react with SARS-CoV-1 and MERS, while they did not share homology with the common cold coronaviruses (Habel et al., 2020). Rha et al. reported that the S 269–277 epitope was specific to SARS-CoV-2, whereas the S 1220–1228 epitope was conserved in SARS-CoV-1 (Rha et al., 2021). In the study of Gangaev, of the 9 CD8 T cell epitopes they identified, 5 were unique for SARS-CoV-2 and 4 were shared between SARS-CoV-2 and SARS-CoV-1 (Gangaev et al., 2020). Prakash et al. also studied conserved pan-species epitope sequences for all coronaviruses, including those responsible for zoonotic infections (Prakash et al., 2020).
Potential for immune escape by SARS-CoV-2 variants
Another topic of relevance is the effect of naturally occurring mutations on epitope recognition. SARS-CoV-2 does mutate, and a key question, particularly for vaccine programs, is whether it will mutate to escape T cell responses. The large breadth of T cell epitopes recognized, and the fact that each individual tends to recognize their own unique sets of epitopes, depending on their HLA polymorphisms, has profound implications in terms of immune escape. A recent study showed that SARS-CoV-2 mutations predicted to have a negative impact on epitope binding to HLA were indeed associated with reduced T cell activity (Agerer et al., 2021). Other analyses of mutations associated with several variants of concern (VOCs) suggest that the vast majority of defined epitopes are conserved in SARS-CoV-2 variants (Tarke et al., 2021b; Redd et al., 2021).
The topic of potential immune escape by variants has been elevated by the observation that several recent SARS-CoV-2 VOCs have accumulated unusually large numbers of mutations and exhibit significant evidence of escape from neutralizing antibodies (Tegally et al., 2021; Wang et al., 2021; Thomson et al., 2021). This evolution appears to be due to the virus’s extended replication in immunocompromised individuals, at least in some cases (Avanzato et al., 2020). Given that immunity against COVID-19 consists of both antibody and T cell responses, there has been concern as to whether these variants escape T cell immunity.
The study of sequence variation and epitope recognition is of particular importance in the context of several well-described VOCs. Two independent studies (Tarke et al., 2021b; Redd et al., 2021) have shown that most of the epitopes defined by Tarke et al. (Tarke et al., 2021a) or Kared et al., (2021) are conserved within VOCs. Consistent with these observations, it has been shown that the antigens containing the sequence variations pertaining to the B.1.1.7, B.1.351, P.1, and CAL.20C variants are cross-recognized by individuals previously infected with the SARS-CoV-2 ancestral strain or that received COVID-19 vaccination. While the frequency of response across the different variants is kept, a decrease in magnitude of 30% or less is observed in terms of T cell reactivity for specific VOCs/assay combinations, suggesting an overall negligible impact of the VOCs in in the context of the T cell responses in the groups of vaccinated and convalescent donors tested thus far (Tarke et al., 2021b; Redd et al., 2021). Because of the high number of different epitopes reported, as noted above, and because of the large breadth of epitopes recognized in any given individual (estimated to be an average of 19 class II and 17 class I epitopes per person, genome-wide, and 9 if only the spike protein is considered), as suggested by one study (Tarke et al., 2021a), it appears unlikely that the new variants will escape T cell recognition at either the population or individual level.
In light of the data that indicate that T cell escape is not occurring (Tarke et al., 2021b), it is also relevant to consider the immunological and virological features that make T cell escape by SARS-CoV-2 unlikely. First, as noted, the broader the T cell response, in terms of epitopes, the less likely viral escape becomes, because any individual epitope that can escape through viral mutation would represent a small fraction of the overall immunity and thus represent a small selective pressure. Given that SARS-CoV-2 is a large RNA virus, the breadth of the CD4 and CD8 T cell responses is not surprising, per se.
Second, there are few examples in the literature of T cell epitope escape in humans for a virus that causes acute infections. In contrast, viruses that cause chronic viral infections, such as HIV and HCV, are well known to escape T cell epitope recognition. This is due to a fundamental difference in selective pressure. Within a single person, there is strong selective pressure for a chronic viral infection to escape T cell responses over time. In contrast, in a population of people, the diversity of HLA alleles presents a fundamental challenge for viral escape. This phenomenon is a basic premise in the evolutionary value of human HLA diversity. The escape of one or more T cell epitopes in one individual is unlikely to give the virus a selection advantage in the next host; indeed, escape mutations are more likely to be disadvantageous, because the original viral protein sequence was selected for functionality. However, in the influenza virus context (Rimmelzwaan et al., 2005), multiple compensatory co-mutations in the nucleoprotein have been observed to restore viral fitness. It remains possible that SARS-CoV-2 cytotoxic T-lymphocyte escape mutants might survive by a similar mechanism. The potential selection of viral T cell escape variants will depend on how well the spread of SARS-CoV-2 is controlled and, although selection for T cell escape variants may be highly restricted (owing to the factors discussed above), it cannot be ruled out at this time.
Third, a cornerstone feature of SARS-CoV-2 is the rapidity of replication and transmission within the human upper respiratory tract. Approximately half of SARS-CoV-2 transmissions occur in the pre-symptomatic phase of infection, before a T cell response has been mounted (in a previously unexposed or unvaccinated individual). The kinetics of SARS-CoV-2 replication and transmission are inconsistent with T cell pressure being a major component of intra-host selection in most individuals nor an evolutionarily relevant pressure, even though viral escape mutations may arise quickly, in acute infection, during the viremic phase. Combined, these virological, immunological, and epidemiological factors make it unlikely that SARS-CoV-2 will escape human T cell responses at the population level. Nevertheless, it is still possible that escape from T cell epitope recognition could occur in immunocompromised patients, some of whom have high levels of viral replication for > 120 days; therefore, it could be speculated that SARS-CoV-2 could/can undergo extensive mutation in such individuals during this time.
As mentioned above, it is important to evaluate SARS-CoV-2 epitope recognition in convalescents over time. Indeed, Bilich et al. (Bilich et al., 2021) published a recent study (which just missed the analysis time-point of March 15, 2021) in which they evaluated the T cell recognition of specific SARS-CoV-2 epitopes in a six-month follow-up of 51 convalescent individuals after mild or moderate SARS-CoV-2 infection. They detected epitopes capable of mediating long-term T cell responses, while responses to other T cell epitopes got lost over time.
Studies addressing TCR repertoires
Several studies have also investigated TCR repertoires and attempted to establish a link between epitope recognition and particular TCR sequences. More specifically, a seminal study by Gittelman et al. (Gittelman et al., 2021) obtained TCR sequence information from the entire municipality of Vò (Italy) during the initial surge of SARS-CoV-2 infections and detected notable correlations with disease severity and other characteristics. Snyder et al. (Snyder et al., 2020) expanded these findings by inferring several epitopes that may be able to be recognized by specific TCRs. They also built a classifier to diagnose infection based solely on TCR sequencing from blood samples. Along the same lines, Shomuradova et al. (Shomuradova et al., 2020) observed specific TCR motifs in the subjects they analyzed, in some cases shared across multiple donors, and Ferretti et al. (Ferretti et al., 2020) sorted epitope-specific T cells and used single-cell sequencing to define paired TCR α and TCR β chains expressed by these T cells. Gangaev et al. have also reported TCR sequences that recognize a defined SARS-CoV-2 epitope (Gangaev et al., 2020).
In conclusion, given the large number of different epitopes recognized in the context of a myriad of different HLA types, it will be necessary to compile an extensive catalog of TCR sequences to completely capture the TCR repertoire associated with SARS-CoV-2 responses in humans. Early reports indicate that the study of TCR repertoires might lead to interesting diagnostic applications and could yield additional insights into the pathogenesis of SARS-CoV-2, particularly given the recent Emergency Use Authorization of a TCR-based diagnostic developed by Adaptive Biotech (see: https://www.fda.gov/media/146478/download).
Conclusions
Here we reviewed 25 different studies describing the identification of over 1,400 different unique epitopes (382 for CD4 and 1052 for CD8) SARS-CoV-2 epitopes recognized by human T cells, herein annotated in terms of available metadata. This review highlights several key findings and also raises outstanding questions for future SARS-COV-2 research to address.
First, the epitope data described here derives in aggregate, from studies with 1,197 human subjects (870 COVID-19 and 327 unexposed controls). These cohorts represent considerable heterogeneity as a function of age, gender, disease severity (with severe disease less represented), and time since symptoms onset. However, different ethnicities were not broadly represented; this will be an important knowledge gap to be addressed in future investigations.
Second, and related to the above issue, HLA-restricted epitopes were identified for 30 class I and 45 class II molecules, which provides good coverage of a number of different loci and alleles. However, while the median number of epitopes per allele is 15, it ranged from 1 to 219, with a large bias toward the HLA alleles that are more frequently encountered in the general population.
Third, we note that while twenty studies defined class I/CD8 epitopes, only 9 defined class II/CD4 epitopes. Also, given the prominent role of CD4 responses in immune responses to SARS CoV2 in the context of natural infection and vaccination, this observation suggests that a more balanced study of both CD4 and CD8 epitopes remains an outstanding issue for future research.
Fourth, in terms of the antigens targeted by epitope identification studies, ten studies screened peptides derived from the entire proteome but fifteen studies concentrated on specific subsets of antigens, mostly based on the fact that the main SARS-CoV-2 T cell antigenic targets have been independently defined utilizing pools of overlapping peptides. Structural proteins (S, M, and N) are dominant targets of T cell responses, but ORF3, ORF8, nsp3, nsp4, and nsp12 are also frequently targeted. Within the main antigens, immunodominant regions are typically pronounced in the case of CD4 recognition but less so in the case of CD8 responses, which are more evenly distributed across the dominant antigens. The precise identification of immunodominant antigens and regions is of interest also for its potential in the context of the identification of immunogenic regions of the SARS CoV2 proteome, conserved in different coronavirus species of pandemic potential.
Finally, the fact that already more than 1,400 epitopes have been identified—also considering that many HLA alleles and regions of the SARS CoV2 proteome are relatively less studied—highlights that a large breadth of epitopes are recognized in human populations, making it unlikely that SARS CoV2 variants might escape T cell recognition at the population level.
Acknowledgments
This work has been supported by NIH contracts 75N93019C00001(A.S. and B.P.) and 75N9301900065 (A.S. and D.W.).
Author contributions
Conceptualization, D.W. and A.S.; Investigation, D.W., A.G., J.S., and A.S.; Resources, R.V., B.P., and A.S.; Data curation, D.W., A.G., J.S., R.V., and A.S.; Writing–original draft, D.W., J.S., and A.S.; Writing–review & editing, D.W., A.G., J.S., R.V., B.P., S.C., and A.S.; Data visualization, D.W., A.G., and J.S.; Supervision, D.W. and A.S.; Funding acquisition, D.W., B.P., and A.S.
Declaration of interests
A.S. is a consultant for Gritstone, Flow Pharma, Merck, Epitogenesis, Gilead, and Avalia. S.C. is a consultant for Avalia. L.J.I. has filed for patent protection for various aspects of T cell epitope and vaccine design work.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.chom.2021.05.010.
Supplemental information
The table is sorted by CD4 and then CD8 epitopes, and then by antigen and starting position. The listing includes CD4 epitopes of 12-25 residues. Individual epitopes have been assigned to unique clusters when they are nested within, or largely overlap (>70% homology) with other epitopes. CD8 epitopes include all epitopes of 8-14 residues. Reported epitopes without minimally defined class I or class II restriction are not included. Respective columns tabulate for each epitope the number of subjects tested and the number that had positive responses, as tabulated by the IEDB’s Immunome Browser. The number of studies describing each epitope is also tabulated, as well as the specific references; the number of references shown may exceed the number of studies tallied, as some studies are cited for both pre-print and final peer-reviewed versions. Epitopes that have been found to elicit positive responses in three or more donors are designated as dominant, and are highlighted accordingly. HLA class I and II restricting alleles, as reported in various studies, are also compiled for each epitope when known.
References
- Abbas A.K., Lichtman A.H., Pillai S. Saunders Elsevier; 2007. Cellular and Molecular Immunology. [Google Scholar]
- Agerer B., Koblischke M., Gudipati V., Montaño-Gutierrez L.F., Smyth M., Popa A., Genger J.W., Endler L., Florian D.M., Mühlgrabner V., et al. SARS-CoV-2 mutations in MHC-I-restricted epitopes evade CD8+ T cell responses. Sci. Immunol. 2021;6:6. doi: 10.1126/sciimmunol.abg6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmann D.M., Boyton R.J. SARS-CoV-2 T cell immunity: Specificity, function, durability, and role in protection. Sci. Immunol. 2020;5:5. doi: 10.1126/sciimmunol.abd6160. [DOI] [PubMed] [Google Scholar]
- Assarsson E., Sidney J., Oseroff C., Pasquetto V., Bui H.H., Frahm N., Brander C., Peters B., Grey H., Sette A. A quantitative analysis of the variables affecting the repertoire of T cell specificities recognized after vaccinia virus infection. J. Immunol. 2007;178:7890–7901. doi: 10.4049/jimmunol.178.12.7890. [DOI] [PubMed] [Google Scholar]
- Avanzato V.A., Matson M.J., Seifert S.N., Pryce R., Williamson B.N., Anzick S.L., Barbian K., Judson S.D., Fischer E.R., Martens C., et al. Case Study: Prolonged Infectious SARS-CoV-2 Shedding from an Asymptomatic Immunocompromised Individual with Cancer. Cell. 2020;183:1901–1912.e9. doi: 10.1016/j.cell.2020.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacher P., Rosati E., Esser D., Martini G.R., Saggau C., Schiminsky E., Dargvainiene J., Schröder I., Wieters I., Khodamoradi Y., et al. Low-Avidity CD4+ T Cell Responses to SARS-CoV-2 in Unexposed Individuals and Humans with Severe COVID-19. Immunity. 2020;53:1258–1271C.e5. doi: 10.1016/j.immuni.2020.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilich T., Nelde A., Heitmann J.S., Maringer Y., Roerden M., Bauer J., Rieth J., Wacker M., Peter A., Hörber S., et al. T cell and antibody kinetics delineate SARS-CoV-2 peptides mediating long-term immune responses in COVID-19 convalescent individuals. Sci. Transl. Med. 2021;13:eabf7517. doi: 10.1126/scitranslmed.abf7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Ruan P., Wang L., Nie X., Ma X., Tan Y. T and B cell Epitope analysis of SARS-CoV-2 S protein based on immunoinformatics and experimental research. J. Cell. Mol. Med. 2021;25:1274–1289. doi: 10.1111/jcmm.16200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft N.P., Smith S.A., Pickering J., Sidney J., Peters B., Faridi P., Witney M.J., Sebastian P., Flesch I.E.A., Heading S.L., et al. Most viral peptides displayed by class I MHC on infected cells are immunogenic. Proc. Natl. Acad. Sci. USA. 2019;116:3112–3117. doi: 10.1073/pnas.1815239116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan J.M., Lindestam Arlehamn C.S., Weiskopf D., da Silva Antunes R., Havenar-Daughton C., Reiss S.M., Brigger M., Bothwell M., Sette A., Crotty S. A Cytokine-Independent Approach To Identify Antigen-Specific Human Germinal Center T Follicular Helper Cells and Rare Antigen-Specific CD4+ T Cells in Blood. J. Immunol. 2016;197:983–993. doi: 10.4049/jimmunol.1600318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash P., Fiore-Gartland A.J., Hertz T., Wang G.C., Sharma S., Souquette A., Crawford J.C., Clemens E.B., Nguyen T.H.O., Kedzierska K., et al. Quantifiable predictive features define epitope-specific T cell receptor repertoires. Nature. 2017;547:89–93. doi: 10.1038/nature22383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demotz S., Grey H.M., Sette A. The minimal number of class II MHC-antigen complexes needed for T cell activation. Science. 1990;249:1028–1030. doi: 10.1126/science.2118680. [DOI] [PubMed] [Google Scholar]
- Dhanda S.K., Vaughan K., Schulten V., Grifoni A., Weiskopf D., Sidney J., Peters B., Sette A. Development of a novel clustering tool for linear peptide sequences. Immunology. 2018;155:331–345. doi: 10.1111/imm.12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanda S.K., Vita R., Ha B., Grifoni A., Peters B., Sette A. ImmunomeBrowser: a tool to aggregate and visualize complex and heterogeneous epitopes in reference proteins. Bioinformatics. 2018;34:3931–3933. doi: 10.1093/bioinformatics/bty463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiPiazza A.T., Graham B.S., Ruckwardt T.J. T cell immunity to SARS-CoV-2 following natural infection and vaccination. Biochem. Biophys. Res. Commun. 2020;538:211–217. doi: 10.1016/j.bbrc.2020.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti A.P., Kula T., Wang Y., Nguyen D.M.V., Weinheimer A., Dunlap G.S., Xu Q., Nabilsi N., Perullo C.R., Cristofaro A.W., et al. Unbiased Screens Show CD8+ T Cells of COVID-19 Patients Recognize Shared Epitopes in SARS-CoV-2 that Largely Reside outside the Spike Protein. Immunity. 2020;53:1095–1107.e3. doi: 10.1016/j.immuni.2020.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleri W., Vaughan K., Salimi N., Vita R., Peters B., Sette A. The Immune Epitope Database: How Data Are Entered and Retrieved. J. Immunol. Res. 2017;2017:5974574. doi: 10.1155/2017/5974574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangaev A., Ketelaars S.L.C., Isaeva O.I., Patiwael S., Dopler A., Hoefakker K.D.E., Biasi S., Gibellini L., Mussini C., Guaraldi G., et al. Identification and characterization of an immunodominant SARS-CoV-2-specific CD8 T cell response. J. Immunother. Cancer. 2020;8:A513–A514. doi: 10.1038/s41467-021-22811-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gfeller D., Guillaume P., Michaux J., Pak H.S., Daniel R.T., Racle J., Coukos G., Bassani-Sternberg M. The Length Distribution and Multiple Specificity of Naturally Presented HLA-I Ligands. J. Immunol. 2018;201:3705–3716. doi: 10.4049/jimmunol.1800914. [DOI] [PubMed] [Google Scholar]
- Gittelman R.M., Lavezzo E., Snyder T.M., Zahid H.J., Elyanow R., Dalai S., Kirsch I., Baldo L., Manuto L., Franchin E., et al. Diagnosis and Tracking of SARS-CoV-2 Infection By T-Cell Receptor Sequencing. medRxiv. 2021 doi: 10.1101/2020.11.09.20228023. [DOI] [Google Scholar]
- Glanville J., Huang H., Nau A., Hatton O., Wagar L.E., Rubelt F., Ji X., Han A., Krams S.M., Pettus C., et al. Identifying specificity groups in the T cell receptor repertoire. Nature. 2017;547:94–98. doi: 10.1038/nature22976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Galarza F.F., McCabe A., Santos E.J.M.D., Jones J., Takeshita L., Ortega-Rivera N.D., Cid-Pavon G.M.D., Ramsbottom K., Ghattaoraya G., Alfirevic A., et al. Allele frequency net database (AFND) 2020 update: gold-standard data classification, open access genotype data and new query tools. Nucleic Acids Res. 2020;48:D783–D788. doi: 10.1093/nar/gkz1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulder P.J., Walker B.D. HIV and HLA class I: an evolving relationship. Immunity. 2012;37:426–440. doi: 10.1016/j.immuni.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum J., Sidney J., Chung J., Brander C., Peters B., Sette A. Functional classification of class II human leukocyte antigen (HLA) molecules reveals seven different supertypes and a surprising degree of repertoire sharing across supertypes. Immunogenetics. 2011;63:325–335. doi: 10.1007/s00251-011-0513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habel J.R., Nguyen T.H.O., van de Sandt C.E., Juno J.A., Chaurasia P., Wragg K., Koutsakos M., Hensen L., Jia X., Chua B., et al. Suboptimal SARS-CoV-2-specific CD8+ T cell response associated with the prominent HLA-A∗02:01 phenotype. Proc. Natl. Acad. Sci. USA. 2020;117:24384–24391. doi: 10.1073/pnas.2015486117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassert M., Geerling E., Stone E.T., Steffen T.L., Feldman M.S., Dickson A.L., Class J., Richner J.M., Brien J.D., Pinto A.K. mRNA induced expression of human angiotensin-converting enzyme 2 in mice for the study of the adaptive immune response to severe acute respiratory syndrome coronavirus 2. PLoS Pathog. 2020;16:e1009163. doi: 10.1371/journal.ppat.1009163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hataye J., Moon J.J., Khoruts A., Reilly C., Jenkins M.K. Naive and memory CD4+ T cell survival controlled by clonal abundance. Science. 2006;312:114–116. doi: 10.1126/science.1124228. [DOI] [PubMed] [Google Scholar]
- Joag V., Wijeyesinghe S., Stolley J.M., Quarnstrom C.F., Dileepan T., Soerens A.G., Sangala J.A., O’Flanagan S.D., Gavil N.V., Hong S.W., et al. Cutting Edge: Mouse SARS-CoV-2 Epitope Reveals Infection and Vaccine-Elicited CD8 T Cell Responses. J Immunol. 2021;206:931–935. doi: 10.4049/jimmunol.2001400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan S.C. Innate and adaptive immune responses to SARS-CoV-2 in humans: relevance to acquired immunity and vaccine responses. Clin. Exp. Immunol. 2021;204:310–320. doi: 10.1111/cei.13582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kared H., Redd A.D., Bloch E.M., Bonny T.S., Sumatoh H., Kairi F., Carbajo D., Abel B., Newell E.W., Bettinotti M.P., et al. SARS-CoV-2-specific CD8+ T cell responses in convalescent COVID-19 individuals. J. Clin. Invest. 2021;131:145476. doi: 10.1172/JCI145476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson A.C., Humbert M., Buggert M. The known unknowns of T cell immunity to COVID-19. Sci. Immunol. 2020;5:5. doi: 10.1126/sciimmunol.abe8063. [DOI] [PubMed] [Google Scholar]
- Keller M.D., Harris K.M., Jensen-Wachspress M.A., Kankate V., Lang H., Lazarski C.A., Durkee-Shock J.R., Lee P.H., Chaudhry K., Webber K., et al. SARS-CoV-2 specific T-cells Are Rapidly Expanded for Therapeutic Use and Target Conserved Regions of Membrane Protein. Blood. 2020 doi: 10.1182/blood.2020008488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilgus J., Jardetzky T., Gorga J.C., Trzeciak A., Gillessen D., Sinigaglia F. Analysis of the permissive association of a malaria T cell epitope with DR molecules. J. Immunol. 1991;146:307–315. [PubMed] [Google Scholar]
- Knierman M.D., Lannan M.B., Spindler L.J., McMillian C.L., Konrad R.J., Siegel R.W. The Human Leukocyte Antigen Class II Immunopeptidome of the SARS-CoV-2 Spike Glycoprotein. Cell Rep. 2020;33:108454. doi: 10.1016/j.celrep.2020.108454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotturi M.F., Peters B., Buendia-Laysa F., Jr., Sidney J., Oseroff C., Botten J., Grey H., Buchmeier M.J., Sette A. The CD8+ T-cell response to lymphocytic choriomeningitis virus involves the L antigen: uncovering new tricks for an old virus. J. Virol. 2007;81:4928–4940. doi: 10.1128/JVI.02632-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotturi M.F., Scott I., Wolfe T., Peters B., Sidney J., Cheroutre H., von Herrath M.G., Buchmeier M.J., Grey H., Sette A. Naive precursor frequencies and MHC binding rather than the degree of epitope diversity shape CD8+ T cell immunodominance. J. Immunol. 2008;181:2124–2133. doi: 10.4049/jimmunol.181.3.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., Chng M.H.Y., Lin M., Tan N., Linster M., et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- Le Bert N., Clapham H.E., Tan A.T., Chia W.N., Tham C.Y.L., Lim J.M., Kunasegaran K., Tan L.W.L., Dutertre C.A., Shankar N., et al. Highly functional virus-specific cellular immune response in asymptomatic SARS-CoV-2 infection. J. Exp. Med. 2021;218:e20202617. doi: 10.1084/jem.20202617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E., Sandgren K., Duette G., Stylianou V.V., Khanna R., Eden J.S., Blyth E., Gottlieb D., Cunningham A.L., Palmer S. Identification of SARS-CoV-2 Nucleocapsid and Spike T-cell Epitopes for Assessing T-cell Immunity. J Virol. 2020 doi: 10.1128/JVI.02002-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsitch M., Grad Y.H., Sette A., Crotty S. Cross-reactive memory T cells and herd immunity to SARS-CoV-2. Nat. Rev. Immunol. 2020;20:709–713. doi: 10.1038/s41577-020-00460-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locci M., Havenar-Daughton C., Landais E., Wu J., Kroenke M.A., Arlehamn C.L., Su L.F., Cubas R., Davis M.M., International AIDS Vaccine Initiative Protocol C Principal Investigators, et al. Human circulating PD-1+CXCR3-CXCR5+ memory Tfh cells are highly functional and correlate with broadly neutralizing HIV antibody responses. Immunity. 2013;39:758–769. doi: 10.1016/j.immuni.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan S., Kode V., Bhojak K., Magdalene C.M., Lee K., Manoharan M., Ramesh A., Sudheendra H., Srivastava A., Sathian R., et al. Immunodominant T-cell epitopes from the SARS-CoV-2 spike antigen reveal robust pre-existing T-cell immunity in unexposed individuals. bioRxiv. 2020 doi: 10.1038/s41598-021-92521-4. 2020.11.03.367375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masopust D., Soerens A.G. Tissue-Resident T Cells and Other Resident Leukocytes. Annu. Rev. Immunol. 2019;37:521–546. doi: 10.1146/annurev-immunol-042617-053214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateus J., Grifoni A., Tarke A., Sidney J., Ramirez S.I., Dan J.M., Burger Z.C., Rawlings S.A., Smith D.M., Phillips E., et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370:89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney D.M., Southwood S., Hinz D., Oseroff C., Arlehamn C.S., Schulten V., Taplitz R., Broide D., Hanekom W.A., Scriba T.J., et al. A strategy to determine HLA class II restriction broadly covering the DR, DP, and DQ allelic variants most commonly expressed in the general population. Immunogenetics. 2013;65:357–370. doi: 10.1007/s00251-013-0684-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMichael A.J., Gotch F.M., Santos-Aguado J., Strominger J.L. Effect of mutations and variations of HLA-A2 on recognition of a virus peptide epitope by cytotoxic T lymphocytes. Proc. Natl. Acad. Sci. USA. 1988;85:9194–9198. doi: 10.1073/pnas.85.23.9194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphey K., Travers P., Walport M., Janeway C. Garland Science; 2012. Janeway’s Immunobiology. [Google Scholar]
- Nelde A., Bilich T., Heitmann J.S., Maringer Y., Salih H.R., Roerden M., Lübke M., Bauer J., Rieth J., Wacker M., et al. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat. Immunol. 2021;22:74–85. doi: 10.1038/s41590-020-00808-x. [DOI] [PubMed] [Google Scholar]
- Nielsen S.S., Vibholm L.K., Monrad I., Olesen R., Frattari G.S., Pahus M.H., Højen J.F., Gunst J.D., Erikstrup C., Holleufer A., et al. SARS-CoV-2 Elicits Robust Adaptive Immune Responses Regardless of Disease Severity. bioRxiv. 2020 doi: 10.1016/j.ebiom.2021.103410. 2020.10.08.331645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien C., Flower D.R., Feighery C. Peptide length significantly influences in vitro affinity for MHC class II molecules. Immunome Res. 2008;4:6. doi: 10.1186/1745-7580-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan D., Arrhenius T., Sidney J., Del Guercio M.F., Albertson M., Wall M., Oseroff C., Southwood S., Colón S.M., Gaeta F.C., et al. On the interaction of promiscuous antigenic peptides with different DR alleles. Identification of common structural motifs. J. Immunol. 1991;147:2663–2669. [PubMed] [Google Scholar]
- Oseroff C., Peters B., Pasquetto V., Moutaftsi M., Sidney J., Panchanathan V., Tscharke D.C., Maillere B., Grey H., Sette A. Dissociation between epitope hierarchy and immunoprevalence in CD8 responses to vaccinia virus western reserve. J. Immunol. 2008;180:7193–7202. doi: 10.4049/jimmunol.180.11.7193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panina-Bordignon P., Tan A., Termijtelen A., Demotz S., Corradin G., Lanzavecchia A. Universally immunogenic T cell epitopes: promiscuous binding to human MHC class II and promiscuous recognition by T cells. Eur. J. Immunol. 1989;19:2237–2242. doi: 10.1002/eji.1830191209. [DOI] [PubMed] [Google Scholar]
- Parker R., Partridge T., Wormald C., Kawahara R., Stalls V., Aggelakopoulou M., Parker J., Doherty R.P., Morejon Y.A., Lee E., et al. Mapping the SARS-CoV-2 spike glycoprotein-derived peptidome presented by HLA class II on dendritic cells. bioRxiv. 2020 doi: 10.1016/j.celrep.2021.109179. 2020.08.19.255901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S., Karosiene E., Dhanda S.K., Jurtz V., Edwards L., Nielsen M., Sette A., Peters B. Determination of a Predictive Cleavage Motif for Eluted Major Histocompatibility Complex Class II Ligands. Front. Immunol. 2018;9:1795. doi: 10.3389/fimmu.2018.01795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S., Croft N.P., Purcell A.W., Tscharke D.C., Sette A., Nielsen M., Peters B. Benchmarking predictions of MHC class I restricted T cell epitopes in a comprehensively studied model system. PLoS Comput. Biol. 2020;16:e1007757. doi: 10.1371/journal.pcbi.1007757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Mentzer A.J., Liu G., Yao X., Yin Z., Dong D., Dejnirattisai W., Rostron T., Supasa P., Liu C., et al. Oxford Immunology Network Covid-19 Response T cell Consortium; ISARIC4C Investigators Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020;21:1336–1345. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters B., Nielsen M., Sette A. T Cell Epitope Predictions. Annu. Rev. Immunol. 2020;38:123–145. doi: 10.1146/annurev-immunol-082119-124838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen J., Purcell A.W., Rossjohn J. Post-translationally modified T cell epitopes: immune recognition and immunotherapy. J. Mol. Med. (Berl.) 2009;87:1045–1051. doi: 10.1007/s00109-009-0526-4. [DOI] [PubMed] [Google Scholar]
- Poran A., Harjanto D., Malloy M., Arieta C.M., Rothenberg D.A., Lenkala D., van Buuren M.M., Addona T.A., Rooney M.S., Srinivasan L., Gaynor R.B. Sequence-based prediction of SARS-CoV-2 vaccine targets using a mass spectrometry-based bioinformatics predictor identifies immunogenic T cell epitopes. Genome Med. 2020;12:70. doi: 10.1186/s13073-020-00767-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poran A., Harjanto D., Malloy M., Rooney M.S., Srinivasan L., Gaynor R.B. Sequence-based prediction of vaccine targets for inducing T cell responses to SARS-CoV-2 utilizing the bioinformatics predictor RECON. bioRxiv. 2020 doi: 10.1186/s13073-020-00767-w. 2020.04.06.027805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prachar M., Justesen S., Steen-Jensen D.B., Thorgrimsen S., Jurgons E., Winther O., Bagger F.O. Identification and validation of 174 COVID-19 vaccine candidate epitopes reveals low performance of common epitope prediction tools. Sci. Rep. 2020;10:20465. doi: 10.1038/s41598-020-77466-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S., Srivastava R., Coulon P.-G., Dhanushkodi N.R., Chentoufi A.A., Tifrea D.F., Edwards R.A., Figueroa C.J., Schubl S.D., Hsieh L., et al. Genome-Wide Asymptomatic B-Cell, CD4+ and CD8+ T-Cell Epitopes, that are Highly Conserved Between Human and Animal Coronaviruses, Identified from SARS-CoV-2 as Immune Targets for Pre-Emptive Pan-Coronavirus Vaccines. bioRxiv, 2020 doi: 10.4049/jimmunol.2001438. 2020.09.27.316018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redd A.D., Nardin A., Kared H., Bloch E.M., Pekosz A., Laeyendecker O., Abel B., Fehlings M., Quinn T.C., Tobian A.A. CD8+ T cell responses in COVID-19 convalescent individuals target conserved epitopes from multiple prominent SARS-CoV-2 circulating variants. medRxiv. 2021 doi: 10.1093/ofid/ofab143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss S., Baxter A.E., Cirelli K.M., Dan J.M., Morou A., Daigneault A., Brassard N., Silvestri G., Routy J.P., Havenar-Daughton C., et al. Comparative analysis of activation induced marker (AIM) assays for sensitive identification of antigen-specific CD4 T cells. PLoS One. 2017;12:e0186998. doi: 10.1371/journal.pone.0186998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rha M.S., Jeong H.W., Ko J.H., Choi S.J., Seo I.H., Lee J.S., Sa M., Kim A.R., Joo E.J., Ahn J.Y., et al. PD-1-Expressing SARS-CoV-2-Specific CD8+ T Cells Are Not Exhausted, but Functional in Patients with COVID-19. Immunity. 2021;54:44–52.e3. doi: 10.1016/j.immuni.2020.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddell S.R., Greenberg P.D. Principles for adoptive T cell therapy of human viral diseases. Annu. Rev. Immunol. 1995;13:545–586. doi: 10.1146/annurev.iy.13.040195.002553. [DOI] [PubMed] [Google Scholar]
- Rimmelzwaan G.F., Berkhoff E.G.M., Nieuwkoop N.J., Smith D.J., Fouchier R.A.M., Osterhaus A.D.M.E. Full restoration of viral fitness by multiple compensatory co-mutations in the nucleoprotein of influenza A virus cytotoxic T-lymphocyte escape mutants. J. Gen. Virol. 2005;86:1801–1805. doi: 10.1099/vir.0.80867-0. [DOI] [PubMed] [Google Scholar]
- Robinson J., Barker D.J., Georgiou X., Cooper M.A., Flicek P., Marsh S.G.E. IPD-IMGT/HLA Database. Nucleic Acids Res. 2020;48(D1):D948–D955. doi: 10.1093/nar/gkz950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagar M., Reifler K., Rossi M., Miller N.S., Sinha P., White L.F., Mizgerd J.P. Recent endemic coronavirus infection is associated with less-severe COVID-19. J. Clin. Invest. 2021;131:131. doi: 10.1172/JCI143380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin U., Muik A., Vogler I., Derhovanessian E., Kranz L.M., Vormehr M., Quandt J., Bidmon N., Ulges A., Baum A., et al. BNT162b2 induces SARS-CoV-2-neutralising antibodies and T cells in humans. medRxiv. 2020 [Google Scholar]
- Saini S.K., Hersby D.S., Tamhane T., Povlsen H.R., Hernandez S.P.A., Nielsen M., Gang A.O., Hadrup S.R. SARS-CoV-2 genome-wide mapping of CD8 T cell recognition reveals strong immunodominance and substantial CD8 T cell activation in COVID-19 patients. bioRxiv. 2020 doi: 10.1126/sciimmunol.abf7550. 2020.10.19.344911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini S.K., Hersby D.S., Tamhane T., Povlsen H.R., Amaya Hernandez S.P., Nielsen M., Gang A.O., Hadrup S.R. SARS-CoV-2 genome-wide T cell epitope mapping reveals immunodominance and substantial CD8+ T cell activation in COVID-19 patients. Sci. Immunol. 2021;6:6. doi: 10.1126/sciimmunol.abf7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salimi N., Fleri W., Peters B., Sette A. The immune epitope database: a historical retrospective of the first decade. Immunology. 2012;137:117–123. doi: 10.1111/j.1365-2567.2012.03611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulien I., Kemming J., Oberhardt V., Wild K., Seidel L.M., Killmer S., Sagar F.D., Daul F., Salvat Lago M., Decker A., et al. Characterization of pre-existing and induced SARS-CoV-2-specific CD8+ T cells. Nat. Med. 2021;27:78–85. doi: 10.1038/s41591-020-01143-2. [DOI] [PubMed] [Google Scholar]
- Sekine T., Perez-Potti A., Rivera-Ballesteros O., Strålin K., Gorin J.B., Olsson A., Llewellyn-Lacey S., Kamal H., Bogdanovic G., Muschiol S., et al. Karolinska COVID-19 Study Group Robust T Cell Immunity in Convalescent Individuals with Asymptomatic or Mild COVID-19. Cell. 2020;183:158–168.e14. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A., Crotty S. Author Correction: Pre-existing immunity to SARS-CoV-2: the knowns and unknowns. Nat. Rev. Immunol. 2020;20:644. doi: 10.1038/s41577-020-00430-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A., Crotty S. Pre-existing immunity to SARS-CoV-2: the knowns and unknowns. Nat. Rev. Immunol. 2020;20:457–458. doi: 10.1038/s41577-020-0389-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A., Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021 doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A., Sidney J. Nine major HLA class I supertypes account for the vast preponderance of HLA-A and -B polymorphism. Immunogenetics. 1999;50:201–212. doi: 10.1007/s002510050594. [DOI] [PubMed] [Google Scholar]
- Shaan Lakshmanappa Y., Elizaldi S.R., Roh J.W., Schmidt B.A., Carroll T.D., Weaver K.D., Smith J.C., Verma A., Deere J.D., Dutra J., et al. SARS-CoV-2 induces robust germinal center CD4 T follicular helper cell responses in rhesus macaques. Nat. Commun. 2021;12:541. doi: 10.1038/s41467-020-20642-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shomuradova A.S., Vagida M.S., Sheetikov S.A., Zornikova K.V., Kiryukhin D., Titov A., Peshkova I.O., Khmelevskaya A., Dianov D.V., Malasheva M., et al. SARS-CoV-2 Epitopes Are Recognized by a Public and Diverse Repertoire of Human T Cell Receptors. Immunity. 2020;53:1245–1257.e5. doi: 10.1016/j.immuni.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidney J., Peters B., Frahm N., Brander C., Sette A. HLA class I supertypes: a revised and updated classification. BMC Immunol. 2008;9:1. doi: 10.1186/1471-2172-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidney J., Peters B., Sette A. Epitope prediction and identification- adaptive T cell responses in humans. Semin. Immunol. 2020;50:101418. doi: 10.1016/j.smim.2020.101418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder T.M., Gittelman R.M., Klinger M., May D.H., Osborne E.J., Taniguchi R., Zahid H.J., Kaplan I.M., Dines J.N., Noakes M.N., et al. Magnitude and Dynamics of the T-Cell Response to SARS-CoV-2 Infection at Both Individual and Population Levels. medRxiv. 2020 2020.07.31.20165647. [Google Scholar]
- Sun L., Middleton D.R., Wantuch P.L., Ozdilek A., Avci F.Y. Carbohydrates as T-cell antigens with implications in health and disease. Glycobiology. 2016;26:1029–1040. doi: 10.1093/glycob/cww062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swadling L., Maini M.K. T cells in COVID-19 - united in diversity. Nat. Immunol. 2020;21:1307–1308. doi: 10.1038/s41590-020-0798-y. [DOI] [PubMed] [Google Scholar]
- Sykulev Y., Joo M., Vturina I., Tsomides T.J., Eisen H.N. Evidence that a single peptide-MHC complex on a target cell can elicit a cytolytic T cell response. Immunity. 1996;4:565–571. doi: 10.1016/s1074-7613(00)80483-5. [DOI] [PubMed] [Google Scholar]
- Takagi A., Matsui M. Identification of HLA-A∗02:01-Restricted Candidate Epitopes Derived from the Nonstructural Polyprotein 1a of SARS-CoV-2 That May Be Natural Targets of CD8+ T Cell Recognition <em>In Vivo</em>. J. Virol. 2021;95:e01837-e20. doi: 10.1128/JVI.01837-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan A.T., Sodsai P., Chia A., Moreau E., Chng M.H., Tham C.Y., Ho Z.Z., Banu N., Hirankarn N., Bertoletti A. Immunoprevalence and immunodominance of HLA-Cw∗0801-restricted T cell response targeting the hepatitis B virus envelope transmembrane region. J. Virol. 2014;88:1332–1341. doi: 10.1128/JVI.02600-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarke A., Sidney J., Kidd C.K., Dan J.M., Ramirez S.I., Yu E.D., Mateus J., Da Silva Antunes R., Moore E., Rubiro P., Methot N., et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep Med. 2021;2:100204. doi: 10.1016/j.xcrm.2021.100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarke A., Sidney J., Methot N., Zhang Y., Dan J.M., Goodwin B., Rubiro P., Sutherland A., da Silva Antunes R., Frazier A., et al. Negligible impact of SARS-CoV-2 variants on CD4 + and CD8 + T cell reactivity in COVID-19 exposed donors and vaccinees. bioRxiv. 2021 doi: 10.1101/2021.02.27.433180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N., et al. Emergence of a SARS-CoV-2 variant of concern with mutations in spike glycoprotein. Nature. 2021;592 doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- Thomson E.C., Rosen L.E., Shepherd J.G., Spreafico R., da Silva Filipe A., Wojcechowskyj J.A., Davis C., Piccoli L., Pascall D.J., Dillen J., et al. ISARIC4C Investigators. COVID-19 Genomics UK (COG-UK) Consortium Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell. 2021;184:1171–1187.e20. doi: 10.1016/j.cell.2021.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trolle T., McMurtrey C.P., Sidney J., Bardet W., Osborn S.C., Kaever T., Sette A., Hildebrand W.H., Nielsen M., Peters B. The Length Distribution of Class I-Restricted T Cell Epitopes Is Determined by Both Peptide Supply and MHC Allele-Specific Binding Preference. J. Immunol. 2016;196:1480–1487. doi: 10.4049/jimmunol.1501721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vita R., Mahajan S., Overton J.A., Dhanda S.K., Martini S., Cantrell J.R., Wheeler D.K., Sette A., Peters B. The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res. 2019;47:D339–D343. doi: 10.1093/nar/gky1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Sidney J., Dow C., Mothé B., Sette A., Peters B. A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput. Biol. 2008;4:e1000048. doi: 10.1371/journal.pcbi.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.F., Kerzerho J., Adotevi O., Nuyttens H., Badoual C., Munier G., Oudard S., Tu S., Tartour E., Maillère B. Comprehensive analysis of HLA-DR- and HLA-DP4-restricted CD4+ T cell response specific for the tumor-shared antigen survivin in healthy donors and cancer patients. J. Immunol. 2008;181:431–439. doi: 10.4049/jimmunol.181.1.431. [DOI] [PubMed] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- Weingarten-Gabbay S., Klaeger S., Sarkizova S., Pearlman L.R., Chen D.Y., Bauer M.R., Taylor H.B., Conway H.L., Tomkins-Tinch C.H., Finkel Y., et al. SARS-CoV-2 infected cells present HLA-I peptides from canonical and out-of-frame ORFs. bioRxiv. 2020 2020.10.02.324145. [Google Scholar]
- Weiskopf D., Angelo M.A., de Azeredo E.L., Sidney J., Greenbaum J.A., Fernando A.N., Broadwater A., Kolla R.V., De Silva A.D., de Silva A.M., et al. Comprehensive analysis of dengue virus-specific responses supports an HLA-linked protective role for CD8+ T cells. Proc. Natl. Acad. Sci. USA. 2013;110:E2046–E2053. doi: 10.1073/pnas.1305227110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yewdell J.W. Confronting complexity: real-world immunodominance in antiviral CD8+ T cell responses. Immunity. 2006;25:533–543. doi: 10.1016/j.immuni.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Yewdell J.W., Bennink J.R. Immunodominance in major histocompatibility complex class I-restricted T lymphocyte responses. Annu. Rev. Immunol. 1999;17:51–88. doi: 10.1146/annurev.immunol.17.1.51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The table is sorted by CD4 and then CD8 epitopes, and then by antigen and starting position. The listing includes CD4 epitopes of 12-25 residues. Individual epitopes have been assigned to unique clusters when they are nested within, or largely overlap (>70% homology) with other epitopes. CD8 epitopes include all epitopes of 8-14 residues. Reported epitopes without minimally defined class I or class II restriction are not included. Respective columns tabulate for each epitope the number of subjects tested and the number that had positive responses, as tabulated by the IEDB’s Immunome Browser. The number of studies describing each epitope is also tabulated, as well as the specific references; the number of references shown may exceed the number of studies tallied, as some studies are cited for both pre-print and final peer-reviewed versions. Epitopes that have been found to elicit positive responses in three or more donors are designated as dominant, and are highlighted accordingly. HLA class I and II restricting alleles, as reported in various studies, are also compiled for each epitope when known.