Human cytomegalovirus (HCMV) is a herpesvirus that, like all herpesvirus, establishes a lifelong infection. HCMV remains a significant cause of morbidity and mortality in immunocompromised individuals and HCMV seropositivity is associated with age-related pathology.
KEYWORDS: cytomegalovirus, endothelial cells, herpesviruses, multivesicular body, secretory autophagy, vesicular trafficking
ABSTRACT
Human cytomegalovirus (HCMV), while highly restricted for the human species, infects a diverse array of cell types in the host. Patterns of infection are dictated by the cell type infected, but cell type-specific factors and how they impact tropism for specific cell types is poorly understood. Previous studies in primary endothelial cells showed that HCMV infection induces large multivesicular-like bodies (MVBs) that incorporate viral products, including dense bodies (DBs) and virions. Here, we define the nature of these large vesicles using a recombinant virus where UL32, encoding the pp150 tegument protein, is fused in frame with green fluorescent protein (GFP, TB40/E-UL32-GFP). In fibroblasts, UL32-GFP-positive vesicles were marked with classical markers of MVBs, including CD63 and lysobisphosphatidic acid (LBPA), both classical MVB markers, as well as clathrin and LAMP1. Unexpectedly, UL32-GFP-positive vesicles in primary human microvascular endothelial cells (HMVECs) were not labeled by CD63, and LBPA was completely lost from infected cells. We defined these UL32-positive vesicles in endothelial cells using markers for the cis-Golgi (GM130), the lysosome (LAMP1), and for autophagy (LC3B). These findings suggest that UL32-GFP-containing MVBs in fibroblasts are derived from the canonical endocytic pathway and take over the classical exosomal release pathway. In contrast, UL32-GFP-containing MVBs in HMVECs are derived from the early biosynthetic pathway and exploit a less-well-characterized early Golgi-LAMP1-associated noncanonical secretory autophagy pathway. These results reveal striking cell type-specific membrane trafficking differences in host pathways that are exploited by HCMV, which may reflect distinct pathways for virus egress.
IMPORTANCE Human cytomegalovirus (HCMV) is a herpesvirus that, like all herpesvirus, establishes a lifelong infection. HCMV remains a significant cause of morbidity and mortality in immunocompromised individuals and HCMV seropositivity is associated with age-related pathology. HCMV infects many cells in the human host and the biology underlying the different patterns of infection in different cell types is poorly understood. Endothelial cells are an important target of infection that contribute to hematogenous spread of the virus to tissues. Here, we define striking differences in the biogenesis of large vesicles that incorporate virions in fibroblasts and endothelial cells. In fibroblasts, HCMV is incorporated into canonical MVBs derived from an endocytic pathway, whereas HCMV matures through vesicles derived from the biosynthetic pathway in endothelial cells. This work defines basic biological differences between these cell types that may impact how progeny virus is trafficked out of infected cells.
INTRODUCTION
Human cytomegalovirus (HCMV) is a betaherpesvirus that is characterized by its ability to establish a lifelong latent infection in humans with the potential for reactivation (1). HCMV is prevalent worldwide, with a seroprevalence ranging from 45% to 99%, depending upon geographic location and socioeconomic factors (2). In healthy individuals, HCMV infection is typically asymptomatic (2, 3). However, in immunocompromised individuals, such as stem cell or organ transplant recipients, HCMV reactivation or primary infection can result in high morbidity and mortality (2, 4). HCMV is vertically transmitted to developing fetuses, and approximately 1 in 150 children are born with congenital HCMV infection in the United States (5), which can result in hearing impairment, microcephaly, and neurodevelopmental delays (6). Asymptomatic seropositivity has also been linked to increased risk for age-related, chronic inflammatory pathologies, including vascular disease, frailty, and immune dysfunction (7–11). Currently, there is no vaccine for HCMV (12). Understanding HCMV biology and the mechanisms by which the virus replicates is important for developing strategies to control virus-related pathology and disease.
While HCMV is highly restricted in its tropism for the human species, a wide variety of cells are susceptible to infection within the human host, including fibroblasts, hematopoietic progenitor cells, myeloid-lineage hematopoietic cells, smooth muscle cells, epithelial cells, and endothelial cells. Fibroblasts, endothelial cells, and epithelial cells are major targets of the virus as these cells play important roles in productive infection in the host (13). Fibroblasts have been the primary model for studying HCMV replication because of the ability of these cells to support robust productive replication. However, HCMV establishes a chronic, low-level persistence in endothelial cells. Viral gene products encoded by the ULb’ region of the HCMV genome, which is lost during serial passage of the virus in fibroblasts (14–16), are required for efficient entry and replication (17–25). While productive HCMV infection in fibroblasts is well understood, we understand much less about the biology of infection in endothelial cells. Endothelial cells are important targets of infection that undoubtedly contribute to HCMV hematogenous dissemination and pathogenesis, as endothelial cells comprise the interface between the circulating blood and organs. Infection of the endothelium increases the recruitment and extravasation of monocytes and decreases vascular permeability (19, 26–30). Further, proinflammatory signaling from the infected endothelium has been postulated to contribute to vascular disease (31–33).
In HCMV-infected human microvascular endothelial cells (HMVECs), we have observed the formation of large vesicles resembling multivesicular bodies (MVBs) that contain both virions and dense bodies (DBs), vesicles of viral tegument proteins (17). In this study, we investigated the origin and identity of the MVB-like vesicles that incorporate viral products in HMVECs and fibroblasts. Using a virus where the pp150 tegument protein, encoded by UL32, is fused to the green fluorescent protein (UL32-GFP), we characterized the large vesicles that incorporate virus products containing UL32-GFP, such as virions and DBs. Interestingly, these large vesicles that contain virions in HMVECs do not have classical markers of MVBs, but do contain the lysosomal marker LAMP1 and the cis-Golgi marker GM130 on the limiting membrane, and the autophagy marker LC3B in the lumen of the vesicles. In contrast, in fibroblasts, UL32-GFP virions were incorporated into MVBs containing classical MVB markers. This unexpected result indicates that HCMV accesses distinct trafficking pathways in HMVECs and fibroblasts and suggests the possibility of distinct routes of egress in these cell types. Indeed, further characterization of these vesicles indicates that infection of HMVECs exploits early biogenesis/exocytic pathways, whereas infection of fibroblasts exploits endocytic pathways.
RESULTS
UL32-GFP-positive vesicles in HMVECs are not classic MVBs.
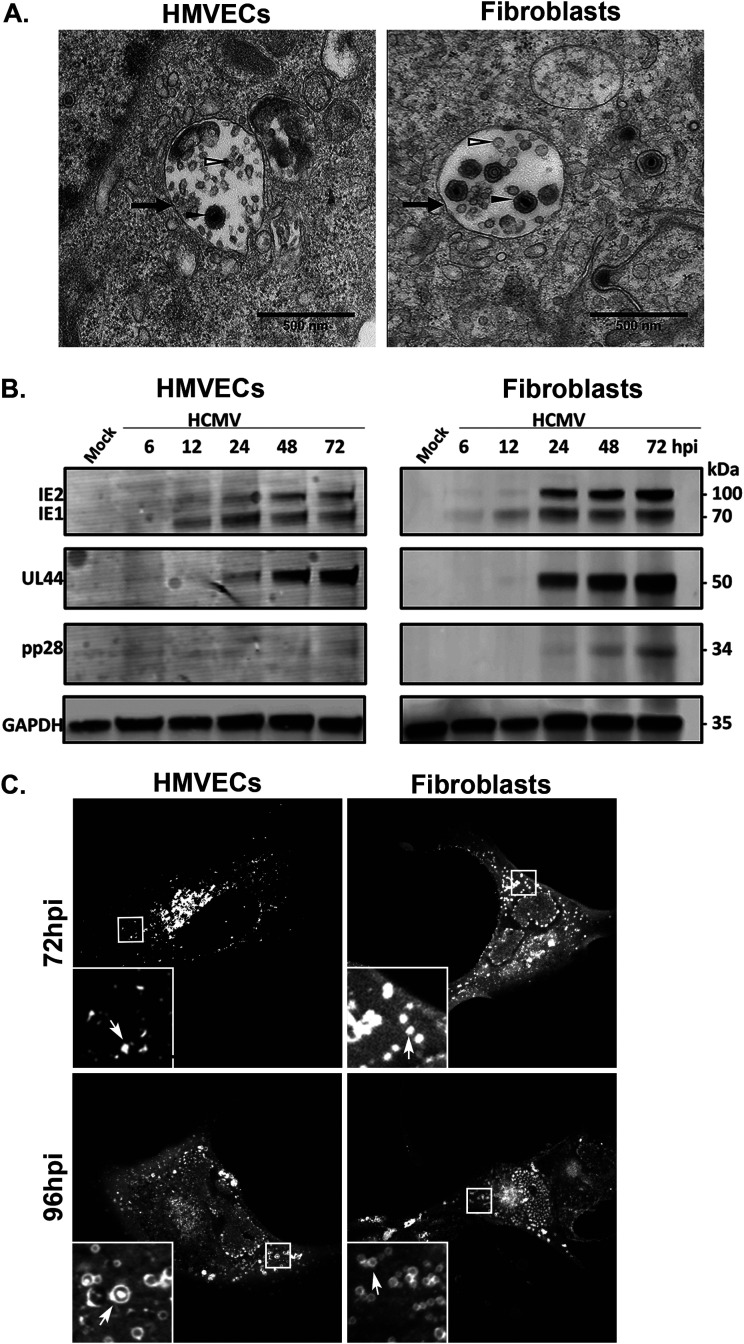
HCMV infection in HMVECs and fibroblasts induces the formation of large vesicles containing intraluminal vesicles that resemble multivesicular bodies (MVBs). By electron microscopy, we observed that virions are incorporated into these MVBs (Fig. 1A). Previous studies have shown that MVB biogenesis and incorporation of viral products are controlled by different viral proteins in different cell types, such as pUL135 in endothelial cells and pUL71 in fibroblasts (17, 34). The requirement of viral proteins in the incorporation of virus into MVBs suggested that this is a virus-directed outcome. However, the biochemical composition and biogenesis of MVBs induced by HCMV infection, and whether or not they differ in different cell types, has not been characterized.
FIG 1.
Virions are incorporated into MVBs. (A) TB40/E-infected HMVECs (MOI = 4) or fibroblasts (MOI = 2) were fixed, embedded, and sectioned for imaging by transmission electron microscopy at 96 and 72 hpi, respectively. Multivesicular bodies (black arrows) are present in the cytoplasm. The lumens of the MVBs contain virions (filled arrowheads) and ILVs (open arrowheads). Scale bars, 500 nm. (B) Kinetics of IE, E, and L proteins were analyzed over a time course. Lysates from HMVECs or fibroblasts were infected at an MOI of 4 or 2, respectively, were collected over the time course shown, and viral IE1/IE2, UL44 (early), and late (pp28) proteins were detected by immunoblotting, with GAPDH as a loading control. (C) UL32-GFP vesicle formation at 72 and 96 hpi in HMVECs and fibroblasts. Cells were imaged by confocal microscopy. UL32-GFP-positive vesicles are indicated by the arrows (insets).
To better define the MVBs induced by HCMV infection using both HMVECs and fibroblasts, we determined the conditions to equivalently infect each cell type and compare the progression of the viral program in each cell type. Multiplicities of infection (MOIs) were chosen to infect ∼60 to 70% of the cells for each cell type. We then analyzed progression of the viral program in each cell type by following the accumulation of viral proteins representing the immediate early (IE), early (UL44), and late (pp28) phases of infection (Fig. 1B). HMVECs accumulate peak levels of early- and late-phase proteins with a ∼24 h delay relative to their accumulation during infection in fibroblasts. Therefore, we chose to space experimental end points for each cell type by 24 h and perform all experiments at 72 hours postinfection (hpi) for fibroblasts and at 96 hpi for HMVECs, respectively. At these time points, the viral assembly compartment (VAC) is apparent in the majority of infected cells and the development of severe cytopathic effect (CPE) will not occur for another 24 h. Using these conditions, we infected HMVECs and fibroblasts with a recombinant TB40/E strain engineered to express a variant of the UL32/pp150 tegument protein that has been fused to green fluorescent protein (GFP) (35). To further validate the time points chosen for comparative analysis, we analyzed the formation of large UL32-positive vesicles in each cell type at each time point. UL32-GFP vesicles are well formed in fibroblasts by 72 hpi, but are not as apparent in HMVECs at that same time point (Fig. 1C). UL32-GFP vesicles become prominent by 96 hpi in HMVECs, at which time infected fibroblasts begin suffering severe virus-induced CPE. Therefore, the kinetics of UL32-positive large vesicle formation is delayed ∼24 h in HMVECs relative to fibroblasts, further supporting our choice of 72 and 96 hpi as time points for comparing infection in fibroblasts and HMVECs, respectively.
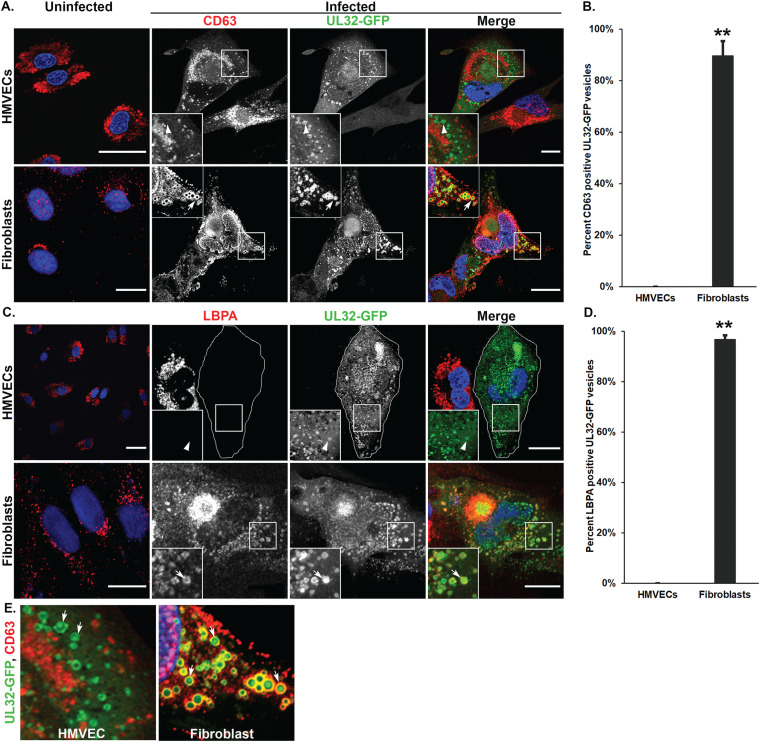
To define the composition of MVB-like vesicles that incorporate viral cargo, we labeled cells with the classic MVB markers CD63 and LBPA. CD63 is a tetraspanin-group protein that is present in MVBs or late endosomes (LEs) and at the cell surface (36). Lysobisphosphatidic acid (LBPA) is present on the membrane of the intraluminal vesicles (ILVs) and is used as marker of ILVs in MVBs (37). In uninfected HMVECs (Fig. 2A and C, top panels) and fibroblasts (Fig. 2A and C, bottom panels), CD63 and LBPA are distributed on punctate perinuclear structures. In infected HMVECs, CD63 (Fig. 2A, top) did not colocalize with UL32-GFP. Strikingly, LBPA was undetectable in infected HMVECs, although adjacent uninfected cells contained LBPA (Fig. 2C). In contrast, in infected fibroblasts, both CD63 (Fig. 2A, bottom) and LBPA (Fig. 2C, bottom) colocalized to large UL32-GFP-positive vesicles. Quantification of the vesicles in fibroblasts shows that 90% and 97% of UL32-GFP colocalized with CD63 and LBPA, respectively, compared to no colocalization of UL32-GFP with either marker in HMVECs (Fig. 2B and D). UL32-GFP labeled the membrane of CD63-positive vesicles in fibroblasts, and also accumulated in the lumen of vesicles, hence likely represents the incorporation of UL32-containing products of virus infection, including virions and dense bodies (Fig. 2E) observed by electron microscopy (Fig. 1). These results suggest that, while UL32-GFP-containing vesicles in fibroblasts are classical MVBs, vesicles in HMVECs are atypical or nonclassical.
FIG 2.
CD63 and LBPA show differential association with UL32-GFP-positive vesicles in HMVECs and fibroblasts. Uninfected or TB40/E-UL32-GFP-infected HMVECs (MOI = 4) or fibroblasts (MOI = 2) were fixed at 96 and 72 hpi, respectively. (A) Cells were labeled with mouse anti-CD63 (red) and imaged by confocal microscopy. Nuclei are stained with DAPI (blue). The large UL32-GFP-positive vesicles (green) do not colocalize with CD63 in HMVECs (arrowheads, insets) but do colocalize with CD63 in fibroblasts (arrows, insets). (B) Quantification of the percentage of vesicles that contain the CD63 that are also positive for UL32-GFP; **, P < 0.01. (C) Uninfected and infected cells were labeled with anti-LBPA (red) and imaged by confocal microscopy. Infection results in loss of LBPA from infected HMVECs, and the infected HMVEC is outlined due to lack of LBPA staining. UL32-GFP-positive vesicles (green) are indicated by the arrowhead (inset). In fibroblasts, UL32-GFP-positive vesicles colocalize with LBPA (arrows). (D) Quantification of the percentage of vesicles that contain the LBPA that are also positive for UL32-GFP; **, P < 0.01. For each quantification, 700 to 900 vesicles were counted for each marker. (E) UL32-GFP accumulates in the lumen of the large vesicles in both HMVEC and fibroblasts (arrows). Scale bars, 20 μm.
Clathrin heavy chain associates with UL32-GFP-positive vesicles.
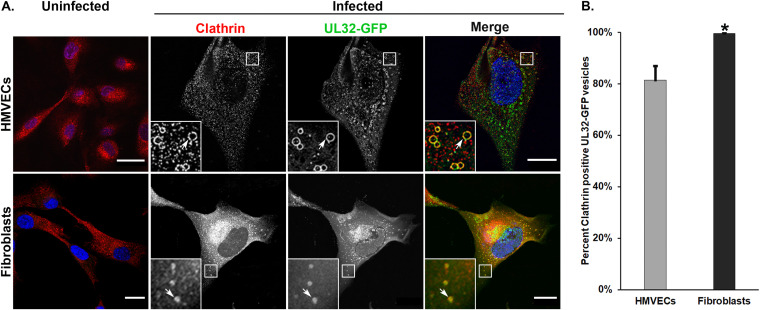
Clathrin acts as an integral component of both endocytic and biosynthetic cargo trafficking (38, 39). Clathrin domains on MVBs receive ubiquitinated cargo for transfer to endosomal sorting complexes required for transport (ESCRT)-I and incorporation into the MVB (40–42). Previous studies have shown accumulation of clathrin near the VAC in infected fibroblasts and this accumulation was decreased by the inhibition of endocytosis (43). However, the localization of clathrin to MVB-like vesicles in the context of HCMV infection has not been examined. In uninfected HMVECs, clathrin was widely distributed on punctate cytoplasmic structures and is tightly localized to the perinuclear region in fibroblasts (Fig. 3). However, in infected HMVECs (Fig. 3, top panels) or fibroblasts (Fig. 3, bottom panels), clathrin was localized on the large (average size of 0.6 to 1.0 μm) UL32-GFP-positive peripheral vesicles, (Fig. 3A, insets). Perinuclear clathrin accumulation in infected fibroblasts is consistent with previous findings that clathrin is relocalized to the VAC during infection (43, 44). However, the relocalization of clathrin to the VAC in infected HMVECs is less prominent. Quantification of the vesicles showed 81% of UL32-GFP-positive vesicles in infected HMVECs and 99% of UL32-GFP-positive vesicles in infected fibroblasts contained clathrin (Fig. 3B). These results demonstrate the accumulation of UL32-GFP on and within large, clathrin-positive, peripheral vesicles in both fibroblasts and HMVECs.
FIG 3.
Clathrin heavy chain colocalizes with UL32-GFP-positive vesicles in HMVECs and fibroblasts. (A) Uninfected or TB40/E-UL32-GFP-infected HMVECs (96 hpi, MOI 4) or fibroblasts (72 hpi, MOI 2) labeled with anti-clathrin heavy chain (red). In uninfected cells, clathrin is distributed in diffuse puncta throughout the cell. There is substantial colocalization of clathrin heavy chain and UL 32-GFP vesicles (arrows, insets) in both HMVECs and fibroblasts. In fibroblasts, clathrin also accumulates in the VAC (lower middle panel). (B) Quantification of clathrin-positive, UL32-GFP positive vesicles. Thirteen hundred vesicles were counted; *, P < 0.05. Scale bars, 20 μm.
UL32-GFP-positive vesicles lack common endosomal and biosynthetic compartment markers in HMVECs.
Our results show that the UL32-GFP-positive vesicles in HMVECs, but not fibroblasts, lack the classical MVB markers CD63 and LBPA. These findings suggest that UL32-positive vesicles in fibroblasts derive from the conventional endocytic pathway, whereas the vesicles in HMVECs may originate from a distinct pathway. To further investigate this, we examined the localization of early and late endosomal, and biosynthetic compartment markers (Fig. 4).
FIG 4.
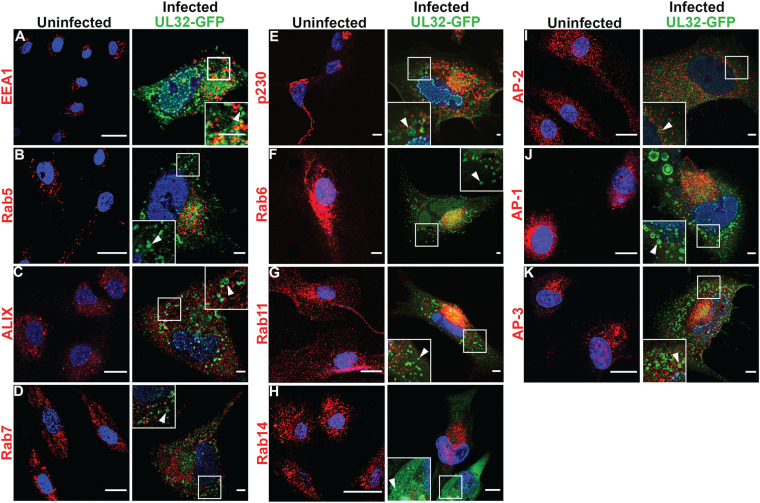
UL32-GFP-positive vesicles do not contain typical endocytic and biosynthetic trafficking markers in HMVECs. (A to K) Uninfected or TB40/E-UL32-GFP-infected HMVECs (96 hpi, MOI 4). Cells were labeled with anti-EEA1 (early endosomes) (A), anti-Rab5 (early endosomes) (B), anti-ALIX (late endosomes) (C), anti-Rab7 (late endosomes) (D), anti-p230 (trans-Golgi) (E), anti-Rab6 (trans-Golgi) (F), anti-Rab11 (recycling endosomes) (G), anti-Rab14 (endosomes) (H), anti-AP-2 (plasma membrane) (I), anti-AP-1 (trans-Golgi) (J), and anti-AP-3 (trans-Golgi) (K) antibodies and secondary antibodies conjugated to Alexa Fluor 647. The UL 32-GFP-positive vesicles (arrowheads) do not colocalize with any of these markers. Nuclei, blue. Scale bars, 20 μm.
The early endosome (EE) is often marked by the presence of early endosomal antigen (EEA) 1 and the small GTPase Rab5, which mediates early endosome docking and fusion in association with phosphatidylinositol 3-phosphate (PI3P) (45). Previous studies have shown that HCMV infection in fibroblasts and HMVECs redistributes EEA1 to the VAC, suggesting a possible role of EEA1 in viral maturation (46, 47). In uninfected HMVECs, EEA1 and Rab5 are distributed on puncta in the cytoplasm (Fig. 4A and B), and neither colocalized with the UL32-GFP vesicles in infected HMVECs (Fig. 4B). These results indicate that UL32-GFP-positive vesicles in HMVECs do not derive from or maintain markers of the early endosomal pathway.
ALG-2 interacting protein-X (ALIX) binds LBPA, and CHMP4B, a component of the ESCRT-III complex, which is involved in membrane curvature and fission (48). A previous study established that downregulation of ALIX reduces the LBPA labeling by 50% (49). As LBPA was undetectable in infected HMVECs, we asked if ALIX localization was affected in infected HMVECs. In uninfected HMVECs, ALIX localized to small cytoplasmic granules (Fig. 4C). In infected HMVECs, ALIX localization was unaltered, and did not colocalize with UL32-GFP vesicles (arrowhead, inset). We further tested for colocalization of the late endosome-associated small GTPase, Rab7. Rab7 regulates MVB biogenesis and trafficking from early endosomes to late endosomes and lysosomes (50). Uninfected HMVECs showed Rab7 labeling distributed throughout the cytoplasm and we observed no colocalization of Rab7 with UL32-GFP vesicles (Fig. 4D). In sum, these results indicate that none of the EE- or LE-associated markers colocalized with UL32-GFP-positive vesicles in HMVECs and suggest that these vesicles do not derive from the endosomal pathway.
Based on these results, we next tested if the UL32-GFP-positive vesicles in HMVECs could be derived from the biosynthetic pathway. We labeled infected HMVECs with the trans-Golgi marker p230. A tubular network of p230 was observed in the perinuclear region of uninfected HMVECs, but this trans-Golgi network (TGN) marker did not colocalize with UL32-GFP-positive vesicles (Fig. 4E), suggesting the UL32-GFP-positive vesicles do not derive from the trans-Golgi network.
Next, we tested the small GTPase Rab6, which regulates retrograde transport from the endosomal compartment via the trans-Golgi to the endoplasmic reticulum (51). A previous study found that Rab6 recruits UL32 to the viral assembly compartment by binding to dynein, a microtubule motor protein, in infected fibroblasts (52). Rab6 was scattered through the cytoplasm in uninfected cells and did not colocalize with UL32-GFP vesicles in infected HMVECs (Fig. 4F). These data suggest that endosomal and trans-Golgi membrane traffic is not involved in the biogenesis of these vesicles in HMVECs.
A previous study showed that murine CMV (MCMV) assembly compartment formation alters the recycling endosomal Rab cascade marked by the small GTPase Rab11 in infected fibroblasts (53). Rab11 was distributed as small punctate structures in the cytoplasm of uninfected cells and did not colocalize with UL32-GFP-positive vesicles in infected HMVECs (Fig. 4G). The small GTPase Rab14, which is involved in the biosynthetic trafficking between Golgi and endosomes and the plasma membrane (54), also did not colocalize with UL32-GFP-positive vesicles in HMVECs (Fig. 4H). These data suggest that UL32-positive vesicles are not derived from endosomal recycling compartments.
Due to our observation of the presence of clathrin on the membrane of UL32-GFP-positive vesicles in HMVECs, we next asked if UL32-GFP-positive vesicles contained the clathrin-associated adaptor proteins AP-2, AP-1, or AP-3. AP-2 binds to phosphatidylinositol 2-phosphate (PIP2) in the plasma membrane and the cargo in clathrin-mediated endocytosis (38). AP-2 labeled small puncta in uninfected HMVECs, and this distribution did not change with infection in infected HMVECs (Fig. 4I). However, AP-2 was somewhat less intense and more diffuse, possibly due to an infection-induced increase in cell size. AP-2 did not colocalize with UL32-GFP vesicles (Fig. 4I). AP-1 recruits clathrin to the TGN and contributes to the biogenesis of vesicles from the TGN (55). Uninfected HMVECs had small puncta of AP-1 in the perinuclear region (Fig. 4J), similar in appearance to p230 (see Fig. 4E). AP-1, like AP-2, also did not colocalize with UL32-GFP vesicles (Fig. 4J). AP-3 mediates the transport from the TGN to the LE or lysosome/lysosome-related organelles (56). As with all other adaptor proteins, AP-3 also did not colocalize with UL32-GFP-positive vesicles in infected HMVECs (Fig. 4K). Together, these findings demonstrate a striking lack of colocalization of UL32-GFP with common endosomal and biosynthetic markers and support the idea that these vesicles originate from a nonclassical membrane trafficking pathway in HMVECs.
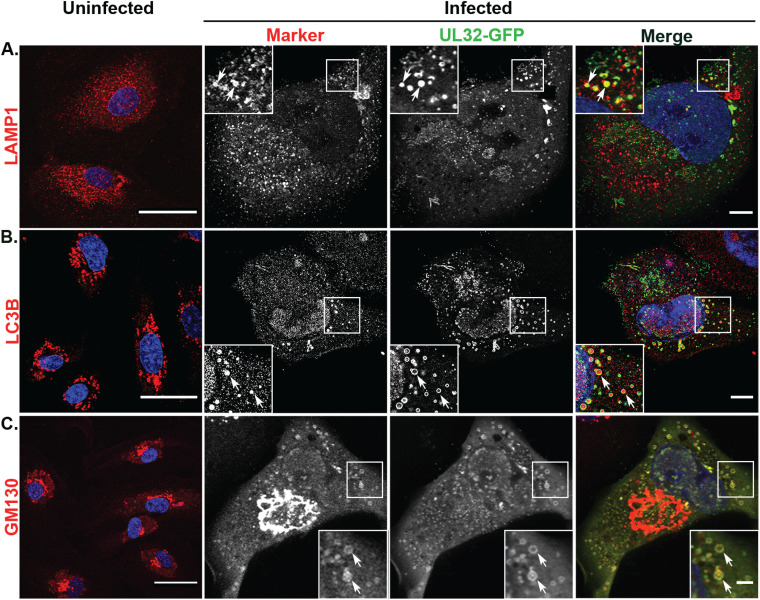
UL32-GFP-positive vesicles associate with lysosomal, autophagic, and early biosynthetic (cis-Golgi) markers in HMVECs.
MVB-associated cargoes have two primary fates: (i) fusion with the lysosomal compartment for degradation or (ii) transport to the plasma membrane for exosomal release (17, 57). Our results show that UL32-GFP-containing vesicles colocalized with classical MVB markers in fibroblasts but not in infected HMVECs. To further characterize these vesicles, we asked if UL32-GFP vesicles colocalize with the lysosomal compartment marker LAMP1. Uninfected HMVECs showed elongated vesicular labeling of LAMP1 throughout the cytoplasm (Fig. 5A). In infected HMVECs, LAMP1 labeling was not substantially altered. However, the UL32-GFP-containing vesicles colocalized with LAMP1 (Fig. 5A).
FIG 5.
LAMP1, GM130, and LC3B localize to UL32-GFP-positive vesicles in HMVECs. Uninfected or TB40/E-UL32-GFP-infected HMVECs were labeled with anti-LAMP1 (late endosomes-lysosomes) (A), anti-LC3B (autophagosomes) (B), and anti-GM130 (cis-Golgi) (C) antibodies and secondary antibodies conjugated to Alexa Fluor 647 (red) at 96 hpi (MOI 4). Nuclei, blue. In uninfected cells, LAMP1 and LC3B are distributed throughout the cytoplasm; GM130 is localized to the perinuclear region. In infected cells, LAMP1, LC3B, and GM130 all colocalize on UL32-GFP-positive vesicles (arrows). Scale bars, 20 μm.
A recent study reported that HCMV hijacks the autophagic component LC3B for envelopment of infectious virus particles, and knockdown of LC3B by shRNA demonstrated reduced viral production (58). Next, we asked if those nonclassical MVBs in infected HMVECs contained LC3B. In uninfected HMVECs, LC3B was distributed in spherical cytoplasmic structures (Fig. 5B). In infected HMVECs, we observed LC3B in the lumen of the UL32-GFP vesicles (Fig. 5B). Luminal localization of LC3B within nonclassical MVBs marked by UL32-GFP in HMVECs suggests that the biogenesis of MVBs in HMVECs may follow a nonclassical secretory autophagy pathway, like LC3-dependent EV loading and secretion (LDELS).
Lysosomal storage vesicles have been described that are labeled by LAMP1, clathrin, and the cis-Golgi marker GM130, but are negative for LE markers (59). These vesicles sometimes also contain LC3B (60). To determine if the large vesicles observed in HMVECs could be related to these structures, we next examined if the UL32-GFP vesicles contained GM130. In infected HMVECs, GM130 labeling was detected as a well-defined and characteristic ring structure around the VAC (Fig. 5C). Further, GM130 colocalizes with the UL32-GFP vesicles in HMVECs (Fig. 5C). These findings indicate that the UL32-GFP vesicles may derive from an early biosynthetic, Golgi-mediated pathway.
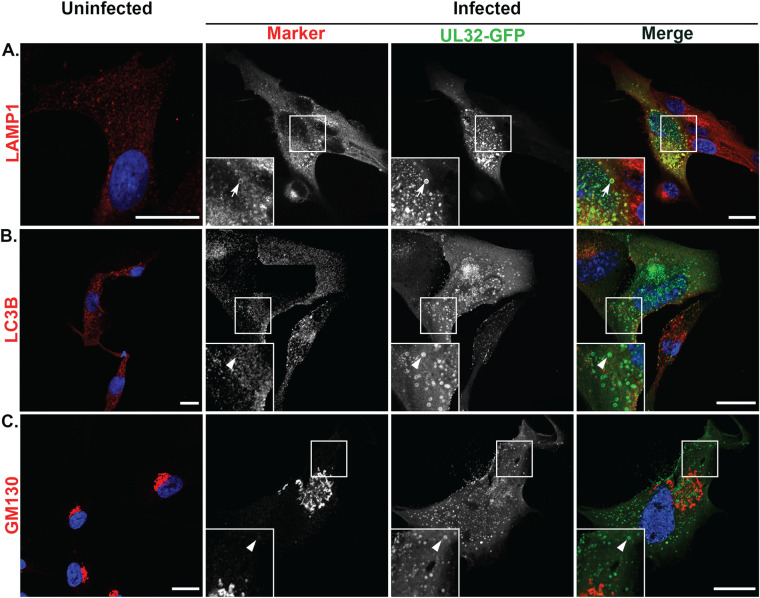
UL32-GFP-containing vesicles are marked by LAMP1, but not by LC3B and GM130, in fibroblasts.
We next sought to analyze the association of LAMP1, LC3B, and GM130 with UL32-GFP-positive vesicles in infected fibroblasts. LAMP1 labeling in uninfected fibroblasts appeared as vesicular structures in the cytoplasm (Fig. 6A). Infected fibroblasts demonstrated a scattered and fine granule labeling of LAMP1 around the VAC, consistent with previous observations (Fig. 6A) (46). However, UL32-GFP-containing vesicles also colocalized with LAMP1 (Fig. 6A). LAMP1 has been reported to localize in the late endosomes (LEs) apart from their primary localization at the lysosomal compartments (61–63). This finding indicates that colocalization of UL32-GFP-positive vesicles with LAMP1 in infected fibroblasts may derive from either LEs or from the lysosomes.
FIG 6.
LAMP1, but not GM130 or LC3B, colocalize with UL32-GFP-positive vesicles in fibroblasts. Uninfected or TB40/E-UL32-GFP-infected fibroblasts were labeled with anti-LAMP1 (late endosomes-lysosomes) (A), anti-LC3B (autophagosomes) (B), and anti-GM130 (cis-Golgi) (C) antibodies and secondary antibodies conjugated to Alexa Fluor 647 (red) at 72 hpi (MOI 2). Nuclei, blue. In uninfected cells, LAMP1 and LC3B are distributed in the cytoplasm; GM130 localizes to the perinuclear region. In infected cells, UL32-GFP vesicles colocalize with LAMP1 (arrows, A), but not with GM130 or LC3B (arrowheads, B, C). Scale bars, 20 μm.
LC3B was present on vesicle structures throughout the cytoplasm in uninfected fibroblasts (Fig. 6B). However, there was no colocalization with UL32-GFP vesicles in infected fibroblasts (Fig. 6B). In uninfected fibroblasts, GM130 localized in the perinuclear region, typical of the cis-medial Golgi (Fig. 6C). In infected fibroblasts, GM130 was localized to the classic ring-like structure of the VAC and showed no colocalization with UL32-GFP vesicles (Fig. 6C).
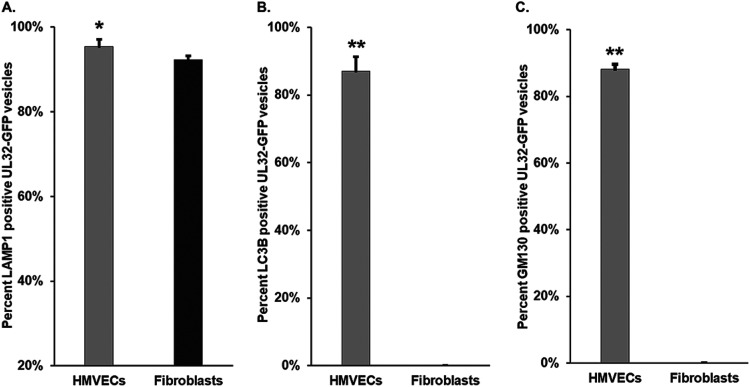
We quantified the colocalization of LAMP1, LC3B, and GM130 with UL32-GFP in both HMVECs and fibroblasts (Fig. 7). In HMVECs and fibroblasts, 92% and 99% of the UL32-GFP vesicles colocalized with LAMP1, respectively (Fig. 7A). However, LC3B and GM130 showed strong discordance between HMVECs and fibroblasts. In infected HMVECs, 87% of UL32-GFP-positive vesicles colocalized with LC3B, compared to none in infected fibroblasts (Fig. 7B). Similarly, 88% of UL32-GFP vesicles localized with GM130 in HMVECs, compared to none in infected fibroblasts (Fig. 7C). The absence of classical MVB markers on UL32-GFP vesicles in HCMV-infected HMVECs, coupled with the presence of LC3B and GM130, suggest that these vesicles are derived from a distinct pathway from that associated with infection in fibroblasts.
FIG 7.
Quantification of LAMP1-, LC3B-, and GM130-positive UL32-GFP vesicles. The percentage of LAMP1 (A), LC3B (B), and GM130 (C) positive UL32-GFP vesicles on both cell types are illustrated. Two hundred to four hundred vesicles were counted for each marker. Most vesicles in both cell types contain LAMP1. However, fibroblasts do not contain LC3B or GM130. *, P < 0.05; **, P < 0.01.
DISCUSSION
HCMV establishes distinct patterns of infection depending upon the cell type infected. HCMV infection in fibroblasts results in a robust productive infection, while vascular endothelium supports a smoldering, chronic infection that serves as a gateway to hematogenous dissemination to distant organs (13, 64, 65). However, the cell biology that distinguishes these patterns of infection and regulates post-entry tropism remains ill defined. Defining the molecular and cellular pathways that distinguish these patterns of infection is fundamental to understanding the basic biology of HCMV infection in diverse host tissues and supports efforts to identify antiviral targets for controlling hematogenous HCMV spread.
We previously observed the incorporation of virus products of replication, virions and dense bodies, into MVB-like bodies in endothelial cells (17). The incorporation of HCMV products into these MVB-like vesicles suggests they are either a means of egress for the virus or a host response, resulting in a dead-end for infection if their contents are targeted for lysosomal destruction. Viruses containing disruptions in the HCMV gene UL135 incorporate fewer virions and DBs into the MVB-like vesicles, suggesting that pUL135 may direct incorporation of virus and DBs into these vesicles, possibly for egress. The existence of a viral gene required for the incorporation of virus products into vesicles suggests this may not represent an unfortunate dead-end cellular response to infection, but a direct goal. Further, the copurification of host exosomal markers with HCMV progeny supports a role for MVBs in viral maturation and egress (92). Remarkably, we find in this study that HCMV exploits a specialized trafficking pathway in endothelial cells that is associated with a LAMP1-mediated biosynthetic and atypical secretory autophagy trafficking pathway. In contrast, infection in fibroblasts routes viral components and progeny through the endocytic pathway to classical MVBs.
Membrane markers present on MVBs in infected fibroblasts represent classical MVBs that include CD63, LBPA, and LAMP1 (36, 37, 57, 60, 66–69). It has been well established that other viruses, such as hepatitis C and hepatitis A, use the ESCRT component Hrs (hepatocyte growth factor receptor substrate required for formation of ILVs in MVB), as well as the VPS4 protein (AAA ATPase and master regulator of MVB sorting) and ALIX, for incorporation into MVBs for export (70, 71). The presence of these markers on virus-containing MVBs in fibroblasts suggests these vesicles originate from the canonical endocytic trafficking pathway. In the canonical endocytic pathway, the early endosome (EE) is the sorting station for cargo and determines its fate (72). The EE matures to form late endosome (LE) by replacing EE resident Rab5 with Rab7 (72). Also during maturation, LEs incorporate endosomal membrane to generate ILVs containing MVBs (36, 72). Depending on the cargo, MVBs can then transport cargo either to the degradative lysosomes or for fusion with the plasma membrane for release of contents from the cell (36, 57, 72).
Unexpectedly, the virus-containing MVBs in HMVECs lack classical MVB markers, indicating their biogenesis from a pathway distinct from endocytic trafficking pathways. The loss of LBPA in the infected HMVECs also implies the generation of altered ILVs and altered lipid metabolism. This may be due to viral interference with LBPA-synthesizing enzymes or their precursors, e.g., phosphatidyl glycerol (70). Altered ILV generation is supported by the lack of colocalization of these vesicles with the exosomal marker ALIX. It has been previously reported that ILVs lacking ALIX have altered biogenesis and shape, thus affecting the vesicle release pathway (49). ALIX-independent generation of ILVs has also been observed with other viruses, e.g., herpes simplex virus (HSV-1) and HIV (73, 74). Moreover, loss of LBPA results in reduced retention of cholesterol (75) and blocks classical exosomal release (76). Furthermore, other lipids, such as ceramides and their metabolite sphingosine-1 phosphate (S1P), have been implicated in MVB release (70). Thus, in microvascular endothelial cells, HCMV likely uses a distinct mechanism for egress, although a definitive role for these vesicles in egress remains to be determined.
In cells from individuals with lysosomal storage disorders, membrane vesicles marked by clathrin, LAMP1, GM130, and LC3B exist that are not directed to lysosomal compartments (77). These vesicles are thought to be exaggerations of normal membrane trafficking pathways. The colocalization of LAMP1, GM130, and the cluster of LC3B in the lumen of the UL32-GFP vesicles supports the idea that vesicles induced by HCMV infection in endothelial cells are similar and are not targeted for degradation; hence, they could be directed to fuse with the plasma membrane for release. This possibility remains to be tested.
Many viruses thwart canoncial degradative autophagy to evade immune response to infection and ensure survival of progeny virions, while other viruses hijack autophagic pathways for their replication or egress from the cell (78). HCMV in fibroblasts also hijacks autophagy during viral maturation (58). Secretory autophagy is a means by which cells secrete proteins; nucelic acids and lipids are part of extracellular vesicles. Secretory autophagy has been adopted by several viruses, including poliovirus, human rhinovirus 2, coxsackievirus, Zika virus, and Epstein Barr virus (EBV), as a route to exit host cells (79–84). These viruses utilize lipidated LC3B-rich membrane-bound vesicles, derived from the autophagosome, to transport virus particles out of the cells. The luminal localization of LC3B within the vesicles of HMVECs suggests that HCMV also uses a secretory autophagy pathway in this cell type. While little is known about luminal LC3B-dependent secretory autophagy, LC3-dependent EV loading and secretion (LDELS) has been reported (85). Interestingly, these vesicles accumulate ceramide, not LBPA, to facilitate membrane scission in an ESCRT-independent manner (85), which supports our finding of loss of LBPA in virus-containing vesicles. Most recently, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been shown to egress using a lysosomal pathway where the virus likely traffics to lysosomes from the Golgi/trans-Golgi network or the ER/ER-Golgi intermediate compartment (ERGIC) (86). This egress pathway requires deacidification of the lysosomes so the incorporated virus is not destroyed. HMVECs may use a similar egress pathway. However, the luminal LC3B in HCMV-containing vesicles also suggests the association with secretory autophagy, distinct from SARS-CoV-2 egress.
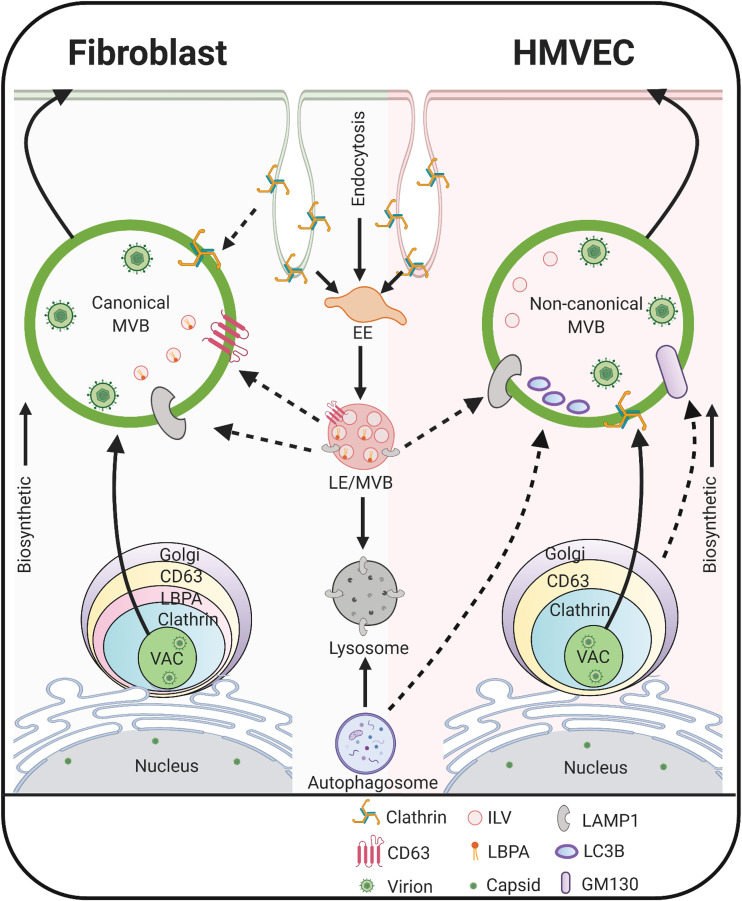
Based on our findings, we propose a model in which HCMV controls maturation and egress from distinct host pathways in infected HMVECs and fibroblasts (Fig. 8). The vesicles in infected fibroblasts contain CD63, LBPA, LAMP1, and clathrin, and thus may originate from canonical endocytic trafficking pathways and represent the classical exosomal release pathway (87). In contrast, vesicles in HMVECs contain clathrin, GM130, LAMP1, and luminal LC3B. This suggests these vesicles derive from the early biosynthetic route. Luminal LC3B may promote virion incorporation and recruit ceramide to facilitate membrane scission and release as EVs. Our results suggest that HCMV has evolved to differentially utilize two distinct vesicle trafficking pathways in fibroblasts and endothelial cells. The virus of host factors driving the redirection of trafficking have yet to be identified. Further, beyond egress and dissemination, EVs released from MVBs or secretory autophagosomes impact the biology of uninfected cells. For example, EVs derived from Kaposi sarcoma-associated herpesvirus lymphoma induce long-term reprogramming of endothelial cells to activate MEK/ERK signaling (93). The possibility that HCMV remodels EV release to impact host biology remains to be thoroughly investigated.
FIG 8.
Proposed model for UL32-postitive MVB biogenesis and egress in HCMV-infected fibroblasts and endothelial cells. In fibroblasts, HCMV infection induces classical MVBs, suggesting the virus exploits membrane trafficking in the endocytic pathway to promote viral incorporation into these vesicles, possibly for release via the exosomal pathway. In endothelial cells, HCMV infection generates MVBs that contain noncanonical markers, including Golgi and lysosomal markers and intraluminal LC3B, suggesting the MVBs in these cells originate from the early biosynthetic pathway and viral infection expands the noncanonical secretory autophagy pathway. The solid arrows denote the known cellular pathways; dotted arrows denote pathways expanded by HCMV infection. The image was created with BioRender.
MATERIALS AND METHODS
Cells.
Primary HMVECs (purchased from Lonza, Walkersville, MD) were cultured in EGM-2 MV Bulletkit medium (microvascular endothelial cell growth medium-2, Lonza) with 5% fetal bovine serum (FBS), 0.2 ml hydrocortisone, 2 ml human fibroblast growth factor (hFGF), 0.5 ml vascular endothelial growth factor (VEGF), 0.5 ml R3-insulin-like growth factor-1 (IGF-1), 0.5 ml ascorbic acid, 0.5 ml human epidermal growth factor (hEGF), and 100 U/ml penicillin. Human primary embryonic lung fibroblasts (MRC-5) (purchased from ATCC; Manassas, VA) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 10 mM HEPES, 1 mM sodium pyruvate, 2 mM l-alanyl glutamine, 0.1 mM nonessential amino acids, 100 U/ml penicillin, and 100 μg/ml streptomycin. All cells were cultured at 37°C in 5% CO2.
Viruses.
The low-passage strain of human cytomegalovirus TB40/E recombinant expressing UL32 fused to GFP, TB40/E-UL32GFP (88), was a generous gift from Christian Sinzger. Virus stocks were propagated by electroporation of infectious bacterial artificial chromosome (BAC) DNA into MRC5 cells and purified by density gradient centrifugation through a 20% D-sorbitol cushion at 20,000 RPM in an SW28 rotor (Beckman Coulter, Fullerton CA) for 80 min at 22°C. Virions were resuspended in Iscove’s modified Dulbecco’s medium (IMDM) containing 2% bovine serum albumin (BSA) and stored at −80°C. Infectious virus yields were determined by 50% tissue culture infectious dose (TCID50) on MRC5 fibroblasts. Virus was not serially propagated in fibroblasts. Virus titers were determined by TCID50 on fibroblasts, as previously described (89).
Transmission electron microscopy.
HMVECs or MRC-5 fibroblasts were mock infected or infected at a multiplicity of infection (MOI) of 4 or 2, respectively, with centrifugal enhancement. Infection medium was replaced at 24 h postinfection for HMVECs and at 6 h postinfection for MRC-5 with the normal growth medium for each respective cell type. Fibroblasts were harvested at 72 hpi and HMVECs at 96 h postinfection (hpi) and fixed in 2.5% glutaraldehyde and 0.1 M PIPES (piperazine-N, N′-bis [2-ethanesulfonic acid]) for 20 min. The fixed cell pellet was postfixed with osmium tetroxide in 0.1 M PIPES and dehydrated in a graded series of alcohol. Pellets were infiltrated with resin and cut into 100-nm sections. The sections were floated onto copper grids and imaged using a Phillips CM-12s transmission electron microscope. Cells were embedded and sectioned by the Arizona Research Laboratories, Arizona Health Sciences Center Core Facility.
Immunofluorescence imaging.
HMVEC and MRC-5 cells were seeded onto 12-mm glass coverslips in 24-well dishes 1 day prior to infection at an MOI of 4 or 2, respectively. Cells were processed for indirect immunofluorescence at 96 hpi for HMVECs and 72 hpi for MRC-5s. Cells were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) (except for LC3B staining, where we used ice-cold 100% methanol as a fixative) for 20 min. After washing with PBS, cells were incubated with 50 mM ammonium chloride (NH4Cl) for 10 min to quench free aldehydes. Cells were blocked and permeabilized with 0.2% saponin in 10% FBS-containing 1× PBS for 30 min (Table 1). After blocking, the cells were incubated with primary antibodies for at least 2 h. Primary antibodies were prepared by using antibody dilution buffer according to the manufacturer’s instructions. After rinsing with 1× PBS for at least three times, cells were incubated with secondary antibodies (Table 1) diluted in antibody dilution buffer, as per the antibody data sheet, for 1 h in a dark chamber. For methanol fixation of cells, we used anti-GFP secondary antibody since methanol quenches the fluorescence of GFP (90). After staining, DNA was stained with DAPI (4′,6-diamidino-2-phenylindole) according to the manufacturer's instructions (Molecular Probes). Coverslips were mounted using Prolong Diamond antifade mounting agent without DAPI (Invitrogen) according to manufacturer's instructions. For superresolution-structured illumination microscopy (SR-SIM) imaging, cells were cured for 60 h in a dark chamber before imaging. Confocal images were obtained with a Zeiss LSM880 inverted confocal microscope (Zeiss, Jena, Germany) with a 63× Plan Apo 1.4 NA oil immersion objective. All images were further processed using NIH-ImageJ (91). Representative single-plane images with 0.5 μm thickness were adjusted for brightness and contrast. Image galleries were created with Adobe Photoshop software (Adobe, San Jose, CA). SR-SIM images were obtained using a Zeiss ELYRA S.1 (SR-SIM) superresolution microscope with a 63× Plan-Apochromat 1.4 NA objective. SIM processing and channel alignment were rendered using ZEN imaging software. Quantification of the vesicles was conducted using Image J. In brief, invert look-up tables (LUTs) of single-plane images in two channels were processed and counted for coincidence of vesicles using point tools. At least one hundred vesicles were counted for each marker. Statistical analysis was performed using the Student’s t test and the error is indicated as standard error of the mean (SEM).
TABLE 1.
Antibodies used in this study
| Antibody | Speciesa | Source | Clone | Dilution | Cellular compartment |
|---|---|---|---|---|---|
| CD63 | M | DSHB | H5C6 | 3 μg/mlb | MVB |
| LBPA | M | Echelon | ML062915-21 | 1:100 | MVB |
| Clathrin | R | Cell signaling | 4796S | 1:50 | Endocytic vesicles and cell surface |
| EEA1 | R | Cell signaling | 3288 | 1:100 | Early endosome |
| Rab5 | M | BD transduction | 610725 | 1:50 | Early endosome |
| ALIX | M | Invitrogen | MA1-83977 | 1:200 | MVB |
| Rab7 | R | Cell signaling | 9367 | 1:100 | Late endosome |
| p230c | M | BD transduction | 611280 | 1:400 | Trans-Golgi |
| Rab6 | R | Cell signaling | 9625 | 1:400 | Golgi-ER |
| Rab11 | R | Cell signaling | 5589 | 1:100 | Recycling endosomes |
| Rab14 | R | Sigma-Aldrich | R0656 | 1:200 | Endosome-Golgi |
| AP-2 | M | Abcam | ab2807 | 1:100 | Endosome |
| AP-1d | M | Sigma-Aldrich | A4200 | 1:100 | Trans-Golgi network |
| AP-3 | M | DSHB | SA4 | 5 μg/mle | Trans-Golgi network |
| LAMP1 | R | Abcam | ab62562 | 1:500 | Late endosome-Lysosome |
| LC3Bf | R | Cell signaling | 3868 | 1:200 | Autophagosome |
| GM130 | M | BD transduction | 610822 | 1:100 | cis-Golgi |
| Anti-GFP | M | DSHB | G1-c 2ea | 5 μg/mlg | |
| Alexa Fluor 647 | NA | Thermo Fisher | 1:1000 | NA | |
| Alexa Fluor 568 | NA | Thermo Fisher | 1:1000 | NA | |
| IE1/2 | M | Gift from Thomas Shenk, Princeton University | 3H4 | 1:1,000 | NA |
| UL44 | M | Virusys | 10D8 | 1:2,500 | NA |
| pp28 | M | Gift from Thomas Shenk, Princeton University | 10B4-29 | 1:50 | NA |
| GAPDH | M | Abcam | 1:15,000 | NA |
R, Rabbit; M, Mouse.
Original concentration 71 μg/ml.
A generous gift from Samuel Campos, University of Arizona.
Original concentration 52 μg/ml.
Fixed with ice-cold 100% methanol.
Original concentration 239 μg/ml.
Immunoblotting.
Lysates were collected over a time course and separated by electrophoresis on precast 4 to 12% Tris-Bis SDS-PAGE gels (GenScript). Gels were transferred onto Immobilon-P PVDF membrane (EMD Millipore). Antibodies were incubated with blocking solution, either 5% milk in Tris-buffered saline with Tween 20 (TBST) or 5% BSA in TBST for 1 h at room temperature at the concentrations indicated in Table 1. After antibody staining, blots were incubated with fluorescent secondary antibodies and imaged using a Li-Cor Odyssey CLx infrared scanner with Image Studio software.
ACKNOWLEDGMENTS
We acknowledge Christian Sinzger (Ulm University, Germany) for the kind gift of the UL32-GFP virus and Thomas Shenk (Princeton University) for the gift of antibodies. We acknowledge Patricia Jansma, William Day, and Douglas Cromey of the University of Arizona Imaging Cores for assistance with confocal and transmission electron microscopy.
This work was funded by National Institutes of Health/National Institute of Allergy and Infectious Diseases grant AI131598 to F.G. and J.M.W.
REFERENCES
- 1.Schottstedt V, Blumel J, Burger R, Drosten C, Groner A, Gurtler L, Heiden M, Hildebrandt M, Jansen B, Montag-Lessing T, Offergeld R, Pauli G, Seitz R, Schlenkrich U, Strobel J, Willkommen H, von KC. 2010. Human cytomegalovirus (HCMV)—revised. Transfus Med Hemother 37:365–375. 10.1159/000322141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cannon MJ, Schmid DS, Hyde TB. 2010. Review of cytomegalovirus seroprevalence and demographic characteristics associated with infection. Rev Med Virol 20:202–213. 10.1002/rmv.655. [DOI] [PubMed] [Google Scholar]
- 3.Riou R, Bressollette-Bodin C, Boutoille D, Gagne K, Rodallec A, Lefebvre M, Raffi F, Senitzer D, Imbert-Marcille BM, Retiere C. 2017. Severe symptomatic primary human cytomegalovirus infection despite effective innate and adaptive immune responses. J Virol 91:e02245-16. 10.1128/JVI.02245-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nogalski MT, Collins-McMillen D, Yurochko AD. 2014. Overview of human cytomegalovirus pathogenesis. Methods Mol Biol 1119:15–28. 10.1007/978-1-62703-788-4_2. [DOI] [PubMed] [Google Scholar]
- 5.Britt WJ. 2018. Maternal immunity and the natural history of congenital human cytomegalovirus infection. Viruses 10:405. 10.3390/v10080405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Permar SR, Schleiss MR, Plotkin SA. 2018. Advancing our understanding of protective maternal immunity as a guide for development of vaccines to reduce congenital cytomegalovirus infections. J Virol 92:e00030-18. 10.1128/JVI.00030-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lebedeva AM, Shpektor AV, Vasilieva EY, Margolis LB. 2018. Cytomegalovirus infection in cardiovascular diseases. Biochemistry (Mosc) 83:1437–1447. 10.1134/S0006297918120027. [DOI] [PubMed] [Google Scholar]
- 8.Leng SX, Margolick JB. 2020. Aging, sex, inflammation, frailty, and CMV and HIV infections. Cell Immunol 348:104024. 10.1016/j.cellimm.2019.104024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Y, Liu S, Leng SX. 2019. Chronic low-grade inflammatory phenotype (CLIP) and senescent immune dysregulation. Clin Ther 41:400–409. 10.1016/j.clinthera.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 10.Sansoni P, Vescovini R, Fagnoni FF, Akbar A, Arens R, Chiu Y-L, Čičin-Šain L, Dechanet-Merville J, Derhovanessian E, Ferrando-Martinez S, Franceschi C, Frasca D, Fulöp T, Furman D, Gkrania-Klotsas E, Goodrum F, Grubeck-Loebenstein B, Hurme M, Kern F, Lilleri D, López-Botet M, Maier AB, Marandu T, Marchant A, Matheï C, Moss P, Muntasell A, Remmerswaal EBM, Riddell NE, Rothe K, Sauce D, Shin E-C, Simanek AM, Smithey MJ, Söderberg-Nauclér C, Solana R, Thomas PG, van Lier R, Pawelec G, Nikolich-Zugich J. 2014. New advances in CMV and immunosenescence. Exp Gerontol 55:54–62. 10.1016/j.exger.2014.03.020. [DOI] [PubMed] [Google Scholar]
- 11.Nikolich-Zugich J, Goodrum F, Knox K, Smithey MJ. 2017. Known unknowns: how might the persistent herpesvirome shape immunity and aging? Curr Opin Immunol 48:23–30. 10.1016/j.coi.2017.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xia L, Su R, An Z, Fu T-M, Luo W. 2018. Human cytomegalovirus vaccine development: immune responses to look into vaccine strategy. Hum Vaccin Immunother 14:292–303. 10.1080/21645515.2017.1391433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sinzger C, Grefte A, Plachter B, Gouw ASH, The TH, Jahn G. 1995. Fibroblasts, epithelial cells, endothelial cells and smooth muscle cells are major targets of human cytomegalovirus infection in lung and gastrointestinal tissues. J General Virology 76:741–750. 10.1099/0022-1317-76-4-741. [DOI] [PubMed] [Google Scholar]
- 14.Cha TA, Tom E, Kemble GW, Duke GM, Mocarski ES, Spaete RR. 1996. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J Virol 70:78–83. 10.1128/JVI.70.1.78-83.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davison AJ. 2002. Evolution of the herpesviruses. Vet Microbiol 86:69–88. 10.1016/s0378-1135(01)00492-8. [DOI] [PubMed] [Google Scholar]
- 16.Murphy E, Yu D, Grimwood J, Schmutz J, Dickson M, Jarvis MA, Hahn G, Nelson JA, Myers RM, Shenk TE. 2003. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc Natl Acad Sci U S A 100:14976–14981. 10.1073/pnas.2136652100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bughio F, Umashankar M, Wilson J, Goodrum F. 2015. Human cytomegalovirus UL135 and UL136 genes are required for postentry tropism in endothelial cells. J Virol 89:6536–6550. 10.1128/JVI.00284-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bughio F, Elliott DA, Goodrum F. 2013. An endothelial cell-specific requirement for the UL133-UL138 locus of human cytomegalovirus for efficient virus maturation. J Virol 87:3062–3075. 10.1128/JVI.02510-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hahn G, Revello MG, Patrone M, Percivalle E, Campanini G, Sarasini A, Wagner M, Gallina A, Milanesi G, Koszinowski U, Baldanti F, Gerna G. 2004. Human cytomegalovirus UL131-128 genes are indispensable for virus growth in endothelial cells and virus transfer to leukocytes. J Virol 78:10023–10033. 10.1128/JVI.78.18.10023-10033.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adler B, Scrivano L, Ruzcics Z, Rupp B, Sinzger C, Koszinowski U. 2006. Role of human cytomegalovirus UL131A in cell type-specific virus entry and release. J Gen Virol 87:2451–2460. 10.1099/vir.0.81921-0. [DOI] [PubMed] [Google Scholar]
- 21.Ryckman BJ, Chase MC, Johnson DC. 2008. HCMV gH/gL/UL128-131 interferes with virus entry into epithelial cells: evidence for cell type-specific receptors. Proc Natl Acad Sci U S A 105:14118–14123. 10.1073/pnas.0804365105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ryckman BJ, Jarvis MA, Drummond DD, Nelson JA, Johnson DC. 2006. Human cytomegalovirus entry into epithelial and endothelial cells depends on genes UL128 to UL150 and occurs by endocytosis and low-pH fusion. J Virol 80:710–722. 10.1128/JVI.80.2.710-722.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scrivano L, Sinzger C, Nitschko H, Koszinowski UH, Adler B. 2011. HCMV spread and cell tropism are determined by distinct virus populations. PLoS Pathog 7:e1001256. 10.1371/journal.ppat.1001256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang D, Shenk T. 2005. Human cytomegalovirus UL131 open reading frame is required for epithelial cell tropism. J Virol 79:10330–10338. 10.1128/JVI.79.16.10330-10338.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang D, Shenk T. 2005. Human cytomegalovirus virion protein complex required for epithelial and endothelial cell tropism. Proc Natl Acad Sci U S A 102:18153–18158. 10.1073/pnas.0509201102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Britt W. 2008. Manifestations of human cytomegalovirus infection: proposed mechanisms of acute and chronic disease. Curr Top Microbiol Immunol 325:417–470. 10.1007/978-3-540-77349-8_23. [DOI] [PubMed] [Google Scholar]
- 27.Grundy JE, Lawson KM, MacCormac LP, Fletcher JM, Yong KL. 1998. Cytomegalovirus-infected endothelial cells recruit neutrophils by the secretion of C-X-C chemokines and transmit virus by direct neutrophil-endothelial cell contact and during neutrophil transendothelial migration. J Infect Dis 177:1465–1474. 10.1086/515300. [DOI] [PubMed] [Google Scholar]
- 28.Revello MG, Gerna G. 2010. Human cytomegalovirus tropism for endothelial/epithelial cells: scientific background and clinical implications. Rev Med Virol 20:136–155. 10.1002/rmv.645. [DOI] [PubMed] [Google Scholar]
- 29.Waldman WJ, Knight DA, Huang EH, Sedmak DD. 1995. Bidirectional transmission of infectious cytomegalovirus between monocytes and vascular endothelial cells: an in vitro model. J Infect Dis 171:263–272. 10.1093/infdis/171.2.263. [DOI] [PubMed] [Google Scholar]
- 30.Bentz GL, Jarquin-Pardo M, Chan G, Smith MS, Sinzger C, Yurochko AD. 2006. Human cytomegalovirus (HCMV) infection of endothelial cells promotes naïve monocyte extravasation and transfer of productive virus to enhance hematogenous dissemination of HCMV. J Virol 80:11539–11555. 10.1128/JVI.01016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Streblow DN, Dumortier J, Moses AV, Orloff SL, Nelson JA. 2008. Mechanisms of cytomegalovirus-accelerated vascular disease: induction of paracrine factors that promote angiogenesis and wound healing. Curr Top Microbiol Immunol 325:397–415. 10.1007/978-3-540-77349-8_22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Botto S, Streblow DN, DeFilippis V, White L, Kreklywich CN, Smith PP, Caposio P. 2011. IL-6 in human cytomegalovirus secretome promotes angiogenesis and survival of endothelial cells through the stimulation of survivin. Blood 117:352–361. 10.1182/blood-2010-06-291245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caposio P, Orloff SL, Streblow DN. 2011. The role of cytomegalovirus in angiogenesis. Virus Res 157:204–211. 10.1016/j.virusres.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schauflinger M, Fischer D, Schreiber A, Chevillotte M, Walther P, Mertens T, von Einem J. 2011. The tegument protein UL71 of human cytomegalovirus is involved in late envelopment and affects multivesicular bodies. J Virol 85:3821–3832. 10.1128/JVI.01540-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sampaio KL, Cavignac Y, Stierhof Y-D, Sinzger C. 2005. Human cytomegalovirus labeled with green fluorescent protein for live analysis of intracellular particle movements. J Virol 79:2754–2767. 10.1128/JVI.79.5.2754-2767.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pols MS, Klumperman J. 2009. Trafficking and function of the tetraspanin CD63. Exp Cell Res 315:1584–1592. 10.1016/j.yexcr.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 37.Bissig C, Gruenberg J. 2013. Lipid sorting and multivesicular endosome biogenesis. Cold Spring Harb Perspect Biol 5:a016816. 10.1101/cshperspect.a016816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaksonen M, Roux A. 2018. Mechanisms of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol 19:313–326. 10.1038/nrm.2017.132. [DOI] [PubMed] [Google Scholar]
- 39.Jaiswal JK, Rivera VM, Simon SM. 2009. Exocytosis of post-Golgi vesicles is regulated by components of the endocytic machinery. Cell 137:1308–1319. 10.1016/j.cell.2009.04.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bache KG, Brech A, Mehlum A, Stenmark H. 2003. Hrs regulates multivesicular body formation via ESCRT recruitment to endosomes. J Cell Biol 162:435–442. 10.1083/jcb.200302131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sachse M, Urbé S, Oorschot V, Strous GJ, Klumperman J. 2002. Bilayered clathrin coats on endosomal vacuoles are involved in protein sorting toward lysosomes. Mol Biol Cell 13:1313–1328. 10.1091/mbc.01-10-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raiborg C, Bache KG, Gillooly DJ, Madshus IH, Stang E, Stenmark H. 2002. Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat Cell Biol 4:394–398. 10.1038/ncb791. [DOI] [PubMed] [Google Scholar]
- 43.Archer MA, Brechtel TM, Davis LE, Parmar RC, Hasan MH, Tandon R. 2017. Inhibition of endocytic pathways impacts cytomegalovirus maturation. Sci Rep 7:46069. 10.1038/srep46069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hasan MH, Davis LE, Bollavarapu RK, Mitra D, Parmar R, Tandon R. 2018. Dynamin is required for efficient cytomegalovirus maturation and envelopment. J Virol 92:e01418-18. 10.1128/JVI.01418-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McBride HM, Rybin V, Murphy C, Giner A, Teasdale R, Zerial M. 1999. Oligomeric complexes link Rab5 effectors with NSF and drive membrane fusion via interactions between EEA1 and syntaxin 13. Cell 98:377–386. 10.1016/s0092-8674(00)81966-2. [DOI] [PubMed] [Google Scholar]
- 46.Das S, Pellett PE. 2011. Spatial relationships between markers for secretory and endosomal machinery in human cytomegalovirus-infected cells versus those in uninfected cells. J Virol 85:5864–5879. 10.1128/JVI.00155-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Das S, Vasanji A, Pellett PE. 2007. Three-dimensional structure of the human cytomegalovirus cytoplasmic virion assembly complex includes a reoriented secretory apparatus. J Virol 81:11861–11869. 10.1128/JVI.01077-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mercier V, Laporte MH, Destaing O, Blot B, Blouin CM, Pernet-Gallay K, Chatellard C, Saoudi Y, Albiges-Rizo C, Lamaze C, Fraboulet S, Petiot A, Sadoul R. 2016. ALG-2 interacting protein-X (Alix) is essential for clathrin-independent endocytosis and signaling. Sci Rep 6:26986. 10.1038/srep26986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matsuo H, Chevallier J, Mayran N, Le Blanc I, Ferguson C, Faure J, Blanc NS, Matile S, Dubochet J, Sadoul R, Parton RG, Vilbois F, Gruenberg J. 2004. Role of LBPA and Alix in multivesicular liposome formation and endosome organization. Science 303:531–534. 10.1126/science.1092425. [DOI] [PubMed] [Google Scholar]
- 50.Vanlandingham PA, Ceresa BP. 2009. Rab7 regulates late endocytic trafficking downstream of multivesicular body biogenesis and cargo sequestration. J Biol Chem 284:12110–12124. 10.1074/jbc.M809277200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wanschers B, van de Vorstenbosch R, Wijers M, Wieringa B, King SM, Fransen J. 2008. Rab6 family proteins interact with the dynein light chain protein DYNLRB1. Cell Motil Cytoskeleton 65:183–196. 10.1002/cm.20254. [DOI] [PubMed] [Google Scholar]
- 52.Indran SV, Britt WJ. 2011. A role for the small GTPase Rab6 in assembly of human cytomegalovirus. J Virol 85:5213–5219. 10.1128/JVI.02605-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lučin P, Kareluša L, Blagojević Zagorac G, Mahmutefendić Lučin H, Pavišić V, Jug Vučko N, Lukanović Jurić S, Marcelić M, Lisnić B, Jonjić S. 2018. Cytomegaloviruses exploit recycling Rab proteins in the sequential establishment of the assembly compartment. Front Cell Dev Biol 6:165–165. 10.3389/fcell.2018.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Junutula JR, De Maziere AM, Peden AA, Ervin KE, Advani RJ, van Dijk SM, Klumperman J, Scheller RH. 2004. Rab14 is involved in membrane trafficking between the Golgi complex and endosomes. Mol Biol Cell 15:2218–2229. 10.1091/mbc.e03-10-0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burgess J, Jauregui M, Tan J, Rollins J, Lallet S, Leventis PA, Boulianne GL, Chang HC, Le Borgne R, Kramer H, Brill JA. 2011. AP-1 and clathrin are essential for secretory granule biogenesis in Drosophila. Mol Biol Cell 22:2094–2105. 10.1091/mbc.E11-01-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park SY, Guo X. 2014. Adaptor protein complexes and intracellular transport. Biosci Rep 34:e00123. 10.1042/BSR20140069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vacca F, Scott C, Gruenberg J. 2016. The late endosome, p 201–210. In Bradshaw RA, Stahl PD (ed), Encyclopedia of cell biology. 10.1016/B978-0-12-394447-4.20017-5. Academic Press, Waltham, MA. [DOI] [Google Scholar]
- 58.Taisne C, Lussignol M, Hernandez E, Moris A, Mouna L, Esclatine A. 2019. Human cytomegalovirus hijacks the autophagic machinery and LC3 homologs in order to optimize cytoplasmic envelopment of mature infectious particles. Sci Rep 9:4560. 10.1038/s41598-019-41029-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vitry S, Bruyère J, Hocquemiller M, Bigou S, Ausseil J, Colle M-A, Prévost M-C, Heard JM. 2010. Storage vesicles in neurons are related to Golgi complex alterations in mucopolysaccharidosis IIIB. Am J Pathol 177:2984–2999. 10.2353/ajpath.2010.100447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Piper RC, Katzmann DJ. 2007. Biogenesis and function of multivesicular bodies. Annu Rev Cell Dev Biol 23:519–547. 10.1146/annurev.cellbio.23.090506.123319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baba K, Kuwada S, Nakao A, Li X, Okuda N, Nishida A, Mitsuda S, Fukuoka N, Kakeya H, Kataoka T. 2020. Different localization of lysosomal-associated membrane protein 1 (LAMP1) in mammalian cultured cell lines. Histochem Cell Biol 153:199–213. 10.1007/s00418-019-01842-z. [DOI] [PubMed] [Google Scholar]
- 62.Eskelinen EL, Tanaka Y, Saftig P. 2003. At the acidic edge: emerging functions for lysosomal membrane proteins. Trends Cell Biol 13:137–145. 10.1016/s0962-8924(03)00005-9. [DOI] [PubMed] [Google Scholar]
- 63.Saftig P, Klumperman J. 2009. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat Rev Mol Cell Biol 10:623–635. 10.1038/nrm2745. [DOI] [PubMed] [Google Scholar]
- 64.Goodrum F, Bughio F. 2015. Viral infection at the endothelium. Oncotarget 6:26541–26542. 10.18632/oncotarget.5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sahni SK. 2007. Endothelial cell infection and hemostasis. Thromb Res 119:531–549. 10.1016/j.thromres.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 66.Bucci C, Stasi M. 2016. Endosome to lysosome transport, p 408–417. In Bradshaw RA, Stahl PD (ed), Encyclopedia of cell biology. 10.1016/B978-0-12-394447-4.20041-2. Academic Press, Waltham, MA. [DOI] [Google Scholar]
- 67.Piper RC. 2013. Multivesicular bodies: biogenesis and function. 10.1036/1097-8542.YB130004. [DOI] [PMC free article] [PubMed]
- 68.Hessvik NP, Llorente A. 2018. Current knowledge on exosome biogenesis and release. Cell Mol Life Sci 75:193–208. 10.1007/s00018-017-2595-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kobayashi T, Stang E, Fang KS, de Moerloose P, Parton RG, Gruenberg J. 1998. A lipid associated with the antiphospholipid syndrome regulates endosome structure and function. Nature 392:193–197. 10.1038/32440. [DOI] [PubMed] [Google Scholar]
- 70.Gruenberg J. 2020. Life in the lumen: the multivesicular endosome. Traffic 21:76–93. 10.1111/tra.12715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ramakrishnaiah V, Thumann C, Fofana I, Habersetzer F, Pan Q, de Ruiter PE, Willemsen R, Demmers JAA, Stalin Raj V, Jenster G, Kwekkeboom J, Tilanus HW, Haagmans BL, Baumert TF, van der Laan LJW. 2013. Exosome-mediated transmission of hepatitis C virus between human hepatoma Huh7.5 cells. Proc Natl Acad Sci U S A 110:13109–13113. 10.1073/pnas.1221899110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Naslavsky N, Caplan S. 2018. The enigmatic endosome—sorting the ins and outs of endocytic trafficking. J Cell Sci 131:jcs216499. 10.1242/jcs.216499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pawliczek T, Crump CM. 2009. Herpes simplex virus type 1 production requires a functional ESCRT-III complex but is independent of TSG101 and ALIX expression. J Virol 83:11254–11264. 10.1128/JVI.00574-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Martin-Serrano J, Neil SJ. 2011. Host factors involved in retroviral budding and release. Nat Rev Microbiol 9:519–531. 10.1038/nrmicro2596. [DOI] [PubMed] [Google Scholar]
- 75.Chevallier J, Chamoun Z, Jiang G, Prestwich G, Sakai N, Matile S, Parton RG, Gruenberg J. 2008. Lysobisphosphatidic acid controls endosomal cholesterol levels. J Biol Chem 283:27871–27880. 10.1074/jbc.M801463200. [DOI] [PubMed] [Google Scholar]
- 76.Sobo K, Le Blanc I, Luyet PP, Fivaz M, Ferguson C, Parton RG, Gruenberg J, van der Goot FG. 2007. Late endosomal cholesterol accumulation leads to impaired intra-endosomal trafficking. PLoS One 2:e851. 10.1371/journal.pone.0000851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lieberman AP, Puertollano R, Raben N, Slaugenhaupt S, Walkley SU, Ballabio A. 2012. Autophagy in lysosomal storage disorders. Autophagy 8:719–730. 10.4161/auto.19469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pleet ML, Branscome H, DeMarino C, Pinto DO, Zadeh MA, Rodriguez M, Sariyer IK, El-Hage N, Kashanchi F. 2018. Autophagy, EVs, and infections: a perfect question for a perfect time. Front Cell Infect Microbiol 8:362–362. 10.3389/fcimb.2018.00362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Münz C. 2017. The autophagic machinery in viral exocytosis. Front Microbiol 8:269. 10.3389/fmicb.2017.00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mutsafi Y, Altan-Bonnet N. 2018. Enterovirus transmission by secretory autophagy. Viruses 10:139. 10.3390/v10030139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang Z-W, Li Z-L, Yuan S. 2016. The role of secretory autophagy in Zika virus transfer through the placental barrier. Front Cell Infect Microbiol 6:206. 10.3389/fcimb.2016.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bird SW, Maynard ND, Covert MW, Kirkegaard K. 2014. Nonlytic viral spread enhanced by autophagy components. Proc Natl Acad Sci U S A 111:13081–13086. 10.1073/pnas.1401437111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nowag H, Münz C. 2015. Diverting autophagic membranes for exocytosis. Autophagy 11:425–427. 10.1080/15548627.2015.1009793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen YH, Du W, Hagemeijer MC, Takvorian PM, Pau C, Cali A, Brantner CA, Stempinski ES, Connelly PS, Ma HC, Jiang P, Wimmer E, Altan-Bonnet G, Altan-Bonnet N. 2015. Phosphatidylserine vesicles enable efficient en bloc transmission of enteroviruses. Cell 160:619–630. 10.1016/j.cell.2015.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Leidal AM, Huang HH, Marsh T, Solvik T, Zhang D, Ye J, Kai F, Goldsmith J, Liu JY, Huang YH, Monkkonen T, Vlahakis A, Huang EJ, Goodarzi H, Yu L, Wiita AP, Debnath J. 2020. The LC3-conjugation machinery specifies the loading of RNA-binding proteins into extracellular vesicles. Nat Cell Biol 22:187–199. 10.1038/s41556-019-0450-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ghosh S, Dellibovi-Ragheb TA, Kerviel A, Pak E, Qiu Q, Fisher M, Takvorian PM, Bleck C, Hsu VW, Fehr AR, Perlman S, Achar SR, Straus MR, Whittaker GR, de Haan CAM, Kehrl J, Altan-Bonnet G, Altan-Bonnet N. 2020. β-Coronaviruses use lysosomes for egress instead of the biosynthetic secretory pathway. Cell 183:1520–1535.e14. 10.1016/j.cell.2020.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mori Y, Koike M, Moriishi E, Kawabata A, Tang H, Oyaizu H, Uchiyama Y, Yamanishi K. 2008. Human herpesvirus-6 induces MVB formation, and virus egress occurs by an exosomal release pathway. Traffic 9:1728–1742. 10.1111/j.1600-0854.2008.00796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sinzger C, Hahn G, Digel M, Katona R, Sampaio KL, Messerle M, Hengel H, Koszinowski U, Brune W, Adler B. 2008. Cloning and sequencing of a highly productive, endotheliotropic virus strain derived from human cytomegalovirus TB40/E. J Gen Virol 89:359–368. 10.1099/vir.0.83286-0. [DOI] [PubMed] [Google Scholar]
- 89.Umashankar M, Petrucelli A, Cicchini L, Caposio P, Kreklywich CN, Rak M, Bughio F, Goldman DC, Hamlin KL, Nelson JA, Fleming WH, Streblow DN, Goodrum F. 2011. A novel human cytomegalovirus locus modulates cell type-specific outcomes of infection. PLoS Pathog 7:e1002444. 10.1371/journal.ppat.1002444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhao J, Zhong F, Yu H, Chen Z, Wang M, Chen J. 2018. Human cytomegalovirus infection-induced autophagy was associated with the biological behavioral changes of human umbilical vein endothelial cell (HUVEC). Biomed Pharmacother 102:938–946. 10.1016/j.biopha.2018.03.156. [DOI] [PubMed] [Google Scholar]
- 91.Schindelin J, Rueden CT, Hiner MC, Eliceiri KW. 2015. The ImageJ ecosystem: an open platform for biomedical image analysis. Mol Reprod Dev 82:518–529. 10.1002/mrd.22489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Turner DL, Korneev DV, Purdy JG, deMarco A, Mathias RA. 2020. The host exosome pathway underpins biogenesis of the human cytomegalovirus virion. Elife 9:e58288. 10.7554/eLife.58288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.McNamara RP, Chugh PE, Bailey A, Costantini LM, Ma Z, Bigi R, Cheves A, Eason AB, Landis JT, Host KM, Xiong J, Griffith JD, Damania B, Dittmer DP. 2019. Extracellular vesicles from Kaposi sarcoma-associated herpesvirus lymphoma induce long-term endothelial cell reprogramming. PLoS Pathog 15:e1007536. 10.1371/journal.ppat.1007536. [DOI] [PMC free article] [PubMed] [Google Scholar]