Abstract
Background
The recent outbreak of the novel SARS-CoV-2 across the globe and the absence of specific drug against this virus lead the scientific community to look into some alternative indigenous treatments. India as a hub of Ayurvedic and medicinal plants can shed light on its treatment using specific active bio-molecules from these plants.
Objectives
Keeping our herbal resources in mind, we were interested to inquire whether some phytochemicals from Indian spices and medicinal plants can be used as alternative therapeutic agents in contrast to synthetic drugs.
Materials and methods
We used insilico molecular docking approach to test whether bioactive molecules of herbal origin such as hyperoside, nimbaflavone, ursolic acid, 6-gingerol, 6-shogaol and 6-paradol, curcumin, catechins and epigallocatechin, α-Hederin, piperine could bind and potentially block the Mproenzyme of the SARS-CoV-2 virus.
Results
Ursolic acid showed the highest docking score (−8.7 kcal/mol) followed by hyperoside (−8.6 kcal/mol), α-Hederin (−8.5 kcal/mol) and nimbaflavone (−8.0 kcal/mol). epigallocatechin, catechins, and curcumin also exhibited high binding affinity (Docking score −7.3, −7.1 and −7.1 kcal/mol) with the Mpro. The remaining tested phytochemicals exhibited moderate binding and inhibitory effects.
Conclusion
This finding provides a basis for biochemical assay of tested bioactive molecules on SARS-CoV-2 virus.
Keywords: SARS CoV-2 virus, Mpro, Insilico molecular docking, Phytochemical, Indian spices, Medicinal plants
1. Introduction
The Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) is a highly infectious virus for novel coronavirus disease -19 (COVID-19) disease that caused the recent outbreak in China in December 2019 and rapidly spread to the other parts of the globe owing to its extreme contagious nature. Its initial symptoms include fever, dry cough, tiredness, aches or pains, diarrhoea, difficulty in breathing, etc. In the human body, it probably settles through the angiotensin-converting enzyme 2 (ACE2) [1] receptor for entry into the host cell and the transmembrane protease, serine 2 (TMPRSS2) for viral spike protein priming [2,3]. The infection gradually took the shape of a pandemic with extremely high mortality [4,5] owing to lack of definite treatment regimen and medications against the virus as well as due to the presence of co-morbidities. Even the so-called developed nations like U.S and several European countries failed to control the infection with their cutting-edge medical technologies at a very early phase. As a result, the World Health Organisation declared COVID-19 as a public health emergency of international concern [6].
The previous name of this betacoronavirus was 2019-nCoV. It was renamed as SARS-CoV-2 by the International Committee on Taxonomy of Viruses (ICTV) [7]. The genome of SARS-CoV-2 has been sequenced [8]. The whole genome sequence analysis of SARS-CoV-2 shows 96.2% similarity with bat coronavirus (SARSr-Co) [9,10], while it shows low sequence identity with that of SARS-CoV (about 79%) or MERS-CoV (about 50%) [11,12].
Since, the onset of the COVID-19 pandemic, globally, researchers are involved in the rapid development of drugs and specific antiviral treatment strategies. Among all the SARS-CoV-2 targets the main protease (Mpro, 3CLpro, nsp5) of the virus received major attention [13,14]. Some alternative targets like spike protein (S), RNA-dependent RNA-polymerase (RdRp, nsp12), NTPase/helicase (nsp13) and papain-like protease (PLpro, part of nsp3) have also been reported in some literature [15,16]. The SARS-CoV-2 Mpro is a 33.8 kDa enzyme which plays a pivotal role in the cleavage of viral polyproteins (pp1a and pp1ab) in a site-specific (L-Q (S, A, G)) manner [17], resulting in the release of functional replicase enzyme which is crucial for transcription and replication of the virus [[18], [19], [20]]. Other essential enzymes which are involved in the replication process such as RdRp or nsp13 cannot fully function without this proteolytic action [13], making Mpro a key enzyme in viral replication cycle. As a result, the inhibition of Mpro could stop viral replication process and thus alleviate disease symptoms [21,22]. For drug discovery against SARS-CoV-2 virus, Mpro is one of the most attractive viral targets. Some studies have already reported synthetic competitive inhibitors against SARS-CoV-2 Mpro [17,23,24]; however, increase in substrate concentration often reduces the effectiveness of such inhibitors. Natural phytochemicals can provide safe and effective treatment by alleviating this limitation.
Although there are no approved drugs for COVID-19, a number of clinical trials are in progress [25]. Lopinavir and ritonavir, combined with Chinese herbal medicines, were used in preliminary clinical studies [26].
Indian medicinal plants and spices are a rich hub of ingredients which can be utilized for drug designing because of their high therapeutic values [27]. Previous docking study already reported that phytochemicals such as hyperoside and nimbaflavone are good candidate drugs against influenza virus strains [28]. A recent study has suggested high binding efficacy of ursolic acid (Tulsi) against surface spike glycoprotein and RNA polymerase of SARS-CoV-2 virus [29]. Ajoene and allicin (Garlic) shows strong virucidal activity against selected viruses including, herpes simplex virus type 1, herpes simplex virus type 2, parainfluenza virus type 3, vaccinia virus, vesicular stomatitis virus, and human rhinovirus type 2 [30]. Curcumin has diverse antiviral activity against dengue virus, herpes simplex virus, Zika and chikungunya virus [[31], [32], [33]]. Previous study reported that catechins and epigallocatechin form green tea leaves have profound antiviral effects [34]. A very recent in silico molecular docking study revealed that piperine from black pepper can act as a potent inhibitor of the antiviral enzymes of dengue and Ebola viruses [35].
Molecular docking is a computational technique, widely used for the study of molecular recognition, prediction of binding mode and binding affinity of complexes formed by two or more known structures. It has become a widely accepted tool for drug discovery [[36], [37], [38], [39], [40]]. This high throughput technique can screen a variety of available drugs to identify potential drugs for novel diseases as well as to predict the adverse effects of novel drugs in a very short time [[41], [42], [43], [44]]. Development of novel drugs is a time-consuming process and generally several years of work is required for clinical approval [45]. Drug repositioning, also known as repurposing, is an effective strategy to combat novel diseases caused by infectious agents that spread rapidly [[46], [47], [48]]. Drugs that have been approved for some disease, are safe for human use [49], and only their effectiveness against the disease of interest needs to be established [50]. In life-threatening cases, where there is no alternative medicine or vaccine, such drug repurposing strategy is particularly attractive. However, clinical trials are necessary to ensure that such treatment is better than a placebo [51,52].
An in silico screening of herbal medicines for treatment of COVID-19 has also been reported [53]. Although several clinical trials are in progress to assess the potential effects of putative therapeutic agents, very limited data is available publicly regarding the in vitro and in vivo activities of the drugs that are currently involved in clinical trials for treatment of COVID-19 [25]. It has been reported that chloroquine phosphate shows anti-COVID-19 activity [54]. Several clinical trials are assessing the potential of protease inhibitors such as lopinavir and ritonavir that have been approved for treatment of other viral infections. Lopinavir and ritonavir were identified in earlier studies to target the Mpro of the SARS virus.
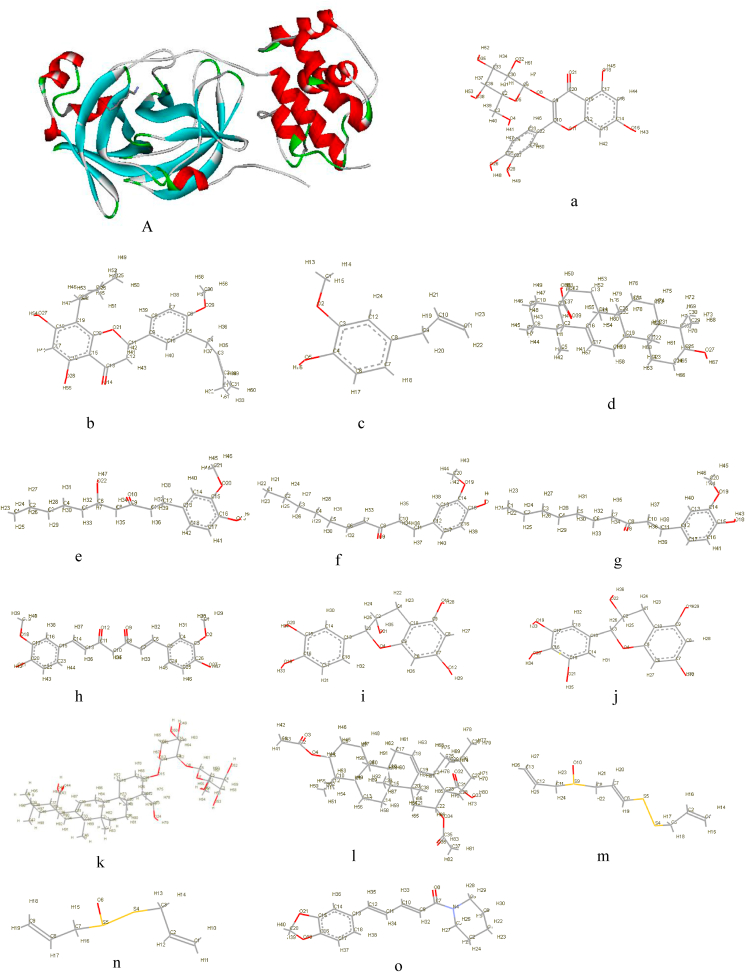
In the present study, we utilized the recently available high resolution experimental structure of the main protease (Mpro) of SARS-CoV-2 (Fig. 1) [55], as the target for molecular docking-based virtual screening. The predictions of this study will provide information that can be utilized for choice of candidate drugs for in vitro, in vivo and clinical trials.
Fig. 1.
3D cartoon structure of SARS-CoV-2 main protease and all 15 ligands in stick form. A) Main protease a) Hyperoside b) Nimbaflavone c) Eugenol d) Ursolic acid ;e) 6-gingerol f) 6-shogaol g) 6-paradol h) Curcumin i) Catechins j) Epigallocatechin k) α-Hederin l) Echinocystic acid diacetate m) Ajoene n) Allicin o) Piperine.
2. Materials and methods
2.1. Protein and chemical compounds
The X-ray crystal structure of COVID-19 Mpro in complex with an inhibitor N3 (PDB ID: 6LU7) [56], having 2.16 Å resolution was downloaded from RCSB Protein Data Bank (PDB) [57]. We collected Simplified Molecular Input Line Entry System (SMILES) code for 15 ligand molecules and then 3D structures were generated and downloaded in PDB format from CORINAClassic (https://www.mn-am.com/online_demos/corina_demo).
For hyperoside and nimbaflavone, SMILE codes were downloaded from Neem Metabolites Structure Database [58]. For rest of the ligands, SMILE codes were collected from DrugBank (https://www.drugbank.ca/) and PubChem (https://pubchem.ncbi.nlm.nih.gov/).
This in silico study was carried out by using AutoDock suite 4.2.6 for protein and ligand preparations, PyRx 0.8 for grid generation and protein-ligand docking, PyMol V1.7.4 (Educational use only) and Discovery Studio 2.5.5 for visualization of docking result and Ligplot + version 1.4.5 for generation of 2D Ligplot. The entire computational study was done in Windows 8.1 operating system.
2.2. Protein preparation
SARS-CoV-2 Mpro was prepared for molecular docking study by using AutoDock suite 4.2.6 software. Protein preparation wizard has the following steps: removal of water molecules, removal of inhibitor N3 from protein structure to obtained fresh protein for docking, addition of polar hydrogens, and addition of Kollman charges. After preparation protein molecule was exported in Protein Data Bank, Partial Charge (Q), & Atom Type (T) (PDBQT) format from AutoDock.
2.3. Ligand preparation
All 15 ligands (Fig. 1) were prepared for docking using AutoDock suite 4.2.6 software. Ligand preparation has the following steps: addition of hydrogen atoms, removal of unwanted molecules, addition of all hydrogens, computation of Gasteiger charges, merging of non-polar hydrogens, generating ionization states at pH 7, tautomers, geometric characteristics, and low-energy ring conformations. After preparation, ligand molecules were exported in PDBQT format for docking in PyRx 0.8 software.
2.4. Grid generation
The grid generation process was done in PyRx 0.8 software. It provided a square block at the active site of the protein for the accurate binding score with thermodynamic optimal energy. A grid box size of x = 27.9382928446, y = 28.2467551684, and z = 30.0038760533 Å points was generated to cover active amino acid residues that are important for docking [55]. The grid was centered at x,y,z coordinates of −13.9467660792, 12.664485092, 68.4908850063.
2.5. Molecular docking
Molecular docking was conducted with PyRx 0.8 software which uses AutodockVina wizard at the backend. 15 ligands were docked with generated grid of prepared protein. The exhaustiveness parameter that controls the extent of the search was chosen as 8, and 9 modes were generated for each ligand. The best ligand pose selection for the receptor was done based on the docking score and lowest Root Mean Square Deviation (RMSD) value.
2.6. Validation of docking score
For validation of docking results, we used another web server Webina 1.0.2 by Durrant Lab [59] with exactly same parameters. The best ligand pose selection for the receptor was done based on the docking score and lowest RMSD value.
2.7. Calculation of inhibition constant [Ki] from binding energy
After docking, best ligand poses for all 15 ligands were selected on the basis of binding energy ΔGb (kcal/mol) and RMSD values. Then inhibition constant (Ki) (nM) were calculated from binding energy.
2.8. Molecular dynamics simulation study
The protein and docked protein-ligand complexes were subjected to Molecular Dynamics (MD) Simulation using MDWeb web portal [60]. GROMACS FULL MD setup was performed using AMBER-99SB∗ force field. Simple Box Solvent Molecular Dynamics (NPT) operations were performed using the following settings: Total time 2 fs, Temperature 300 K, Total time 10 ns, Output frequency 500 steps and Total 10,000 snapshots, to obtained MD trajectory file. Water molecules and ions were removed from trajectories to obtained dry trajectory. The RMSD and B-factor fluctuations along the residues were calculated for the protein and all docked protein-ligand complexes and plotted to compare the protein backbone stability. Due to limitation in computing power, no further analyses were performed.
3. Results
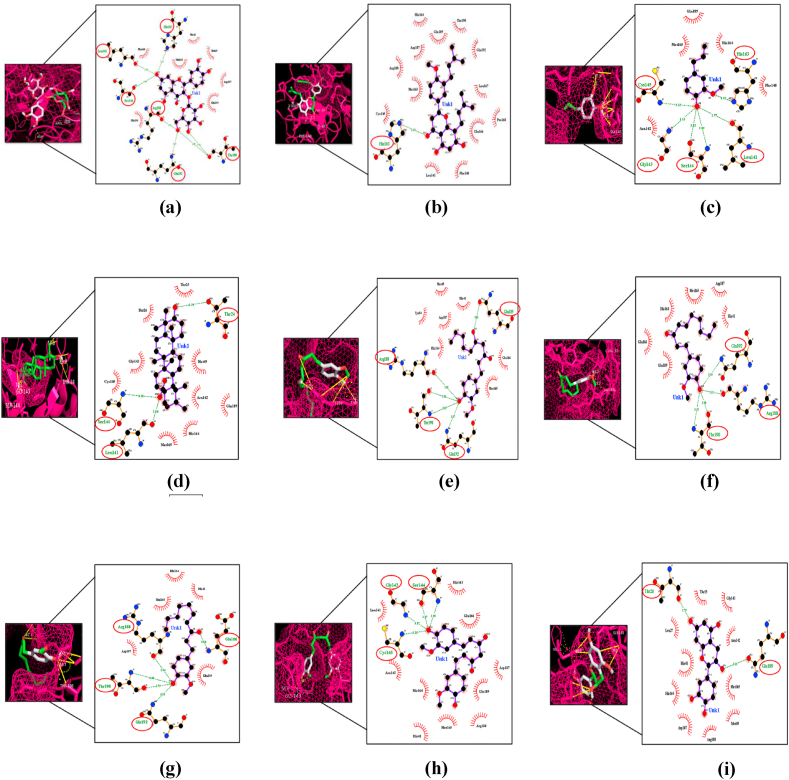
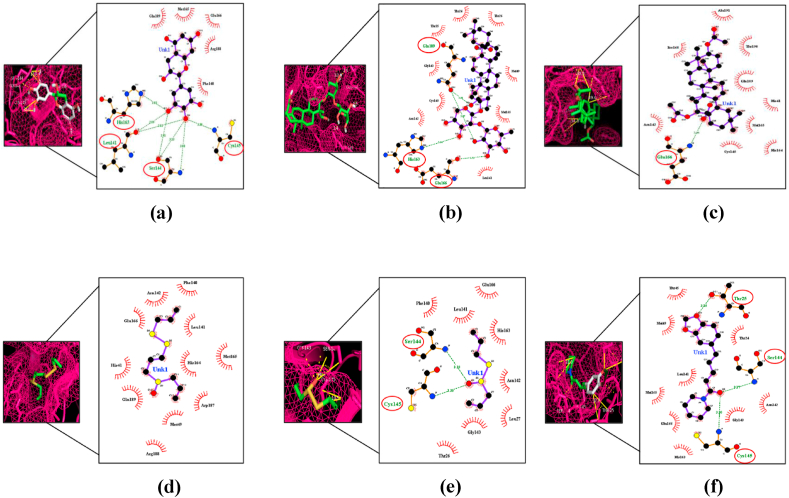
This study was done to identify possible compounds that can bind to the Mpro, which may be used as a potential drug target for SARS-CoV-2. We tested 15 bioactive compounds from Indian spices and medicinal plants that have been previously reported for their antiviral activity [28,31,34,[61], [62], [63], [64], [65]]. These compounds can bind with the Mpro with a docking score of −8.7 to −3.6 kcal/mol (Table 1). Ursolic acid (Drugbank ID DB15588), a compound of Tulsi, reported to have antiviral activity [29], had highest docking score −8.7 (kcal/mol) than others (Table 1), and formed three hydrogen bonds (H-bonds) with Thr24, Leu141∗, Ser144∗ residues of Mpro (Fig. 2). Hyperoside and nimbaflavone were predicted to have a docking score of −8.6 and −8.0 kcal/mol (Table 1). Hyperoside forms six H-bonds with Leu141∗, Ser144∗, His163∗, Arg188∗, Thr190∗, Gln192 residues whereas nimbaflavone exhibited single H-bond with His163∗ residue of SARS-CoV-2 Mpro (Fig. 2). Eugenol (DrugBank ID DB09086), the principal bioactive compound of clove, exhibited a docking score of −4.9 kcal/mol (Table 1), and formed six H-bonds with Leu141∗, Gly143, Ser144∗, Cys145, His163∗ residues of viral Mpro (Fig. 2). Natural compounds of ginger i.e., 6-gingerol (PubChem ID 442793), 6-shogaol (PubChem ID 5281794) and 6-paradol (PubChem ID 94378) were predicted to have a docking score of −5.8, −5.8 and −5.7 kcal/mol (Table 1). 6-Gingerol forms five interacting H-bonds with Arg188∗, Gln189, Thr190∗, Gln192 residues, 6-shogaol forms four interacting H-bonds with three residues of SARS-CoV-2 Mpro (Arg188∗, Thr190∗, Gln192), whereas 6-paradol exhibited five interacting H-bonds with Glu166, Arg188∗, Thr190∗, Gln192 residues of the viral Mpro (Fig. 2). Curcumin (DrugBank ID DB11672), the active compound of turmeric, had a docking score of −7.1 kcal/mol (Table 1) and formed four H-bonds with two interacting residues of Mpro (Gly143∗, Ser144∗) (Fig. 2). Two main naturally occurring compounds of tea plant, catechins (PubChem ID 1203) and epigallocatechin (DrugBank ID DB03823), predicted to have a docking score of −7.1 kcal/mol and −7.3 kcal/mol respectively (Table 1). Catechins formed two H-bonds with Thr26, Gln189 residues (Fig. 2) of viral Mpro whereas epigallocatechin exhibited seven interacting H-bonds with Leu141∗, Ser144∗, Cys145, His163∗ residues of Mpro (Fig. 3). α-Hederin (PubChemID73296), had a significantly higher docking score of −8.5 kcal/mol (Table 1), and formed four interacting H-bonds with His163∗, Glu166, Gln189 residues of SARS-CoV-2 Mpro (Fig. 3). Echinocystic acid diacetate (PubChem ID 476534), a triterpenoid saponin compound, had a docking score prediction of −6.7 kcal/mol (Table 1), and exhibited single interacting H-bond with Glu166 residue of Mpro (Fig. 3). Two main natural compounds of garlic i.e, ajoene (PubChem ID 5386591) and allicin (DrugBank ID DB11780) were predicted to have a docking score of −4.1 kcal/mol and −3.6 kcal/mol (Table 1). Allicin formed two interacting H-bonds with Ser144∗, Cys145 residues of Mpro whereas ajoene did not form any H-bond with SARS-CoV-2 Mpro (Fig. 3). Piperine (DrugBank ID DB12582), a naturally occurring bioactive compound of black pepper, had a docking score prediction −6.8 kcal/mol (Table 1). Piperine formed three interacting H-bonds with Thr25, Ser144∗, Cys145 residues of viral Mpro (Fig. 3). Docking results were validated by Webina 1.0.2 web server using the same docking parameters. Not much significant difference was observed between binding affinity predictions by PyRx 0.8 and Webina 1.0.2 web server by Durrant lab (Table 3).
Table 1.
Summary of all 15 bioactive compounds screened against SARS CoV-2 Mpro with their respective source, binding energy, interacting residues and inhibition constant.
| Compounds | Source | SMILE code collected from | AutoDock Binding Energy ΔGb (kcal/mol) |
No. of H-bonds | Interacting Residues | Inhibition constant (Ki) (nM) |
|---|---|---|---|---|---|---|
| Hyperoside | Neem(Azadirachtaindica) | Neem Metabolite Structure Database | −8.6 | 6 | Leu141a, Ser144a, His163a, Arg188a, Thr190a, Gln192 |
494.36 |
| Nimbaflavone | Neem(Azadirachtaindica) | Neem Metabolite Structure Database | −8 | 1 | His163a | 1370.95 |
| Eugenol | Clove (Syzygiumaromaticum) | DrugBank, DB09086 |
−4.9 | 6 | Leu141a, Gly143, Ser144a, Cys145, His163a |
256085.29 |
| Ursolic acid | Tulsi or holy basil (Ocimum sanctum) | Drugbank, DB15588 |
−8.7 | 3 | Thr24, Leu141a, Ser144a |
421.27 |
| 6-gingerol | Ginger (Zingiberofficinale) | PubChem, 442793 |
−5.8 | 5 | Arg188a, Gln189, Thr190a, Gln192 |
56008.89 |
| 6-shogaol | Ginger (Zingiberofficinale) | PubChem, 5281794 |
−5.8 | 4 | Arg188a, Thr190a, Gln192 |
56008.89 |
| 6-paradol | Ginger (Zingiberofficinale) | PubChem, 94378 |
−5.7 | 5 | Glu166, Arg188, Thr190a, Gln192 |
66387.61 |
| Curcumin | Turmeric (Curcuma longa) | DrugBank, DB11672 |
−7.1 | 4 | Gly143, Ser144a |
6268.33 |
| Catechins | Tea plant (Camellia sinensis) | PubChem, 1203 |
−7.1 | 2 | Thr26, Gln189 |
6268.33 |
| Epigallocatechin | Tea plant (Camellia sinensis) | DrugBank, DB03823 (EXPT01331) |
−7.3 | 7 | Leu141a, Ser144a, Cys145, His163a |
4461.61 |
| α-Hederin | Black cumin (Nigella sativa) |
PubChem, 73296 |
−8.5 | 4 | His163a, Glu166a, Gln189 |
585.97 |
| Echinocystic acid diacetate | Sponge gourd Luffa cylindrica |
PubChem, 476534 |
−6.7 | 1 | Glu166 | 12249.81 |
| Ajoene | Garlic (Allium sativum) | PubChem, 5386591 |
−4.1 | – | – | 987829.94 |
| Allicin | Garlic (Allium sativum) | DrugBank, DB11780 | −3.6 | 2 | Ser144a, Cys145 |
2288176.65 |
| Piperine | Black pepper (Piper nigrum) | DrugBank, DB12582 | −6.8 | 3 | Thr25, Ser144a, Cys145 |
10334.73 |
-: Nil.
Hotspot residue.
Fig. 2.
The binding site of SARS CoV-2 Mpro, represented as a mesh, shows ligand interactions. Amino acid residues that formed polar H-bonds with ligands are highlighted with red circle in Ligplot. (a) Hyperoside (b) Nimbaflavone (c) Eugenol (d) Ursolic acid (e) 6-gingerol (f) 6-shogaol (g) 6-paradol (h) Curcumin (i) Catechins.
Fig. 3.
The binding site of SARS CoV-2 Mpro, represented as a mesh, shows ligand interactions. Amino acid residues that formed polar H-bonds with Ligands are highlighted with red circle in Ligplot. (a) Epigallocatechin (b) α-Hederin (c) Echinocystic acid diacetate (d) Ajoene (e) Allicin (f) Piperine.
Table 3.
Predicted binding energies (Kcal/mol) of all 15 phytochemicals and 3 compounds in PyRx 0.8 and Webina 1.0.2
| Compounds | Binding Energy ΔGb (kcal/mol) |
|
|---|---|---|
| Docking with PyRx 0.8 | Docking with Webina 1.0.2 | |
| Hyperoside | −8.6 | −8.6 |
| Nimbaflavone | −8 | −8 |
| Eugenol | −4.9 | −4.9 |
| Ursolic acid | −8.7 | −8.7 |
| 6-gingerol | −5.8 | −5.6 |
| 6-shogaol | −5.8 | −5.8 |
| 6-paradol | −5.7 | −5.3 |
| Curcumin | −7.1 | −6.9 |
| Catechins | −7.1 | −7.1 |
| Epigallocatechin | −7.3 | −7.3 |
| α-Hederin | −8.5 | −8.6 |
| Echinocystic acid diacetate | −6.7 | −6.9 |
| 6-gingerol | −4.1 | −4.2 |
| 6-shogaol | −3.6 | −3.5 |
| 6-paradol | −6.8 | −7 |
| Ajoene | −4.9 | −4.9 |
| Allicin | −4.1 | −4.1 |
| Piperine | −3.6 | −3.7 |
| quercetin | −7.4 | −7.4 |
| Remdesivir | −7.7 | −7.6 |
| PF-00835231 | −8.4 | −8.4 |
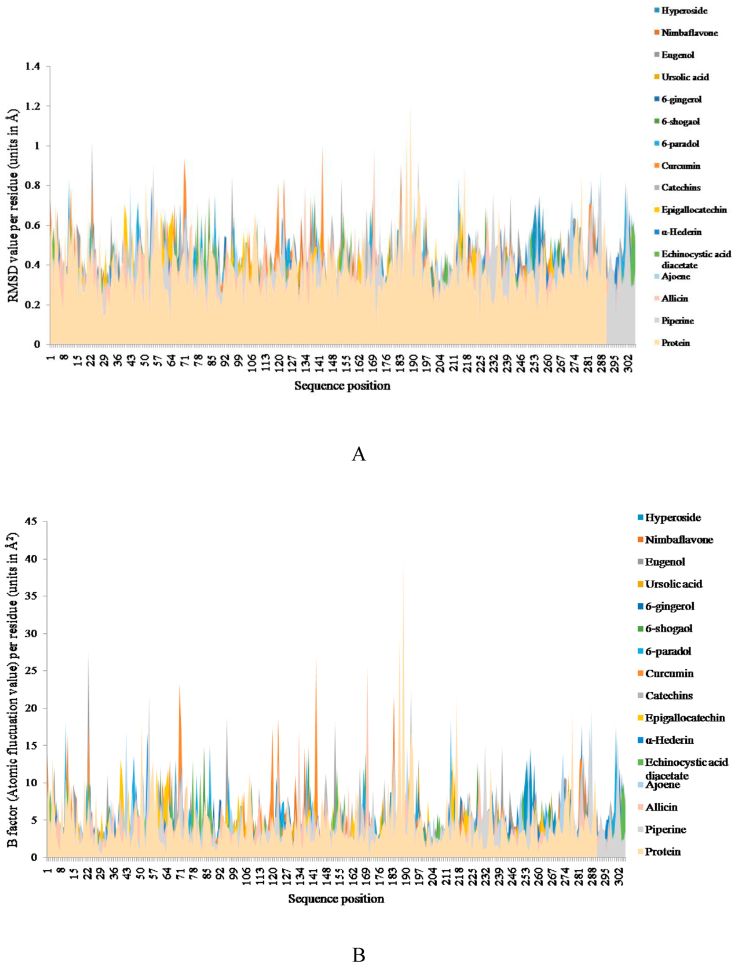
We selected all 15 ligands to simulate using MDWeb web portal [60]. To evaluate the stability of protein-ligand complexes, the RMSD of the protein backbone and protein-ligand complex were calculated during a 10 ns MD trajectory. The RMSD fluctuation per residue showed that protein-ligand complexes were stable (Fig. 4A) which indicates that the protein-ligand complexes remain intact during simulation process. As shown in Fig. 4A most of the values of RMSD were fluctuated between 0.4 Å and 1.2 Å, with an average of 0.7 Å.
Fig. 4.
A) Plots of Root Mean Square Deviation (RMSD) fluctuations per residue in Protein and all 15 protein-ligand complexes. B) Plots of B-factor fluctuations per residue in Protein and all 15 protein-ligand complexes.
For further analysis of residual atomic flexibility, an isotropic temperature factor (B-factor) was calculated. B-factor generally reflects the mobility of each residue around its mean position. It is an important tool for analysing the dynamic stability during MD simulation process. Fig. 4B shows B-factor fluctuation per residue of protein and all protein-ligand complexes. Based on the general trend of change, B-factor fluctuation of protein-ligand complexes are nearly similar to that of protein backbone; however, some residues show higher flexibility. As shown in Fig. 4B, B-factor values were fluctuated between 5 Å2and 15 Å2 with an average of 9 Å2. Detailed information regarding the same has been provided in the Supplementary Tables S1 and S2.
3.1. Predicted binding affinity of all 15 phytochemicals compared to quercetin, remdesivir and PF-00835231 against SARS-CoV-2 Mpro
We compared docking results of all 15 phytochemicals with three well-known drugs viz., quercetin (DrugBank ID DB04216), remdesivir (DrugBank ID DB14761), and PF-00835231 (PubChem CID 11561899) that can target SARS-CoV-2 Mpro (Table 2). Quercetin had a docking score prediction −7.4 kcal/mol. Remdesivir was predicted to have a docking score of −7.7 kcal/mol. PF-00835231 compound exhibited high binding affinity for Mpro (−8.4 kcal/mol). Comparison of the docking results of all 15 phytochemicals showed that ursolic acid (−8.7 kcal/mol), hyperoside (−8.6 kcal/mol) and α-Hederin (−8.5 kcal/mol) have greater binding affinity with Mpro of SARS-CoV-2 virus than quercetin, remdesivir and PF-00835231, which are currently being used in COVID-19 treatment (Table 2).
Table 2.
Predicted binding affinity of all 15 tested phytochemicals and Quercetin, Remdesivir, PF-00835231 against SARS-CoV-2 Mpro.
| Compound | AutoDock Binding Energy ΔGb (kcal/mol) |
No. of H-bonds | Interacting Residues | Inhibition constant (Ki) (nM) |
|---|---|---|---|---|
| Ursolic acid | −8.7 | 3 | Thr24, Leu141a, Ser144a | 421.27 |
| Hyperoside | −8.6 | 6 | Leu141a, Ser144a, His163a, Arg188a, Thr190a, Gln192 |
494.36 |
| α-Hederin | −8.5 | 4 | His163a, Glu166, Gln189 | 585.97 |
| PF-00835231 | −8.4 | 8 | His41, Leu141a, Gly143a, Ser144a, Cys145, Gln189 | 3764.107 |
| Nimbaflavone | −8 | 1 | His163a | 1370.95 |
| Remdesivir | −7.7 | 5 | Phe140, Leu141a, Gly143a, Ser144a, His163a | 2260.33 |
| quercetin | −7.4 | 8 | Leu141a, Gly143a, Ser144a, Cys145, His163a | 694.55 |
| Epigallocatechin | −7.3 | 7 | Leu141a, Ser144a, Cys145, His163a |
4461.61 |
| Curcumin | −7.1 | 4 | Gly143a, Ser144a | 6268.33 |
| Catechins | −7.1 | 2 | Thr26, Gln189 | 6268.33 |
| Piperine | −6.8 | 3 | Thr25, Ser144a, Cys145 | 10334.73 |
| Echinocystic acid diacetate | −6.7 | 1 | Glu166 | 12249.81 |
| 6-gingerol | −5.8 | 5 | Arg188a, Gln189, Thr190a, Gln192 |
56008.89 |
| 6-shogaol | −5.8 | 4 | Arg188a, Thr190a, Gln192 | 56008.89 |
| 6-paradol | −5.7 | 5 | Glu166, Arg188a, Thr190a, Gln192 |
66387.61 |
| Eugenol | −4.9 | 6 | Leu141a, Gly143, Ser144a, Cys145, His163a |
256085.29 |
| Ajoene | −4.1 | Nil | Nil | 987829.94 |
| Allicin | −3.6 | 2 | Ser144a, Cys145 | 2288176.65 |
Hotspot residue.
4. Discussion
In this study, we evaluated the efficacy of 15 bioactive compounds from Indian spices and medicinal plants as potential inhibitors of SARS-CoV-2 Mpro. All these compound have some antiviral properties as reported in published literature. Hyperoside, a neem secondary metabolite, has potential effects against influenza virus nucleoprotein [28]. Ursolic acid exhibits strong antiviral activity against rotavirus [66]. 6-Gingerol and 6-paradol show high efficacy against hepatitis C virus [67]. Green tea catechins and epigallocatechin have been reported to have antiviral effects against numerous viruses such as herpes simplex virus, hepatitis B virus, hepatitis C virus, etc. [34]. Previous study reported that eugenol can inhibit human herpes virus in vitro and in vivo [62]. Curcumin can inhibit Zika and chikungunya viruses [31]. Garlic compound allicin is highly effective against human cytomegalovirus, influenza B, herpes simplex virus type 1, herpes simplex virus type 2 [68].
The molecular docking approach provides an opportunity to test different drugs against SARS-CoV-2 Mpro in combination with N3 inhibitor. Recent docking study showed that the inhibitor N3 can bind to the substrate binding pockets of new COVID-19 Mpro [56]. This substrate binding pockets are located within a cleft between domain I and II and are highly conserved among all SARS-CoV-2 Mpros which make it a good target for designing drugs for anti-COVID-19 activity [56]. Similarly, we analysed the above mentioned bioactive compounds from Indian spices and medicinal plants which may act as potential drug targets for SARS-CoV-2 Mpro. Out of the 15 bioactive compounds, 9 exhibited very high docking scores (scores > −6.5 kcal/mol) (Table 1). Highest docking score was exhibited by ursolic acid (−8.7 kcal/mol) which is the principal bioactive compound of Tulsi leaf extract. The α-Hederin exhibited second highest docking score of −8.5 kcal/mol (Table 1). Neem secondary metabolites, hyperoside and nimbaflavone exhibited docking score as −8.6 and −8.0 kcal/mol (Table 1). Curcumin exhibited docking score of −7.1 kcal/mol (Table 1). Compounds of tea plant i.e., catechins and epigallocatechin exhibited a docking score of −7.1 kcal/mol and −7.3 kcal/mol respectively (Table 1). Piperine, the bioactive compound from black pepper yielded docking score −6.8 kcal/mol (Table 1). Echinocystic acid diacetate yielded docking score −6.7 kcal/mol (Table 1). We identified five hotspot residues namely Leu141, Ser144, His163, Arg188, Thr190 (Table 1) on the sequence of SARS-CoV-2 Mprowhich exhibited effective interaction with all the tested bioactive compounds. These residues can be targeted for potential drug designing to block SARS-CoV-2 Mpro. In addition, we compared (Table 2) the binding capacity of these tested bioactive compounds with widely used popular drugs against SARS-CoV-2 infection across the world. These widely used drugs are quercetin, remdesivir, PF-00835231. Interestingly, three of our tested compounds namely ursolic acid, hyperoside, α-hederin exhibited stronger binding and inhibitory potential against SARS-CoV-2 Mpro compared to quercetin, remdesivir, PF-00835231.
Our study suffers from some potential limitations. It would have been better to use GROMACS full package for entire molecular dynamics simulation analyses. We did not use MM/PBSA and MM/GBSA program for calculating free binding energy calculation of each bioactive compound. We could not determine the inhibitory effects in term of IC50 value for each of these tested drug. Moreover, our finding needs wet lab experimental validation.
5. Conclusion
In the present study, we selected and tested 15 bioactive compounds against SARS-CoV-2 Mpro. These compounds are found among Indian spices and medicinal plants and exhibit antiviral properties. Among them, nine compounds namely ursolic acid, α-Hederin, hyperoside, nimbaflavone, curcumin, catechins, epigallocatechin, piperine, and echinocystic acid diacetate exhibited very high docking score against SARS-CoV-2 Mpro. We also identified a set of hotspot residues on the peptide chain of the viral protease which are important for protein-ligand interactions and can be targeted for designing novel drugs against SARS-CoV-2 Mpro. Moreover, through comparative analyses we demonstrated that three bioactive agents have higher potential to inhibit the SARS-CoV-2 Mpro, than the drugs that are being used widely across the globe in treatment of COVID19. This comparative analyses makes us optimistic to develop herbal drugs without any side-effects in near future. This is particularly important as many cases have been reported from across the globe regarding secondary drug complications among the patients following recovery from SARC-CoV-2 infection. Our study will help researchers to carry similar analyses for other drugs and Ayurvedic bioactive agents and all these would contribute effectively to win the battle against SARS-CoV-2 infections.
Source(s) of funding
The work is financially supported by Department of Science and Technology, Government of West Bengal, India, Grant no. SG/WBDST/S&T 1000114/2016.
Conflict of interest
None.
Acknowledgement
The infrastructural support and instrumental facilities were provided by UGC-UPE II, DST-FIST, UGC-PURSE program at University of Calcutta. Pinku Halder is thankful to University Grants Commission, Government of India for providing him NET-JRF fellowship for his research.
Footnotes
Peer review under responsibility of Transdisciplinary University, Bangalore.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jaim.2021.05.003.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matsuyama S., Nagata N., Shirato K., Kawase M., Takeda M., Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J Virol. 2010;84(24):12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glowacka I., Bertram S., Müller M.A., Allen P., Soilleux E., Pfefferle S., et al. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J Virol. 2011;85(9):4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao S., Lin Q., Ran J., Musa S.S., Yang G., Wang W., et al. Preliminary estimation of the basic reproduction number of novel coronavirus (2019-nCoV) in China, from 2019 to 2020: a data-driven analysis in the early phase of the outbreak. Int J Infect Dis. 2020;92:214–217. doi: 10.1016/j.ijid.2020.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lai C.-C., Shih T.-P., Ko W.-C., Tang H.-J., Hsueh P.-R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int J Antimicrob Agents. 2020;55(3):105924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5(4):536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yadav P.D., Potdar V.A., Choudhary M.L., Nyayanit D.A., Agrawal M., Jadhav S.M., et al. Full-genome sequences of the first two SARS-CoV-2 viruses from India. Indian J Med Res. 2020;151(2 & 3):200–209. doi: 10.4103/ijmr.IJMR_663_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paraskevis D., Kostaki E.G., Magiorkinis G., Panayiotakopoulos G., Sourvinos G., Tsiodras S. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect Genet Evol. 2020;79:104212. doi: 10.1016/j.meegid.2020.104212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang X., Wu C., Li X., Song Y., Yao X., Wu X., et al. On the origin and continuing evolution of SARS-CoV-2. Natl Sci Rev. 2020;7(6):1012–1023. doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thiel V., Ivanov K.A., Putics Á., Hertzig T., Schelle B., Bayer S., et al. Mechanisms and enzymes involved in SARS coronavirus genome expression. J Gen Virol. 2003;84(Pt 9):2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- 14.Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z., et al. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc Natl AcadSci USA. 2003;100(23):13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hilgenfeld R., Peiris M. From SARS to MERS: 10 years of research on highly pathogenic human coronaviruses. Antivir Res. 2013;100(1):286–295. doi: 10.1016/j.antiviral.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020;10(5):766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma C., Sacco M.D., Hurst B., Townsend J.A., Hu Y., Szeto T., et al. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020;30(8):678–692. doi: 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ziebuhr J., Snijder E.J., Gorbalenya A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J Gen Virol. 2000;81(Pt 4):853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- 19.Hegyi A., Ziebuhr J. Conservation of substrate specificities among coronavirus main proteases. J Gen Virol. 2002;83(Pt 3):595–599. doi: 10.1099/0022-1317-83-3-595. [DOI] [PubMed] [Google Scholar]
- 20.Du Q.-S., Wang S.-Q., Zhu Y., Wei D.-Q., Guo H., Sirois S., et al. Polyprotein cleavage mechanism of SARS CoVMpro and chemical modification of the octapeptide. Peptides. 2004;25(11):1857–1864. doi: 10.1016/j.peptides.2004.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang P.-H. Characterization and inhibition of SARS-coronavirus main protease. Curr Top Med Chem. 2006;6(4):361–376. doi: 10.2174/156802606776287090. [DOI] [PubMed] [Google Scholar]
- 22.Yang H., Bartlam M., Rao Z. Drug design targeting the main protease, the Achilles' heel of coronaviruses. Curr Pharmaceut Des. 2006;12(35):4573–4590. doi: 10.2174/138161206779010369. [DOI] [PubMed] [Google Scholar]
- 23.Zhu L., George S., Schmidt M.F., Al-Gharabli S.I., Rademann J., Hilgenfeld R. Peptide aldehyde inhibitors challenge the substrate specificity of the SARS-coronavirus main protease. Antivir Res. 2011;92(2):204–212. doi: 10.1016/j.antiviral.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368(6489):409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu H. Drug treatment options for the 2019-new coronavirus (2019-nCoV) Biosci Trends. 2020;14(1):69–71. doi: 10.5582/bst.2020.01020. [DOI] [PubMed] [Google Scholar]
- 26.Wang Z., Chen X., Lu Y., Chen F., Zhang W. Clinical characteristics and therapeutic procedure for four cases with 2019 novel coronavirus pneumonia receiving combined Chinese and Western medicine treatment. Biosci Trends. 2020;14(1):64–68. doi: 10.5582/bst.2020.01030. [DOI] [PubMed] [Google Scholar]
- 27.Introduction and importance of medicinal plants and herbs National Health Portal of India. https://www.nhp.gov.in/introduction-and-importance-of-medicinal-plants-and-herbs-mtl Available from:
- 28.Ahmad A., Javed M.R., Rao A.Q., Husnain T. Designing and screening of universal drug from neem (Azadirachtaindica) and standard drug chemicals against influenza virus nucleoprotein. BMC Compl Alternative Med. 2016;16(1):519. doi: 10.1186/s12906-016-1469-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar A.H. Molecular docking of natural compounds from Tulsi (ocimum sanctum) and neem (azadirachtaindica) against SARS-CoV-2 protein targets. BEMS Reports. 2020;6(1):11–13. [Google Scholar]
- 30.Weber N.D., Andersen D.O., North J.A., Murray B.K., Lawson L.D., Hughes B.G. In vitro virucidal effects of Allium sativum (garlic) extract and compounds. Planta Med. 1992;58(5):417–423. doi: 10.1055/s-2006-961504. [DOI] [PubMed] [Google Scholar]
- 31.Mounce B.C., Cesaro T., Carrau L., Vallet T., Vignuzzi M. Curcumin inhibits Zika and chikungunya virus infection by inhibiting cell binding. Antivir Res. 2017;142:148–157. doi: 10.1016/j.antiviral.2017.03.014. [DOI] [PubMed] [Google Scholar]
- 32.Padilla-S L., Rodríguez A., Gonzales M.M., Gallego G.J.C., Castaño O.J.C. Inhibitory effects of curcumin on dengue virus type 2-infected cells in vitro. Arch Virol. 2014;159(3):573–579. doi: 10.1007/s00705-013-1849-6. [DOI] [PubMed] [Google Scholar]
- 33.Kutluay S.B., Doroghazi J., Roemer M.E., Triezenberg S.J. Curcumin inhibits herpes simplex virus immediate-early gene expression by a mechanism independent of p300/CBP histone acetyltransferase activity. Virology. 2008;373(2):239–247. doi: 10.1016/j.virol.2007.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu J., Xu Z., Zheng W. A review of the antiviral role of green tea catechins. Molecules. 2017;22(8):1337. doi: 10.3390/molecules22081337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nag A., Chowdhury R.R. Piperine, an alkaloid of black pepper seeds can effectively inhibit the antiviral enzymes of Dengue and Ebola viruses, an in silico molecular docking study. Virusdisease. 2020;31(3):308–315. doi: 10.1007/s13337-020-00619-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marshall G.R. Computer-aided drug design. Annu Rev Pharmacol Toxicol. 1987;27:193–213. doi: 10.1146/annurev.pa.27.040187.001205. [DOI] [PubMed] [Google Scholar]
- 37.Leelananda S.P., Lindert S. Computational methods in drug discovery. Beilstein J Org Chem. 2016;12:2694–2718. doi: 10.3762/bjoc.12.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuntz I.D. Structure-based strategies for drug design and discovery. Science. 1992;257(5073):1078–1082. doi: 10.1126/science.257.5073.1078. [DOI] [PubMed] [Google Scholar]
- 39.Macalino S.J.Y., Gosu V., Hong S., Choi S. Role of computer-aided drug design in modern drug discovery. Arch Pharm Res (Seoul) 2015;38(9):1686–1701. doi: 10.1007/s12272-015-0640-5. [DOI] [PubMed] [Google Scholar]
- 40.Talluri S. Computational protein design of bacteriocins based on structural scaffold of aureocin A53. Int J Bioinf Res Appl. 2019;15(2):129. [Google Scholar]
- 41.Jones G., Willett P., Glen R.C., Leach A.R., Taylor R. Development and validation of a genetic algorithm for flexible docking. J Mol Biol. 1997;267(3):727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- 42.Friesner R.A., Banks J.L., Murphy R.B., Halgren T.A., Klicic J.J., Mainz D.T., et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47(7):1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 43.Irwin J.J., Shoichet B.K. Docking screens for novel ligands conferring new biology. J Med Chem. 2016;59(9):4103–4120. doi: 10.1021/acs.jmedchem.5b02008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mohanasundaram N., Sekhar T. Computational studies of molecular targets regarding the adverse effects of isoniazid drug for tuberculosis. Curr Pharmacogenomics Personalized Med (CPPM) 2019;16(3):210–218. [Google Scholar]
- 45.Fons N. Focus: drug development: textbook of drug design and discovery, fifth edition. Yale J Biol Med. 2017;90(1):160. [Google Scholar]
- 46.Ashburn T.T., Thor K.B. Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov. 2004;3(8):673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 47.Li J., Zheng S., Chen B., Butte A.J., Swamidass S.J., Lu Z. A survey of current trends in computational drug repositioning. Briefings Bioinf. 2016;17(1):2–12. doi: 10.1093/bib/bbv020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chopra G., Samudrala R. Exploring polypharmacology in drug discovery and repurposing using the CANDO platform. Curr Pharmaceut Des. 2016;22(21):3109–3123. doi: 10.2174/1381612822666160325121943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H., Wang Y.M., Xu J.Y., Cao B. [Potential antiviral therapeutics for 2019 novel coronavirus] Zhonghua Jiehe He Huxi Zazhi. 2020;43(3):170–172. doi: 10.3760/cma.j.issn.1001-0939.2020.03.004. [DOI] [PubMed] [Google Scholar]
- 50.Hurle M.R., Yang L., Xie Q., Rajpal D.K., Sanseau P., Agarwal P. Computational drug repositioning: from data to therapeutics. Clin Pharmacol Ther. 2013;93(4):335–341. doi: 10.1038/clpt.2013.1. [DOI] [PubMed] [Google Scholar]
- 51.Ding X. Drug screening: drug repositioning needs a rethink. Nature. 2016;535(7612):355. doi: 10.1038/535355d. [DOI] [PubMed] [Google Scholar]
- 52.Novac N. Challenges and opportunities of drug repositioning. Trends Pharmacol Sci. 2013;34(5):267–272. doi: 10.1016/j.tips.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 53.Zhang D.-H., Wu K.-L., Zhang X., Deng S.-Q., Peng B. In silico screening of Chinese herbal medicines with the potential to directly inhibit 2019 novel coronavirus. J Integr Med. 2020;18(2):152–158. doi: 10.1016/j.joim.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gao J., Tian Z., Breakthrough Yang X. Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci Trends. 2020;14(1):72–73. doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- 55.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., et al. Structure of Mpro from COVID-19 virus and discovery of its inhibitors. BioRxiv. 2020 doi: 10.1101/2020.02.26.964882. [DOI] [Google Scholar]
- 56.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582(7811):289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 57.Burley S.K., Berman H.M., Kleywegt G.J., Markley J.L., Nakamura H., Velankar S. Protein data bank (PDB): the single global macromolecular structure archive. Methods Mol Biol. 2017;1607:627–641. doi: 10.1007/978-1-4939-7000-1_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hatti K.S., Muralitharan L., Hegde R., Kush A. Neemdb: convenient database for neem secondary metabolites. Bioinformation. 2014;10(5):314–315. doi: 10.6026/97320630010314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kochnev Y., Hellemann E., Cassidy K.C., Durrant J.D. Webina: an open-source library and web app that runs AutoDockVina entirely in the web browser. Bioinformatics. 2020;36(16):4513–4515. doi: 10.1093/bioinformatics/btaa579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hospital A., Andrio P., Fenollosa C., Cicin-Sain D., Orozco M., Gelpí J.L. MDWeb and MDMoby: an integrated web-based platform for molecular dynamics simulations. Bioinformatics. 2012;28(9):1278–1279. doi: 10.1093/bioinformatics/bts139. [DOI] [PubMed] [Google Scholar]
- 61.Chang J.S., Wang K.C., Yeh C.F., Shieh D.E., Chiang L.C. Fresh ginger (Zingiberofficinale) has anti-viral activity against human respiratory syncytial virus in human respiratory tract cell lines. J Ethnopharmacol. 2013;145(1):146–151. doi: 10.1016/j.jep.2012.10.043. [DOI] [PubMed] [Google Scholar]
- 62.Cortés-Rojas D.F., de Souza C.R.F., Oliveira W.P. Clove (Syzygiumaromaticum): a precious spice. Asian Pac J Trop Biomed. 2014;4(2):90–96. doi: 10.1016/S2221-1691(14)60215-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Astani A., Reichling J., Schnitzler P. Screening for antiviral activities of isolated compounds from essential oils. Evid Based Complement Alternat Med. 2011;2011:253643. doi: 10.1093/ecam/nep187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mikaili P., Maadirad S., Moloudizargari M., Aghajanshakeri S., Sarahroodi S. Therapeutic uses and pharmacological properties of garlic, shallot, and their biologically active compounds. Iran J Basic Med Sci. 2013;16(10):1031–1048. [PMC free article] [PubMed] [Google Scholar]
- 65.Prasad S., Tyagi A.K. Ginger and its constituents: role in prevention and treatment of gastrointestinal cancer. Gastroenterol Res Pract. 2015;2015:142979. doi: 10.1155/2015/142979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tohmé M.J., Giménez M.C., Peralta A., Colombo M.I., Delgui L.R. Ursolic acid: a novel antiviral compound inhibiting rotavirus infection in vitro. Int J Antimicrob Agents. 2019;54(5):601–609. doi: 10.1016/j.ijantimicag.2019.07.015. [DOI] [PubMed] [Google Scholar]
- 67.Abd El-Wahab A., El-Adawi H., El-Demellawy M. In vitro study of the antiviral activity of Zingiberofficinale. Planta Med. 2009;75(9) doi: 10.1055/s-0029-1234649. [DOI] [Google Scholar]
- 68.Ankri S., Mirelman D. Antimicrobial properties of allicin from garlic. Microb Infect. 1999;1(2):125–129. doi: 10.1016/s1286-4579(99)80003-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.