Andes orthohantavirus (ANDV) is a New World hantavirus that is a major cause of hantavirus cardiopulmonary syndrome (HCPS; also referred to as hantavirus pulmonary syndrome) in South America, particularly in Chile and Argentina. ANDV is one of the few hantaviruses for which there is a reliable animal model, the Syrian hamster model, which recapitulates important aspects of human disease.
KEYWORDS: orthohantaviruses, disease modeling, pathogenesis
ABSTRACT
Hantavirus cardiopulmonary syndrome (HCPS) is a severe respiratory disease caused by orthohantaviruses in the Americas with a fatality rate as high as 35%. In South America, Andes orthohantavirus (Hantaviridae, Orthohantavirus; ANDV) is a major cause of HCPS, particularly in Chile and Argentina, where thousands of cases have been reported since the virus was discovered. Two strains of ANDV that are classically used for experimental studies of the virus are Chile-9717869, isolated from the natural reservoir, the long-tailed pygmy rice rat, and CHI-7913, an isolate from a lethal human case of HCPS. An important animal model for studying pathogenesis of HCPS is the lethal Syrian golden hamster model of ANDV infection. In this model, ANDV strain Chile-9717869 is uniformly lethal and has been used extensively for pathogenesis, vaccination, and therapeutic studies. Here, we show that the CHI-7913 strain, despite having high sequence similarity with Chile-9717869, does not cause lethal disease in Syrian hamsters. CHI-7913, while being able to infect hamsters and replicate to moderate levels, showed a reduced ability to replicate within the tissues compared with Chile-9717869. Hamsters infected with CHI-7913 had reduced expression of cytokines interleukin-4 (IL-4), IL-6, and gamma interferon compared with Chile-9717869-infected animals, suggesting potentially limited immune-mediated pathology. These results demonstrate that certain ANDV strains may not be lethal in the classical Syrian hamster model of infection, and further exploration into the differences between lethal and nonlethal strains should provide important insights into molecular determinants of pathogenic hantavirus infection.
IMPORTANCE Andes orthohantavirus (ANDV) is a New World hantavirus that is a major cause of hantavirus cardiopulmonary syndrome (HCPS; also referred to as hantavirus pulmonary syndrome) in South America, particularly in Chile and Argentina. ANDV is one of the few hantaviruses for which there is a reliable animal model, the Syrian hamster model, which recapitulates important aspects of human disease. Here, we infected hamsters with a human isolate of ANDV, CHI-7913, to assess its pathogenicity compared with the classical lethal Chile-9717869 strain. CHI-7913 had 22 amino acid differences from Chile-9717869, did not cause lethal disease in hamsters, and showed reduced ability to replicate in vivo. Our data indicate potentially important molecular signatures for the pathogenesis of ANDV infection in hamsters and may lead to insights into what drives the pathogenesis of certain hantaviruses in humans.
INTRODUCTION
Orthohantaviruses (herein referred to as hantaviruses) are a group of viruses in the order Bunyavirales, family Hantaviridae, subfamily Mammantavirinae that are enveloped and contain trisegmented, negative-sense RNA genomes (1). The three genomic segments include a small segment, S, a medium segment, M, and a large segment, L, which encode the nucleocapsid (N) protein and, in some species, a nonstructural protein (NSs), surface glycoproteins (Gn and Gc), and an RNA-dependent RNA polymerase, respectively (1). Hantaviruses are zoonotic pathogens with a worldwide distribution and have been found to be carried mainly by cricetid and murid rodents, including mice, rats, and voles, but they have also been found to be carried by bats, shrews, and moles (2). Those that cause human disease are rodent-borne viruses, and humans become infected following exposure to aerosolized virus in contaminated rodent excreta and/or secreta. Hantaviruses found in Europe and Asia are referred to as Old World hantaviruses, while those found in the Americas are called New World hantaviruses.
Classically, Old and New World hantaviruses are responsible for two distinct diseases in humans; however, more recent evidence suggests significant overlap in the stages and symptoms of disease caused by both groups of viruses. Hemorrhagic fever with renal syndrome (HFRS), caused by Old World viruses, is typified by acute kidney involvement and hemorrhaging (3). While HFRS is generally mild, it can have a fatality rate as high as 15% depending on the causative pathogen, and HFRS cases number in the thousands each year, highlighting a significant health burden in these areas (1). Hantavirus cardiopulmonary syndrome (HCPS), caused by New World hantaviruses in the Americas, is a severe respiratory illness that has a lower incidence than HFRS but has a higher case fatality rate. HCPS is characterized by vascular leakage in the lungs, pulmonary edema, respiratory failure, and cardiogenic shock and can have a fatality rate of up to 35% (4). HCPS disease typically starts with a mild febrile stage following an incubation period of around 2 to 3 weeks. The disease course is rapid after the onset of symptoms, with respiratory symptoms beginning within days of illness onset. Cough, dyspnea, and low blood pressure occur, followed by cyanosis and respiratory failure as early as 5 to 6 days following symptom onset. Patients are typically hospitalized at this stage and often require mechanical ventilation within days of hospital admittance (1). While considered a respiratory disease, renal involvement and hemorrhaging can occur in HCPS patients, highlighting the overlap between HCPS and HFRS disease.
Two of the main causative agents of HCPS in the Americas include Sin Nombre virus (SNV), found in North America, and Andes virus (ANDV), found in South America (5, 6). ANDV is carried by the long-tailed pygmy rice rat (Oligoryzomys longicaudatus) and is responsible for the majority of HCPS cases in Chile and Argentina (7). These countries experience hundreds of HCPS cases, and there are currently no approved vaccines or therapeutics for prevention or treatment of HCPS and ANDV infection. Additionally, ANDV is the only hantavirus for which there is documented evidence of person-to-person transmission, which has been responsible for outbreaks of the virus in South America (8, 9).
The Syrian golden hamster model of HCPS is currently the most well-characterized animal model of hantavirus infection and leads to a uniform and reproducible lethal outcome that recapitulates several aspects of human disease (10, 11). This model has been used extensively to attempt to characterize HCPS as well as test a number of different vaccines and therapeutics (12–17). This classical model relies on ANDV strain Chile-9717869 (herein referred to as Chile-9717869), which was isolated from the rodent reservoir during an outbreak in Chile. Infection of Syrian hamsters with Maporal virus (a closely related New World hantavirus) can also cause lethal disease, albeit in only approximately 30% of infected hamsters (18, 19). Both ANDV Chile-9717869 and Maporal viruses remain lethal in Syrian hamsters following passage on VeroE6 cells. Other strains of ANDV have been isolated and characterized and share high sequence and amino acid identity with Chile-9717869 but have not been assessed in the hamster model for infection/disease characteristics (20). The ANDV isolate CHI-7913 (herein referred to as CHI-7913) was first isolated from a human case of HCPS. The virus was isolated from the serum of a 10-year-old boy who had become infected with ANDV following infection of his grandmother (21). This strain has been used for characterization of antibodies from human cases and for structural analyses of hantaviral glycoproteins, including epitope-mapping studies (22, 23). Pathogenesis studies utilizing the CHI-7913 strain of ANDV in the Syrian hamster model have not been reported.
The exact mechanisms of pathogenesis of HCPS are not well understood, but vascular leakage occurs and is accompanied by an excessive inflammatory immune response, which likely contributes to disease. It is thought that immune-mediated pathology contributes to disease based on T cell phenotypes seen during HCPS as well as an infiltration of antigen-presenting cells into the lungs during infection (24, 25). In the Syrian hamster model of HCPS, an upregulation of proinflammatory cytokines occurs immediately before the onset of clinical signs and deterioration of the infected animals (26). During infection of rodent reservoirs, predominant regulatory T cell responses appear to drive persistent infections characterized by an absence of overt illness, suggesting that inflammatory responses are deleterious during hantavirus infection (27, 28). Interestingly, however, in the Syrian hamster model of HCPS, T cells appear to be dispensable for lethal disease, as T cell depletion does not affect infection outcome (29, 30), and immunosuppression of Syrian hamsters renders them susceptible to lethal SNV infection (31, 32). Detailed immunological analyses between lethal and nonlethal HCPS infections in an animal model have not been reported, and such studies could lead to insights into how the proinflammatory response during infection contributes to disease course.
Molecular determinants of pathogenesis for hantavirus infections have been difficult to study due to the lack of a developed reverse genetics system to reliably produce viruses harboring specific mutations. While ANDV is lethal in Syrian hamsters, wild-type SNV does not cause disease, revealing that differences between the proteins of these viruses are key to pathogenesis in this species. Successful reassortment of ANDV and SNV revealed that the ANDV M segment was not sufficient to produce a lethal outcome in Syrian hamsters; however, more detailed studies have been elusive (33). Furthermore, comparative studies of different strains of ANDV in the Syrian hamster model to attempt to identify key regions of the ANDV genome for pathogenesis have not been performed.
Here, we show that the human isolate of ANDV CHI-7913, which was isolated from a lethal case of HCPS, can infect Syrian golden hamsters but does not cause lethal disease. We compared the sequences of our classically used strain Chile-9717869 and CHI-7913 to attempt to shed light on regions of the ANDV genome that are important for pathogenesis in the Syrian hamster model of HCPS. We also monitored how the immune response develops in hamsters infected with either strain of virus to attempt to determine the immunological determinants of pathogenic ANDV infection.
RESULTS
Genetic comparison of ANDV strains.
We used next-generation sequencing to identify genetic differences between ANDV strains Chile-9717869 and CHI-7913. The sequences obtained from our stocks of both strains of ANDV were greater than 99% similar, but not 100% identical, to the type strain sequences in GenBank, likely due to mutations accumulated through serial passage in tissue culture. We wanted to directly compare the sequences of Chile-9717869 and CHI-7913 to attempt to identify differences in the ANDV genome that may be important for pathogenesis in Syrian hamsters. Following sequencing of both viruses, at the nucleotide level they had a sequence identity of 92.8% in the S segment, 94.6% in the M segment, and 94.0% in the L segment (GenBank accession numbers MT956618 [CHI-7913 S], MT956619 [CHI-9713 M], MT956620 [CHI-9713 L], MT956622 [Chile-9717869 S], MT956623 [Chile-9717869 M], and MT956621 [Chile-9717869 L]). As seen in Table 1, we identified 23 amino acid changes between the two viruses. There were none in the N protein, eight in the coding region of the glycoprotein precursor, and 15 in the coding region of the RNA-dependent RNA polymerase.
TABLE 1.
Single-nucleotide polymorphisms causing amino acid changes observed between Andes virus strains Chile-9717869 and CHI-7913
| Region (n) | Nucleotide position(s) | Sequence for:a |
Amino acid |
||
|---|---|---|---|---|---|
| Chile-9717869 | CHI-7913 | Change | Position | ||
| S segment (0) | |||||
| M segment (8) | 23 | GTT | GCT | V→A | 8 |
| 31 | GTC | ATC | V→I | 11 | |
| 880 | CAC | TAC | H→Y | 294 | |
| 1036 | GTA | ATA | V→I | 346 | |
| 1057, 1058 | ACA | GTA | T→V | 353 | |
| 1609, 1611 | ATT | GTC | I→V | 537 | |
| 2812 | ACA | GCA | T→A | 938 | |
| 3067 | ACA | GCA | T→A | 1023 | |
| L segment (15) | 422 | ACT | ATT | T→I | 141 |
| 541 | ATT | GTT | I→V | 181 | |
| 830 | TTA | TCA | L→S | 277 | |
| 1012 | TCT | GCT | S→A | 338 | |
| 1037 | AGG | AAG | R→K | 346 | |
| 1204 | ATC | GTC | I→V | 402 | |
| 2338 | GAT | AAT | D→N | 780 | |
| 3097 | AAT | GAT | N→D | 1033 | |
| 3754 | GCA | ACA | A→T | 1252 | |
| 3909 | GAT | GAA | D→E | 1303 | |
| 4993 | GTA | ATA | V→I | 1665 | |
| 5249 | AAA | AGA | K→R | 1750 | |
| 5480 | ACA | CCA | T→P | 1828 | |
| 6325 | ATT | GTT | I→V | 2109 | |
| 6337 | ACA | GCA | T→A | 2113 | |
Nucleotides in bold and underlined represent the polymorphismn observed between the two ANDV isolates.
In vitro growth kinetics.
Triplicate analysis of baby hamster kidney (BHK) cells infected at a low multiplicity of infection (MOI) with ANDV strain Chile-9717869 or CHI-7913 revealed an average of 2-log-greater titers in the supernatant of Chile-9717869 infected cells on days 3, 5, and 7 postinfection (Fig. 1).
FIG 1.
In vitro growth kinetics of ANDV strains Chile-971789 and CHI-7913. Nearly confluent baby hamster kidney (BHK) cells were infected in 24-well dishes with either ANDV Chile-9717869 or CHI-7913 at an MOI of 0.001 (100 μl total volume). Inoculum was removed after 1 h, and cells were washed and cultured for 3, 5, or 7 days postinfection, at which point supernatants were collected and analyzed for the presence of ANDV-RNA by real-time RT-qPCR methodologies. Data shown represent average genomic copies per ml with standard errors of the means (SEM).
Pathogenesis of Chile-9717869 and CHI-7913 in Syrian hamsters.
The differing sources of each strain of ANDV brought about the question of whether these strains are equally pathogenic in the Syrian hamster model of HCPS. We sought to determine whether infection of Syrian hamsters with the ANDV strain CHI-7913 produces a similar lethal outcome and disease course as the classically used Chile-9717869 strain. Infection of Syrian hamsters with ANDV via the intranasal (i.n.) route results in a longer disease course compared to intraperitoneal injection while maintaining similar pathogenesis (26). We chose to infect hamsters intranasally with both strains in an attempt to better characterize the kinetics of infection and the immune response generated by hamsters during infection. Interestingly, infection of hamsters with CHI-7913 did not result in lethal disease (Fig. 2A). Infection of hamsters with Chile-9717869 resulted in 100% lethality, with all hamsters succumbing to infection by 12 days postinfection (dpi) (Fig. 2A). Animals infected with CHI-7913 also did not show any clinical signs of disease that are noticed during HCPS in hamsters, such as hunched posture, ruffled fur, inactivity, and labored breathing.
FIG 2.
Infection of hamsters with ANDV strains Chile-9717869 and CHI-7913. Groups of Syrian hamsters were infected with either ANDV strain CHI-7913 or Chile-9717869 or DMEM placebo control. (A) Survival of animals infected with either virus (n = 6/strain). (B) Weights of hamsters infected with ANDV (n = 6/strain) or DMEM control (n = 2) throughout the course of infection. The exception is the day 13 ANDV 9717869 infection group, which only had two survivors remaining. (C) Lung weights of hamsters infected with ANDV or DMEM (n = 6/infection or 2 for DMEM controls) throughout the course of infection. (D) Lung/body weight ratios of hamsters infected with ANDV throughout the course of infection. Reported are data means with SEM. Significance in panel A was assessed using the log rank (Mantel Cox) test.
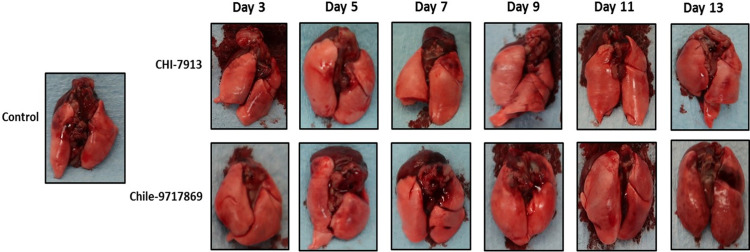
To attempt to better characterize the disease course in both groups, we performed a serial sample study to examine both groups of animals at various time points throughout the disease course. Groups of six animals were euthanized on days 3, 5, 7, 9, 11, and 13 with an anticipated mean time to death of 12 to 14 days. Hamsters infected with ANDV typically do not suffer from weight loss throughout the disease course, and animals from both groups in our experiment did not lose weight until those in the Chile-9717869 group lost weight perimortem (Fig. 2B). Our group has also previously shown that the weight of the lungs of infected hamsters can be an important indicator of disease outcome, specifically the lung/body weight ratio (26). This ratio is affected by cellular lung infiltration and fluid build-up as a part of the pathology of HCPS. Here, the weight of sets of lungs from hamsters in each group did not differ from each other or control animals given Dulbecco’s modified Eagle medium (DMEM) (Fig. 2C). Interestingly, the lung/body weight ratio also did not differ between groups or between either group and control animals (Fig. 2D). Therefore, it remains to be seen whether this is a predictive indicator of disease severity. We also examined the heart and lungs from each group for pathology throughout infection. Noticeable signs of pathology were present in the heart and lungs of hamsters in the Chile-9717869 group, including focally extensive areas of consolidation and hyperemia (Fig. 3). Overall, the number of animals that had visible lesions or damage to the lungs did not appear to differ significantly between the groups until day 13, when the extent of gross pathological lung damage appeared to be significantly higher in the Chile-9717869 animals (Fig. 3). Additionally, we performed histological examinations of the lungs in each group of hamsters. Hematoxylin and eosin staining did not reveal any significantly noticeable signs of pathology in either group and no clear differences between groups that could distinguish between infections with either virus (Fig. 4).
FIG 3.
Gross pathology in hamsters infected with ANDV strains Chile-9717869 and CHI-7913. Following infection with ANDV strain CHI-7913 or Chile-9717869, hamsters were serially euthanized beginning on day 3 postinfection. Macroscopic identification of lesions in the lungs of infected hamsters was performed. Representative heart and lungs from infected hamsters are depicted.
FIG 4.
Histopathology of lungs in ANDV-infected hamsters. Hamsters were infected with ANDV Chile 9717869 or CHI-7913 via the intranasal route. Hematoxylin and eosin staining was performed on the lungs of Chile-9717869- and CHI-7913-infected hamsters taken on day 11 (n = 6/group) and day 13 (n = 6 for CHI-7913 or 2 for Chile-9717869) postinfection. Representative sections are shown. Scale bar, 200 μM.
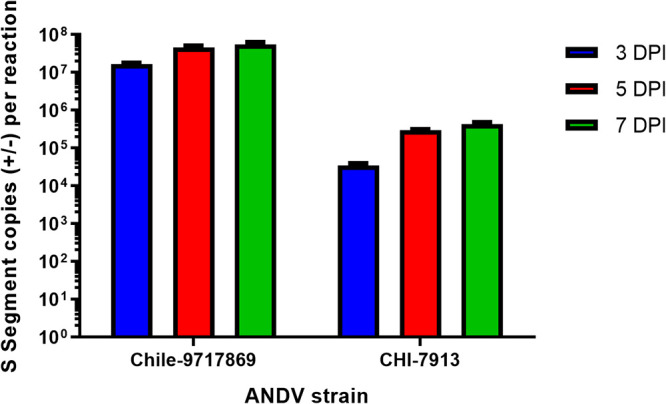
Viral RNA levels during Chile-9717869 and CHI-7913 infection.
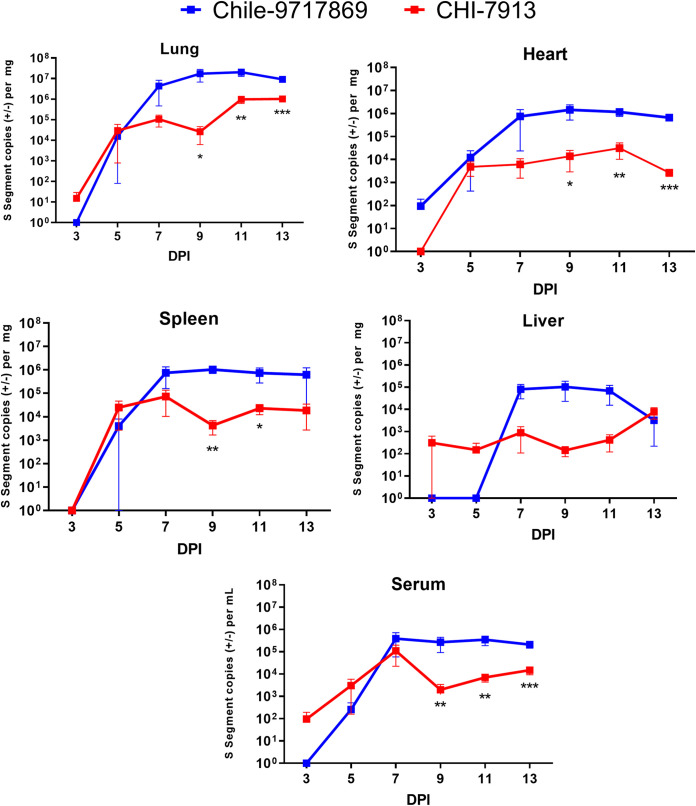
We wanted to determine whether both viruses replicate to similar levels during infection. We collected serum, lungs, heart, liver, and spleen from infected animals and extracted viral RNA for real-time quantitative PCR (RT-qPCR) analyses of viral S segment copy number. Early during infection, up to day 5, both viruses replicated to similar levels in all tissues examined, except for the liver, which did not have detectable Chile-9717869 RNA until day 7 (Fig. 5). Following day 5, however, Chile-9717869 RNA levels began to replicate to consistently higher levels than CHI-7913 in all tissues. During the later stages of infection from 9 dpi onward, animals infected with Chile-9717869 had viral RNA copy numbers that were consistently two logs higher than those infected with CHI-7913 (Fig. 5). Significantly higher levels of Chile-9717869 viral RNA were seen in all tissues examined after day 7 with the exception of the liver, which appeared to also have higher levels of RNA, but these differences were not statistically significant (Fig. 5). Differences in viral RNA copy number between the two viruses reflect some replicative attenuation of the CHI-7913 strain in Syrian hamsters. Surprisingly, however, CHI-7913 replicated to high levels in the lungs of infected hamsters, reaching greater than 106 copies/mg 11 dpi onward in half the animals tested and just below that number in the remaining animals (Fig. 5). That the virus is able to replicate to high levels in the primary affected organ despite little pathology or disease suggests a varied host response to CHI-7913 infection compared to Chile-9717869.
FIG 5.
Viral RNA levels in hamsters throughout ANDV infection. Following infection with ANDV strain CHI-7913 or Chile-9717869, groups of hamsters were serially euthanized beginning on day 3 postinfection. Serum and the indicated tissues were collected to examine the amount of ANDV RNA present. Shown are data means with SEM (n = 6/group, except for Chile-9717869 at day 13, which had only two surviving animals). Significance was determined at each time point by Mann-Whitney test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Host immune response to infection.
As noted above, while we observed lower levels of CHI-7913 RNA in the lungs after day 5 postinfection, significant levels of viral RNA were present despite a lack of noticeable signs of disease or pathology. Previous work has characterized the innate and adaptive immune response made by Syrian hamsters infected with ANDV by the i.n. route via RT-qPCR of genes within affected tissues (26). Here, we took a similar approach and analyzed the expression of a number of genes that were shown to be either upregulated or downregulated during lethal ANDV infection in an attempt to determine whether differences in host immune responses are critical for the development of disease during infection with these two strains of ANDV.
There is evidence that an excessive inflammatory response can be deleterious during hantavirus infection, and several cytokines have been shown to be correlated with severe disease and a lethal outcome both in human and animal models (34–36). Here, cytokine mRNA over time during infection with Chile-9717869 and CHI-7913 showed similar levels of expression in most cytokines examined (Fig. 6). However, there were differences in some cytokines that may play a role in infection outcome. Animals infected with CHI-7913 had lower mean expression levels of gamma interferon (IFN-γ), interleukin-4 (IL-4), IL-6, and IL-10 seen for at least one time point during peak illness, although there were fluctuations in expression levels throughout the course of infection (Fig. 6). We also performed a clustering analysis based on the expression patterns of each cytokine seen in each animal throughout infection. The expression of cytokines such as IL-6, IFN-γ, and IL-10 clustered closely together along with expression of IL-2 and the transcription factor FoxP3. In terms of the expression profiles of individual animals, higher expression levels of a number of genes were seen in the early to mid-stages of infection (3 to 7 dpi). Animals from both groups had individuals that showed these higher expression levels, with more CHI-7913-infected hamsters within this cluster. Of those animals taken on day 13 in both groups, most showed lower overall expression and clustered closely together along with some animals from both groups in the mid-stages of disease.
FIG 6.
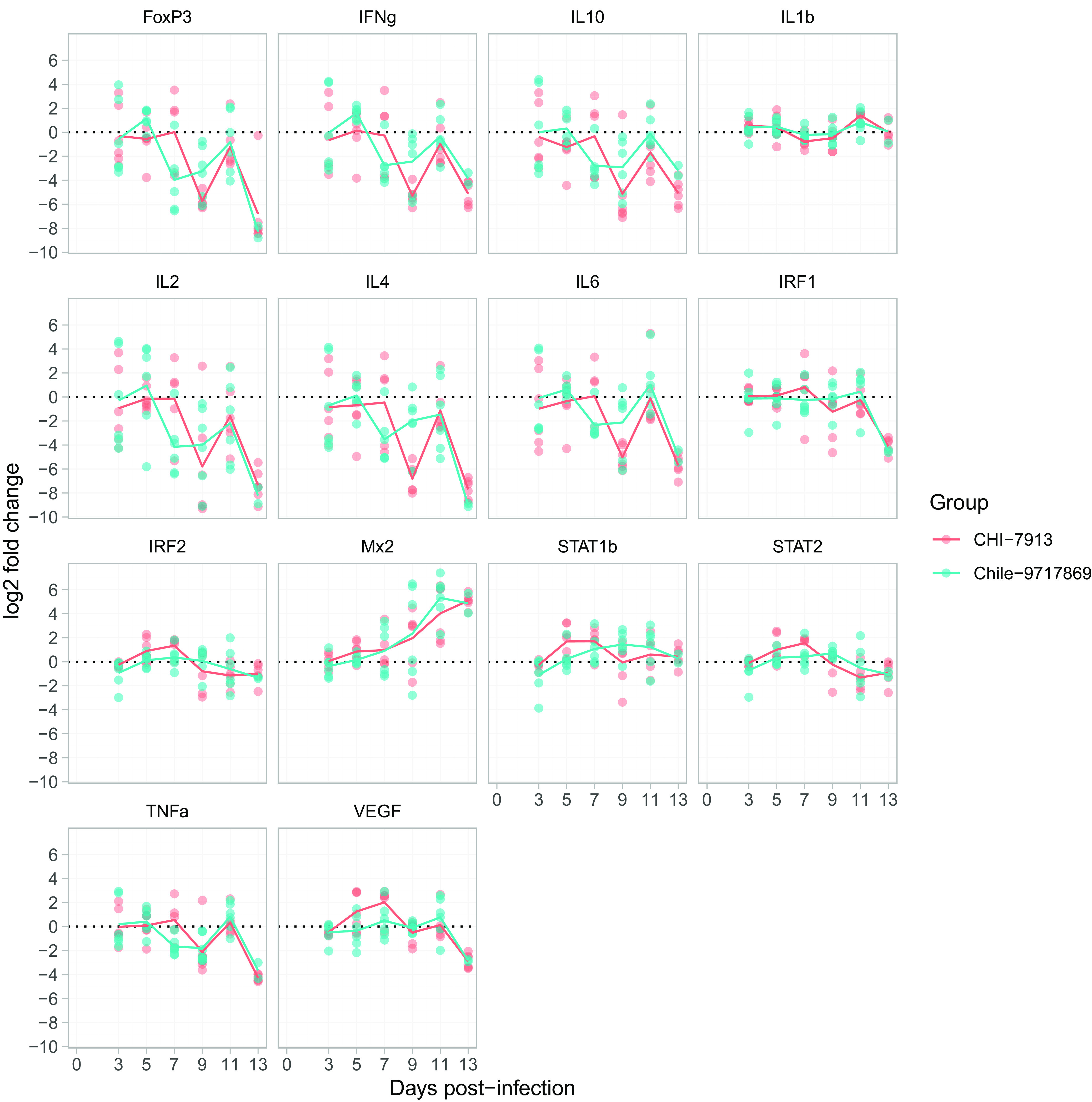
Changes in cytokine mRNA levels during ANDV infection. Following infection with ANDV strain CHI-7913 or Chile-9717869, groups of hamsters were serially euthanized beginning on day 3 postinfection. Lung samples were taken to determine the mRNA expression of various cytokines and immune genes compared with control animals given DMEM only. Reported are the log2 fold change in gene expression compared with DMEM animals. Shown are all data points (n = 6/group, except for Chile-9717869 at day 13, which had only two surviving animals). Lines represent means of all data points from each time point.
Humoral immune responses.
Serum samples from hamsters collected at experimental time points or at the time of euthanasia were examined for anti-SNV N and neutralizing antibodies using enzyme-linked immunosorbent assay (ELISA) and plaque reduction neutralization test (PRNT) methods. Sera collected at days 3, 5, 7, 9, 11, and 13 (corresponding to terminal disease in Chile-9717869-infected hamsters) postinfection were mostly negative (titers, <100) for the presence of anti-N antibodies, with the exception of a terminal hamster infected with Chile-9717869 that had a titer of 400. In addition, all six survivors in the CHI-7913 infection group seroconverted with ELISA titers of ≥6,400 in serum collected at the end of the experiment (42 dpi). Neutralizing antibody responses (80% plaque reduction titers [PRNT80]) were not detected in any of the hamster serum samples collected at a defined experimental time point or at the time of euthanasia. Survivors in the CHI-7913 infection group all had detectable neutralizing antibodies with PRNT80 titers ranging from 40 to 160 (Table 2).
TABLE 2.
Anti-N ELISA and PRNT80 titers in hamster sera at defined time points after Andes virus infection
| Collection day postinfection | ANDV strain | No. of samples | Titer |
|
|---|---|---|---|---|
| ELISA anti-N | PRNT80 | |||
| 3 | Chile-9717869 | 6 | <100 | Negative |
| CHI-7913 | 6 | <100 | Negative | |
| 5 | Chile-9717869 | 6 | <100 | Negative |
| CHI-7913 | 6 | <100 | Negative | |
| 7 | Chile-9717869 | 6 | <100 | Negative |
| CHI-7913 | 6 | <100 | Negative | |
| 9 | Chile-9717869 | 6 | <100 | Negative |
| CHI-7913 | 6 | <100 | Negative | |
| 11 | Chile-9717869 | 6 | <100 | Negative |
| CHI-7913 | 6 | <100 | Negative | |
| 13 | Chile-9717869 | 2 | <100 (n = 1), 400 (n = 1) | Negative |
| CHI-7913 | 6 | <100 | Negative | |
| Survivors (42 dpi) | Chile-9717869 | 0 | ||
| CHI-7913 | 6 | ≥6,400 | 40 (n = 1); 80 (n = 4); 160 (n = 1) | |
DISCUSSION
ANDV has been the cause of thousands of HCPS cases in South America since the discovery of New World hantaviruses in the early 1990s. It is one of several New World hantaviruses that have been discovered that can cause human disease, while others appear to be nonpathogenic in humans. While there have been some studies on the molecular determinants of pathogenesis with regard to hantavirus infections in humans, specifically with regard to receptor usage, it has been difficult to assess what determines pathogenesis of given hantaviruses in humans and in various animal models (37, 38). Different hantavirus species show various levels of replicative abilities and pathogenesis in different species. ANDV and, to a lesser extent, Maporal virus, is lethal in Syrian hamsters, while SNV does not cause disease. A passaged SNV increases replication in hamsters, particularly in the pulmonary endothelium, without clinical signs of disease (39). Neither SNV nor ANDV can cause disease in nonhuman primates unless, with SNV, a virus passaged only in the rodent reservoir (deer mouse) is used as the challenge virus (11, 34, 40). Despite these differences, without a reliable reverse genetics system, it has been difficult to study which genetic components are important for the development of disease in animal models. An early study using a reassortant virus that included the S and L segments of SNV and the M segment of ANDV showed that the M segment alone was not sufficient for lethal disease in Syrian hamsters (33). Evidence that the accumulation of very few mutations could alter the infectivity of both Puumala virus (PUUV) and SNV suggest that even minor genetic differences could have a significant effect on pathogenesis in animal models (34, 41, 42). Since these studies, no further characterization of molecular determinants of hantavirus pathogenesis has been done. Here, we attempted to characterize infection of Syrian hamsters with two distinct ANDV isolates, Chile-9717869, a reservoir host isolate that has been used extensively in hamsters for pathogenesis studies, and CHI-7913, which is a human isolate.
Sequencing of both viruses from our working stocks revealed a high similarity; however, there were 23 amino acid differences in the coding regions between the two viruses (Table 1). Eight amino acid changes were detected in the glycoproteins between the two viruses, while 15 were detected in the L protein, the RNA-dependent RNA polymerase. In terms of the locations of the different amino acid residues between the two viruses in the glycoproteins, CHI-7913 had two substitutions in the leader sequence of Gn as well as four in the ectodomain of Gn and two in the ectodomain of Gc (43–46). These changes may play a role in intracellular trafficking of the glycoprotein or in viral attachment, which may be partly responsible for the differences in replication seen between the two viruses. It is possible that differences seen in pathology are due to differences in replication due to changes in the L protein, as supplementation of a nonlethal virus, SNV, with the M segment of the lethal Chile-9717869 strain in hamsters does not result in clinical signs or disease pathology (33). None of the amino acid changes between the two viruses fall within any of the motifs that are conserved in RNA-dependent RNA polymerases across the Hantaviridae, from premotif A through motif E (46). The substitutions between these viruses may affect folding or tertiary structures of the protein. How these changes alter replication ability remains to be investigated.
In the absence of a developed reverse genetics system for hantaviruses, it will be difficult to determine which mutations are key drivers of pathogenic infection. In terms of ANDV glycoproteins, there is little data on the interaction of New World hantavirus Gn and Gc with specific hantavirus receptors. There is some evidence that mutations in the replication complexes of hantaviruses such as SNV and PUUV can lead to reduced infectivity of rodent hosts following adaptation to tissue culture (34, 41).
Infection of hamsters with Chile-9717869 resulted in uniform lethality, as expected. Surprisingly, infection with CHI-7913 did not cause lethal disease in any of the hamsters included in the survival group despite being isolated from a lethal human case (Fig. 2). Importantly, this vast difference in lethality could not be attributed to an increased or suppressed host adaptive immune response following infection. In all but one hamster, no appreciable anti-N antibody or neutralizing antibody responses were documented in either group during the acute stages of infection (Table 2).
A difficulty of the hamster model of HCPS is the lack of clinical signs of disease throughout infection. We typically do not detect signs of disease until the end stages of infection during respiratory failure (approximately 24 to 36 h before death); however, we wanted to monitor groups of animals infected with both viruses through a serial sample study to attempt to better characterize infection. Hamsters in both groups did not see any changes in weight compared with one another or with DMEM control animals aside from a steep drop in weight in the Chile-9717869 group at the final time point, day 13 (Fig. 2B). This was not unexpected based on previous studies and our experience with this model. Previous experiments indicated that lung weight during disease course and, more specifically, the lung/body weight ratio is an important indicator of disease severity (26). Therefore, we weighed the set of lungs of each animal at the time points throughout the study and compared them to total body weight as described above. Surprisingly, we did not notice any significant differences between the groups of virus-infected animals compared with control animals throughout infection. Aside from the expected clinical signs of infection seen in the Chile-9717869 group on the day before death, no obvious differences in infection were noticeable.
Throughout the serial sample study, gross pathological examination of the lungs was performed following euthanasia and subsequent tissue processing. Upon macroscopic examination of the lungs in each group, there were very few signs of noticeable lung pathology until late in disease, when areas of consolidation and hyperemia were seen in the Chile-9717869 group (Fig. 3). We also determined the amount of viral RNA in the tissues throughout infection. Chile-9717869 RNA levels were higher in every tissue tested beginning on day 7 postinfection, indicating that CHI-7913 does not replicate as efficiently in hamsters in vivo. Similar observations were made in vitro using BHK cells (Fig. 1). These data suggest that specific differences in viral sequence are important for replication in hamsters and in humans, as this isolate was from a lethal human case. This difference may be due in part to differences in viral entry ability or to altered capacity of the polymerase to initiate either viral replication or transcription. It is likely that the differences in infection outcome are due to changes in the RNA-dependent RNA polymerase, as the M segment from Chile-9717869 alone is not sufficient to cause disease (33). However, it is possible that infection outcome is multifactorial, and that a recombinant between SNV and ANDV does not fully represent what is occurring during ANDV infection. Similarly, passage of SNV in hamsters (HA-SNV) demonstrated increased titers of SNV in hamsters with no discernible clinical signs of disease, even after 20 passages (39).
Previous work also reported on the expression of various immune genes during ANDV infection of Syrian hamsters (26). We sought to determine if differences in the immune response generated during infection play a role in reducing viral replication, limiting disease pathology, and ultimately clearing infection. Ultimately, we did not detect any clear and obvious differences in the expression levels of the genes examined that might hint at the importance of either for keeping viral replication in check or for exacerbating pathogenesis. Similar to previous reports, hamsters showed high expression of Mx-2 and STAT-1 upon infection in both groups. There were differences in mean expression levels between hamsters infected with both viruses, including expression of IL-4, IL-6, IFN-γ, and IL-10 (Fig. 6). Of these, IL-10 and IL-6 have been shown to be significantly upregulated during HCPS, with IL-6 potentially playing an important role in whether patients survive infection (35, 36). Lower levels of IL-6 have been correlated with increased survival in HCPS patients, suggesting a reduction in the inflammatory cascade is beneficial during infection (36). However, the fluctuation in expression levels and variability within and between groups makes it difficult to determine the importance of individual genes on the outcome of infection. Animals that were taken down at the same time point also generally had similar gene expression profiles regardless of which virus they were infected with. However, several genes that we assessed in our study differed in their expression patterns compared with previously reported experiments (26).
In this study, we examined the pathogenesis of a human isolate of ANDV, CHI-7913, in Syrian hamsters. While ANDV (strain Chile-9717869) typically causes lethal infection of hamsters at low doses, the CHI-7913 isolate did not result in lethal infection or noticeable signs of pathology. We showed that relatively few amino acid changes between two hantavirus isolates could have significant implications in infection outcome. While a reverse genetics system for hantaviruses is currently a tool that has not been developed, our study highlights the importance of developing such a system, which could help identify components of hantaviruses that are critical for causing disease. This could have implications not only for pathogenic infection in animal models but also for infections of reservoir hosts, for which hantaviruses are highly adapted and fail to infect following very few amino acid changes (41, 42). Geographic disparities in the sequences of hantaviruses or their hosts can affect their ability to infect rodent populations in geographically distinct areas. The identification of specific changes that mediate this inability could lead to important insights regarding transmission to rodents and humans. Ultimately, here we showed that the lethal ANDV model of HCPS is strain specific and that an isolate that was lethal in humans, CHI-7913, does not cause lethal infection in hamsters. Further study of specific genomic and amino acid differences between hantavirus species and strains is warranted and could help determine key molecular determinants of hantavirus infectivity and pathogenesis.
MATERIALS AND METHODS
Ethics statement.
The animal experiments described were carried out at the National Microbiology Laboratory (NML) of the Public Health Agency of Canada. All experiments were approved by an institutional animal care committee at the Canadian Science Center for Human and Animal Health in accordance with guidelines provided by the Canadian Council on Animal Care. All animals were acclimated for at least 1 week prior to experimental manipulations. All infectious ANDV work was performed under biosafety level 4 conditions at the NML. The animals were given food and water ad libitum and monitored daily throughout the course of the experiments.
Generation of cDNA.
Total nucleic acids from both ANDV stocks were subjected to DNase treatment using the DNA-free DNA removal kit (AM1906; Ambion). Briefly, a 0.1× volume of 10× DNase I buffer was added to each sample, followed by 1 μl of rDNase I enzyme, and incubated at 37°C for 20 min. The reaction was supplemented with an additional 1 μl of rDNase I enzyme and incubated at 37°C for an additional 20 min. The rDNase I was inactivated by adding a 0.2× volume of DNase inactivation reagent to each sample and incubated at room temperature for 2 min with periodic mixing. The samples were centrifuged at 10,000 × g for 2 min to pellet the DNase inactivation reagent, and the supernatants were transferred to fresh microcentrifuge tubes. First-strand cDNA was generated using the SuperScript IV first-strand synthesis system (18091050; Invitrogen). Briefly, a total of 8 μl of RNA was combined with 1 μl of 10 mM deoxynucleoside triphosphate mix, 3 μl of laboratory-grade water (W3513; Sigma-Aldrich), and 1 μl of random hexamers. The samples were incubated at 65°C for 5 min and then cooled on ice for at least 1 min. A total of 7 μl of a master mix comprising 4 μl of 5× Superscript IV buffer, 1 μl of 100 mM dithiothreitol (DTT), 1 μl of RNase inhibitor, and 1 μl of Superscript IV reverse transcriptase was added to each sample. The samples were subjected to a series of incubations: room temperature for 10 min, 50°C for 20 min, and 80°C for 10 min. Upon completion of the final incubation, 1 μl of E. coli RNase H was added to each sample and incubated at 37°C for 20 min. All incubations were conducted using a ThermoMixer C (Eppendorf) instrument without mixing. The second strand of cDNA was generated using the NEBNext mRNA second-strand synthesis module (E6111L; NEB) according to the manufacturer’s specifications, followed by purification using the Qiagen QIAquick PCR purification kit (28104; Qiagen). Ten microliters of 3 M NaHCO3 (S0296; Teknova) was added to each sample to ensure the appropriate pH for column binding, and samples were eluted off the columns with 25 μl of buffer EB (10 Mm Tris-Cl, pH 8.5). The ds-cDNA from each sample was quantified on the Qubit 3 fluorometer using the dsDNA high-sensitivity kit.
High-throughput sequencing.
Sequencing libraries were generated using the Nextera XT library prep kit (FC-131-1024; Illumina) according to the manufacturer’s protocol, except for the bead-based library normalization, which was omitted in favor of manual library quantification and pooling. Briefly, the libraries were quantified on the Qubit 3 fluorometer using the dsDNA high-sensitivity kit and then pooled manually such that the total mass of each sample within the pool was equivalent. The fragment size distribution and concentration of the pool were assessed using a Bioanalyzer 2100 (Agilent) with the DNA HS kit (5067-4626; Agilent) and Qubit 3 fluorometer with the dsDNA high-sensitivity kit, respectively. The pool was diluted to 4 nM with laboratory-grade water, denatured with freshly prepared 0.2 N NaOH (H022431J1601; Teknova), and then diluted to a final loading concentration of 12 pM according to the manufacturer’s specifications. A total of 6 μl of 12.5 pM PhiX control V3 (FC-110-3001; Illumina) was added to the pool at a final concentration of 1%. Paired-end sequencing was conducted on a MiSeq (Illumina) using a reagent kit v2 (300-cycle) cartridge and Micro flow cell (MS-103-1002; Illumina).
Bioinformatics.
The BBDuk module within BBMap v 38.22 (https://sourceforge.net/projects/bbmap/) was used to remove reads associated with Illumina’s PhiX control V3, followed by read adaptor and quality trimming using fastp v 0.19.7 (47). Subsequently, reads were taxonomically classified with Kraken2 v 2.0.7 beta using a database, current as of 22 March 2019, comprising all complete genomes in NCBI’s Refseq for viruses, bacteria, and archaea, as well as a collection of known vectors (UniVec_Core), the human genome GRCh38.p13 (GCF_000001405.39), and Centrifuge v 1.0.4 beta using a database comprising NCBI’s nucleotide nonredundant sequences, current as of 3 March 2018 (48, 49). Reads were filtered such that only those classified as virus or unclassified were retained for further analysis. The filtered reads were subjected to de novo assembly using Shovill v 1.0.4 (https://github.com/tseemann/shovill) and Unicycler v 0.4.7 and were mapped against reference genomes in GenBank (Andes virus strain Chile-9717869, AF291702 to AF291704; Andes virus strain CHI-7913, AY228237 to AY228239) using Snippy v 4.4.5 (https://github.com/tseemann/snippy) (50). The de novo contigs generated by Shovill and Unicycler were aligned with the consensus sequences (with all variants instantiated) generated by Snippy using MAFFT v 7.309 (50). Any discrepancies (i.e., single-nucleotide variants) between the de novo and templated assemblies were resolved by interrogating the .bam files generated by Snippy in Geneious v 9.1.8 (Biomatters). Corrections to the final consensus sequence were made based on the result best supported by the underlying read data.
Viruses.
Virus stocks of ANDV strains Chile-9717869 and CHI-7913 were propagated in VeroE6 cells as described previously (26, 51).
In vitro growth kinetics.
Baby hamster kidney fibroblast (BHK) cells were used to seed 24-well cell culture plates and maintained in Eagle’s minimal essential medium (Lonza) supplemented with tryptose phosphate broth (Sigma-Aldrich), 5% fetal bovine serum (FBS; Fisher Scientific), and 1% l-glutamine (HyClone). At 90% confluence, cells were infected in triplicate with ANDV strain Chile-9717869 or CHI-7913 at a multiplicity of infection (MOI) of 0.001 (100 μl total volume). After 1 h, the inoculum was removed and replaced with maintenance medium with reduced (2%) FBS. Supernatant samples were collected from representative wells on days 0 (immediately after inoculum was removed), 3, 5, and 7 postinfection and analyzed by quantitative real-time PCR (RT-qPCR) as outlined below.
Animals and infections.
Five- to 6-week-old female Syrian golden hamsters (Mesocricetus auratus) were anesthetized with inhalational isoflurane and given an intranasal (i.n.) inoculation of 150 focus-forming units of ANDV strain Chile-9717869 (n = 42) or CHI-7913 (n = 42) or plain medium (DMEM; n = 12) (50 μl per nare) using a p200 pipette and sterile tip. Animals were monitored daily for clinical signs of disease, including hunched posture, labored breathing, and lethargy according to an approved scoring sheet. Beginning on day 3 postinfection and continuing every second day until terminal disease, groups of 6 hamsters per isolate and 2 DMEM controls were euthanized and samples collected for virological, immunological, and histological analysis. The remaining six hamsters per infection group were included for clinical observations, disease manifestations, and survival. Animals requiring euthanasia due to clinical score or a predetermined experimental time point were exsanguinated via cardiac puncture after induction of deep anesthesia.
Detection of ANDV RNA.
Viral RNA levels were detected as described previously (17). Briefly, at the indicated time points, groups of hamsters were anesthetized with inhalational isoflurane and euthanized for collection of blood and tissues for analysis of viral RNA levels. RT-qPCR detection of ANDV S segment RNA was performed on a Quant Studio 3 instrument (Applied Biosystems) using a one-step protocol with a QuantiTect probe RT-PCR kit (Qiagen) and ANDV-specific primers and probe (ANDVforw, AGGCAGTGGAGGTGGAC; ANDVrev, CCCTGTTGGATCAACTGGTT; ANDVProbe, FAM-ACGGGCAGCTGTGTCTACATTGGA-TAMRA) according to the manufacturer’s instructions. RT-qPCR stages consisted of reverse transcription (50°C for 30 min), Taq activation (95°C for 15 min), and amplification (40 cycles of 94°C for 15 s and 60°C for 60 s). Data acquisition occurred at the end of the annealing/extension stage (60°C for 60 s) of each amplification cycle. Samples were quantified against a standard curve of ANDV S segment in vitro-transcribed RNA ranging from 5 × 107 to five S segment copies.
Detection of N-specific antibodies.
Antibodies cross-react against both ANDV and SNV N protein; therefore, to confirm seroconversion in ANDV-infected animals, an anti-SNV N ELISA was performed. Briefly, 96-well, half-area, high-binding polystyrene plates (Corning) were coated with recombinant SNV N protein at 15 ng per well and incubated overnight at 4°C. The following day, the plates were washed with PBS-T five times and then blocked with 5% skim milk in PBS-T (PBS plus 0.1% Tween 20) for 1 h at 37°C. Serum samples were diluted (1:100 to 1:6,400) in blocking buffer and added to the plates for 1 h. The plates were washed with PBS-T another five times, and secondary horseradish peroxidase-conjugated polyclonal rabbit anti-Syrian hamster IgG secondary antibody (1:1,000 dilution; LSBio) was added to the plates for 1 h at 37°C. The plates were washed with PBS-T and ABTS substrate (ThermoFisher) was added, and plates were incubated for 30 min at room temperature before reading the optical density (OD) values at 405 nm. Positive samples were those that had an OD greater than the mean OD plus three standard deviations seen in the negative-control wells.
PRNT.
A surrogate virus neutralization assay was used based on a recombinant vesicular stomatitis virus (VSV) expressing the glycoproteins of ANDV Chile 9717869 in place of its own (VSV-ANDV GP). Hamster serum samples collected as outlined above were inactivated by incubation at 56°C for 30 min. VeroE6 cells were seeded in 12-well culture plates and maintained in DMEM (HyClone) supplemented with 10% FBS, 1% penicillin-streptomycin (HyClone), and 2 mM glutamine (HyClone). The following day, 2-fold serial dilutions of the sera diluted in DMEM were incubated with 50 plaque-forming units (PFU) of VSV-ANDV GP for 1 h at 37°C and subsequently applied to a monolayer of confluent VeroE6 cells for 1 h at 37°C. After removing the inoculum, cells were overlaid with minimal essential medium (Fisher) containing 2% FBS and 1% low-melting-point agarose (Fisher). Plaques were visualized using a 0.2% (wt/vol) crystal violet solution containing 4% formaldehyde and counted after incubation for 5 days at 37°C and 5% CO2. Eighty percent plaque reduction (PRNT80) was calculated and data were fitted into a four-parameter logistic curve in GraphPad Prism (GraphPad Software, San Diego, CA; www.graphpad.com).
Cytokine mRNA analysis.
RNA was extracted from lung samples using an RNeasy plus minikit (Qiagen), which includes a genomic DNA elimination step, per the manufacturer’s instructions. Lung RNA was quantified using a Qubit 4 fluorometer and the Qubit RNA assay kit (Invitrogen). Host responses, including the expression of forkhead box P3 (FoxP3), IFN-γ, IL-1b, -2, -4, -6, and -10, interferon regulatory factor 1 (IRF1) and IRF2, myxovirus resistance protein 2 (Mx2), signal transducer activator of transcription 1b (STAT1b) and STAT2, transforming growth factor alpha (TNF-α), and vascular endothelial growth factor (VEGF), were analyzed on 20 ng of lung RNA using the one-step QuantiTect probe RT-qPCR kit using the primer/probe sets described previously (26). Ribosomal protein L18 (RPL18) was used as an internal control. All RT-qPCR assays were performed on a QuantStudio 3 instrument (Applied Biosystems).
Histology.
Hematoxylin and eosin staining was performed as described previously (17). Briefly, formalin-fixed tissues were embedded in paraffin wax to make paraffin blocks. Five-millimeter sections were cut and mounted on Superfrost microscope slides (Fisher). Following an overnight incubation at 37°C, sections were deparaffinized with three 5-min changes of xylene. Slides were then immersed three times in 100%, twice in 95%, and once in 70% ethanol for 3 min each. They were then washed with distilled water for 2 min and then stained for 2 min with hematoxylin (7211; Richard Allen Scientific). A water rinse was performed for 2 min, followed by differentiation in 1% acid alcohol treatment (8 to 12 washes) and a second rinse in Scott's tap water substitute (Sigma-Aldrich) for 1 min, followed by a rinse for one min. A 2-min counterstain was then performed in eosin Y (Surgipath). Sections were dehydrated with two washes of 95% ethanol for 3 min each. A second set of washes was then performed three times in 100% ethanol for 3 min each and then cleared with three changes of xylene for 5 min each. Slides were mounted with Permount (Fisher) for viewing and scanned with a Zeiss Mirax Midi. Histological slides and gross pathology examinations were performed by certified veterinary pathologists.
Statistical analysis.
All results were analyzed and graphed using GraphPad Prism 8 software or R software (www.r-project.org). Immune gene expression data were analyzed in R 3.6.1; the checkpoint package was used to set all packages to their version on 11 November 2019. Calculation of delta-delta threshold cycle was performed using dplyr and tidyr (www.r-project.org); graphing was performed using ggplot2 (www.ggplot2.tidyverse.org). Statistical significance between groups was determined using a Mann-Whitney test or Kaplan-Meier analysis with log-rank test where applicable.
Data availability.
Sequences of the S, M, and L segments of CHI-7913 and Chile-9717869 are available in GenBank under accession numbers MT956618 (CHI-7913 S), MT956619 (CHI-9713 M), MT956620 (CHI-9713 L), MT956622 (Chile-9717869 S), MT956623 (Chile-9717869 M), and MT956621 (Chile-9717869 L).
REFERENCES
- 1.Jonsson CB, Figueiredo LT, Vapalahti O. 2010. A global perspective on hantavirus ecology, epidemiology, and disease. Clin Microbiol Rev 23:412–441. 10.1128/CMR.00062-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schlegel M, Jacob J, Krüger DH, Rang A, Ulrich RG. 2014. Hantavirus emergence in rodents, insectivores and bats: what comes next, p 235–292. In The role of animals in emerging viral diseases. Academic Press, New York, NY. [Google Scholar]
- 3.Jiang H, Du H, Wang LM, Wang PZ, Bai XF. 2016. Hemorrhagic fever with renal syndrome: pathogenesis and clinical picture. Front Cell Infect Microbiol 6:1. 10.3389/fcimb.2016.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borges AA, Campos GM, Moreli ML, Souza RL, Aquino VH, Saggioro FP, Figueiredo LT. 2006. Hantavirus cardiopulmonary syndrome: immune response and pathogenesis. Microbes Infect 8:2324–2330. 10.1016/j.micinf.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 5.Drebot MA, Jones S, Grolla A, Safronetz D, Strong JE, Kobinger G, Lindsay RL. 2015. Hantavirus pulmonary syndrome in Canada: an overview of clinical features, diagnostics, epidemiology and prevention. Can Commun Dis Rep 41:124–131. 10.14745/ccdr.v41i06a02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Figueiredo LT, Souza WM, Ferres M, Enria DA. 2014. Hantaviruses and cardiopulmonary syndrome in South America. Virus Res 187:43–54. 10.1016/j.virusres.2014.01.015. [DOI] [PubMed] [Google Scholar]
- 7.Firth C, Tokarz R, Simith DB, Nunes MR, Bhat M, Rosa ES, Medeiros DB, Palacios G, Vasconcelos PF, Lipkin WI. 2012. Diversity and distribution of hantaviruses in South America. J Virol 86:13756–13766. 10.1128/JVI.02341-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martinez-Valdebenito C, Calvo M, Vial C, Mansilla R, Marco C, Palma RE, Vial PA, Valdivieso F, Mertz G, Ferres M. 2014. Person-to-person household and nosocomial transmission of Andes hantavirus, Southern Chile, 2011. Emerg Infect Dis 20:1629–1636. 10.3201/eid2010.140353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martinez VP, Bellomo C, San Juan J, Pinna D, Forlenza R, Elder M, Padula PJ. 2005. Person-to-person transmission of Andes virus. Emerg Infect Dis 11:1848–1853. 10.3201/eid1112.050501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Safronetz D, Ebihara H, Feldmann H, Hooper JW. 2012. The Syrian hamster model of hantavirus pulmonary syndrome. Antiviral Res 95:282–292. 10.1016/j.antiviral.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hooper JW, Larsen T, Custer DM, Schmaljohn CS. 2001. A lethal disease model for hantavirus pulmonary syndrome. Virology 289:6–14. 10.1006/viro.2001.1133. [DOI] [PubMed] [Google Scholar]
- 12.Brown KS, Safronetz D, Marzi A, Ebihara H, Feldmann H. 2011. Vesicular stomatitis virus-based vaccine protects hamsters against lethal challenge with Andes virus. J Virol 85:12781–12791. 10.1128/JVI.00794-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hooper JW, Ferro AM, Wahl-Jensen V. 2008. Immune serum produced by DNA vaccination protects hamsters against lethal respiratory challenge with Andes virus. J Virol 82:1332–1338. 10.1128/JVI.01822-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hooper JW, Brocato RL, Kwilas SA, Hammerbeck CD, Josleyn MD, Royals M, Ballantyne J, Wu H, Jiao JA, Matsushita H, Sullivan EJ. 2014. DNA vaccine-derived human IgG produced in transchromosomal bovines protect in lethal models of hantavirus pulmonary syndrome. Sci Transl Med 6:264ra162. 10.1126/scitranslmed.3010082. [DOI] [PubMed] [Google Scholar]
- 15.Safronetz D, Haddock E, Feldmann F, Ebihara H, Feldmann H. 2011. In vitro and in vivo activity of ribavirin against Andes virus infection. PLoS One 6:e23560. 10.1371/journal.pone.0023560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Safronetz D, Falzarano D, Scott DP, Furuta Y, Feldmann H, Gowen BB. 2013. Antiviral efficacy of favipiravir against two prominent etiological agents of hantavirus pulmonary syndrome. Antimicrob Agents Chemother 57:4673–4680. 10.1128/AAC.00886-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warner BM, Stein DR, Jangra RK, Slough MM, Sroga P, Sloan A, Frost KL, Booth S, Chandran K, Safronetz D. 2019. Vesicular stomatitis virus-based vaccines provide cross-protection against Andes and Sin Nombre viruses. Viruses 11:645. 10.3390/v11070645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meissner JD, Rowe JE, Borucki MK, St Jeor SC. 2002. Complete nucleotide sequence of a Chilean hantavirus. Virus Res 89:131–143. 10.1016/S0168-1702(02)00129-6. [DOI] [PubMed] [Google Scholar]
- 19.Milazzo ML, Eyzaguirre EJ, Molina CP, Fulhorst CF. 2002. Maporal viral infection in the Syrian golden hamster: a model of hantavirus pulmonary syndrome. J Infect Dis 186:1390–1395. 10.1086/344735. [DOI] [PubMed] [Google Scholar]
- 20.Cruz CD, Forshey BM, Vallejo E, Agudo R, Vargas J, Blazes DL, Guevara C, Laguna-Torres VA, Halsey ES, Kochel TJ. 2012. Novel strain of Andes virus associated with fatal human infection, central Bolivia. Emerg Infect Dis 18:750–757. 10.3201/eid1805.111111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galeno H, Mora J, Villagra E, Fernandez J, Hernandez J, Mertz GJ, Ramirez E. 2002. First human isolate of Hantavirus (Andes virus) in the Americas. Emerg Infect Dis 8:657–661. 10.3201/eid0807.010277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tischler ND, Galeno H, Rosemblatt M, Valenzuela PD. 2005. Human and rodent humoral immune responses to Andes virus structural proteins. Virology 334:319–326. 10.1016/j.virol.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 23.Valdivieso F, Vial P, Ferres M, Ye C, Goade D, Cuiza A, Hjelle B. 2006. Neutralizing antibodies in survivors of Sin Nombre and Andes hantavirus infection. Emerg Infect Dis 12:166–168. 10.3201/eid1201.050930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Terajima M, Ennis FA. 2011. T cells and pathogenesis of hantavirus cardiopulmonary syndrome and hemorrhagic fever with renal syndrome. Viruses 3:1059–1073. 10.3390/v3071059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kilpatrick ED, Terajima M, Koster KT, Catalina MD, Cruz J, Ennis FA. 2004. Role of specific CD8+ T cells in the severity of a fulminant zoonotic viral hemorrhagic fever, hantavirus pulmonary syndrome. J Immunol 172:3297–3304. 10.4049/jimmunol.172.5.3297. [DOI] [PubMed] [Google Scholar]
- 26.Safronetz D, Zivcec M, Lacasse R, Feldmann F, Rosenke R, Long D, Haddock E, Brining D, Gardner D, Feldmann H, Ebihara H. 2011. Pathogenesis and host response in Syrian hamsters following intranasal infection with Andes virus. PLoS Pathog 7:e1002426. 10.1371/journal.ppat.1002426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schountz T, Prescott J, Cogswell AC, Oko L, Mirowsky-Garcia K, Galvez AP, Hjelle B. 2007. Regulatory T cell-like responses in deer mice persistently infected with Sin Nombre virus. Proc Natl Acad Sci U S A 104:15496–15501. 10.1073/pnas.0707454104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Easterbrook JD, Zink MC, Klein SL. 2007. Regulatory T cells enhance persistence of the zoonotic pathogen Seoul virus in its reservoir host. Proc Natl Acad Sci U S A 104:15502–15507. 10.1073/pnas.0707453104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hammerbeck CD, Hooper JW. 2011. T cells are not required for pathogenesis in the Syrian hamster model of hantavirus pulmonary syndrome. J Virol 85:9929–9944. 10.1128/JVI.05356-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prescott J, Safronetz D, Haddock E, Robertson S, Scott D, Feldmann H. 2013. The adaptive immune response does not influence hantavirus disease or persistence in the Syrian hamster. Immunology 140:168–178. 10.1111/imm.12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brocato RL, Hammerbeck CD, Bell TM, Wells JB, Queen LA, Hooper JW. 2014. A lethal disease model for hantavirus pulmonary syndrome in immunosuppressed Syrian hamsters infected with Sin Nombre virus. J Virol 88:811–819. 10.1128/JVI.02906-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vergote V, Laenen L, Vanmechelen B, Ranst MV, Verbeken E, Hooper JW, Maes P. 2017. A lethal disease model for New World hantaviruses using immunosuppressed Syrian hamsters. PLoS Negl Trop Dis 11:e0006042. 10.1371/journal.pntd.0006042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McElroy AK, Smith JM, Hooper JW, Schmaljohn CS. 2004. Andes virus M genome segment is not sufficient to confer the virulence associated with Andes virus in Syrian hamsters. Virology 326:130–139. 10.1016/j.virol.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 34.Safronetz D, Prescott J, Feldmann F, Haddock E, Rosenke R, Okumura A, Brining D, Dahlstrom E, Porcella SF, Ebihara H, Scott DP, Hjelle B, Feldmann H. 2014. Pathophysiology of hantavirus pulmonary syndrome in rhesus macaques. Proc Natl Acad Sci U S A 111:7114–7119. 10.1073/pnas.1401998111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morzunov SP, Khaiboullina SF, St Jeor S, Rizvanov AA, Lombardi VC. 2015. Multiplex Analysis of serum cytokines in humans with hantavirus pulmonary syndrome. Front Immunol 6:432. 10.3389/fimmu.2015.00432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borges AA, Campos GM, Moreli ML, Moro Souza RL, Saggioro FP, Figueiredo GG, Livonesi MC, Moraes Figueiredo LT. 2008. Role of mixed Th1 and Th2 serum cytokines on pathogenesis and prognosis of hantavirus pulmonary syndrome. Microbes Infect 10:1150–1157. 10.1016/j.micinf.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 37.Geimonen E, Neff S, Raymond T, Kocer SS, Gavrilovskaya IN, Mackow ER. 2002. Pathogenic and nonpathogenic hantaviruses differentially regulate endothelial cell responses. Proc Natl Acad Sci U S A 99:13837–13842. 10.1073/pnas.192298899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gavrilovskaya IN, Shepley M, Shaw R, Ginsberg MH, Mackow ER. 1998. beta3 integrins mediate the cellular entry of hantaviruses that cause respiratory failure. Proc Natl Acad Sci U S A 95:7074–7079. 10.1073/pnas.95.12.7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Safronetz D, Prescott J, Haddock E, Scott DP, Feldmann H, Ebihara H. 2013. Hamster-adapted Sin Nombre virus causes disseminated infection and efficiently replicates in pulmonary endothelial cells without signs of disease. J Virol 87:4778–4782. 10.1128/JVI.03291-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McElroy AK, Bray M, Reed DS, Schmaljohn CS. 2002. Andes virus infection of cynomolgus macaques. J Infect Dis 186:1706–1712. 10.1086/345768. [DOI] [PubMed] [Google Scholar]
- 41.Lundkvist A, Cheng Y, Sjolander KB, Niklasson B, Vaheri A, Plyusnin A. 1997. Cell culture adaptation of Puumala hantavirus changes the infectivity for its natural reservoir, Clethrionomys glareolus, and leads to accumulation of mutants with altered genomic RNA S segment. J Virol 71:9515–9523. 10.1128/JVI.71.12.9515-9523.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Warner BM, Stein DR, Griffin BD, Tierney K, Leung A, Sloan A, Kobasa D, Poliquin G, Kobinger GP, Safronetz D. 2019. Development and characterization of a Sin Nombre virus transmission model in Peromyscus maniculatus. Viruses 11:183. 10.3390/v11020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hepojoki J, Strandin T, Lankinen H, Vaheri A. 2012. Hantavirus structure–molecular interactions behind the scene. J Gen Virol 93:1631–1644. 10.1099/vir.0.042218-0. [DOI] [PubMed] [Google Scholar]
- 44.Cifuentes-Munoz N, Salazar-Quiroz N, Tischler ND. 2014. Hantavirus Gn and Gc envelope glycoproteins: key structural units for virus cell entry and virus assembly. Viruses 6:1801–1822. 10.3390/v6041801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Willensky S, Bar-Rogovsky H, Bignon EA, Tischler ND, Modis Y, Dessau M. 2016. Crystal structure of glycoprotein C from a hantavirus in the post-fusion conformation. PLoS Pathog 12:e1005948. 10.1371/journal.ppat.1005948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kukkonen SK, Vaheri A, Plyusnin A. 2005. L protein, the RNA-dependent RNA polymerase of hantaviruses. Arch Virol 150:533–556. 10.1007/s00705-004-0414-8. [DOI] [PubMed] [Google Scholar]
- 47.Chen S, Zhou Y, Chen Y, Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884–i890. 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wood DE, Lu J, Langmead B. 2019. Improved metagenomic analysis with Kraken 2. bioRxiv 10.1101/762302. [DOI] [PMC free article] [PubMed]
- 49.Kim D, Song L, Breitwieser LP, Salzberg SL. 2016. Centrifuge: rapid and sensitive classification of metagenomic sequences. Genome Res 26:1721–1729. 10.1101/gr.210641.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Safronetz D, Hegde NR, Ebihara H, Denton M, Kobinger GP, St Jeor S, Feldmann H, Johnson DC. 2009. Adenovirus vectors expressing hantavirus proteins protect hamsters against lethal challenge with Andes virus. J Virol 83:7285–7295. 10.1128/JVI.00373-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequences of the S, M, and L segments of CHI-7913 and Chile-9717869 are available in GenBank under accession numbers MT956618 (CHI-7913 S), MT956619 (CHI-9713 M), MT956620 (CHI-9713 L), MT956622 (Chile-9717869 S), MT956623 (Chile-9717869 M), and MT956621 (Chile-9717869 L).