Summary
Knowledge of the epitopes of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) targeted by T cells in recovered (convalescent) individuals is important for understanding T cell immunity against coronavirus disease 2019 (COVID-19). This information can aid development and assessment of COVID-19 vaccines and inform novel diagnostic technologies. Here, we provide a unified description and meta-analysis of SARS-CoV-2 T cell epitopes compiled from 18 studies of cohorts of individuals recovered from COVID-19 (852 individuals in total). Our analysis demonstrates the broad diversity of T cell epitopes that have been recorded for SARS-CoV-2. A large majority are seemingly unaffected by current variants of concern. We identify a set of 20 immunoprevalent epitopes that induced T cell responses in multiple cohorts and in a large fraction of tested individuals. The landscape of SARS-CoV-2 T cell epitopes we describe can help guide immunological studies, including those related to vaccines and diagnostics. A web-based platform has been developed to help complement these efforts.
Keywords: SARS-CoV-2, T cells, epitopes, CD4, CD8, convalescent patients, immunodominant epitopes, immunoprevalent epitopes, population coverage, variants of concern
Graphical abstract
Highlights
-
•
Meta-analysis of T cell epitopes from 18 studies of individuals recovered from COVID-19
-
•
20 immunoprevalent SARS-CoV-2 T cell epitopes identified across multiple cohorts
-
•
Large majority of epitopes appear unaffected by current variants of concern
-
•
Web dashboard for reporting and analyzing SARS-CoV-2 T cell epitope data
Quadeer et al. provide a meta-analysis of SARS-CoV-2 T cell epitope data from 18 studies involving 852 individuals recovered from COVID-19. Their analysis highlights the characteristics of the reported epitopes and identifies 20 immunoprevalent epitopes targeted in multiple cohorts and in a majority of tested individuals. An associated web-platform is reported.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of coronavirus disease 2019 (COVID-19), has led to a global public health crisis. Development of COVID-19 vaccines and diagnostic tests are aided by an understanding of the natural protective immune responses to SARS-CoV-2. This includes humoral and cellular immunity, mediated by antibodies and T cells, respectively. A significant amount of COVID-19 research has focused on understanding antibody responses,1, 2, 3 but studies informing on the role of T cells have also started to emerge.
Initial results suggest a potential key role of T cells in protecting against COVID-19.4,5 Studies of individuals recovered from COVID-19 have detected SARS-CoV-2-specific T cells 9 months after infection,6 showing promising signs for the potential of T cells to provide lasting immunity. Longevity may be a concern for antibody responses, which have been reported to decline within few months after infection.7, 8, 9 Observations consistent with this have also been reported for the most closely related human CoV, SARS-CoV, for which T cells have been shown to persist up to 17 years after infection,10 whereas antibody responses waned after a few years.11
Characterizing SARS-CoV-2 T cell epitopes as well as their human leukocyte antigen (HLA) association is important for multiple reasons. It informs on the expected SARS-CoV-2 natural or vaccine-induced T cell responses in a population of specific ethnicity or a specific geographical region, which is tied to the composition of HLA alleles prevalent in that population. It can help with assessing the T cell responses that may be induced by COVID-19 vaccines (which currently mainly focus on the spike [S] protein12) and provide possible directions for boosting the T cell response by including specific immunodominant epitopes. It can guide vaccine assessment studies probing whether a vaccine induces T cell responses similar to those commonly generated during natural infection. It can also aid with monitoring potential viral escape from T cell responses via genetic mutations and can facilitate development of T cell-based diagnostics for distinguishing recovered from unexposed individuals. T cell-based diagnostics may have advantages over serological assays, given the uncertainties related to the appearance and persistence of SARS-CoV-2-specific antibody responses in infected individuals.7, 8, 9,13
Here we present a unified account of the current knowledge (as of April 20, 2021) of SARS-CoV-2 T cell epitopes associated with individuals recovered from COVID-19. We collate and analyze data of T cell epitopes that have been identified experimentally in independent studies of blood samples from different cohorts. These data are compiled from 18 studies (15 published, 3 preprints) of T cell responses in a total of 852 individuals recovered from COVID-19 (Table 1). Our analysis highlights the different characteristics of epitopes reported for SARS-CoV-2 and identifies a specific set of epitopes that appear to induce T cell responses in multiple cohorts and in a large fraction of tested individuals. This information regarding SARS-CoV-2 T cell epitopes can provide directions for future immunological studies. We report a web dashboard we developed to support these ongoing scientific efforts.
Table 1.
Summary of immunological studies reporting SARS-CoV-2 T cell epitopes targeted in individuals recovered from COVID-19 (as of April 20, 2021)
| No. | Studya | Geography (Country) | Total Individuals | Gender (Female/Male) | Median Age (Range) | Disease Severityb (Asymptomatic or Mild) | Disease Severityb (Moderate or Severe) | Blood Collection Time (Days)c | Initial Peptide Selection Procedured | Proteins | Total Peptides Tested | T cell Assay | Total Distinct Epitopes Identified | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Saini et al.14 | Denmark | 18 | 6/12 | 43.5 (29–82) | 7 | 11 | As close as possible to the first positive test | NetMHCpan-4.1 | all | 2,204 | multimer qualitative binding | 122 | |
| 2 | Kared et al.15 | Baltimore/ Washington, USA | 30 | 12/18 | 42.5 (19–77) | N/A | N/A | 27–62 after symptom resolution | Grifoni et al.;16 Prachar et al.17 | all | 408 | multimer qualitative binding | 46 | |
| 3 | Schulien et al.18 | Germany | 26 | 14/12 | 32.5 (24–56) | 26 | 0 | 24 (14–70) after symptom onset | ANN-4.0, SARS-CoV epitopes16 | S, N, M, ORF1ab, ORF3a | 66 | multimer qualitative binding | 37 | |
| 4 | Poran et al.19 | N/A | 3 | N/A | N/A | N/A | N/A | N/A | HLAthena | all | 23 | multimer qualitative binding | 11 | |
| 5 | Shomuradova et al.20 | Moscow, Russia | 31 | 16/15 | 35 (17–59) | 21 | 10 | 33 (17–49) after positive test/after disease onset | NetMHCpan-4.0 and identity with SARS-CoV > 60% | S | 13 | multimer qualitative binding | 10 | |
| 6 | Nielsen et al.21 | Denmark | 203 | 92/111 | 47 (21–79) | 17 | 186 | 44 (14–129) after symptom onset | N/A | S, N, M | 9 | multimer qualitative binding | 9 | |
| 7 | Chour et al.22 | USA | 2 | 0/2 | 50 (30–70) | 0 | 2 | 6.5 (2–13) post symptom onset | NetMHC-4.0 | S | 96 | multimer qualitative binding | 6 | |
| 8 | Sekine et al.23 | Sweden | 66 | 14/41 | 51 | 40 | 26 | 53.5 (IQR: 45.5–61) after symptom onset | NetMHCpan-4.1 | all | 13 | multimer qualitative binding | 4 | |
| 9 | Nguyen et al.24 | Melbourne, Australia | 7 | 1/6 | 32 (19–74) | 6 | 1 | 90 (5–145) after disease onset | overlapping peptide pools and 5 N-specific immunogenic peptides25,18,26,27 | S, N, M | N/A | ICS IFN-γ release; multimer qualitative binding | 3 | |
| 10 | Rha et al.28 | South Korea | 37 | 18/19 | 46 (21–83) | 23 | 14 | 46 (19–125) after symptom onset | N/A | S, N, M | 8 | multimer qualitative binding | 2 | |
| 11 | Ferretti et al.26 | New Jersey/Louisiana, USA | 78 | 55/23 | 19.5 (0–80) | 55 | 23 | 49 (11–111) post diagnosis | identification of 20-mer peptides by TScan screen29 followed by NetMHC-4.0 | all | ~240 | Single-allele ELISA IFN-γ; multimer qualitative bindinge | 28 | |
| 12 | Nelde et al.30 | Germany | 116 | 55/61 | 44 (18–75) | 36 | 80 | 37.7 (19–52) after positive test | SYFPEITHI-1.0, NetMHCpan-4.0 | all | 120 | ICS IFN-γ release; ELISPOT IFN-γ release | 47 | |
| 13 | Hu et al.31 | Chongqing, China | 37 | 16/21 | 47 (20–67) | 34 | 3 | N/A | NetMHCpan-4.0, SARS-CoV epitopes32,16 | S, N | 78 | ELISPOT IFN-γ release | 15 | |
| 14 | Habel et al.25 | Melbourne, Australia | 18 | 10/8 | 54 (22–76) | 11 | 7 | 47 (36–102) after disease onset | NetCTLpan, NetMHCpan | S, N, M, ORF1ab | 14 | ICS IFN-γ release | 14 | |
| 15 | Lineburg et al.33 | Queensland, Australia | 37 | 23/14 | 51 (20–75) | 7 | 5 | 62 (46–124) after positive test | overlapping peptide pools followed by peptide matrix analysis | all | N/A | ICS IFN-γ release | 4 | |
| 16 | Lee et al.34 | New South Wales, Australia | 2 | N/A | N/A | 0 | 2 | 30–60 after symptom resolution | Network analysis35 followed by NetMHCpan-4.1 and NetMHCIIpan-4.0 | S, N | 2 | ICS IFN-γ release | 2 | |
| 17 | Tarke et al.36 | San Diego, USA | 99 | 58/41 | 41 (19–91) | 90 | 9 | 67 (3–184) after symptom onset | NetMHCpan-4.0 | all | 7,525 | AIM assay37 | 803 | |
| 18 | Peng et al.27 | UK | 42 | 16/26 | 57 (20–95) | 28 | 14 | 42 (30–62) after symptom onset | 15- to 18-mer peptides overlapping by 10 residues, SARS-CoV epitopes32 | all except ORF1 | 450 | ELISPOT IFN-γ release | 46f | |
Studies reporting precise epitopes along with the cognate HLA information.
Definition of disease severity varies among studies.
IQR, interquartile range.
NetCTLpan,38 NetMHC-4.0,39 NetMHCpan,40 NetMHCpan-4.0,41 NetMHCpan-4.1,42 SYFPEITHI-1.0,43 ANN-4.0,44 and HLAthena.45
Six (of 28) epitopes were identified using multimer qualitative binding.
Only two (of 46) epitopes that were reported as precise epitopes with the cognate HLA information were considered.
Results and discussion
In the 18 studies we considered, the set of recovered individuals covers a population across four continents and well-distributed age, gender, disease severity, and blood collection time within and across studies (Table 1). Half of the studies have characterized T cell responses against the whole proteome, whereas the others have focused on responses mounted against subsets of SARS-CoV-2 proteins, typically involving the S and nucleocapsid [N] proteins. In the majority of cases, the T cell response was measured in blood samples of individuals against a set of peptides predicted by bioinformatics tools or earlier bioinformatics-based analyses16,17,32 (reviewed in Sohail et al.46), whereas a few studies employed overlapping peptide pools spanning the SARS-CoV-2 proteome. All 18 studies reported optimal epitopes along with cognate HLA information. Of these, 10 studies (1–10 in Table 1) experimentally determined HLA restrictions of the reported epitopes by using multimer qualitative binding. Of the remaining eight studies, one study (11 in Table 1) employed mono-allelic cell line assays to identify specific HLA-restricted epitopes, whereas others (12–18 in Table 1) inferred HLA restrictions using standard functional assays (such as activation-induced markers [AIMs] or intracellular cytokine staining [ICS]/enzyme-linked immunospot [ELISPOT] interferon γ [IFN-γ] release) and HLA haplotype information for individuals. Although the latter set of studies involved some predictive element in deconvolving the HLA allele responsible for the observed response from the individual’s haplotype, the reported HLA associations were supported by HLA binding assays or use of accurate peptide-HLA binding prediction methods.
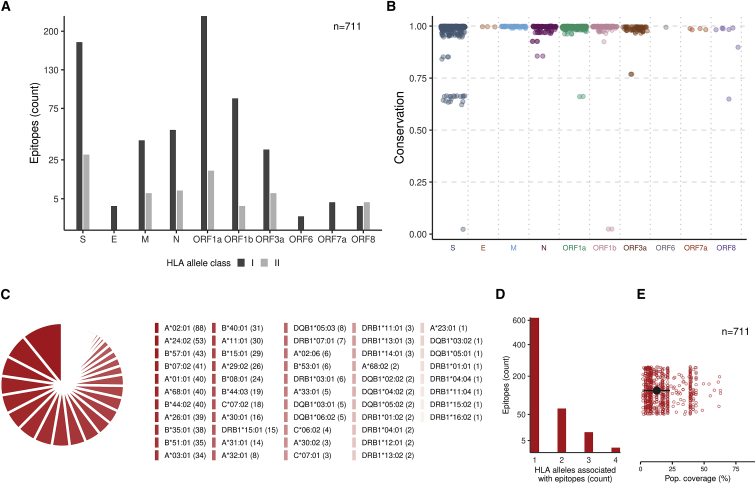
A total of 711 unique T cell epitopes with information of a cognate HLA allele have been reported in the 18 immunological studies we considered (see STAR Methods for details). Of these, 635 are CD8+ (HLA class I restricted), and 76 are CD4+ (HLA class II restricted) (Figure 1A). The set of epitopes covers each protein from the canonical reading frames of SARS-CoV-2 except open reading frame 7b [ORF7b] and ORF10. The largest numbers of epitopes fall within S and ORF1a—the most exposed protein and the longest SARS-CoV-2 ORF, respectively. These epitopes broadly cover almost the entire S protein, including the receptor-binding domain, whereas the majority of epitopes in ORF1a lie within the non-structural protein [nsp3] (PL2-PROPapain-like proteinase) and nsp4 proteins (File S1). These nsp3 and nsp4 proteins participate in assembly of virally induced cytoplasmic double-membrane vesicles, which are necessary for viral replication.47
Figure 1.
Features of SARS-CoV-2-specific T cell epitopes reported to elicit an immune response in blood samples of individuals recovered from COVID-19
(A) The number of epitopes (n = 711) according to HLA class restriction. S, spike; E, envelope; M, membrane; N, nucleocapsid.
(B) The conservation of each epitope among the global SARS-CoV-2 genetic sequences (as of April 20, 2021). Further details of the S-derived epitopes (n = 16) with low genetic conservation (<0.9) and their association with current VOCs are provided in Table S1.
(C) Diversity of HLA associations reported for SARS-CoV-2 epitopes. The number of epitopes associated with a particular HLA allele is shown in brackets.
(D) The number of HLA alleles associated with each epitope.
(E) Estimate of the global population coverage of each epitope (STAR Methods). The median is shown as a black circle and the median absolute deviation as an error bar.
All identified epitopes have high genetic conservation (>0.9) among the ∼860,000 SARS-CoV-2 sequences (as of April 20, 2021), except for 24 epitopes (Figure 1B). Of these, three epitopes (612YQDVNCTEV620 in S and 305NVLFSTVFPPTSFGP319 and 309STVFPPTSF317, in ORF1b) have very low conservation (∼0.02) because they encompass mutations (S:D614G and ORF1b:P314L, underlined) that are now dominant globally, with the former mutation reported to have increased virus infectivity and transmission.48, 49, 50 Two-thirds of the 24 epitopes (16 of 24) with low genetic conservation (<0.9) belong to S. These make up ∼8% (16 of 208) of all unique T cell epitopes derived from S—4-fold higher than the fraction of epitopes with low conservation derived from all other proteins (∼2%, 8 of 503). This can be of interest in the context of T cell responses against current COVID-19 vaccines that focus largely on the S protein, assuming that the T cell epitopes targeted in response to vaccination are similar to those seen in recovered individuals. Although limited data are currently available about epitope-specific T cell responses following vaccination, this assumption is partially supported by a study of eight S-derived epitopes reported to elicit T cell responses in vaccinated persons,51 of which we observe seven to be targeted in recovered individuals (File S1). Of the 16 S-derived T cell epitopes with low genetic conservation (<0.9), all but one encompass sites harboring mutations that define the variants of concern (VOCs) (Table S1).52,53 For example, the epitope 495YGFQPTNGV503 encompasses the N501Y mutation associated with the three VOCs B.1.1.7, B.1.351, and P.1, and the epitopes 142GVYYHKNNK150 and 144YYHKNNKSW152 encompass the deletion at Y144 associated with two variants, B.1.1.7 and B.1.525. The higher genetic variation observed in S as opposed to other proteins is, at least in part, likely to be driven by escape from neutralizing antibodies.54 Despite the potential of S-derived epitopes to escape T cells elicited to target the non-mutated epitopes (by natural infection or vaccination), responses against the significant majority of S-derived epitopes we studied are not expected to be affected by VOCs (Figure 1B). Moreover, at the level of HLA alleles, for each of the 36 HLA alleles associated with S-derived epitopes, each of them is associated with at least one conserved (>0.9) S-derived epitope (File S1). Collectively, the observed heterogeneity of T cell responses against SARS-CoV-2 provides little evidence to suggest that the observed genetic variation in the S protein may significantly affect T cell immunity, in line with a recent report.55 Escape from T cell pressure may become an important evolutionary factor when strong and diverse selective pressure is imposed by widespread vaccination, and this requires close monitoring.
The overall set of reported epitopes was associated with 52 HLA class I and II alleles (Figure 1C). Most HLA alleles are associated with multiple epitopes, with each of 21 alleles having an association with 14 epitopes or more. The same epitope may be presented by multiple HLA alleles, as evidenced by studies of the related SARS-CoV56 as well as other viruses.57 However, for the majority of SARS-CoV-2 epitopes, only a single associated HLA allele has been reported so far (Figure 1D). This appears in part to be due to the limited number of recovered COVID-19 individuals who have been studied (Table 1). The limited number of associated HLA alleles translates to a median global population coverage estimate per epitope of only 12% (Figure 1E). Thus, investigation of additional HLA alleles associated with the identified SARS-CoV-2 epitopes is required to providing a more accurate indication of their individual population coverage. An expanded list of likely HLA associations may be predicted for some of the reported epitopes by using prior knowledge of genetically matched experimentally determined T cell epitopes of SARS-CoV and their associated HLA alleles (for details, see File S1 and Figure S1). These predictions, when confirmed, would provide an increase in the median population coverage of the selected SARS-CoV-2 epitopes from 16.8% to 40%, with a few epitopes having around 60% coverage (Figure S1).
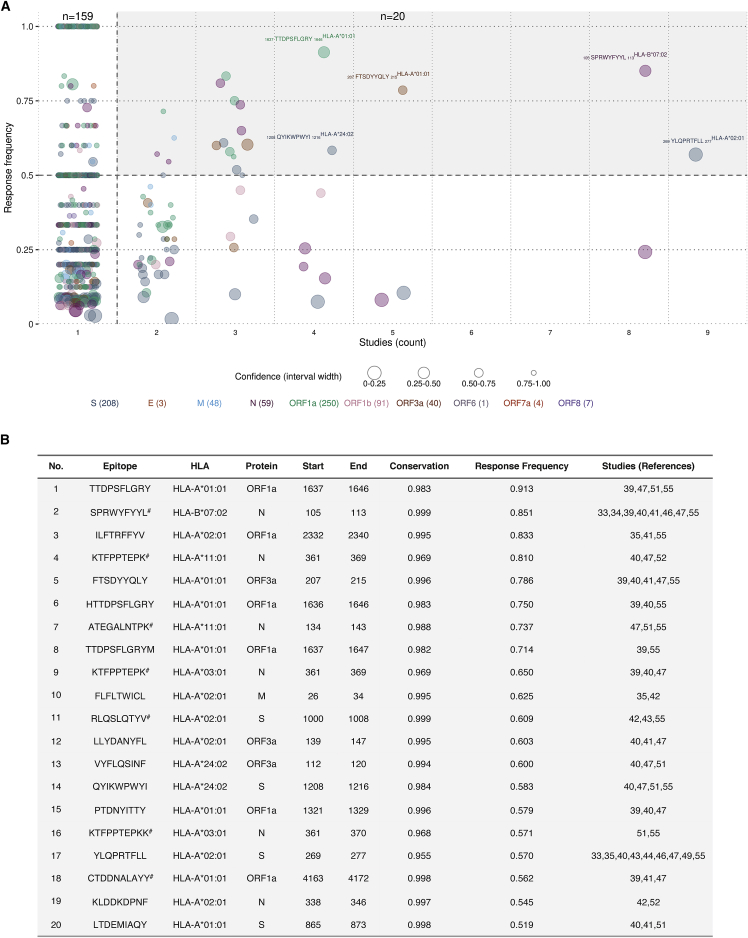
Quantifying the immunodominance of each reported epitope-HLA pair using the standard response frequency (RF) metric58, 59, 60 (STAR Methods) revealed that 179 of the reported (812) epitope-HLA pairs had an RF score exceeding 0.5, indicating that they induced T cell responses in over half of the subjects tested across studies (Figure 2A). Confidence in the estimated RF values varies with the number of tested subjects, with higher confidence being attributed to epitope-HLA pairs with larger numbers of tested subjects (STAR Methods). The majority (159 of 179) of epitope-HLA pairs with an RF exceeding 0.5 had been reported in a single study only. Among these, pairs with high confidence appear promising, but responses against them should be investigated in different cohorts to further confirm their immunodominance. Responses against the remaining (20 of 179) epitope-HLA pairs were registered in more than one study, and although per-study variation was observed in the results (Figure S2), these pairs appear to be immunoprevalent.61 This is because responses against each epitope-HLA pair were recorded for more than half of the tested recovered individuals collectively across multiple studies despite differences in characteristics of the donor cohorts (age, gender, geographical location, and disease severity), blood collection time, and methodology used to determine the epitopes (initial peptide selection procedure and T cell assay) (Table 1).
Figure 2.
Identification of immunoprevalent SARS-CoV-2 T cell epitopes
(A) Response frequency (RF) of unique epitope-HLA pairs versus the number of immunological studies reporting a T cell response against them. The size of each circle represents the confidence in the respective RF value (STAR Methods). The 20 immunoprevalent epitope-HLA pairs (having an RF exceeding 0.5 and reported in more than one study) are shown with a shaded background, and five highly immunoprevalent epitope-HLA pairs (having an RF exceeding 0.5 and reported in at least four studies) are labeled.
(B) Details of the identified 20 immunoprevalent epitope-HLA pairs (ordered according to decreasing RF). Epitope-HLA pairs matched genetically to those determined experimentally for SARS-CoV are marked (#).
All of the 20 identified immunoprevalent epitopes have high genetic conservation (>0.9) (Figure 2B), and none of them encompass mutations associated with the current VOCs. Interestingly, 35% of the immunoprevalent epitope-HLA pairs (n = 20) had an identical match to experimentally determined SARS-CoV epitope-HLA pairs (Figure 2B). This fraction of matched epitope-HLA pairs is over three times higher than that (11%) in the complete set of SARS-CoV-2 epitope-HLA pairs (p < 10−3, Fisher’s exact test), suggesting that many of the immunoprevalent T cell epitopes of SARS-CoV-2 are also cross-reactive to SARS-CoV. Of the 20 identified immunoprevalent epitope-HLA pairs, six each belonged to N and ORF1a, four to S, three to ORF3a, and one to the membrane [M] protein (Figure 2B). This indicates that ∼80% of the immunoprevalent epitopes lie in proteins other than S, suggesting that vaccine candidates targeting these proteins may have benefits in terms of T cell immunity. Of the identified immunoprevalent epitopes, five epitopes (HLA-A∗02:01-restricted 269YLQPRTFLL277 in S, HLA-B∗07:02-restricted 105SPRWYFYYL113 in N, HLA-A∗01:01-restricted 207FTSDYYQLY215 in ORF3a and 1637TTDPSFLGRY1646 in ORF1a, and HLA-A∗24:02-restricted 1208QYIKWPWYI1216 in S) appeared to be highly immunoprevalent, eliciting T cell responses in more than ∼60% of the tested individuals recovered from COVID-19 in four immunological studies or more. Collectively, around 71% of the global population is estimated to carry the associated HLA alleles and, hence, may generate a T cell response against at least one of these five epitopes. Two of these highly immunoprevalent epitopes (105SPRWYFYYL113 in N and 269YLQPRTFLL277 in S) are attracting considerable attention, and detailed analyses of T cell responses against them have been reported.24,25,33
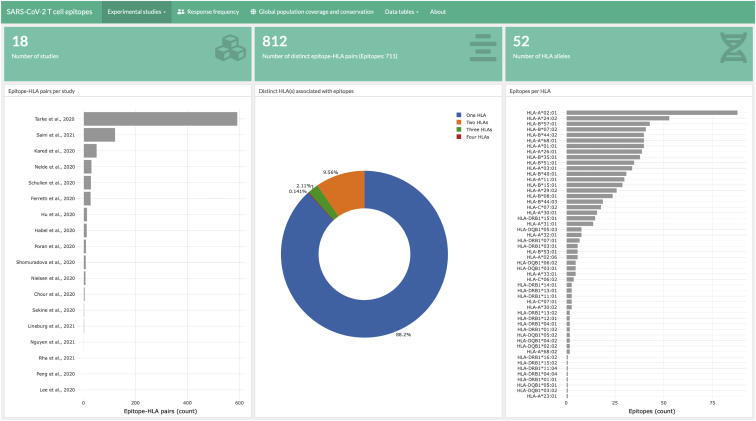
The SARS-CoV-2 T cell epitope data we compiled and report here were integrated into a web-based dashboard (Figure 3).62 This dashboard provides exportable data tables listing the SARS-CoV-2 epitopes and graphic displays to summarize different characteristics of the epitopes, including aggregate information as well as specific details of the individual epitopes. We plan to update the dashboard with new experimental information as it becomes available, with the goal of aiding further research to understand T cell responses against SARS-CoV-2 and guide studies related to COVID-19 vaccines and diagnostics. Although we focused the current study on SARS-CoV-2-specific T cell responses recorded in recovered individuals, knowledge of T cell epitopes targeted in animal studies63,64 is also informative. These may inform potential immune targets in COVID-19-infected individuals and can help guide further immunological experiments that seek to probe T cell responses that arise because of natural infection or those elicited by vaccination. The SARS-CoV-2 epitopes reported to be targeted in animal models were also incorporated into the web dashboard.
Figure 3.
Snapshot of the web dashboard developed for reporting and analyzing SARS-CoV-2 T cell epitope data (as of April 20, 2021)
The web dashboard62 provides aggregated information regarding the T cell epitopes and their HLA associations. Exportable data tables are provided to aid further research.
Overall, the data we described, based on recent experimental studies, demonstrates an impressive and diverse list of SARS-CoV-2 T cell epitopes targeted by individuals recovered from COVID-19. Subsets of these epitopes exhibit desirable properties, including high genetic conservation and high RF across multiple cohorts, and they appear to have the potential to collectively induce a T cell response in a large fraction of the population. Current knowledge of the landscape of T cell epitopes for SARS-CoV-2 is still evolving, and further studies of different cohorts of recovered individuals, encompassing a broad diversity of HLA profiles, are required to provide a more comprehensive understanding. Moreover, further systematic studies are required to ascertain possible correlates between the responses against T cell epitopes and disease protection. Knowledge of SARS-CoV-2 T cell epitopes could play an important role in contributing to the fight against COVID-19 by guiding diverse applications and novel technologies, including development, assessment, and monitoring of vaccines and development of improved diagnostic assays.
Limitations of the study
There are multiple limitations of our study. Our analysis is unable to make associations between epitope-specific T cell responses and levels of disease severity, nor can it capture differences between T cell responses according to age or gender. The majority of the immunological studies we investigate do not report epitope-specific T cell responses at this level of detail, and, hence, such an analysis could not be performed. In terms of the HLA associations of the reported SARS-CoV-2 epitopes, our study predicted additional associations based on homology with experimental T cell epitopes of SARS-CoV and their associated HLA alleles (File S1). These additional HLA associations, although promising, still need to be confirmed for SARS-CoV-2 by further immunological studies. Moreover, our analysis mainly involved CD8+ T cell epitopes. This was due to the limited number of CD4+ T cell epitopes that have been reported so far with a unique HLA allele association. Further experimental studies are needed to precisely identify CD4+ T cell epitopes with distinct HLA alleles. This would help to determine potential immunoprevalent CD4+ epitopes in the global population, similar to those reported for CD8+ T cells in this work, and would contribute to providing a more comprehensive understanding of the relationship between T cell responses and convalescence for COVID-19.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| T cell epitopes from convalescent COVID-19 patients | Chour et al.22 | Figure 3 |
| T cell epitopes from convalescent COVID-19 patients | Shomuradova et al.20 | Table 1 |
| T cell epitopes from convalescent COVID-19 patients | Nelde et al.30 | Extended Data Tables 2 and 3 |
| T cell epitopes from convalescent COVID-19 patients | Poran et al.19 | Figure 3 |
| T cell epitopes from convalescent COVID-19 patients | Ferretti et al.26 | Table 1 |
| T cell epitopes from convalescent COVID-19 patients | Peng et al.27 | Tables 1 and 2 |
| T cell epitopes from convalescent COVID-19 patients | Kared et al.15 | Figure S2 |
| T cell epitopes from convalescent COVID-19 patients | Schulien et al.18 | Table S1 |
| T cell epitopes from convalescent COVID-19 patients | Habel et al.25 | Figure 2B |
| T cell epitopes from convalescent COVID-19 patients | Hu et al.31 | Table 1 |
| T cell epitopes from convalescent COVID-19 patients | Nielsen et al.21 | Figure 5B |
| T cell epitopes from convalescent COVID-19 patients | Tarke et al.36 | Tables S3 and S5 |
| T cell epitopes from convalescent COVID-19 patients | Sekine et al.23 | Table S2 |
| T cell epitopes from convalescent COVID-19 patients | Saini et al.14 | Table S5 |
| T cell epitopes from convalescent COVID-19 patients | Lee et al.34 | Figure 13 |
| T cell epitopes from convalescent COVID-19 patients | Rha et al.28 | Figure 1C |
| T cell epitopes from convalescent COVID-19 patients | Lineburg et al.33 | Table S3 |
| T cell epitopes from convalescent COVID-19 patients | Nguyen et al.24 | Figure 3C |
| Response frequencies of SARS-CoV-2 T cell epitopes | This paper | File S1; Mendeley data: https://dx.doi.org/10.17632/fwn3kbbh6y.1 |
| Genetic conservation of SARS-CoV-2 T cell epitopes among SARS-CoV-2 sequences (as of 20 April 2021) | This paper | File S1; Mendeley data: https://dx.doi.org/10.17632/fwn3kbbh6y.1 |
| Total subjects tested for each SARS-CoV-2 T cell epitope across studies | This paper | File S1; Mendeley data: https://dx.doi.org/10.17632/fwn3kbbh6y.1 |
| Total subjects responded to each SARS-CoV-2 T cell epitope across studies | This paper | File S1; Mendeley data: https://dx.doi.org/10.17632/fwn3kbbh6y.1 |
| Additional HLA alleles predicted for SARS-CoV-2 T cell epitopes | This paper | File S1; Mendeley data: https://dx.doi.org/10.17632/fwn3kbbh6y.1 |
| SARS-CoV-2 genome sequence data for conservation analysis | GISAID (https://www.gisaid.org) | All full genome and high coverage sequences available as of 20 April 2021 |
| SARS-CoV-2 reference genome for aligning the sequences | https://www.ncbi.nlm.nih.gov/nuccore/NC_045512.2 | GenBank: NC_045512.2 |
| SARS-CoV T cell epitope data | IEDB; https://www.iedb.org/ | IEDB was queried for epitopes with positive MHC binding and positive T cell assays using “Severe acute respiratory syndrome-related coronavirus” as “Organism” on 21 February 2020. |
| Software and algorithms | ||
| SARS-CoV-2 T cell epitope web-dashboard | This paper | https://www.mckayspcb.com/SARS2TcellEpitopes/ |
| MAFFT | Katoh and Standley65 | https://mafft.cbrc.jp/alignment/software/ |
| Estimated population coverage of SARS-CoV-2 T cell epitopes based on associated HLA alleles | IEDB Analysis Resource - Population coverage tool | http://tools.iedb.org/population/download/ |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact Matthew R. McKay (m.mckay@ust.hk).
Materials availability
This study did not generate new materials.
Data and code availability
Compiled data of the SARS-CoV-2 T cell epitopes is available to download from the web dashboard, https://www.mckayspcb.com/SARS2TcellEpitopes/. File S1 has been deposited to Mendeley Data: http://dx.doi.org/10.17632/fwn3kbbh6y.1.
Experimental model and subject details
We compiled data from 20 immunological studies that reported SARS-CoV-2 epitopes targeted by T cells in individuals recovered from COVID-19. Two studies7,10 reported responses against synthetic peptide pool libraries using functional or molecular assays, and identified long immunogenic peptides. As complete information of epitopes was not available in these studies, we focused on the remaining 18 studies that provided precise epitopes along with their HLA restriction (Table 1). All statistics related to the patients that participated in each of these 18 studies (age, gender, geographical location, disease severity, blood collection time) are summarized in Table 1. Across the considered 18 studies, the total number of recovered COVID-19 individuals was 852. A total of 1,209 epitopes were obtained from these immunological studies (Table 1). Removing the epitopes with no HLA allele information and those for which the number of tested and responded patients was not reported at a distinct epitope-HLA level resulted in a total of 711 unique epitopes (812 unique epitope-HLA pairs; listed in File S1).
Method details
Sequence data, epitope conservation and coverage
SARS-CoV-2 genomic sequences were obtained from the GISAID database (https://www.gisaid.org/) on 20 April 2021. We downloaded only the complete (full-genome) sequences derived from human hosts with high coverage using the options provided on the GISAID database. All of the 859,233 downloaded sequences were aligned to the SARS-CoV-2 reference genome (GenBank: NC_045512.2) using MAFFT.65 The genomic MSA was translated using an in-house code to obtain the protein MSAs. The positions of the open reading frames provided with the reference sequence were used to identify the respective protein regions of the full genome.
The conservation of each SARS-CoV-2 T cell epitope was calculated as the fraction of SARS-CoV-2 sequences that encompassed the precise epitope sequence. The coverage of SARS-CoV-2 T cell epitopes at any position of a protein was calculated by counting the number of epitopes that included that position.
Response frequency (RF)
RF score58 was used to quantify the immunodominance of the SARS-CoV-2 epitopes reported to be recognized by T cells in recovered COVID-19 individuals. The RF score of an epitope is defined as follows:
where is the number of subjects responding to the epitope in study , is the number of subjects tested for a response against the epitope in study , and is the total number of studies. An RF score calculated using a large number of tested subjects would be more reliable than one calculated using relatively few subjects. To account for this, we computed the 95% confidence interval for the RF score of each epitope using the binomial cumulative distribution function58. In Figure 2, we defined the confidence in RF value of an epitope as the inverse of the length of the corresponding 95% confidence interval. That is, values of RF with a smaller 95% confidence interval have higher confidence, and vice versa.
Estimating global population coverage of epitopes
The global population coverage of an epitope refers to the percentage of individuals in the world population that is expected to mount a T cell response against that epitope. The population coverage of a T cell epitope was calculated based on the HLA alleles associated with it using the tool downloaded from the IEDB Analysis Resource (http://tools.iedb.org/population/download/). This tool employs global HLA allele frequency data obtained from the Allele Frequency Net Database (http://www.allelefrequencies.net/) to estimate the population coverage.
Quantification and statistical analysis
Statistical analyses were performed using MATLAB (R2019b) and the R language (version 3.6) using the RStudio server (version 1.3). The web-based platform was developed using the open source R Shiny (version 1.5) development framework. Fisher’s exact test was used to compute the statistical significance associated with enrichment of SARS-CoV epitopes among immunoprevalent SARS-CoV-2 epitopes. The 95% confidence interval for the RF score of each epitope in Figure 2 was determined using the binomial cumulative distribution function.58
Acknowledgments
The authors were supported by the General Research Fund of the Hong Kong Research Grants Council (RGC) (grants 16204519 and 16201620). S.F.A. was additionally supported by the Hong Kong Ph.D. Fellowship Scheme (HKPFS).
Author contributions
A.A.Q. and M.R.M. conceptualized the study and wrote the manuscript. A.A.Q. compiled the data from the literature. A.A.Q., S.F.A., and M.R.M. analyzed the data. A.A.Q. and S.F.A. performed the computations. S.F.A. generated the figures and developed the web dashboard.
Declaration of interests
The authors declare no competing interests.
Published: May 21, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2021.100312.
Contributor Information
Ahmed Abdul Quadeer, Email: eeaaquadeer@ust.hk.
Matthew R. McKay, Email: m.mckay@ust.hk.
Supplemental information
It includes, for each epitope, the associated HLA allele, location within the proteome, genetic conservation in global SARS-CoV-2 sequences (as of 20 April 2021), and the computed response frequency across studies (as reported in Figure 2). Number of total subjects tested, number of responding subjects, and response frequency per study for each epitope-HLA pair are also listed. For (n = 90) SARS-CoV-2 epitopes having a genetic match with SARS-CoV known epitopes, the IEDB IDs of the corresponding SARS-CoV epitopes as well as the associated HLAs are listed. This list of epitopes is compiled from both published studies and non-peer-reviewed studies available so far only on preprint servers. Results from non-peer-reviewed studies should be interpreted with caution. This table is also available from Mendeley Data at https://dx.doi.org/10.17632/fwn3kbbh6y.1.
References
- 1.Yong S.E.F., Anderson D.E., Wei W.E., Pang J., Chia W.N., Tan C.W., Teoh Y.L., Rajendram P., Toh M.P.H.S., Poh C. Connecting clusters of COVID-19: an epidemiological and serological investigation. Lancet Infect. Dis. 2020;20:809–815. doi: 10.1016/S1473-3099(20)30273-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zohar T., Alter G. Dissecting antibody-mediated protection against SARS-CoV-2. Nat. Rev. Immunol. 2020;20:392–394. doi: 10.1038/s41577-020-0359-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu F., Wang A., Liu M., Wang Q., Chen J., Xia S., Ling Y., Zhang Y., Xun J., Lu L. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. medRxiv. 2020 doi: 10.1101/2020.03.30.20047365. [DOI] [Google Scholar]
- 4.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weiskopf D., Schmitz K.S., Raadsen M.P., Grifoni A., Okba N.M.A., Endeman H., van den Akker J.P.C., Molenkamp R., Koopmans M.P.G., van Gorp E.C.M. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020;5:eabd2071. doi: 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Z., Liu J., Deng H., Yang X., Wang H., Feng X., Zelinskyy G., Trilling M., Sutter K., Lu M. SARS-CoV-2-specific T cell memory is long-lasting in the majority of convalescent COVID-19 individuals. bioRxiv. 2020 doi: 10.1101/2020.11.15.383463. [DOI] [Google Scholar]
- 7.Snyder T.M., Gittelman R.M., Klinger M., May D.H., Osborne E.J., Taniguchi R., Zahid H.J., Kaplan I.M., Dines J.N., Noakes M.N. Magnitude and dynamics of the T-cell response to SARS-CoV-2 infection at both individual and population levels. medRxiv. 2020 doi: 10.1101/2020.07.31.20165647. [DOI] [Google Scholar]
- 8.Long Q.-X., Tang X.-J., Shi Q.-L., Li Q., Deng H.-J., Yuan J., Hu J.-L., Xu W., Zhang Y., Lv F.-J. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020;26:1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- 9.Seow J., Graham C., Merrick B., Acors S., Steel K.J.A., Hemmings O., O’Bryne A., Kouphou N., Pickering S., Galao R. Longitudinal evaluation and decline of antibody responses in SARS-CoV-2 infection. medRxiv. 2020 doi: 10.1101/2020.07.09.20148429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., Chng M.H.Y., Lin M., Tan N., Linster M. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 11.Tang F., Quan Y., Xin Z.-T., Wrammert J., Ma M.-J., Lv H., Wang T.-B., Yang H., Richardus J.H., Liu W., Cao W.C. Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: a six-year follow-up study. J. Immunol. 2011;186:7264–7268. doi: 10.4049/jimmunol.0903490. [DOI] [PubMed] [Google Scholar]
- 12.Le T.T., Cramer J.P., Chen R., Mayhew S. Evolution of the COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020;19:667–668. doi: 10.1038/d41573-020-00151-8. [DOI] [PubMed] [Google Scholar]
- 13.Liu Z.-L., Liu Y., Wan L.-G., Xiang T.-X., Le A.-P., Liu P., Peiris M., Poon L.L.M., Zhang W. Antibody profiles in mild and severe cases of COVID-19. Clin. Chem. 2020;66:1102–1104. doi: 10.1093/clinchem/hvaa137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saini S.K., Hersby D.S., Tamhane T., Povlsen H.R., Amaya Hernandez S.P., Nielsen M., Gang A.O., Hadrup S.R. SARS-CoV-2 genome-wide T cell epitope mapping reveals immunodominance and substantial CD8+ T cell activation in COVID-19 patients. Sci. Immunol. 2021;6:eabf7550. doi: 10.1126/sciimmunol.abf7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kared H., Redd A.D., Bloch E.M., Bonny T.S., Sumatoh H., Kairi F., Carbajo D., Abel B., Newell E.W., Bettinotti M.P. SARS-CoV-2-specific CD8+ T cell responses in convalescent COVID-19 individuals. J. Clin. Invest. 2021;131:e145476. doi: 10.1172/JCI145476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grifoni A., Sidney J., Zhang Y., Scheuermann R.H., Peters B., Sette A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe. 2020;27:671–680.e2. doi: 10.1016/j.chom.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prachar M., Justesen S., Steen-Jensen D.B., Thorgrimsen S., Jurgons E., Winther O., Bagger F.O. Identification and validation of 174 COVID-19 vaccine candidate epitopes reveals low performance of common epitope prediction tools. Sci. Rep. 2020;10:20465. doi: 10.1038/s41598-020-77466-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schulien I., Kemming J., Oberhardt V., Wild K., Seidel L.M., Killmer S., Sagar, Daul F., Salvat Lago M., Decker A. Characterization of pre-existing and induced SARS-CoV-2-specific CD8+ T cells. Nat. Med. 2021;27:78–85. doi: 10.1038/s41591-020-01143-2. [DOI] [PubMed] [Google Scholar]
- 19.Poran A., Harjanto D., Malloy M., Arieta C.M., Rothenberg D.A., Lenkala D., van Buuren M.M., Addona T.A., Rooney M.S., Srinivasan L., Gaynor R.B. Sequence-based prediction of SARS-CoV-2 vaccine targets using a mass spectrometry-based bioinformatics predictor identifies immunogenic T cell epitopes. Genome Med. 2020;12:70. doi: 10.1186/s13073-020-00767-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shomuradova A.S., Vagida M.S., Sheetikov S.A., Zornikova K.V., Kiryukhin D., Titov A., Peshkova I.O., Khmelevskaya A., Dianov D.V., Malasheva M. SARS-CoV-2 epitopes are recognized by a public and diverse repertoire of human T cell receptors. Immunity. 2020;53:1245–1257.e5. doi: 10.1016/j.immuni.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nielsen S.S.F., Vibholm L.K., Monrad I., Olesen R., Frattari G.S., Pahus M.H., Højen J.F., Gunst J.D., Erikstrup C., Holleufer A. SARS-CoV-2 elicits robust adaptive immune responses regardless of disease severity. bioRxiv. 2020 doi: 10.1101/2020.10.08.331645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chour W., Xu A.M., Ng A.H.C., Choi J., Xie J., Yuan D., Lee J.K., Delucia D.C., Edmark R., Jones L. Shared antigen-specific CD8+ T cell responses against the SARS-COV-2 spike protein in HLA A∗02:01 COVID-19 participants. medRxiv. 2020 doi: 10.1101/2020.05.04.20085779. [DOI] [Google Scholar]
- 23.Sekine T., Perez-Potti A., Rivera-Ballesteros O., Strålin K., Gorin J.-B., Olsson A., Llewellyn-Lacey S., Kamal H., Bogdanovic G., Muschiol S., Karolinska COVID-19 Study Group Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183:158–168.e14. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nguyen T.H.O., Rowntree L.C., Petersen J., Chua B.Y., Hensen L., Kedzierski L., van de Sandt C.E., Chaurasia P., Tan H.-X., Habel J.R. CD8+ T cells specific for an immunodominant SARS-CoV-2 nucleocapsid epitope display high naive precursor frequency and TCR promiscuity. Immunity. 2021;54:1066–1082.e5. doi: 10.1016/j.immuni.2021.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Habel J.R., Nguyen T.H.O., van de Sandt C.E., Juno J.A., Chaurasia P., Wragg K., Koutsakos M., Hensen L., Jia X., Chua B. Suboptimal SARS-CoV-2-specific CD8+ T cell response associated with the prominent HLA-A∗02:01 phenotype. Proc. Natl. Acad. Sci. USA. 2020;117:24384–24391. doi: 10.1073/pnas.2015486117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferretti A.P., Kula T., Wang Y., Nguyen D.M.V., Weinheimer A., Dunlap G.S., Xu Q., Nabilsi N., Perullo C.R., Cristofaro A.W. Unbiased screens show CD8+ T cells of COVID-19 patients recognize shared epitopes in SARS-CoV-2 that largely reside outside the spike Protein. Immunity. 2020;53:1095–1107.e3. doi: 10.1016/j.immuni.2020.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng Y., Mentzer A.J., Liu G., Yao X., Yin Z., Dong D., Dejnirattisai W., Rostron T., Supasa P., Liu C., Oxford Immunology Network Covid-19 Response T cell Consortium. ISARIC4C Investigators Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020;21:1336–1345. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rha M.-S., Jeong H.W., Ko J.-H., Choi S.J., Seo I.-H., Lee J.S., Sa M., Kim A.R., Joo E.-J., Ahn J.Y. PD-1-expressing SARS-CoV-2-specific CD8+ T cells are not exhausted, but functional in patients with COVID-19. Immunity. 2021;54:44–52.e3. doi: 10.1016/j.immuni.2020.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kula T., Dezfulian M.H., Wang C.I., Abdelfattah N.S., Hartman Z.C., Wucherpfennig K.W., Lyerly H.K., Elledge S.J. T-Scan: A genome-wide method for the systematic discovery of T cell epitopes. Cell. 2019;178:1016–1028.e13. doi: 10.1016/j.cell.2019.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nelde A., Bilich T., Heitmann J.S., Maringer Y., Salih H.R., Roerden M., Lübke M., Bauer J., Rieth J., Wacker M. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat. Immunol. 2021;22:74–85. doi: 10.1038/s41590-020-00808-x. [DOI] [PubMed] [Google Scholar]
- 31.Hu C., Shen M., Han X., Chen Q., Li L., Chen S., Zhang J., Gao F., Wang W., Wang Y. Identification of Cross-Reactive CD8+ T Cell Receptors with High Functional Avidity to a SARS-CoV-2 Immunodominant Epitope and Its Natural Mutant Variants. bioRxiv. 2020 doi: 10.1101/2020.11.02.364729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020;12:254. doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lineburg K.E., Grant E.J., Swaminathan S., Chatzileontiadou D.S.M., Szeto C., Sloane H., Panikkar A., Raju J., Crooks P., Rehan S. CD8+ T cells specific for an immunodominant SARS-CoV-2 nucleocapsid epitope cross-react with selective seasonal coronaviruses. Immunity. 2021;54:1055–1065.e5. doi: 10.1016/j.immuni.2021.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee E., Sandgren K., Duette G., Stylianou V.V., Khanna R., Eden J.-S., Blyth E., Gottlieb D., Cunningham A.L., Palmer S. Identification of SARS-CoV-2 nucleocapsid and spike T-cell epitopes for assessing T-cell immunity. J. Virol. 2020;95:e02002-20. doi: 10.1128/JVI.02002-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gaiha G.D., Rossin E.J., Urbach J., Landeros C., Collins D.R., Nwonu C., Muzhingi I., Anahtar M.N., Waring O.M., Piechocka-Trocha A. Structural topology defines protective CD8+ T cell epitopes in the HIV proteome. Science. 2019;364:480–484. doi: 10.1126/science.aav5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tarke A., Sidney J., Kidd C.K., Dan J.M., Ramirez S.I., Yu E.D., Mateus J., da Silva Antunes R., Moore E., Rubiro P. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep Med. 2021;2:100204. doi: 10.1016/j.xcrm.2021.100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reiss S., Baxter A.E., Cirelli K.M., Dan J.M., Morou A., Daigneault A., Brassard N., Silvestri G., Routy J.-P., Havenar-Daughton C. Comparative analysis of activation induced marker (AIM) assays for sensitive identification of antigen-specific CD4 T cells. PLoS ONE. 2017;12:e0186998. doi: 10.1371/journal.pone.0186998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stranzl T., Larsen M.V., Lundegaard C., Nielsen M. NetCTLpan: pan-specific MHC class I pathway epitope predictions. Immunogenetics. 2010;62:357–368. doi: 10.1007/s00251-010-0441-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andreatta M., Nielsen M. Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics. 2016;32:511–517. doi: 10.1093/bioinformatics/btv639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nielsen M., Lundegaard C., Blicher T., Lamberth K., Harndahl M., Justesen S., Røder G., Peters B., Sette A., Lund O., Buus S. NetMHCpan, a method for quantitative predictions of peptide binding to any HLA-A and -B locus protein of known sequence. PLoS ONE. 2007;2:e796. doi: 10.1371/journal.pone.0000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jurtz V., Paul S., Andreatta M., Marcatili P., Peters B., Nielsen M. NetMHCpan-4.0: Improved peptide–MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. J. Immunol. 2017;199:3360–3368. doi: 10.4049/jimmunol.1700893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reynisson B., Alvarez B., Paul S., Peters B., Nielsen M. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020;48(W1):W449–W454. doi: 10.1093/nar/gkaa379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rammensee H., Bachmann J., Emmerich N.P.N., Bachor O.A., Stevanović S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 44.Nielsen M., Lundegaard C., Worning P., Lauemøller S.L., Lamberth K., Buus S., Brunak S., Lund O. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. 2003;12:1007–1017. doi: 10.1110/ps.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abelin J.G., Keskin D.B., Sarkizova S., Hartigan C.R., Zhang W., Sidney J., Stevens J., Lane W., Zhang G.L., Eisenhaure T.M. Mass spectrometry profiling of HLA-associated peptidomes in mono-allelic cells enables more accurate epitope prediction. Immunity. 2017;46:315–326. doi: 10.1016/j.immuni.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sohail M.S., Ahmed S.F., Quadeer A.A., McKay M.R. In silico T cell epitope identification for SARS-CoV-2: Progress and perspectives. Adv. Drug Deliv. Rev. 2021;171:29–47. doi: 10.1016/j.addr.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.UniProt . 2020. SARS-CoV-2 replicase polyprotein 1ab.https://covid-19.uniprot.org/uniprotkb/P0DTD1 [Google Scholar]
- 48.Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., Zhang X., Muruato A.E., Zou J., Fontes-Garfias C.R. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2021;592:116–121. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Sheffield COVID-19 Genomics Group Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hou Y.J., Chiba S., Halfmann P., Ehre C., Kuroda M., Dinnon K.H., 3rd, Leist S.R., Schäfer A., Nakajima N., Takahashi K. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020;370:1464–1468. doi: 10.1126/science.abe8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sahin U. BNT162b2 induces SARS-CoV-2-neutralising antibodies and T cells in humans. medRxiv. 2020 2020.12.09.20245175. [Google Scholar]
- 52.Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 53.Wang P., Casner R.G., Nair M.S., Wang M., Yu J., Cerutti G., Liu L., Kwong P.D., Huang Y., Shapiro L., Ho D.D. Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. Cell Host Microbe. 2021;29:747–751.e4. doi: 10.1016/j.chom.2021.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Greaney A.J., Starr T.N., Gilchuk P., Zost S.J., Binshtein E., Loes A.N., Hilton S.K., Huddleston J., Eguia R., Crawford K.H.D. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe. 2021;29:44–57.e9. doi: 10.1016/j.chom.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tarke A., Sidney J., Methot N., Zhang Y., Dan J.M., Goodwin B., Rubiro P., Sutherland A., da Silva Antunes R., Frazier A. Negligible impact of SARS-CoV-2 variants on CD4+ and CD8+ T cell reactivity in COVID-19 exposed donors and vaccinees. bioRxiv. 2021 doi: 10.1101/2021.02.27.433180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ahmed S.F., Quadeer A.A., McKay M.R. COVIDep: a web-based platform for real-time reporting of vaccine target recommendations for SARS-CoV-2. Nat. Protoc. 2020;15:2141–2142. doi: 10.1038/s41596-020-0358-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vita R., Mahajan S., Overton J.A., Dhanda S.K., Martini S., Cantrell J.R., Wheeler D.K., Sette A., Peters B. The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res. 2019;47(D1):D339–D343. doi: 10.1093/nar/gky1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dhanda S.K., Vita R., Ha B., Grifoni A., Peters B., Sette A. ImmunomeBrowser: a tool to aggregate and visualize complex and heterogeneous epitopes in reference proteins. Bioinformatics. 2018;34:3931–3933. doi: 10.1093/bioinformatics/bty463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tian Y., Grifoni A., Sette A., Weiskopf D. Human T cell response to dengue virus infection. Front. Immunol. 2019;10:2125. doi: 10.3389/fimmu.2019.02125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weiskopf D., Angelo M.A., de Azeredo E.L., Sidney J., Greenbaum J.A., Fernando A.N., Broadwater A., Kolla R.V., De Silva A.D., de Silva A.M. Comprehensive analysis of dengue virus-specific responses supports an HLA-linked protective role for CD8+ T cells. Proc. Natl. Acad. Sci. USA. 2013;110:E2046–E2053. doi: 10.1073/pnas.1305227110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sidney J., Peters B., Sette A. Epitope prediction and identification- adaptive T cell responses in humans. Semin. Immunol. 2020;50:101418. doi: 10.1016/j.smim.2020.101418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ahmed S.F., Quadeer A.A., McKay M.R. 2020. A web-based dashboard for reporting epitopes targeted by T cells in convalescent COVID-19 patients.https://www.mckayspcb.com/SARS2TcellEpitopes/ [Google Scholar]
- 63.Corbett K.S., Edwards D.K., Leist S.R., Abiona O.M., Boyoglu-Barnum S., Gillespie R.A., Himansu S., Schäfer A., Ziwawo C.T., DiPiazza A.T. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586:567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smith T.R.F., Patel A., Ramos S., Elwood D., Zhu X., Yan J., Gary E.N., Walker S.N., Schultheis K., Purwar M. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat. Commun. 2020;11:2601. doi: 10.1038/s41467-020-16505-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
It includes, for each epitope, the associated HLA allele, location within the proteome, genetic conservation in global SARS-CoV-2 sequences (as of 20 April 2021), and the computed response frequency across studies (as reported in Figure 2). Number of total subjects tested, number of responding subjects, and response frequency per study for each epitope-HLA pair are also listed. For (n = 90) SARS-CoV-2 epitopes having a genetic match with SARS-CoV known epitopes, the IEDB IDs of the corresponding SARS-CoV epitopes as well as the associated HLAs are listed. This list of epitopes is compiled from both published studies and non-peer-reviewed studies available so far only on preprint servers. Results from non-peer-reviewed studies should be interpreted with caution. This table is also available from Mendeley Data at https://dx.doi.org/10.17632/fwn3kbbh6y.1.
Data Availability Statement
Compiled data of the SARS-CoV-2 T cell epitopes is available to download from the web dashboard, https://www.mckayspcb.com/SARS2TcellEpitopes/. File S1 has been deposited to Mendeley Data: http://dx.doi.org/10.17632/fwn3kbbh6y.1.