Bacterial plasmids are extrachromosomal genetic elements that often carry antimicrobial resistance (AMR) genes and genes encoding increased virulence and can be transmissible among bacteria by conjugation. One key group of plasmids is the incompatibility group I1 (IncI1) plasmids that have been isolated from multiple Enterobacteriaceae of food animal origin and clinically ill human patients.
KEYWORDS: incompatibility group I1 plasmids, plasmid replication, plasmid maintenance, plasmid transfer, virulence, antimicrobial resistance, plasmid biology, plasmid genetics, public health
SUMMARY
Bacterial plasmids are extrachromosomal genetic elements that often carry antimicrobial resistance (AMR) genes and genes encoding increased virulence and can be transmissible among bacteria by conjugation. One key group of plasmids is the incompatibility group I1 (IncI1) plasmids, which have been isolated from multiple Enterobacteriaceae of food animal origin and clinically ill human patients. The IncI group of plasmids were initially characterized due to their sensitivity to the filamentous bacteriophage If1. Two prototypical IncI1 plasmids, R64 and pColIb-P9, have been extensively studied, and the plasmids consist of unique regions associated with plasmid replication, plasmid stability/maintenance, transfer machinery apparatus, single-stranded DNA transfer, and antimicrobial resistance. IncI1 plasmids are somewhat unique in that they encode two types of sex pili, a thick, rigid pilus necessary for mating and a thin, flexible pilus that helps stabilize bacteria for plasmid transfer in liquid environments. A key public health concern with IncI1 plasmids is their ability to carry antimicrobial resistance genes, including those associated with critically important antimicrobials used to treat severe cases of enteric infections, including the third-generation cephalosporins. Because of the potential importance of these plasmids, this review focuses on the distribution of the plasmids, their phenotypic characteristics associated with antimicrobial resistance and virulence, and their replication, maintenance, and transfer.
INTRODUCTION
Bacterial plasmids are extrachromosomal genetic elements that are linear or circular DNA molecules that exist independently of the host chromosome in microbial cells and can replicate autonomously (1). Plasmids are seen most often in bacteria but have also been detected in archaea and eukaryotic organisms, where they are typically associated with the mitochondria (2). Plasmids have their own replication origin and can be stably inherited. Despite some similarities with chromosomal elements, plasmids do differ from bacterial chromosomes in several key characteristics (3, 4). Compared to bacterial chromosomes, plasmids typically contain fewer genes, are not essential for host survival, and most of the time have multiple copies in a cell (4). Many plasmids, including several that carry antimicrobial resistance (AMR) genes and genes encoding increased virulence, are transmissible by conjugation (5). While most infections caused by pathogens such as Salmonella enterica, Escherichia coli, and related organisms are self-limiting, some are more severe due to a variety of factors, including the infectious dose, route of inoculation, host immunity, and virulence characteristics of the infecting organisms (6). Severe manifestations of disease often require the use of antimicrobial agents to manage the infection. One set of challenges that has arisen is that many bacteria have developed resistance to antimicrobial agents used to control them.
Historically, plasmids were classified based on their compatibility for coexistence with one another in a single strain (7). With this typing approach, plasmids are assigned to different incompatibility (Inc) groups based on their incompatibility to coexist in the same cell (8–10). These incompatibility typing methods have been used to study the dissemination of plasmid-mediated antimicrobial resistance and the corresponding evolution of plasmids, with some of the more common examples among enteric bacteria being the IncF, IncI1, IncA/C, and IncX groups (11). Among these groups, the IncI1 plasmids are isolated from bacteria from human patients and food animals and are often associated with clinically relevant strains, although their host range appears to be relatively limited to a few enteric species, including E. coli and S. enterica (12). Several representative plasmids have been identified for their potential to carry and disseminate antimicrobial resistance among enteric pathogens (9, 13). Along with IncA/C plasmids, the IncI1 plasmids are the most common plasmid types associated with the dissemination of genes encoding resistance to extended-spectrum cephalosporins, which are the antimicrobial agents used in management of severe Salmonella infections (14).
IncI1 plasmids are characterized by the following distinguishing traits. They encode two types of sex pili, thin, flexible pili to aid in liquid matrix mating and thick, rigid pili needed for mating in both liquid and surface environments (15). In addition, IncI1 plasmids carry sog, which encodes a DNA primase that is vital for the establishment of plasmids following DNA transfer into recipient cells (16, 17). IncI1 plasmids can also carry genes responsible for antimicrobial resistance, attachment, and virulence and those that contribute to stable inheritance during cell division and plasmid maintenance (14, 18). Some IncI1 plasmids are bacteriocinogenic plasmids, which are capable of synthesizing bacteriocins, i.e., toxic compounds produced by host bacteria to antagonize other bacteria (19–21).
The IncI group of plasmids were initially characterized due to their sensitivity to the filamentous bacteriophage If1 (22). The receptors for the phage were determined to be on the tips of the thin flexible pili encoded on the IncI1 plasmid (23). There are two prototypical IncI1 plasmids that have been extensively studied, namely, R64 and pColIb-P9, that share functional similarity in plasmid replication, stability, and conjugal transfer mechanisms (24–26). More recently, isolated IncI1 plasmids have been characterized that share many of these characteristics; however, there are often differences in genes that encode antimicrobial resistance and/or increased virulence which are highlighted throughout the review (27, 28). The sequence of the R64 plasmid can be broken down into 5 different regions associated with replication, antimicrobial resistance, plasmid stability/maintenance, leading (first sequence transferred during conjugation to establish stable plasmid in the recipient), and transfer sequences (25). More detailed discussion of the regions will be presented in the following sections and are highlighted in Fig. 1. This review will focus on IncI1 plasmids, their phenotypic characteristics associated with antimicrobial resistance and virulence, and their transfer and genetics.
FIG 1.
Overview of the major regions that generally make up IncI1-type plasmids as described by Sampei et al. (25). These regions include the plasmid replication and control regions, variable regions encoding antimicrobial resistance and/or virulence-associated genes, genes associated with plasmid stability and partitioning, the leading region, which may play a role in conjugal transfer, and regions associated with conjugal transfer (25). The plasmid represented is pSH1148_107, GenBank accession number JN983049 (43).
DISTRIBUTION
With the enhanced capabilities of DNA sequencing, an increasing number of whole plasmid sequences are available to researchers. When GenBank was searched using microbial nucleotide BLAST by querying the reference replicon sequences for IncI1 plasmid types using a reference sequence (29) against the “complete plasmids” genome database, a total of 133 IncI1 complete plasmid sequences were identified. When the resultant plasmid GenBank information was extracted using FeatureExtract 1.2 program (Danish Technical University), a total of 16,636 IncI1 loci were cataloged. In an attempt to develop a nonredundant data set, extracted duplicate sequences were removed in Excel and then manually reviewed to generate a draft nonredundant data set, which contained approximately 1,400 unique IncI1 coding sequences, including variants of genes. Half of these sequences were identified as encoding “hypothetical proteins,” while other genes were predicted to be associated with antimicrobial resistance, biocide/heavy metal resistance, virulence, horizontal gene transfer elements, and plasmid transfer (Table 1). IncI1 plasmid sequences were detected in isolates from the following bacterial species in GenBank: Escherichia albertii, E. coli, Klebsiella pneumoniae, Salmonella enterica, Shigella dysenteriae, Shigella flexneri, and Shigella sonnei, and thus the host range appears to be limited to the Enterobacteriaceae.
TABLE 1.
Summary of gene products associated with antimicrobial resistance, metal/biocide resistance, virulence, partition/maintenance, conjugal transfer, and gene transfer detected in fully sequenced IncI1 plasmids
| Function | Product |
|---|---|
| Antimicrobial resistance | Aminoglycoside N-acetyltransferase AAC(3)-I |
| Aminoglycoside N-acetyltransferase AAC(3)-II | |
| Aminoglycoside acetyltransferase AacC | |
| Aminoglycoside adenyltransferase AadA1 | |
| Aminoglycoside adenyltransferase AadA2 | |
| Aminoglycoside 3′-phosphotransferase APH(3′)-I | |
| Aminoglycoside N-acetyltransferase III ACC(3)-III | |
| Aminoglycoside N-acetyltransferase AAC(3)-VIa | |
| Aminoglycoside O-phosphotransferase APH(3′)-II | |
| Aminoglycoside O-phosphotransferase APH(6)-Ic | |
| Hygromycin resistance protein Hrp | |
| Streptomycin phosphotransferase protein StrA | |
| Streptomycin phosphotransferase protein StrB | |
| Chloramphenicol efflux MFS transporter CmlA | |
| Chloramphenicol/florfenicol efflux MFS transporter FloR | |
| Extended-spectrum beta-lactamase SHV-12 | |
| Extended-spectrum beta-lactamase CTX-M-1 | |
| Extended-spectrum beta-lactamase CTX-M-3 | |
| Extended-spectrum beta-lactamase CTX-M-8 | |
| Extended-spectrum beta-lactamase CTX-M-14 | |
| Extended-spectrum beta-lactamase CTX-M-15 | |
| Extended-spectrum beta-lactamase CTX-M-55 | |
| Extended-spectrum beta-lactamase CTX-M-123 | |
| Class C beta-lactamase CMY-2 | |
| Class C beta-lactamase CMY-4 | |
| Class C beta-lactamase CMY-42 | |
| Class C beta-lactamase CMY-111 | |
| Beta-lactamase, TEM-1 | |
| Beta-lactamase, TEM-20 | |
| Beta-lactamase, TEM-52 | |
| Beta-lactamase, TEM-57 | |
| Beta-lactamase, TEM-210 | |
| Oxacillin-hydrolyzing class D beta-lactamase OXA-2 | |
| Dihydrofolate reductase DhfrA1 | |
| Dihydrofolate reductase DfrA17 | |
| Dihydropteroate synthase type-1 Sul1 | |
| Dihydropteroate synthase type-2 Sul2 | |
| Fosfomycin resistance glutathione transferase FosA3 | |
| Macrolide ABC transporter permease/ATP-binding protein MacB | |
| Macrolide 2′-phosphotransferase MphB | |
| Tetracycline resistance MFS efflux pump TetA | |
| Tetracycline efflux MFS transporter TetC | |
| Tetracycline resistance ribosomal protection protein TetM | |
| Tetracycline resistance transcriptional regulator TetR | |
| Metal/biocide resistance | Arsenical pump-driving ATPase ArsA |
| Arsenical efflux pump membrane protein ArsB | |
| Arsenate reductase ArsC | |
| Arsenical resistance operon transcriptional repressor ArsD | |
| Arsenical resistance operon transcriptional regulator ArsR | |
| Mercuric ion reductase MerA | |
| Organomercurial lyase MerB | |
| Mercuric transport protein MerC | |
| Mercuric resistance operon coregulator MerD | |
| Mercuric transporter protein MerE | |
| Mercuric transcriptional regulator MerR | |
| Quaternary ammonium compound efflux SMR transporter QacE | |
| Quaternary ammonium compound resistance protein QacH | |
| Quaternary ammonium compound resistance protein SugE | |
| Silver- or copper-binding protein SilE | |
| Virulence | Colicin 1B Cib |
| Colicin 1B immunity protein Cbi | |
| Colicin Ia immunity protein Cia | |
| Colicin Ia protein Cai | |
| Colicin M activity protein Cma | |
| Colicin M immunity protein Cmi | |
| CS1 fimbrial subunit A CfaA | |
| CS1 fimbrial subunit B CfaB | |
| Plasmid-encoded fimbriae Pef | |
| Partition/maintenance | Stable plasmid inheritance protein A ParA |
| Stable plasmid inheritance protein B ParB | |
| Plasmid maintenance protein CcdA | |
| Plasmid maintenance protein CcdB | |
| Plasmid maintenance protein VagD | |
| Plasmid maintenance protein VagC | |
| Plasmid maintenance protein PndA | |
| Plasmid maintenance protein PndB | |
| Plasmid maintenance protein PndC | |
| Plasmid maintenance protein RelB | |
| Plasmid maintenance protein RelE | |
| Conjugal transfer | Conjugal transfer protein TraA |
| Conjugal transfer transcription antiterminator TraB | |
| Conjugal transfer protein TraC | |
| Conjugal transfer system coupling protein TraD | |
| Conjugal transfer protein PilI | |
| Conjugal transfer protein PilJ | |
| Conjugal transfer protein PilK | |
| Conjugal transfer outer membrane protein PilL | |
| Conjugal transfer protein PilM | |
| Conjugal transfer protein PilN | |
| Conjugal transfer protein PilO | |
| Conjugal transfer pilus biogenesis protein PilP | |
| Conjugal transfer protein PilQ | |
| Conjugal transfer pilus biogenesis protein PilR | |
| Conjugal transfer pilus biogenesis protein PilS | |
| Conjugal transfer lytic transglycosylase PilT | |
| Conjugal transfer peptidase PilU | |
| Conjugal transfer pilus-tip adhesin protein PilV | |
| Conjugal transfer pilus assembly protein PilX | |
| Conjugal transfer pilus assembly protein TraE | |
| Conjugal transfer protein TraF | |
| Conjugal transfer protein TraG | |
| Conjugal transfer pilus assembly protein TraH | |
| Conjugal transfer lipoprotein TraI | |
| Conjugal transfer protein TraJ | |
| Conjugal transfer protein TraK | |
| Conjugal transfer pilus assembly protein TraL | |
| Conjugal transfer protein TraM | |
| Conjugal transfer protein TraN | |
| Conjugal transfer protein TraO | |
| Conjugal transfer protein TraP | |
| Conjugal transfer protein TraQ | |
| Conjugal transfer protein TraR | |
| Conjugal transfer protein TraS | |
| Conjugal transfer protein TraT | |
| Conjugal transfer pilus assembly protein TraU | |
| Conjugal transfer protein TraV | |
| Conjugal transfer pilus assembly protein TraW | |
| Conjugal transfer pilus acetylation protein TraX | |
| Conjugal transfer integral membrane protein TraY | |
| Conjugal transfer protein TrbA | |
| Conjugal transfer protein TrbB | |
| Conjugal transfer protein TrbC | |
| Conjugal transfer oriT-specific DNA-binding protein NikA | |
| Conjugal transfer relaxase protein NikB | |
| Conjugal transfer protein FinQ | |
| Gene transfer | Insertion element IS1 protein InsA |
| Insertion element IS1 protein InsB | |
| IS629 element | |
| IS100 element | |
| IS4321 element | |
| IS200 element | |
| IS256 element | |
| IS26 element | |
| IS3 element | |
| IS4 element | |
| IS481 element | |
| ISEcp1 element | |
| IS1294 element | |
| IS5 element | |
| IS6 element | |
| IS630 element | |
| IS66 element | |
| IS91 element | |
| ISKra4 element | |
| ISL3 element | |
| Class 1 integron integrase IntI1 | |
| Tn21 protein Urf2 | |
| Tn3 transposase |
A factor in the persistence of IncI1 plasmids in the Enterobacteriaceae is that they contribute traits such as antimicrobial resistance and colicin production yet appear to convey minimal metabolic burden on the host strains. Johnson et al. demonstrated that the acquisition of an IncI1 plasmid did not significantly affect the fitness of the host bacterium, and in some cases the fitness cost associated with the acquisition of an IncI1 plasmid was negative (i.e., beneficial) (30). For example, in E. coli strains carrying IncA/C plasmids, the fitness cost of the acquisition of an IncI1 was no greater than that of carrying the IncA/C plasmid alone or, in some cases, was a negative cost (30). Similarly, Kaldhone et al. found that many IncI1-positive Salmonella isolates with the greatest ability to multiply and persist in intestinal epithelial cells carried additional large plasmids, including IncA/C, IncHI2, IncFIB, and IncB/O, which in theory should have high metabolic costs (31). Freire Martín et al. cured an IncI1 plasmid to evaluate mechanical burden of carrying the plasmid and determined that there was not a burden associated with IncI1 plasmids in S. enterica serotype 4,5,12:i:- (32). Conversely, an IncI1 CTX-M1 plasmid imparted a growth disadvantage upon K. pneumoniae (33). Potential reasons for this disparity in findings across studies are that the K. pneumoniae plasmid may have been recently acquired and not gone through coevolution far enough to compensate for the growth and fitness costs or that there were differences in the plasmids that led to variable costs, since the IncI1 plasmid multilocus sequence typing (pMLST) classifications of the plasmids in K. pneumoniae and S. enterica 4,5,12:i:- were different (32).
When examined from a host source range perspective, IncI1 plasmids have been isolated from environmental sources and several different animal species, including cats, cattle, chickens, dog, fish, goats, horses, rabbits, sheep, swine, and turkeys (31, 34–36). When factors such as the host, geographical origin of isolation of the IncI1 plasmids present in GenBank (described above), and the literature were assessed, isolates were found to originate from South America, North America, Europe, Asia, Australia, and Africa (28, 37–42). The sequenced strains were isolated from as far back as 1969 (GenBank accession: NZ_CP018638) and came from a wide range of animal species (as noted above), retail foods (retail beef, chicken, pork, and turkey products), and forest soil (GenBank accession: NZ_CP010233). IncI1 plasmids have been detected in isolates collected from several human patients. Among the sequenced plasmids present in GenBank, E. coli, S. enterica, S. sonnei, and K. pneumoniae were isolated from a variety of specimens, including stool, blood, urine, wound sites, and peritoneal fluid; these findings correspond well to the published literature (14, 21, 38, 43–45). IncI1 plasmids have also been isolated from the stools of mothers during pregnancy and lead to subsequent transmission to their newborns (46, 47).
PLASMID REPLICATION
The process of plasmid replication and partitioning during bacterial cell division is a multistep process which is often encoded by a rep operon (48, 49). Because replication of plasmids can be metabolically costly to bacteria, there are regulatory mechanisms that limit the copy numbers of plasmids in strains to allow enough copies to ensure that daughter cells retain plasmids and avoid postsegregation killing yet not too many copies to be overly metabolically taxing (49, 50). The IncI1 plasmids are low copy number plasmids, whose replication is tightly controlled by negative regulation of replication initiation (51).
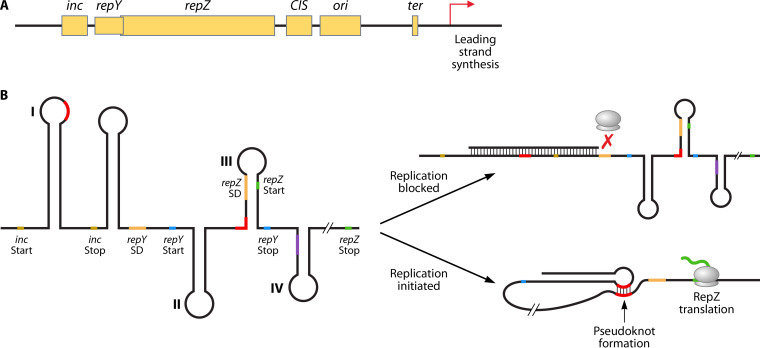
The control of IncI1 plasmid replication is likely best studied in the plasmid ColIb-P9 (52, 53). ColIb-P9 and other IncI1 plasmids, such as R64, have a 3-kb replication control region that encodes the initiation, control, and termination of unidirectional replication of the IncI1 plasmids (upper left portion of Fig. 2) (25, 51). This replication control region is generally conserved across sequenced IncI1 plasmids (31, 45, 54). The main replication initiation protein is the 39-kDa RepZ protein. RepZ interacts with the origin of replication (ori) which is near repZ to initiate replication of the plasmid sequence. Termination of plasmid replication occurs at CIS, which is located between repZ and ori (Fig. 3A). Control of repZ expression and translation, and subsequently control of plasmid replication and copy number, is associated with the negative regulator inc and positive regulator repY (55). The inc gene encodes an antisense RNA of approximately 70 bp in length, which contains nucleotides (5′-rGCCA-3′) that bind to the conserved 5′-rUUGGCG-3′ motif in the “structure I” stem-loop configuration of the Rep mRNA repressing translation of RepZ (56) (Fig. 3B). The interaction of the Inc RNA and Rep mRNA prevents the formation of a pseudoknot structure that leads to RepZ translation (57).
FIG 2.
Genetic map of IncI1 plasmid pSH1148_107, GenBank accession number JN983049 (43). The gene names of known genes are included along with color coding of the predicted functions of each of the genes (25).
FIG 3.
(A) Diagram of the replication control region for IncI1 plasmids. RepZ is the main replication initiation protein and interacts with the origin of replication (ori), which is near repZ, to initiate replication of the plasmid sequence. Termination of plasmid replication occurs at CIS, which is located between repZ and ori (57). (B) Predicted RNA structure of the replication control (Rep) region of the IncI1 plasmid and predicted mechanisms of replication control. Control of repZ translation, and subsequently control of plasmid replication and copy number, is associated with the negative regulator inc and the positive regulator repY. To control replication, inc mRNA binds to the inc sequence and blocks the ribosomal binding site to inhibit RepY translation. To activate replication, inc mRNA is unbound from inc, allowing translation of RepY, which facilitates pseudoknot formation (binding of structure I to structure III at the binding sites indicated in red) that opens the ribosomal binding site to facilitate RepZ expression (based on data from reference 55).
For RepZ mRNA to be translated, RepY, a short protein of 29 amino acids, must first be translated. The 3′ of repY overlaps the 5′ repZ and is located adjacent to the repZ start codon. RepY expression is also under the control of inc, such that the binding of the Inc RNA to structure I causes steric hinderance of the ribosomal binding site (RBS) for translation of repY (55) (Fig. 3B). Translation of repY leads to the formation of the activated pseudoknot, which is initiated during the termination step of repY translation, opening access to the repZ RBS and facilitating base pairing between nucleotides in structure III and structure I of the RepZ mRNA (55) (Fig. 3B). The pseudoknot structure allows for translation of RepZ that is needed for plasmid replication. Following initiation of RepZ translation, the Inc RNA rapidly binds to structure I of the RepZ mRNA, inhibiting further translation of RepZ, thus keeping the plasmid replication in check (55).
HOST ADDICTION SYSTEMS
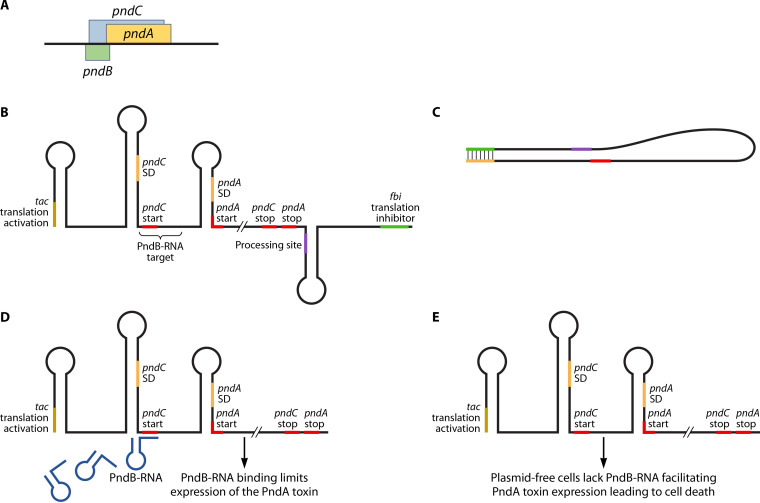
Many plasmids encode host addiction systems consisting of long-acting toxins and shorter-acting antitoxins, which, if the plasmids were lost (cured), would be lethal to the bacterium through a process known as postsegregation killing (58, 59). The following toxin/antitoxin (TA) systems have been identified in IncI1 plasmids: ccdAB, relBE, and pndBCA (59, 60). PndCA is part of the Hok/Sok TA family, in which there is the stable mRNA encoding a toxin (e.g., Hok) and the more unstable antisense RNA (e.g., Sok) that limits toxin translation (61). Pnd is named due to its promotion of nucleic acid degradation (62). The pnd TA genes are located within the transfer region of plasmid R64 and pSH1148_107 (43, 63) (Fig. 2). pndC overlaps pndA in the plasmid sequence, while pndB is located on the opposite DNA strand (Fig. 4). Studies indicate that the presence of the Pnd toxin leads to degradation of RNA following the addition of the antibiotic rifampin to pnd-positive E. coli (64, 65). Rifampin is a drug that targets and inactivates DNA-dependent bacterial RNA polymerase (66). This inhibition of RNA synthesis can lead to a reduction of the pndB transcription and subsequent degradation of the inhibitory RNA molecules. This PndB RNA degradation subsequently leads to an increase in PndCA translation (65, 67). PndA serves as a toxin that functions by damaging the bacterial cell membrane, while PndC acts by promoting the translation of pndA. The pndCA mRNA is very stable in bacteria; however, its levels are negatively regulated through an unstable complementary pndB-encoded RNA molecule that prevents the toxin translation (61, 63). In cells that lose plasmids carrying the TA system, the unstable RNA degrades, allowing for translation of the residual pndCA mRNA, thereby killing the plasmid-free host cells (63).
FIG 4.
Diagram of the Pnd toxin-antitoxin system. The system can encode the PndA toxin that causes lethal damage to the bacterial cell membrane when expressed. (A) The Pnd operon is made up of pndC that overlaps pndA in the plasmid sequence, and pndB is located on the opposite DNA strand. pndC modulates pndA expression, and pndB encodes an RNA molecule that suppresses expression of pndA and toxin formation. (B) Transcription of the pnd operon leads to the formation of a complex mRNA molecule whose translation is regulated by multiple mechanisms. The 5′ end of the inactive RNA molecule contains a translational activation element (tac), and the 3′ end contains a fold-back inhibition element (fbi). Between these elements are the nucleotides for the translation of PndC and PndA and a processing site for the formation of the functional mRNA molecules. (C) The fbi and tac sites are complementary to one another and bind to prevent translation of the unprocessed RNA molecule. (D) Following processing (cleavage at the processing site and removal of the fbi element), the regulation of the translation of the Pnd toxin in the activated mRNA is due to the binding of the small, very short-lived PndB RNA molecules that bind to the more stable PndCA mRNA overlapping the translation start site for PndC. (E) In cases where the plasmids are lost, all of the short-lived PndB are degraded, allowing the translation of the long-lived PndA toxin leading to cell membrane damage and cell death.
The regulation of the expression of the Pnd toxin is due to the binding of the small pndB-encoded RNA molecule that binds to the more stable PndCA mRNA (68). The antisense PndB RNA is complementary to both the translation initiation sequence of pndC and the leader region of PndA mRNA, thus regulating PndA translation (61, 69). Based on the model Hok/Sok TA system, the translation of pndC would be tightly controlled by the antisense PndB RNA (Fig. 4). The expression of pndA is coupled to the expression of pndC; thus, the regulation of PndA levels by pndB likely occurs indirectly though pndC (69). For translation of PndA to occur, the PndA mRNA is cleaved at its 3′ end, which converts the inactive form of the mRNA to the form that is translated. The presence of the PndB RNA binding likely limits this cleavage and keeps the translation in check when present at adequate levels. In strains where the plasmids are lost, the antisense RNA is degraded and the expression of the PndA is able to be initiated from the more stable mRNA molecules, leading to cell death of the plasmid cured bacteria (61).
CONJUGAL TRANSFER
Bacterial conjugation is the transfer of genetic material between bacterial cells by direct cell-to-cell contact or by a bridge-like connection (pilus) between two cells. Conjugation facilitates the horizontal transfer of different genes among different bacteria (70) and potentially allows for the rapid adaptation and evolution observed among bacteria to respond to varied stresses, such as antibiotic exposure, and an increase in their ability to colonize hosts and cause disease (71). The ability of IncI1 plasmids to disseminate among enteric bacteria has been well studied to develop an understanding of their plasmid conjugation strategies. One of the best-studied conjugative transfer systems is that of R64, which, like many other IncI1 plasmids, is a relatively complex system compared to most other plasmid types (15). The transfer region of IncI1 plasmids is larger than those of most other plasmid types at approximately 54 kb in size (Fig. 1); in comparison, the transfer regions of most IncF plasmids are generally around 33 kb (72). The large IncI1 transfer region typically contains 48 open reading frames (ORFs) (Fig. 2), which encode both a thick, rigid conjugal pilus and a thin, flexible pilus (15).
Much of the early work elucidating the mechanisms of IncI1-associated conjugation mechanisms originated from studies by Komano and colleagues in Japan (for examples see references 15, 25, and 73–76). The transfer region typically consists of genes for the thick conjugal pilus encoded by the tra/trb genes and a thin flexible pilus encoded by pil genes that plays a key role in facilitating conjugation in liquid environments (15, 77) (Fig. 2). Based on the analyses by Sampei et al. (25), the transfer region of the prototypical IncI1 plasmid R64 could be separated into four major functional groupings, including those with regulatory functions (traABCD), the relaxation complex for conjugation initiation (oriT and nikAB), the type IV pilus (T4P) formation (pil gene cluster), and the tra/trb general conjugal apparatus. The IncI1 Tra and Trb proteins share amino acid sequence similarity with the Dot/Icm virulence plasmids from Legionella pneumophila and the tumor-inducing (Ti) VirB/D4 T4SS of Agrobacterium tumefaciens, rather than with the more widely studied IncF plasmids (76, 78, 79). Likewise, the IncI1 T4P is ancestrally related to the toxin co-regulated pilus (TCP) from Vibrio cholerae and the bundle-forming pilus (BFP) of enteropathogenic E. coli (EPEC) and is classified as a type IVb pilus (T4bP) based on its physical structures (80–83).
In the sequence of R64 and similar plasmids, the transfer region is adjacent to the replication initiation sequence, with the traABCD just upstream of the inc replication regulatory sequence and downstream of the pil operon (25). Both traB and traC encode proteins that are essential for conjugative transfer of IncI1 plasmids in both liquid and solid media (73). TraB is homologous to NusG, a protein that interacts with RNA polymerase, increases the rate of transcription, and affects transcription termination (84, 85). TraC is also predicted to be a positive regulator for expression of transfer-associated genes (73). TraA is predicted to have a helix-turn-helix domain that is characteristic of a DNA-binding domain and may affect the regulation of transcription (86). In the IncI1 plasmid pESBL-EA11, when the region adjacent to traA was disrupted by a transposon, it led to an elevated (>10-fold) transfer efficiency; thus, this region has been termed the high frequency of transfer (Hft) region (87, 88). This disruption of the Hft region led to overexpression of TraA, which led to the observed increased rate of the conjugal transfer. The expression of TraA likely facilitates the activation of TraBC in pESBL-EA11, which had previously been reported to be key to the transcription of the downstream transfer-associated genes (87). The function of TraD is currently unknown, and several sequenced IncI1 plasmids appear to lack the traD gene annotated in R64 (25, 84, 88).
The tra and trb genes encode the thick, rigid conjugal pilus for both solid and liquid media mating (76). As noted above, the tra and trb genes have been sequenced and their proteins possess predicted structural similarities to the Dot/Icm and Ti T4SS (Table 2 and Fig. 5). Early studies mapping the plasmid gene function in R64 indicated that the traEFG genes likely form an operon that is not essential for conjugation to occur (74). TraF and TraG do not appear to have protein homologs within the Dot/Icm and Ti T4SS (76, 78, 79, 89–92). The functions of these gene products are not well understood; TraG likely functions as a histidine phosphatase (76), and more recent studies have indicated that the expression of traF is significantly upregulated in E. coli strains carrying plasmid pTF2. These strains demonstrated increased conjugation rates following exposure to cephalosporin antibiotics (93). TraE is predicted to be homologous to the VirE1 of the Ti T4SS (76). VirE1 functions in a chaperone-like fashion to facilitate the export of VirE2, a nonspecific, single-stranded DNA-binding protein that was shown to transfer tumor DNA from A. tumefaciens to plant cells (94, 95).
TABLE 2.
Genes in the IncI1 type 4 secretion system (T4SS) and homologs of the proteins from other better-characterized T4SS
| IncI1 gene | Predicted function/product | Protein homolog(s)a | Reference(s) |
|---|---|---|---|
| trbC | Type 4 coupling protein | DotL (VirD4) | 76, 103 |
| trbB | Protein disulfide isomerase | TrbC(F) | 25, 91 |
| trbA | T4SS coupling complex protein | DotM | 76, 103 |
| traY | Integral membrane protein | DotA | 76, 79 |
| traX | Inner membrane protein | 25, 76 | |
| traW | Outer membrane lipoprotein | 25, 76 | |
| traV | Cytoplasmic transfer protein | 25, 76 | |
| traU | ATPase, nucleotide binding protein | IcmB/DotO (VirB4) | 76, 79, 91 |
| traT | T4SS coupling complex protein | IcmJ/DotN | 102, 103 |
| traS | Cytoplasmic transfer protein | 25, 76 | |
| traR | Inner membrane protein | IcmD/DotP, IcmC/DotE, DotV | 102 |
| traQ | Inner membrane protein | IcmD/DotP, IcmC/DotE, DotV | 25, 102 |
| traP | Inner membrane protein-T4SS core complex | DotF (VirB3) | 25, 79 |
| traO | Inner membrane protein-T4SS core complex | DotG (VirB10) | 76, 79, 91 |
| traN | Outer membrane lipoprotein-T4SS core complex | DotH (VirB9) | 76, 91 |
| traM | Inner membrane protein-T4SS core complex | DotI (VirB8) | 76, 78, 79, 89 |
| traL | Outer membrane/periplasmic protein-signal peptide | 25, 76 | |
| traK | Inner membrane protein | IcmT | 76 |
| traJ | Outer membrane lipoprotein-T4SS core complex | DotB (VirB11) | 76, 101 |
| traI | Outer membrane lipoprotein-T4SS core complex | DotC (VirB7) | 76, 92 |
| traH | Outer membrane lipoprotein | DotD | 76, 102 |
| traG | Outer membrane/periplasmic protein-signal peptide | 25, 76 | |
| traF | Outer membrane/periplasmic protein-signal peptide | 25, 76 | |
| traE | Cytoplasmic transfer protein | (VirE1) | 25, 76 |
Proteins from the Dot/ICM T4SS and the VirB/D4 Ti T4SS (in parentheses).
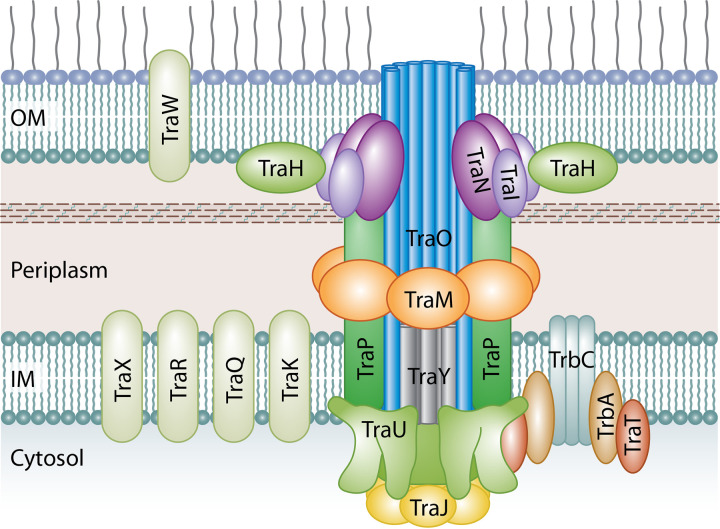
FIG 5.
Predicted structure of the Tra/Trb T4SS of IncI1 plasmids based on homologs from the better-characterized Dot/Icm and Ti T4SSs. The inner membrane portion of the T4SS is predicted to be made up of TraJ, TraM, TraO, TraP, TraU, and TraY proteins. TraP forms a multimer with TraO and TraY through the inner membrane and into the periplasmic space where the complex appears to interact with the outer membrane complex. TraU has homology to DotO of the Dot/Icm T4SS, which forms a hexamer that sits at the base of the main pore channel of the secretion system where it interacts with the TraJ multimer. TraJ homologs are predicted to form hexamers that are on the cytoplasmic side of the secretion system and form a transitory complex with the TraU multimer in line with the core complex. TrbC is a T4CP that forms a complex with TrbA and TraT and functions to help deliver macromolecules from the cytoplasm to the T4SS section apparatus to traverse the cell membranes. The outer membrane complex made up of 13 subunits comprised of TraI, TraH, and TraN surrounds the TraO secretion channel consisting of 18 subunits. OM, outer membrane; IM, inner membrane.
The gene products of traH through traY likely form a distinct operon from that of traEFG (76). When Komano et al. (76) disrupted the various genes in the tra region, they found that traMNPQRUVWY were essential for conjugal transfer of R64, whereas disruption of the traIJKLOTX genes significantly affected plasmid transfer efficiency but did not completely eliminate it (76). The predicted homologs for several of the genes are shown in Table 2; many of the genes are associated with the formation of the pilus structure (Fig. 5). Even though there are overlapping homologies with many of the proteins in the more extensively studied Dot/Icm and VirB/D4 T4SSs, there are some protein homologs that are not apparent in the IncI1 Tra/Trb T4SS, which may affect extrapolation of the IncI1 T4SS structure. The inner membrane portion of the T4SS is predicted to be made up of TraJ, TraM, TraO, TraP, TraU, and TraY proteins, which are homologous to VirB11 (DotB), VirB8 (DotI), VirB10 (DotG), VirB3 (DotF), VirB4 (DotO), and DotA, respectively (76, 78, 79, 96, 97). The IncI1 T4SS lacks a homologous protein to DotJ of Legionella Dot/Icm T4SS membrane complex (92). DotJ forms a heterooctomer complex with DotI, while TraM, the DotI homolog, forms a homohexamer, which is similar to the VirB8 structure of the Agrobacterium Ti T4SS (92). Based on homology to DotG, TraO is predicted to form an 18-mer that serves as a periplasmic channel for the secretion system (98). The TraO multimer interacts with an outer membrane complex that is made up of 13 subunits comprised of TraI, TraH, and TraN (98–100) (Fig. 5). The TraO multimer appears to extend from the inner membrane complex to, or through, the outer membrane complex, forming the channel for the plasmid DNA to be transferred out of the cytoplasm during conjugation (101). Interestingly, inactivation of traH, nuc, and traS did not inhibit conjugation in R64, and thus they may not be required for conjugation (76). The TraR and TraQ proteins were identified as having indirect homology with VirB2, which forms a multiprotein complex serving as the pilus extending from the bacterial cells of the VirB/D4 T4SSs (79). In the Dot/Icm T4SS, there does not appear to be an analogous protein polymer to VirB2 (78, 99, 101). Because of the lack of direct homology to VirB2 and the lack of a homolog in the Dot/Icm T4SS, it is not clear whether or not TraR and/or TraQ forms the pilus appendage. Other investigators have predicted that TraR and TraQ are likely inner membrane proteins (76, 102). TraP forms a multimer with TraO through the inner membrane and into the periplasmic space where the complex appears to interact with the TraM, TraI, and TraH multimer (99, 101). TraY was shown to encode transmembrane helices that are homologous to those of DotA of the Dot/Icm T4SS (76, 102) and analogous to VirB6 of the VirB/D4 T4SS (79). These proteins have been predicted to be part of the inner membrane complex and interact with the homologs of TraO and TraP in the secretion system (100). DotA, the TraY homolog, was also shown to be essential for Legionella pathogenicity (78, 102, 103).
Several other tra genes encode proteins that are predicted to be associated with the inner membrane based on their homology to the Dot/Icm T4SS; these include TraK, TraQ, TraR, and TraX (76, 79, 102). TraK is predicted to be an ortholog of IcmT, an integral membrane protein that was shown to be essential for the T4SS function; however, its specific role in the process is currently not known (102). One of the membrane proteins for which little is known about its function is TraW, an outer membrane lipoprotein that has been shown to be essential for conjugal transfer in R64 (76).
T4SSs are characterized by the presence of ATPases that energize the secretion functions. In the Ti T4SS, these ATPases are VirB4, VirB11, and VirD4, while in the Dot/Icm T4SS the corresponding proteins are DotO (IcmB), DotB, and DotL, respectively (104). In R64 and most other IncI1 plasmids, the corresponding ATPases are TraU, TraJ, and TrbC, respectively (76, 79, 102). Each of these ATPases is predicted to form hexamers that help drive the molecules being secreted across the T4SS (79). TrbC is a type 4 coupling protein (T4CP) that forms a complex with TrbA and TraT, which are DotM and DotN (IcmJ) homologs, respectively (76, 103, 105, 106). A predicted structure of the T4CP based on recent findings for the Dot/ICM system shows that the TrbC hexamer forms a central channel and each monomer interacts with a TrbA subunit that is bound to a TraT monomer (105, 106) (Fig. 5). The Dot/ICM system has additional cytoplasmic components (IcmS, IcmW, and LygA) likely involved in substrate recognition that appear to be absent in the IncI1 T4CP complex (106). T4CP complexes function by helping deliver macromolecules from the bacterial cytoplasm to the T4SS section apparatus to traverse the cell membranes (100–103). TraU has homology to DotO (IcmB) and VirB4 and plays key roles in type 4 secretion (70, 102). The related proteins, such as DotO, form hexamers that sit at the base of the main pore channel of the secretion system (Fig. 5) and likely lead to conformation changes in the T4SS core complex facilitating transfer of macromolecules across the cell membranes (96, 97, 107). TraJ homologs, such as DotB, are predicted to form hexamers that are on the cytoplasmic side of the secretion system and form a transitory complex with the DotO (TraU) in line with the core complex (96, 97). In this model, the TraJ hexamer docks with the TraU complex to facilitate the transfer of macromolecules from the T4CPs on the cytoplasm to the base of the T4SS channel, where the TraJ hexamer facilitates the loading of the macromolecules into the core complex for secretion across the membranes (107, 108).
Upstream of traY are excAB and pndCA, which encode surface exclusion and host addiction, respectively (25, 109). Adjacent to pndCA is the trbABC operon, where the trbA and trbC genes were found to be essential for conjugal transfer of IncI1 plasmids (63). As noted above, TrbC and TrbA are key elements of the T4CP complex that likely plays a key role in the delivery of macromolecules to the T4SS (78, 89). trbB deletion mutants in R64, while significantly less efficient in conjugal transfer, maintain the ability to transfer plasmids (63); TrbB is a homolog to the IncF pilus protein TrbCF, where TrbCF resides in the periplasm and appears to function in pilus stabilization and pore formation (91).
During the conjugal transfer of DNA across the pili, the processing of plasmid DNA transfer is initiated through a relaxation complex or relaxosome encoded by an operon that includes nikA and nikB, along with oriT (110, 111) (Fig. 2). The NikA and NikB proteins of the relaxosome function as a nickase that recognizes a short DNA motif (5′-YATCCTG*Y-3′) in oriT where the double-stranded DNA is nicked (* marks the nick site) (111). The nicked DNA strand is transferred to the recipient cell during conjugation. oriT also contains two inverted repeats, one that is 8 bases and another that is 17 bases. The 17-bp inverted repeats have a single nucleotide difference between the repeat sequences. To form the active relaxosome, NikA binds specifically to one of the 17-bp repeat sequences, leading to a change in the bending of the oriT DNA (112). The three-dimensional structure of NikA is similar to that of known transcriptional repressors and when bound to oriT interacts with the relaxase protein NikB to form the active relaxosome. The bending of the oriT DNA orients the nick site and NikB, which in turn introduces a nick in oriT to initiate transfer (111, 112). Following nicking, the single-stranded DNA molecule is transferred along with NikB, which remains attached to the single-stranded DNA (ssDNA), into the recipient cells. The second 8-bp inverted repeat in oriT plays a role in termination of DNA transfer into the recipient cell, after which the transferred ssDNA molecules are recircularized and a complementary strand synthesized (25). The sog gene, located between traL and nuc in the tra region (Fig. 2), encodes two proteins, SogS and SogL. SogL functions both as a primase to help initiate synthesis of the complementary strand of the newly transferred plasmid and for suppression of E. coli dnaG mutations which can negatively affect DNA synthesis and, subsequently, conjugation (16, 113, 114). Both SogL and SogS are transferred into the recipient cell as part of the conjugal transfer of ColIb-P9; however, SogS lacks the primase activity of SogL, and its function is less well understood (16, 113).
An operon encoding the T4P is located just upstream of traD in R64 (traC in pESBL-EA11 and R621a) and is made up of 14 genes (pilI to pilV) (115). The T4P is a thin, flexible pilus that early studies indicated was required for conjugal transfer of R64 and ColIb-P9 in liquid media but not on solid media (116). More recent studies have indicated that this observation is not universal among IncI1 plasmids, in that pilRSTUV are also required for transfer of pESBL-EA11 on solid media (88). Of the 14 pil genes, 12 appear to be required for the formation of the conjugal pilus (pilK to pilV) in R64 and ColIb-P9, while the functions of pilI and pilJ remain unknown (15). The T4P encoded on the pSERB1 plasmid of enteroaggregative E. coli was also shown to contribute to conjugal transfer of the IncI1 plasmid and to aid adherence of the bacteria to epithelial cells and surfaces, facilitating biofilm formation (77). Similar contributions of T4P to biofilm formation have also been noted in several other species (77, 83). The function of the T4P in conjugation in liquid media appears to be stabilization of the mating bacteria through the formation of aggregates of the donor and recipient strains (116).
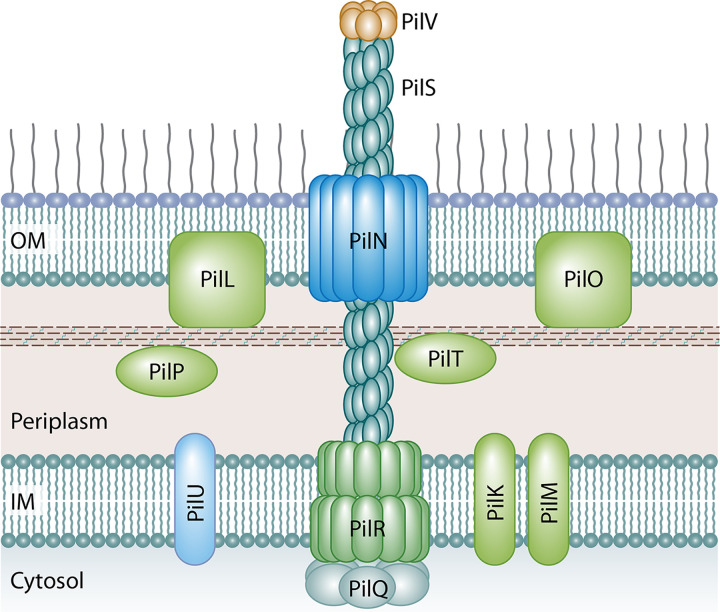
The pil genes that encode the T4P structure extending from the surface of the bacterial cell are pilS, which encodes the major prepilin, and pilV, which encodes the minor prepilin. Other structural genes include pilR, which encodes the inner membrane spanning protein, and pilN, which encodes the outer membrane secretin through which the PilS polymer extends (76, 84, 115) (Fig. 6). The PilS prepilin is initially translated as a 22-kDa precursor that is cleaved by the PilU prepilin peptidase to form a 19-kDa monomer that is assembled into a polymer that is secreted to form the extending T4P structure (117) (Fig. 6). At the terminal tip of the pilus is the PilV adhesin, which has been shown to interact with lipopolysaccharide (LPS) on a recipient cell. Through these interactions, PilV functions in recognition of the recipient that facilitates conjugation in liquid environments. Different PilV variants recognize specific oligosaccharide moieties in the LPS core on the surface of recipient cells, and this affects the range of recipients that the donor strains can conjugate with in liquid media (118–120). Control of recipient recognition is mediated by a shufflon, which is a multi-inversion system that is located at the 3′ end of pilV and functions as a biological switch that mediates variable expression of the PilV protein (118, 121, 122). Shufflons are generally composed of four DNA segments, three of which (segments A, B, and C) are divided into two open reading frames that are subject to inversion and are separated by seven recombination sites. The combination of the DNA segments and recombination sites allows for the potential formation of 7 different PilV adhesion variants (118). The recombination of the shufflon elements is mediated by Rci, which is a site-specific recombinase whose gene is located just upstream of the shufflon region (25, 123). Recent next-generation sequencing experiments have identified that the shufflon region may display even greater variability due to deletions of pilV segments or insertion sequence (IS) elements inserted at recombination sites (124, 125). Because of the location of the shufflon sequences at the C-terminal region of pilV, the proteins formed have a conserved N-terminal region of approximately 361 amino acids and variable C-terminal regions that can vary in size between 69 and 113 amino acids (118).
FIG 6.
Predicted structure of the T4P of IncI1 plasmids based on homologs from the better-characterized TCP and BFG pili. PilS forms the major prepilin polymer complex that extends from the cell to form the T4P and is capped by PilV subunits that make up the minor prepilin multimer that interacts with specific oligosaccharide in LPS on a recipient cell. Other structural elements include PilR, which is the integral inner membrane spanning protein, and PilN, which forms the outer membrane secretin through which the PilS polymer extends. PilT is predicted to be a lytic transglycosidase that may function to create a pore through the peptidoglycan layer to allow elongation of the pilus structure. PilR proteins are predicted to be platform proteins that transfer energy from the system ATPases, such as PilQ, to the T4P structure.
PilT is predicted to be a lytic transglycosidase that is localized to the periplasm, and it has been suggested that it functions to create a pore through the peptidoglycan layer to allow elongation of the pilus structure (126). PilR is predicted to be an inner membrane protein whose amino acid sequence has similarity to those of BfgE and TcpE of the BFP and TCP T4bP (115, 126). These proteins are predicted to be platform proteins that transfer energy from the system ATPases, such as PilQ, to the T4P structure (127). The other proteins identified as essential for facilitating R64 and ColIb-P9 transfer include PilK and PilM, which are inner membrane-associated pilus biogenesis proteins, PilL outer membrane lipoprotein, PilN outer membrane pilus secretin protein, PilO outer membrane-associated pilus biogenesis protein, PilP periplasmic-associated pilus biogenesis protein, and PilQ cytoplasmic ATPase (25, 115, 128). The PilN outer membrane monomers form a ring structure that is predicted to serve as the channel for passage of extending T4P structure across the outer membrane of R64 during elongation, and the PilQ complex is a cytoplasmic ATPase that powers the assembly of the pilus structure (115, 126–128).
ANTIMICROBIAL RESISTANCE
A key reason that IncI1 plasmids have drawn attention by the public health community is their ability to carry antimicrobial resistance genes, including those associated with crucial antimicrobials used to treat severe cases of enteric infections such as the third-generation cephalosporins, fluoroquinolones, and macrolides (129–131). IncI1 plasmids are known to carry a variety of different resistance genes (Table 1), and hence they possess the potential to encode multidrug resistance (MDR) in bacterial pathogens (37, 132, 133).
The best-studied antimicrobial resistance associated with the IncI1 plasmids is that of those genes that encode β-lactam resistance. β-Lactamases are enzymes that cause hydrolysis of oxyimino-β-lactam antimicrobial agents (134). There are multiple generations of β-lactam antimicrobial compounds that exhibit a spectrum of activity levels, and they are widely used in clinical practice (129, 134). Extended-spectrum β-lactamases (ESBLs) are enzymes that can inhibit a broader range of β-lactam antibiotics (135, 136). Numerous researchers have investigated the prevalence of these ESBL genes in IncI1 plasmids (9, 18, 27, 38, 137–157). Many variants of ESBL enzymes have evolved over a period of time, largely due to mutations with the genes encoding the enzymes (158). There are some broad classes of ESBLs, including the TEM and SHV families of β-lactamases that were prevalent in the 1980s and 1990s, respectively, and CTX-M β-lactamases that have been prominent since the early 2000s (159–161). The blaTEM-1, blaTEM-20, blaTEM-52, and blaSHV-12 genes are those most commonly associated with IncI1 plasmids (14, 37, 133, 151, 162–165).
The blaCTX-M variants are unique from blaTEM and blaSHV enzyme types (166). The blaCTX-M family contains multiple subtypes, and many have been reported to be associated with IncI1 plasmids (37). The blaCTX-M variants are present in enteric organisms, including E. coli, Salmonella spp., and Klebsiella spp. from around the world, and have been detected in both nosocomial and community settings (37, 160). The global presence of blaCTX-M variants led to in-depth studies to understand them (143, 167–172). Some blaCTX-M variants are associated with insertion sequences (IS) present on the plasmids. The ISEcp1 is associated with blaCTX-M-5 and blaCTX-M-15, while ISCR1 is linked to blaCTX-M-2 and blaCTX-M-9 (27, 146). These IS elements are hypothesized to carry outward reading promoters that confer high level expression of blaCTX-M. The blaCTX-M-15 is the most universal ESBL among E. coli strains carrying blaCTX-M. These strains often belong to ST131 and are resistant to quinolones in addition to cephalosporins (173). The increasing prevalence of E. coli in community-associated and nosocomial infections make this broadening of antimicrobial resistance a public health concern.
Additionally, IncI1 plasmids have been identified that carry the blaCMY gene that encodes AmpC β-lactamase (141, 162, 174–179) leading to resistance to several β-lactam antibiotics, including ampicillin, cefoxitin, ceftriaxone, and amoxicillin clavulanate (180). The primary blaCMY variant associated with IncI1 plasmids is CMY-2. The blaCMY-2 gene has been found in a diverse range of IncI1 plasmids based on pMLST, with the gene being detected in plasmids representing a variety of sequence types and clonal clusters (37). Within the IncI1 plasmids, the blaCMY-2 genes are typically found in IS elements, including ISEsp1 and IS1294, that facilitate their mobilization and insertion to different regions of the plasmids (14, 181, 182). Other blaCMY genes that have been identified in IncI1 plasmids include blaCMY-4 (183), blaCMY-42 (164, 176, 184), and blaCMY-111 (183).
In addition to the β-lactam resistance genes, IncI1 plasmids have also been characterized that carry resistance genes for several other antimicrobial agents, including sulfonamides, trimethoprim, chloramphenicol, aminoglycosides, and tetracyclines (28, 43, 185) (Table 1). For chloramphenicol resistance, the genes cmlA and floR have been identified and are typically associated with integrons and IS elements, such as IS26 and ISCR2 (18, 28, 186). Many of the chloramphenicol resistance genes are colocated with the sulfonamide resistance gene sul2 (28, 186, 187). In addition, sul1 has also been detected in several IncI1 plasmids of organisms collected from a variety of animal sources and diverse geographical locations (43, 187, 188). Trimethoprim resistance genes, including dfrA1 and dfrA17, have been identified in plasmids that were isolated from E. coli in human patients, food animals, and wild birds in Europe (162, 187). The most commonly detected tetracycline resistance gene in IncI1 plasmids appears to be tetA (21, 188, 189). There is greater diversity of aminoglycoside resistance genes that have been detected on the plasmids, including those associated with gentamicin [aacC and aac(3)-IV] (28, 43), kanamycin [aph(4)-1a] (18), and streptomycin (aadA1, aadA2, aadA5, strA, and strB) (28, 43, 188). This wide variety of genes encoding resistance to clinically relevant antimicrobials is concerning, especially in light of the ability of several IncI1 plasmids to conjugally transfer among different bacteria (21, 43). Many of the resistance genes are associated with IS elements, integrons, and transposons which may further facilitate their transmission among plasmids that are coresident within bacteria and potentially the host chromosome (21, 43, 190). Therefore, the distribution of antimicrobial resistance genes to and from IncI1 plasmids is an important area for public health surveillance.
VIRULENCE AND COLICIN PRODUCTION
To cause infection, Salmonella must traverse the upper gastrointestinal tract, compete with commensal intestinal bacteria, and invade and persist within the intestinal epithelia (6). The potential contribution of IncI1 plasmids to virulence has not been well understood, as much of the research has focused on their contributions to antimicrobial resistance (21, 31). There have been several efforts to sequence IncI1-positive bacteria to examine the genetics of IncI1 plasmids (25, 45, 54, 84, 88), and several have identified genes that may be associated with virulence (191). Virulence factors allow the bacterium to have an increased ability to colonize a host niche, provide entry into, survive within, and exit from a host, evade or suppress the host’s immune response, or obtain required nutrients that are limited in the host environment (192, 193). Among these factors are biofilm formation that increases colonization ability, bacteriocin production which limits niche competition, nutrient acquisition, such as for iron, bacterial uptake systems that facilitate invasion and improve intracellular survival in host cells, and regulatory factors that mediate the expression of virulence genes (192).
Many of the sequenced IncI1 plasmids carry genes, such as cib, that encode the production of bacteriocins which can provide a competitive advantage for the host against members of the microbiota that they may be competing against for niche colonization (19, 194). The cib gene, which encodes colicin Ib, has been reported to be commonly carried on IncI1 plasmids (21, 195). Most of these strains positive for cib also carry a colicin immunity gene (imm) that protects the strain from the toxin (14, 21). The spectrum of inhibition of colicin Ib appears to be quite narrow; in characterization studies by Kaldhone et al., strains expressing the colicin were able to inhibit a limited number of E. coli strains and none of the non-E. coli species tested, including S. enterica, K. pneumoniae, Enterobacter cloacae, Pseudomonas aeruginosa, and Enterococcus faecalis (31).
In functional studies examining the role of colicin in pathogenesis, Nedialkova et al. demonstrated that colicin Ib produced by Salmonella enterica serotype Typhimurium strains could inhibit the growth of other enteric organisms in a murine model system, especially when there was inflammation in the gastrointestinal (GI) tract, while colicin-negative strains lacked the competitive colonization advantage (196). The expression of colicins appears to be dependent on external factors, including during periods of iron limitation, as is observed during the GI inflammatory response, where cib expression becomes altered (197). The impact of intestinal inflammation and iron limitation on bacteriocin activity is likely due to enterobactin siderophore receptors on the surface of bacterial cells that can also serve as colicin Ib receptors (19, 198). Periods of iron limitation can lead to increased expression of siderophore receptors (also known as colicin receptors) by commensal organisms and to increased susceptibility to colicin Ib. In Salmonella, this competitive colonization advantage coupled with an increased ability to acquire iron from the environment helps facilitate Salmonella uptake into the intestinal epithelial cells (194). These potential contributions to virulence coupled with the apparent minimal metabolic costs of IncI1 plasmid carriage, especially in the presence of other plasmids, may explain why a high percentage of IncI1 plasmid-carrying isolates contain multiple large plasmids, including those of the IncA/C, IncHI2, IncFIB, and IncX replicon types (31, 45, 54).
Several IncI1 plasmids carry genes that encode DNA repair mechanisms following DNA damage due to UV light exposure and DNA-damaging compounds (199–201). The I group mutation and protection (imp) operon contains three genes, impA, impB, and impC, that are functionally similar to the chromosomally encoded umuCD genes of the error-prone DNA repair system (201). DNA damage can initiate an SOS response in bacteria. Expression SOS response in genes is generally held in check by a LexA repressor binding to the SOS box in the umuCD promoter region (202, 203). During the SOS response, single-stranded (damaged) DNA initially interacts with and activates RecA, which then leads to autoproteolysis of the LexA repressor and expression of a cascade of genes, including umuC and umuD, due to derepression of the LexA repressor (202). Additionally, activated RecA triggers the autocleavage of UmuD to its active form associated with its error-prone DNA repair capabilities (204). The imp system has been best characterized in the IncI1 plasmid TP110 and is found in both R64 and ColI1-P9 (25, 199–201). When the IncI1 positive Salmonella strains characterized by Kaldhone et al. were assessed, 24/43 (56%) of the isolates contained the full impCAB operon, while 4 (9%) additional isolates carried all but the 3′ end of the impB sequence, which was a similar phenomenon to that reported previously in Shigella (25, 31, 202, 205).
In the impCAB operon, impC overlaps the impA start codon by 2 nucleotides and impA overlaps the impB translation initiation sequence by 1 nucleotide (202). The imp promoter region, which is upstream of impC, contains the sequence of an SOS box, where a LexA repressor could potentially bind. Based on homology to UmuCD, ImpA and ImpB likely serve as an error-prone DNA polymerase (206). ImpC may function in a regulatory role for the expression of impA and impB, as it has homology to regulatory proteins, including DinI, which can inhibit the LexA and UmuD cleavage functions of RecA (207). Shigella strains that lost the impCAB-containing plasmid or had a mutated impB gene exhibited reduced ability to survive following UV irradiation compared to that of the wild-type strains, indicating the importance of these genes for UV resistance (202, 208–210). The process of error-prone DNA repair leads to increased rates of mutagenesis in the strains, potentially facilitating compensatory mutations associated with survival during high-stress periods (208). Some of the mutations may manifest as increased levels of resistance to certain clinically relevant antimicrobials, including the fluoroquinolones (208, 211, 212), and the ability to survive in the GI tract following exposure to bile salts, such as sodium deoxycholate, that can damage bacterial DNA (208, 210).
CONCLUSIONS
This review focused on the genetic and phenotypic characterization of IncI1 plasmids. IncI1 plasmids have garnered significant attention due to the carriage and dissemination of a wide range of antimicrobial resistance genes, including those encoding resistance to critically important antimicrobials such at the third-generation cephalosporins (37, 132, 133). The widespread carriage of ESBL- and AmpC β-lactamase-encoding genes is very problematic, as these genes in IncI1 plasmids have been isolated globally from a wide range of animal species and patients (37, 160). Also concerning is the fact that these plasmids have been shown to carry genes encoding resistance for several other antimicrobial agents, including aminoglycosides, chloramphenicol, sulfonamides, trimethoprim, and tetracyclines (28, 43, 185). This carriage of the wide variety of resistance genes is critical, especially in light of the ability of several IncI1 plasmids to conjugally transfer among different bacteria (21, 43). Additionally, many of the resistance genes are associated with IS elements, integrons, and transposons that contribute to their transmission among plasmids and the chromosome and other plasmids that are coresident within bacteria, leading to a further potential to spread. Because of the multiple transfer mechanisms carried on the plasmids, their host addiction systems, and their relatively low fitness costs, the IncI1 plasmids will likely remain a concern for the maintenance and dissemination of antimicrobial resistance transfer in the future.
ACKNOWLEDGMENTS
Pravin Kaldhone’s graduate assistantship was provided by the Center for Advanced Surface Engineering under the National Science Foundation grant number OIA-1457888 and by the Arkansas EPSCoR Program, ASSET III.
The material described in the manuscript is not a formal dissemination of information by the FDA and does not represent agency position or policy.
Biographies

Steven L. Foley is a member of the Senior Biomedical Research and Biomedical Product Assessment Service and Deputy Director of the Division of Microbiology with the U.S. Food and Drug Administration’s National Center for Toxicological Research. Dr. Foley earned his Ph.D. in Cellular and Molecular Biology from North Dakota State University and completed a postdoctoral fellowship with FDA’s Center for Veterinary Medicine. His research program focuses on understanding the role of plasmids in Salmonella virulence and antimicrobial resistance and factors that contribute to their spread. He also has an interest in the integration of laboratory and bioinformatics approaches to predict potential public health risks associated with enteric pathogens.

Pravin R. Kaldhone received his M.B.B.S. from the University of Mumbai, his M.S. from the University of Central Arkansas, and his Ph.D. from the University of Arkansas and is an ORISE postdoctoral fellow at National Center for Toxicological Research (NCTR/FDA). His graduate school work emphasized on antimicrobial resistance and virulence of Salmonella serovars relevant to public health and the role of dietary supplements such as prebiotics on gut microbiome in food animals. He was a research associate at Marshfield Clinic where he investigated the role of mobile genetic elements from methicillin-resistant Staphylococcus aureus in survival and evolution. His current research focuses on exploring genetic toxicity of nanomaterials using whole-genome sequencing of mammalian cells expanded from single-cell clones and testing opioid compounds for maternal and perinatal toxicity using multiple approaches to enhance oversight of FDA-regulated products.

Steven C. Ricke received his B.S. from the University of Illinois and his Ph.D. from the University of Wisconsin and was a USDA-ARS postdoctoral fellow at North Carolina State University. He was a professor at Texas A&M University until 2005, when he became the Wray Endowed Chair and Director of the Center for Food Safety at the University of Arkansas. In 2020 he became the Director for Meat Science and Animal Biologics Science Discovery Program in the Department of Animal and Dairy Sciences at the University of Wisconsin-Madison. His research program is focused primarily on virulence and pathogenic characteristics of foodborne pathogens with emphasis on their growth, survival, and pathogenesis under conditions encountered during food animal production and processing. He is also conducting studies on the gastrointestinal tract microbiota of food animals and the impact of feed additives such as prebiotics on the microbial composition and functionality.

Jing Han received her B.S. from Shandong Agricultural University, her M.S. from China Agricultural University, and her Ph.D. from Iowa State University and was a postdoctoral fellow at National Center for Toxicological Research (NCTR/FDA). She worked at the Public Health Laboratory of Arkansas Department of Health as a microbiologist in 2015, then rejoined NCTR as a research microbiologist where she served as a leading investigator on a project focusing on smokeless tobacco-associated microbiology. Currently her research is focused primarily on the characterization of antimicrobial resistance and associated genetic factors in Salmonella serovars associated with food animals and invasive human infections and the development of improved databases to identify Salmonella-virulence genes and develop the analytical tools for whole-genome-sequencing (WGS) data analyses.
REFERENCES
- 1.Johnson TJ, Nolan LK. 2009. Plasmid replicon typing. Methods Mol Biol 551:27–35. 10.1007/978-1-60327-999-4_3. [DOI] [PubMed] [Google Scholar]
- 2.Gualberto JM, Kuhn K. 2014. DNA-binding proteins in plant mitochondria: implications for transcription. Mitochondrion 19(Pt B):323–328. 10.1016/j.mito.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 3.Johnson TJ, Lang KS. 2012. IncA/C plasmids: an emerging threat to human and animal health? Mob Genet Elements 2:55–58. 10.4161/mge.19626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson TJ, Nolan LK. 2009. Pathogenomics of the virulence plasmids of Escherichia coli. Microbiol Mol Biol Rev 73:750–774. 10.1128/MMBR.00015-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frost LS, Koraimann G. 2010. Regulation of bacterial conjugation: balancing opportunity with adversity. Future Microbiol 5:1057–1071. 10.2217/fmb.10.70. [DOI] [PubMed] [Google Scholar]
- 6.Foley SL, Lynne AM. 2008. Food animal-associated Salmonella challenges: pathogenicity and antimicrobial resistance. J Anim Sci 86:E173–187. 10.2527/jas.2007-0447. [DOI] [PubMed] [Google Scholar]
- 7.Novick RP. 1987. Plasmid incompatibility. Microbiol Rev 51:381–395. 10.1128/MR.51.4.381-395.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanad YM, Johnson K, Park SH, Han J, Deck J, Foley SL, Kenney B, Ricke S, Nayak R. 2016. Molecular characterization of Salmonella enterica serovars isolated from a turkey production facility in the absence of selective antimicrobial pressure. Foodborne Pathog Dis 13:80–87. 10.1089/fpd.2015.2002. [DOI] [PubMed] [Google Scholar]
- 9.Wong MH, Kan B, Chan EW, Yan M, Chen S. 2016. IncI1 plasmids carrying various blaCTX-M genes contribute to ceftriaxone resistance in Salmonella enterica serovar Enteritidis in China. Antimicrob Agents Chemother 60:982–989. 10.1128/AAC.02746-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Couturier M, Bex F, Bergquist PL, Maas WK. 1988. Identification and classification of bacterial plasmids. Microbiol Rev 52:375–395. 10.1128/MR.52.3.375-395.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Douarre PE, Mallet L, Radomski N, Felten A, Mistou MY. 2020. Analysis of COMPASS, a new comprehensive plasmid database revealed prevalence of multireplicon and extensive diversity of IncF plasmids. Front Microbiol 11:483. 10.3389/fmicb.2020.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Fernandez A, Chiaretto G, Bertini A, Villa L, Fortini D, Ricci A, Carattoli A. 2008. Multilocus sequence typing of IncI1 plasmids carrying extended-spectrum beta-lactamases in Escherichia coli and Salmonella of human and animal origin. J Antimicrob Chemother 61:1229–1233. 10.1093/jac/dkn131. [DOI] [PubMed] [Google Scholar]
- 13.Mo SS, Sunde M, Ilag HK, Langsrud S, Heir E. 2017. Transfer potential of plasmids conferring extended-spectrum-cephalosporin resistance in Escherichia coli from poultry. Appl Environ Microbiol 83:e00654-17. 10.1128/AEM.00654-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith H, Bossers A, Harders F, Wu G, Woodford N, Schwarz S, Guerra B, Rodriguez I, van Essen-Zandbergen A, Brouwer M, Mevius D. 2015. Characterization of epidemic IncI1-Iγ plasmids harboring ambler class A and C genes in Escherichia coli and Salmonella enterica from animals and humans. Antimicrob Agents Chemother 59:5357–5365. 10.1128/AAC.05006-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshida T, Kim SR, Komano T. 1999. Twelve pil genes are required for biogenesis of the R64 thin pilus. J Bacteriol 181:2038–2043. 10.1128/JB.181.7.2038-2043.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilkins BM, Thomas AT. 2000. DNA-independent transport of plasmid primase protein between bacteria by the I1 conjugation system. Mol Microbiol 38:650–657. 10.1046/j.1365-2958.2000.02164.x. [DOI] [PubMed] [Google Scholar]
- 17.Komano T, Kim SR, Yoshida T. 1995. Mating variation by DNA inversions of shufflon in plasmid R64. Adv Biophys 31:181–193. 10.1016/0065-227x(95)99391-2. [DOI] [PubMed] [Google Scholar]
- 18.Riccobono E, Di Pilato V, Di Maggio T, Revollo C, Bartoloni A, Pallecchi L, Rossolini GM. 2015. Characterization of IncI1 sequence type 71 epidemic plasmid lineage responsible for the recent dissemination of CTX-M-65 extended-spectrum beta-lactamase in the Bolivian Chaco region. Antimicrob Agents Chemother 59:5340–5347. 10.1128/AAC.00589-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cascales E, Buchanan SK, Duche D, Kleanthous C, Lloubes R, Postle K, Riley M, Slatin S, Cavard D. 2007. Colicin biology. Microbiol Mol Biol Rev 71:158–229. 10.1128/MMBR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luria SE, Suit JL, Neidhardt FC. 1987. Colicins and col plasmids, p 1615–1624. In Escherichia coli and Salmonella Typhimurium: cellular and Molecular Biology. American Society for Microbiology, Washington, DC. [Google Scholar]
- 21.Kaldhone PR, Han J, Deck J, Khajanchi B, Nayak R, Foley SL, Ricke SC. 2018. Evaluation of the genetics and functionality of plasmids in incompatibility group I1-positive Salmonella enterica. Foodborne Pathog Dis 15:168–176. 10.1089/fpd.2017.2332. [DOI] [PubMed] [Google Scholar]
- 22.Meynell GG, Lawn AM. 1968. Filamentous phages specific for the I sex factor. Nature 217:1184–1186. 10.1038/2171184a0. [DOI] [PubMed] [Google Scholar]
- 23.Bradley DE. 1984. Characteristics and function of thick and thin conjugative pili determined by transfer-derepressed plasmids of incompatibility groups I1, I2, I5, B, K and Z. J Gen Microbiol 130:1489–1502. 10.1099/00221287-130-6-1489. [DOI] [PubMed] [Google Scholar]
- 24.Meynell E, Datta N. 1966. The relation of resistance transfer factors to the F-factor (sex-factor) of Escherichia coli K12. Genet Res 7:134–140. 10.1017/s0016672300009538. [DOI] [PubMed] [Google Scholar]
- 25.Sampei G, Furuya N, Tachibana K, Saitou Y, Suzuki T, Mizobuchi K, Komano T. 2010. Complete genome sequence of the incompatibility group I1 plasmid R64. Plasmid 64:92–103. 10.1016/j.plasmid.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 26.Hama C, Takizawa T, Moriwaki H, Urasaki Y, Mizobuchi K. 1990. Organization of the replication control region of plasmid ColIb-P9. J Bacteriol 172:1983–1991. 10.1128/jb.172.4.1983-1991.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zong Z, Ginn AN, Dobiasova H, Iredell JR, Partridge SR. 2015. Different IncI1 plasmids from Escherichia coli carry ISEcp1-blaCTX-M-15 associated with different Tn2-derived elements. Plasmid 80:118–126. 10.1016/j.plasmid.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 28.Johnson TJ, Shepard SM, Rivet B, Danzeisen JL, Carattoli A. 2011. Comparative genomics and phylogeny of the IncI1 plasmids: a common plasmid type among porcine enterotoxigenic Escherichia coli. Plasmid 66:144–151. 10.1016/j.plasmid.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 29.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 30.Johnson TJ, Singer RS, Isaacson RE, Danzeisen JL, Lang K, Kobluk K, Rivet B, Borewicz K, Frye JG, Englen M, Anderson J, Davies PR. 2015. In vivo transmission of an IncA/C plasmid in Escherichia coli depends on tetracycline concentration, and acquisition of the plasmid results in a variable cost of fitness. Appl Environ Microbiol 81:3561–3570. 10.1128/AEM.04193-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaldhone PR, Carlton A, Aljahdali N, Khajanchi B, Sanad YM, Han J, Deck J, Ricke SC, Foley SL. 2019. Evaluation of incompatibility group I1 (IncI1) plasmid-containing Salmonella enterica and assessment of the plasmids in bacteriocin production and biofilm development. Front Vet Med 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Freire Martín I, Thomas CM, Laing E, AbuOun M, La Ragione RM, Woodward MJ. 2016. Curing vector for IncI1 plasmids and its use to provide evidence for a metabolic burden of IncI1 CTX-M-1 plasmid pIFM3791 on Klebsiella pneumoniae. J Med Microbiol 65:611–618. 10.1099/jmm.0.000271. [DOI] [PubMed] [Google Scholar]
- 33.Lee SW, Edlin G. 1985. Expression of tetracycline resistance in pBR322 derivatives reduces the reproductive fitness of plasmid-containing Escherichia coli. Gene 39:173–180. 10.1016/0378-1119(85)90311-7. [DOI] [PubMed] [Google Scholar]
- 34.Moremi N, Manda EV, Falgenhauer L, Ghosh H, Imirzalioglu C, Matee M, Chakraborty T, Mshana SE. 2016. Predominance of CTX-M-15 among ESBL producers from environment and fish gut from the shores of Lake Victoria in Mwanza. Tanzania Front Microbiol 7:1862. 10.3389/fmicb.2016.01862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dahmen S, Haenni M, Madec JY. 2012. IncI1/ST3 plasmids contribute to the dissemination of the blaCTX-M-1 gene in Escherichia coli from several animal species in France. J Antimicrob Chemother 67:3011–3012. 10.1093/jac/dks308. [DOI] [PubMed] [Google Scholar]
- 36.Giles M, Cawthraw SA, AbuOun M, Thomas CM, Munera D, Waldor MK, La Ragione RM, Ritchie JM. 2018. Host-specific differences in the contribution of an ESBL IncI1 plasmid to intestinal colonization by Escherichia coli O104:H4. J Antimicrob Chemother 73:1579–1585. 10.1093/jac/dky037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carattoli A, Villa L, Fortini D, Garcia-Fernandez A. 2018. Contemporary IncI1 plasmids involved in the transmission and spread of antimicrobial resistance in Enterobacteriaceae. Plasmid 102392. 10.1016/j.plasmid.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 38.Brouwer MS, Bossers A, Harders F, van Essen-Zandbergen A, Mevius DJ, Smith HE. 2014. Complete genome sequences of IncI1 plasmids carrying extended-spectrum beta-lactamase genes. Genome Announc 2:e00859-14. 10.1128/genomeA.00859-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J, Stephan R, Power K, Yan Q, Hachler H, Fanning S. 2014. Nucleotide sequences of 16 transmissible plasmids identified in nine multidrug-resistant Escherichia coli isolates expressing an ESBL phenotype isolated from food-producing animals and healthy humans. J Antimicrob Chemother 69:2658–2668. 10.1093/jac/dku206. [DOI] [PubMed] [Google Scholar]
- 40.Bleicher A, Schofl G, Rodicio MR, Saluz HP. 2013. The plasmidome of a Salmonella enterica serovar Derby isolated from pork meat. Plasmid 69:202–210. 10.1016/j.plasmid.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 41.Castellanos LR, Donado-Godoy P, Leon M, Clavijo V, Arevalo A, Bernal JF, Timmerman AJ, Mevius DJ, Wagenaar JA, Hordijk J. 2017. High heterogeneity of Escherichia coli sequence types harbouring ESBL/AmpC genes on IncI1 plasmids in the Colombian poultry chain. PLoS One 12:e0170777. 10.1371/journal.pone.0170777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castellanos LR, van der Graaf-van Bloois L, Donado-Godoy P, Mevius DJ, Wagenaar JA, Hordijk J, Zomer AL. 2019. Phylogenomic investigation of IncI1-Iγ plasmids harboring blaCMY-2 and blaSHV-12 in Salmonella enterica and Escherichia coli in multiple countries. Antimicrob Agents Chemother 63. 10.1128/AAC.02546-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han J, Lynne AM, David DE, Tang H, Xu J, Nayak R, Kaldhone P, Logue CM, Foley SL. 2012. DNA sequence analysis of plasmids from multidrug resistant Salmonella enterica serotype Heidelberg isolates. PLoS One 7:e51160. 10.1371/journal.pone.0051160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang D, Zhao Y, Feng J, Hu L, Jiang X, Zhan Z, Yang H, Yang W, Gao B, Wang J, Li J, Yin Z, Zhou D. 2019. Replicon-based typing of inci-complex plasmids, and comparative genomics analysis of IncIγ/K1 plasmids. Front Microbiol 10:48. 10.3389/fmicb.2019.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaldhone PR, Khajanchi BK, Han J, Nayak R, Ricke SC, Foley SL. 2017. Draft genome sequences of Salmonella enterica isolates containing incompatibility group I1 plasmids from swine, poultry, and human sources. Genome Announc 5. 10.1128/genomeA.01056-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hagbo M, Ravi A, Angell IL, Sunde M, Ludvigsen J, Diep DB, Foley SL, Vento M, Collado MC, Perez-Martinez G, Rudi K. 2020. Experimental support for multidrug resistance transfer potential in the preterm infant gut microbiota. Pediatr Res 88:57–65. 10.1038/s41390-019-0491-8. [DOI] [PubMed] [Google Scholar]
- 47.Ravi A, Avershina E, Foley SL, Ludvigsen J, Storro O, Oien T, Johnsen R, McCartney AL, L'Abee-Lund TM, Rudi K. 2015. The commensal infant gut meta-mobilome as a potential reservoir for persistent multidrug resistance integrons. Sci Rep 5:15317. 10.1038/srep15317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pinto UM, Pappas KM, Winans SC. 2012. The ABCs of plasmid replication and segregation. Nat Rev Microbiol 10:755–765. 10.1038/nrmicro2882. [DOI] [PubMed] [Google Scholar]
- 49.Brantl S. 2014. Plasmid replication control by antisense RNAs. Microbiol Spectr 2:PLAS-0001-2013. 10.1128/microbiolspec.PLAS-0001-2013. [DOI] [PubMed] [Google Scholar]
- 50.Scott JR. 1984. Regulation of plasmid replication. Microbiol Rev 48:1–23. 10.1016/b978-0-12-048850-6.50006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Asano K, Mizobuchi K. 1998. Copy number control of IncIα plasmid ColIb-P9 by competition between pseudoknot formation and antisense RNA binding at a specific RNA site. EMBO J 17:5201–5213. 10.1093/emboj/17.17.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Asano K, Mizobuchi K. 2000. Structural analysis of late intermediate complex formed between plasmid ColIb-P9 Inc RNA and its target RNA. How does a single antisense RNA repress translation of two genes at different rates? J Biol Chem 275:1269–1274. 10.1074/jbc.275.2.1269. [DOI] [PubMed] [Google Scholar]
- 53.Clewell DB, Helinski DE. 1970. Existence of the colicinogenic factor-sex factor ColI-b-P9 as a supercoiled circular DNA-protein relaxation complex. Biochem Biophys Res Commun 41:150–156. 10.1016/0006-291x(70)90481-x. [DOI] [PubMed] [Google Scholar]
- 54.Aljahdali NH, Kaldhone PR, Foley SL, Khajanchi BK. 2019. Whole genome sequences of 35 incompatibility group I1 plasmid-carrying Salmonella enterica isolates from food animal and clinical sources. Microbiol Resour Announc 8. 10.1128/MRA.00831-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Asano K, Hama C, Inoue S, Moriwaki H, Mizobuchi K. 1999. The plasmid ColIb-P9 antisense Inc RNA controls expression of the RepZ replication protein and its positive regulator repY with different mechanisms. J Biol Chem 274:17924–17933. 10.1074/jbc.274.25.17924. [DOI] [PubMed] [Google Scholar]
- 56.Asano K, Niimi T, Yokoyama S, Mizobuchi K. 1998. Structural basis for binding of the plasmid ColIb-P9 antisense Inc RNA to its target RNA with the 5′-rUUGGCG-3′ motif in the loop sequence. J Biol Chem 273:11826–11838. 10.1074/jbc.273.19.11826. [DOI] [PubMed] [Google Scholar]
- 57.Asano K, Mizobuchi K. 1998. An RNA pseudoknot as the molecular switch for translation of the repZ gene encoding the replication initiator of IncIalpha plasmid ColIb-P9. J Biol Chem 273:11815–11825. 10.1074/jbc.273.19.11815. [DOI] [PubMed] [Google Scholar]
- 58.Harms A, Brodersen DE, Mitarai N, Gerdes K. 2018. Toxins, targets, and triggers: an overview of toxin-antitoxin biology. Mol Cell 70:768–784. 10.1016/j.molcel.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 59.Unterholzner SJ, Poppenberger B, Rozhon W. 2013. Toxin-antitoxin systems: biology, identification, and application. Mob Genet Elements 3:e26219. 10.4161/mge.26219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kamruzzaman M, Shoma S, Thomas CM, Partridge SR, Iredell JR. 2017. Plasmid interference for curing antibiotic resistance plasmids in vivo. PLoS One 12:e0172913. 10.1371/journal.pone.0172913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thisted T, Nielsen AK, Gerdes K. 1994. Mechanism of post-segregational killing: translation of Hok, SrnB and Pnd mRNAs of plasmids R1, F and R483 is activated by 3'-end processing. EMBO J 13:1950–1959. 10.1002/j.1460-2075.1994.tb06464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Akimoto S, Ohnishi Y. 1982. R483 and F plasmid genes promoting RNA degradation: comparative restriction mapping. Microbiol Immunol 26:779–793. 10.1111/j.1348-0421.1982.tb00224.x. [DOI] [PubMed] [Google Scholar]
- 63.Furuya N, Komano T. 1996. Nucleotide sequence and characterization of the trbABC region of the IncI1 Plasmid R64: existence of the pnd gene for plasmid maintenance within the transfer region. J Bacteriol 178:1491–1497. 10.1128/jb.178.6.1491-1497.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sakikawa T, Akimoto S, Ohnishi Y. 1985. Cloning and expression of the pnd gene of R16: determination of transcriptional direction and evolutionary analysis. Microbiol Immunol 29:791–801. 10.1111/j.1348-0421.1985.tb00882.x. [DOI] [PubMed] [Google Scholar]
- 65.Nielsen AK, Thorsted P, Thisted T, Wagner EG, Gerdes K. 1991. The rifampicin-inducible genes srnB from F and pnd from R483 are regulated by antisense RNAs and mediate plasmid maintenance by killing of plasmid-free segregants. Mol Microbiol 5:1961–1973. 10.1111/j.1365-2958.1991.tb00818.x. [DOI] [PubMed] [Google Scholar]
- 66.Finch RG. 2003. Antibiotic and chemotherapy: anti-infective agents and their use in therapy. J Antimicrob Chemother 52:740–741. [Google Scholar]
- 67.Saramago M, Barria C, Arraiano CM, Domingues S. 2015. Ribonucleases, antisense RNAs and the control of bacterial plasmids. Plasmid 78:26–36. 10.1016/j.plasmid.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 68.Nielsen AK, Gerdes K. 1995. Mechanism of post-segregational killing by hok-homologue pnd of plasmid R483: two translational control elements in the pnd mRNA. J Mol Biol 249:270–282. 10.1006/jmbi.1995.0296. [DOI] [PubMed] [Google Scholar]
- 69.Thisted T, Sorensen NS, Wagner EG, Gerdes K. 1994. Mechanism of post-segregational killing: Sok antisense RNA interacts with Hok mRNA via its 5′-end single-stranded leader and competes with the 3′-end of Hok mRNA for binding to the mok translational initiation region. EMBO J 13:1960–1968. 10.1002/j.1460-2075.1994.tb06465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smillie C, Garcillan-Barcia MP, Francia MV, Rocha EP, de la Cruz F. 2010. Mobility of plasmids. Microbiol Mol Biol Rev 74:434–452. 10.1128/MMBR.00020-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aminov RI. 2011. Horizontal gene exchange in environmental microbiota. Front Microbiol 2:158. 10.3389/fmicb.2011.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Frost LS, Ippen-Ihler K, Skurray RA. 1994. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol Rev 58:162–210. 10.1128/MMBR.58.2.162-210.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim SR, Funayama N, Komano T. 1993. Nucleotide sequence and characterization of the traABCD region of IncI1 plasmid R64. J Bacteriol 175:5035–5042. 10.1128/jb.175.16.5035-5042.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Komano T, Funayama N, Kim SR, Nisioka T. 1990. Transfer region of IncI1 plasmid R64 and role of shufflon in R64 transfer. J Bacteriol 172:2230–2235. 10.1128/jb.172.5.2230-2235.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Komano T, Kim SR, Nisioka T. 1987. Distribution of shufflon among IncI plasmids. J Bacteriol 169:5317–5319. 10.1128/jb.169.11.5317-5319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Komano T, Yoshida T, Narahara K, Furuya N. 2000. The transfer region of IncI1 plasmid R64: similarities between R64 tra and legionella icm/dot genes. Mol Microbiol 35:1348–1359. 10.1046/j.1365-2958.2000.01769.x. [DOI] [PubMed] [Google Scholar]
- 77.Dudley EG, Abe C, Ghigo JM, Latour-Lambert P, Hormazabal JC, Nataro JP. 2006. An IncI1 plasmid contributes to the adherence of the atypical enteroaggregative Escherichia coli strain C1096 to cultured cells and abiotic surfaces. Infect Immun 74:2102–2114. 10.1128/IAI.74.4.2102-2114.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Christie PJ, Gomez-Valero L, Buchrieser C. 2017. Biological diversity and evolution of type IV secretion systems. Curr Top Microbiol Immunol 413:1–30. 10.1007/978-3-319-75241-9_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guglielmini J, Neron B, Abby SS, Garcillan-Barcia MP, de la Cruz F, Rocha EP. 2014. Key components of the eight classes of type IV secretion systems involved in bacterial conjugation or protein secretion. Nucleic Acids Res 42:5715–5727. 10.1093/nar/gku194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Altier C. 2005. Genetic and environmental control of Salmonella invasion. J Microbiol 43 Spec No:85–92. [PubMed] [Google Scholar]
- 81.Anantha RP, Stone KD, Donnenberg MS. 2000. Effects of bfp mutations on biogenesis of functional enteropathogenic Escherichia coli type IV pili. J Bacteriol 182:2498–2506. 10.1128/jb.182.9.2498-2506.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kumar A, Das B, Kumar N. 2020. Vibrio pathogenicity island-1: the master determinant of cholera pathogenesis. Front Cell Infect Microbiol 10:561296. 10.3389/fcimb.2020.561296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Giltner CL, Nguyen Y, Burrows LL. 2012. Type IV pilin proteins: versatile molecular modules. Microbiol Mol Biol Rev 76:740–772. 10.1128/MMBR.00035-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Takahashi H, Shao M, Furuya N, Komano T. 2011. The genome sequence of the incompatibility group Iγ plasmid R621a: evolution of IncI plasmids. Plasmid 66:112–121. 10.1016/j.plasmid.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 85.Richardson LV, Richardson JP. 2005. Identification of a structural element that is essential for two functions of transcription factor NusG. Biochim Biophys Acta 1729:135–140. 10.1016/j.bbaexp.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 86.Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM. 2005. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol Rev 29:231–262. 10.1016/j.femsre.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 87.Poidevin M, Sato M, Altinoglu I, Delaplace M, Sato C, Yamaichi Y. 2018. Mutation in ESBL plasmid from Escherichia coli O104:H4 leads autoagglutination and enhanced plasmid dissemination. Front Microbiol 9:130. 10.3389/fmicb.2018.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yamaichi Y, Chao MC, Sasabe J, Clark L, Davis BM, Yamamoto N, Mori H, Kurokawa K, Waldor MK. 2015. High-resolution genetic analysis of the requirements for horizontal transmission of the ESBL plasmid from Escherichia coli O104:H4. Nucleic Acids Res 43:348–360. 10.1093/nar/gku1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Christie PJ, Whitaker N, Gonzalez-Rivera C. 2014. Mechanism and structure of the bacterial type IV secretion systems. Biochim Biophys Acta 1843:1578–1591. 10.1016/j.bbamcr.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Guglielmini J, de la Cruz F, Rocha EP. 2013. Evolution of conjugation and type IV secretion systems. Mol Biol Evol 30:315–331. 10.1093/molbev/mss221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lawley TD, Klimke WA, Gubbins MJ, Frost LS. 2003. F factor conjugation is a true type IV secretion system. FEMS Microbiol Lett 224:1–15. 10.1016/S0378-1097(03)00430-0. [DOI] [PubMed] [Google Scholar]
- 92.Kuroda T, Kubori T, Thanh Bui X, Hyakutake A, Uchida Y, Imada K, Nagai H. 2015. Molecular and structural analysis of Legionella DotI gives insights into an inner membrane complex essential for type IV secretion. Sci Rep 5:10912. 10.1038/srep10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Moller TSB, Liu G, Boysen A, Thomsen LE, Luthje FL, Mortensen S, Moller-Jensen J, Olsen JE. 2017. Treatment with cefotaxime affects expression of conjugation associated proteins and conjugation transfer frequency of an inci1 plasmid in Escherichia coli. Front Microbiol 8:2365. 10.3389/fmicb.2017.02365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dym O, Albeck S, Unger T, Jacobovitch J, Branzburg A, Michael Y, Frenkiel-Krispin D, Wolf SG, Elbaum M. 2008. Crystal structure of the Agrobacterium virulence complex VirE1-VirE2 reveals a flexible protein that can accommodate different partners. Proc Natl Acad Sci U S A 105:11170–11175. 10.1073/pnas.0801525105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sundberg C, Meek L, Carroll K, Das A, Ream W. 1996. VirE1 protein mediates export of the single-stranded DNA-binding protein VirE2 from Agrobacterium tumefaciens into plant cells. J Bacteriol 178:1207–1212. 10.1128/jb.178.4.1207-1212.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Park D, Chetrit D, Hu B, Roy CR, Liu J. 2020. Analysis of Dot/Icm type IVB secretion system subassemblies by cryoelectron tomography reveals conformational changes induced by DotB binding. mBio 11. 10.1128/mBio.03328-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chetrit D, Hu B, Christie PJ, Roy CR, Liu J. 2018. A unique cytoplasmic ATPase complex defines the Legionella pneumophila type IV secretion channel. Nat Microbiol 3:678–686. 10.1038/s41564-018-0165-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Durie CL, Sheedlo MJ, Chung JM, Byrne BG, Su M, Knight T, Swanson M, Lacy DB, Ohi MD. 2020. Structural analysis of the Legionella pneumophila Dot/Icm type IV secretion system core complex. Elife 9. 10.7554/eLife.59530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kubori T, Nagai H. 2016. The Type IVB secretion system: an enigmatic chimera. Curr Opin Microbiol 29:22–29. 10.1016/j.mib.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 100.Ghosal D, Jeong KC, Chang YW, Gyore J, Teng L, Gardner A, Vogel JP, Jensen GJ. 2019. Molecular architecture, polar targeting and biogenesis of the Legionella Dot/Icm T4SS. Nat Microbiol 4:1173–1182. 10.1038/s41564-019-0427-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang S, Wang D, Du D, Li S, Yan W. 2018. Advances in the assembly model of bacterial type IVB secretions systems. Appl Sci 8:2368. 10.3390/app8122368. [DOI] [Google Scholar]
- 102.Nagai H, Kubori T. 2011. Type IVB secretion systems of Legionella and other Gram-negative bacteria. Front Microbiol 2:136. 10.3389/fmicb.2011.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gomez-Valero L, Chiner-Oms A, Comas I, Buchrieser C. 2019. Evolutionary dissection of the Dot/Icm system based on comparative genomics of 58 Legionella species. Genome Biol Evol 11:2619–2632. 10.1093/gbe/evz186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ghosal D, Chang YW, Jeong KC, Vogel JP, Jensen GJ. 2017. In situ structure of the Legionella Dot/Icm type IV secretion system by electron cryotomography. EMBO Rep 18:726–732. 10.15252/embr.201643598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kim H, Kubori T, Yamazaki K, Kwak MJ, Park SY, Nagai H, Vogel JP, Oh BH. 2020. Structural basis for effector protein recognition by the Dot/Icm type IVB coupling protein complex. Nat Commun 11:2623. 10.1038/s41467-020-16397-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Meir A, Mace K, Lukoyanova N, Chetrit D, Hospenthal MK, Redzej A, Roy C, Waksman G. 2020. Mechanism of effector capture and delivery by the type IV secretion system from Legionella pneumophila. Nat Commun 11:2864. 10.1038/s41467-020-16681-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li YG, Hu B, Christie PJ. 2019. Biological and structural diversity of type iv secretion systems. Microbiol Spectr 7. 10.1128/microbiolspec.PSIB-0012-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wallden K, Williams R, Yan J, Lian PW, Wang L, Thalassinos K, Orlova EV, Waksman G. 2012. Structure of the VirB4 ATPase, alone and bound to the core complex of a type IV secretion system. Proc Natl Acad Sci U S A 109:11348–11353. 10.1073/pnas.1201428109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Furuya N, Komano T. 1994. Surface exclusion gene of IncI1 plasmid R64: nucleotide sequence and analysis of deletion mutants. Plasmid 32:80–84. 10.1006/plas.1994.1047. [DOI] [PubMed] [Google Scholar]
- 110.Furuya N, Komano T. 1991. Determination of the nick site at oriT of IncI1 plasmid R64: global similarity of oriT structures of IncI1 and IncP plasmids. J Bacteriol 173:6612–6617. 10.1128/jb.173.20.6612-6617.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Furuya N, Komano T. 1995. Specific binding of the NikA protein to one arm of 17-base-pair inverted repeat sequences within the oriT region of plasmid R64. J Bacteriol 177:46–51. 10.1128/jb.177.1.46-51.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yoshida H, Furuya N, Lin YJ, Guntert P, Komano T, Kainosho M. 2008. Structural basis of the role of the NikA ribbon-helix-helix domain in initiating bacterial conjugation. J Mol Biol 384:690–701. 10.1016/j.jmb.2008.09.067. [DOI] [PubMed] [Google Scholar]
- 113.Narahara K, Rahman E, Furuya N, Komano T. 1997. Requirement of a limited segment of the sog gene for plasmid R64 conjugation. Plasmid 38:1–11. 10.1006/plas.1997.1297. [DOI] [PubMed] [Google Scholar]
- 114.Merryweather A, Rees CE, Smith NM, Wilkins BM. 1986. Role of sog polypeptides specified by plasmid ColIb-P9 and their transfer between conjugating bacteria. EMBO J 5:3007–3012. 10.1002/j.1460-2075.1986.tb04599.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kim SR, Komano T. 1997. The plasmid R64 thin pilus identified as a type IV pilus. J Bacteriol 179:3594–3603. 10.1128/jb.179.11.3594-3603.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yoshida T, Furuya N, Ishikura M, Isobe T, Haino-Fukushima K, Ogawa T, Komano T. 1998. Purification and characterization of thin pili of IncI1 plasmids ColIb-P9 and R64: formation of PilV-specific cell aggregates by type IV pili. J Bacteriol 180:2842–2848. 10.1128/JB.180.11.2842-2848.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shimoda E, Muto T, Horiuchi T, Furuya N, Komano T. 2008. Novel class of mutations of pilS mutants, encoding plasmid R64 type IV prepilin: interface of PilS-PilV interactions. J Bacteriol 190:1202–1208. 10.1128/JB.01204-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gyohda A, Furuya N, Ishiwa A, Zhu S, Komano T. 2004. Structure and function of the shufflon in plasmid R64. Adv Biophys 38:183–213. 10.1016/S0065-227X(04)80166-7. [DOI] [PubMed] [Google Scholar]
- 119.Ishiwa A, Komano T. 2000. The lipopolysaccharide of recipient cells is a specific receptor for PilV proteins, selected by shufflon DNA rearrangement, in liquid matings with donors bearing the R64 plasmid. Mol Gen Genet 263:159–164. 10.1007/s004380050043. [DOI] [PubMed] [Google Scholar]
- 120.Ishiwa A, Komano T. 2003. Thin pilus PilV adhesins of plasmid R64 recognize specific structures of the lipopolysaccharide molecules of recipient cells. J Bacteriol 185:5192–5199. 10.1128/jb.185.17.5192-5199.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Komano T. 1999. Shufflons: multiple inversion systems and integrons. Annu Rev Genet 33:171–191. 10.1146/annurev.genet.33.1.171. [DOI] [PubMed] [Google Scholar]
- 122.Komano T, Kubo A, Kayanuma T, Furuichi T, Nisioka T. 1986. Highly mobile DNA segment of IncI alpha plasmid R64: a clustered inversion region. J Bacteriol 165:94–100. 10.1128/jb.165.1.94-100.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kubo A, Kusukawa A, Komano T. 1988. Nucleotide sequence of the rci gene encoding shufflon-specific DNA recombinase in the IncI1 plasmid R64: homology to the site-specific recombinases of integrase family. Mol Gen Genet 213:30–35. 10.1007/BF00333394. [DOI] [PubMed] [Google Scholar]
- 124.Brouwer MS, Tagg KA, Mevius DJ, Iredell JR, Bossers A, Smith HE, Partridge SR. 2015. IncI shufflons: assembly issues in the next-generation sequencing era. Plasmid 80:111–117. 10.1016/j.plasmid.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 125.Brouwer MSM, Jurburg SD, Harders F, Kant A, Mevius DJ, Roberts AP, Bossers A. 2019. The shufflon of IncI1 plasmids is rearranged constantly during different growth conditions. Plasmid 102:51–55. 10.1016/j.plasmid.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 126.Sakai D, Komano T. 2002. Genes required for plasmid R64 thin-pilus biogenesis: identification and localization of products of the pilK, pilM, pilO, pilP, pilR, and pilT genes. J Bacteriol 184:444–451. 10.1128/jb.184.2.444-451.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Craig L, Forest KT, Maier B. 2019. Type IV pili: dynamics, biophysics and functional consequences. Nat Rev Microbiol 17:429–440. 10.1038/s41579-019-0195-4. [DOI] [PubMed] [Google Scholar]
- 128.Sakai D, Komano T. 2000. The pilL and pilN genes of IncI1 plasmids R64 and ColIb-P9 encode outer membrane lipoproteins responsible for thin pilus biogenesis. Plasmid 43:149–152. 10.1006/plas.1999.1434. [DOI] [PubMed] [Google Scholar]
- 129.Gilbert DN, Chambers HF, Eliopoulos GM, Saag MS, Pavia AT. 2019. The Sanford guide to antimicrobial therapy 2019. 50 years: 1969–2019. Antimicrobial Therapy, Inc, Hyde Park, VT. [Google Scholar]
- 130.DuPont HL. 2015. Bacillary dysentery: Shigella and enteroinvasive Escherichia coli, p 2569–2574. In Bennett JE, Dolin R, Blaser MJ (), Mandell, Douglas, and Bennett’s principles and practice of infectious diseases, 8th ed. Elsevier Saunders, Philadelphia, PA. [Google Scholar]
- 131.Pegues DA, Miller SI. 2015. Salmonella Species, p 2559–2568. In Bennett JE, Dolin R, Blaser MJ (), Mandell, Douglas, and Bennett’s principles and practice of infectious diseases, 8th ed. Elsevier Saunders, Philadelphia, PA. [Google Scholar]
- 132.Folster JP, Grass JE, Bicknese A, Taylor J, Friedman CR, Whichard JM. 2017. Characterization of resistance genes and plasmids from outbreaks and illness clusters caused by Salmonella resistant to ceftriaxone in the United States, 2011–2012. Microb Drug Resist 23:188–193. 10.1089/mdr.2016.0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Cloeckaert A, Praud K, Doublet B, Bertini A, Carattoli A, Butaye P, Imberechts H, Bertrand S, Collard JM, Arlet G, Weill FX. 2007. Dissemination of an extended-spectrum-beta-lactamase blaTEM-52 gene-carrying IncI1 plasmid in various Salmonella enterica serovars isolated from poultry and humans in Belgium and France between 2001 and 2005. Antimicrob Agents Chemother 51:1872–1875. 10.1128/AAC.01514-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bush K. 2003. B-lactam antibiotics: penicillins, p 224–259. In Greenwood D, Whitley RJ, Finch RG, Greenwood D, Norrby SR, Whitley RJ (), Antibiotic and chemotherapy anti-infective agents and their use in therapy, vol 8. Churchill Livingstone, London. [Google Scholar]
- 135.Jacoby GA, Han P. 1996. Detection of extended-spectrum beta-lactamases in clinical isolates of Klebsiella pneumoniae and Escherichia coli. J Clin Microbiol 34:908–911. 10.1128/JCM.34.4.908-911.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Doumith M, Dhanji H, Ellington MJ, Hawkey P, Woodford N. 2012. Characterization of plasmids encoding extended-spectrum beta-lactamases and their addiction systems circulating among Escherichia coli clinical isolates in the UK. J Antimicrob Chemother 67:878–885. 10.1093/jac/dkr553. [DOI] [PubMed] [Google Scholar]
- 137.Abgottspon H, Stephan R, Bagutti C, Brodmann P, Hachler H, Zurfluh K. 2014. Characteristics of extended-spectrum cephalosporin-resistant Escherichia coli isolated from Swiss and imported poultry meat. J Food Prot 77:112–115. 10.4315/0362-028X.JFP-13-120. [DOI] [PubMed] [Google Scholar]
- 138.Abraham S, Kirkwood RN, Laird T, Saputra S, Mitchell T, Singh M, Linn B, Abraham RJ, Pang S, Gordon DM, Trott DJ, O’Dea M. 2018. Dissemination and persistence of extended-spectrum cephalosporin-resistance encoding IncI1-blaCTXM-1 plasmid among Escherichia coli in pigs. ISME J 12:2352–2362. 10.1038/s41396-018-0200-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Agerso Y, Jensen JD, Hasman H, Pedersen K. 2014. Spread of extended spectrum cephalosporinase-producing Escherichia coli clones and plasmids from parent animals to broilers and to broiler meat in a production without use of cephalosporins. Foodborne Pathog Dis 11:740–746. 10.1089/fpd.2014.1742. [DOI] [PubMed] [Google Scholar]
- 140.Arvand M, Bettge-Weller G, Fruth A, Uphoff H, Pfeifer Y. 2015. Extended-spectrum beta-lactamase-producing Shiga toxin gene (stx1)-positive Escherichia coli O91:H14 carrying blaCTX-M-15 on an IncI1-ST31 plasmid isolated from a human patient in Germany. Int J Med Microbiol 305:404–407. 10.1016/j.ijmm.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 141.Ben Sallem R, Ben Slama K, Rojo-Bezares B, Porres-Osante N, Jouini A, Klibi N, Boudabous A, Saenz Y, Torres C. 2014. IncI1 plasmids carrying bla(CTX-M-1) or bla(CMY-2) genes in Escherichia coli from healthy humans and animals in Tunisia. Microb Drug Resist 20:495–500. 10.1089/mdr.2013.0224. [DOI] [PubMed] [Google Scholar]
- 142.Berger M, Berger P, Denamur E, Mellmann A, Dobrindt U. 2018. Core elements of the vegetative replication control of the Inc1 plasmid pO104_90 of Escherichia coli O104:H4 also regulate its transfer frequency. Int J Med Microbiol 308:962–968. 10.1016/j.ijmm.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 143.Cloeckaert A, Praud K, Lefevre M, Doublet B, Pardos de la Gandara M, Granier SA, Brisabois A, Weill FX. 2010. IncI1 plasmid carrying extended-spectrum-beta-lactamase gene blaCTX-M-1 in Salmonella enterica isolates from poultry and humans in France, 2003 to 2008. Antimicrob Agents Chemother 54:4484–4486. 10.1128/AAC.00460-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Cortes-Cortes G, Lozano-Zarain P, Torres C, Castaneda M, Sanchez GM, Alonso CA, Lopez-Pliego L, Mayen MG, Martinez-Laguna Y, Rocha-Gracia RC. 2016. Detection and molecular characterization of Escherichia coli strains producers of extended-spectrum and CMY-2 type beta-lactamases, isolated from turtles in Mexico. Vector Borne Zoonotic Dis 16:595–603. 10.1089/vbz.2014.1725. [DOI] [PubMed] [Google Scholar]
- 145.Curiao T, Canton R, Garcillan-Barcia MP, de la Cruz F, Baquero F, Coque TM. 2011. Association of composite IS26-sul3 elements with highly transmissible IncI1 plasmids in extended-spectrum-beta-lactamase-producing Escherichia coli clones from humans. Antimicrob Agents Chemother 55:2451–2457. 10.1128/AAC.01448-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Dhanji H, Doumith M, Hope R, Livermore DM, Woodford N. 2011. ISEcp1-mediated transposition of linked blaCTX-M-3 and blaTEM-1b from the IncI1 plasmid pEK204 found in clinical isolates of Escherichia coli from Belfast, UK. J Antimicrob Chemother 66:2263–2265. 10.1093/jac/dkr310. [DOI] [PubMed] [Google Scholar]
- 147.Ferreira JC, Penha Filho RA, Andrade LN, Berchieri A, Jr, Darini AL. 2014. IncI1/ST113 and IncI1/ST114 conjugative plasmids carrying blaCTX-M-8 in Escherichia coli isolated from poultry in Brazil. Diagn Microbiol Infect Dis 80:304–306. 10.1016/j.diagmicrobio.2014.09.012. [DOI] [PubMed] [Google Scholar]
- 148.Fischer EA, Dierikx CM, van Essen-Zandbergen A, van Roermund HJ, Mevius DJ, Stegeman A, Klinkenberg D. 2014. The IncI1 plasmid carrying the blaCTX-M-1 gene persists in in vitro culture of a Escherichia coli strain from broilers. BMC Microbiol 14:77. 10.1186/1471-2180-14-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Handel N, Otte S, Jonker M, Brul S, ter Kuile BH. 2015. Factors that affect transfer of the IncI1 beta-lactam resistance plasmid pESBL-283 between E. coli strains. PLoS One 10:e0123039. 10.1371/journal.pone.0123039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Irrgang A, Hammerl JA, Falgenhauer L, Guiral E, Schmoger S, Imirzalioglu C, Fischer J, Guerra B, Chakraborty T, Kasbohrer A. 2018. Diversity of CTX-M-1-producing E. coli from German food samples and genetic diversity of the blaCTX-M-1 region on IncI1 ST3 plasmids. Vet Microbiol 221:98–104. 10.1016/j.vetmic.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 151.Jones-Dias D, Manageiro V, Canica M. 2016. Influence of agricultural practice on mobile bla genes: IncI1-bearing CTX-M, SHV, CMY and TEM in Escherichia coli from intensive farming soils. Environ Microbiol 18:260–272. 10.1111/1462-2920.13021. [DOI] [PubMed] [Google Scholar]
- 152.Kameyama M, Chuma T, Yokoi T, Yabata J, Tominaga K, Miyasako D, Iwata H, Okamoto K. 2012. Emergence of Salmonella enterica serovar infantis harboring IncI1 plasmid with bla(CTX-M-14) in a broiler farm in Japan. J Vet Med Sci 74:1213–1216. 10.1292/jvms.11-0488. [DOI] [PubMed] [Google Scholar]
- 153.Kim JS, Kim J, Jeon SE, Kim SJ, Kim NO, Hong S, Kang YH, Han S, Chung GT. 2014. Complete nucleotide sequence of the IncI1 plasmid pSH4469 encoding CTX-M-15 extended-spectrum beta-lactamase in a clinical isolate of Shigella sonnei from an outbreak in the Republic of Korea. Int J Antimicrob Agents 44:533–537. 10.1016/j.ijantimicag.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 154.Madec JY, Doublet B, Ponsin C, Cloeckaert A, Haenni M. 2011. Extended-spectrum beta-lactamase blaCTX-M-1 gene carried on an IncI1 plasmid in multidrug-resistant Salmonella enterica serovar Typhimurium DT104 in cattle in France. J Antimicrob Chemother 66:942–944. 10.1093/jac/dkr014. [DOI] [PubMed] [Google Scholar]
- 155.Madec JY, Haenni M, Ponsin C, Kieffer N, Rion E, Gassilloud B. 2016. Sequence type 48 Escherichia coli carrying the blaCTX-M-1 IncI1/ST3 plasmid in drinking water in France. Antimicrob Agents Chemother 60:6430–6432. 10.1128/AAC.01135-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Pan YS, Zong ZY, Yuan L, Du XD, Huang H, Zhong XH, Hu GZ. 2016. Complete sequence of pEC012, a multidrug-resistant IncI1 ST71 plasmid carrying blaCTX-M-65, rmtB, fosA3, floR, and oqxAB in an avian Escherichia coli ST117 strain. Front Microbiol 7:1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zurfluh K, Wang J, Klumpp J, Nuesch-Inderbinen M, Fanning S, Stephan R. 2014. Vertical transmission of highly similar blaCTX-M-1-harboring IncI1 plasmids in Escherichia coli with different MLST types in the poultry production pyramid. Front Microbiol 5:519. 10.3389/fmicb.2014.00519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Nordmann P. 1998. Trends in beta-lactam resistance among Enterobacteriaceae. Clin Infect Dis 27 Suppl 1:S100–S106. 10.1086/514905. [DOI] [PubMed] [Google Scholar]
- 159.Chong Y, Ito Y, Kamimura T. 2011. Genetic evolution and clinical impact in extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Infect Genet Evol 11:1499–1504. 10.1016/j.meegid.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 160.Chong Y, Shimoda S, Shimono N. 2018. Current epidemiology, genetic evolution and clinical impact of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Infect Genet Evol 61:185–188. 10.1016/j.meegid.2018.04.005. [DOI] [PubMed] [Google Scholar]
- 161.Lahlaoui H, Ben Haj Khalifa A, Ben Moussa M. 2014. Epidemiology of Enterobacteriaceae producing CTX-M type extended spectrum beta-lactamase (ESBL). Med Mal Infect 44:400–404. 10.1016/j.medmal.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 162.Accogli M, Fortini D, Giufre M, Graziani C, Dolejska M, Carattoli A, Cerquetti M. 2013. IncI1 plasmids associated with the spread of CMY-2, CTX-M-1 and SHV-12 in Escherichia coli of animal and human origin. Clin Microbiol Infect 19:E238–240. 10.1111/1469-0691.12128. [DOI] [PubMed] [Google Scholar]
- 163.Alonso CA, Michael GB, Li J, Somalo S, Simon C, Wang Y, Kaspar H, Kadlec K, Torres C, Schwarz S. 2017. Analysis of blaSHV-12-carrying Escherichia coli clones and plasmids from human, animal and food sources. J Antimicrob Chemother 72:1589–1596. 10.1093/jac/dkx024. [DOI] [PubMed] [Google Scholar]
- 164.Feng Y, Yang P, Xie Y, Wang X, McNally A, Zong Z. 2015. Escherichia coli of sequence type 3835 carrying blaNDM-1, blaCTX-M-15, blaCMY-42 and blaSHV-12. Sci Rep 5:12275. 10.1038/srep12275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Haenni M, Saras E, Metayer V, Doublet B, Cloeckaert A, Madec JY. 2012. Spread of the blaTEM-52 gene is mainly ensured by IncI1/ST36 plasmids in Escherichia coli isolated from cattle in France. J Antimicrob Chemother 67:2774–2776. 10.1093/jac/dks282. [DOI] [PubMed] [Google Scholar]
- 166.Bauernfeind A, Grimm H, Schweighart S. 1990. A new plasmidic cefotaximase in a clinical isolate of Escherichia coli. Infection 18:294–298. 10.1007/BF01647010. [DOI] [PubMed] [Google Scholar]
- 167.Rodriguez-Bano J, Navarro MD, Romero L, Muniain MA, de Cueto M, Rios MJ, Hernandez JR, Pascual A. 2006. Bacteremia due to extended-spectrum β-lactamase-producing Escherichia coli in the CTX-M era: a new clinical challenge. Clin Infect Dis 43:1407–1414. 10.1086/508877. [DOI] [PubMed] [Google Scholar]
- 168.Hopkins KL, Liebana E, Villa L, Batchelor M, Threlfall EJ, Carattoli A. 2006. Replicon typing of plasmids carrying CTX-M or CMY beta-lactamases circulating among Salmonella and Escherichia coli isolates. Antimicrob Agents Chemother 50:3203–3206. 10.1128/AAC.00149-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Rodriguez I, Barownick W, Helmuth R, Mendoza MC, Rodicio MR, Schroeter A, Guerra B. 2009. Extended-spectrum {beta}-lactamases and AmpC {beta}-lactamases in ceftiofur-resistant Salmonella enterica isolates from food and livestock obtained in Germany during 2003–07. J Antimicrob Chemother 64:301–309. 10.1093/jac/dkp195. [DOI] [PubMed] [Google Scholar]
- 170.Garcia-Fernandez A, Cloeckaert A, Bertini A, Praud K, Doublet B, Weill FX, Carattoli A. 2007. Comparative analysis of IncHI2 plasmids carrying blaCTX-M-2 or blaCTX-M-9 from Escherichia coli and Salmonella enterica strains isolated from poultry and humans. Antimicrob Agents Chemother 51:4177–4180. 10.1128/AAC.00603-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Marcade G, Deschamps C, Boyd A, Gautier V, Picard B, Branger C, Denamur E, Arlet G. 2009. Replicon typing of plasmids in Escherichia coli producing extended-spectrum beta-lactamases. J Antimicrob Chemother 63:67–71. 10.1093/jac/dkn428. [DOI] [PubMed] [Google Scholar]
- 172.Chouchani C, El Salabi A, Marrakchi R, Ferchichi L, Walsh TR. 2012. Characterization of IncA/C conjugative plasmid harboring blaTEM-52 and blaCTX-M-15 extended-spectrum beta-lactamases in clinical isolates of Escherichia coli in Tunisia. Eur J Clin Microbiol Infect Dis 31:1081–1087. 10.1007/s10096-011-1410-z. [DOI] [PubMed] [Google Scholar]
- 173.Zhang L, Lu X, Zong Z. 2013. The emergence of blaCTX-M-15-carrying Escherichia coli of ST131 and new sequence types in Western China. Ann Clin Microbiol Antimicrob 12:35. 10.1186/1476-0711-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Cao G, Allard M, Hoffmann M, Muruvanda T, Luo Y, Payne J, Meng K, Zhao S, McDermott P, Brown E, Meng J. 2018. Sequence analysis of IncA/C and IncI1 plasmids isolated from multidrug-resistant Salmonella Newport using single-molecule real-time sequencing. Foodborne Pathog Dis 15:361–371. 10.1089/fpd.2017.2385. [DOI] [PubMed] [Google Scholar]
- 175.Folster JP, Tolar B, Pecic G, Sheehan D, Rickert R, Hise K, Zhao S, Fedorka-Cray PJ, McDermott P, Whichard JM. 2014. Characterization of blaCMY plasmids and their possible role in source attribution of Salmonella enterica serotype Typhimurium infections. Foodborne Pathog Dis 11:301–306. 10.1089/fpd.2013.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ingti B, Saikia P, Paul D, Maurya AP, Dhar Chanda D, Chakravarty A, Deshamukhya C, Bhattacharjee A. 2018. Occurrence of blaCMY-42 on an IncI1 plasmid in multidrug-resistant Escherichia coli from a tertiary referral hospital in India. J Glob Antimicrob Resist 14:78–82. 10.1016/j.jgar.2018.02.021. [DOI] [PubMed] [Google Scholar]
- 177.Sellera FP, Fernandes MR, Moura Q, Lopes RB, Souza TA, Cerdeira L, Lincopan N. 2018. Draft genome sequence of a blaCMY-2/IncI1-harbouring Escherichia coli D:ST457 isolated from coastal benthic organisms. J Glob Antimicrob Resist 14:83–84. 10.1016/j.jgar.2018.06.010. [DOI] [PubMed] [Google Scholar]
- 178.Sidjabat HE, Seah KY, Coleman L, Sartor A, Derrington P, Heney C, Faoagali J, Nimmo GR, Paterson DL. 2014. Expansive spread of IncI1 plasmids carrying blaCMY-2 amongst Escherichia coli. Int J Antimicrob Agents 44:203–208. 10.1016/j.ijantimicag.2014.04.016. [DOI] [PubMed] [Google Scholar]
- 179.Tiba-Casas MR, Camargo CH, Soares FB, Doi Y, Fernandes SA. 2018. Emergence of CMY-2-producing Salmonella Heidelberg associated with IncI1 plasmids isolated from poultry in Brazil. Microb Drug Resist 25:271–276. 10.1089/mdr.2018.0044. [DOI] [PubMed] [Google Scholar]
- 180.Liebana E, Carattoli A, Coque TM, Hasman H, Magiorakos AP, Mevius D, Peixe L, Poirel L, Schuepbach-Regula G, Torneke K, Torren-Edo J, Torres C, Threlfall J. 2013. Public health risks of enterobacterial isolates producing extended-spectrum beta-lactamases or AmpC beta-lactamases in food and food-producing animals: an EU perspective of epidemiology, analytical methods, risk factors, and control options. Clin Infect Dis 56:1030–1037. 10.1093/cid/cis1043. [DOI] [PubMed] [Google Scholar]
- 181.Tagg KA, Iredell JR, Partridge SR. 2014. Complete sequencing of IncI1 sequence type 2 plasmid pJIE512b indicates mobilization of blaCMY-2 from an IncA/C plasmid. Antimicrob Agents Chemother 58:4949–4952. 10.1128/AAC.02773-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Yassine H, Bientz L, Cros J, Goret J, Bebear C, Quentin C, Arpin C. 2015. Experimental evidence for IS1294b-mediated transposition of the blaCMY-2 cephalosporinase gene in Enterobacteriaceae. J Antimicrob Chemother 70:697–700. 10.1093/jac/dku472. [DOI] [PubMed] [Google Scholar]
- 183.Kao CY, Chen JW, Liu TL, Yan JJ, Wu JJ. 2018. Comparative genomics of Escherichia coli sequence type 219 clones from the same patient: evolution of the IncI1 blaCMY-carrying plasmid in vivo. Front Microbiol 9:1518. 10.3389/fmicb.2018.01518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Piazza A, Comandatore F, Romeri F, Pagani C, Mattioni Marchetti V, Brilli M, Panelli S, Migliavacca R, Ridolfo A, Olivieri P, Gismondo MR, Bandi C, Rimoldi SG. 2018. Detection of ST1702 Escherichia coli blaNDM-5 and blaCMY-42 genes positive isolates from a Northern Italian hospital. New Microbiol 41:230–231. [PubMed] [Google Scholar]
- 185.Wu S, Dalsgaard A, Hammerum AM, Porsbo LJ, Jensen LB. 2010. Prevalence and characterization of plasmids carrying sulfonamide resistance genes among Escherichia coli from pigs, pig carcasses and human. Acta Vet Scand 52:47. 10.1186/1751-0147-52-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Freire Martin I, AbuOun M, Reichel R, La Ragione RM, Woodward MJ. 2014. Sequence analysis of a CTX-M-1 IncI1 plasmid found in Salmonella 4,5,12:i:-, Escherichia coli and Klebsiella pneumoniae on a UK pig farm. J Antimicrob Chemother 69:2098–2101. 10.1093/jac/dku098. [DOI] [PubMed] [Google Scholar]
- 187.Valcek A, Roer L, Overballe-Petersen S, Hansen F, Bortolaia V, Leekitcharoenphon P, Korsgaard HB, Seyfarth AM, Hendriksen RS, Hasman H, Hammerum AM. 2019. IncI1 ST3 and IncI1 ST7 plasmids from CTX-M-1-producing Escherichia coli obtained from patients with bloodstream infections are closely related to plasmids from E. coli of animal origin. J Antimicrob Chemother 74:2171–2175. 10.1093/jac/dkz199. [DOI] [PubMed] [Google Scholar]
- 188.Dolejska M, Papagiannitsis CC. 2018. Plasmid-mediated resistance is going wild. Plasmid 99:99–111. 10.1016/j.plasmid.2018.09.010. [DOI] [PubMed] [Google Scholar]
- 189.Szmolka A, Lestar B, Paszti J, Fekete P, Nagy B. 2015. Conjugative IncF and IncI1 plasmids with tet(A) and class 1 integron conferring multidrug resistance in F18(+) porcine enterotoxigenic E. coli. Acta Vet Hung 63:425–443. 10.1556/004.2015.040. [DOI] [PubMed] [Google Scholar]
- 190.Kaldhone P, Nayak R, Lynne AM, David DE, McDermott PF, Logue CM, Foley SL. 2008. Characterization of Salmonella enterica serovar Heidelberg from Turkey-associated sources. Appl Environ Microbiol 74:5038–5046. 10.1128/AEM.00409-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Johnson TJ, Logue CM, Johnson JR, Kuskowski MA, Sherwood JS, Barnes HJ, DebRoy C, Wannemuehler YM, Obata-Yasuoka M, Spanjaard L, Nolan LK. 2012. Associations between multidrug resistance, plasmid content, and virulence potential among extraintestinal pathogenic and commensal Escherichia coli from humans and poultry. Foodborne Pathog Dis 9:37–46. 10.1089/fpd.2011.0961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Webb SA, Kahler CM. 2008. Bench-to-bedside review: bacterial virulence and subversion of host defences. Crit Care 12:234. 10.1186/cc7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Cross AS. 2008. What is a virulence factor? Crit Care 12:196. 10.1186/cc7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Baumler AJ, Sperandio V. 2016. Interactions between the microbiota and pathogenic bacteria in the gut. Nature 535:85–93. 10.1038/nature18849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Ayala FJ, Krane DE, Hartl DL. 1994. Genetic variation in IncI1-ColIb plasmids. J Mol Evol 39:129–133. 10.1007/BF00163801. [DOI] [PubMed] [Google Scholar]
- 196.Nedialkova LP, Denzler R, Koeppel MB, Diehl M, Ring D, Wille T, Gerlach RG, Stecher B. 2014. Inflammation fuels colicin Ib-dependent competition of Salmonella serovar Typhimurium and E. coli in enterobacterial blooms. PLoS Pathog 10:e1003844. 10.1371/journal.ppat.1003844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Spriewald S, Glaser J, Beutler M, Koeppel MB, Stecher B. 2015. Reporters for single-cell analysis of colicin Ib expression in Salmonella enterica serovar Typhimurium. PLoS One 10:e0144647. 10.1371/journal.pone.0144647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Guterman SK. 1973. Colicin B: mode of action and inhibition by enterochelin. J Bacteriol 114:1217–1224. 10.1128/JB.114.3.1217-1224.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Dowden SB, Glazebrook JA, Strike P. 1984. UV inducible UV protection and mutation functions on the I group plasmid TP110. Mol Gen Genet 193:316–321. 10.1007/BF00330687. [DOI] [PubMed] [Google Scholar]
- 200.Sedgwick SG, Thomas SM, Hughes VM, Lodwick D, Strike P. 1989. Mutagenic DNA repair genes on plasmids from the 'pre-antibiotic era'. Mol Gen Genet 218:323–329. 10.1007/BF00331285. [DOI] [PubMed] [Google Scholar]
- 201.Strike P, Lodwick D. 1987. Plasmid genes affecting DNA repair and mutation. J Cell Sci Suppl 6:303–321. 10.1242/jcs.1984.supplement_6.20. [DOI] [PubMed] [Google Scholar]
- 202.Runyen-Janecky LJ, Hong M, Payne SM. 1999. The virulence plasmid-encoded impCAB operon enhances survival and induced mutagenesis in Shigella flexneri after exposure to UV radiation. Infect Immun 67:1415–1423. 10.1128/IAI.67.3.1415-1423.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Walker GC. 1984. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Rev 48:60–93. 10.1128/MR.48.1.60-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Shinagawa H, Iwasaki H, Kato T, Nakata A. 1988. RecA protein-dependent cleavage of UmuD protein and SOS mutagenesis. Proc Natl Acad Sci U S A 85:1806–1810. 10.1073/pnas.85.6.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Lodwick D, Owen D, Strike P. 1990. DNA sequence analysis of the imp UV protection and mutation operon of the plasmid TP110: identification of a third gene. Nucleic Acids Res 18:5045–5050. 10.1093/nar/18.17.5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Glazebrook JA, Grewal KK, Strike P. 1986. Molecular analysis of the UV protection and mutation genes carried by the I incompatibility group plasmid TP110. J Bacteriol 168:251–256. 10.1128/jb.168.1.251-256.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Yasuda T, Morimatsu K, Horii T, Nagata T, Ohmori H. 1998. Inhibition of Escherichia coli RecA coprotease activities by DinI. EMBO J 17:3207–3216. 10.1093/emboj/17.11.3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Joo LM, Macfarlane-Smith LR, Okeke IN. 2007. Error-prone DNA repair system in enteroaggregative Escherichia coli identified by subtractive hybridization. J Bacteriol 189:3793–3803. 10.1128/JB.01764-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Bernstein C, Bernstein H, Payne CM, Beard SE, Schneider J. 1999. Bile salt activation of stress response promoters in Escherichia coli. Curr Microbiol 39:68–72. 10.1007/s002849900420. [DOI] [PubMed] [Google Scholar]
- 210.Prieto AI, Ramos-Morales F, Casadesus J. 2004. Bile-induced DNA damage in Salmonella enterica. Genetics 168:1787–1794. 10.1534/genetics.104.031062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Guerrant RL, Steiner TS. 2005. Principles and syndromes of enteric infections, p 1215–1230. In Mandell GL, Bennett JE, Dolin R (), Principles and practice of infectious diseases, 6th ed, vol 1. Elsevier Churchill Livingstone, Philadelphia, PA. [Google Scholar]
- 212.Guerrant RL, Van Gilder T, Steiner TS, Thielman NM, Slutsker L, Tauxe RV, Hennessy T, Griffin PM, DuPont H, Sack RB, Tarr P, Neill M, Nachamkin I, Reller LB, Osterholm MT, Bennish ML, Pickering LK, Infectious Diseases Society of A.. 2001. Practice guidelines for the management of infectious diarrhea. Clin Infect Dis 32:331–351. 10.1086/318514. [DOI] [PubMed] [Google Scholar]