Abstract
Objective
To estimate the burden of active infection and anti-SARS-CoV-2 IgG antibodies in Karnataka, India, and to assess variation across geographical regions and risk groups.
Methods
A cross-sectional survey of 16,416 people covering three risk groups was conducted between 3–16 September 2020 using the state of Karnataka’s infrastructure of 290 healthcare facilities across all 30 districts. Participants were further classified into risk subgroups and sampled using stratified sampling. All participants were subjected to simultaneous detection of SARS-CoV-2 IgG using a commercial ELISA kit, SARS-CoV-2 antigen using a rapid antigen detection test (RAT) and reverse transcription-polymerase chain reaction (RT-PCR) for RNA detection. Maximum-likelihood estimation was used for joint estimation of the adjusted IgG, active and total prevalence (either IgG or active or both), while multinomial regression identified predictors.
Results
The overall adjusted total prevalence of COVID-19 in Karnataka was 27.7% (95% CI 26.1–29.3), IgG 16.8% (15.5–18.1) and active infection fraction 12.6% (11.5–13.8). The case-to-infection ratio was 1:40 and the infection fatality rate was 0.05%. Influenza-like symptoms or contact with a COVID-19-positive patient were good predictors of active infection. RAT kits had higher sensitivity (68%) in symptomatic people compared with 47% in asymptomatic people.
Conclusion
This sentinel-based population survey was the first comprehensive survey in India to provide accurate estimates of the COVID-19 burden. The findings provide a reasonable approximation of the population immunity threshold levels. Using existing surveillance platforms coupled with a syndromic approach and sampling framework enabled this model to be replicable.
Keywords: Sentinel survey, COVID-19, Antibody testing, Karnataka
Introduction
The global pandemic of SARS-CoV-2 causing coronavirus disease 2019 (COVID-19) raged across the world within a few months. India has the second-highest burden of COVID-19, with 8.8 million infected and 130,070 deaths as of 16 November 2020 (Ministry of Health and Family Welfare Services, 2020). Currently, India has only case-based reporting as the prime strategy for estimating disease burden and trend through all the epidemic phases. Case-based reporting has the advantages of rationalizing testing, isolating cases, and tracing and quarantining contacts (Hellewell et al., 2020); however, it does not provide an estimate of the true burden of the disease, as it mostly identifies sicker people seeking care or those who have better access to healthcare. Hence, the reported case counts of COVID-19 have grossly underestimated the true prevalence or disease burden of the pandemic. The two rounds of national seroprevalence surveys conducted by the Indian Council of Medical Research (ICMR) indicated that 81–130 infections were missed for every reported case in the initial survey conducted in May 2020 (Murhekar et al., 2020), which improved to missing nearly 26–32 infections per reported case by August 2020. Serological surveys, such as those conducted by the ICMR, can help to understand the burden of past infections. It is also important to detect active infections during a pandemic; however, this is challenging since 45% of the infected people have mild or no symptoms (Chatterjee et al., 2020). Furthermore, the estimation is affected by poor in-person testing due to inaccessibility, stigma and supply-side inadequacies.
Effective public health measures require reliable understanding of the existing burden of disease through epidemiological investigations. Joint estimation of IgG prevalence and active SARS-CoV-2 infections can help to detect, manage and control the disease outbreak. Seroprevalence estimates from around the world show varying numbers ranging from 0.07% in hospital patients to 54.1% in slum inhabitants (Malani et al., 2021, Herzog et al., 2020, Meyerowitz-Katz and Merone, 2020, Korth et al., 2020, Stringhini et al., 2020, Hallal et al., 2020, Noh et al., 2020, Financial Express, 2021, Aarti et al., 2020). The ICMR survey results reported 0.73% prevalence across India (May–June 2020), which increased to 7% by the end of September (Murhekar et al., 2020). The surveys in slums and non-slums of Mumbai (Malani et al., 2021) showed considerable variation: 54.1% (95% CI 52.7–55.6) and 16.1% (95% CI 14.9–17.4) prevalence, respectively. Serosurveys in a healthcare setting of North India showed prevalence increasing from 2.3% in April to 50.6% in July (Siddiqui et al., 2020). However, there are concerns about using only IgG prevalence as a marker of population immunity threshold and for estimation of period prevalence of infection. These include inability to detect IgG antibodies over time, varying sampling methods, the unreliable nature of the predictive value of positive antibody tests with varying sensitivity and specificity of different tests affecting the tests’ reliability, and the presence of other types of immune response (La Marca et al., 2020, To et al., 2020, Zou et al., 2020). Some of these concerns could be mitigated by understanding the burden of active infections concurrently with IgG estimation.
Karnataka has an estimated population of 70.7 million (Statistics DoEa, 2013) spread over 191,791 km2. The first confirmed COVID-19 case was reported on 09 March 2020. As of 16 November 2020, there were 861,647 cumulative cases, 27,146 active cases and 11,529 deaths (Ministry of Health and Family Welfare Services, 2020). This study aimed to estimate the combined proportion of the population with past or active SARS-CoV-2 infections in Karnataka and assess the variation across geographical regions and risk groups. The results of what is perhaps the first comprehensive statewide sentinel-based survey in India are presented here. The design and analysis methodologies of this survey can serve as the blueprint for other similar surveys.
Methods
Study setting, design and sample size
Setting
This was the first round of the proposed serial cross-sectional surveys across the districts of Karnataka. This state has 30 administrative districts. The capital district Bengaluru has approximately 13.6 million residents. The study was conducted during 03–16 September 2020.
Design
Each district was a unit of the survey, except Bengaluru, which was subdivided into nine units. From the resulting 38 units, geographically representative healthcare facilities (district hospitals or community healthcare centres or primary healthcare centres) with the expertise to conduct the survey were selected for this sentinel-based population survey (Figure 1 and Appendix D). The participants included only adults aged ≥18 years. The survey excluded those already diagnosed with SARS-CoV-2 infection or those who did not agree to provide informed consent to participate in the survey. The population was stratified into three risk groups based on community exposure and vulnerability to COVID-19: the low-risk group comprised pregnant women presenting for a periodic antenatal check-up, people attending the outpatient departments for common ailments and their attendees; the moderate-risk group comprised people with high contact in the community such as bus conductors and autorickshaw drivers, vendors at vegetable markets, healthcare workers, individuals in containment zones, people in congregate settings (markets, malls, retail stores, bus stops, railway stations), and waste collectors (The Lancet, 2020); the high-risk group comprised the elderly (aged ≥60 years) and people with comorbid conditions such as chronic liver, lung or renal disease, diabetes, heart disease, hypertension, immunocompromised conditions, and malignancy. The protocol mandated that the elderly be systematically sampled from a population register, and those with comorbidities be systematically sampled from a list maintained by the facility on those suffering from non-communicable diseases. In each of the other risk subgroups, participants were also systematically sampled.
Figure 1.
Sites (blue dots) of the survey representing geographical spread across Karnataka.
The inset picture shows the sites across Bengaluru (multi-coloured dots).
Sample size
A 10% prevalence and design effect of 3 were assumed. A margin of error of 5% was required. Under normal error assumption, a standard calculation for a 95% CI required that the minimum sample size be 432 per unit; this was then equally divided among the risk groups (144 per risk group). In each risk group, the number of samples was further equally divided among the risk subgroups. This led to a total sample size of 16,416 across the 38 units (Supplementary Table 4/Appendix D).
Sample collection and laboratory testing
From participants in the low-risk group, both nasopharyngeal and oropharyngeal swab samples were collected for the RT-PCR test following the ICMR protocol and 4 ml of venous blood for the IgG antibody test. In the moderate-risk and high-risk groups, two swab samples in different media were collected for the antigen and RT-PCR tests and 4 ml of venous blood.
The rapid antigen detection test (RAT) was performed using the Antigen Standard Q COVID-19 Ag detection kit, a rapid chromatographic immunoassay for the qualitative detection of antigens specific to SARS-CoV-2. The RT-PCR test was performed for all low-risk participants and those who tested negative on the RAT through the current ICMR-approved testing network. For antibody testing, the collected venous blood sample was left undisturbed at room temperature for 30 min for clotting, then centrifuged at 3,000 rpm, and the serum was transported to the laboratory by maintaining a cold chain. SARS-CoV-2-specific IgG antibodies were detected using a commercially available, validated and ICMR-approved kit (Covid Kavach Anti SARS-CoV-2 IgG antibody detection ELISA, Zydus Cadila, India) (Sapkal et al., 2020); the test was performed as per the manufacturer’s instructions. The results were interpreted as positive or negative for SARS-CoV-2 IgG antibodies based on the cut-off value of optical densities obtained with positive and negative samples provided in the kit.
Data collection
After obtaining written informed consent, information on basic demographic details, exposure history to laboratory-confirmed COVID-19 cases, symptoms suggestive of COVID-19 in the preceding month, and clinical history were recorded on a web-based application designed specifically for the study and were linked to the samples using the ICMR Specimen Referral Forms for COVID-19. The category, symptoms, contact, and comorbidity information for participants were gathered using the web-based application. RAT/RT-PCR results were entered into the ICMR test-data portal. The IgG antibody test results were retrieved directly from the labs. A consolidated line-list of all the participants was then created. From this, subsets of participants in risk categories, subcategories, age groups, sex, and geographical units were used to jointly estimate IgG prevalence, active infection fraction, and total burden in the respective categories. Symptoms and comorbidity data, which were part of the consolidated line-list, were used in the regression. Only anonymised data with no personal identifiers were used for the analysis.
Ethical considerations
The Institutional Ethics Committee (IEC) of the Indian Institute of Public Health-Bengaluru campus reviewed and approved the study (vide. IIPHHB/TRCIEC/174/2020). The participants were informed of the purpose of the survey, the samples that would be taken and were requested to respond to some screening questions. Those who were aged <18 years, already diagnosed with SARS-CoV-2 infection, unwilling to provide samples for the test, and who did not agree to provide informed consent were excluded. After obtaining informed consent, information on basic demographic details, exposure history, symptoms observed in the previous month, and clinical history were noted. Participants’ test results were available and shared with them by the concerned healthcare facility.
Statistical analysis
IgG prevalence was defined as the fraction of the sampled population with IgG antibodies. Active infection fraction was defined as the fraction of the sampled population who tested positive on the RT-PCR/RAT test. Total prevalence of COVID-19 was defined as the fraction of the sampled population with either IgG or active infection.
To enable joint estimation of IgG prevalence, active infection fraction and total prevalence, it was first modelled that an individual be in one of four disease states: having active infection but no IgG antibodies, having IgG antibodies but no evidence of active infection, having both IgG antibodies and active infection, and having neither active infection nor IgG antibodies. The disease state of the individual is, however, hidden and can only be inferred from the RAT, RT-PCR and IgG antibody test outcomes. This leads to a parametric model for the probabilities of test outcomes (observations), given the disease-state probabilities (parameters of the model), after taking the sensitivities and specificities of the tests into account.
To obtain the joint estimates of the parameters in a stratum, maximum likelihood estimation was used, which ipso facto provided estimates already adjusted for the sensitivities and specificities of the tests. The joint estimation was an extension of the Rogan-Gladen formula (Rogan and Gladen, 1978). The procedure also accounted for the protocol-induced variation of test types across participants. Confidence intervals were obtained by invoking asymptotic normality of the maximum likelihood estimates, with their covariance matrix being approximated by the inverse of the Fisher information matrix of the parametric model. Further, weighted adjusted estimates for Karnataka were obtained after weighing each district’s prevalence estimates by the population fraction in that district. Facility-level weighting was not used because some of the healthcare facilities served the entire district population. Also, the estimates for prevalence across risk subcategories were unweighted. Odds ratios across risk subcategories (with respect to one reference subcategory) were calculated by restricting attention to the relevant subcategories.
To identify the weights on various independent/explanatory variables (symptoms, comorbidities, etc.) for predicting past infection and active infection, multinomial regressions were used to regress the test outcomes on two disjoint sets of independent variables. The procedure could be embedded within the framework of the generalised linear model with multinomial logit functions along with a custom link function that accounted for the test-type variability across participants, and also the tests’ sensitivities and specificities. Important explanatory variables were captured using the Wald test.
The details are given in the Supplementary material provided.
Results
This was the first statewide sentinel-based population survey conducted in India. It was carried out in 290 healthcare facilities spread across the state of Karnataka. Of the 16,585 people surveyed in the different risk categories, this study presents the results for 15,624 individuals whose RAT plus RT-PCR and COVID Kavach ELISA antibody test results were matched (Appendix C). A total of 16,585 IgG results were provided. The results of 513 were not considered due to missing information and inability to match the participant in the database; 448 entries were further unmapped to the line list because of manual data-entry errors or because data were not retrievable from the ICMR portal. Also, 18 IgG samples were inconclusive (Figure 1 in Supplementary material/Appendix C).
IgG prevalence
The overall weighted adjusted seroprevalence of IgG was 16.8% (95% CI 15.5–18.1); this was as of 03 September 2020 and at the state level, obtained after adjusting for the serial sensitivities and specificities of all tests (Table 1 ).
Table 1.
Seroprevalence of IgG antibodies against SARS-CoV-2 and active infection in Karnataka.
| Category | Type | Samplesa | % IgG against SARS-CoV-2b |
% active infection with COVID-19 b |
% prevalence of COVID-19b |
Odds ratio | |
|---|---|---|---|---|---|---|---|
| State | Karnataka | Crude | 15,939 | 2,565/15,939 = 16.1% | 2,363/14,132 = 16.7% | 4,582/15,939 = 28.7% | |
| Adjusted | 15,624 | 15.7 | 12 | 26.3 | |||
| Weighted adjusted | 15,624 | 16.8 (15.5–18.1) | 12.6 (11.5–13.8) | 27.7 (26.1–29.3) | |||
| Demography | Sex | Male | 8,165 | 15.8 (14.3–17.4) | 15.5 (13.9–17.2) | 29.8 (27.7–31.8) | 1.51 (1.23–1.88) |
| Female | 7,445 | 14.8 (13.2–16.4) | 8.4 (7–9.8) | 21.9 (19.9–23.8) | 1 | ||
| Age, years | 18–29 | 5,184 | 12.5 (10.7–14.3) | 7.1 (5.6–8.6) | 19 (16.8–21.3) | 1 | |
| 30–39 | 3,353 | 16 (13.6–18.4) | 11.2 (9–13.5) | 25.7 (22.7–28.7) | 1.47 (1.09–1.99) | ||
| 40–49 | 2,447 | 15.4 (12.6–18.2) | 15.8 (12.8–18.8) | 29.3 (25.6–33) | 1.77 (1.27–2.44) | ||
| 50–59 | 1,792 | 17.6 (14.2–21) | 17.3 (13.7–20.9) | 33.3 (28.9–37.7) | 2.13 (1.5–3) | ||
| ≥60 | 2,848 | 18.1 (15.3–20.8) | 15.9 (13.2–18.7) | 31.6 (28.1–35) | 1.97 (1.44–2.67) | ||
| Region | Urban | 14,107 | 15.8 (14.6–17) | 12.4 (11.3–13.6) | 26.7 (25.2–28.2) | 1.54 (1.11–2.23) | |
| Rural | 1,517 | 10.6 (7.4–13.7) | 9 (5.9–12.1) | 19.1 (15–23.3) | 1 | ||
| Risk category | High-risk | 5,322 | 17.9 (15.9–19.9) | 15.9 (13.8–17.9) | 31.7 (29.1–34.2) | 1.78 (1.37–2.31) | |
| Moderate-risk | 5,253 | 14.3 (12.4–16.2) | 12.3 (10.4–14.1) | 25.4 (23–27.8) | 1.3 (1–1.71) | ||
| Low-risk | 5,049 | 13.6 (11.8–15.5) | 8.1 (6.5–9.8) | 20.7 (18.4–23) | 1 | ||
| Risk sub-category d |
High-risk | Elderly | 2,445 | 17.7 (14.7–20.6) | 16.8 (13.7–19.8) | 32.4 (28.6–36.2) | 2.5 (1.7–3.76) |
| People with comorbidities | 2,455 | 18.2 (15.2–21.2) | 14.7 (11.8–17.6) | 30.5 (26.8–34.2) | 2.29 (1.55–3.45) | ||
| Moderate-risk | Containment zones | 1,138 | 16.2 (12–20.4) | 16.3 (11.9–20.7) | 31 (25.5–36.5) | 2.34 (1.45–3.81) | |
| Bus conductors/auto drivers | 1,008 | 16.1 (11.7–20.6) | 13.9 (9.5–18.3) | 28.9 (23.2–34.6) | 2.12 (1.28–3.51) | ||
| Vendors at vegetable markets | 1,025 | 15.4 (11.1–19.8) | 13.5 (9.2–17.8) | 27.9 (22.3–33.5) | 2.02 (1.22–3.34) | ||
| Congregate settingsc | 1,259 | 13.6 (9.8–17.3) | 13.5 (9.6–17.4) | 25.8 (20.9–30.8) | 1.81 (1.12–2.95) | ||
| Healthcare workers | 1,107 | 11.8 (8–15.6) | 4.9 (1.9–7.9) | 16 (11.4–20.6) | 0.99 (0.54–1.72) | ||
| Low-risk | Outpatient department | 2,632 | 14.8 (12.1–17.5) | 13 (10.3–15.6) | 26 (22.6–29.5) | 1.83 (1.24–2.78) | |
| Pregnant women | 2,555 | 12.4 (9.8–14.9) | 4.1 (2.2–5.9) | 16.1 (13.1–19.1) | 1 | ||
| Pre-existing medical conditions | More than one | 529 | 18.3 (11.9–24.7) | 15.8 (9.3–22.3) | 31.3 (23.2–39.4) | 1.38 (0.85–2.14) | |
| One | 1,941 | 17.1 (13.8–20.4) | 17.5 (14–20.9) | 32.4 (28.2–36.7) | 1.45 (1.1–1.91) | ||
| None | 13,154 | 14.9 (13.7–16.1) | 11.2 (10–12.3) | 24.8 (23.3–26.3) | 1 | ||
| Symptoms | More than one | 803 | 15.7 (10.7–20.6) | 35.6 (28.7–42.5) | 48.9 (41.6–56.2) | 3.39 (2.32–5.01) | |
| One | 3,423 | 15.9 (13.5–18.4) | 20.6 (17.8–23.4) | 34.4 (31.1–37.7) | 1.86 (1.47–2.36) | ||
| None | 11,398 | 15.1 (13.8–16.4) | 8 (7–9.1) | 22 (20.4–23.5) | 1 | ||
Included only samples that were mapped to individuals.
All estimates were adjusted for sensitivities and specificities of the RAT, RT-PCR and antibody testing kits and procedures. The assumed values were RAT sensitivity 0.5, specificity 0.975; RT-PCR sensitivity 0.95, specificity 0.97; IgG ELISA kit sensitivity 0.921, specificity 0.977 Weighted estimates for Karnataka estimated the prevalence in each unit and then weights according to population.
Markets, malls, retail stores, bus stops, railway stations, waste collectors.
Some individuals recruited in the moderate-risk and low-risk categories were moved to high-risk because of age or comorbidities.
Active infection
It was estimated that 12.6% (95% CI 11.5–13.8) of the seemingly unsuspecting participants in the general population or an estimated 89,17,539 (95% CI 81,39,023–97,66,828) people had active infection (on 16 September 2020). This was based on the numbers that tested positive on RT-PCR/RAT and after taking into account the IgG outcomes and the serial sensitivities and specificities of all tests (Table 1).
Total prevalence of COVID-19
The adjusted total prevalence of COVID-19 at state level was 27.7% (95% CI 26.1–29.3) as of 16 September 2020 (combined IgG and active infection (Table 1).
Stratifications
The seroprevalence of IgG among males and females was similar, but the active infection was higher in males than females (15.5% vs. 8.4%) (Table 1). Thus, the total prevalence was higher in males than in females (29.8% vs. 21.9%). Estimates of both seroprevalence and total prevalence were higher in the elderly population and lower among those aged <30 years. The high-risk population had a higher prevalence (31.7%; 95% CI 29.1–34.2), followed by the moderate-risk population (25.4%; 95% CI 23.0–27.8) and then the low-risk population (20.7; 95% CI 18.4–23.0) (Table 1).
Case-to-infection ratio (CIR)
At the state level, it was estimated that there were 40 infected individuals for every RT-PCR-confirmed case detected as of 16 September 2020 (Table 2 ). This was estimated by using 484,954 reported number of cases in Karnataka (Ministry of Health and Family Welfare Services, 2020) and the adjusted prevalence of COVID-19 (27.7%) against SARS-CoV-2. The cases-to-infections ratio ranged between 11–112 across units.
Table 2.
Seroprevalence of IgG antibodies against SARS-CoV-2 and active infection in districts of Karnataka state (N = 15,624).
| Unit | Samplesa | % IgG against SARS-CoV2b | % active infectionb | % prevalence of COVID-19b | CIR | IFR |
|---|---|---|---|---|---|---|
| Karnataka | 15,624 | 16.8 (15.5–18.1) | 12.6 (11.5–13.8) | 27.7 (26.1–29.3) | 1 : 40 | 0.05% |
| Ballari | 406 | 22.7 (14.9–30.4) | 34.3 (25.2–43.3) | 43.5 (34–53) | 1 : 49 | 0.04% |
| Davanagere | 412 | 16.8 (9.8–23.7) | 29.1 (20.2–38) | 40.9 (31.3–50.5) | 1 : 62 | 0.06% |
| Udupi | 439 | 17 (10.3–23.7) | 22.6 (15–30.2) | 37 (28.2–45.9) | 1 : 34 | 0.05% |
| Vijayapura | 381 | 24.1 (15.9–32.4) | 13.8 (6.6–21.1) | 35.6 (25.9–45.3) | 1 : 112 | 0.02% |
| Raichur | 404 | 23.1 (15.2–30.9) | 12.1 (5.4–18.7) | 34.3 (25–43.6) | 1 : 76 | 0.02% |
| Chikmagalur | 436 | 12.3 (6.3–18.4) | 20.9 (13.1–28.7) | 32.1 (23.1–41.1) | 1 : 54 | 0.06% |
| Yadgir | 422 | 15.7 (9–22.5) | 18.5 (11.2–25.9) | 31.9 (23–40.8) | 1 : 62 | 0.02% |
| Hassan | 410 | 13.6 (7.1–20.1) | 21.1 (12.8–29.4) | 31 (21.7–40.3) | 1 : 44 | 0.08% |
| Belgaum | 430 | 23.9 (16.2–31.7) | 6.4 (1.4–11.4) | 30.3 (21.6–39) | 1 : 95 | 0.02% |
| Kalaburagi | 425 | 17.4 (10.5–24.4) | 14.4 (7.8–21) | 30.1 (21.4–38.7) | 1 : 60 | 0.04% |
| Bengaluru Urban Conglomerate | 3617 | 22.4 (19.6–25.3) | 9.2 (7.1–11.2) | 30.1 (26.9–33.3) | 1 : 23 | 0.07% |
| Ramanagar | 408 | 14.2 (7.5–20.8) | 16.1 (8.7–23.6) | 29.6 (20.4–38.7) | 1 : 76 | 0.02% |
| Tumakuru | 429 | 7 (1.9–12) | 25.1 (16.2–34.1) | 29.5 (20–39) | 1 : 82 | 0.08% |
| Bengaluru Rural | 432 | 15.6 (9–22.2) | 16.4 (9–23.8) | 29 (20.2–37.9) | 1 : 46 | 0.02% |
| Haveri | 417 | 15 (8.3–21.7) | 14.5 (7.8–21.3) | 28.8 (20.1–37.6) | 1 : 72 | 0.04% |
| Mysuru | 402 | 19.2 (11.9–26.6) | 8.4 (2.7–14) | 27.6 (18.9–36.3) | 1 : 34 | 0.07% |
| Dakshina Kannada | 430 | 15 (8.5–21.6) | 13.5 (6.9–20) | 27.4 (19–35.9) | 1 : 34 | 0.11% |
| Chitradurga | 411 | 10.3 (4.4–16.3) | 16 (8.5–23.4) | 26 (17.2–34.9) | 1 : 85 | 0.01% |
| Mandya | 414 | 18.8 (11.4–26.1) | 6.7 (1.3–12.1) | 25.5 (16.8–34.2) | 1 : 54 | 0.02% |
| Koppal | 427 | 20 (12.7–27.2) | 2.6 (0–6.2) | 22.6 (14.7–30.5) | 1 : 38 | 0.04% |
| Shivamogga | 426 | 8.1 (2.9–13.4) | 13.7 (6.8–20.5) | 21.8 (13.5–30) | 1 : 31 | 0.09% |
| Chamarajanagar | 383 | 16 (8.8–23.1) | 6.6 (1.1–12.1) | 21.3 (12.9–29.7) | 1 : 72 | 0.02% |
| Kodagu | 412 | 12.3 (6–18.5) | 8.7 (2.8–14.5) | 20.7 (12.6–28.9) | 1 : 56 | 0.03% |
| Bidar | 407 | 18.2 (10.9–25.4) | 0.7 (0–3.3) | 18.9 (11.2–26.5) | 1 : 64 | 0.04% |
| Uttara Kannada | 419 | 8.4 (3–13.8) | 8.7 (3–14.4) | 16.6 (9.1–24.1) | 1 : 33 | 0.04% |
| Kolar | 431 | 10.3 (4.5–16.1) | 6.7 (1.6–11.9) | 16.3 (9–23.6) | 1 : 59 | 0.04% |
| Chikkaballapur | 412 | 6.7 (1.6–11.8) | 5.8 (0–11.8) | 12.4 (4.8–20) | 1 : 28 | 0.07% |
| Bagalkot | 401 | 4.4 (0–8.9) | 9.7 (3.6–15.8) | 12.3 (5.3–19.4) | 1 : 31 | 0.08% |
| Gadag | 341 | 6.8 (1.3–12.3) | 2.7 (0–8.5) | 9.5 (1.6–17.4) | 1 : 14 | 0.11% |
| Dharwad | 440 | 7.6 (2.5–12.6) | 2 (0–5.5) | 9.2 (3.2–15.2) | 1 : 13 | 0.21% |
Included only samples that were mapped to individuals; CIR: case-to-infection ratio; IFR: infection fatality rate.
Adjusted for sensitivities and specificities of RAT, RT-PCR, and antibody testing kits and procedure.
Infection fatality rate (IFR)
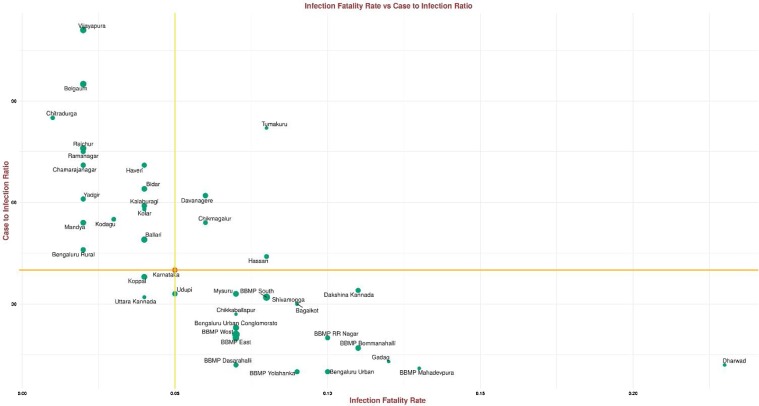
As of 03 September 2020, the IFR due to COVID-19 in Karnataka was estimated as 0.05%, with more than half of the units (21 of 38) above state IFR; the highest was estimated in the Dharwad district (0.21%) (Table 2 and Figure 2 ).
Figure 2.
Scatter plot of CIR versus IFR. The size of the point indicates the IgG prevalence in the units.
The horizontal and the vertical lines intersect at Karnataka’s IFR and CIR. Moving clockwise from the upper-left quadrant: a unit with a larger green disk had high IgG antibody prevalence, low IFR and high CIR, such a unit was missing cases and deaths; a unit with a larger green disk in the upper-right quadrant had high IgG antibody prevalence, high IFR and high CIR, such a unit was also likely missing cases but death reporting was better than average; a unit with a larger green disk in the bottom-right quadrant had high IgG antibody prevalence, high IFR and low CIR, such a unit did well in identifying cases and had better-than-average reporting of deaths; a unit with a larger green disk in the bottom left had low IFR and low CIR, such a unit saw a surge in cases but did well in identifying cases and had low fatality rates, perhaps due to good clinical practices that could be studied and replicated elsewhere.
District/unit variations across the state
The IgG prevalence was highest in Vijayapura district (24.1%) and lowest in Bagalkot district (4.4%). The state capital Bengaluru had an IgG prevalence of 22.4% (95% CI 19.6–25.3). The active infection fraction was highest in Ballari (34.3%) and lowest in Bidar (0.7%). Bengaluru’s active infection fraction was an estimated 9.2% (95% CI 7.1–11.2). The overall COVID-19 prevalence was lowest in Dharwad district (9.2%) and highest in Ballari district (43.5%) (Table 2). The total COVID-19 prevalence in Bengaluru was estimated to be 30.1% (95% CI 26.9–33.3). Within Bengaluru itself (3,617 samples), it was estimated that BBMP West had the highest IgG against SARS-CoV-2 and total prevalence of COVID-19. In contrast, BBMP Mahadevapura had the least (Supplementary Table 1). Again, BBMP RR Nagar had the highest active infection fraction within Bengaluru and BBMP East had the lowest (Supplementary Table 1). Districts with high case infection ratios (>40) were Vijayapura, Belgaum, Chitradurga, Tumakuru, Raichur, Ramanagar, Haveri, Chamarajanagar, Bidar, Davanagere, Yadgir, Kalaburagi, Kolar, Kodagu, Mandya, Chikmagalur, Ballari, Bengaluru Rural, and Hassan (Table 2). To summarise: there was differential exposure to the disease across the state.
Explanatory variables
A generalised linear model-based multinomial regression for symptoms indicated that headache, chest pain, wheezing, rhinorrhoea, cough, sore throat, muscle ache, fatigue, chills, and fever were significant variables that predicted active infection, with fever being the most significant. Diarrhoea, chest-pain, rhinorrhoea, fatigue, and fever predicted the presence of IgG antibodies to some extent, with diarrhoea having the highest weight. A second generalised linear model-based multinomial regression yielded additional variables that predicted active infection. These were attendance at the outpatient department of the healthcare facilities and contact with COVID-19-positive patients. Additional variables that predicted the presence of IgG were professions who had more contact with the public, residence in containment zones and the urbanisation level of the district (Tables 3 and 4 ).
Table 3.
Generalized linear model: prediction of active, IgG and simultaneous IgG and active infection.
| Predictor | Active p-value@ | IgG p-value@ |
Active and IgG p-value@ |
Logistic p-value@ |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Intercept | −2.4 | 0.044 | *** | −1.7 | 0.031 | *** | −4.1 | 0.12 | *** | −1.1 | 0.021 | *** |
| Diarrhoea | 0.54 | 0.48 | 1 | 0.32 | *** | 1.4 | 0.85 | . | 0.79 | 0.25 | ** | |
| Abdominal pain | −3 | 3.8 | −0.05 | 0.25 | −0.11 | 0.85 | −0.23 | 0.18 | ||||
| Vomiting | 0.56 | 0.4 | . | −0.023 | 0.39 | −6.2 | 24 | 0.18 | 0.24 | |||
| Headache | 0.56 | 0.16 | *** | −0.032 | 0.18 | 0.34 | 0.46 | 0.23 | 0.1 | * | ||
| Other respiratory symptoms | 0.37 | 0.52 | 0.33 | 0.39 | −6.4 | 33 | 0.27 | 0.28 | ||||
| Chest pain | 0.68 | 0.23 | ** | 0.55 | 0.21 | ** | −0.51 | 1.3 | 0.46 | 0.14 | ** | |
| Wheezing | 1 | 0.46 | * | −0.089 | 0.58 | 1.3 | 0.86 | . | 0.37 | 0.31 | ||
| Shortness of breath | 0.62 | 0.49 | 0.12 | 0.59 | 0.37 | 1.5 | 0.48 | 0.34 | ||||
| Runny nose | 0.95 | 0.27 | *** | 0.55 | 0.28 | * | −7.4 | 32 | 0.56 | 0.19 | ** | |
| Cough | 0.84 | 0.086 | *** | 0.13 | 0.09 | . | 0.3 | 0.28 | 0.41 | 0.054 | *** | |
| Sore throat | 0.71 | 0.37 | * | −0.078 | 0.46 | 1 | 0.8 | . | 0.32 | 0.25 | ||
| Muscle ache | 0.67 | 0.18 | *** | 0.033 | 0.2 | 0.25 | 0.57 | 0.25 | 0.12 | * | ||
| Fatigue | 0.68 | 0.25 | ** | 0.42 | 0.23 | * | −2.4 | 7.8 | 0.42 | 0.16 | ** | |
| Chills | 0.77 | 0.21 | *** | −0.35 | 0.3 | 0.47 | 0.61 | 0.19 | 0.15 | |||
| Fever | 1.5 | 0.085 | *** | 0.21 | 0.11 | * | 1.4 | 0.23 | *** | 0.8 | 0.058 | *** |
@ *** indicates p < 0.001; ** indicates p < 0.01; * indicates p < 0.05;. indicates p < 0.1.
Table 4.
Generalized linear model: prediction of active, IgG and simultaneous IgG and active infection.
| Predictor | Active p-value@ | IgG p-value@ |
Active/IgG p-value@ |
Logistic p-value@ |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Intercept | −3.3 | 0.19 | *** | −2.6 | 0.13 | *** | −6.6 | 0.76 | *** | −1.8 | 0.081 | *** |
| Chronic liver disease | −0.59 | 1 | 0.38 | 0.66 | 0.54 | 1.5 | −0.012 | 0.47 | ||||
| Chronic renal disease | −7.3 | 82 | −0.03 | 0.56 | −5.6 | 22 | −0.53 | 0.47 | ||||
| Diabetes | 0.075 | 0.12 | −0.033 | 0.11 | 0.034 | 0.29 | 0.013 | 0.068 | ||||
| Heart disease | −0.17 | 0.4 | 0.42 | 0.27 | . | −0.21 | 1.1 | 0.15 | 0.2 | |||
| Hypertension | 0.14 | 0.12 | −0.072 | 0.11 | 0.13 | 0.3 | 0.04 | 0.071 | ||||
| Immunocompromised condition | −0.44 | 0.45 | −0.6 | 0.4 | . | −1.3 | 2.2 | −0.46 | 0.23 | * | ||
| Malignancy | 1 | 0.85 | 0.46 | 0.93 | −4.6 | 32 | 0.53 | 0.61 | ||||
| High-risk | 0.59 | 0.29 | * | −0.11 | 0.33 | −1 | 1.3 | 0.14 | 0.19 | |||
| Moderate-risk | 0.41 | 0.35 | −0.56 | 0.37 | . | −0.22 | 1.9 | −0.075 | 0.23 | |||
| OPD attendee | 0.67 | 0.19 | *** | 0.078 | 0.11 | 0.93 | 0.57 | . | 0.23 | 0.075 | ** | |
| Bus conductor or auto driver | 0.28 | 0.41 | 0.74 | 0.4 | * | 0.6 | 1.9 | 0.33 | 0.24 | |||
| In containment zone | 0.5 | 0.39 | 0.74 | 0.39 | * | 0.79 | 1.9 | 0.43 | 0.24 | . | ||
| Healthcare worker | −1 | 0.45 | * | 0.27 | 0.39 | −0.29 | 1.9 | −0.25 | 0.24 | |||
| In congregate setting | 0.32 | 0.4 | 0.52 | 0.39 | . | 0.59 | 1.9 | 0.29 | 0.24 | |||
| Comorbidity | 0.04 | 0.33 | 0.37 | 0.34 | 1.8 | 1.4 | . | 0.17 | 0.2 | |||
| Elderly | 0.35 | 0.34 | 0.34 | 0.34 | 1.4 | 1.4 | 0.21 | 0.2 | ||||
| Vegetable vendor | 0.33 | 0.4 | 0.7 | 0.39 | * | 0.37 | 2 | 0.33 | 0.24 | |||
| Age 30–39 | 0.21 | 0.11 | * | 0.24 | 0.084 | ** | 0.95 | 0.42 | * | 0.2 | 0.055 | *** |
| Age 40–49 | 0.53 | 0.11 | *** | 0.17 | 0.098 | * | 1.2 | 0.44 | ** | 0.3 | 0.062 | *** |
| Age 50–59 | 0.64 | 0.13 | *** | 0.34 | 0.11 | *** | 1 | 0.48 | * | 0.42 | 0.07 | *** |
| Age 60+ | 0.31 | 0.15 | * | 0.26 | 0.13 | * | 1.5 | 0.51 | ** | 0.29 | 0.083 | *** |
| Male | 0.45 | 0.078 | *** | 0.052 | 0.063 | 0.02 | 0.2 | 0.19 | 0.041 | *** | ||
| Other | −0.86 | 1.8 | −28 | 640000 | −14 | 2300 | −1.6 | 1 | ||||
| Urban or rural | 0.39 | 0.12 | *** | 0.36 | 0.11 | *** | 1.2 | 0.63 | * | 0.32 | 0.065 | *** |
| Contact with a positive patient | 0.75 | 0.096 | *** | 0.11 | 0.11 | 0.77 | 0.27 | ** | 0.39 | 0.063 | *** | |
| Urbanization | −0.56 | 0.15 | *** | 0.68 | 0.1 | *** | 0.24 | 0.37 | 0.14 | 0.073 | . |
@ *** indicates p < 0.001; ** indicates p < 0.01; * indicates p < 0.05;. indicates p < 0.1.
RAT versus RT-PCR sensitivity for symptomatic and asymptomatic people
As of September 2020, it was noted that RAT was more sensitive for symptomatic individuals. For participants with symptoms, 543 RAT were positive out of the 798 RT-PCR-confirmed positive participants. This yielded a sensitivity of 68.0% for those with symptoms. In contrast, for participants without symptoms, 348 RAT were positive out of 742 RT-PCR-confirmed positive participants. This yielded a sensitivity of 46.9% for those without symptoms.
Discussion
The study involved healthcare workers and several state and central agencies across the entire state of Karnataka and was based around 290 healthcare facilities. It addressed the important question of measuring past and active infection of COVID-19 in Karnataka. Further, this was the first study in India, and probably elsewhere, that jointly estimated the proportion of people who already had the SARS-CoV-2 infection (IgG antibody positive) and who currently had an active infection (RT-PCR/RAT positive). The combined estimates could lead to timely, informed and evidence-based public health responses.
The survey, although sentinel-based, sampled from the general population in the moderate-risk and high-risk groups. The low-risk group sampled from the facilities were restricted to healthcare workers and patients coming for periodic care or visiting outpatient departments. Further, the survey ensured good geographical spread across Karnataka. The design was thus aimed at capturing as representative a population as possible. The sampling frame serves as a reference standard and could be used for population-representative surveillance in the future. A serological test for IgG with high sensitivity (0.921) and specificity (0.977) was used, thereby yielding a better predictive value for a positive test.
An estimation of the IgG prevalence alone would have assessed the state’s burden at 16.8% prevalence. In contrast, the dual assessment of viral markers and antibodies gave the IgG prevalence and also the active infection fraction of 12.6% and a total COVID-19 burden of 27.7%. This significantly larger estimate calls for an entirely different response from the state and highlights this survey’s benefits. Moreover, 2.1% of the population showed both viral RNA and IgG antibodies. The correlates and implications of the simultaneous presence of viral RNA and IgG antibodies might require further examination in future studies.
The statewide CIR was 40 infections for every case. The number of reported cases up to 03 September 2020 was 361,305; another 123,649 reported cases were added between 03 September and 16 September. Clearly there was a surge during this 2-week period and 34% of reported cases were added. This suggests that the active infection fraction during this period should have been 34% of 16.8% estimated IgG level (i.e., 5.7%); however, the active infection fraction from the survey was as high as 12.6%. The CIR for the period prior to 03 September 2020 was 32 versus a CIR of 72 during the survey period, suggesting that a greater number of cases were missed during the surge in the first half of September 2020. The overall CIR of 40 is to be understood as a weighted average of the estimates for the two periods.
Public health responses and control measures in India have been largely shaped by case-based evidence, namely: data from the line list and contact tracing. For example, in a study from Tamil Nadu and Andhra Pradesh, Laxminarayan et al. showed that the infection probabilities ranged from 4.7–10.7% for low-risk and high-risk contacts (Laxminarayan et al., 2020). Kumar et al. showed that both asymptomatic and symptomatic SARS-CoV-2 cases transmitted the infection, with the symptomatic cases being the main driving force within Karnataka state (Kumar et al., 2021). However, the current seroprevalence-based estimates did not suffer from selection bias, as the sampling frame included the general population and went beyond reported cases and contact tracing. Therefore, the current study complements the understanding by providing better estimates of the actual burden of infection in the community.
Across risk groups, the elderly and those with comorbidities had a higher prevalence of COVID-19 (Hu et al., 2020, CDC COVID-19 Response Team, 2020, Guan et al., 2020), suggesting that they are at higher risk of contracting the infection. Despite the expected lower exposure, higher prevalence in them offers the possibility of infection with lower viral dose or that younger age groups have a protective immune response.
The reported IFR due to COVID-19 of 0.05% was likely an underestimate and a function of how well each district reported death data. Studies worldwide have found that the IFR of COVID-19 ranged from 0.17–4.16% (Rinaldi and Paradisi, 2020, Bendavid et al., 2021, Lewis and Torgerson, 2012). A high estimated IFR of 0.21% was seen in Dharwad. Comparing Dharwad (population 1.8 million, IgG level 7.6%, 113 days to survey start date since 50 cases, urbanisation index = 0.56, deaths = 337) with Shivamogga (population 1.7 million, estimated IgG 8.1%, 106 days to survey start date since 50 cases, urbanisation index = 0.30, deaths = 134), 200 more deaths were seen in Dharwad for roughly similar populations, IgG levels and days since 50 cases. The higher deaths could have been due to reporting differences or issues related to clinical practice or travel from neighbouring units to avail critical or tertiary health care facilities at Dharwad. Further research is required to test these hypotheses.
This regression analysis determined which symptoms accurately predict active and past infections. Among symptoms, diarrhoea, chest pain, rhinorrhoea, fatigue, and fever predict the presence of IgG antibodies. This suggests that COVID-19 may have consequences that last beyond the active infection period. Diarrhoea suggests that the gastrointestinal tract manifestations might stay longer, which may have implications to explore with oral vaccines. Influenza-like-illness symptoms and history of contact with a COVID-19-positive person are strong predictors of active infection.
Recruitment of the low risk participants from the facilities may suggest a bias in the estimate. This was mitigated by systematically sampling only among pregnant women and attendees of outpatient clinics. A design effect of 3 was used to account for any possible bias. The low-risk participants were not administered the RAT due to the test’s low sensitivity. The statistical methodology handled such partial test administrations. The number of samples per unit of 432 was based solely on a nominal 10% IgG prevalence assumption and a requirement of a 5% margin of error. The actual 95% CI depends on the true prevalence. The reported 95% CIs were based on the inferred estimates and made use of all the available test data (IgG, RT-PCR and RAT outcomes).
This sentinel-based state-wide comprehensive population survey and the associated comparative analysis gave insight into the state of the pandemic in the different districts of Karnataka and the varying levels of prevalence across the different stratifications based on age, sex and risk. Figure 2 shows important epidemiological metrics such as IFR, CIR and their variation across geographical regions and population strata. This shows which units were likely to be missing cases but had better-than-average death reporting, which units did well in identifying cases and had better-than-average reporting of deaths, or which units had not seen a surge in cases, yet had done well in identifying cases and had low IFR. Testing strategies could be tuned to local variation, based on each unit’s total prevalence and CIR. Thus, establishing district-level sentinel-based surveillance to systematically monitor the trend of infection in the long term, to inform local decision-making at a district level, would facilitate and augment the necessary public health response towards the COVID-19 epidemic in Karnataka. It would also help to identify regions with high severity of the disease, identify at-risk populations and enable evidence-based intervention and resource allocation to effectively manage the pandemic. Repetition of the survey can better inform changes in the extent and speed of transmission and help evaluate the potential impact of containment strategies over time in different parts of the state.
The question of designing facility-level public health responses for COVID-19, rather than district-level, immediately arises. As the districts are the first-level administrative units for COVID-19 management, the survey was designed to be tight for a 5% margin of error and a 95% CI at district level. Facility-level responses can be designed if there is sufficient reliability on the estimates at each facility.
The sentinel-based population survey strategy for monitoring COVID-19 through healthcare facilities has great relevance for global public health. Sentinel surveillance systems enable identification of trends over time and are also easier to manage in terms of planning logistics, using existing human resources and are less cost-intensive. These enable timely implementation of serological studies. Team et al. reported a sentinel survey conducted by the Indian Council of Medical Research in the early stages of the pandemic (ICMR COVID Study Group et al., 2020). Such methodologies could be included by the WHO in its Solidarity II global serological study protocol (World Health Organization, 2020). This study’s findings hold significant potential to improve clinical management and guide public health interventions to reduce the burden of COVID-19 in India and other lower and middle-income countries.
Contributors
The survey was a collaborative effort of the Department of Health and Family Welfare, National Institute of Mental Health and Neuro-Sciences, Indian Institute of Public Health - Bangalore, Indian Institute of Science, Indian Statistical Institute (Bangalore Centre), UNICEF, MS Ramaiah Medical College, Bangalore Medical College, and others. Giridhara R Babu, R Lalitha, Lalitha Krishnappa, Pradeep Banandur, and Kumar DE Vasanth designed the protocol. Mysore Kalappa Sudarshan, CN Manjunath, Gopalkrishna Gururaj, Timmanahalli Sobagaiah Ranganath, Asish Satapathy, Lokesh Alahari, and Anita Desai reviewed and provided feedback on the design, implementation of the survey, analyses, and helped articulate the findings. Jawaid Akhtar and Pankaj Kumar Pandey reviewed the protocol, led the implementation of the survey and identified district-level public health responses. M Rajagopal Padma, Mohammed Shariff and Parimala S. Maroor coordinated the implementation across the state and reviewed the manuscript. Deepa Ravi, Shilpa Shiju, Prameela Dinesh, Vinitha Thakar, and Eunice Lobo developed the detailed protocol and standard operating procedures (protocol manual), implemented the study and reviewed the manuscript. Giridhara R. Babu, Siva Athreya and Rajesh Sundaresan planned and executed the data analysis, arrived at the initial findings and drafted the manuscript. Ambica Rangaiah, Ashok Munivenkatappa, Krishna S, Shantala Gowdara Basawarajappa, HG Sreedhara, Siddesh KC, Amrutha Kumari B, Nawaz Umar, Mythri BA developed the lab protocols and provided the test results. Ravi Vasanthapuram designed and developed the protocol and revised the manuscript. All authors reviewed and approved the final manuscript.
Conflict of interest
We declare no competing interests.
Data sharing
The data are accessible to researchers upon formal request for data addressed to the Commissioner, Health and Family Welfare Services, Government of Karnataka.
Role of funding source
The study was supported by the National Health Mission Karnataka. This work was also supported by the Wellcome Trust/D BT India Alliance Fellowship [Grant number: IA/CPHI/14/1/501499] awarded to Dr Giridhara R Babu, the SERB-MATRICS grant awarded to Siva R. Athreya, and the Centre for Networked Intelligence(Indian Institute of Science) grant given to Rajesh Sundaresan. The funders of the study had no role in the study design, data collection, data analysis, data interpretation, or the writing of the report. They did not participate in the decision to submit the manuscript for publication. The principal investigator (GRB) and key investigators had full access to the data. The corresponding author had final responsibility for the decision to submit for publication.
Acknowledgments
We would like to express our thanks to: Dr Arundathi, IAS, MD – NHM, Dr Patil Om Prakash R Director – DHFWS, State Surveillance Unit for their support; DSOs, DAPCU officers, AMOs and medical officers, district microbiologists and district epidemiologists for coordinating and implementing the survey and providing guidance for sample collection as per sample size to healthcare facility lab staff and coordinating sample transportation to mapped RT-PCR and antibody testing ICMR labs; Lab Nodal Officer and staff of ICMR labs for RT-PCR testing and IgG antibody testing; Mr. Ramesh and team for providing a robust web platform for data collection; lab technicians, counsellors – ICTC and NCDC, staff nurses, and healthcare workers for filling data in the survey App, collection of samples and sending samples to higher labs; Ms. Manjushree, entomologist, DHFWS for helping in fetching RAT/RT-PCR results from ICMR Portal; Ms. Maithili Karthik and Ms. Sindhu ND, PHFI, for help with the line list matching; Mr. Nihesh Rathod, Indian Institute of Science, for the generation of Karnataka and Bengaluru Urban Conglomerate maps; Nitya Gadhiwala and Abhiti Mishra of the Indian Statistical Institute for help in collation of COVID-19 data from Karnataka state bulletins and R graphics; all study participants for providing their consent to be part of this survey.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.ijid.2021.05.043.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Aarti N., Aurnab G., LS S . 2020. Epidemiological and serological surveillance of COVID-19 in Pune city organizations; pp. 1–4. India; Pune. [Google Scholar]
- Bendavid E., Mulaney B., Sood N., Shah S., Bromley-Dulfano R., Lai C., et al. COVID-19 antibody seroprevalence in Santa Clara County, California. Int J Epidemiol. 2021 doi: 10.1093/ije/dyab010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC COVID-19 Response Team Preliminary estimates of the prevalence of selected underlying health conditions among patients with coronavirus disease 2019—United States, February 12–March 28, 2020. MMWR Morb Mortal Wkly Rep. 2020;69(13):382. doi: 10.15585/mmwr.mm6913e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S., Sarkar A., Karmakar M., Chatterjee S., Paul R. How the asymptomatic population is influencing the COVID-19 outbreak in India? arXiv preprint arXiv. 2020:1–12. 2006.03034. [Google Scholar]
- Financial Express . 2021. Sero-prevalence survey Delhi: here’s why survey result in the capital is’ remarkable’. [Google Scholar]
- Guan W.-J., Liang W.-H., Zhao Y., Liang H.-R., Chen Z.-S., Li Y.-M., et al. Comorbidity and its impact on 1590 patients with Covid-19 in China: a nationwide analysis. Eur Respir J. 2020;55(5) doi: 10.1183/13993003.00547-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallal P.C., Hartwig F.P., Horta B.L., Silveira M.F., Struchiner C.J., Vidaletti L.P., et al. SARS-CoV-2 antibody prevalence in Brazil: results from two successive nationwide serological household surveys. Lancet Glob Health. 2020;8(11) doi: 10.1016/S2214-109X(20)30387-9. e1390-e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellewell J., Abbott S., Gimma A., Bosse N.I., Jarvis C.I., Russell T.W., et al. Feasibility of controlling COVID-19 outbreaks by isolation of cases and contacts. Lancet Glob Health. 2020:e488–e496. doi: 10.1016/S2214-109X(20)30074-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog S., De Bie J., Abrams S., Wouters I., Ekinci E., Patteet L., et al. Seroprevalence of IgG antibodies against SARS coronavirus 2 in Belgium: a prospective cross-sectional study of residual samples. medRxiv. 2020:1–15. doi: 10.2807/1560-7917.ES.2022.27.9.2100419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Sun J., Dai Z., Deng H., Li X., Huang Q., et al. Prevalence and severity of corona virus disease 2019 (COVID-19): a systematic review and meta-analysis. J Clin Virol. 2020 doi: 10.1016/j.jcv.2020.104371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ICMR COVID Study Group, COVID Epidemiology & Data Management Team, COVID Laboratory Team, VRDLN Team Laboratory surveillance for SARS-CoV-2 in India: performance of testing & descriptive epidemiology of detected COVID-19, January 22-April 30, 2020. Ind J Med Res. 2020;151(May (5)):424. doi: 10.4103/ijmr.IJMR_1896_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korth J., Wilde B., Dolff S., Anastasiou O.E., Krawczyk A., Jahn M., et al. SARS-CoV-2-specific antibody detection in healthcare workers in Germany with direct contact to COVID-19 patients. J Clin Virol. 2020 doi: 10.1016/j.jcv.2020.104437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N., Hameed S.K.S., Babu G.R., Venkataswamy M.M., Dinesh P., Bg P.K., et al. Descriptive epidemiology of SARS-CoV-2 infection in Karnataka state, South India: transmission dynamics of symptomatic vs. asymptomatic infections. EClinicalMedicine. 2021;32 doi: 10.1016/j.eclinm.2020.100717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Marca A., Capuzzo M., Paglia T., Roli L., Trenti T., Nelson S.M. Testing for SARS-CoV-2 (COVID-19): a systematic review and clinical guide to molecular and serological in-vitro diagnostic assays. Reprod Biomed Online. 2020:483–499. doi: 10.1016/j.rbmo.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxminarayan R., Wahl B., Dudala S.R., Gopal K., Neelima S., Reddy K.J., et al. Epidemiology and transmission dynamics of COVID-19 in two Indian states. Science. 2020;370(6517):691–697. doi: 10.1126/science.abd7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis F.I., Torgerson P.R. A tutorial in estimating the prevalence of disease in humans and animals in the absence of a gold standard diagnostic. Emerg Themes Epidemiol. 2012;9(1):1–8. doi: 10.1186/1742-7622-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malani A., Shah D., Kang G., Lobo G.N., Shastri J., Mohanan M., et al. Seroprevalence of SARS-CoV-2 in slums versus non-slums in Mumbai, India. Lancet Glob Health. 2021;9(2):e110–e111. doi: 10.1016/S2214-109X(20)30467-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerowitz-Katz G., Merone L. A systematic review and meta-analysis of published research data on COVID-19 infection-fatality rates. Int J Infect Dis. 2020:138–148. doi: 10.1016/j.ijid.2020.09.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ministry of Health and Family Welfare Services . 2020. #IndiaFightsCorona COVID-19 in India, Corona Virus Tracker. COVID-19 Dashbaord.https://www.mygov.in/covid-19 [Google Scholar]
- Murhekar M.V., Bhatnagar T., Selvaraju S., Rade K., Saravanakumar V., Thangaraj J.W.V., et al. Prevalence of SARS-CoV-2 infection in India: Findings from the national serosurvey, May-June 2020. Ind J Med Res. 2020;152(1):48. doi: 10.4103/ijmr.IJMR_3290_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh J.Y., Seo Y.B., Yoon J.G., Seong H., Hyun H., Lee J., et al. Seroprevalence of anti-SARS-CoV-2 antibodies among outpatients in southwestern Seoul, Korea. J Korean Med Sci. 2020;35(33) doi: 10.3346/jkms.2020.35.e311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi G., Paradisi M. An empirical estimate of the infection fatality rate of COVID-19 from the first Italian outbreak. medRxiv. 2020:1–18. [Google Scholar]
- Rogan W.J., Gladen B. Estimating prevalence from the results of a screening test. Am J Epidemiol. 1978;107(1):71–76. doi: 10.1093/oxfordjournals.aje.a112510. [DOI] [PubMed] [Google Scholar]
- Sapkal G., Shete-Aich A., Jain R., Yadav P.D., Sarkale P., Lakra R., et al. Development of indigenous IgG ELISA for the detection of anti-SARS-CoV-2 IgG. Ind J Med Res. 2020;151(5):444. doi: 10.4103/ijmr.IJMR_2232_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui S., Naushin S., Pradhan S., Misra A., Tyagi A., Looma M., et al. 2020. SARS-CoV-2 antibody seroprevalence and stability in a tertiary care hospital-setting. Available at SSRN 3696827. [Google Scholar]
- Statistics DoEa . Government of Karnataka; 2013. Projected population of Karnataka (2012-2021) p. 60. [Google Scholar]
- Stringhini S., Wisniak A., Piumatti G., Azman A.S., Lauer S.A., Baysson H., et al. Seroprevalence of anti-SARS-CoV-2 IgG antibodies in Geneva, Switzerland (SEROCoV-POP): a population-based study. Lancet. 2020:313–319. doi: 10.1016/S0140-6736(20)31304-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Lancet The plight of essential workers during the COVID-19 pandemic. Lancet (London, England) 2020;395(10237):1587. doi: 10.1016/S0140-6736(20)31200-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K.K.-W., Tsang O.T.-Y., Leung W.-S., Tam A.R., Wu T.-C., Lung D.C., et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . 2020. Solidarity II global serologic study for COVID-19. Accessed from: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/global-research-on-novel-coronavirus-2019-ncov/solidarity-2-global-serologic-study-for-covid-19. [Google Scholar]
- Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382(12):1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.