T cells are crucial for the control of persistent EBV infection and the development of EBV-associated diseases. The EBV gB protein is essential for virus entry into B cells and epithelial cells and is thus a target antigen for vaccine development.
KEYWORDS: EBV, glycoprotein B, T cell epitope
ABSTRACT
Glycoprotein B (gB) is an essential fusion protein for Epstein-Barr virus (EBV) infection of both B cells and epithelial cells and is thus a promising target antigen for a prophylactic vaccine to prevent or reduce EBV-associated disease. T cell responses play key roles in the control of persistent EBV infection and the efficacy of a vaccine. However, to date, T cell responses to gB have been characterized for only a limited number of human leukocyte antigen (HLA) alleles. Here, we screened gB T cell epitopes in 23 healthy EBV carriers and 10 patients with nasopharyngeal cancer (NPC) using a peptide library spanning the entire gB sequence. We identified 12 novel epitopes in the context of seven new HLA restrictions that are common in Asian populations. Two epitopes, gB214–223 and gB840–849, restricted by HLA-B*58:01 and -B*38:02, respectively, elicited specific CD8+ T cell responses to inhibit EBV-driven B cell transformation. Interestingly, gB-specific CD8+ T cells were more frequent in healthy viral carriers with EBV reactivation than in those without EBV reactivation, indicating that EBV reactivation in vivo stimulates both humoral (VCA-gp125-IgA) and cellular responses to gB. We further found that most gB epitopes are conserved among different EBV strains. Our study broadens the diversity and HLA restrictions of gB epitopes and suggests that gB is a common target of T cell responses in healthy viral carriers with EBV reactivation. In particular, the precisely mapped and conserved gB epitopes provide valuable information for prophylactic vaccine development.
IMPORTANCE T cells are crucial for the control of persistent EBV infection and the development of EBV-associated diseases. The EBV gB protein is essential for virus entry into B cells and epithelial cells and is thus a target antigen for vaccine development. Understanding T cell responses to gB is important for subunit vaccine design. Here, we comprehensively characterized T cell responses to full-length gB. Our results expand the available gB epitopes and HLA restrictions, particularly those common in Asian populations. Furthermore, we showed that gB-specific CD8+ T cells inhibit B cell transformation ex vivo and that gB-specific CD8+ T cell responses in vivo may be associated with intermittent EBV reactivation in asymptomatic viral carriers. These gB epitopes are highly conserved among geographically separated EBV strains. Precisely mapped and conserved T cell epitopes may contribute to immune monitoring and the development of a gB subunit vaccine.
INTRODUCTION
Epstein-Barr virus (EBV) is a gammaherpesvirus that targets both B cells and epithelial cells and establishes persistent infection in over 90% of adults worldwide (1). Although most individuals are asymptomatic after primary infection, EBV is a direct causative agent of life-threatening B-lymphoproliferative disorders in immunocompromised or T cell-suppressed patients (2), indicating that T cell responses play a crucial role in controlling EBV. Primary infection with EBV is the cause of infectious mononucleosis (IM), and persistent infection is causally associated with a number of lymphomas and epithelial malignancies, including Burkitt lymphoma, Hodgkin lymphoma, NK/T cell lymphoma, nasopharyngeal cancer (NPC), and a subset of gastric cancers, accounting for approximately 200,000 new cancer cases each year. Thus, there is a strong need for the development of a prophylactic vaccine to prevent or reduce EBV-associated diseases (3).
Prior efforts to develop a vaccine to prevent EBV infection and/or EBV-associated diseases have focused on the viral surface antigen gp350 (4, 5). However, gp350 is required for viral entry into only B cells but not epithelial cells (6–8). Consistently, the majority of serum neutralizing antibodies using gp350 as the sole immunogen are capable of preventing only infection of B cells but promote infection of epithelial cells (9). A vaccine candidate that can protect both B and epithelial cells from infection should be further evaluated. Among EBV envelope glycoproteins, glycoprotein B (gB) is a fusogen responsible for the fusion of viral and host membranes and is required for EBV infection of both B cells and epithelial cells (10). Furthermore, gB has shown its potential in human cytomegalovirus (CMV) subunit vaccines, preventing ∼50% of primary infections in postpartum healthy women (11). Similar protective effects have also been observed for a guinea pig CMV gB subunit vaccine (12). Therefore, the gB protein is a promising target antigen for vaccine development against EBV.
T cell responses play a key role in controlling the reactivation of herpesviruses, including EBV. In addition to eliciting neutralizing antibodies, a vaccine that can induce an effective T cell response to EBV-infected cells may have an advantage in providing long-term prophylactic protection and/or reducing symptomatic diseases by restricting viral reactivation. The only successful herpesvirus vaccine, Shingrix, appears to act by this type of mechanism to prevent varicella-zoster virus (VZV)-induced zoster (13). The key determinants of the success of this VZV vaccine are that the viral glycoprotein E (gE) antigen, which contains immunodominant epitopes, strongly induces gE-specific CD4+ T cell responses and the adoption of an adjuvant that can boost the long-term T cell response (13, 14). To date, gB epitopes have been characterized in only a limited number of human leukocyte antigen (HLA) molecules in Caucasian populations (15–18). As a vaccine candidate antigen, the EBV gB protein would be useful for a gB subunit vaccine design upon the definition of critical T cell responses.
In this study, we aimed to determine T cell responses against EBV gB in healthy viral carriers and patients with NPC using a peptide library spanning the entire gB sequence and to characterize their target epitopes and HLA-restricting alleles. We further revealed the inhibition of EBV-driven B cell transformation by gB-specific CD8+ T cells and compared the differences in gB-specific CD8+ T cell responses in healthy viral carriers with or without EBV reactivation and patients with NPC. Our study broadens the diversity and HLA restrictions of gB epitopes and suggests that gB is a common target of T cell responses when EBV is reactivated in healthy donors.
RESULTS
Screening gB-specific T cell epitopes in healthy EBV carriers and patients with NPC.
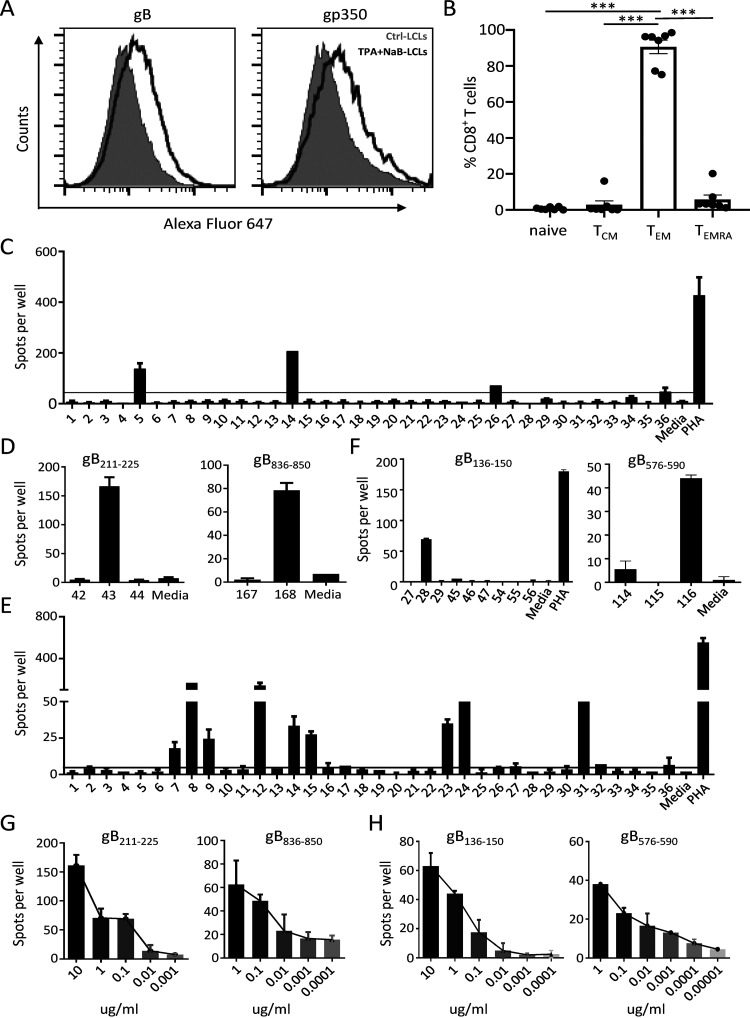
To efficiently generate EBV-specific effector T cells enriched in gB specificity, lymphoblastoid cell lines (LCLs) established from 23 healthy EBV carriers and 10 patients with NPC were treated with tetradecanoyl phorbol acetate (TPA) and sodium butyrate (NaB) (TPA+NaB-LCLs) to trigger EBV lytic replication and to upregulate the expression of lytic antigens, including gB. Flow cytometry analyses revealed increased expression of late lytic proteins, gB and gp350, in LCLs induced by TPA+NaB (Fig. 1A). Analysis of the CD8+ T cell differentiation profile revealed that more than 90% of CD8+ T cells displayed an effector memory (TEM) phenotype (CCR7− CD45RA−) (Fig. 1B). These data indicate that TPA+NaB-LCLs effectively stimulated the expansion of EBV-specific T cells, which are likely to contain lytic antigen-specific T cells.
FIG 1.
Screening gB epitopes using a gB peptide library. (A) Flow cytometry for determining gB and gp350 expression on Ctrl-LCLs and TPA+NaB-LCLs. (B) Distribution of subsets of EBV-specific CD8+ T cells, including naive (CCR7+ CD45RA+), central memory (TCM) (CCR7+ CD45RA−), effector memory (TEM) (CCR7− CD45RA−), and terminally differentiated effector (TEMRA) (CCR7− CD45RA+) T cells. The means ± standard errors of the means (SEMs) are shown (n = 7). ***, P < 0.001 (by a two-tailed Mann-Whitney test). (C) TPA+NaB-LCL-reactivated EBV-specific T cells (5 × 104 cells/well) from donor 1 (D1) were stimulated with a gB peptide library pooled into 36 pools. Responses were measured by IFN-γ ELISPOT assays. The black horizontal line indicates the threshold level used to determine positive results (5 times the number of spot-forming cells [SFCs]/5 × 104 unstimulated EBV-specific T cells). (D) Confirmation of the two peptides 43 (gB211–225) and 168 (gB836–850) eliciting T cell responses using single peptides by ELISPOT assays. (E) TPA+NaB-LCL-reactivated EBV-specific T cells (5 × 104 cells/well) from patient 4 (P4) were stimulated with a gB peptide library pooled into 36 pools. Responses were measured by IFN-γ ELISPOT assays. The black horizontal line indicates the threshold level used to determine positive results (5 times the number of SFCs/5 × 104 unstimulated T cells). (F) Confirmation of the two peptides 28 (gB136–150) and 116 (gB576–590) eliciting T cell responses using single peptides by ELISPOT assays. (G) Dose-dependent T cell responses to peptide 43 (gB211–225) (left) and peptide 168 (gB836–850) (right) at the indicated concentrations as determined by ELISPOT assays. (H) Dose-dependent T cell responses stimulated against peptide 28 (gB136–150) (left) and peptide 116 (gB576–590) (right) at the indicated concentrations as determined by ELISPOT assays. In panels C to H, the means ± standard deviations (SDs) are shown (n = 2 biological replicates).
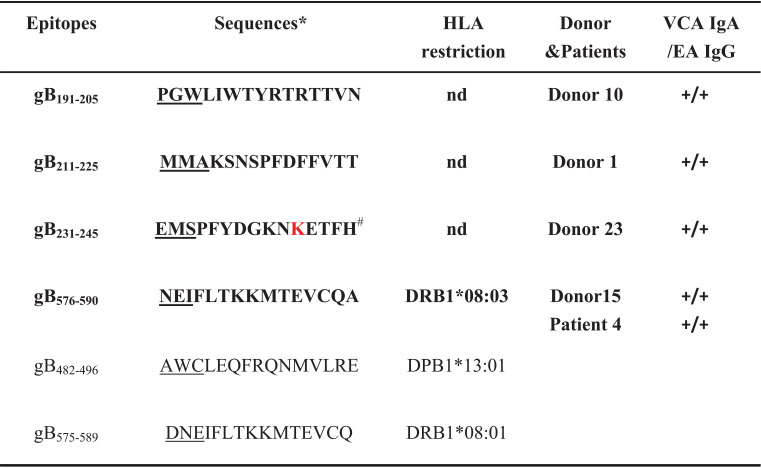
The gB-specific T cells within healthy donor- and patient-derived EBV-specific T cells were screened based on interferon gamma (IFN-γ) production in response to a gB peptide pool that covered full-length gB by overlapping 15-residue peptides using enzyme-linked immunospot (ELISPOT) assays. Figure 1C and D illustrates a screening strategy for gB-specific responses from donor 1 (D1). First, EBV-specific T cells from D1 were screened against 36 peptide subpools representing all 857 residues of gB (B95.8 strain); positive responses were detected against peptides within pools 5, 14, 26, and 36 (Fig. 1C). Subsequent screening of individual 15-mer peptides from pools 5 and 14 or pools 26 and 36 identified two peptides, 43 (gB211–225) and 168 (gB836–850), that were recognized by EBV-specific T cells (Fig. 1D). Similarly, another example of EBV-specific T cells from a patient with NPC (patient 4 [P4]) in response to gB peptides is shown in Fig. 1E and F. Two peptides, 28 (gB136–150) and 116 (gB576–590), were identified to have the ability to stimulate the T cell response. Dose-dependent responses to peptides detected by IFN-γ ELISPOT assays further confirmed that the four peptides gB211–225, gB136–150, gB576–590, and gB836–850 were able to stimulate EBV-specific T cell responses (Fig. 1G and H). Altogether, using these methods, gB-specific T cell responses were detected against 13 15-mer epitopes in EBV-specific T cell lines expanded by TPA+NaB-treated autologous LCLs from a total of nine healthy viral carriers and one patient with NPC (Tables 1 and 2).
TABLE 1.
gB CD8+ T cell epitopesa
| Epitope | Sequenceb | HLA restriction | Donor or patient | Presence of VCA-IgA/EA-IgG |
|---|---|---|---|---|
| gB101–115 | KIVTNILIYNGWYADc | A*24:02 | Donor 4 | −/− |
| gB139–147 | YQCYNAVKM | B*13:01 | Patient 4 | +/+ |
| gB214–223 | KSNSPFDFFV | B*58:01 | Donor 1 | +/+ |
| gB221–231 | FFVTTTGQTVE | C*03:04 | Donor 11 | +/+ |
| gB276–284 | FLDKGTYTL | A*02:01 | Donor 23 | +/+ |
| gB511–525 | GKAVAAKRLGDVISVc | ND | Donor 22 | +/− |
| gB606–614 | NDYHHFKTI | ND | Donor 16 | +/+ |
| gB786–796 | INPISKTELQA | B*55:02 | Donor 8 | +/+ |
| gB840–849 | YHDPETAAAL | B*38:02 | Donor 1 | +/+ |
| gB106–114 | ILIYNGWYA | A*02:01 | ||
| gB190–198 | APGWLIWTY | B*35:08 | ||
| gB544–552 | VPGSETMCY | B*35:01 | ||
| gB827–835 | AARDRFPGL | B*07 |
Listed are the amino acid sequences of newly identified (in boldface type) and previously described gB epitopes. ND, not determined.
Epitopes are also indicated by the first 3 amino acids (underlined) in the text.
Optimal sequences have not been determined.
TABLE 2.
gB CD4+ T cell epitopesa
Listed are the amino acid sequences of newly identified (in boldface type) and previously described gB epitopes. nd, not determined. *Epitopes are also indicated by the first 3 amino acids (underlined) in the text. #One variation in epitope gB231–245 is marked in red. The amino acid sequence of this epitope in the B95.8, Akata, GD1, AG876, and Mutu EBV strains is EMSPFYDGKNKETFH; that in the YCCLE1 and SNU-719 strains is EMSPFYDGKNEETFH; and that in the M81 and C666 strains is EMSPFYDGKNTETFH.
Characterization of HLA restrictions and optimal CD8 epitopes.
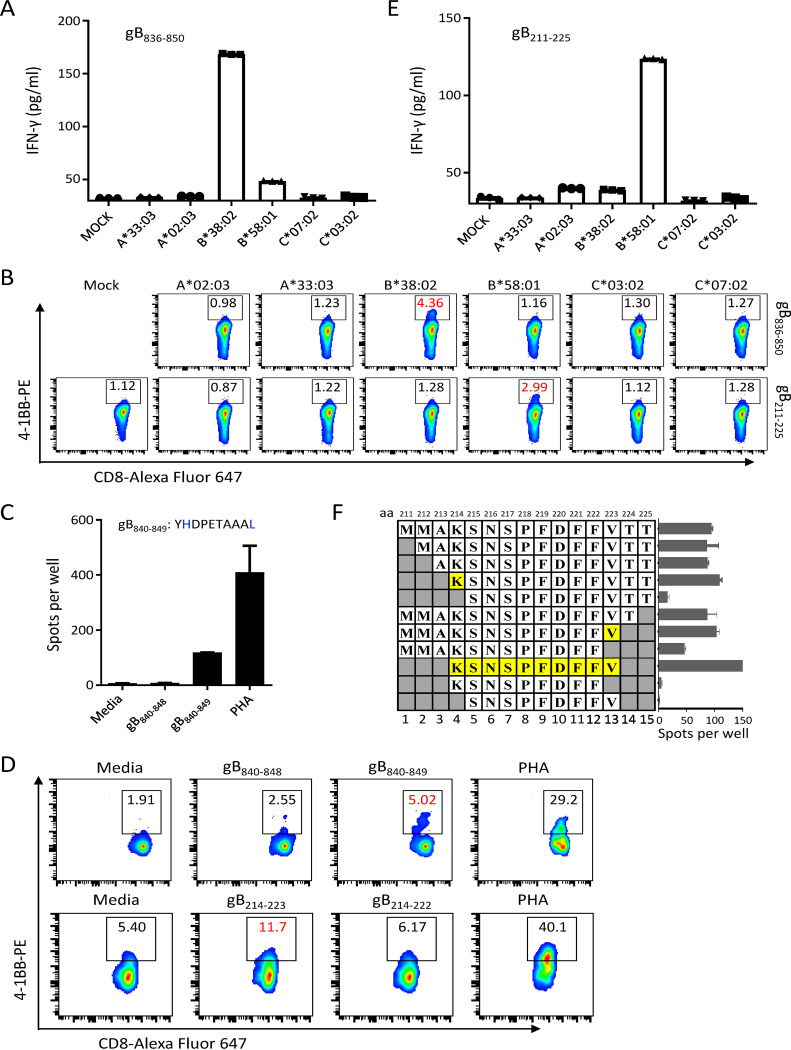
We next sought to determine the HLA restrictions and optimal peptides of the newly identified gB CD8+ T cell epitopes. We first sequenced the HLA class I (HLA-I) alleles in each subject and constructed a panel of COS7 cell lines that individually expressed each of these host HLA-I alleles (19). These HLA-I-overexpressing COS7 cells were then used to present the 15-mer peptides to gB peptide-specific CD8+ T cells. HLA-B*38:02-overexpressing COS7 cells preloaded with the peptide gB836–850 (RRRRYHDPETAAALL) activated gB836–850-specific CD8+ T cells, which displayed increased IFN-γ secretion (Fig. 2A) and upregulated expression of the T cell activation surface marker 4-1BB (Fig. 2B, top row). We further determined that gB840–849 represents the optimal peptide recognized by peptide-specific CD8+ T cells (Fig. 2C and D, top row). The sequence of gB840–849 (YHDPETAAAL) is concordant with the canonical anchor residues of 10-mer HLA-B*38:02 ligands, with histidine at P2 and leucine or phenylalanine at the C terminus (20). Similarly, we identified the HLA-B*58:01 restriction for the peptide gB211–225 (MMAKSNSPFDFFVTT) (Fig. 2B, bottom row, and Fig. 2E). Peptide-specific CD8+ T cells recognized the optimal gB214–223 sequence (KSNSPFDFFV) (Fig. 2D, bottom row, and Fig. 2F), which is consistent with the canonical anchor residues of 10-mer HLA-B*58:01 ligands with serine at P2 and tryptophan or phenylalanine at the C terminus (20). Using the same strategy, four other HLA restrictions, A*24:02, B*13:01, B*55:02, and C*03:04, were determined for the newly identified gB epitopes gB101–115, gB139–147, gB786–796, and gB221–231, respectively (Table 1). In total, we characterized 9 CD8+ T cell epitopes of the 13 novel 15-mer peptide epitopes by IFN-γ enzyme-linked immunosorbent assays (ELISAs) or ELISPOT assays (Table 1). Among these CD8+ T cell epitopes, eight were newly identified, while one epitope, FLDKGTYTL, has been previously reported (15). Six HLA class I restrictions and the optimal sequences were determined for six novel epitopes (Table 1).
FIG 2.
Identification of HLA restrictions and optimal peptides from donor 1. (A) IFN-γ production by gB836–850-specific CD8+ T cells was evaluated by ELISAs after coculture with COS7 cells overexpressing the indicated HLA-I alleles pulsed with the peptide gB836–850. (B) The expression of 4-1BB on gB836–850- andgB211–225-specific CD8+ T cells was evaluated by flow cytometry after coculture with COS7 cells overexpressing the indicated HLA-I alleles pulsed with peptides gB836–850 and gB211–225, respectively. (C) IFN-γ ELISPOT assays of the 9-mer and 10-mer candidate peptides within the 15-mer peptide gB836–850 identified gB840–849 as the optimal epitope. (D) The expression of 4-1BB on gB836–850-specific CD8+ T cells in response to gB840–848 and gB840–849 or on gB211–225-specific CD8+ T cells in response to gB214–222 and gB214–223 was measured by flow cytometry. (E) IFN-γ production by gB211–225-specific CD8+ T cells was evaluated by ELISAs after coculture with COS7 cells overexpressing the indicated HLA-I alleles pulsed with the peptide gB211–225. (F) Truncation analysis of the peptide gB211–225 identified gB214–223 as the optimal epitope as determined by IFN-γ ELISPOT assays. In panels A and E, the means ± SEMs are shown (n = 3 technical replicates). In panels C and F, the means ± SEMs are shown (n = 2 biological replicates).
Identification of gB CD4 epitopes.
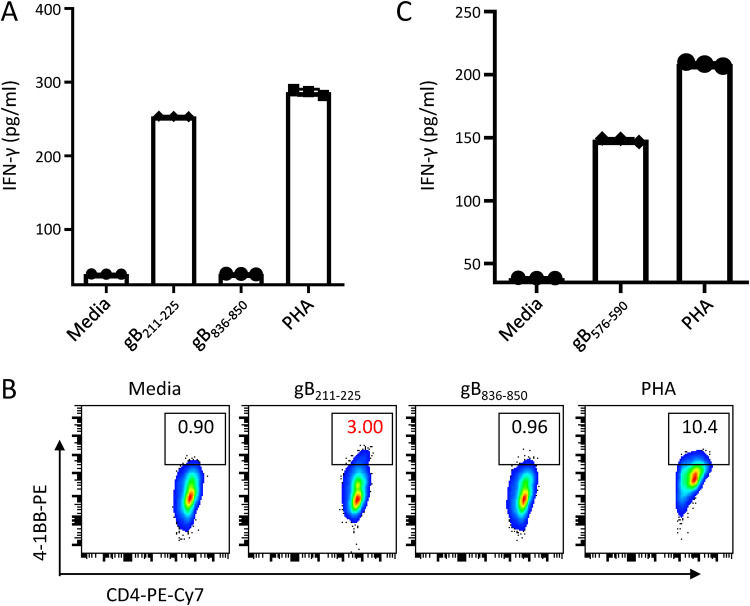
In parallel with the CD8+ T cell epitopes, 4 HLA class II-restricted epitopes were confirmed out of the 13 IFN-γ ELISPOT-positive peptides. The peptide-specific CD4-enriched T cell population from D1 secreted IFN-γ after pulsation with the peptide gB211–225 (Fig. 3A). Upregulation of 4-1BB also confirmed that the CD4+ T cells from D1 recognized the peptide gB211–225 (Fig. 3B). The peptide-specific CD4-enriched T cell population from P4 recognized the peptide gB576–590 (Fig. 3C). Using these assays, we identified two other new CD4+ T cell epitopes, gB191–205 and gB231–245, from D10 and D23, respectively (Table 2). The epitope gB576–590 (NEIFLTKKMTEVCQA) identified from two individuals with HLA-DRB1*08:03 almost coincided with the known CD4+ T cell epitope gB575–589 (DNEIFLTKKMTEVCQ) restricted by HLA-DRB1*08:01 (18, 21). Another novel CD4+ T cell epitope, gB191–205 (PGWLIWTYRTRTTVN), partially overlapped the previously described CD8+ T cell epitope gB190–198 (APGWLIWTY). We did not identify HLA class II restriction of the remaining three 15-mer epitopes due to the limited number of CD4-enriched T cells.
FIG 3.
Identification of CD4+ T cell epitopes from donor 1 and patient 4. (A and B) Peptide-specific CD4-enriched cells from D1 were stimulated with the indicated peptides. The secretion of IFN-γ was measured by an IFN-γ ELISA (A), and the expression of 4-1BB on peptide-specific CD4+ T cells was assessed by flow cytometry (B). (C) Peptide-specific CD4-enriched cells from P4 were stimulated with the peptide gB576–590. The secretion of IFN-γ was measured by an IFN-γ ELISA. In panels A and C, the means ± SEMs are shown (n = 3 technical replicates).
gB-specific CD8+ T cells restricted EBV-driven B cell transformation.
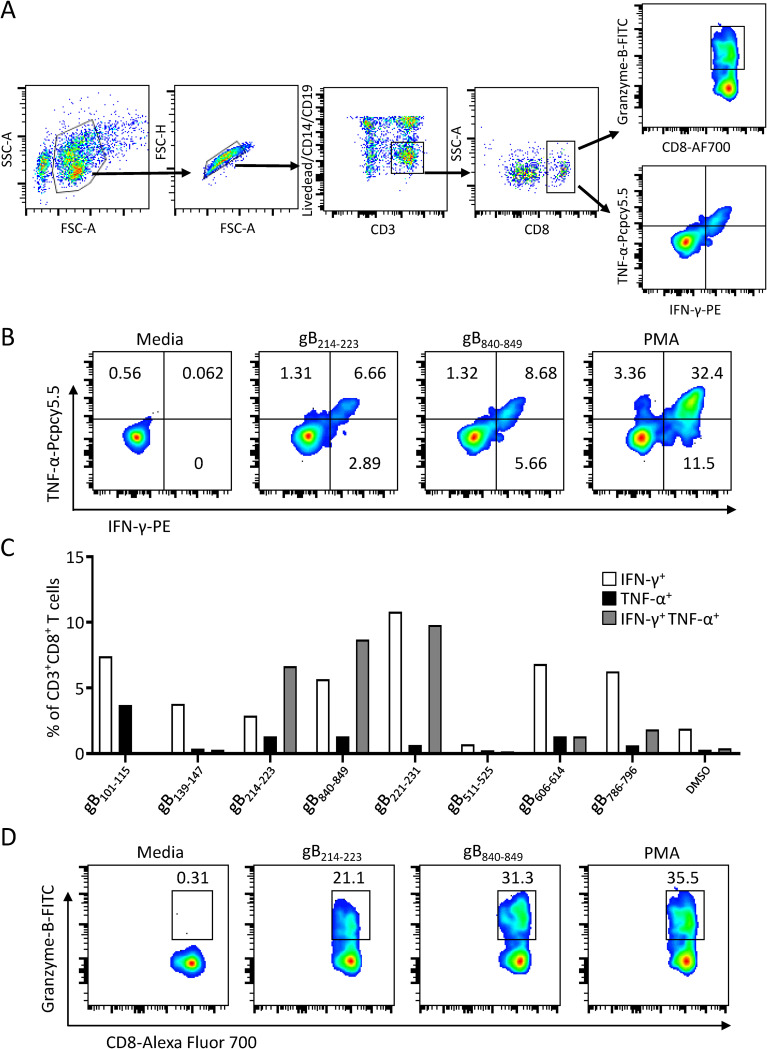
To further characterize cytokine production, gB peptide-specific T cells were stimulated with eight novel single CD8+ T cell epitopes identified in the present study, and the frequencies of IFN-γ- and/or tumor necrosis factor alpha (TNF-α)-producing and granzyme-B-producing CD3+ CD8+ T cells were determined by intracellular cytokine staining (ICS) (Fig. 4A). Three epitopes, gB214–223, gB221–231, and gB840–849, stimulated more than 5% of the CD8+ T cells simultaneously to secrete both IFN-γ and TNF-α (Fig. 4B and C). To a lesser extent, two other epitopes, gB606–614 and gB786–796, stimulated approximately 1% of the CD8+ T cells to secrete both IFN-γ and TNF-α. Only IFN-γ or TNF-α single-producing CD8+ T cells were detected for peptide gB101–115 (Fig. 4C). Additionally, CD8+ T cells from D1 showed a high level of granzyme B expression in response to two epitopes, gB214–223 and gB840–849, indicating that these two peptide-specific CD8+ T cells may be able to directly lyse gB-expressed target cells (Fig. 4D). Granzyme B-producing CD3+ CD8+ T cells were not detected in response to the other six epitopes.
FIG 4.
Cytokine secretion of CD8+ T cells in response to gB epitopes. (A) Gating strategy for the flow cytometric analysis of cytokine production in peptide-specific CD8+ T cells. CD8+ T cells were gated as CD8+ CD3+ CD14– CD19– live single lymphocytes. FSC, forward scatter; SSC, side scatter; AF700, Alexa Fluor 700. (B) Flow cytometric analysis of the proportions of IFN-γ-producing (IFN-γ+) and/or TNF-α-producing CD8+ gB214–223- or gB840–849-specific T cells after coculture with the corresponding peptides for 6 h. (C) Bar chart showing the percentages of IFN-γ+ and/or TNF-α+ peptide-specific CD8+ T cells in response to eight gB epitopes. Peptide-specific CD8+ T cells stimulated with DMSO were used as negative controls, and the means of the proportions of IFN-γ+ and/or TNF-α+ CD8+ T cells in the control groups are shown. (D) The percentages of granzyme B-producing gB214–223- or gB840–849-specific CD8+ T cells in response to the corresponding epitopes for 6 h were examined by flow cytometry.
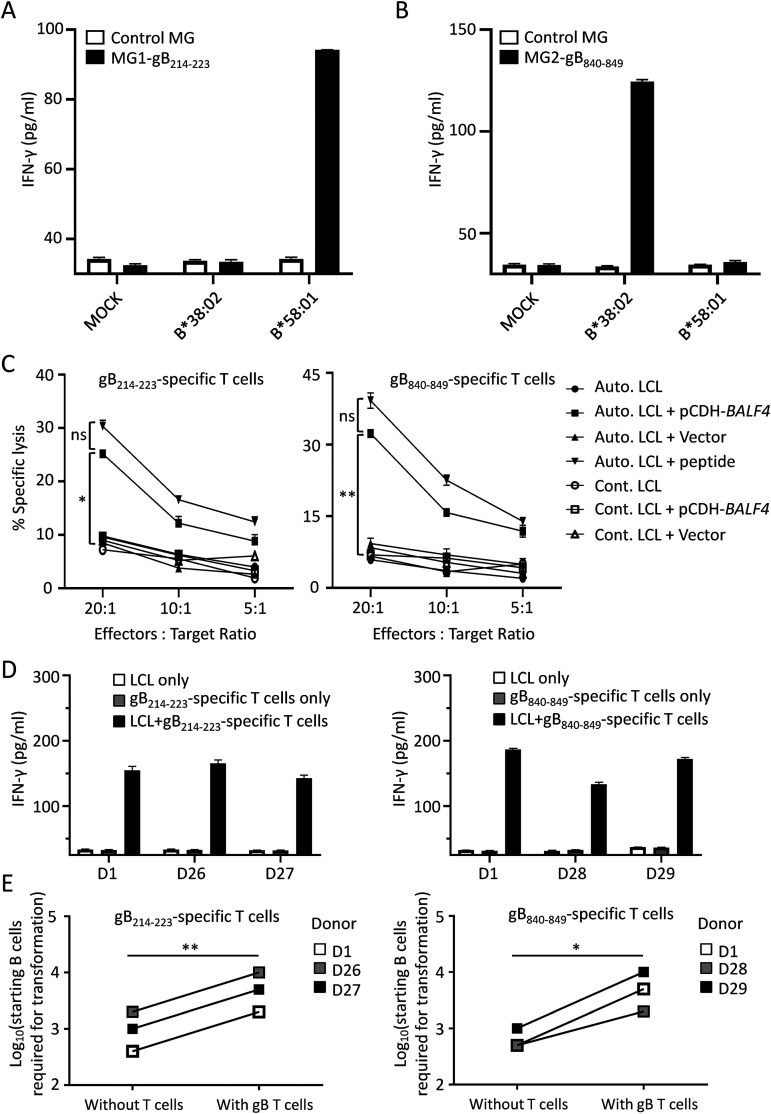
To test whether the two epitopes gB214–223 and gB840–849 could be endogenously processed, target cells (COS7 cells) overexpressing the corresponding epitope-encoding minigenes and HLA molecules were exposed to gB214–223 or gB840–849 epitope-specific T cells. COS7 cells expressing the minigene gB214–223 and HLA-B*58:01 stimulated gB214–223-specific CD8+ T cells to secrete IFN-γ (Fig. 5A). Similarly, COS7 cells expressing both HLA-B*38:02 and the minigene gB840–849 were recognized by gB840–849-specific CD8+ T cells (Fig. 5B). Additionally, LCLs were exposed to gB214–223 or gB840–849 peptide-specific T cells for 4 h, and specific lysis was examined by lactate dehydrogenase (LDH) cytotoxicity assays. Only autologous LCLs overexpressing the full-length gB protein or preloaded with target peptides were specifically recognized and lysed by these two types of peptide-specific T cells, whereas specific lysis could not be observed for autologous LCLs (Fig. 5C). EBV establishes a latency III program in LCLs that express a limited number of latent antigens, and only a small population of LCLs, <5%, are positive for gB expression (15). The low frequency of gB-positive LCLs may explain why no specific lysis of autologous LCLs by gB214–223- and gB840–849-specific T cells was observed after 4 h of coculture in the LDH cytotoxicity assays. We then cocultured gB214–223- and gB840–849-specific CD8+ T cells with LCLs derived from HLA-matched individuals for a prolonged time overnight and detected markedly elevated secretion of IFN-γ in the supernatant (Fig. 5D), indicating that gB214–223- and gB840–849-specific CD8+ T cells in fact recognized HLA-matched LCLs.
FIG 5.
gB peptide-specific CD8+ T cells recognized the endogenously processed epitopes gB214–223 and gB840–849 and limited B cell transformation after EBV infection. (A and B) The secretion of IFN-γ by peptide-specific CD8+ T cells was measured by an IFN-γ ELISA after coculture with COS7 cells coexpressing the HLA-I alleles indicated and minigenes expressing gB214–223 (MG1-gB214–223) (A) or gB840–849 (MG2-gB840–849) (B) and the control minigene (control MG). The means ± SEMs are shown (n = 3 technical replicates). (C) Specific lysis of LCLs after coculture with gB214–223- or gB840–849-specific CD8+ T cells was evaluated by the LDH release assay. Autologous LCLs (Auto. LCLs) or HLA-mismatched LCLs (Cont. LCLs) were infected with retrovirus expressing gB (pCDH-BALF4) or an empty vector and cocultured with gB214–223- or gB840–849-specific T cells at different effector-to-target cell ratios. Autologous LCLs pulsed with the corresponding peptides were used as the positive control. The means ± SDs are shown (n = 2 biological replicates). * and ** indicate significant differences observed between the groups (Auto. LCL and pCDH-BALF4) and other groups. *, P < 0.05; **, P < 0.005; ns, no significance (by one-way analysis of variance [ANOVA]). (D) The LCLs established from HLA-matched donors D1, D26, and D27 (HLA-A*58:01) and donors D1, D28, and D29 (HLA-A*38:02) were cocultured with gB214–223 (left)- and gB840–849 (right)-specific CD8+ T cells overnight, and the secretion of IFN-γ in the supernatant was measured by an IFN-γ ELISA. (E) gB214–223 (left)- and gB840–849 (right)-specific CD8+ T cells inhibited primary B cell transformation after EBV infection. The log10-transformed numbers of starting B cells required for transformation by EBV are indicated. Primary B cells enriched from the same HLA-matched donors as the ones described above for panel D were infected with the B95.8 virus and cocultured with or without a constant number of peptide-specific CD8+ T cells (10,000 T cells/well) at different effector-to-target cell ratios of between 1,000:1 and 1:3. LCL outgrowth was monitored by microscopy, and the B cell identity of the outgrowing cells was confirmed by flow cytometry. The data are shown as the means ± SDs (n = 3 biological replicates). *, P < 0.05, **, P < 0.005 (by a paired t test).
Delivered as EBV virion components, gB and other antigens are presented by recently infected B cells, and these de novo-infected B cells can be the targets of CD8+ T cells during the initial phase of infection (15, 22). Therefore, we next investigated whether gB-specific CD8+ T cells could inhibit B cell transformation after EBV infection. Primary B cells were sorted from donors whose HLA matched the HLA restrictions of gB214–223- and gB840–849-specific CD8+ T cells (HLA-B*58:01 and -B*38:02, respectively) and were infected by the B95.8 virus. A gradient number of starting B cells was cocultured with or without a fixed number of gB peptide-specific CD8+ T cells. B cell transformation by EBV was evaluated by flow cytometry. B cells sorted from three donors, D1, D26, and D27, were cocultured with gB214–223-specific CD8+ T cells, and five times the number of starting B cells was required for the successful transformation of B cells (Fig. 5E, left). In the presence of gB840–849-specific CD8+ T cells, we observed a 4- to 10-fold increase in the number of starting B cells from donors D1, D28, and D29 for successful EBV-driven transformation (Fig. 5E, right). These results indicate that gB-specific CD8+ T cells efficiently inhibited the EBV transformation of B cells.
gB-specific T cell responses in healthy EBV carriers and patients with NPC.
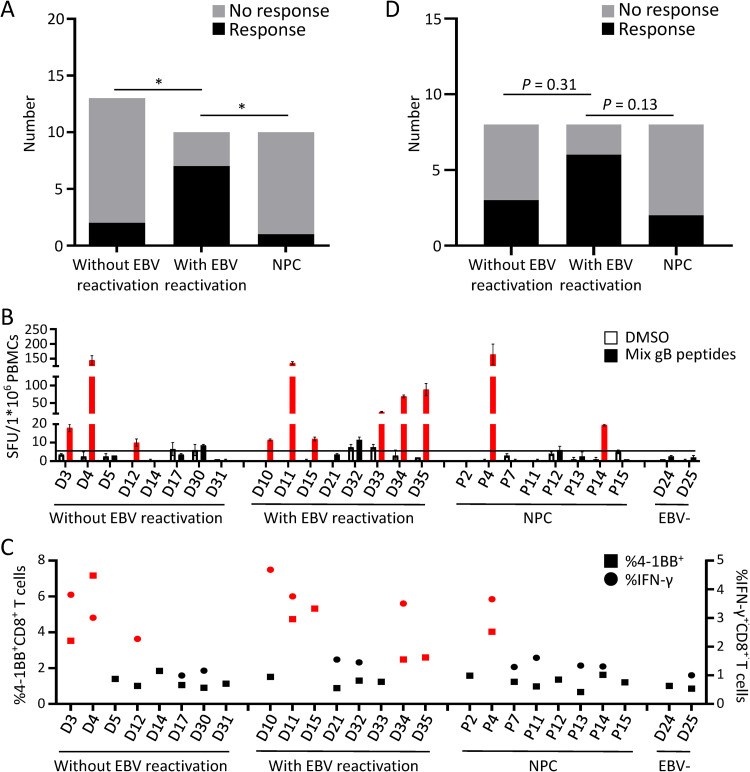
To understand the correlation between gB-specific T cell responses and the EBV infection status, we used VCA-gp125-IgA and EA-IgG to differentiate donors with EBV reactivation from latent viral carriers (see Materials and Methods for details). Except for two control EBV-seronegative donors, D24 and D25, all the healthy donors included in this study were positive for EBNA-1–IgG and VCA-IgG but negative for VCA-IgM, indicating that they were EBV carriers and excluding acute EBV infection. In contrast to most healthy EBV carriers who are VCA-gp125-IgA and EA-IgG negative, donors with EBV reactivation are expected to be positive for VCA-gp125-IgA and EA-IgG (23–25). Out of the 10 individuals whose EBV-specific T cell lines displayed a positive reaction to gB epitopes, 7 displayed asymptomatic EBV reactivation with positivity for VCA-gp125-IgA and EA-IgG (Tables 1 and 2). Interestingly, the EBV-specific T cell lines expanded from 7 of 10 donors with EBV reactivation (70%) displayed a positive reaction to the gB peptide, whereas the EBV-specific T cell lines from only 15% of donors without EBV reactivation (2 of 13) and 10% of patients with NPC (1 of 10) responded to the gB peptides (Fig. 6A). Taken together, these results indicate that gB-specific T cells are more likely to be present in individuals with EBV reactivation than in those without EBV reactivation and patients with NPC.
FIG 6.
gB-specific T cell responses in healthy viral carriers with and without EBV reactivation and patients with NPC. (A) Summary of the gB-specific T cell responses from the 33 EBV-specific T cell lines expanded by autologous LCLs. Two of 13 T cell lines expanded from healthy donors without EBV reactivation (15.4%), 7 of 10 from healthy donors with EBV reactivation (70%), and 1 of 10 from patients with NPC (10%) displayed positive T cell responses to gB peptides. *, P < 0.05 (by Fisher’s exact test). (B) The frequency of gB-specific CD8+ T cell responses in PBMCs from eight healthy viral carriers without EBV reactivation, eight donors with EBV reactivation, and eight patients with NPC was directly evaluated by the IFN-γ ELISPOT assay. The black horizontal line indicates the threshold level used to determine positive results (2 times the number of SFCs/1 × 106 unstimulated CD4-depleted PBMCs). The means ± SEMs are shown (n = 2 biological replicates). SFU, spot-forming units. (C) The frequencies of IFN-γ+ or 4-1BB+ CD8+ T cells in response to gB epitopes from eight healthy viral carriers without EBV reactivation, eight donors with EBV reactivation, and eight patients with NPC were measured by ICS and flow cytometry. IFN-γ ICS assays were not performed in six healthy donors and three patients due to the limited numbers of PBMCs. More than 2-fold increases in IFN-γ+ or 4-1BB+ cells compared with the DMSO background group are indicated in red in panels B and C. ELISPOT and flow cytometry assays were performed with CD4-depleted PBMCs stimulated with the gB CD8 epitope mixture or DMSO for 16 h. (D) Summary of the positive gB-specific CD8+ T cell responses detected from healthy viral carriers with and without EBV reactivation and patients with NPC directly ex vivo as shown in panels B and C. The CD8+ T cells from three of the eight donors without EBV reactivation (37.5%), six of the eight donors with EBV reactivation (75%), and two of the eight patients with NPC (25%) responded to the gB peptides. P values were yielded by Fisher’s exact test.
To further explore the frequency of gB-specific T cells in healthy EBV carriers with different EBV infection stages and patients with NPC, we examined gB-specific CD8+ T cell responses in peripheral blood mononuclear cells (PBMCs) from 16 donors with or without EBV reactivation and 8 patients with NPC directly ex vivo. CD8+ T cell responses after gB peptide stimulation were evaluated by IFN-γ ELISPOT assays and ICS, and the upregulation of the T cell activation marker 4-1BB on the CD8+ T cell surface was measured by flow cytometry. Compared to EBV-seronegative healthy donors (D24 and D25), six of eight donors with EBV reactivation (75%) exhibited at least a 2-fold upregulation of IFN-γ secretion, and four of them also showed increased cell surface expression of 4-1BB in response to gB peptide stimulation (Fig. 6B and C). In comparison, three of eight donors without EBV reactivation (37.5%) and only two of eight patients with NPC (25%) responded to the gB peptides (Fig. 6B and C). Although not significant at the level of a P value of <0.05, these P values indicate the trend in the higher frequency of gB-specific CD8+ T cells observed among the individuals with EBV reactivation than in those without reactivation and patients with NPC (Fig. 6D). Consistent with the results of the gB-specific reactivity in EBV-specific T cells expanded by autologous LCLs (Fig. 6A), these data further confirmed that gB-specific CD8+ T cells were more common in the peripheral blood from healthy donors with EBV reactivation than in individuals without EBV reactivation and patients with NPC.
Most gB epitopes are conserved in different EBV strains.
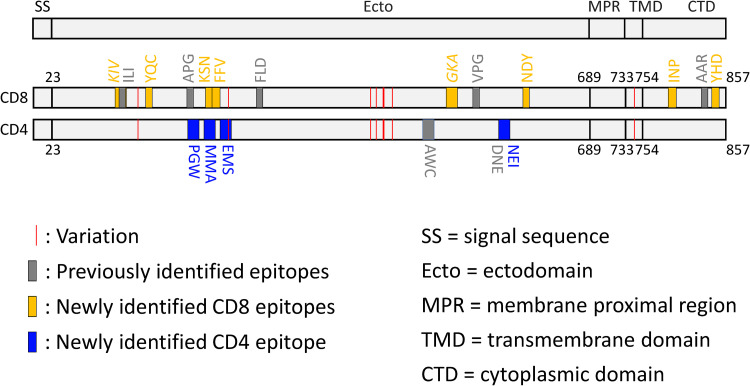
In this study, the gB peptide library was designed according to EBV strain B95.8. Since different EBV strains have been shown to have distinct geographical and population distributions (26, 27), we compared the amino acid sequences of 19 gB epitopes identified thus far among nine different EBV strains. All CD8+ T cell epitopes were fully conserved. One variation was present in a CD4+ T cell epitope (Fig. 7 and Table 2). T cell epitopes were distributed in the ectodomain, specifically domains I, III, and IV, as well as in the intracellular domain of gB, whereas we failed to identify any T cell epitopes in domain II (Fig. 7). These data indicate that full-length gB contains the most abundant T cell epitopes.
FIG 7.
Schematic of gB CD8 and CD4 epitopes. The newly identified gB CD8 and CD4 epitopes are shown as orange and blue vertical bars, respectively, and are indicated by the first 3 amino acids of each peptide. The gray bars highlight previously identified T cell epitopes. The red vertical lines represent gB amino acid variations among nine EBV strains. The CD8 epitopes for which the optimal sequences have not been defined are shown in italics. Details of all of the gB CD8 and CD4 epitopes are shown in Tables 1 and 2.
DISCUSSION
The characterization of T cell responses during natural human infections with VZV, HIV, and influenza virus has defined critical epitopes on target antigens and advanced the development of subunit vaccines (28–30). In this study, we comprehensively explored T cell responses to gB in healthy EBV carriers with and without viral reactivation and in patients with NPC using an overlapping peptide library covering the entire gB sequence. In addition to a previously identified FLD epitope, we identified 12 novel T cell epitopes in the context of seven new HLA restrictions that are common in Asian populations. Interestingly, we found that the gB-specific CD8+ T cells in healthy donors with EBV reactivation are more frequent than those in healthy donors without EBV reactivation and patients with NPC, suggesting that gB is a common target of CD8+ T cells in healthy donors with EBV reactivation. Our data have broadened the availability of gB T cell epitopes and HLA restrictions. In addition, because 18 of the 19 gB epitopes identified thus far are conserved among different EBV strains, they can be used to track and evaluate T cell responses to the gB subunit vaccine.
Of the five previously defined gB CD8+ T cell epitopes (Table 1), restriction was limited to four HLA alleles, including both HLA-A and HLA-B alleles but not HLA-C alleles. In the present study, we identified an additional eight gB CD8+ T cell epitopes restricted by HLA-A and HLA-B as well as HLA-C alleles (Table 1). Six of the eight new CD8+ T cell epitopes were restricted by common Asian HLA class I alleles, including HLA-A*24:02, HLA-B*13:01, HLA-B*38:02, HLA-B*55:02, HLA-B*58:01, and HLA-C*03:04. These HLA alleles cover approximately one-third of the population in South China and Southeast Asia (http://www.allelefrequencies.net/default.asp), significantly increasing the population coverage of the HLA restrictions of gB epitopes. Our study revealed that the defined epitopes were mainly restricted by HLA-B alleles, in accordance with a previous study (31). Of particular interest, among the newly identified HLA class I alleles, HLA-B*13:01 and HLA-B*55:02 are protective alleles, while HLA-B*38:02 and HLA-B*58:01 are susceptible alleles for the development of NPC, a disease endemic in South China and Southeast Asia and potentially linked to impaired T cell surveillance (32). Our study suggests that the gB epitopes can elicit a CD8+ T cell response restricted by common susceptible and protective HLA alleles for NPC, adding more value to the development of a gB subunit vaccine to prevent EBV infection or reactivation in populations in South China and Southeast Asia where NPC is endemic. In addition to CD8+ T cell epitopes, four CD4+ T cell epitopes were identified, although the precise HLA-II restrictions of the four CD4+ T cell epitopes require further investigation. Knowledge of gB T cell epitopes and their HLA restrictions will facilitate the generation of key reagents, for example, peptide-specific tetramers, to evaluate the T cell response to gB.
Except for one CD4+ T cell epitope, gB231–245, all of the other gB T cell epitopes were conserved among different geographically separated EBV strains (Fig. 7). We also found that T cell responses were highly enriched in one region from residues 190 to 245. The epitope-enriched region, located at ectodomain domain I of gB, is not only able to elicit both CD4+ and CD8+ T cell responses in Asian and Caucasian populations but also critical for gB-mediated membrane fusion with the two fusion loops located in domain I (10). In addition to epitope-enriched domain I, the C-terminal domain (CTD) also harbors CD8+ T cell epitopes in the context of several common HLA restrictions, including HLA-B*07, HLA-B*38:02, and HLA-B*55:02. Therefore, gB subunit vaccines containing an ectodomain and a CTD may elicit more T cell responses.
EBV establishes persistent infection through its latency in B cells, from which it intermittently reactivates the lytic cycle and shuttles between B cells and epithelial cells to produce progeny virus particles for transmission. In healthy carriers, the detection of virus in throat wash samples and the simultaneous appearance of VCA-gp125-IgA, EA-IgG, and EBNA-1–IgG clearly indicate EBV reactivation, in contrast to donors without reactivation, who are negative for VCA-gp125-IgA and EA-IgG (23–25). We found that healthy virus carriers with EBV reactivation showed a significantly higher rate of T cell response to the gB peptides than donors without EBV reactivation and patients with NPC. These results indicated that intermittent EBV reactivation may elicit gB-specific T cell responses in vivo. In addition to the specific lysis of gB-overexpressing LCLs, we showed that gB peptide-specific CD8+ T cells expanded from donors with EBV reactivation were also able to inhibit EBV-driven B cell transformation. Our study suggests that EBV reactivation elicits gB-specific T cell responses, which may in turn play a role in the control of EBV lytic activation.
In this study, we made a very interesting observation about the relatively low levels of gB-specific T cell responses in patients with NPC. Unlike healthy individuals with intermittent EBV reactivation, which elicits both gB-specific antibody (VCA-gp125-IgA) and T cell immune responses, despite an aberrantly strong IgA response to EBV antigens, T cell responses to gB in patients with NPC are less common than those in healthy donors with EBV reactivation. Currently, the mechanisms underlying this inconsistency remain unclear and may indicate that the form or origin of EBV reactivation involved in NPC tumorigenesis differs from that governing the EBV-positive serological response in healthy individuals. Elevated serum EBV IgA antibody levels can be detected 3 to 5 years before the onset of NPC, and the association between IgA antibody titers and NPC risk is strong, indicating that EBV reactivation plays a role in the early or premalignant stages of NPC (33). However, whether EBV reactivation occurs in submucosal lymphoid cells or sporadic malignant epithelial cells remains unclear since a cellular niche of lytic producer cells has not been identified so far in NPC tumors via histochemical staining (34). Furthermore, we failed to detect the gB protein using an immunohistochemistry (IHC) assay (data not shown) despite the abundant BALF4 (gB) mRNA levels in NPC tumors (35).
In agreement with the relatively low frequency of gB-specific T cell responses, most studies have reported only normal or suppressed T cell responses to EBV-encoded antigens in patients with NPC (36, 37), which differs from the highly elevated humoral immune responses observed for NPC. In contrast, in healthy seropositive individuals, the expression of a variety of EBV antigens during lytic viral replication gives rise to significant CD4 and CD8 T cell responses (38). We hypothesize that the loss of EBV-specific T cell immunity may contribute to the development of NPC. With a limited number of antigens being expressed, only latent or abortive lytic EBV infection, rather than full lytic infection, has been observed in NPC (34). This is probably a key mechanism underlying the immune escape of EBV-infected tumor cells. In addition to reduced antigen expression, multiple viral gene products, the expression of which has been observed in NPC tumors (39), including BNLF2a, BGLF5, and BILF1, inhibit the antigen processing machinery and major histocompatibility complex (MHC) molecule expression of infected cells (40–42). A viral interleukin-10 (vIL-10) homolog encoded by the BCRF1 gene also interferes with the immune recognition of infected cells by effector T cells (43, 44). Moreover, in NPC tumors, an immunosuppressive tumor microenvironment caused by persistent exposure to EBV antigens and the recruitment of immunosuppressive cells, such as myeloid-derived suppressor cells (MDSCs) and regulatory T cells (Tregs), leads to the functional impairment or exhaustion of effector T cells (36, 45). Altogether, these studies suggest that EBV has evolved various strategies to evade immune recognition and T cell surveillance in NPC.
In summary, we report the screening of gB epitopes from healthy EBV carriers and patients with NPC and show that gB-specific T cells potently restrict EBV-driven B cell transformation. The frequent detection of gB-specific T cell responses in EBV IgA serology-positive healthy individuals suggests that gB is a common target of T cells in vivo when EBV is reactivated. This study increases the understanding of the T cell immune responses to gB and suggests that a gB subunit vaccine may elicit T cell responses in the context of broad HLA restrictions. Precisely mapped gB T cell epitopes could contribute to immune monitoring and the development of a gB subunit vaccine.
MATERIALS AND METHODS
Ethics statement.
This study was approved by the institutional ethical review boards of the Sun Yat-sen University Cancer Center, and written informed consent was obtained from all participants.
Establishment of lymphoblastoid cell lines.
Heparinized peripheral blood (20 to 40 ml) was collected from 35 EBV-seropositive healthy donors, 2 EBV-seronegative control donors, and 15 patients with NPC. Human PBMCs were isolated by Ficoll density gradient centrifugation. A total of 2 × 106 PBMCs were used to establish EBV-infected LCLs using the B95.8 EBV strain (46). Briefly, PBMCs were incubated with the virus-containing supernatant and cultured in RF10 medium (RPMI 1640 with 10% fetal bovine serum [FBS], 1 mM minimal essential medium [MEM] sodium pyruvate, 2 mM l-glutamine, 5 mM HEPES buffer solution, 55 mM 2-mercaptoethanol, 100 mM MEM nonessential amino acids, 100 U/ml penicillin, and 100 mg/ml streptomycin; purchased from Gibco [Thermo Fisher Scientific]) that contained 0.4 μg/ml cyclosporine. After approximately 5 or 6 weeks, the LCLs were stable and could be used for subsequent experiments.
ELISA of EBV VCA-IgM, VCA-IgG, VCA-IgA, EA-IgG, and EBNA-1–IgG.
The titers of serum antibodies against EBV antigens were measured by an enzyme-linked immunosorbent assay (ELISA) using commercial kits from Euroimmun (Germany) for VCA-IgM and VCA-IgG and from IBL (Hamburg, Germany) for VCA-IgA, EA-IgG, and EBNA-1–IgG according to the manufacturers’ instructions. Briefly, the levels of seromarkers were standardized by calculating the ratio of the optical density (OD) of the sample to that of a reference control (rOD). The positive cutoff values for the rOD were indicated by the manufacturer’s instructions. Reactivated EBV infection is expected to be negative for VCA-IgM but positive for VCA-IgG, VCA-IgA, EA-IgG, and EBNA-1–IgG. A viral carrier without EBV reactivation is expected to be positive for VCA-IgG and EBNA-1–IgG but negative for VCA-IgM and EA-IgG. The positive serological marker for acute EBV infection is VCA-IgM.
Induction of EBV lytic production in LCLs.
LCLs were seeded in RF10 medium (5 × 105 cells/ml) and treated with 20 ng/ml 12-O-tetradecanoyl-phorbol-1-acetate (TPA; Sigma) and 5 μmol/liter sodium butyrate (NaB; Sigma) for 8 h. After treatment, LCLs were washed twice and cultured in fresh RF10 medium for an additional 72 h (TPA+NaB-LCLs). LCLs without any stimulation were used as controls (Ctrl-LCLs). After incubation, cells were collected for flow cytometric analysis.
Generation of EBV-specific T cell lines.
Donor- and patient-derived EBV-specific T cells were generated from cryopreserved PBMCs using autologous LCLs treated with TPA+NaB (TPA+NaB-LCLs) as antigen-presenting cells (APCs), as described previously (47). Briefly, PBMCs (2 × 106 cells/2-ml 24-well plate) were stimulated with irradiated (40 Gy) autologous TPA+NaB-LCLs at an effector-to-APC ratio of 40:1 in 50/50 medium (RF10 medium mixed 1:1 with X-Vivo-15 medium). Cultures were supplemented with recombinant human IL-2 (Sigma) (10 U/ml) on day 4, and 50% of the culture medium was changed every 2 days thereafter. After 10 to 12 days, viable cells were restimulated with irradiated (40 Gy) autologous TPA+NaB-LCLs at an effector-to-APC ratio of 4:1 in 50/50 medium. The IL-2 concentration in the culture was successively increased to 100 U/ml. Cells were analyzed by ELISPOT assays on days 21 to 25. For CD8+ T cell differentiation profile analysis, cells were monitored by flow cytometry on day 21.
Expansion of peptide-specific T cells.
T cells specific for the newly identified peptide epitopes were established using cryopreserved PBMCs (∼1 × 107), as previously described (48). Briefly, one-third of the PBMCs were preloaded with a single peptide (10 μg/ml) in X-Vivo-15 medium (Lonza) for 2 h at 37°C, washed twice, and mixed with the remaining autologous PBMCs in 50/50 medium. After 3 days, cultures with IL-2 (10 U/ml) were added, and half of the medium was replaced every 2 days thereafter. Cells were restimulated on day 7 with irradiated (50 Gy) autologous PBMCs preloaded with peptide. After 10 to 12 days, peptide-specific CD8+ T cells were purified by magnetic beads (Miltenyi Biotec) for HLA restriction analysis. In cytotoxicity assays, peptide-specific CD8+ T cells were used as effectors.
Cell sorting.
Peptide-specific CD8+ T cells and peptide-specific CD8-deleted (CD4-encriched) cells were isolated from peptide-specific T cells using microbeads (Miltenyi Biotec) according to the manufacturer’s instructions. The purity of CD8+ T cells was above 95%.
Peptides.
A total of 163 overlapping peptides (15-mers overlapping by 10 amino acids [aa]) covering the complete sequence of BALF4 (B95.8 strain; UniProtKB accession number P03188) and truncated peptides within 15-mer peptides were purchased from Sangon Biotech (Shanghai, China), with a purity of >95%. Seven peptides in the gB peptide library failed to be synthesized: peptides 2, 20, 118, 124, 126, 147, and 148. Synthetic peptides were dissolved in dimethyl sulfoxide (DMSO) at 20 mg/ml. As described previously, 15-mer overlapping peptides were pooled in 36 pools such that each peptide was represented in 2 pools (47). All peptides were represented by the position of their inclusive amino acids from the N terminus (e.g., gBxx–yy), and epitopes were also identified by the first 3 amino acids of the gB sequence. Consecutive 15-mer peptides were designated by their position starting from the N-terminal 15-mers [e.g., gB1–15(1) is peptide 1, and gB6–20(2) is peptide 2, etc.]. Aliquots of peptides were stored at −80°C.
IFN-γ ELISPOT assays.
IFN-γ ELISPOT assays were performed according to the manufacturer’s instructions (human IFN-γ ELISPOT set, BD ELISPOT). Briefly, ELISPOT plates were precoated overnight with an anti-IFN-γ monoclonal antibody (mAb). A total of 5 × 104 EBV-specific T cells were plated and stimulated with gB peptide pools (1 μg/ml each peptide) or individual peptides (0.01 to 10,000 ng/ml, as indicated). Equivalent volumes of DMSO and 1 μg/ml phytohemagglutinin (PHA) were used as negative and positive controls, respectively. After incubation for 16 to 20 h at 37°C in 5% CO2, the secondary biotinylated anti-IFN-γ antibody was added to the plates. Following incubation with streptavidin-horseradish peroxidase (HRP), the reactions were developed with the 3-amino-9-ethylcarbazole (AEC) substrate (AEC substrate set, BD ELISPOT). Spots were quantified on the plates using Immunospot (Cellular Technology Ltd., Shaker Heights, OH). The results are shown as spots per well and were considered positive if the magnitude of the response exceeded 2 or 5 times the background level of the negative control, as noted in the figure legends, and if more than 5 spots per well were observed. Two EBV-specific T cell lines from EBV-seronegative (VCA-IgM/G/A-negative) healthy donors were used as negative controls. In some experiments, cells were collected for flow cytometric analysis.
Retroviral infection of the COS7 tumor cell line and LCLs.
Briefly, 293T cells (8 × 106) were plated in a 100-mm plate overnight at 37°C. Ten micrograms of cloned pCDH-EF1α-puro containing individual HLA-I alleles or BALF4 (B95.8 strain) and packaging plasmids (6 μg psPAX2 and 4 μg pMD2.G) were transfected using 30 μl of polyethyleneimine (PEI) (Polysciences). Empty vectors were used as negative controls. Forty-eight hours later, lentiviruses were harvested and concentrated. COS7 cells (5 × 105) were plated in 6-well plates in RF10 medium overnight at 37°C, and lentiviruses encoding HLA-I alleles were added to the plates. LCLs (5 × 106) were mixed with lentiviruses encoding gB, and centrifugation was performed at 800 × g for 1 h at 37°C with 8 μg/ml Polybrene (Sigma). Next, LCLs were plated in 6-well plates. After incubation for 6 to 8 h at 37°C in 5% CO2, the medium was replaced. Transfectants were maintained by puromycin selection and confirmed to express HLA-I alleles or gB by staining with a pan-HLA-I antibody (clone W6/32; eBioscience) or anti-EBV gB mouse mAb (5B2; Santa Cruz Biotechnology).
HLA restriction analysis and definition of optimal epitopes.
COS7 cells overexpressing specific HLA-I alleles (2.5 × 104 cells) were plated in flat-bottom 96-well plates in RF10 medium and incubated overnight at 37°C. The following day, candidate peptides (10 μg/ml) were added and pulsed for 4 h, and the cells were washed three times with 50/50 medium. Peptide-specific CD8+ T cells (50,000 to 150,000 cells/well) were added to the wells and cocultured overnight at 37°C. Peptide-specific CD4-enriched T cell populations (100,000 to 500,000 cells/well) were plated and stimulated with the candidate peptides overnight at 37°C. After incubation, the supernatant was collected for the detection of IFN-γ (BioLegend), and cells were collected for flow cytometric analysis. The Immune Epitope Database and Analysis Resource (IEDB) (http://www.iedb.org) and SYFPEITHI (http://www.syfpeithi.de) algorithms were used to predict the optimal peptides used in the validation assays.
Flow cytometry.
Single-cell suspensions were prepared and incubated with anti-human FcR block (BioLegend) before incubation for 30 min at 4°C with fluorochrome-conjugated antibodies. Subsets of EBV-specific CD8+ T cells were detected using LiveDead-NIR (Thermo Fisher), CD3-BV650 (clone UCHT1; BD Biosciences), CD8-Alexa Fluor 647 (SK1; BioLegend), CD19-allophycocyanin (APC)-Cy7 (HIB19; BioLegend), CCR7-phycoerythrin (PE) (3D12; Thermo Fisher), and CD45RA-fluorescein isothiocyanate (FITC) (HI100; Thermo Fisher); gB peptide-specific CD8 T cells were assessed using LiveDead-NIR (Thermo Fisher), CD3-Brilliant Violet 650 (UCHT1; BD Biosciences), CD4-PE-Cy7 (RPA-T4; BioLegend), CD14-APC-Cy7 (63D3; BioLegend), CD19-APC-Cy7 (HIB19; BioLegend), CD137-PE (4B4-1; BioLegend), CD8-Alexa Fluor 647 (SK1; BioLegend), or CD8-Alexa Fluor 700 (RPA-T8; BioLegend). gB and gp350 on LCLs were stained with gB (5B2; Santa Cruz) and gp350 (72A1), respectively, and the Alexa Fluor 647 secondary antibody (Invitrogen) was then added at a 1:500 dilution. All antibodies were diluted at 1:100 unless otherwise noted. For intracellular molecule staining, peptide-specific T cells were plated in 96-well round-bottom plates and stimulated with individual peptides (10 μg per well) for 6 h in the presence of 5 μg/ml brefeldin A (BioLegend) and 2 μM monensin (BioLegend). Cells with no stimulation were used as a negative control, while cells stimulated with phorbol myristate acetate (PMA)-ionomycin (Sigma) were used as a positive control. Following incubation, the cells were subsequently surface stained, fixed and permeabilized with 1× fixation buffer/permeabilization wash buffer (BioLegend), and stained with the following antibodies at a 1:50 dilution: IFN-γ–PE (4S.B3; BioLegend), TNF-α–PerCP-Cy5.5 (MAb11; BioLegend), and granzyme B-FITC (GB11; BioLegend). Cells were acquired on a CytoFLEX S instrument (Beckman Coulter), and the data were analyzed using FlowJo software X 10.0.7 (TreeStar).
In vitro cytotoxicity assays.
The cytotoxicity of gB214–223- and gB840–849-specific T cells was analyzed using the Pierce LDH cytotoxicity assay kit (Thermo Scientific) according to the manufacturer’s instructions. Briefly, 1 × 104 target cells/well were incubated with effector cells (peptide-specific CD8+ T cells) in a 96-well round-bottom plate for 4 h at different effector-to-target cell ratios. After incubation, 50 μl of the sample medium was transferred to a new 96-well flat-bottom plate, and 50 μl of the reaction mixture was added. Following incubation at room temperature for 30 min, 50 μl of stop solution was added to the plate, and the absorbance was measured at 490 nm and 680 nm using a plate reader (MD SpectraMax Plus 384).
Minigene design.
Minigenes inducing gB peptide-specific CD8+ T cell responses were designed to encode candidate T cell epitopes (49). The NetChop 3.1 cleavage prediction tool was used to analyze whether the epitopes could be endogenously cleaved (50). Linker sequences K were included in the minigene gB214–223 to allow cleavage. Minigenes, including the Kozak sequence and stop codon, were codon optimized and synthesized by General Biosystems (Anhui, China). The minigenes were then cloned into the pCDH-EF1α-puro vector and yielded the following peptide products: MK KSNSPFDFFV for minigene gB214–223, M YHDPETAAAL for minigene gB840–849, and M FFVTTTGQTVE for minigene gB221–231 (control minigene).
gB epitope conservation analysis.
The full-length gB amino acid sequences of the nine EBV strains, including B95.8 (GenBank accession number V01555.2; NCBI protein accession number CAA24806.1), Akata (GenBank accession number KC207813.1; protein accession number AFY97902.1), GD1 (GenBank accession number AY961628.3; protein accession number AAY41155.1), M81 (GenBank accession number KF373730; protein accession number AGZ95225.1), C666 (GenBank accession number MK540243.1; protein accession number QCF47661.1), AG876 (GenBank accession number DQ279927.1; protein accession number ABB89287.1), Mutu (GenBank accession number KC207814.1; protein accession number AFY97983.1), YCCLE1 (GenBank accession number AP015016.1; protein accession number BAU51603.1), and SNU-719 (GenBank accession number AP015015; protein accession number ASU89799.1), were downloaded from the National Center for Biotechnology Information database. The sequences were aligned and analyzed using ClustalX 2.0 (51) and Jalview 2 (52).
CD8+ T cell inhibition of B cell outgrowth.
Primary B cells enriched from PBMCs were infected with wild-type EBV (B95.8) at a multiplicity of infection of 1. After 2 h of incubation at 37°C, primary B cells were seeded in 96-well round-bottom plates in RF10 medium containing 50 IU/ml IL-2 and cocultured with or without a constant number of gB-specific CD8+ T cells (10,000 T cells/well) at different effector-to-target cell ratios of between 1,000:1 and 1:3 in three replicates. Half of the medium in these cocultures was replaced once a week with fresh RF10 medium without cytokine supplementation. After 4 weeks of incubation, outgrowth was monitored by microscopy, and the B cell identity of the outgrowing cells was confirmed by flow cytometry.
Statistical analysis.
All statistical analyses were performed with GraphPad Prism 8.0 software and are indicated in the figure legends. The sample numbers (n) and replicates in each experiment are indicated in the figure legends. All statistical tests were two sided, and P values of <0.05 were considered statistically significant.
ACKNOWLEDGMENTS
This work was supported by grants from the National Natural Science Foundation of China (81872228 to M.X.), the Guangdong Basic and Applied Basic Research Foundation (2020B1515020002 to M.X.), and the Fundamental Research Funds for the Central Universities to M.X.
We have no competing interests to declare.
REFERENCES
- 1.Cohen JI. 2000. Epstein-Barr virus infection. N Engl J Med 343:481–492. 10.1056/NEJM200008173430707. [DOI] [PubMed] [Google Scholar]
- 2.Taylor GS, Long HM, Brooks JM, Rickinson AB, Hislop AD. 2015. The immunology of Epstein-Barr virus-induced disease. Annu Rev Immunol 33:787–821. 10.1146/annurev-immunol-032414-112326. [DOI] [PubMed] [Google Scholar]
- 3.Cohen JI, Fauci AS, Varmus H, Nabel GJ. 2011. Epstein-Barr virus: an important vaccine target for cancer prevention. Sci Transl Med 3:107fs7. 10.1126/scitranslmed.3002878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sokal EM, Hoppenbrouwers K, Vandermeulen C, Moutschen M, Leonard P, Moreels A, Haumont M, Bollen A, Smets F, Denis M. 2007. Recombinant gp350 vaccine for infectious mononucleosis: a phase 2, randomized, double-blind, placebo-controlled trial to evaluate the safety, immunogenicity, and efficacy of an Epstein-Barr virus vaccine in healthy young adults. J Infect Dis 196:1749–1753. 10.1086/523813. [DOI] [PubMed] [Google Scholar]
- 5.Gu SY, Huang TM, Ruan L, Miao YH, Lu H, Chu CM, Motz M, Wolf H. 1995. First EBV vaccine trial in humans using recombinant vaccinia virus expressing the major membrane antigen. Dev Biol Stand 84:171–177. [PubMed] [Google Scholar]
- 6.Chen J, Sathiyamoorthy K, Zhang X, Schaller S, Perez White BE, Jardetzky TS, Longnecker R. 2018. Ephrin receptor A2 is a functional entry receptor for Epstein-Barr virus. Nat Microbiol 3:172–180. 10.1038/s41564-017-0081-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang H, Li Y, Wang H-B, Zhang A, Chen M-L, Fang Z-X, Dong X-D, Li S-B, Du Y, Xiong D, He J-Y, Li M-Z, Liu Y-M, Zhou A-J, Zhong Q, Zeng Y-X, Kieff E, Zhang Z, Gewurz BE, Zhao B, Zeng M-S. 2018. Ephrin receptor A2 is an epithelial cell receptor for Epstein-Barr virus entry. Nat Microbiol 3:1–8. 10.1038/s41564-017-0080-8. [DOI] [PubMed] [Google Scholar]
- 8.Hutt-Fletcher LM. 2007. Epstein-Barr virus entry. J Virol 81:7825–7832. 10.1128/JVI.00445-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turk SM, Jiang R, Chesnokova LS, Hutt-Fletcher LM. 2006. Antibodies to gp350/220 enhance the ability of Epstein-Barr virus to infect epithelial cells. J Virol 80:9628–9633. 10.1128/JVI.00622-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Backovic M, Longnecker R, Jardetzky TS. 2009. Structure of a trimeric variant of the Epstein-Barr virus glycoprotein B. Proc Natl Acad Sci U S A 106:2880–2885. 10.1073/pnas.0810530106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pass RF. 2009. Development and evidence for efficacy of CMV glycoprotein B vaccine with MF59 adjuvant. J Clin Virol 46(Suppl 4):S73–S76. 10.1016/j.jcv.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schleiss MR, Choi KY, Anderson J, Mash JG, Wettendorff M, Mossman S, Van Damme M. 2014. Glycoprotein B (gB) vaccines adjuvanted with AS01 or AS02 protect female guinea pigs against cytomegalovirus (CMV) viremia and offspring mortality in a CMV-challenge model. Vaccine 32:2756–2762. 10.1016/j.vaccine.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L, Zhu L, Zhu H. 2016. Efficacy of varicella (VZV) vaccination: an update for the clinician. Ther Adv Vaccines 4:20–31. 10.1177/2051013616655980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malavige GN, Jones L, Black AP, Ogg GS. 2008. Varicella zoster virus glycoprotein E-specific CD4+ T cells show evidence of recent activation and effector differentiation, consistent with frequent exposure to replicative cycle antigens in healthy immune donors. Clin Exp Immunol 152:522–531. 10.1111/j.1365-2249.2008.03633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pudney VA, Leese AM, Rickinson AB, Hislop AD. 2005. CD8+ immunodominance among Epstein-Barr virus lytic cycle antigens directly reflects the efficiency of antigen presentation in lytically infected cells. J Exp Med 201:349–360. 10.1084/jem.20041542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turcanova V, Hollsberg P. 2004. Sustained CD8+ T-cell immune response to a novel immunodominant HLA-B*0702-associated epitope derived from an Epstein-Barr virus helicase-primase-associated protein. J Med Virol 72:635–645. 10.1002/jmv.20023. [DOI] [PubMed] [Google Scholar]
- 17.Khanna R, Burrows SR. 2000. Role of cytotoxic T lymphocytes in Epstein-Barr virus-associated diseases. Annu Rev Microbiol 54:19–48. 10.1146/annurev.micro.54.1.19. [DOI] [PubMed] [Google Scholar]
- 18.Adhikary D, Behrends U, Moosmann A, Witter K, Bornkamm GW, Mautner J. 2006. Control of Epstein-Barr virus infection in vitro by T helper cells specific for virion glycoproteins. J Exp Med 203:995–1006. 10.1084/jem.20051287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malekzadeh P, Pasetto A, Robbins PF, Parkhurst MR, Paria BC, Jia L, Gartner JJ, Hill V, Yu Z, Restifo NP, Sachs A, Tran E, Lo W, Somerville RP, Rosenberg SA, Deniger DC. 2019. Neoantigen screening identifies broad TP53 mutant immunogenicity in patients with epithelial cancers. J Clin Invest 129:1109–1114. 10.1172/JCI123791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarkizova S, Klaeger S, Le PM, Li LW, Oliveira G, Keshishian H, Hartigan CR, Zhang W, Braun DA, Ligon KL, Bachireddy P, Zervantonakis IK, Rosenbluth JM, Ouspenskaia T, Law T, Justesen S, Stevens J, Lane WJ, Eisenhaure T, Lan Zhang G, Clauser KR, Hacohen N, Carr SA, Wu CJ, Keskin DB. 2020. A large peptidome dataset improves HLA class I epitope prediction across most of the human population. Nat Biotechnol 38:199–209. 10.1038/s41587-019-0322-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Long HM, Leese AM, Chagoury OL, Connerty SR, Quarcoopome J, Quinn LL, Shannon-Lowe C, Rickinson AB. 2011. Cytotoxic CD4+ T cell responses to EBV contrast with CD8 responses in breadth of lytic cycle antigen choice and in lytic cycle recognition. J Immunol 187:92–101. 10.4049/jimmunol.1100590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abbott RJ, Quinn LL, Leese AM, Scholes HM, Pachnio A, Rickinson AB. 2013. CD8+ T cell responses to lytic EBV infection: late antigen specificities as subdominant components of the total response. J Immunol 191:5398–5409. 10.4049/jimmunol.1301629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maurmann S, Fricke L, Wagner HJ, Schlenke P, Hennig H, Steinhoff J, Jabs WJ. 2003. Molecular parameters for precise diagnosis of asymptomatic Epstein-Barr virus reactivation in healthy carriers. J Clin Microbiol 41:5419–5428. 10.1128/jcm.41.12.5419-5428.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Paschale M, Clerici P. 2012. Serological diagnosis of Epstein-Barr virus infection: problems and solutions. World J Virol 1:31–43. 10.5501/wjv.v1.i1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Obel N, Hoier-Madsen M, Kangro H. 1996. Serological and clinical findings in patients with serological evidence of reactivated Epstein-Barr virus infection. APMIS 104:424–428. 10.1111/j.1699-0463.1996.tb00737.x. [DOI] [PubMed] [Google Scholar]
- 26.Xu M, Yao Y, Chen H, Zhang S, Cao SM, Zhang Z, Luo B, Liu Z, Li Z, Xiang T, He G, Feng QS, Chen LZ, Guo X, Jia WH, Chen MY, Zhang X, Xie SH, Peng R, Chang ET, Pedergnana V, Feng L, Bei JX, Xu RH, Zeng MS, Ye W, Adami HO, Lin X, Zhai W, Zeng YX, Liu J. 2019. Genome sequencing analysis identifies Epstein-Barr virus subtypes associated with high risk of nasopharyngeal carcinoma. Nat Genet 51:1131–1136. 10.1038/s41588-019-0436-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Correia S, Palser A, Elgueta Karstegl C, Middeldorp JM, Ramayanti O, Cohen JI, Hildesheim A, Fellner MD, Wiels J, White RE, Kellam P, Farrell PJ. 2017. Natural variation of Epstein-Barr virus genes, proteins, and primary microRNA. J Virol 91:e00375-17. 10.1128/JVI.00375-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Doorn E, Liu H, Ben-Yedidia T, Hassin S, Visontai I, Norley S, Frijlink HW, Hak E. 2017. Evaluating the immunogenicity and safety of a BiondVax-developed universal influenza vaccine (Multimeric-001) either as a standalone vaccine or as a primer to H5N1 influenza vaccine: phase IIb study protocol. Medicine (Baltimore) 96:e6339. 10.1097/MD.0000000000006339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brosio F, Masetti G, Matteo G, Stefanati A, Gabutti G. 2018. A novel nonlive, adjuvanted herpes zoster subunit vaccine: a report on the emerging clinical data and safety profile. Infect Drug Resist 11:1401–1411. 10.2147/IDR.S148303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen H, Ndhlovu ZM, Liu D, Porter LC, Fang JW, Darko S, Brockman MA, Miura T, Brumme ZL, Schneidewind A, Piechocka-Trocha A, Cesa KT, Sela J, Cung TD, Toth I, Pereyra F, Yu XG, Douek DC, Kaufmann DE, Allen TM, Walker BD. 2012. TCR clonotypes modulate the protective effect of HLA class I molecules in HIV-1 infection. Nat Immunol 13:691–700. 10.1038/ni.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Forrest C, Hislop AD, Rickinson AB, Zuo J. 2018. Proteome-wide analysis of CD8+ T cell responses to EBV reveals differences between primary and persistent infection. PLoS Pathog 14:e1007110. 10.1371/journal.ppat.1007110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang M, Lautenberger JA, Gao X, Sezgin E, Hendrickson SL, Troyer JL, David VA, Guan L, McIntosh CE, Guo X, Zheng Y, Liao J, Deng H, Malasky M, Kessing B, Winkler CA, Carrington M, De The G, Zeng Y, O’Brien SJ. 2012. The principal genetic determinants for nasopharyngeal carcinoma in China involve the HLA class I antigen recognition groove. PLoS Genet 8:e1003103. 10.1371/journal.pgen.1003103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ji MF, Wang DK, Yu YL, Guo YQ, Liang JS, Cheng WM, Zong YS, Chan KH, Ng SP, Wei WI, Chua DT, Sham JS, Ng MH. 2007. Sustained elevation of Epstein-Barr virus antibody levels preceding clinical onset of nasopharyngeal carcinoma. Br J Cancer 96:623–630. 10.1038/sj.bjc.6603609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsao SW, Tsang CM, Lo KW. 2017. Epstein-Barr virus infection and nasopharyngeal carcinoma. Philos Trans R Soc Lond B Biol Sci 372:20160270. 10.1098/rstb.2016.0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu L, Lin Z, Wu Y, Dong J, Zhao B, Cheng Y, Huang P, Xu L, Xia T, Xiong D, Wang H, Li M, Guo L, Kieff E, Zeng Y, Zhong Q, Zeng M. 2016. Comprehensive profiling of EBV gene expression in nasopharyngeal carcinoma through paired-end transcriptome sequencing. Front Med 10:61–75. 10.1007/s11684-016-0436-0. [DOI] [PubMed] [Google Scholar]
- 36.Fogg M, Murphy JR, Lorch J, Posner M, Wang F. 2013. Therapeutic targeting of regulatory T cells enhances tumor-specific CD8+ T cell responses in Epstein-Barr virus associated nasopharyngeal carcinoma. Virology 441:107–113. 10.1016/j.virol.2013.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin X, Gudgeon NH, Hui EP, Jia H, Qun X, Taylor GS, Barnardo MCNM, Lin CK, Rickinson AB, Chan ATC. 2008. CD4 and CD8 T cell responses to tumour-associated Epstein-Barr virus antigens in nasopharyngeal carcinoma patients. Cancer Immunol Immunother 57:963–975. 10.1007/s00262-007-0427-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moss DJ, Burrows SR, Khanna R. 2007. EBV: immunobiology and host response, p 904–914. In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom. [PubMed] [Google Scholar]
- 39.Jin S, Li R, Chen MY, Yu C, Tang LQ, Liu YM, Li JP, Liu YN, Luo YL, Zhao Y, Zhang Y, Xia TL, Liu SX, Liu Q, Wang GN, You R, Peng JY, Li J, Han F, Wang J, Chen QY, Zhang L, Mai HQ, Gewurz BE, Zhao B, Young LS, Zhong Q, Bai F, Zeng MS. 2020. Single-cell transcriptomic analysis defines the interplay between tumor cells, viral infection, and the microenvironment in nasopharyngeal carcinoma. Cell Res 30:950–965. 10.1038/s41422-020-00402-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zuo J, Quinn LL, Tamblyn J, Thomas WA, Feederle R, Delecluse HJ, Hislop AD, Rowe M. 2011. The Epstein-Barr virus-encoded BILF1 protein modulates immune recognition of endogenously processed antigen by targeting major histocompatibility complex class I molecules trafficking on both the exocytic and endocytic pathways. J Virol 85:1604–1614. 10.1128/JVI.01608-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rowe M, Glaunsinger B, van Leeuwen D, Zuo J, Sweetman D, Ganem D, Middeldorp J, Wiertz EJ, Ressing ME. 2007. Host shutoff during productive Epstein-Barr virus infection is mediated by BGLF5 and may contribute to immune evasion. Proc Natl Acad Sci U S A 104:3366–3371. 10.1073/pnas.0611128104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Croft NP, Shannon-Lowe C, Bell AI, Horst D, Kremmer E, Ressing ME, Wiertz EJ, Middeldorp JM, Rowe M, Rickinson AB, Hislop AD. 2009. Stage-specific inhibition of MHC class I presentation by the Epstein-Barr virus BNLF2a protein during virus lytic cycle. PLoS Pathog 5:e1000490. 10.1371/journal.ppat.1000490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niiro H, Otsuka T, Abe M, Satoh H, Ogo T, Nakano T, Furukawa Y, Niho Y. 1992. Epstein-Barr virus BCRF1 gene product (viral interleukin 10) inhibits superoxide anion production by human monocytes. Lymphokine Cytokine Res 11:209–214. [PubMed] [Google Scholar]
- 44.Lee CH, Yeh TH, Lai HC, Wu SY, Su IJ, Takada K, Chang Y. 2011. Epstein-Barr virus Zta-induced immunomodulators from nasopharyngeal carcinoma cells upregulate interleukin-10 production from monocytes. J Virol 85:7333–7342. 10.1128/JVI.00182-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cai TT, Ye SB, Liu YN, He J, Chen QY, Mai HQ, Zhang CX, Cui J, Zhang XS, Busson P, Zeng YX, Li J. 2017. LMP1-mediated glycolysis induces myeloid-derived suppressor cell expansion in nasopharyngeal carcinoma. PLoS Pathog 13:e1006503. 10.1371/journal.ppat.1006503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nagy N. 2017. Establishment of EBV-infected lymphoblastoid cell lines. Methods Mol Biol 1532:57–64. 10.1007/978-1-4939-6655-4_3. [DOI] [PubMed] [Google Scholar]
- 47.Straathof KC, Leen AM, Buza EL, Taylor G, Huls MH, Heslop HE, Rooney CM, Bollard CM. 2005. Characterization of latent membrane protein 2 specificity in CTL lines from patients with EBV-positive nasopharyngeal carcinoma and lymphoma. J Immunol 175:4137–4147. 10.4049/jimmunol.175.6.4137. [DOI] [PubMed] [Google Scholar]
- 48.Koutsakos M, Illing PT, Nguyen THO, Mifsud NA, Crawford JC, Rizzetto S, Eltahla AA, Clemens EB, Sant S, Chua BY, Wong CY, Allen EK, Teng D, Dash P, Boyd DF, Grzelak L, Zeng W, Hurt AC, Barr I, Rockman S, Jackson DC, Kotsimbos TC, Cheng AC, Richards M, Westall GP, Loudovaris T, Mannering SI, Elliott M, Tangye SG, Wakim LM, Rossjohn J, Vijaykrishna D, Luciani F, Thomas PG, Gras S, Purcell AW, Kedzierska K. 2019. Human CD8(+) T cell cross-reactivity across influenza A, B and C viruses. Nat Immunol 20:613–625. 10.1038/s41590-019-0320-6. [DOI] [PubMed] [Google Scholar]
- 49.Stronen E, Toebes M, Kelderman S, van Buuren MM, Yang W, van Rooij N, Donia M, Boschen ML, Lund-Johansen F, Olweus J, Schumacher TN. 2016. Targeting of cancer neoantigens with donor-derived T cell receptor repertoires. Science 352:1337–1341. 10.1126/science.aaf2288. [DOI] [PubMed] [Google Scholar]
- 50.Nielsen M, Lundegaard C, Lund O, Keşmir C. 2005. The role of the proteasome in generating cytotoxic T-cell epitopes: insights obtained from improved predictions of proteasomal cleavage. Immunogenetics 57:33–41. 10.1007/s00251-005-0781-7. [DOI] [PubMed] [Google Scholar]
- 51.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 52.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. 2009. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]