Abstract
COVID-19 pandemic has resulted in an unprecedented global public health crisis. It is obvious that SARS-CoV-2 vaccine is needed to control the global COVID-19 public health crisis. Since obvious advantages including fast manufacturing speed, potent immunogenicity and good safety profile, six mRNA vaccines have been used to prevent SARS-CoV-2 infections in clinic with lipid nanoparticles (LNP) formulation via intramuscular injection. In this work, we first constructed RBD-encoding mRNA (RBD-mRNA) formulated in liposomes (LPX/RBD-mRNA) and investigated the influence of administration routes on the immunogenicity. LPX/RBD-mRNA can express RBD in vivo and successfully induced SARS-CoV-2 RBD specific antibodies in the vaccinated mice, which efficiently neutralized SARS-CoV-2 pseudotyped virus. Moreover, the administration routes were found to affect the virus neutralizing capacity of sera derived from the immunized mice and the types (Th1-type and Th2-type) of cellular immune responses. This study indicated that liposome-based RBD-mRNA vaccine with optimal administration route might be a potential candidate against SARS-CoV-2 infection with good efficacy and safety.
Keywords: SARS-CoV-2, mRNA vaccines, Receptor-binding domain, Liposomes, Administration route
Graphical abstract
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has extended throughout the world at the speed of exponential [1]. And it has resulted in an unprecedented global public health crisis that was declared “the coronavirus disease 2019 (COVID-19) pandemic” by the World Health Organization (WHO) [2]. The emergency shutdown of public life in many countries has caused unparalleled social and economic grief [3]. Since it happened nearly one year ago, the virus evil has infected over 70 million innocent people and killed about 1.6 million people in the whole world (https://covid19.who.int/). It is obvious that there is an urgent need for intervention before it is too late, especially advantageous treatments or vaccines against the COVID-19 pandemic [4].
As a novel beta-coronavirus, SARS-CoV-2 shares 79% genome sequence identity with SARS-CoV and 50% with MERS-CoV [5,6]. The six functional open reading frames (ORFs) of genome organization are arranged in replicase (ORF1a/ORF1b), spike (S), envelope (E), membrane (M), and nucleocapsid (N) [7,8]. Together with the other two highly pathogenic human coronaviruses, SARS-CoV and MERS-CoV, SARS-CoV-2 invades host cells by binding to its receptor [9], angiotensin-converting enzyme 2 (ACE2), through the receptor-binding domain (RBD) of S protein [10]. Based on our knowledge of the infection mechanism of SARS-CoV-2, utilizing RBD as immunogen to induce specific antibodies may block this recognition and prevent the invasion efficiently [11]. Recent studies also demonstrated that the use of RBD to produce neutralizing antibodies may reduce the risk of antibody-dependent enhancement (ADE) of infection [12].
Messenger RNA (mRNA)-based vaccines have been used to prevent viral infections with obvious advantages including fast manufacturing speed, potent immunogenic, and good safety profile [13,14]. Six mRNA vaccines formulated in lipid nanoparticles (LNP) against COVID-19 have been in clinical trials or granted a conditional marketing authorization [15,16]. However, all of these mRNA vaccines were formulated in LNP and administrated by intramuscular injection [17]. Other formulations like liposome (LP) and mode of administration have not been evaluated in the limited timeframe. On the other hand, their effectiveness and safety have not yet been proven completely [18]. Therefore, we developed RBD-encoding mRNA (RBD-mRNA) formulated in liposomes and investigated the influence of routes of administration on the immunogenicity. The LP-formulated mRNA vaccine was finally found to have good immunogenicity in mice treated by multiple routes of administration and protected host cells from SARS-CoV-2 pseudovirus infection, which supports further research for clinical development in humans.
2. Materials and methods
2.1. Materials
1,2-dioleoyloxy-3-(trimethylammonium)propane (DOTAP) chloride and cholesterol were purchased from A.V.T. Pharmaceutical Co., Ltd. T7 mScript™ Standard mRNA Production System was obtained from CELLSCRIPT. All the plasmid templates were provided by Convenience Biology. Restriction enzyme was supplied from New England Biolabs. Luciferase substrate was purchased from BioVision. Dulbecco's Modified Eagle's medium (DMEM), trypsin-EDTA, 1% penicillin-streptomycin (P/S) and fetal bovine serum (FBS) were obtained from Gibco. HEK293T cell line, derived from human embryonic kidney 293 cells, was purchased from American Type Culture Collection (ATCC). Human ACE2-overexpressing cells (HEK293T-hACE2), created by transducing HEK293T cells with lentiviral vector expressing ACE2, were provided sincerely by Prof. Guangwen Lu and Aiping Tong in our lab. The immortalized murine dendritic cells (DC2.4 cells) were kindly provided by Prof. Xun Sun from West China School of Pharmacy, Sichuan University (Chengdu, China). Recombinant SARS-CoV-2 spike RBD-His protein was obtained from Navoprotein. HRP-conjugated anti-mouse IgG, IgM, IgG1, and IgG2a were purchased from Abcam. RNase-free water was prepared using a Milli-Q Synthesis System. All other solvents and reagents were used as received.
2.2. Cell culture
HEK293T, HEK293T-hACE2 and DC2.4 cells were maintained in DMEM containing 10% FBS (v/v) and 1% P/S (v/v) in a humidified atmosphere containing 5% CO2 at 37 °C.
2.3. Animals
C57BL/6 male mice (8 weeks old) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd.. All animals received care in compliance with the guidelines outlined in the Guide for the Care and Use of Laboratory Animals. The procedures on animals were approved by the ‘Animal Care and Use Committee’ of Sichuan University.
2.4. mRNA synthesis
SARS-CoV-2 isolate Wuhan-Hu-1 (GenBank MN908947.3) was used as the reference amino acid sequence alignment of RBD. The mRNA was produced in vitro using T7 RNA polymerase-mediated transcription from a linearized DNA template containing the optimal codons of RBD region and the untranslated regions [19].
2.5. Encapsulation of mRNA into LP
The liposomes were prepared using a modified thin-film dispersion method [20]. DOTAP chloride and cholesterol were dissolved in ethanol with the molar ratio of 1:1. The organic solvents were subsequently removed using a rotary evaporator to produce a thin film of lipid at 37 °C. The lipid film was hydrated with 4 mL RNase-free water for 1 h at 60 °C. The final LP preparation was acquired after the above suspension was sonicated for 3 min at 100 W in an ice bath and filtered through a 0.22 μm syringe filter. The LP was mixed with mRNA at an N/P ratio of 3:1 to form mRNA encapsulated liposome (LPX). LPX was supplemented with glucose to form an isotonic solution before use.
2.6. Determination of particle size and size distribution
The size distribution of LPX in 5% glucose solution was measured by a Malvern Zetasizer Nano ZS90 with a He—Ne laser (633 nm) at 90° collecting optics. The data were analyzed by Malvern Dispersion Technology Program 7.13.
2.7. Storage stability
LPX/RBD-mRNA containing 10% sucrose was stored at 4, −20 and -80 °C with multipartite for about one month. On day 0, 1, 8, 13 and 36, samples under different Storage conditions were thawed at room temperature. The size distribution of LPX was measured by DLS.
2.8. RBD-mRNA expression in vitro
For western blotting analysis, HEK293T cells were seeded in 24-well plates at 2 × 105 cells/well. After 12 h incubation, the cells were transfected with RBD-mRNA (2 μg/mL) using Lipofectamine 2000 Transfection Reagent (Thermo Fisher Scientific). 24 h later, the cells were collected and then lysed in RIPA lysis buffer. The expression of RBD protein was then detected by western blotting.
For flow cytometry analysis, HEK293T and DC2.4 cells were seeded in 24-well plates at 2 × 105 cells/well. After 12 h incubation, the cells were transfected with RBD-EGFP-mRNA encoding EGFP-tag fused RBD at C-terminal (2 μg/mL) using LPX. 24 h later, the cells were collected and then analyzed by flow cytometry.
For enzyme-linked immunosorbent assay (ELISA) analysis, HEK293T cells were seeded in 24-well plates at 2 × 105 cells/well. After 12 h incubation, the cells were transfected with RBD-mRNA-loaded LP (LPX/RBD-mRNA) at a dose of 2 μg/mL mRNA. 12 and 24 h later, the cells and supernatant were collected. The cells were lysed with ultrasonication. The cell lysate and supernatant were clarified by centrifugation at 13000 rpm and then analyzed by SARS-CoV-2 spike RBD ELISA kit (Sino Biological) according to the product specification.
2.9. RBD-mRNA expression in vivo
For the analysis of RBD-mRNA expression in major tissue organs, C57BL/6 male mice were intravenously administered with LPX/RBD-mRNA, LPX encapsulating firefly luciferase reporter-encoding mRNA (FLuc-mRNA), via tail vein at a dose of 30 μg mRNA per mouse. 6 h later, the mice were injected intraperitoneally (i.p.) with luciferase substrate. 15 min later, all the mice were sacrificed and the liver, spleen, lung, and kidney were excised. Fluorescence signals were collected by In Vivo Imaging System (IVIS) Spectrum instrument (PerkinElmer) for 60 s.
For the detection of mRNA-translated RBD secreted into blood, C57BL/6 male mice were administered with LPX/RBD-mRNA via different routes of administration at a dose of 30 μg mRNA per mouse. After predetermined time point 0.5, 1, 2, 3 and 4 days, 200 μL of blood samples were collected from the retro-orbital plexus of mice. The blood was then centrifuged at 4 °C (3000 g, 10 min) to collect the sera. The content of the RBD protein in sera was measured with SARS-CoV-2 spike RBD ELISA kit (Sino Biological) according to the product specification.
2.10. Vaccination in mice
C57BL/6 mice were randomly divided into six groups. 30 μg mRNA/150 μL of LPX/RBD-mRNA was administered to mice on day 0, day 10, and day 20 by intravenous (i.v.), intramuscular (i.m.), hypodermic (i.h.), intradermal (i.d.), or intraperitoneal (i.p.) injection, respectively. Specifically, i.v. and i.m. injections were administered in the lateral tail vein and the right caudal thigh muscles, respectively. The i.h. and i.d. injections were performed in the neck of the mice and the dermis of skin on the mouse back, which is located between the epidermis and the hypodermis. The i.p. injection was made in the lower right quadrant of the mouse's abdomen. Blood samples were collected by a sterile capillary from eye sockets on day 15 and day 30. After centrifuged at 5000 g for 15 min at 4 °C, sera samples were acquired and heated at 56 °C for 30 min for further use.
2.11. Antibody detection
Epitope-specific antibody titers were detected by a semi-quantitative enzyme-linked immunosorbent assay (ELISA). Briefly, recombinant SARS-CoV-2 RBD protein was coated on 96-well assay plate (High Binding polystyrene, Corning) with a final concentration of 2.0 μg/mL. After incubated overnight at 4 °C, plates were blocked with 2% Bull Serum Albumin (BSA) for 4 h at room temperature and washed three times with washing buffer. Subsequently, serial dilutions of the sera samples were added and incubated overnight at 4 °C. After washed five times with washing buffer, HPR-conjugated goat anti-mouse IgG (1:50000 dilution), IgM (1:50000 dilution), IgG1 (1:100000 dilution), and IgG2a (1:2000 dilution) were added to the plate and incubated for 2 h at 37 °C. The trimethyl borane (TMB) solution (Solarbio) was then added and the plates were incubated for 30 min without light. Finally, 2 M H2SO4 was applied to stop the reactions, and the following measurement was performed on an ELISA reader at 450 nm.
2.12. Pseudovirus-based neutralization assay
HEK293T-hACE2 cells were seeded in 96-well plates at 2 × 104 cells/well and incubated for 24 h. Serial 3-fold diluted sera with DMEM, starting at 1:50, were incubated with SARS-CoV-2 pseudovirus (Genewiz) at a final concentration of 2 × 105 TU/mL for 1 h at 37 °C and then used to treat HEK293T-hACE2 cells. After another incubation for 48 h, the cells were collected for Green Fluorescent Proteins (GFP) positive rate analysis using flow cytometry. The virus control wells were only treated by SARS-CoV-2 pseudovirus without pre-incubation with any serum at the same concentration. The 50% neutralization titer (NT50) was defined as the serum dilution at which the GFP positive rates were reduced by 50% compared with the virus control wells.
2.13. Toxicity assay
Male C57BL/6 mice were treated with 30 μg mRNA/150 μL of LPX/RBD-mRNA by different injection routes. The blood was drawn and serum was used for cytokine analysis 12 h post-injection. All the mice were then sacrificed 14 days after boost vaccination. The heart, liver, spleen, lung and kidney were collected. Tissues were finally sectioned and stained with H&E. All slides images were taken by an Olympus BX 43 fluorescence microscope (Olympus Corp, Tokyo, Japan).
2.14. Statistical analysis
Statistical analyses were performed using GraphPad Prism 8.01 software. One-way analysis of variance (ANOVA) was used to assess the significance of differences among groups. The significance was defined as follows: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
3. Results
3.1. Preparation and characterization of LPX/RBD-mRNA
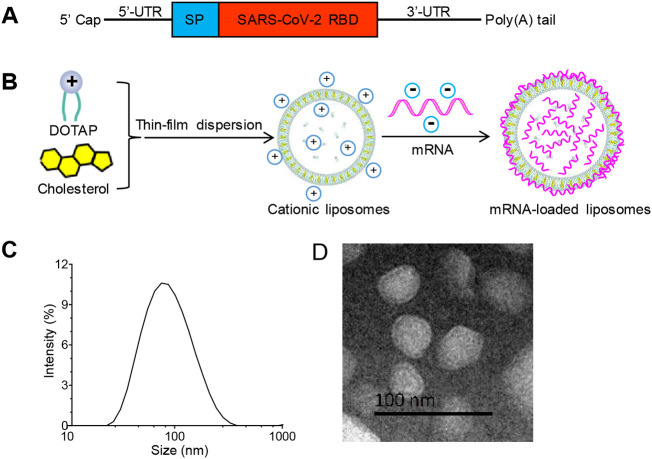
Based on the sequence information of SARS-CoV-2 isolate Wuhan-Hu-1, we generated RBD as the model target antigen from amino acids 319–541 of SARS-CoV-2 spike protein [21]. We designed RBD-mRNA with a type 1 (N7mGpppGm) cap, optimal 5′ and 3′ untranslated sequences, a secretion signal sequence, and a poly(A) tail (Fig. 1A). LP was prepared using a classical thin-film dispersion method [20]. RBD-mRNA was mixed with LP to form LPX/RBD-mRNA (Fig. 1B). Dynamic light scattering (DLS) measurement showed that the average hydrodynamic diameter of the LPX was 74.2 ± 0.8 nm with a narrow distribution (PDI = 0.22 ± 0.03) (Fig. 1C). Meanwhile, the zeta potential was about 40 mV. The typical morphology of LPX/RBD-mRNA was analyzed using transmission electron microscopy (TEM), which showed that the particles were nearly spherical in size about 50–150 nm (Fig. 1D).
Fig. 1.
Preparation and characterization of RBD-mRNA loaded liposome (LPX/RBD-mRNA). (A) Schematic of the RBD-mRNA construct. (B) Sketch map of the preparation of LPX/RBD-mRNA by a thin-film dispersion method. (C) The size distribution of LPX/RBD-mRNA measured by DLS. (D) Typical TEM image of LPX/RBD-mRNA (scale bar 100 nm).
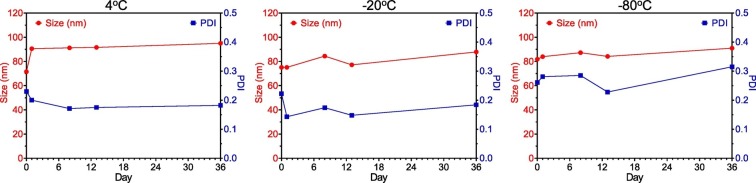
The time dependent DLS measurement indicated that the particle sizes and PDI were not changed over about one month (Fig. 2 ), suggesting the good stability of LPX/RBD-mRNA stored under different conditions.
Fig. 2.
The time dependent size variation of LPX/RBD-mRNA stored under different conditions.
3.2. RBD-mRNA expression
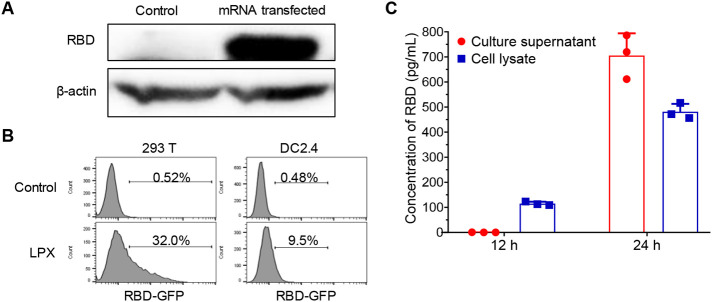
To verify the structure of RBD-mRNA, we transfected HEK293T cells with RBD-mRNA using the commercial transfection reagent and detected RBD protein with western blotting. As shown in Fig. 3A, RBD-mRNA can be successfully translated into RBD protein. Next, we evaluated the in vitro delivery capability of LP for mRNA in HEK293T and DC2.4 cell lines using RBD-EGFP-mRNA. The flow cytometry analysis showed that the two cell lines can efficiently express EGFP after treated by LPX/RBD-EGFP-mRNA. The EGFP+ cells were up to 32.0% and 9.5% in HEK293T and DC2.4 cells (Fig. 3B), respectively. Furthermore, ELISA analysis was carried out to test the RBD expression in 293T cells treated by LPX/RBD-mRNA. Fig. 3C demonstrated that RBD was translated high-efficiently and can also be secreted outside the cells.
Fig. 3.
The efficient expression of mRNA loaded into LP in vitro. (A) The RBD-mRNA can be successfully translated into RBD protein in HEK293T cells at 24 h post-transfection. (B) Flow cytometry analysis of GFP positive rate of HEK293T and DC2.4 cells at 24 h after transfection with LPX/RBD-EGFP-mRNA. (C) The ELISA detection of RBD expressed in HEK293T cell lysate and secreted into the culture supernatant at 12 h and 24 h after LPX/RBD-mRNA transfection.
3.3. LPX translation and tissue distribution in vivo
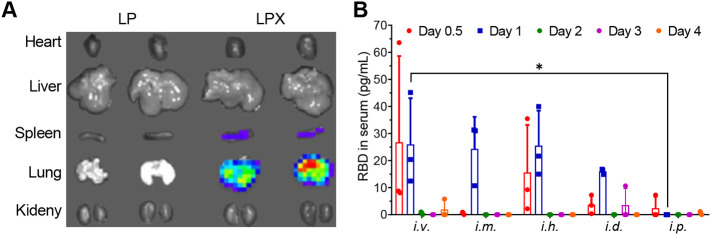
To visualize the translation and tissue distribution in vivo of the LPX formulation, we prepared LPX/FLuc-mRNA and injected it to C57BL/6 mice via intravenous administration. 6 h later, a robust expression of FLuc-mRNA was observed in the lung and a low expression in the spleen, whereas no signal was detected in other tissues (Fig. 4A). Next, we evaluated the secretion of RBD protein into sera of C57BL/6 mice treated by LPX/RBD-mRNA via five different injection routes (i.v., i.m., i.h., i.d. and i.p). As seen in Fig. 4B, the RBD protein was detected in the sera of mice after 12 h (day 0.5) of treatment by i.v., i.h., i.d. and i.p. LPX/RBD-mRNA rather than i.m. LPX/RBD-mRNA. While the RBD protein was found in the sera of all the mice except those treated by i.p. LPX/RBD-mRNA after 24 h (day 1) of treatment. There were almost no RBD proteins found in the sera at day 2, day 3 and day 4 in all the groups.
Fig. 4.
The effective expression of mRNA encapsulated into LPX in vivo (n = 3). (A) The representative image of the expression and biodistribution of FLuc-mRNA at 6 h in C57BL/6 mice treated by LPX/ FLuc-mRNA via intravenous injection. (B) The RBD levels in sera in C57BL/6 mice treated by LPX/RBD-mRNA via different injection routes. * P < 0.05.
3.4. Evaluation of immunogenicity and efficacy of LPX/RBD-mRNA in mice
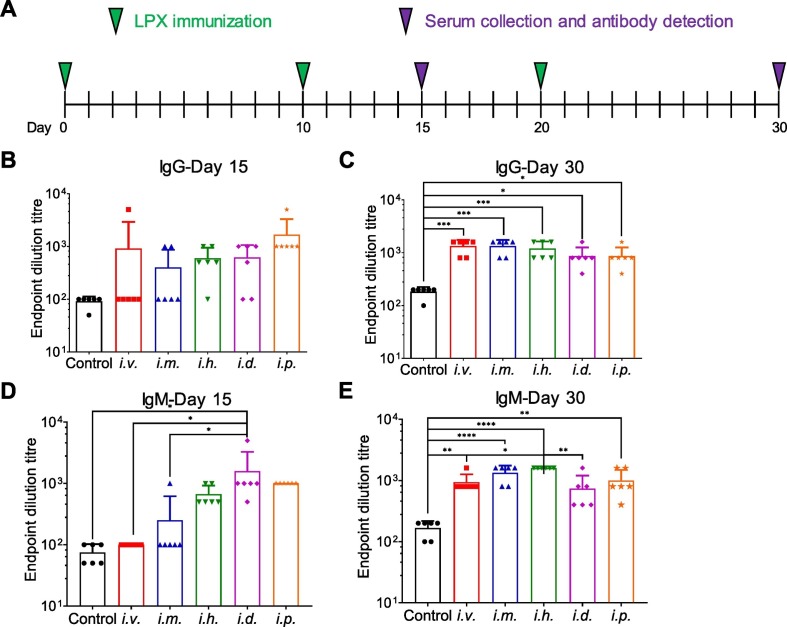
The immune responses induced by LPX/RBD-mRNA were tested in C57BL/6 mice inoculated via five different injection routes (i.v., i.m., i.h., i.d. and i.p) with 30 μg of RBD-mRNA. These mice were administrated LPX/RBD-mRNA on day 0, day 10, and day 20. And serum samples were collected to detect RBD-specific antibodies on day 15 and day 30 (Fig. 5A). After twice immunizations, SARS-CoV-2 RBD-specific IgG and IgM antibodies with 50% endpoint dilution titers approached about 1/1000 on day 15 and day 30 (Fig. 5B–E).
Fig. 5.
Immune responses induced by LPX/RBD-mRNA in vaccinated C57BL/6 mice (n = 6). (A) Schematic diagram of immunization and sample collection. (B-E) The SARS-CoV-2 RBD specific IgG and IgM antibody titers on day 15 and day 30 determined by ELISA. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
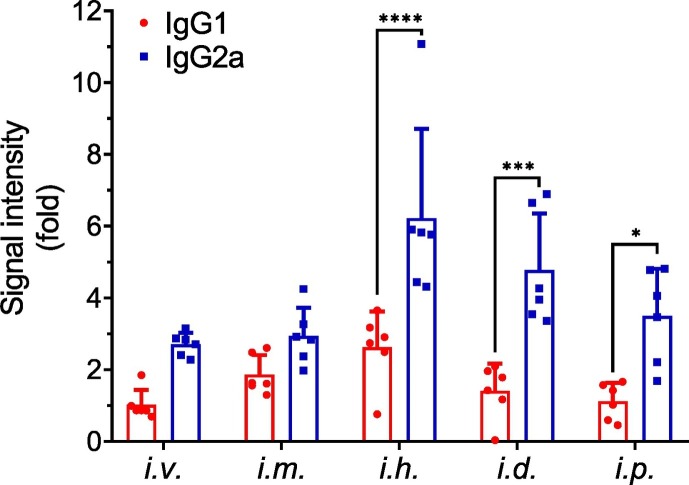
3.5. IgG subtypes determination
IgG1 antibody is a marker for Th2-biased immune response, whereas IgG2a antibodies indicate a favorable Th1-biased immune response in mice [22]. IgG subtype-specific ELISA was performed with the sera collected on day 30 (Fig. 6 ). There was no significant difference of IgG subtypes between the i.v. and i.m. administration, while higher IgG2a responses than IgG1 was induced by the other three injection routes (i.h., i.d. and i.p.).
Fig. 6.
IgG1 and IgG2a levels in C57BL/6 mice vaccinated with LPX/RBD-mRNA via different injection routes (n = 6). The IgG subtypes in serum on day 30 were determined by ELISA. Statistical analyses were performed using GraphPad Prism 8.01 software. One-way analysis of variance (ANOVA) was used to assess the significance of differences among groups.
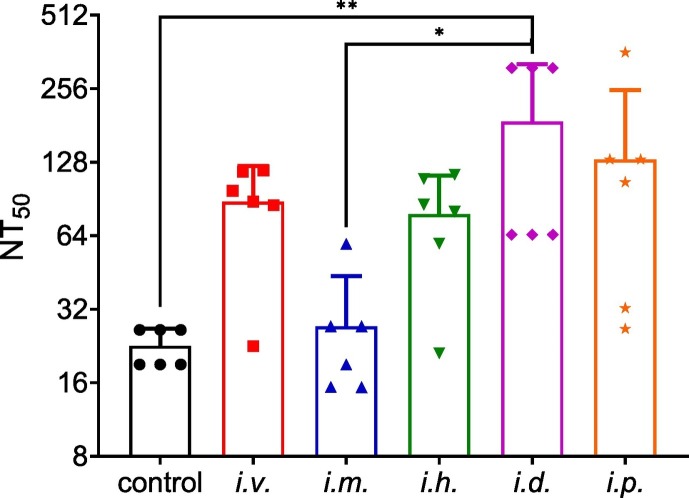
3.6. Pseudovirus neutralization
The SARS-CoV-2 neutralization assay was performed using pseudovirus. The immune sera from the vaccinated mice were tested for the neutralizing activity against SARS-CoV-2 pseudovirus in HEK293T-hACE2 cells. After three immunizations, the sera on day 30 were used to test the pseudovirus neutralization. As seen in Fig. 7 , similar NT50 titers (about 1/100 to 1/200) were approached in the four groups (i.v., i.h., i.d. and i.p.), while a relatively lower NT50 titer (1/30) was found in mice with i.m. administration.
Fig. 7.
SARS-CoV-2 pseudovirus neutralization. The NT50 titers of the sera on day 30 were determined using SARS-CoV-2 pseudovirus infection. Statistical analyses were performed using GraphPad Prism 8.01 software. One-way analysis of variance (ANOVA) was used to assess the significance of differences among groups.
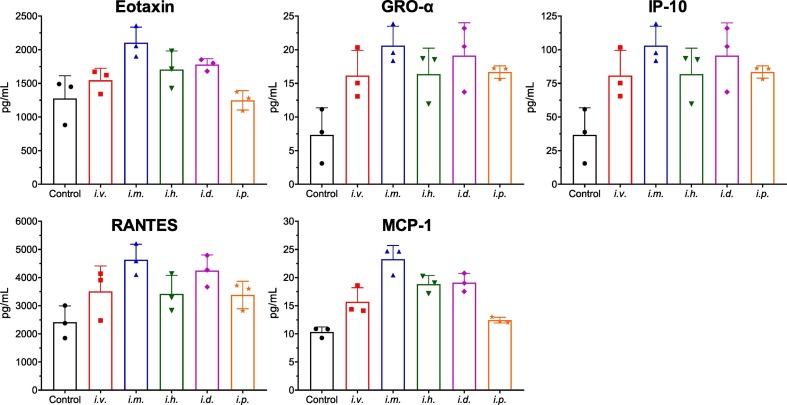
3.7. Toxicity assay
The immune tolerability of LPX/RBD-mRNA were tested in C57BL/6 mice inoculated via five different injection routes (i.v., i.m., i.h., i.d. and i.p) with 30 μg of RBD-mRNA. Changes of eotaxin, GRO-α, IP-10, RANTES and MCP-1 were detected 12 h after administration. All the groups presented increase in these immune function indexes (Fig. 8 ). The i.p. administration group induced the lowest systemic cytokine production. Interestingly, the i.v. administration group induced a lower increase than i.m. administration group.
Fig. 8.
Immune tolerability of LPX/RBD-mRNA in the treated mice. The serum concentrations of cytokines including eotaxin, GRO-α, IP-10, RANTES, and MCP-1 were measured by ELISA 12 h after a single administration of 30 μg LPX/RBD-mRNA (n = 3 per group).
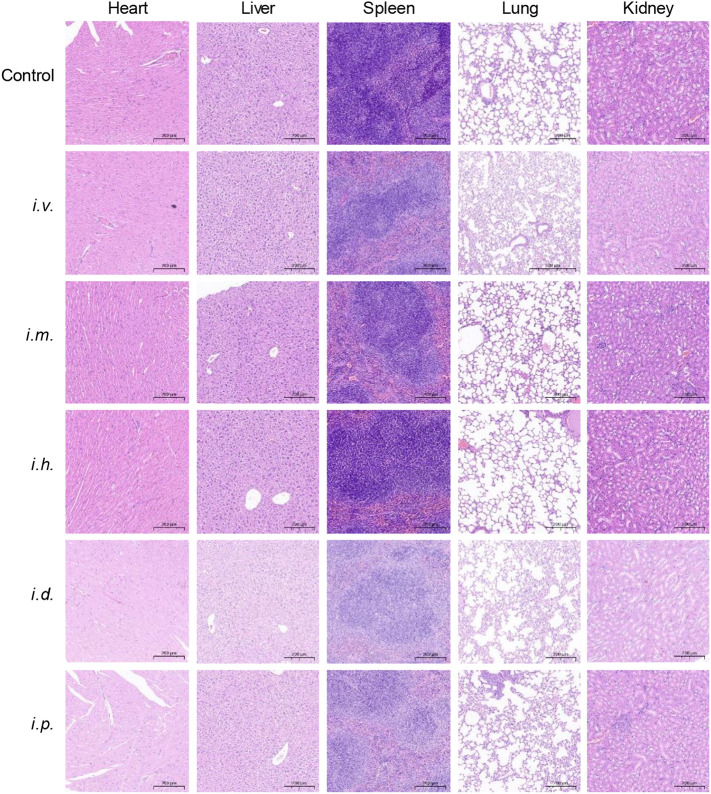
After boost vaccination, all the mice were sacrificed and the major organs were excised and examined via H&E staining (Fig. 9 ). The results showed that there were no obvious histopathologic differences in the main organs between the control and LPX/RBD-mRNA treatment groups. These data demonstrated that the LPX/RBD-mRNA would not cause systemic toxicity.
Fig. 9.
H&E analyses of major organs after boost vaccination with 30 μg LPX/RBD-mRNA (scale bar 200 μm).
4. Discussion
SARS-CoV-2 vaccines seem helpful to control the global COVID-19 public health crisis [23]. In this study, we provided insights into RBD-mRNA encapsulated liposome to fight against SARS-CoV-2 infection and characterized its immunogenicity in different modes of administration. To our best knowledge, LPX/mRNA vaccines are the first example to accomplish this aim. We observed that LPX/RBD-mRNA immunization induced RBD-specific IgG antibodies in mice, which were able to efficiently neutralize SARS-CoV-2 pseudotyped virus. Thus, this liposome-based mRNA vaccine is worthy of being further investigated in non-human primates to evaluate the protection efficacy against the wild-type SARS-CoV-2 virus in the Biosafety Level 3 Laboratory (BSL-3).
mRNA vaccines have not yet produced any formally registered new drug although two COVID-19 mRNA vaccines have been approved for emergency use [24]. All the six COVID-19 mRNA vaccines in clinic are delivered via LNP and administrated via i.m. injection [17]. mRNA vaccines are a relatively neonatal field [25]. More mRNA delivery systems need to be further researched and developed [26]. Safety and efficacy of mRNA-liposomal formulations have been designed and showed promise for cancer immunotherapy and infectious diseases [27]. Although i.m. administration is the most commonly used route for preventive vaccines, i.h. administration also is explored for several new vaccines under development [28,29]. In this study, LPX/RBD-mRNA vaccine was systemically investigated via various routes of administration (i.v., i.m., i.h., i.d. and i.p.). The administration routes showed a slight influence on the concentration of RBD secreted into sera in mice as shown in Fig. 3B. Moreover, the i.d. administration can rapidly induce the specific IgM compared to i.v. and i.m. vaccination routes in the early stage of immunization according to the data determined on day 15 (Fig. 5D). Nevertheless, LPX/RBD-mRNA vaccination with five different administration induced strong immune response with SARS-CoV-2 RBD-specific IgG and IgM antibodies at similar levels. However, i.d. administration achieved stronger efficacy on the SARS-CoV-2 pseudovirus neutralization than i.m. injection route. These data indicated that the administration route of mRNA vaccine might influence the final efficacy of the preventative vaccines. The i.m. administration has been used in the two COVID-19 mRNA vaccines approved for emergency use (mRNA-1273 and BNT162b2). The immune tolerability assay indicated that i.m. administration group have the highest increase on some immune function index (Fig. 8), while i.p. administration induced the lowest systemic cytokine production. Even the i.v. administration group induced a lower increase than i.m. administration group.
Avoiding induction of non-neutralizing antibody and Th2-biased cellular immune responses are important factors for secure SARS-CoV-2 vaccine [30]. mRNA vaccines can efficiently induce key regulators of the Tfh cell program and influence the functional properties of Tfh cells [31]. Th2-biased cellular immune responses have been associated with the enhancement of lung disease following infection [30]. Compared with the full-length S protein, RBD antigen may induce fewer non-neutralizing antibodies, lowering the risk of potential ADE of SARS-CoV-2 infection [12]. In this study, we found that the administration routes would affect the IgG subtypes (Fig. 6). The three injection routes (i.h., i.d. and i.p.) prefer to induce Th1-biased immune responses. Therefore, it's necessary to screen the optimal administration route of mRNA vaccines in order to achieve good preventive efficacy and safety.
5. Conclusion
In summary, we reported a SARS-CoV-2 RBD-mRNA encapsulated liposome to fight against SARS-CoV-2 infection for the first time. The immunogenicity was systemically characterized in different modes of administrations. LPX/RBD-mRNA immunizations induced SARS-CoV-2 RBD specific antibodies in mice vaccinated by 3 doses, which were able to efficiently neutralize SARS-CoV-2 pseudotyped virus. We also observed that the administration routes of LPX/RBD-mRNA vaccine influenced the virus neutralizing capacity of sera and the types (Th1-type and Th2-type) of cellular immune responses. Eventually, our investigation indicated that liposome-based mRNA vaccine is a potential and promising candidate for SARS-CoV-2 vaccines and that the rational design for administration route might be essential to mRNA vaccines in the prevention of infectious diseases.
Author Contribution
Research idea and protocol: Xiangrong Song; Research practical work: Hai Huang, Caili Zhang, Shuping Yang, Wen Xiao, Qian Zheng; Analyses and discussion of results: Hai Huang, Caili Zhang, Xiangrong Song; Research writing and revision: Hai Huang, Caili Zhang, Shuping Yang, Xiangrong Song.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgements
This work was supported by the National Key S&T Special Projects (2018ZX09201018-024), Sichuan Province Science and Technology Support Program (21GJHZ0241 and 2020YFS0008).
References
- 1.Anderson R.M., Heesterbeek H., Klinkenberg D., Hollingsworth T.D.J.T.L. How will country-based mitigation measures influence the course of the COVID-19 epidemic? 2020;395:931–934. doi: 10.1016/S0140-6736(20)30567-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fauci A.S., Lane H.C., Redfield R.R. Covid-19 - navigating the uncharted. N. Engl. J. Med. 2020;382:1268–1269. doi: 10.1056/NEJMe2002387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lurie N., Saville M., Hatchett R., Halton J. Developing Covid-19 vaccines at pandemic speed. 2020;382:1969–1973. doi: 10.1056/NEJMp2005630. [DOI] [PubMed] [Google Scholar]
- 4.Le T.T., Cramer J.P., Chen R., Mayhew S. Evolution of the COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020;19:667–668. doi: 10.1038/d41573-020-00151-8. [DOI] [PubMed] [Google Scholar]
- 5.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rabaan A.A., Al-Ahmed S.H., Haque S., Sah R., Tiwari R., Malik Y.S., Dhama K., Yatoo M.I., Bonilla-Aldana D.K., Rodriguez-Morales A.J. SARS-CoV-2, SARS-CoV, and MERS-COV: a comparative overview. Infez. Med. 2020;28:174–184. [PubMed] [Google Scholar]
- 7.Hu B., Guo H., Zhou P., Shi Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2020;19:141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rangan R., Zheludev I.N., Hagey R.J., Pham E.A., Wayment-Steele H.K., Glenn J.S., Das R. RNA genome conservation and secondary structure in SARS-CoV-2 and SARS-related viruses: a first look. RNA. 2020;26:937–959. doi: 10.1261/rna.076141.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 11.Yang J., Wang W., Chen Z., Lu S., Yang F., Bi Z., Bao L., Mo F., Li X., Huang Y., Hong W., Yang Y., Zhao Y., Ye F., Lin S., Deng W., Chen H., Lei H., Zhang Z., Luo M., Gao H., Zheng Y., Gong Y., Jiang X., Xu Y., Lv Q., Li D., Wang M., Li F., Wang S., Wang G., Yu P., Qu Y., Yang L., Deng H., Tong A., Li J., Wang Z., Yang J., Shen G., Zhao Z., Li Y., Luo J., Liu H., Yu W., Yang M., Xu J., Wang J., Li H., Wang H., Kuang D., Lin P., Hu Z., Guo W., Cheng W., He Y., Song X., Chen C., Xue Z., Yao S., Chen L., Ma X., Chen S., Gou M., Huang W., Wang Y., Fan C., Tian Z., Shi M., Wang F.S., Dai L., Wu M., Li G., Wang G., Peng Y., Qian Z., Huang C., Lau J.Y., Yang Z., Wei Y., Cen X., Peng X., Qin C., Zhang K., Lu G., Wei X. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature. 2020;586:572–577. doi: 10.1038/s41586-020-2599-8. [DOI] [PubMed] [Google Scholar]
- 12.Wu F., Yan R., Liu M., Liu Z., Wang Y., Luan D., Wu K., Song Z., Sun T., Ma Y.J.m. 2020. Antibody-dependent enhancement (ADE) of SARS-CoV-2 infection in recovered COVID-19 patients: studies based on cellular and structural biology analysis. [Google Scholar]
- 13.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines - a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corbett K.S., Edwards D., Leist S.R., Abiona O.M., Boyoglu-Barnum S., Gillespie R.A., Himansu S., Schafer A., Ziwawo C.T., DiPiazza A.T., Dinnon K.H., Elbashir S.M., Shaw C.A., Woods A., Fritch E.J., Martinez D.R., Bock K.W., Minai M., Nagata B.M., Hutchinson G.B., Bahl K., Garcia-Dominguez D., Ma L., Renzi I., Kong W.P., Schmidt S.D., Wang L., Zhang Y., Stevens L.J., Phung E., Chang L.A., Loomis R.J., Altaras N.E., Narayanan E., Metkar M., Presnyak V., Liu C., Louder M.K., Shi W., Leung K., Yang E.S., West A., Gully K.L., Wang N., Wrapp D., Doria-Rose N.A., Stewart-Jones G., Bennett H., Nason M.C., Ruckwardt T.J., McLellan J.S., Denison M.R., Chappell J.D., Moore I.N., Morabito K.M., Mascola J.R., Baric R.S., Carfi A., Graham B.S. SARS-CoV-2 mRNA vaccine development enabled by prototype pathogen preparedness. bioRxiv. 2020 doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Perez Marc G., Moreira E.D., Zerbini C., Bailey R., Swanson K.A., Roychoudhury S., Koury K., Li P., Kalina W.V., Cooper D., Frenck R.W., Jr., Hammitt L.L., Tureci O., Nell H., Schaefer A., Unal S., Tresnan D.B., Mather S., Dormitzer P.R., Sahin U., Jansen K.U., Gruber W.C., C.C.T. Group Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., McGettigan J., Khetan S., Segall N., Solis J., Brosz A., Fierro C., Schwartz H., Neuzil K., Corey L., Gilbert P., Janes H., Follmann D., Marovich M., Mascola J., Polakowski L., Ledgerwood J., Graham B.S., Bennett H., Pajon R., Knightly C., Leav B., Deng W., Zhou H., Han S., Ivarsson M., Miller J., Zaks T., C.S. Group Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong Y., Dai T., Wei Y., Zhang L., Zheng M., Zhou F. A systematic review of SARS-CoV-2 vaccine candidates. Signal Transduct. Target Ther. 2020;5:237. doi: 10.1038/s41392-020-00352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 19.Richner J.M., Himansu S., Dowd K.A., Butler S.L., Salazar V., Fox J.M., Julander J.G., Tang W.W., Shresta S., Pierson T.C., Ciaramella G., Diamond M.S. Modified mRNA vaccines protect against Zika virus infection. Cell. 2017;168 doi: 10.1016/j.cell.2017.02.017. 1114–1125 e1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang F., Xiao W., Elbahnasawy M.A., Bao X., Zheng Q., Gong L., Zhou Y., Yang S., Fang A., Farag M.M.S., Wu J., Song X. Optimization of the linker length of mannose-cholesterol conjugates for enhanced mRNA delivery to dendritic cells by liposomes. Front. Pharmacol. 2018;9:980. doi: 10.3389/fphar.2018.00980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horner C., Schurmann C., Auste A., Ebenig A., Muraleedharan S., Dinnon K.H., 3rd, Scholz T., Herrmann M., Schnierle B.S., Baric R.S., Muhlebach M.D. A highly immunogenic and effective measles virus-based Th1-biased COVID-19 vaccine. Proc. Natl. Acad. Sci. U. S. A. 2020;117:32657–32666. doi: 10.1073/pnas.2014468117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamey G., Schaferhoff M., Hatchett R., Pate M., Zhao F., McDade K.K. Ensuring global access to COVID-19 vaccines. Lancet. 2020;395:1405–1406. doi: 10.1016/S0140-6736(20)30763-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahase E. British Medical Journal Publishing Group. 2020. Covid-19: UK approves Pfizer and BioNTech vaccine with rollout due to start next week. [DOI] [PubMed] [Google Scholar]
- 25.Abbasi J. COVID-19 and mRNA vaccines-first large test for a new approach. JAMA. 2020;324:1125–1127. doi: 10.1001/jama.2020.16866. [DOI] [PubMed] [Google Scholar]
- 26.Pardi N., Hogan M.J., Weissman D. Recent advances in mRNA vaccine technology. Curr. Opin. Immunol. 2020;65:14–20. doi: 10.1016/j.coi.2020.01.008. [DOI] [PubMed] [Google Scholar]
- 27.Tan L., Sun X. Recent advances in mRNA vaccine delivery. Nano Res. 2018;11:5338–5354. [Google Scholar]
- 28.Ols S., Yang L., Thompson E.A., Pushparaj P., Tran K., Liang F., Lin A., Eriksson B., Karlsson Hedestam G.B., Wyatt R.T., Lore K. Route of vaccine administration alters antigen trafficking but not innate or adaptive immunity. Cell Rep. 2020;30:3964–3971. doi: 10.1016/j.celrep.2020.02.111. (e3967) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang N.N., Li X.F., Deng Y.Q., Zhao H., Huang Y.J., Yang G., Huang W.J., Gao P., Zhou C., Zhang R.R., Guo Y., Sun S.H., Fan H., Zu S.L., Chen Q., He Q., Cao T.S., Huang X.Y., Qiu H.Y., Nie J.H., Jiang Y., Yan H.Y., Ye Q., Zhong X., Xue X.L., Zha Z.Y., Zhou D., Yang X., Wang Y.C., Ying B., Qin C.F. A Thermostable mRNA vaccine against COVID-19. Cell. 2020;182:1271–1283. doi: 10.1016/j.cell.2020.07.024. (e1216) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeyanathan M., Afkhami S., Smaill F., Miller M.S., Lichty B.D., Xing Z. Immunological considerations for COVID-19 vaccine strategies. Nat. Rev. Immunol. 2020;20:615–632. doi: 10.1038/s41577-020-00434-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lederer K., Castano D., Gomez Atria D., Oguin T.H., 3rd, Wang S., Manzoni T.B., Muramatsu H., Hogan M.J., Amanat F., Cherubin P., Lundgreen K.A., Tam Y.K., Fan S.H.Y., Eisenlohr L.C., Maillard I., Weissman D., Bates P., Krammer F., Sempowski G.D., Pardi N., Locci M. SARS-CoV-2 mRNA vaccines foster potent antigen-specific germinal center responses associated with neutralizing antibody generation. Immunity. 2020;53:1281–1295. doi: 10.1016/j.immuni.2020.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]