Abstract
Impact of direct heat stress (HS) on genetic parameter estimates, i.e., HS close to the trait recording date, was verified in several previous studies conducted in dairy and beef cattle populations. The aim of the present study was to analyze the impact of time-lagged HS at different recording periods during late pregnancy (a.p.) and postpartum (p.p.) on genetic parameter estimates for birth weight (BWT) and weight gain traits (200 d- and 365 d-weight gain (200dg, 365dg)) in offspring of the dual-purpose cattle breed Rotes Hhenvieh (RHV). Furthermore, we estimated genetic correlations within traits across time-lagged climatic indicators, in order to proof possible genotype by environment interactions (GE). Trait recording included 5,434 observations for BWT, 3,679 observations for 200dg and 2,998 observations for 365dg. Time-lagged climatic descriptors were classes for the mean temperature humidity index (mTHI) and number of HS days (nHS) from the following periods: 7 d-period a.p. (BWT), 56 d-period a.p., and 56 d-period p.p. (200dg and 365dg). Genetic parameters were estimated via 2-trait animal models, i.e., defining the same trait in different climatic environments as different traits. Genetic variances and heritabilities for all traits increased with increasing mTHI- and nHS-classes for all recording periods, indicating pronounced genetic differentiation with regard to time-lagged in utero HS and HS directly after birth. Similarly, in low mTHI- and nHS-classes indicating cold stress, genetic variances, and heritabilities were larger than for temperate climates. Genetic correlations substantially smaller than 0.80 indicating G E were observed when considering same traits from mTHI- and nHS-classes in greater distance. Estimated breeding values (EBV) of the 10 most influential sires with the largest number of offspring records fluctuated across mTHI- and nHS-classes. Correlations between sire EBV for same traits from distant climatic classes confirmed the genetic correlation estimates. Sires displaying stable EBV with climatic alterations were also identified. Selection of those sires might contribute to improved robustness in the RHV outdoor population genetically.
Keywords: dual-purpose cattle, genotypeenvironment interaction, genetic parameters, time-lagged heat stress
Introduction
The observed (Kaspar et al., 2013) and projected (Kreienkamp et al., 2020) climate change in Germany with continuously increasing temperatures has direct impact especially on primary and functional traits of beef and dual-purpose cattle kept in outdoor production systems. Phenotypically, heat stress (HS) hampered daily weight gains in growing crossbred calves (Habeeb et al., 2011) and feedlot heifers (Mitlhner et al., 2001), and impaired meat quality traits in beef cattle (Kadim et al., 2004). Genetically, studies conducted in beef cattle (Guidolin et al., 2012; Williams et al., 2012; Raidan et al., 2015) identified different genetic parameters for performance traits in different environments, indicating possible genotype by environment interactions (GE). A GE indicates that the most appropriate genotype depends on the given environmental conditions. This is especially important for outdoor populations, kept in highly variable environments. In dairy cattle, GE were proven for prompt HS impact on primary and functional traits (e.g., Bohlouli et al., 2013; Santana et al., 2016). A few genetic studies addressed beef or dual purpose cattle. In U.S. Angus cattle, Bradford et al. (2016) reported GE interaction for weaning weight due to HS impact.
Phenotypically, in addition to prompt detrimental HS impact, time-lagged impact of HS in dams on offspring performances was reported for dairy cows (Tao et al., 2012; Guo et al., 2016; Monteiro et al., 2016). Accordingly, in our previous study conducted in an outdoor dual-purpose population, Halli et al. (2020) identified time-lagged effects of intrauterine and direct HS on weight traits record in offspring. However, with regard to genetic parameter estimations in dependency of climatic alterations in cattle populations, time-lagged analyses are lacking. An explanation for time-lagged genetic impact of intra uterine HS addresses epigenetic modifications of the calf genome, inducing alterations of gene expressions (Seong et al., 2011; Daxinger and Whitelaw, 2012; Sakatani, 2017; Skibiel et al., 2018). Epigenetic modifications caused impairments in offspring production and reproduction performances (Gudex et al., 2014; Monteiro et al., 2016) and induced metabolomic changes (Guo et al., 2016).
The aim of this study was to analyze the impact of HS at different recording periods during late pregnancy and postpartum (p.p.) on genetic (co)variance components for birth weight (BWT) and weight gain traits in the dual-purpose cattle breed Rotes Hhenvieh (RHV) from an across-generation perspective. Genetic correlations and estimated breeding values (EBV) among climatic levels within the same trait were studied, in order to infer possible GE due to time-lagged HS impact.
Materials and Methods
The research did not involve any direct physical contact to the animals. No experimental studies were conducted for this project. All performance data were obtained from an existing database. Hence, no additional statement of institutional animal care and use committee is required.
Animal traits
Data were provided by Vereinigte Informationssysteme Tierhaltung w. V. (VIT; Verden, Germany), with permission of all RHV breeding organizations involved. The RHV dataset included the performance traits BWT (from 179 herds), 200 d-weight gain (200dg; from 154 herds) and 365 d-weight gain (365dg; from 184 herds). The traits 200dg and 365dg were calculated as the difference between 200 d weight and BWT, and between 365 d weight and BWT, respectively. The herds were spread over Germany, and located in pasture-based production systems. Data editing was performed using the SAS University Edition (SAS Institute, Cary, NC). All traits were corrected for outliers by excluding values lower or higher than the mean 3 SD in all traits. Accordingly, calves with a BWT 20 kg and 53 kg, a 200dg 19 kg and 371 kg or a 365dg 75 kg and 522 kg, were included in the ongoing analyses. Animals which provided information for both weight gain traits (200dg and 365dg) were only considered when both traits were recorded in the same herd. Data recording spanned a period of 20 yr (2000 to 2019) and included 5,434 observations for BWT, 3,679 observations for 200dg and 2,998 observations for 365dg. Means for BWT, 200dg and 365dg were 36.8 ( 5.28 kg), 194.5 ( 57.5 kg), and 297.6 ( 71.0 kg), respectively.
Lactation numbers of cows were grouped into 8 parity groups, i.e., considering lactations 1 to 7 separately, and all lactations larger than 7 simultaneously in one group. Calving seasons were defined as winter (December, January, and February), spring (March, April, and May), summer (June, July, and August), and autumn (September, October, and November). The pedigree data consisted of 23,518 animals, 6,763 dams, and 1,300 sires.
Meteorological data
According to the postal codes, herds were assigned to geographic regions, and merged with the nearest official weather station. In this regard, longitude and latitude information were considered for the identification of the nearest weather station, using the Geosphere package version 1.5 to 10 in R (Hijmans et al., 2019). For the analyses, 45 different weather stations were allocated to the farms. The mean distance between a farm and a weather station was 22.7 km (minimal distance: 523.4 m; maximal distance: 54.5 km).
The temperature humidity index (THI) as an indicator for climatic impact was calculated using the following equation:
where T is the dry bulb temperature and RH is the relative humidity (NRC, 1971).
A mean daily THI (mTHI) was calculated for different recording periods. For BWT, the 7 d-period before birth of the calf (a.p.) was considered. For 200dg and 365dg, the 56 d-periods a.p. and p.p. of the calf birth date were analyzed. The last week (Muller et al., 1975) and the last 2 mo of gestation (Bauman and Currie, 1980) are defined as periods of high importance for bovine fetus growth, which explains the focus on these specific periods in our study. Furthermore, the trait specific week selection a.p. and p.p. based on the HS impact in our previous phenotypic across-generation analyses (Halli et al., 2020). Four mTHI-classes were defined: class 1: mTHI 39, class 2: mTHI 40 to 49, class 3: mTHI 50 to 59 and class 4: mTHI 60. Furthermore, we counted and classified the numbers of HS days (nHS), i.e., the number of days where mTHI was 60. Classes of nHS within the different recording periods were defined as follows:
7 d-period: class 0: 0 HS days; class 1: 1 to 3 HS days; class 2: 4 to 7 HS days.
56 d-period: class 0: 0 HS days; class 1: 1 to 10 HS days; class 2: 11 to 30 HS days; class 3: 31 to 56 HS days.
The means for the production traits with corresponding SD for mTHI- and nHS-classes are listed in Table 1. Means for BWT increased with increasing mTHI and nHS in the 7 d-period before birth of the calf. Mean values for 200dg and 365dg decreases with increasing mTHI- and nHS-class in the 56 d-period before and after birth.
Table 1.
Means with corresponding SD for BWT, 200dg (200 d-weight gain), and 365dg (365 d-weight gain) in mTHI- and nHS-classes from different recording periods
| Trait1 (Rec. period2) | HS indicator | Mean | SD | Min | Max | n |
|---|---|---|---|---|---|---|
| BWT, kg (7 d a.p.3) | mTHI-class5 1 | 36.07 | 5.19 | 20.00 | 53.00 | 1461 |
| 2 | 36.25 | 5.06 | 20.00 | 52.00 | 1208 | |
| 3 | 37.13 | 5.28 | 20.00 | 53.00 | 1398 | |
| 4 | 37.88 | 5.60 | 20.00 | 53.00 | 998 | |
| nHS-class6 0 | 36.34 | 5.16 | 20.00 | 53.00 | 3396 | |
| 1 | 37.42 | 5.41 | 20.00 | 53.00 | 809 | |
| 2 | 37.80 | 5.62 | 20.00 | 53.00 | 860 | |
| 200dg, kg (56 d a.p.) | mTHI-class 1 | 211.45 | 69.54 | 38.00 | 370.00 | 892 |
| 2 | 192.92 | 53.00 | 38.00 | 368.00 | 960 | |
| 3 | 188.57 | 46.61 | 47.00 | 371.00 | 1221 | |
| 4 | 181.79 | 59.17 | 46.00 | 370.00 | 582 | |
| nHS-class 0 | 205.13 | 65.21 | 38.00 | 370.00 | 1465 | |
| 1 | 191.33 | 43.89 | 47.00 | 368.00 | 978 | |
| 2 | 185.45 | 51.61 | 46.00 | 371.00 | 710 | |
| 3 | 180.40 | 59.15 | 60.00 | 344.00 | 502 | |
| 200dg, kg (56 d p.p.4) | mTHI-class 1 | 179.87 | 63.54 | 38.00 | 368.00 | 814 |
| 2 | 214.45 | 61.35 | 46.00 | 371.00 | 816 | |
| 3 | 202.67 | 56.55 | 53.00 | 370.00 | 923 | |
| 4 | 182.76 | 43.39 | 58.00 | 358.00 | 1102 | |
| nHS-class 0 | 192.40 | 65.69 | 38.00 | 371.00 | 1333 | |
| 1 | 213.78 | 55.48 | 60.00 | 370.00 | 706 | |
| 2 | 195.21 | 54.76 | 53.00 | 370.00 | 637 | |
| 3 | 181.94 | 43.31 | 58.00 | 332.00 | 979 | |
| 365dg, kg (56 d a.p.) | mTHI-class 1 | 308.45 | 73.37 | 98.00 | 522.00 | 886 |
| 2 | 294.57 | 66.18 | 99.00 | 522.00 | 745 | |
| 3 | 286.30 | 66.01 | 99.00 | 512.00 | 959 | |
| 4 | 298.88 | 79.28 | 90.00 | 492.00 | 369 | |
| nHS-class 0 | 304.35 | 70.89 | 98.00 | 522.00 | 1377 | |
| 1 | 292.49 | 59.82 | 118.00 | 512.00 | 700 | |
| 2 | 283.28 | 73.98 | 99.00 | 512.00 | 565 | |
| 3 | 295.61 | 80.44 | 90.00 | 492.00 | 317 | |
| 365dg, kg (56 d p.p.) | mTHI-class 1 | 302.42 | 71.66 | 98.00 | 522.00 | 709 |
| 2 | 316.89 | 69.54 | 119.00 | 522.00 | 702 | |
| 3 | 301.80 | 67.37 | 99.00 | 517.00 | 676 | |
| 4 | 271.45 | 65.78 | 90.00 | 512.00 | 872 | |
| nHS-class 0 | 307.17 | 71.00 | 98.00 | 522.00 | 1196 | |
| 1 | 313.58 | 69.31 | 137.00 | 522.00 | 515 | |
| 2 | 295.33 | 64.70 | 118.00 | 517.00 | 507 | |
| 3 | 268.55 | 66.43 | 90.00 | 481.00 | 741 |
1Traits: BWT, birth weight; 200dg, 200 d-weight gain; 365dg, 365 d-weight gain.
2Rec. period, recording period (7 d, 7 d period; 56 d, 56 d period).
3a.p., ante partum/prenatal.
4p.p., postpartum/postnatal.
5mTHI-class, class for mTHI (class 1: mean THI 39, class 2: mean THI 40 to 49, class 3: mean THI 50 to 59 and class 4: mean THI 60).
6nHS-class, class for number of HS days (7 d-period: class 0: 0 d; class 1: 1 to 3 d; class 2: 4 to 7 d; 56 d-period: class 0: 0 d; class 1: 1 to 10 d; class 2: 11 to 30 d; class 3: 31 to 56 d).
Statistical models
(Co)variance components, genetic correlations and EBV for the traits BWT, 200dg and 365dg were estimated using the AI-REML algorithm as implemented in the DMU software package (Madsen and Jensen, 2013). In this regard, series of 2-trait animal models were defined, considering same traits from the mTHI- and nHS-classes as different traits. Hence, for each trait, 6 two-trait models were run when grouping data according to the mTHI-class. Depending on the climate recording period (7 d or 56 d period), 3 or 6 two-trait models, respectively, were run when the nHS-class was the considered HS indicator.
The statistical bivariate animal model in matrix notation was defined as follows:
where y1 and y2 represented either BWT, 200dg or 365dg in environment 1 or 2. The vectors for the traits 1 (b1) and 2 (b2) included the fixed effects for parity group, herd, calving season and sex; u1 and u2 were vectors of additive genetic animal effects for the 2 traits; and e1 and e2 were vectors for the random residual effects for both traits. X1, X2, Z1, and Z2 were incidence matrices for b1, b2, a1, and a2, respectively.
The variancecovariance structure for random effects was:
where g11 and g22 were additive genetic variances for the 2 traits; g12 and g21 were additive genetic covariances between both traits; A was the additive genetic relationship matrix based on pedigree data considering at least 3 generations back, and r11 and r22 were residual variances for both traits. In our preliminary analyses, we included maternal genetic effects for BWT. We found a quite small maternal genetic variance component (<5% of the total variance). Furthermore, direct heritabilites for BWT as outlined in the results did not change when considering the maternal genetic component, and were in the same range when analyzing different THI intervals. Furthermore, model information criteria based on log-likelihoods were very similar when comparing models with and without maternal genetic effects. Correlations between BWT breeding values from models with and without maternal genetic effects were very close to 1. Hence, due to the small maternal genetic component and very similar estimates from models with and without maternal genetic effects, we studied the RHV farm management in detail. In the farms, we identified cross-fostering as common practice. Hence, we decided to exclude the maternal genetic effect from our models in all bivariate runs. Due to the 2-trait modeling approach in series of bivariate runs, repeated variance components and heritabilities in same traits from same HS indicator classes and intervals were averaged in the ongoing results, presentations and discussions. Nevertheless, variance components and heritabilities for same traits in same HS indicator classes and intervals were very similar.
Furthermore, the series of 2-trait models generated several EBV for the same individual for the same trait, HS indicator and HS interval. Averaged EBV (i.e., the average per trait, HS indicator and HS interval) of the most 10 influential sires with more than 180 phenotyped offspring were selected, and plotted in dependency of HS indicators to study the impact of time lagged HS on genetic evaluations. Furthermore, we considered the EBV from all 1,300 sires, and we calculated correlations between breeding values for same traits recorded in different climatic classes.
Changes of variance components, especially of additive genetic variances, indicate environmental sensitivity (Bohlouli et al., 2019). High heritabilities in same traits from specific environmental levels indicate optimal performance test environments (Schierenbeck et al., 2011). Genetic correlations between same traits from different mTHI- and nHS-classes lower than 0.80 (Robertson, 1959) as well as substantial EBV alterations for different climatic levels were used as indicators for possible G E.
Results
Birth weight
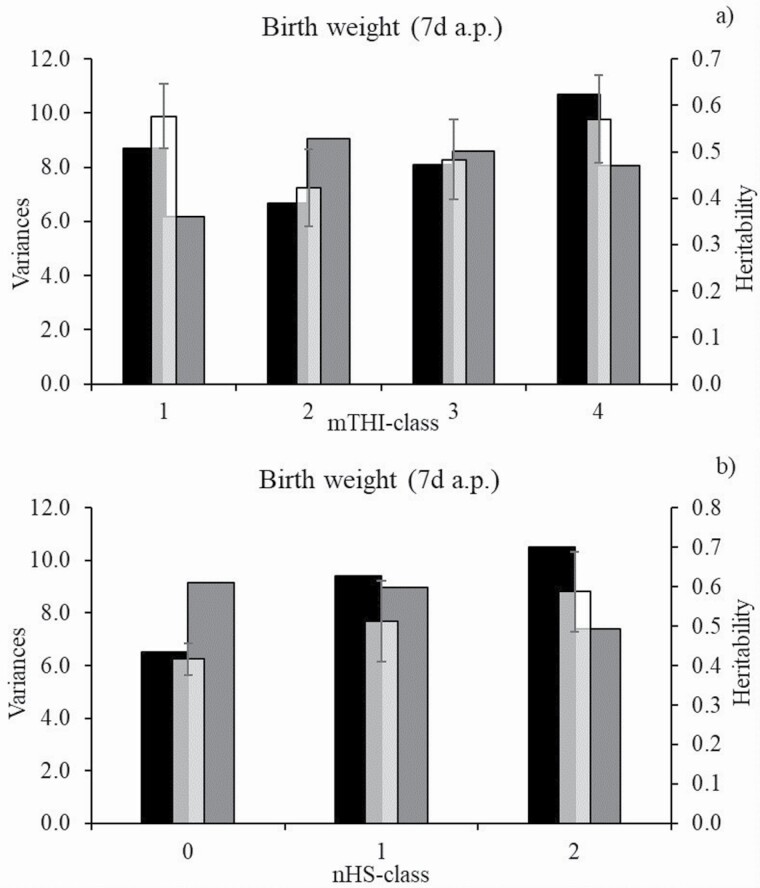
Heritabilities for BWT ranged from 0.42 to 0.58 in dependency of mTHI (Figure 1a), and from 0.42 to 0.59 in dependency of nHS (Figure 1b) recorded in the 7 d period before birth. For both HS indicators, additive genetic variances for BWT were largest in classes indicating HS, i.e., mTHI 60 (mTHI-class 4) and 4 to 7 HS days (nHS-class 2). Furthermore, HS in the observed period was associated with quite small residual variances for BWT, explaining the large heritabilities in the respective mTHI- and nHS-classes. With regard to mTHI, also lowest temperatures and humidities (mTHI 39) contributed to lime-lagged increased additive genetic variances and heritabilities for BWT. Mean THI classes representing the temperate climatic zone implied the smallest BWT heritability estimates, especially due to the increasing residual variance component. Similarly, nHS-classes representing no HS days (nHS-class 0) or only 1 to 3 HS days (nHS-class 1) showed higher residual variances and lower heritability estimates for BWT than 4 to 7 HS days (nHS-class 2).
Figure 1.
Additive genetic variance, residual variance, and heritability with corresponding SE for birth weight (BWT) in dependency of (a) class of mTHI or (b) class of nHS days from the prenatal 7 d-period. Black bars = additive genetic variances, grey bars = residual variances, and white bars = heritability.
Genetic correlations between BWT across different mTHI-classes are shown in Table 2, and across different nHS-classes in Table 3. For BWT, genetic correlations between neighboring mTHI classes were in a narrow range from 0.71 to 0.80. For mTHI-classes in great distance, i.e., classes 1 and 4, the BWT genetic correlation dropped to 0.53. Genetic correlations for BWT from different nHS-classes ranged from 0.70 to 0.86 (Table 3). The highest genetic correlation (0.86) was estimated when considering BWT from 0 HS days (nHS-class 0) and 1 to 3 HS days (nHS-class 1), and the lowest (0.70) between BWT from 0 HS days (nHS-class 0) and 4 to 7 HS days (nHS-class 2).
Table 2.
Heritabilities for BWT, 200dg (200 d-weight gain), and 365dg (365 d-weight gain) with corresponding SE in different mTHI-classes (diagonal), and genetic correlations with corresponding SE between same traits across mTHI-classes (above the diagonal)
| Trait1 Rec. period3 |
mTHI-class2 | 1 | 2 | 3 | 4 |
|---|---|---|---|---|---|
| BWT, kg | 1 | 0.58 (0.07) | 0.71 (0.14) | 0.71 (0.11) | 0.53 (0.15) |
| (7 d a.p.4) | 2 | 0.42 (0.08) | 0.80 (0.11) | 0.58 (0.17) | |
| 3 | 0.48 (0.09) | 0.76 (0.12) | |||
| 4 | 0.57 (0.09) | ||||
| 200dg, kg | 1 | 0.49 (0.10) | 0.51 (0.21) | 0.04 (0.20) | 0.06 (0.27) |
| (56 d a.p.) | 2 | 0.42 (0.10) | 0.38 (0.20) | 0.32 (0.23) | |
| 3 | 0.45 (0.08) | 0.55 (0.16) | |||
| 4 | 0.53 (0.11) | ||||
| 365dg, kg | 1 | 0.53(0.09) | 0.40 (0.19) | 0.34 (0.21) | 0.21 (0.27) |
| (56 d a.p.) | 2 | 0.59 (0.11) | 0.87 (0.13) | 0.60 (0.22) | |
| 3 | 0.38 (0.08) | 0.93 (0.12) | |||
| 4 | 0.72 (0.16) | ||||
| 200dg, kg | 1 | 0.49 (0.10) | 0.43 (0.18) | 0.06 (0.20) | 0.13 (0.22) |
| (56 d p.p.5) | 2 | 0.63 (0.11) | 0.14 (0.16) | 0.09 (0.20) | |
| 3 | 0.59 (0.10) | 0.30 (0.18) | |||
| 4 | 0.60 (0.07) | ||||
| 365dg, kg | 1 | 0.64 (0.11) | 0.48 (0.20) | 0.37 (0.22) | 0.21 (0.27) |
| (56 d p.p.) | 2 | 0.48 (0.11) | 0.43 (0.23) | 0.53 (0.21) | |
| 3 | 0.51 (0.12) | 0.22 (0.23) | |||
| 4 | 0.52 (0.09) |
1Traits: BWT, birth weight; 200dg, 200 d-weight gain; 365dg, 365 d-weight gain.
2mTHI-class, class for mTHI (class 1: mean THI 39, class 2: mean THI 40 to 49, class 3: mean THI 50 to 59 and class 4: mean THI 60).
3Rec. period, recording period (7 d, 7 d period; 56 d, 56 d period).
4a.p., ante partum/prenatal.
5p.p., postpartum/postnatal.
Table 3.
Heritabilities for BWT, 200dg (200 d-weight gain), and 365dg (365 d-weight gain) with corresponding SE in different nHS-classes (diagonal), and genetic correlations with corresponding SE between same traits across nHS-classes (above the diagonal)
| Trait1 Rec. period3 |
nHS-class2 | 0 | 1 | 2 | 3 |
|---|---|---|---|---|---|
| BWT, kg | 0 | 0.42 (0.04) | 0.86 (0.11) | 0.70 (0.12) | |
| (7 d a.p.4) | 1 | 0.51 (0.10) | 0.80 (0.14) | ||
| 2 | 0.59 (0.10) | ||||
| 200dg, kg | 0 | 0.43 (0.07) | 0.38 (0.19) | 0.13 (0.21) | 0.27 (0.22) |
| (56 d a.p.) | 1 | 0.34 (0.10) | 0.43 (0.24) | -0.06 (0.25) | |
| 2 | 0.48 (0.12) | 0.74 (0.19) | |||
| 3 | 0.57 (0.12) | ||||
| 365dg, kg | 0 | 0.43 (0.07) | 0.53 (0.17) | 0.39 (0.22) | 0.42 (0.24) |
| (56 d a.p.) | 1 | 0.52 (0.12) | 0.77 (0.18) | 0.49 (0.26) | |
| 2 | 0.47 (0.13) | 1.00 (0.16) | |||
| 3 | 0.63 (0.18) | ||||
| 200dg, kg | 0 | 0.42 (0.12) | 0.18 (0.20) | 0.11 (0.20) | -0.17 (0.20) |
| (56 d p.p.5) | 1 | 0.45 (0.12) | 0.67 (0.22) | 0.13 (0.24) | |
| 2 | 0.56 (0.13) | 0.58 (0.18) | |||
| 3 | 0.61 (0.12) | ||||
| 365dg, kg | 0 | 0.48 (0.08) | 0.78 (0.20) | 0.06 (0.23) | 0.43 (0.22) |
| (56 d p.p.) | 1 | 0.47 (0.13) | 0.64 (0.26) | 0.46 (0.25) | |
| 2 | 0.48 (0.16) | 0.23 (0.27) | |||
| 3 | 0.51 (0.10) |
1Traits: BWT, birth weight; 200dg, 200 d-weight gain; 365dg, 365 d-weight gain.
2nHS-class, class for number of HS days (7 d-period: class 0: 0 d; class 1: 1 to 3 d; class 2: 4 to 7 d; 56 d-period: class 0: 0 d; class 1: 1 to 10 d; class 2: 11 to 30 d; class 3: 31 to 56 d).
3Rec. period, recording period (7 d, 7 d period; 56 d, 56 d period).
4a.p., ante partum/prenatal.
5p.p., postpartum/postnatal.
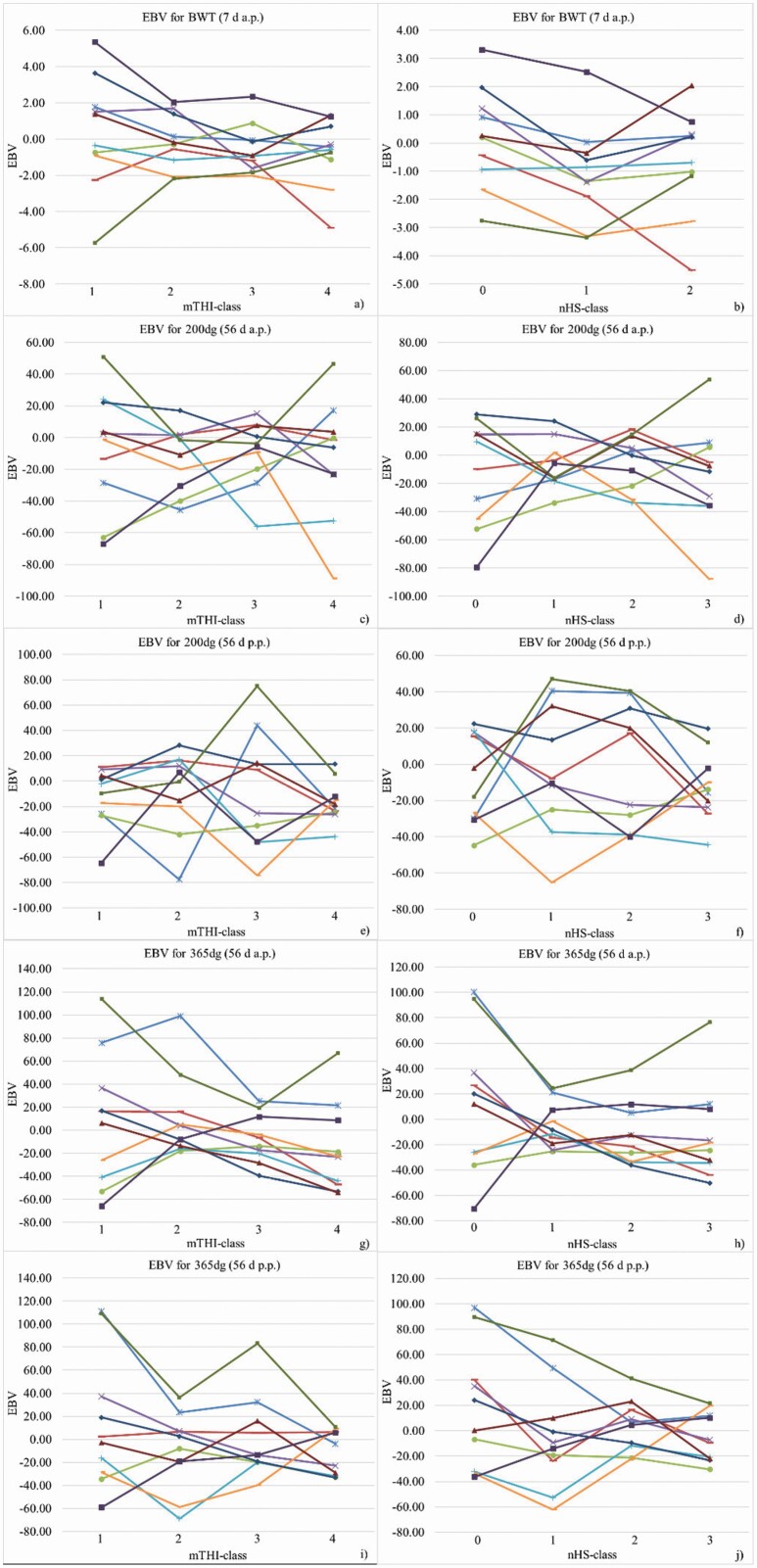
Breeding value alterations of 10 influential sires with more than 180 phenotyped progeny for BWT in dependency of time-lagged mTHI- and nHS-classes are shown in Figure 4a and b, respectively. Sire EBV for BWT varied across mTHI- and nHS-classes. EBV variations of the influential sires did not show a common pattern, and reflected individual particularities. Stable EBV across the mTHI- or nHS-range were found for a small number of individuals, only. Correlations between sire breeding values for same traits from different climatic classes decreased with increasing class distance. The correlation considering THI-classes 1 and 2 was 0.86, 0.80 when considering THI-classes 1 and 3, and dropped to 0.75 when considering THI classes 1 and 4. The correlation when considering THI-classes 2 and 4 was 0.71, but increased to 0.83 for the neighboring THI classes 3 and 4.
Figure 4.
EBV for birth weight (BWT), 200 d-weight gain (200dg), and 365 d-weight gain (365dg) for 10 sires with at least 180 progeny by classes for the mTHI (a), c), e), g), and i)) and by classes for the nHS days (b), d), f), h), and j)) in different recording periods. Individual sires are represented by same colors in all subfigures.
200 d weight gain
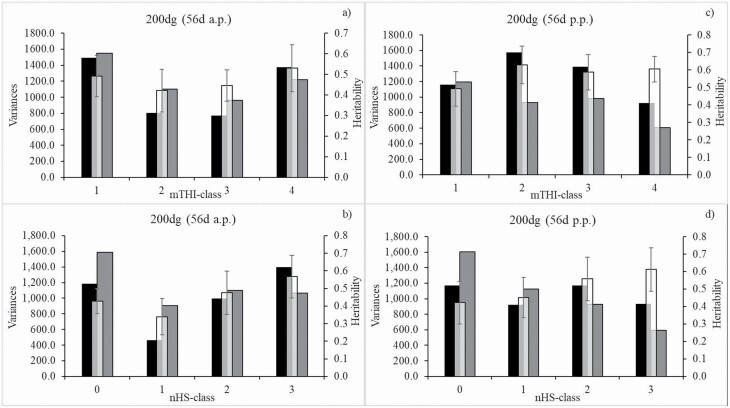
With regard to meteorological data recording in the 56d period before birth, the additive genetic variances and heritabilities for 200dg were highest in mTHI-class 1 (mTHI 39) and in mTHI-class 4 (mTHI 60; Figure 2a). For moderate mTHI (i.e., mTHI 40 to 49 (mTHI-class 2), and mTHI 50 to 59 (mTHI-class 3)), residual variances were considerably larger than the additive genetic variances, contributing to the smaller heritabilities. Accordingly, when evaluating the nHS impact, the additive genetic variance and heritability for 200dg were largest in the highest nHS-class with 31 to 56 HS days before birth (Figure 2b). For a low nHS (i.e., 0 HS days (nHS-class 0) and 1 to 10 HS days (nHS-class 1)), residual variances for 200dg were larger than the additive genetic variances, explaining the smaller heritabilities. Generally, 200dg heritabilities were moderate to large in the range from 0.42 to 0.53 for the different mTHI-classes, and from 0.34 to 0.57 for the different nHS-classes.
Figure 2.
Additive genetic variance, residual variance, and heritability with corresponding SE for 200 d-weight gain (200dg) in dependency of class of mTHI from the prenatal (a)) and postnatal (c)) 56 d-period or class of nHS days from the prenatal (b)) and postnatal (d)) 56 d-period. Black bars = additive genetic variances, grey bars = residual variances, and white bars = heritability.
With regard to the climatic impact from the 56 d a.p.-period, genetic correlations between 200dg from the different mTHI-classes were low (0.06) to moderate (0.55; Table 2). Genetic correlations were stronger between neighboring mTHI-classes (0.38 to 0.55) than for mTHI-classes in greater distance (0.06 to 0.32). Similarly, genetic correlations were low in the range from 0.06 to 0.27 (Table 3) when considering distant nHS-classes, but substantially increased up to 0.74 (200dg from 11 to 30 HS days (nHS-class 2) and 31 to 56 HS days (nHS-class 3)).
EBV for 200dg of 10 selected sires varied widely across mTHI- and nHS-classes (Figure 4c and d), without any consistent direction. None of the 10 sires had positive EBV across all mTHI-classes. Interestingly, for some sires, highest EBVs for 200dg were found in both unfavorable climatic conditions (hot or cold).
With regard to climatic impact from 56 d p.p.-period, the additive genetic variance for 200dg was largest from mTHI-class 2, contributing to the largest heritability with an estimate of 0.63 (Figure 2c). Accordingly, zero HS days (nHS-class 0) were associated with a large additive genetic variance for 200dg. Nevertheless, due to the smallest residual variance, the 200dg heritability was highest (0.61) in nHS-class 3 (31 to 56 HS days; Figure 2d).
Genetic correlations between 200dg from different mTHI-classes were low (0.06) to moderate (0.43) (Table 2). The highest genetic correlation was estimated when considering the adjacent mTHI-classes 1 and 2. With regard to the nHS-classes, genetic correlations between 200dg ranged from 0.17 to 0.67 (Table 3). The negative genetic correlation (0.17) was estimated when considering 200dg records from nHS-classes 0 and 3. The genetic correlation was largest (0.67) based on data records from the neighboring nHS-classes 1 and 2.
Again, the 200dg EBV of the 10 selected sires fluctuated across mTHI- and nHS-classes (Figure 4e and f). Among these influential sires, stable EBV across the THI- or nHS-range were not found. Correlations between sire breeding values (considering all 1,300 sires) for same traits from different climatic classes followed the pattern of genetic correlation estimates.
365 d weight gain
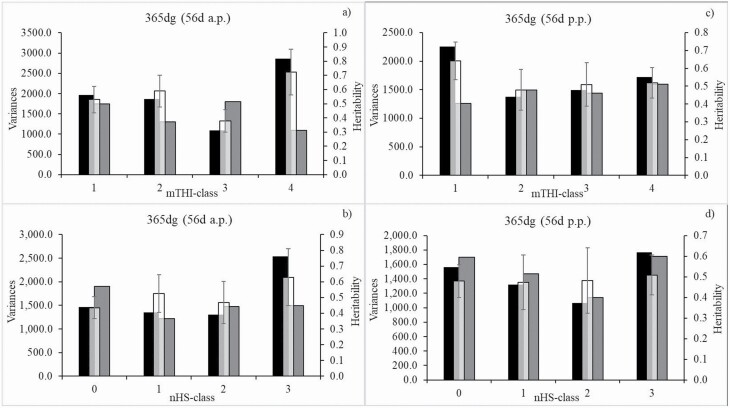
With regard to the climatic impact from the 56 d a.p.-period, the additive genetic variance for 365dg was largest when mTHI was 60 (mTHI-class 4; Figure 3a), contributing to the large heritability of 0.72. In analogy with the trait 200dg, a high nHS (31 to 56 d (nHS-class 5)) implied the highest additive genetic variance and the highest heritability (0.63) for 365dg (Figure 3b). The 365dg heritability range was from 0.43 in nHS-class 0 to 0.63 in nHS-class 3.
Figure 3.
Additive genetic variance, residual variance, and heritability with corresponding SE for 365 d-weight gain (365dg) in dependency of class of mTHI from the prenatal (a)) and postnatal (c)) 56 d-period or class of nHS days from the prenatal (b)) and postnatal (d)) 56 d-period. Black bars = additive genetic variances, grey bars = residual variances, and white bars = heritability.
Genetic correlations between 365dg from different mTHI-classes ranged from 0.21 to 0.93 (Table 2). The highest genetic correlation estimated based on data from the neighboring mTHI-classes 3 and 4 (mTHI 50 to 59 and mTHI 60), and the lowest when considering the distant mTHI-classes 1 and 4 (mTHI 39 and mTHI 60). With regard to nHS-classes, genetic correlations ranged from 0.39 (classes 0 and 2) to 1 (classes 2 and 3).
The variation of 365dg EBV from the 10 selected sires across mTHI- and nHS-classes is shown in Figure 4g and h, respectively. Some sires showed increasing EBV with decreasing mTHI or HS days, and vice versa.
With regard to the climatic impact from the 56 d p.p.-period, the lowest residual variance, the highest additive genetic variance and the highest heritability (0.64) for 365dg was estimated in mTHI-class 1 (mTHI 39; Figure 3c). The 365dg heritability range was from 0.48 (mTHI-class 2) to 0.64 (mTHI-class 1). The highest number of 31 to 56 HS days in nHS-class 3 was associated with the highest additive genetic variance and 365dg heritability of 0.51 (Figure 3d).
Genetic correlations between 365dg from different mTHI-classes ranged from 0.21 (consideration of data from mTHI-classes 1 and 4) to 0.53 (consideration of data from mTHI-classes 2 and 4; Table 2). With regard to the nHS-classes, 365dg genetic correlations ranged from 0.06 (consideration of data from nHS-class 0 and data from nHS-class 2) to 0.78 (consideration of data from nHS-class 0and data from nHS-class1).
The sire EBV for 365dg showed strong fluctuations across mTHI- and nHS-classes (Figure 4i and j, respectively). Only a small number of sires displayed quite stable EBV across the mTHI- or nHS-range.
Discussion
Birth weight
Generally, the BWT heritabilities in the RHV population in dependency of time-lagged climatic impact reflect estimates from previous studies, e.g., a BWT heritability of 0.61 in Zebu crosses (Mackinnon et al., 1991). Lower BW heritabilities were estimated when additionally accounting for maternal-genetic effects as done by Mllenhoff (2008) in Angus and Fleckvieh cattle populations, or by Chud et al. (2014) and Eler et al. (1995) in Nellore beef cattle. In the RHV suckler cow system with cross-fostering as common practice, calves not necessarily receive milk from their biological mother. We exemplary tested the impact of the maternal genetic effect in the model for BWT and found the effect to be very low, supporting estimates by Yin and Knig (2019) in another cattle breed. Hence, we simplified our statistical models and ignored the maternal genetic component as indicated in the methods section. Additive genetic variances and heritabilities increased when dams were kept under climatically stressful conditions in the 7 d period before calving. Accordingly, but with focus on direct stress impact, Schierenbeck et al. (2011) indicated more extreme trait responses in challenging environments, contributing to an increase of additive genetic variances and SD of daughter yield deviations. In contrast to our findings, but addressing direct HS impact, Santana et al. (2013) estimated larger BWT heritabilities in beef cattle from Brazil in favorable compared with harsh environments. Such results are in agreement with the theoretical expectations by Hammond (1947), who suggested progeny testing in superior environments to fully express their genetic potential. As the accuracy of selection depends on trait heritabilities, heritability alterations across different climatic environments indicate impact on selection response, suggesting, when referring to our results, an HS environment during late gestation. In such context, Schafberg et al. (2006) emphasized the increasing heritabilities for diseases resistances in harsh environments. In causality, disease resistances were associated with improved weight gain development in dairy cattle populations (Mahmoud et al., 2018). Hence, directions for the impact of environmental stress on genetic parameter estimates are quite controversial. The substantial climatic differences in different continents or countries, i.e., almost constant tropical conditions in Brazil, but obvious THI variations in Germany, might explain specific climatic impact on identified THI thresholds and on genetic parameter estimates in different cattle populations (Brgemann et al., 2012).
Regarding time-lagged HS impact, Skibiel et al. (2018) found epigenetic modifications (differentially methylated cytosines and genes) when comparing in utero heat stressed with cooled cattle, which may explain influence on morphological characteristics, morbidity and reduced lactation performance of in utero heat stressed offspring. Accordingly, to explain time-lagged HS influences on genetic parameter estimates for BWT from an across-generation perspective, we assume possible alterations of gene expressions due to epigenetics. Such mechanisms on gene expression profile alterations were identified in heat stressed male mice (Cammack et al., 2009).
Genetic correlations between BWT from different mTHI- or nHS-classes were close to 0.80 when considering neighboring classes, but substantially dropped for HS descriptors in great distance. Robertson (1959) and Guidolin et al. (2012) suggested a genetic correlation lower than 0.80 with a corresponding SE lower than 0.20 as an indicator for G E. In the present study, SE for genetic correlation estimates were in the range from 0.11 and 0.17. Possible GE through mTHI or nHS impact from substantially different classes is in agreement with genetic studies focusing on direct HS impact, e.g., Bohlouli et al. (2013), Bohlouli et al., 2019), and Santana et al. (2016). Hence, similar mechanisms might contribute to direct and time-lagged HS impact on genetic (co)variance components.
200 d weight gain
Heritability estimates for 200dg from our study were in the range from 0.34 to 0.63. Most of the other genetic studies in beef cattle analyzed the trait weaning weight, which is comparable with 200dg. Nevertheless, heritabilities for weaning weight were generally lower in the range from 0.11 to 0.28 (Mackinnon et al., 1991; Meyer, 1995; Jeyaruban et al., 2009; Williams et al., 2012; Santana et al., 2013; Bradford et al., 2016) than the 200dg estimates from the present study. An explanation for smaller heritablities in commercial beef breeds compared to dual-purpose RHV may be the effect of intensive selection, with impact of phenotypic uniformity, or, vice versa, heterogeneity in unselected populations likewise RHV (Via and Lande, 1985). In agreement with estimates for BWT, genetic variances and heritabilities for 200dg increased when dams were exposed to heat or cold stress during the recording period (here: 56 d) before calving. Contrary, Bradford et al. (2016) found the highest heritability for weaning weight under no-heat load conditions, and the lowest heritability for intermediate heat load conditions.
With regard to the 56 d period after birth, only HS but not cold stress, was associated with a pronounced 200dg genetic differentiation. A low mTHI of 39 (mTHI-class 1) or zero HS days (nHS-class 0) implied an increase of the residual variance component, indicating impact of further environmental effects on 200dg which were not considered in the statistical models, such as specific feed supply. Ignoring of specific feeding or management groups in genetic evaluations also contributed to increased residual variances for milk production traits in dairy cattle (Knig et al., 2005).
With regard to the climatic impact from the 56 d p.p.-period, enlarged residual variances contributed to lowest 200dg heritabilities in mTHI-class 1 (mTHI 39) and nHS-class 0 (0 HS days). Contrary to our findings, Santana et al. (2013) and Williams et al. (2012) found larger heritabilities for weaning weight in favorable than in harsh environments. In addition to the increased genetic variances under HS conditions, Williams et al. (2012) reported lower additive genetic variances compared with estimates from favorable environments. When referring to the genetic parameters from the present study, selection response in 200dg will be maximized when dams suffered HS in the 56 d-period before or after birth.
With regard to both climate data recording periods 56 d a.p and 56 d p.p., genetic correlations between 200dg from adjacent mTHI- or nHS-classes were substantially larger than genetic correlations from distant climatic descriptors. Nevertheless, all genetic correlation estimates between 200dg from different mTHI- or nHS-classes were <0.80, but SE exceeded 0.20, indicating only a few obvious GE when considering the concept by Robertson (1959) and Guidolin et al. (2012). The partly negative genetic correlations when considering distant climatic classes indicate the initiation of different genetic mechanisms or gene expression due to HS or due to cold stress impact. Sonna et al. (2002) reported ~100 mammalian genes which were affected by HS and ~20 genes which were affected by cold stress and emphasized the similarities and differences in gene expressions response to both climatic stressors. Thermal stress-induced changes in gene expression and in activity of expressed proteins initiated cell stress responses with impact on increased thermotolerance (Lindquist, 1986). From an across-generation perspective, differences in epigenetic modifications are assumed.
365 d weight gain
Also for 365dg, heritabilities in dependency of climatic descriptors were in a moderate-to--large range from 0.38 to 0.64. Instead of weight gain after 1 yr, most of the previous studies conducted in beef cattle considered body weight at the age of 365 d. For 365 d weights in Nellore cattle, Guidolin et al. (2012) estimated heritabilities from 0.24 to 0.62 in different states from Brazil. Bradford et al. (2016) found a heritability of 0.32 for yearling weight in American Angus cattle. As identified for BWT and 200dg, additive genetic variances for 365d and heritabilities were largest in mTHI-class 4 (mTHI 60) and nHS-class 3 (31 to 56 HS days) from the 56 d a.p. period, indicating HS impact. Again, such results are in contrast to findings by Bradford et al. (2016), who reported smaller yearling weight heritabilities for intermediate heat load conditions compared with no-heat load conditions. Regarding different stressors and their impact on epigenetic modifications, Skibiel et al. (2018) highlighted the importance of in utero HS contributing to modifications of the offspring genome, with possible impact on offspring traits recorded later in life.
In agreement with the 56 d period a.p., climatic stress from the 56 d period p.p. was associated with increasing additive genetic variances and heritabilities for 365dg when compared with temperate conditions. However, in contrast to 200dg and climatic impact from the 56 d period p.p., additive genetic variances and heritabilities for 365dg were also comparably large in mTHI-class 1 (mTHI 39) indicating cold stress conditions. As reported by Sonna et al. (2002), cold induces changes in expression of genes with well-established physiological functions and these changes are part of the cellular response to thermal stress. Nevertheless, both climatic descriptors mTHI and nHS indicate large 365dg heritablities when dams suffered HS during the 56 d period a.p. (in utero HS), and when new born calves suffered HS or cold stress directly after birth during the 56 d period p.p. Such results indicate additional selection response for 365dg due to time-lagged stress conditions, contributing to improved genetic differentiation. In dairy cattle, such long-lasting effects through in utero HS were reported by Kipp et al. (2020) for female fertility traits and even longevity.
Again, genetic correlations between 365dg from different mTHI- or nHS-classes were lower when considering distant climatic descriptors. This was observed for both climatic indicator recording periods 56 d a.p. and 56 d p.p. Especially consideration of classes indicating HS and cold stress, e.g., mTHI 39 (mTHI-class 1) and mTHI 60 (mTHI-class 4), indicated obvious GE. Hence, observations made for direct HS impact via random regression model applications with low genetic correlations for THI in great distance (Brgemann et al, 2011; Al-Kanaan et al., 2015) are also relevant from a time-lagged or even an across-generation perspective.
Breeding values
Due to the globalization of dairy cattle breeding and the increasing climatic challenge, it is imperative to select robust sires displaying quite stable favorable breeding values in production as well as in functional traits along the climatic scale. Inclusion of the THI effect in genetic and genomic models contributed to an increase of genomic prediction and EBV accuracies, and inferred obvious G E through re-rankings of sires (Santana et al. 2013; Bernabucci et al., 2014; Bohlouli et al., 2019). For all traits, most of the 10 influential sires showed obvious variations in EBV across mTHI- and nHS-classes, indicating environmental sensitivity (Streit et al., 2012). Only a small number of sires indicated robustness with favorable EBV across all climatic levels. Interestingly, some sires had their highest EBV in both challenging environments representing either HS or cold stress. Regarding sire robustness, Knig and May (2018) identified quite stable sire EBV for traits with moderate-to-large heritbabilties along environmental descriptors, but obvious EBV fluctuations for low heritability functional traits. Environmental sensitivity and indications for G E in dependency of trait heritabilities also were shown by Yin and Knig (2018). Interestingly, Yin and Knig (2018) additionally identified impact of genomic and genetic herd characteristics on estimates for genetic correlations and sire EBVs. Daughters from specific sires, e.g., according to country origin or percentages of Holstein Friesian gene percentages, showed specific trait responses with impact on sire EBVs. Hence, explaining time-lagged-specific daughter response on climatic alterations, offspring from some sires seem to be more susceptible to in utero HS. Such mechanisms including genome modifications need to be studied further on. From a practical breeding perspective, results suggest selection of sires based on their EBV for production traits at time-lagged mTHI- or nHS-classes. For larger datasets, ongoing modelling approaches should focus on random regression approaches, as done in several direct HS-studies (Hammami et al., 2013). Random regression model applications allow the estimation of breeding values and genetic parameters on a continuous THI gradient. Nevertheless, in previous RRM studies (Bohlouli et al., 2019), genetic parameters and EBV had quite large standard errors at the extreme ends of the THI scale (due to the small number of observations). Hence, for small datasets and a limited number of records at extreme THI, Yin and Knig (2018) favored robust multiple-trait model applications.
Our study focused on the time-lagged impact of HS from different recording periods during late pregnancy and postpartum on genetic parameter estimates for BWT and weight gain traits in an local dual-purpose cattle breed, kept under pasture-based conditions. To our knowledge, this is a first study addressing such time-lagged climatic effects on genetic (co)variance components for BWT and weight gain traits. Challenging climatic impact in terms of increasing mTHI and nHS from periods a.p. (in utero climate stress) and p.p. contributed to an increase of additive genetic variances and heritabilities of all traits BWT, 200dg and 365dg. This was the case especially for HS conditions, and partly observed for cold stress environments. Hence, due to the larger heritabilities, selection response in beef cattle traits can be increased when dams suffer HS during late gestation or when calves suffer HS after birth, indicating a pronounced genetic differentiation in challenging environments (also from a time-lagged perspective). From a genetics perspective, such findings may be explained through in utero HS impact on genome modifications (i.e., epigenetics). The impact of in utero HS on (co)variance components implied genetic correlations in same traits from different mTHI- or nHS-levels lower than 0.80, indicating obvious GE. The GE effects were proven by substantial alterations of sire EBVs for production traits in dependency of time-lagged climatic alterations. However, such EBV variations suggest further opportunities with regard to sire selection for improving robustness in outdoor populations such as RHV.
Acknowledgment
This work was financially supported by the German Federal Ministry of Food and Agriculture (BMEL) through the Federal Office for Agriculture and Food (BLE), grant number 2816BM010. The authors gratefully thank for the support.
Glossary
Abbreviations
- EBV
estimated breeding values
- GE
genotypeenvironment interaction
- HS
heat stress
- RH
relative humidity
- RHV
Rotes Hhenvieh
- THI
temperature humidity index
Conflict of Interest Statement
The authors declare no real or perceived conflicts of interest.
Literature Cited
- Al-Kanaan, A., Knig S., and Brgemann K.. . 2015. Effects of heat stress on semen characteristics of Holstein bulls estimated on a continuous phenotypic and genetic scale. Livest. Sci. 177:15–24.. doi: 10.1016/j.livsci.2015.04.003 [DOI] [Google Scholar]
- Bauman, D. E., and Currie W. B.. . 1980. Partitioning of nutrients during pregnancy and lactation: a review of mechanisms involving homeostasis and homeorhesis. J. Dairy Sci. 63:1514–1529.. doi: 10.3168/jds.s0022-0302(80)83111-0. [DOI] [PubMed] [Google Scholar]
- Bernabucci, U., Biffani S., Buggiotti L., Vitali A., Lacetera N., and Nardone A.. . 2014. The effects of heat stress in Italian Holstein dairy cattle. J. Dairy Sci. 97:471–486.. doi: 10.3168/jds.2013-6611. [DOI] [PubMed] [Google Scholar]
- Bohlouli, M., Alijani S., Naderi S., Yin T., and Knig S.. . 2019. Prediction accuracies and genetic parameters for test-day traits from genomic and pedigree-based random regression models with or without heat stress interactions. J. Dairy Sci. 102:488–502.. doi: 10.3168/jds.2018-15329. [DOI] [PubMed] [Google Scholar]
- Bohlouli, M., Shodja J., Alijani S., and Eghbal A.. . 2013. The relationship between temperature-humidity index and test-day milk yield of Iranian Holstein dairy cattle using random regression model. Livest. Sci. 157:414–420.. doi: 10.1016/j.livsci.2013.09.005 [DOI] [Google Scholar]
- Bradford, H. L., Fragomeni B. O., Bertrand J. K., Lourenco D. A., and Misztal I.. . 2016. Genetic evaluations for growth heat tolerance in Angus cattle. J. Anim. Sci. 94:4143–4150.. doi: 10.2527/jas.2016-0707. [DOI] [PubMed] [Google Scholar]
- Brgemann, K., Gernand E., von Borstel U. U., and Knig S.. . 2011. Genetic analyses of protein yield in dairy cows applying random regression models with time-dependent and temperature x humidity-dependent covariates. J. Dairy Sci. 94:4129–4139.. doi: 10.3168/jds.2010-4063. [DOI] [PubMed] [Google Scholar]
- Brgemann, K., Gernand E., Knig von Borstel U., and Knig S.. . 2012. Defining and evaluating heat stress thresholds in different dairy cow production systems. Archiv Tierzucht 55:13–24.. [Google Scholar]
- Cammack, K. M., Antoniou E., Hearne L., and Lamberson W. R.. . 2009. Testicular gene expression in male mice divergent for fertility after heat stress. Theriogenology 71:651–661.. doi: 10.1016/j.theriogenology.2008.09.029. [DOI] [PubMed] [Google Scholar]
- Chud, T. C. S., Caetano S. L., Buzanskas M. E., Grossi D. A., Guidolin D. G. F., Nascimento G. B., Rosa J. O., Lbo R. B., and Munari D. P.. . 2014. Genetic analysis for gestation length, birth weight, weaning weight, and accumulated productivity in Nellore beef cattle. Livest. Sci. 170:16–21.. doi: 10.1016/j.livsci.2014.09.024 [DOI] [Google Scholar]
- Daxinger, L., and Whitelaw E.. . 2012. Understanding transgenerational epigenetic inheritance via the gametes in mammals. Nat. Rev. Genet. 13:153–162.. doi: 10.1038/nrg3188. [DOI] [PubMed] [Google Scholar]
- Eler, J. P., Van Vleck L. D., Ferraz J. B., and Lobo R. B.. . 1995. Estimation of variances due to direct and maternal effects for growth traits of Nellore cattle. J. Anim. Sci. 73:3253–3258.. doi: 10.2527/1995.7311325 [DOI] [PubMed] [Google Scholar]
- Gudex, B., Johnson D., and Singh K.. . 2014. Prenatal maternal and possible transgenerational epigenetic effects on milk production. PLoS One 9:e98928. doi: 10.1371/journal.pone.0098928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidolin, D. G. F., Buzanskas M. E., Ramos S. B., Venturini G. C., Lbo R. B., Paz C. C. P., Munari D. P., and Oliveira J. A.. . 2012. Genotype-environment interaction for post-weaning traits in Nellore beef cattle. Anim. Prod. Sci. 52:975–980.. doi: 10.1071/AN11037 [DOI] [Google Scholar]
- Guo, J. R., Monteiro A. P. A., Weng X. S., Ahmed B. M., Laporta J., Hayen M. J., Dahl G. E., Bernard J. K., and Tao S.. . 2016. Short communication: effect of maternal heat stress in late gestation on blood hormones and metabolites of newborn calves. J. Dairy Sci. 99:6804–6807.. doi: 10.3168/jds.2016-11088. [DOI] [PubMed] [Google Scholar]
- Habeeb, A. A. M., A. E. Gad, A. A. El-Tarabany, and M. A. A. Atta. 2018. Negative effects of heat stress on growth and milk production of farm animals. J. Anim. Hus. & Dairy Sci. 2:1 12. [Google Scholar]
- Halli, K., Brgemann K., Bohlouli M., and Knig S.. . 2020. Time-lagged and acute impact of heat stress on production and fertility traits in the local dual-purpose cattle breed Rotes Hhenvieh under pasture-based conditions. Transl. Anim. Sci. 4:txaa148. doi: 10.1093/tas/txaa148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammami, H., Bormann J., Mhamdi N., Montaldo H. H., and Gengler N.. . 2013. Evaluation of heat stress effects on production traits and somatic cell score of Holsteins in a temperate environment. J. Dairy Sci. 96:1844–1855.. doi: 10.3168/jds.2012-5947. [DOI] [PubMed] [Google Scholar]
- Hammond, J. 1947. Animal breeding in relation to nutrition and environmental conditions. Biol. Rev. Camb. Philos. Soc. 22:195–213.. doi: 10.1111/j.1469-185x.1947.tb00330.x. [DOI] [PubMed] [Google Scholar]
- Hijmans, R. J., Williams E., and Vennes C.. . 2019. Package geosphere. Available from https://cran.r-project.org/web/packages/geosphere/index.html [accessed 25 March, 2020].
- Jeyaruban, M. G., Johnston D. J., and Graser H.-U.. . 2009. Estimation of genotype x environment interactions for growth, fatness and reproductive traits in Australian Angus cattle. Anim. Prod. Sci. 49:1–8.. doi: 10.1071/EA08098 [DOI] [Google Scholar]
- Kadim, T., Mahgoub O., Al-Ajmi D. S., Al-Maqbaly R. S., Al-Mugheiry S. M., and Bartolome D. Y.. . 2004. The influence of season on quality characteristics of hot-boned beef m. longissimus thoracis. Meat SCI. 66:831–836.. doi: 10.1016/j.meatsci.2003.08.001 [DOI] [PubMed] [Google Scholar]
- Kaspar, F., Mller-Westermeier G., Penda E., Mchel H., Zimmermann K., Kaiser-Weiss A., and Deutschlnder T.. . 2013. Monitoring of climate change in Germany data, products and services of Germanys national climate data centre. Adv. Sci. Res. 10:99–106.. doi: 10.5194/asr-10-99-2013 [DOI] [Google Scholar]
- Kipp, C., Brgemann K., Zieger P., Mtze K., Mcklinghoff Wicke S., and Knig S.. . 2020. Across-generation influence of maternal heat stress during late gestation in dairy cows. Proc. EAAP 2020 Virtual Meeting. [DOI] [PubMed] [Google Scholar]
- Knig, S., Dietl G., Raeder I., and Swalve H. H.. . 2005. Genetic relationships for dairy performance between large-scale and small-scale farm conditions. J. Dairy Sci. 88:4087–4096.. doi: 10.3168/jds.S0022-0302(05)73093-9. [DOI] [PubMed] [Google Scholar]
- Knig, S., and May K.. . 2019. Invited review: phenotyping strategies and quantitative-genetic background of resistance, tolerance and resilience associated traits in dairy cattle. Animal 13:897–908.. doi: 10.1017/S1751731118003208. [DOI] [PubMed] [Google Scholar]
- Kreienkamp, F., Lorenz P., and Geiger T.. . 2020. Statistically downscaled CMIP6 projections show stronger warming for Germany. Atmosphere 11:1245. doi: 10.3390/atmos11111245 [DOI] [Google Scholar]
- Lindquist, S. 1986. The heat-shock response. Annu. Rev. Biochem. 55:1151–1191.. [DOI] [PubMed] [Google Scholar]
- Mackinnon, M. J., Meyer K., and Hetzel D. J. S.. . 1991. Genetic variation and covariation for growth, parasite resistance and heat tolerance in tropical cattle. Livest. Prod. Sci. 27:105–122.. doi: 10.1016/0301-6226(91)90090-D [DOI] [Google Scholar]
- Madsen, P., and Jensen J.. . 2013. DMU: a package for analysing multivariate mixed models. Available from http://dmu.agrsci.dk/DMU/DocCurrent/dmuv6guide.5.2.pdf. [accessed November 18, 2020].
- Mahmoud, M., Zeng Y., Shirali M., Yin T., Brgemann K., Knig S., and Haley C.. . 2018. Genome-wide pleiotropy and shared biological pathways for resistance to bovine pathogens. PLoS One 13:e0194374. doi: 10.1371/journal.pone.0194374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, K. 1995. Estimates of genetic parameters and breeding values for New Zealand and Australian Angus cattle. Aust. J. Agric. Res. 46:1219–1229.. doi: 10.1071/AR9951219 [DOI] [Google Scholar]
- Mitlhner, F. M., Morrow J. L., Dailey J. W., Wilson S. C., Galyean M. L., Miller M. F., and McGlone J. J.. . 2001. Shade and water misting effects on behavior, physiology, performance, and carcass traits of heat-stressed feedlot cattle. J. Anim. Sci. 79:2327–2335.. doi: 10.2527/2001.7992327x [DOI] [PubMed] [Google Scholar]
- Monteiro, A. P. A., Tao S., Thompson I. M. T., and Dahl G. E.. . 2016. In utero heat stress decreases calf survival and performance through the first lactation. J. Dairy Sci. 99:8443–8450.. doi: 10.3168/jds.2016-11072. [DOI] [PubMed] [Google Scholar]
- Mllenhoff, A. 2008. Schtzung genetisch-statistischer Parameter bei Fleischrindern der Rassen Deutsche Angus und Deutsches Fleckvieh sowie deren Einfachkreizungen [PhD dissertation]. Giessen, Germany: Justus-Liebig-University. [Google Scholar]
- Muller, L. D., Beardsley G. L., Ellis R. P., Reed D. E., and Owens M. J.. . 1975. Calf response to the initiation of parturition in dairy cows with dexamethasone or dexamethasone with estradiol benzoate. J. Anim. Sci. 41:1711–1716.. doi: 10.2527/jas1975.4161711x. [DOI] [PubMed] [Google Scholar]
- National Research Council (NRC). 1971. A guide to environmental research on animals. Washington, DC: National Academic Science. [Google Scholar]
- Raidan, F. S. S., Passafaro T. L., Fragomeni B. O., Josahkian L. A., Pereira I. G., and Toral F. L. B.. . 2015. Genotype x environment interaction in individual performance and progeny tests in beef cattle. J. Anim. Sci. 93:920–933.. doi: 10.2527/jas2014-798 [DOI] [PubMed] [Google Scholar]
- Robertson, A. 1959. The sampling variance of the genetic correlation coefficient. Biometrics 15:469–485.. doi: 10.2307/2527750 [DOI] [Google Scholar]
- Sakatani, M. 2017. Effects of heat stress on bovine preimplantation embryos produced in vitro. J. Reprod. Dev. 63:347–352.. doi: 10.1262/jrd.2017-045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santana, M. L., Jr, Bignardi A. B., Pereira R. J., Menndez-Buxadera A., and El Faro L.. . 2016. Random regression models to account for the effect of genotype by environment interaction due to heat stress on the milk yield of Holstein cows under tropical conditions. J. Appl. Genet. 57:119–127.. doi: 10.1007/s13353-015-0301-x. [DOI] [PubMed] [Google Scholar]
- Santana, M. L., Eler J. P., Cardoso F. F., Albuquerque L. G., and Ferraz J. B.. . 2013. Phenotypic plasticity of composite beef cattle performance using reaction norms model with unknown covariate. Animal 7:202–210.. doi: 10.1017/S1751731112001711. [DOI] [PubMed] [Google Scholar]
- Schafberg, R., Rosner F., and Swalve H. H.. . 2006. Examinations on intramammary infections in dairy cows based on pathogen-specific data. Proceedings of the 8th World Congress on Genetics Applied to Livestock Production, Belo Horizonte, Minas Gerais, Brazil; p. 15–13.. [Google Scholar]
- Schierenbeck, S., Reinhardt F., Reents R., Simianer H., and Knig S.. . 2011. Identification of informative cooperator herds for progeny testing based on yield deviations. J. Dairy Sci. 94:2071–2082.. doi: 10.3168/jds.2010-3466. [DOI] [PubMed] [Google Scholar]
- Seong, K. H., Li D., Shimizu H., Nakamura R., and Ishii S.. . 2011. Inheritance of stress-induced, ATF-2-dependent epigenetic change. Cell 145:1049–1061.. doi: 10.1016/j.cell.2011.05.029. [DOI] [PubMed] [Google Scholar]
- Skibiel, A. L., Peagaricano F., Amorn R., Ahmed B. M., Dahl G. E., and Laporta J.. . 2018. In utero heat stress alters the offspring epigenome. Sci. Rep. 8:14609. doi: 10.1038/s41598-018-32975-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonna, L. A., Fujita J., Gaffin S. L., and Lilly C. M.. . 2002. Invited review: effects of heat and cold stress on mammalian gene expression. J. Appl. Physiol. (1985). 92:1725–1742.. doi: 10.1152/japplphysiol.01143.2001. [DOI] [PubMed] [Google Scholar]
- Streit, M., Reinhardt F., Thaller G., and Bennewitz J.. . 2012. Reaction norms and genotype-by-environment interaction in the German Holstein dairy cattle. J. Anim. Breed. Genet. 129:380–389.. doi: 10.1111/j.1439-0388.2012.00999.x. [DOI] [PubMed] [Google Scholar]
- Tao, S., Monteiro A. P., Thompson I. M., Hayen M. J., and Dahl G. E.. . 2012. Effect of late-gestation maternal heat stress on growth and immune function of dairy calves. J. Dairy Sci. 95:7128–7136.. doi: 10.3168/jds.2012-5697. [DOI] [PubMed] [Google Scholar]
- Via, S., and Lande R.. . 1985. Genotype-environment interaction and the evolution of phenotypic plasticity. Evolution 39:505–522.. doi: 10.1111/j.1558-5646.1985.tb00391.x. [DOI] [PubMed] [Google Scholar]
- Williams, J. L., Bertrand J. K., Misztal I., and ukaszewicz M.. . 2012. Genotype by environment interaction for growth due to altitude in United States Angus cattle. J. Anim. Sci. 90:2152–2158.. doi: 10.2527/jas.2011-4365. [DOI] [PubMed] [Google Scholar]
- Yin, T., and Knig S.. . 2018. Heritabilities and genetic correlations in the same traits across different strata of herds created according to continuous genomic, genetic, and phenotypic discriptors. J. Dairy Sci. 101:2171–2186.. doi: 10.3168/jds.201713575 [DOI] [PubMed] [Google Scholar]
- Yin, T., and Knig S.. . 2019. Genome-wide associations and detection of potential candidate genes for direct genetic and maternal genetic effects influencing dairy cattle body weight at different ages. Genet. Sel. Evol. 51:4. doi: 10.1186/s12711-018-0444-4. [DOI] [PMC free article] [PubMed] [Google Scholar]