Abstract
Detection, segmentation and classification of nuclei are fundamental analysis operations in digital pathology. Existing state-of-the-art approaches demand extensive amount of supervised training data from pathologists and may still perform poorly in images from unseen tissue types. We propose an unsupervised approach for histopathology image segmentation that synthesizes heterogeneous sets of training image patches, of every tissue type. Although our synthetic patches are not always of high quality, we harness the motley crew of generated samples through a generally applicable importance sampling method. This proposed approach, for the first time, re-weighs the training loss over synthetic data so that the ideal (unbiased) generalization loss over the true data distribution is minimized. This enables us to use a random polygon generator to synthesize approximate cellular structures (i.e., nuclear masks) for which no real examples are given in many tissue types, and hence, GAN-based methods are not suited. In addition, we propose a hybrid synthesis pipeline that utilizes textures in real histopathology patches and GAN models, to tackle heterogeneity in tissue textures. Compared with existing state-of-the-art supervised models, our approach generalizes significantly better on cancer types without training data. Even in cancer types with training data, our approach achieves the same performance without supervision cost. We release code and segmentation results1 on over 5000 Whole Slide Images (WSI) in The Cancer Genome Atlas (TCGA) repository, a dataset that would be orders of magnitude larger than what is available today.
1. Introduction
Existing state-of-the-art supervised image analysis methods [11, 22, 13, 48, 3, 62, 59, 61, 9, 66, 64, 24, 40] largely rely on the availability of large annotated training datasets which requires the involvement of domain experts. This is a time-consuming and expensive process. Moreover, for methods that generalize on various input types, supervised data must be collected for every input type. For example, labeled satellite images from regions such as north Europe and south Africa are all needed to train a robust satellite image analysis method [65, 49]. In pathology image analysis, to achieve optimal performance, the data annotation phase often must be repeated for different tissue types such as different cancer sites, fat tissue, necrotic regions, blood vessels, and glands, because of tissue heterogeneity as well as variations in tissue preparation and image acquisition. The detection, segmentation, and classification of nuclei are core analysis steps in virtually all pathology imaging studies [11, 22, 13, 48, 3, 62, 59, 61, 9, 66, 64, 40, 23, 2, 29] and precision medicine [17, 12]. It is the first step in extracting interpretable features that provide valuable diagnostic and prognostic cancer indicators [14, 15, 1, 43, 20]. Manual generation of nucleus segmentation ground truth data takes a long time. In our experience, a training dataset consisting of 50 image patches (12M pixels) takes 120230 hours of an expert pathologists time. This training dataset is extremely small compared with the volume of data in a large study (e.g. 10k whole slide images, 50T pixels). This is a major impediment to robust nucleus segmentation.
One approach to address this problem is training data synthesis [26, 16, 51]. All existing training data synthesis approaches assume that the distribution of synthetic data is the same as the distribution of real data. However, this is often not the case, especially for synthesis of histopathology images with cellular structure (e.g. nuclear masks), since no real examples of nuclear masks are given for most cancer types. We propose an importance sampling based approach that minimizes the ideal (unbiased) generalization loss over the distribution of real data, even when given a biased distribution (of synthetic data). This allows us to enumerate possible cellar structures for training data synthesis. Our pipeline (see Fig. 2):
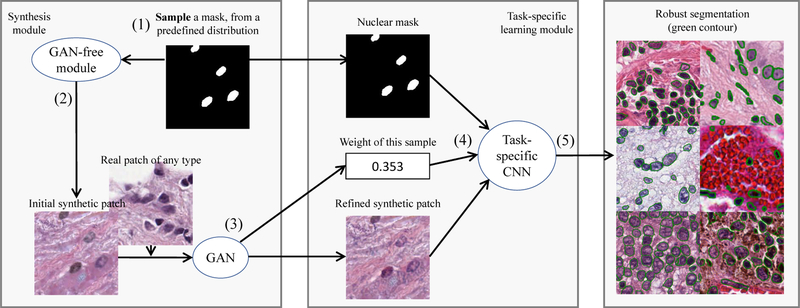
Figure 2.
Overview of our pipeline: we use a GAN-free module to synthesize (sample) an initial synthetic pathology image patch with its nuclear mask. We then refine the initial synthetic patch using a GAN and compute its sample weight. We finally train a task-specific (e.g. segmentation, classification, etc.) CNN on this sampled instance. If a sampled ground truth structure does not produce a realistic synthetic example, the impact of this instance on the training loss is down-weighted.
Samples a nucleus segmentation mask from a predefined, approximate ground truth generator;
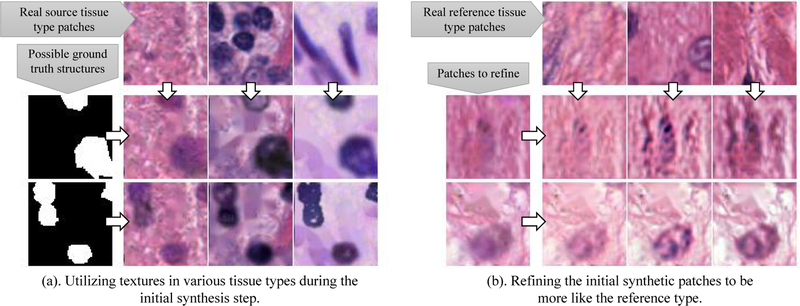
Constructs an initial synthetic patch utilizing real textures (Fig. 3) of the input tissue type;
Uses a GAN model to make the initial synthetic patch more realistic;
Computes an importance weight of this synthetic example, from the discriminators output simply using Bayes theorem; and
Trains a task-specific (e.g. segmentation) CNN using the synthetic patch, mask and importance weight.
Figure 3.
Inside our GAN-free module: synthesizing a histopathology image patch utilizing textures in any given tissue type. This step generates an image patch matches the given mask.
In other words, we enumerate possible ground truth structures during generation of synthetic training patches. If a resulting patch is not realistic, we decrease its impact in the training loss. Similarly, if a resulting patch is not only very realistic, but also rarely synthesized, then we increase its impact in the training loss.
To summarize, our contributions are: (1) Synthesizing perfectly realistic training patches with masks is almost impossible when we are not given any real examples of nuclear masks. We propose an importance sampling based method that reweighs the losses of approximately generated examples, for training a task-specific (e.g. nucleus segmentation) network, minimizing the ideal (unbiased) generalization loss over the real data distribution. (2) We show how to compute importance weights from the outputs of the GAN discriminator by simply using the Bayes theorem, without any computational overhead. (3) We propose a hybrid synthesis pipeline that utilizes textures in real histopathology patches for synthesis of any tissue patches. (4) The proposed method is robust to tissue heterogeneity. When there are no supervised datasets for a test cancer type, our nucleus segmentation CNN significantly outperforms supervised methods in across-cancer generalization. Even for the few tissue types for which supervised data exist, our method matches the performance of supervised methods. (5) We release nucleus segmentation results on over 5000 Whole Slide Images (WSI) of 13 major cancer types in The Cancer Genome Atlas (TCGA) repository. These results are at least four orders of magnitude larger than currently available human annotated datasets. We believe that this large-scale dataset, even though not as accurately annotated, is a useful feature for future pathology image analysis research.
2. Related Work
Detection and segmentation of nuclei is a fundamental analytical step in virtually all pathology imaging studies [11, 22, 13, 48, 3, 62, 59, 61, 9, 66, 64, 40, 23, 2, 29] and precision medicine [17, 12]. Recent works in image analysis have proposed crowd-sourcing or high-level, less accurate annotations, such as scribbles, to generate large training datasets manually [34, 57, 64]. Work by Zhou et al. [68] segments nuclei inside a tissue image and redistributes the segmented nuclei inside the image. The segmentation masks of the redistributed nuclei are assumed to be the predicted segmentation masks. This work requires segmentation masks and does not generate new textures and shapes. Generative Adversarial Networks (GANs) [44] have been proposed for generation of realistic images [16, 6, 4, 51, 8, 67, 42, 25, 46, 38]. For example, an image-to-image translation GAN [26, 16] synthesizes eye fundus images. However, it requires an accurate supervised segmentation network to segment eye vessels out, as part of the synthesis pipeline. The S+U learning framework [51] refines initially synthesized images via a GAN to increase their realism. This method achieves state-of-the-art results in eye gaze and hand pose estimation tasks. Recently, a GAN based approach [37] is able to synthesize realistic pathology images with nuclear masks. It is limited to cancer types with ground truth masks, since it requires real mask examples. GANs are also used to synthesize images of various styles of the same content. Cycle-GAN etc. [35, 69] transfers content of images to target styles without training with paired images. The universal style transfer approach [32, 54] solves this problem by providing a reference style to the generator network. However, to apply any of the GAN models for synthesizing image and masks, examples of both real images and masks are required.
3. Importance Sampling for Loss Estimation
In this section we show how to minimize the ideal (unbiased) task-specific (e.g. segmentation, classification, etc.) generalization loss over the distribution of real data, given an approximate sampling distribution (of synthetic data). We define a random variable X representing an image/patch, with its ground truth T, and the probability density function of real images as p(X, T). In practice, X and T are discrete. The task-specific generalization loss LR(R) with model parameters R is:
| (1) |
where f() is the loss function such as the conventional segmentation loss [36, 41]. To minimize the generalization loss defined by Eq. 1, we sample one example X, T from the distribution defined by p((X, T )), then minimize the loss f(X, T). If there are infinite real samples, the empirical loss converges exactly to Eq. 1. In this work, we synthesize training examples X, T. We define the probability density function of synthetic images as g(X, T). Ideally p(X, T) is equivalent to g(X, T). However, for synthesizing unbiased examples and corresponding ground truth nuclear masks, an unbiased modeling of nuclear masks is needed existing training image synthesis methods [51] heavily depend on unbiased ground truth image structure modeling, such as size of eyeballs, color of iris. This is almost impossible for histopathology images because of the paucity of annotated data and the cellular structure heterogeneity across tissue types.
To estimate the ideal (unbiased) generalization loss with g((X, T )), we formulate the task-specific loss as follows:
| (2) |
Instead of sampling X, T from the real pdf p(X, T), we can now sample X, T from the synthetic pdf g(X, T) and minimize a new loss function F(X, T) = f(X, T)p(X, T)/g(X, T). This is the standard importance sampling approach [7]: when sampling from p(X, T) is expensive, we sample from g(X, T) then re-weight each sample by multiplying its loss with weight p(X, T)/g(X, T). Note that for the resulting generalization loss estimation to be unbiased, for all X, T with p(X, T) > 0, it is required that also g(X, T) > 0.
Given an image X, the underlying ground truth T is fixed. Thus, we can simply drop T in PDFs:
| (3) |
The right hand side of Eq. 3 can be derived from the output of a GAN discriminator. A discriminator trained with cross-entropy (log-likelihood) loss estimates the probability that X is sampled from the real distribution instead of the synthetic distribution: Pr(X ~ p|X). The discriminator is trained with real and synthetic examples. Denote a constant c as the ratio between the numbers of synthetic input samples and real input samples: c = Pr(X ~ g)/Pr(X ~ p). Thus p(X) = Pr(X|X ~ p), g(X) = Pr(X|X ~ g). Using Bayes theorem, we have:
| (4) |
Rearranging Eq. 4 gives us the importance weight formulated by the discriminators output Pr(X ~ p|X):
| (5) |
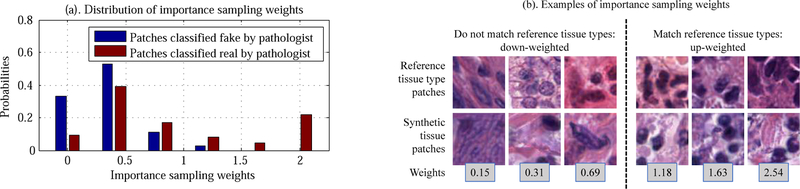
If a synthetic patch is unrealistic (Pr(X ~ p|X) 0.5), it will be down-weighted (contribute less to the loss). If a synthetic patch is realistic and rarely generated, it will be up-weighted (contribute more to the loss). We show the visualization of importance weights in Fig. 7.
Figure 7.
Evaluation and visualization of importance sampling weights. (a). Synthetic patches classified as real by pathologists have higher importance weights than patches classified as fake. (b). Visualization of importance sampling weights.
Optimality of unbiased loss minimization:
Since we learn Pr(X ~ p|X) via training the discriminator on the unbiased dataset (i.e. unlimited samples of X ~ p and X ~ g), we can easily show that this yield unbiased generalization loss minimization: The unbiased generalization loss over the distribution of real data defined by Eq. 1 is equivalent to Eq. 2. Since we can sample from the synthetic data distribution g easily, the only term in Eq. 2 need to learn is the importance weight p(X)/g(X), defined by Eq. 5. Hence, an unbiased discriminator output Pr(X ~ p|X) yields unbiased importance weights, and further, unbiased generalization loss.
4. Heterogeneous Patch Synthesis
We now show how to synthesize (sample) training examples. Fig. 2 shows the overview of our method which learns from unlabeled real histopathology images of heterogeneous texture and cellular structure (e.g. nuclear mask).
4.1. Initial synthesis
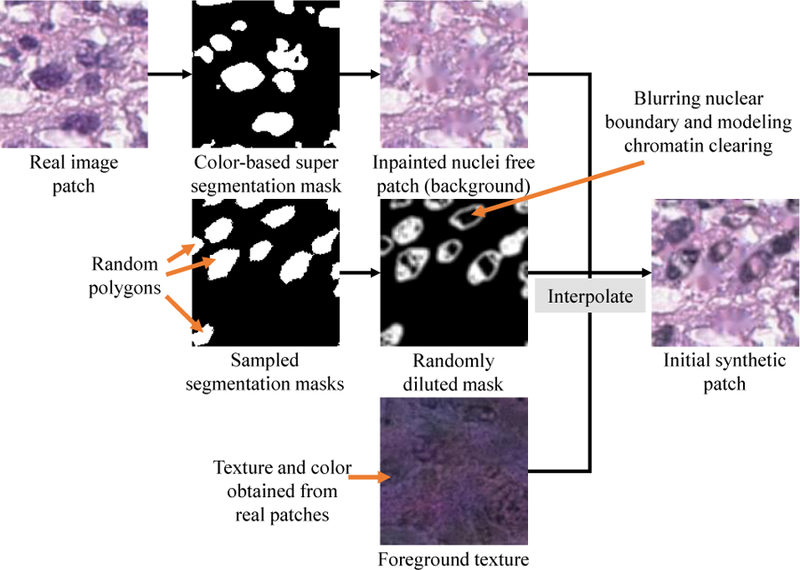
This step generates synthetic patches that are not necessarily realistic for all given target tissue types. Thus, a significant part of this process is predefined regardless of the target tissue type. First, we randomly generate a set of polygons as nuclear masks. In particular, we perturb points on a circle closer/further away from the center according to a random irregularity value. These polygons are of variable sizes and irregularities and are allowed to randomly overlap with each other by a predefined number of pixels. To model the correlation between the shapes of nearby nuclei, all polygons are distorted by a random quadrilateral transform. The purpose of such a mask is to provide a generic representation of the basic structures in tissues and to induce greater variability in the synthetic images. We consider the generated masks as foreground/background masks (nuclei as the foreground and tissue as the background) and utilize textures from real histopathology image patches to generate initial synthetic image patches in a background/foreground manner. This is a fast process; synthesizing a 200200 pixel patch at 40X magnification takes one second using a single CPU core.
Generating Background Patches:
First, we remove the nuclei in a source image patch to create a background patch on which we add the synthetic nuclei. We apply a simple Ostus threshold-based super-segmentation method [33] on the source image patch to determine the nuclear material. In super-segmentation, a segmented region always fully contains the foreground object (nucleus in this case). We replace the pixels corresponding to the segmented nuclear material with color and texture values similar to the background pixels via image inpainting [55]. Super-segmentation may not precisely delineate nucleus boundaries and may include non-nuclear material in segmented nuclei. This is acceptable, because the objective of this step is to guarantee that only background tissue texture and intensity properties are used to synthesize the background patch.
Simulating Foreground Nuclear Textures:
We apply a sub-segmentation method to the source patch to gather nuclear textures from segmented regions. In sub-segmentation, a segmented region is fully contained in the foreground object. This ensures that pixels within real nuclei are used for generating realistic foreground (nuclei) in synthetic images. Since nuclei are generally small and make up a small portion of tissue, sub-segmentation will yield a very limited amount of nuclear material which is not enough for existing reconstruction methods. Thus, our approach utilizes textures in the Eosin channel [19] of a randomly extracted real patch and combines them with nuclear color obtained via sub-segmentation of the source patch to generate nuclear textures.
Combining Foreground and Background:
Let us define Ii,j, Ai,j, Bi,j, Mi,j as pixel values at position i, j in the resulting synthetic patch, the nuclear texture patch, the nucleus free patch, and the nucleus mask patch, respectively. To combine nuclear and non-nuclear textures according to the nucleus mask patch, Ii,j can be set to Ai,jMi,j + Bi,j(1 Mi,j). This may result in significant artifacts, such as obvious nuclear boundaries. Additionally, clear chromatin phenomena in certain types of nuclei are not modeled. Thus, our method randomly clears the interior of the polygons in the nucleus mask patch and blurs their boundaries before applying the above equation.
4.2. Refining the Initial Synthesis
These initial synthetic image patches are refined via adversarial training. We also use the discriminators output to compute the importance sampling weight defined by Eq. 5. For this phase we have implemented a refiner (generator) CNN and a discriminator CNN.
Given an input image patch I and a reference type patch S, the refiner G with trainable parameters G outputs a refined patch X = G(I, S; G). Ideally, an output patch is (1). Regularized: The pixel-wise difference between the initial synthetic patch and the refined patch is small enough so that the synthetic ground truth remains unchanged. (2). Realistic for the given type: It is a realistic representation of the type of the reference patch. (3). Informative and hard: It provides a challenging example for the task-specific CNN so that the trained task-specific CNN will be robust.
We construct three losses: , , and for each of the properties above, respectively. The first two losses, and , are based on the S+U method [51]. The weighted average of these losses is defined as the final loss LG for training the refiner CNN:
| (6) |
We set hyperparameters = 1.0, = 1.0, = 0.0000001 in experiments.
The regularization loss is defined as an elastic net [70]: , where E[] is the expectation function applied on the training set, || ||1and || ||2 are the L-1 and L-2 norms and 1 and 2 are predefined parameters. We use 1 = 0.00001 and 2 = 0.0001 in experiments.
The loss for achieving a realistic representation in the reference type, by training the refiner (generator) G, is where D (X, S; D) is the output of the discriminator D with trainable parameters D given the refined patch X and the same reference type patch S as input. It is the estimated probability by D that input X matches the tissue type of S. The discriminator D has two classes of input: pairs of real patches within the same type S, S and a pair with one synthetic patch X, S. Its loss is the standard class.ification loss .
The generator and discriminator both take a reference patch and refine or classify the other input patch according to textures in the reference patch. This feature is implemented with an asymmetric siamese network [10, 28], as shown in Fig. 4 and Fig. 5.
Figure 4.
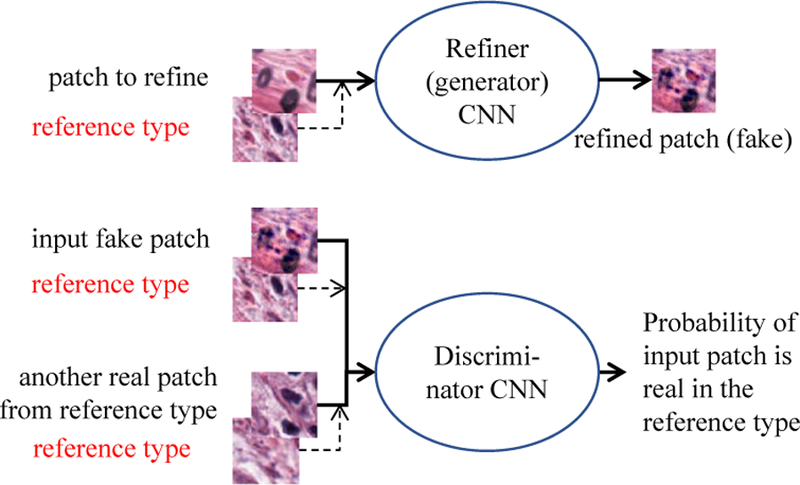
Inside our GAN module: in addition to the input real/fake patches, we provide additional reference type patches extracted from nearby regions of the real patches. If the fake patch is realistic, but does not reflect the same tissue type as the reference type, the discriminator is still able to tell the difference. As a result, the refiner learns to generate patches in the reference style.
Figure 5.
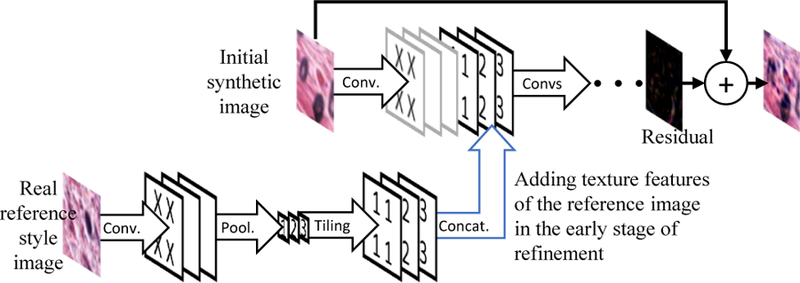
Our refiner (generator) CNN adds information of the reference type patch into the refinement stage, so that the initial synthetic patch will be refined according to the reference type.
It has been shown that GANs are able to generate challenging training examples that yield robust classification/segmentation models [30, 50, 31, 21, 60]. Thus, the refiner is trained with loss to generate challenging training examples (with larger loss) for the task-specific CNN. We simply define as the negative of the task-specific loss: , where LR(R) is the loss of a task-specific model R with trainable parameters R. When training the refiner, we update G to produce refined patches that maximize LR. When training the task-specific CNN, we update R to minimize LR. The underlying segmentation ground truth of the refined patches would change significantly if overpowered . We down-weight by a factor of 0.0001 to minimize the likelihood of this unwanted outcome.
4.3. Visual Evaluation by a Human Expert
Fig. 6, 7, 8 show examples of our initial synthetic and refined patches. To verify that synthetic patches are realistic, we asked a pathologist to distinguish real versus synthetic patches. In particular, we showed the pathologist 100 randomly extracted real patches, 100 randomly selected initial synthetic patches, and 100 randomly selected refined patches. Out of this set, the pathologist selected the patches he thought were real. The pathologist classified almost half of the initial synthetic patches (46%) and most of the refined patches (64%) as real. The pathologist classified (83%) of the real patches as real. This is because many of those real patches are out-of-focus or contain no nuclei. Fig. 7 shows the distributions of weights of the realistic synthetic patches versus the unrealistic synthetic patches. This verifies that the realistic synthetic patches have higher importance sampling weights and vice versa.
Figure 6.
The effect of using different source tissue texture patches and reference type patches. The resulting synthetic patches have the same textures/types as the source/reference patches.
Figure 8.
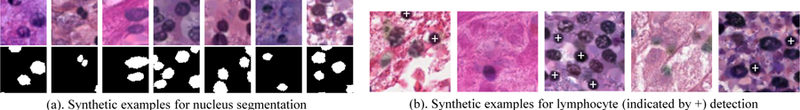
Examples of various kinds of synthetic patches we generated.
5. Experiments
We conducted experiments with datasets from the MIC-CAI18 and MICCAI17 nucleus segmentation challenges [39, 58] and the generalized nucleus segmentation dataset [29] containing seven cancer types. Additionally, we evaluated our method with a lymphocyte detection dataset [23].
We implemented the refiner, outlined in Fig. 5, with 21 convolutional layers and 2 pooling layers. The discriminator has 15 convolutional layers and 3 pooling layers. As the task-specific CNNs, we used U-net [47] and a network with 15 convolutional layers and 2 pooling layers for nucleus detection and segmentation, and a network with 11 convolutional layers for classification. For details, please refer to our source code. We used an open source implementation of GAN [27, 51] as part of our implementation. We initialize all networks randomly (no pretraining). During testing, we normalize the color of input H&E patches [45].
5.1. Nucleus Segmentation Experiments
Supervised methods heavily depend on representative datasets. However, currently only a few cancer types have supervised datasets due to the extensive amount of labor and expert domain knowledge required for histopathology image annotation. For cancer types without labeled data, supervised methods achieve worse performance than on cancer types with labeled data. We verified this argument using the MICCAI18 and MICCAI16/17 nucleus segmentation datasets [39, 58]. The MICCAI18 nucleus segmentation challenge dataset [39] contains 15 training and 18 testing tissue images extracted from whole slide images of two cancer types. The MICCAI17 dataset [58] contains 32 training and 32 testing images, extracted from whole slide images of four cancer types. A typical resolution is 600 600 pixels. In addition, we tested the across dataset generalization ability of our method using the test set of the generalized nucleus segmentation dataset [29]. The test set contains 14 1000 1000 pixel patches in seven cancer types.
Note that annotating one nucleus takes about 2 minutes. It would take about 225 man-hours to generate these training datasets. Unsupervised synthetic image generation and training can result in significant time savings in such cases, while enabling the generation of larger training datasets.
We evaluated several methods in the nucleus segmentation experiments; these methods are listed below. In the following, Universal denotes the proposed method trained with patches extracted from whole slide images for all cancer types in the TCGA repository. More specifically, we randomly extracted a 500 500-pixel tissue patch at 40X (for 20X images, we upsampled the patch to 40X) from each diagnostic whole slide image in the TCGA repository. This generated about 10k tissue patches.
Universal U-net.
The proposed method with U-net [47] as the task-specific CNN. Our U-net has two outputs: one for nucleus detection, and one for class-level nucleus segmentation. We then combined detection and class-level segmentation results to achieve instance-level segmentation using watershed [5, 2].
Universal CNN.
The proposed method with a 15 layer segmentation/detection network.
Universal U-net + real data.
Since U-net is computationally efficient, we train a U-net with both synthetic as well as real data from the MICCAI18 training dataset, as the model we deploy on over 5000 WSIs.
Type-specific U-net / CNN.
We use the semi-supervised U-nets [47] and the 15/11 layer CNN as standalone supervised networks, trained with real, human annotated tissue image patches from up to four cancer types. We augment the real patches by rotation, mirroring, and scaling.
In order to obtain every tissue type for unsupervised learning of our method, we synthesized 75 75-pixel and 200 200-pixel patches according to patches sampled from every TCGA WSI. The GAN-free module generated 100k initial synthetic patches. Then we used GAN for image refinement and importance sampling based task-specific training on those initial synthetic patches.
We tested the supervised methods with the following two setups: (1) Within cancer type. We trained the type-specific, supervised CNNs with the training sets of all two MICCAI18 and four MICCAI17 cancer types. (2) Across cancer types. We excluded the training images of one cancer type, trained a type-specific, supervised CNN with the training images from all of the other cancer types, and evaluated the trained CNN on the images of the excluded type. We repeated this for all two/four cancer types and report performance as the average of all runs.
We used the average of two definitions of DICE coefficients as the performance metric. The first version is the standard DICE coefficient [18, 53]: denote the set of segmented pixels as S and the set of ground truth nuclear pixels as T, . The second is a variant of the original to capture mismatch in the way the segmented objects are split, while the overall segmentation may be very similar. The evaluation results are shown in Tab. 1. Our approach outperforms the supervised methods significantly on testing cancer types without supervised data (across cancer types). Even when supervised data exists for every cancer type (within cancer type), our approach performs as well as the state-of-the-art approaches.
Table 1.
Nucleus segmentation results on the MICCAI18 and MICCAI17 nucleus segmentation datasets. For each of the three network architecture, our approach outperforms the supervised methods significantly on cancer types without supervised data (across cancer). Even when supervised data exists for all cancer types (within cancer), our approach performs as well as state-of-the-art approaches without any supervision cost, due to the large scale of the synthetic dataset. The MICCAI18 winners approach is unknown to us.
| Nucleus segmentation methods | MICCAI18 DICE Avg. |
MICCAI17 DICE Avg. |
|---|---|---|
| Supervised methods tested within cancer types | ||
| Type-specific CNN | 0.8013 | 0.7713 |
| Type-specific U-net | 0.8391 | 0.7645 |
| Contour-aware net [9] | 0.812 | - |
| CSP-CNN [23] | 0.8362 | 0.7681 |
| MICCAI18 winner | 0.870 | - |
| MICCAI17 winner [58] | - | 0.783 |
| Supervised methods tested across cancer types | ||
| Type-specific CNN | 0.7818 | 0.7314 |
| Type-specific U-net | 0.8010 | 0.7179 |
| Proposed unsupervised method for all cancer types | ||
| Universal CNN | 0.8180 | 0.7708 |
| Universal U-net | 0.8401 | 0.7612 |
| Universal U-net + real data | 0.8678 | 0.7863 |
To further verify that our method outperforms baseline methods on tissue types without supervised data, we evaluated nucleus segmentation methods across datasets: we trained supervised method on the MICCAI17 training set and tested it on the test set of the generalized nucleus segmentation dataset [29]. As shown in Tab. 2, our method generalizes significantly better across datasets, than the supervised, type-specific method. Thus, we release segmentation results on 5000 WSIs in the TCGA repository [56]. Existing largest human annotated dataset [29] contains 100 patches of size 1000 1000 pixels. The scale of our segmentation results are larger than 10M such patches. We believe that this large-scale dataset, even though not as accurately annotated, is a useful feature for future pathology image analysis research.
Table 2.
Across dataset evaluation results. The type-specific CNN is trained on the MICCAI17 training set amd evaluated on the test set of the generalized nucleus segmentation dataset [29]. Our unsupervised method generalizes significantly better, than the supervised type-specific method.
| Nucleus segmentation methods | DICE Avg. |
|---|---|
| Type-specific U-net, across dataset | 0.7328 |
| Universal U-net + real data | 0.7713 |
5.2. Ablation Studies
We evaluated the importance of three components of our method: importance weights in the loss function, utilizing a real reference type patch for refinement, and generating hard examples for CNN training. We removed one feature at a time and measured performance for nucleus segmentation on the MICCAI17 dataset. The experimental results using U-net are shown in Tab. 3. The proposed methods reduce the segmentation error by 5.4%, 7.8%, and 3.2%.
Table 3.
Ablation study using the MICCAI17 nucleus segmentation challenge dataset. Proposed methods reduce the segmentation error (1DICE average) by 5.4%, 7.8%, and 3.2%.
| Nucleus segmentation methods | DICE Avg. |
|---|---|
| No hard examples | 0.7476 |
| No reference patch during refinement | 0.7410 |
| No importance weights | 0.7533 |
| Universal CNN (proposed) | 0.7612 |
5.3. Human evaluation on 13 cancer types in TCGA
To evaluate nucleus segmentation methods in an uncontrolled environment, we randomly extracted 133 500 500 pixel patches from 13 major cancer types (that have more than 500 WSIs each) in TCGA [56], applied segmentation methods on those patches, and blindly compared the segmentation quality between our method and the baseline. For segmentation methods, we use the fully supervised U-net (type-specific U-net) trained on the MICCAI18 training set as the baseline, and the U-net trained on both synthetic and real MICCAI18 training data (Universal U-net + real data) as our method. For human evaluation, an expert pathologist blindly compared the segmentation results in terms of TruePositives FalsePositives FalseNegatives in each patch. As a result, out of the 133 patches, in 83 patches our method is better than the baseline, in 46 patches our method is worse, in 4 patches they are similar. We show three failure cases in Fig. 9.
Figure 9.
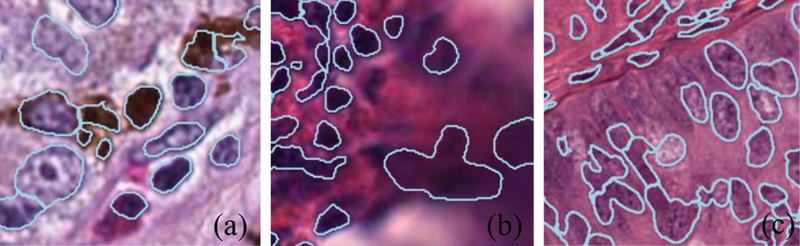
Three failure cases: Dark pigment in melanoma (a) and out-of-focus (b) scenarios are not modeled by our synthesis pipeline. Some light-colored nuclei with clear chromatin (c) are not detected when they are close to dark, easy-to-detect nuclei.
5.4. Lymphocyte Detection Experiments
The lymphocyte detection dataset [23] has 1367 labeled training patches and 418 testing patches cropped from 12 representative lung adenocarcinoma whole slide tissue images. Patches with lymphocytes in the center are labeled positive. Our method synthesized lymphocytes as round and dark objects with around 7 microns in diameter. Some synthetic image examples are shown in Fig. 8. Table 4 shows experimental evaluation of our method against a level set features based method [67] and supervised VGG16 method [52]. We used the Area Under the ROC curve (AU-ROC) measure as the evaluation metric.
Table 4.
Lymphocyte detection on the lymphocyte dataset [23]. Without any supervision cost, our method outperforms all supervised models trained on patches of just one cancer type.
6. Conclusions
Supervised methods rely on large volumes of labeled histopathology data which are expensive to generate. We introduced a method that learns from heterogeneous pathology patches in an unsupervised manner. Our method synthesizes training patches with importance weights, such that the task-specific (e.g. segmentation) CNN is trained to minimize the ideal (unbiased) generalization error over real data. When no supervised data exists for a cancer type, our result is significantly better than across-cancer generalization results by supervised methods. Even when supervised data exists, our approach performs as well as supervised methods, due to the much larger scale of synthetic data. We release segmentation results on over 5000 WSIs, which is orders of magnitude larger than currently available human annotated datasets. In future work we will demonstrate the generality of our importance sampling based loss minimization approach on other tasks such as mixed-quality image classification [63].
Figure 1.
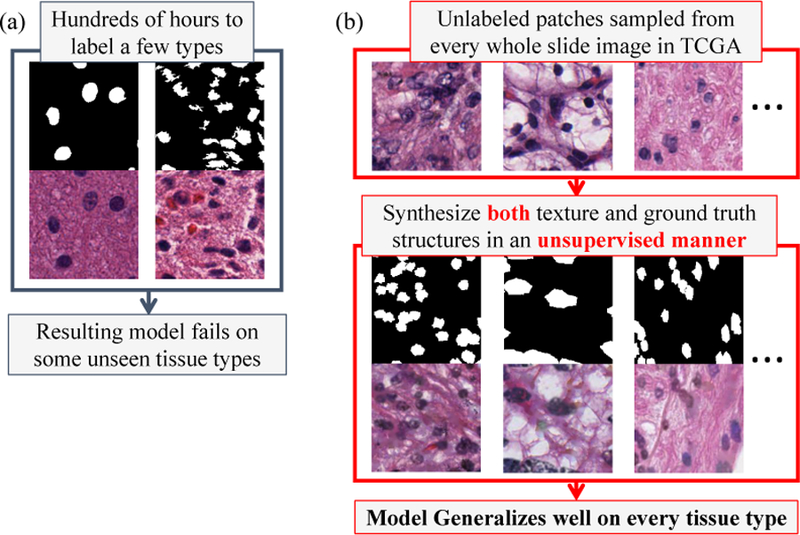
(a). Standard learning methods learn and perform well only with tissue types for which ground truth training data exists. (b). We propose to synthesize both image texture and ground truth structures for training a supervised model, even when no real ground truth structures are given. As a result, our model generalizes well on unseen tissue types.
Acknowledgement
This work was supported in part by 1U24CA180924-01A1, 3U24CA215109-02, and 1UG3CA225021-01 from the National Cancer Institute, R01LM011119-01 and R01LM009239 from the U.S. National Library of Medicine, and a gift from Adobe. This work used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation grant number ACI-1548562. Specifically, it used the Bridges system, which is supported by NSF award number ACI-1445606, at the Pittsburgh Supercomputing Center (PSC).
Footnotes
References
- [1].Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Cavalho S, Bussink J, Monshouwer R, Haibe-Kains B, Rietveld D, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nature communications, 2014. [DOI] [PMC free article] [PubMed]
- [2].Bai M and Urtasun R. Deep watershed transform for instance segmentation. In CVPR, 2017. [Google Scholar]
- [3].Bayramoglu N and Heikkil J. Transfer learning for cell nuclei classification in histopathology images. In ECCV Workshops, 2016. [Google Scholar]
- [4].Bayramoglu N, Kaakinen M, Eklund L, and Heikkila J. Towards virtual h&e staining of hyperspectral lung histology images using conditional generative adversarial networks. In CVPR, 2017. [Google Scholar]
- [5].Beucher S. Watershed, hierarchical segmentation and waterfall algorithm. In Mathematical morphology and its applications to image processing 1994. [Google Scholar]
- [6].Bi L, Kim J, Kumar A, Feng D, and Fulham M. Synthesis of positron emission tomography (pet) images via multichannel generative adversarial networks (gans). In Molecular Imaging, Reconstruction and Analysis of Moving Body Organs, and Stroke Imaging and Treatment 2017. [Google Scholar]
- [7].Bishop CM. Pattern recognition and machine learning 2006.
- [8].Calimeri F, Marzullo A, Stamile C, and Terracina G. Biomedical data augmentation using generative adversarial neural networks. In International Conference on Artificial Neural Networks, 2017. [Google Scholar]
- [9].Chen H, Qi X, Yu L, Dou Q, Qin J, and Heng P-A. Dcan: Deep contour-aware networks for object instance segmentation from histology images. Medical Image Analysis, 2017. [DOI] [PubMed]
- [10].Chopra S, Hadsell R, and LeCun Y. Learning a similarity metric discriminatively, with application to face verification. In CVPR, 2005. [Google Scholar]
- [11].Colen R, Foster I, Gatenby R, Giger ME, Gillies R, Gutman D, Heller M, Jain R, Madabhushi A, Madhavan S, et al. Nci workshop report: clinical and computational requirements for correlating imaging phenotypes with genomics signatures. Translational oncology, 2014. [DOI] [PMC free article] [PubMed]
- [12].Collins FS and Varmus H. A new initiative on precision medicine. New England Journal of Medicine, 2015. [DOI] [PMC free article] [PubMed]
- [13].Cooper LA, Carter AB, Farris AB, Wang F, Kong J, Gutman DA, Widener P, Pan TC, Cholleti SR, Sharma A, et al. Digital pathology: Data-intensive frontier in medical imaging. Proceedings of the IEEE, 2012. [DOI] [PMC free article] [PubMed]
- [14].Cooper LA, Kong J, Gutman DA, Wang F, Cholleti SR, Pan TC, Widener PM, Sharma A, Mikkelsen T, Flanders AE, et al. An integrative approach for in silico glioma research. IEEE Transactions on Biomedical Engineering, 2010. [DOI] [PMC free article] [PubMed]
- [15].Cooper LA, Kong J, Gutman DA, Wang F, Gao J, Appin C, Cholleti S, Pan T, Sharma A, Scarpace L, et al. Integrated morphologic analysis for the identification and characterization of disease subtypes. Journal of the American Medical Informatics Association, 2012. [DOI] [PMC free article] [PubMed]
- [16].Costa P, Galdran A, Meyer MI, Abrmoff MD, Niemeijer M, Mendona AM, and Campilho A. Towards adversarial retinal image synthesis. arXiv, 2017. [DOI] [PubMed]
- [17].Council NR et al. Toward precision medicine: building a knowledge network for biomedical research and a new taxonomy of disease National Academies Press, 2011. [PubMed] [Google Scholar]
- [18].Dice LR. Measures of the amount of ecologic association between species. Ecology, 1945.
- [19].Fischer AH, Jacobson KA, Rose J, and Zeller R. Hematoxylin and eosin staining of tissue and cell sections. Cold Spring Harbor Protocols, 2008. [DOI] [PubMed]
- [20].Gillies RJ, Kinahan PE, and Hricak H. Radiomics: images are more than pictures, they are data. Radiology, 2015. [DOI] [PMC free article] [PubMed]
- [21].Goodfellow IJ, Shlens J, and Szegedy C. Explaining and harnessing adversarial examples. In ICLR, 2015. [Google Scholar]
- [22].Gurcan MN and Madabhushi A. Digital pathology. SPIE, 2013. [DOI] [PMC free article] [PubMed]
- [23].Hou L, Nguyen V, Kanevsky AB, Samaras D, Kurc TM, Zhao T, Gupta RR, Gao Y, Chen W, Foran D, et al. Sparse autoencoder for unsupervised nucleus detection and representation in histopathology images. Pattern Recognition, 86:188–200., 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hou L, Samaras D, Kurc TM, Gao Y, Davis JE, and Saltz JH. Patch-based convolutional neural network for whole slide tissue image classification. In CVPR, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Huang X, Li Y, Poursaeed O, Hopcroft J, and Belongie S. Stacked generative adversarial networks. In CVPR, 2017. [Google Scholar]
- [26].Isola P, Zhu J-Y, Zhou T, and Efros AA. Image-to-image translation with conditional adversarial networks. In CVPR, 2017. [Google Scholar]
- [27].Kim T. Simulated+unsupervised learning in tensorflow https://github.com/carpedm20/simulated-unsupervised-tensorflow.
- [28].Koch G. Siamese neural networks for one-shot image recognition. In ICML workshop, 2015. [Google Scholar]
- [29].Kumar N, Verma R, Sharma S, Bhargava S, Vahadane A, and Sethi A. A dataset and a technique for generalized nuclear segmentation for computational pathology. IEEE transactions on medical imaging, 2017. [DOI] [PubMed]
- [30].Le H, Vicente TFY, Nguyen V, Hoai M, and Samaras D. A+ D net: Training a shadow detector with adversarial shadow attenuation. In ECCV, 2018. [Google Scholar]
- [31].Lemley J, Bazrafkan S, and Corcoran P. Smart augmentation-learning an optimal data augmentation strategy. IEEE Access, 2017.
- [32].Li Y, Fang C, Yang J, Wang Z, Lu X, and Yang M-H. Universal style transfer via feature transforms. In NIPS, 2017. [Google Scholar]
- [33].Liao P-S, Chen T-S, Chung P-C, et al. A fast algorithm for multilevel thresholding. J. Inf. Sci. Eng, 2001.
- [34].Lin D, Dai J, Jia J, He K, and Sun J. Scribble-sup: Scribble-supervised convolutional networks for semantic segmentation. In CVPR, 2016. [Google Scholar]
- [35].Liu M-Y, Breuel T, and Kautz J. Unsupervised image-to-image translation networks. In NIPS, 2017. [Google Scholar]
- [36].Long J, Shelhamer E, and Darrell T. Fully convolutional networks for semantic segmentation. In CVPR, 2015. [DOI] [PubMed] [Google Scholar]
- [37].Mahmood F, Borders D, Chen R, McKay GN, Salimian KJ, Baras A, and Durr NJ. Deep adversarial training for multi-organ nuclei segmentation in histopathology images. arXiv, 2018. [DOI] [PMC free article] [PubMed]
- [38].Mauricio A, Lpez J, Huauya R, and Diaz J. High-resolution generative adversarial neural networks applied to histological images generation. In International Conference on Artificial Neural Networks, 2018. [Google Scholar]
- [39].MICCAI 2018 Challenge. Segmentation of Nuclei in Pathology Images http://miccai.cloudapp.net/competitions/83, 2018.
- [40].Murthy V, Hou L, Samaras D, Kurc TM, and Saltz JH. Center-focusing multi-task CNN with injected features for classification of glioma nuclear images. In Winter Conference on Applications of Computer Vision (WACV), 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Noh H, Hong S, and Han B. Learning deconvolution network for semantic segmentation. In CVPR, 2015. [Google Scholar]
- [42].Osokin A, Chessel A, Salas REC, and Vaggi F. Gans for biological image synthesis. In ICCV, 2017. [Google Scholar]
- [43].Parmar C, Leijenaar RT, Grossmann P, Velazquez ER, Bussink J, Rietveld D, Rietbergen MM, Haibe-Kains B, Lambin P, and Aerts HJ. Radiomic feature clusters and prognostic signatures specific for lung and head & neck cancer. Scientific reports, 2015. [DOI] [PMC free article] [PubMed]
- [44].Radford A, Metz L, and Chintala S. Unsupervised representation learning with deep convolutional generative adversarial networks. In ICLR, 2016. [Google Scholar]
- [45].Reinhard E, Adhikhmin M, Gooch B, and Shirley P. Color transfer between images. IEEE Computer graphics and applications, 21(5):34–41., 2001. [Google Scholar]
- [46].Richter SR, Vineet V, Roth S, and Koltun V. Playing for data: Ground truth from computer games. In ECCV, 2016. [Google Scholar]
- [47].Ronneberger O, Fischer P, and Brox T. U-net: Convolutional networks for biomedical image segmentation. In MIC-CAI, 2015. [Google Scholar]
- [48].Saltz J, Almeida J, Gao Y, Sharma A, Bremer E, DiPrima T, Saltz M, Kalpathy-Cramer J, and Kurc T. Towards generation, management, and exploration of combined radiomics and pathomics datasets for cancer research. AMIA Summits on Translational Science Proceedings, 2017. [PMC free article] [PubMed]
- [49].Sankaran S, Khot LR, Espinoza CZ, Jarolmasjed S, Sathuvalli VR, Vandemark GJ, Miklas PN, Carter AH, Pumphrey MO, Knowles NR, et al. Low-altitude, high-resolution aerial imaging systems for row and field crop phenotyping: A review. European Journal of Agronomy, 2015.
- [50].Shrivastava A, Gupta A, and Girshick R. Training region-based object detectors with online hard example mining. In CVPR, 2016. [Google Scholar]
- [51].Shrivastava A, Pfister T, Tuzel O, Susskind J, Wang W, and Webb R. Learning from simulated and unsupervised images through adversarial training. In CVPR, 2017. [Google Scholar]
- [52].Simonyan K and Zisserman A. Very deep convolutional networks for large-scale image recognition. In ICLR, 2014. [Google Scholar]
- [53].Srensen T. {A method of establishing groups of equal amplitude in plant sociology based on similarity of species and its application to analyses of the vegetation on Danish commons}. Biol. Skr, 1948.
- [54].Taigman Y, Polyak A, and Wolf L. Unsupervised cross-domain image generation. ICLR, 2017.
- [55].Telea A. An image inpainting technique based on the fast marching method. Journal of graphics tools, 2004.
- [56].The TCGA team. The Cancer Genome Atlas https://cancergenome.nih.gov/.
- [57].Vicente T, Hou L, Yu C-P, Hoai M, and Samaras D. Large-scale training of shadow detectors with noisily-annotated shadow examples. In ECCV, 2016. [Google Scholar]
- [58].Vu QD, Graham S, To MNN, Shaban M, Qaiser T, Koohbanani NA, Khurram SA, Kurc T, Farahani K, Zhao T, et al. Methods for segmentation and classification of digital microscopy tissue images. arXiv, 2018. [DOI] [PMC free article] [PubMed]
- [59].Wang S, Yao J, Xu Z, and Huang J. Subtype cell detection with an accelerated deep convolution neural network. In MICCAI, 2016. [Google Scholar]
- [60].Wang X, Shrivastava A, and Gupta A. A-fast-rcnn: Hard positive generation via adversary for object detection. In CVPR, 2017. [Google Scholar]
- [61].Xie Y, Xing F, Kong X, Su H, and Yang L. Beyond classification: structured regression for robust cell detection using convolutional neural network. In MICCAI, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Xu J, Xiang L, Liu Q, Gilmore H, Wu J, Tang J, and Madabhushi A. Stacked sparse autoencoder (ssae) for nuclei detection on breast cancer histopathology images. Medical Imaging, 2016. [DOI] [PMC free article] [PubMed]
- [63].Yang F, Zhang Q, Wang M, and Qiu G. Quality classified image analysis with application to face detection and recognition. arXiv, 2018.
- [64].Yang L, Zhang Y, Chen J, Zhang S, and Chen DZ. Suggestive annotation: A deep active learning framework for biomedical image segmentation. In MICCAI, 2017. [Google Scholar]
- [65].Yuan C, Zhang Y, and Liu Z. A survey on technologies for automatic forest fire monitoring, detection, and fighting using unmanned aerial vehicles and remote sensing techniques. Canadian journal of forest research, 2015.
- [66].Zhang Y, Yang L, Chen J, Fredericksen M, Hughes DP, and Chen DZ. Deep adversarial networks for biomedical image segmentation utilizing unannotated images. In MIC-CAI, 2017.
- [67].Zhao J, Xiong L, Jayashree K, Li J, Zhao F, Wang Z, Pranata S, Shen S, and Feng J. Dual-agent gans for photorealistic and identity preserving profile face synthesis. In NIPS, 2017. [Google Scholar]
- [68].Zhou N, Yu X, Zhao T, Wen S, Wang F, Zhu W, Kurc T, Tannenbaum A, Saltz J, and Gao Y. Evaluation of nucleus segmentation in digital pathology images through large scale image synthesis. SPIE Medical Imaging. International Society for Optics and Photonics, 2017. [DOI] [PMC free article] [PubMed]
- [69].Zhu J-Y, Park T, Isola P, and Efros AA. Unpaired image-to-image translation using cycle-consistent adversarial networks. In CVPR, 2017. [Google Scholar]
- [70].Zou H and Hastie T. Regularization and variable selection via the elastic net. Journal of the Royal Statistical Society: Series B (Statistical Methodology), 2005.