Influenza pandemics occur when an IAV from nonhuman hosts enters the human population and adapts to give rise to a lineage capable of sustained transmission among humans. Despite recurring zoonotic infections involving avian- or swine-adapted IAVs, influenza pandemics occur infrequently because IAVs typically exhibit low fitness in a new host species.
KEYWORDS: fitness, influenza virus, risk assessment, transmission, viral load, zoonotic infection
ABSTRACT
Efforts to estimate the risk posed by potentially pandemic influenza A viruses (IAVs) and to understand the mechanisms governing interspecies transmission have been hampered by a lack of animal models that yield relevant and statistically robust measures of viral fitness. To address this gap, we monitored several quantitative measures of fitness in a guinea pig model: infectivity, magnitude of replication, kinetics of replication, efficiency of transmission, and kinetics of transmission. With the goal of identifying metrics that distinguish human- and nonhuman-adapted IAVs, we compared strains derived from humans to those circulating in swine and canine populations. Influenza A/Panama/2007/99 (H3N2), A/Netherlands/602/2009 (H1N1), A/swine/Kansas/77778/2007 (H1N1), A/swine/Spain/53207/2004 (M1 P41A) (H1N1), and A/canine/Illinois/41915/2015 (H3N2) viruses were evaluated. Our results revealed higher infectivity and faster kinetics of viral replication and transmission for human and canine strains compared to the swine viruses. Conversely, peak viral titers and efficiency of transmission were higher for human strains relative to both swine and canine IAVs. Total viral loads were comparable among all strains tested. When analyzed together, data from all strains point to peak viral load as a principal driver of transmission efficiency and replication kinetics as a major driver of transmission kinetics. While the dose initiating infection did not strongly impact peak viral load, dose was found to modulate the kinetics of viral replication and, in turn, the timing of transmission. Taken together, our results point to peak viral load and transmission efficiency as key metrics differentiating human and nonhuman IAVs and suggest that high peak viral load precipitates robust transmission.
IMPORTANCE Influenza pandemics occur when an IAV from nonhuman hosts enters the human population and adapts to give rise to a lineage capable of sustained transmission among humans. Despite recurring zoonotic infections involving avian- or swine-adapted IAVs, influenza pandemics occur infrequently because IAVs typically exhibit low fitness in a new host species. Anticipating when a zoonosis might lead to a pandemic is both critical for public health preparedness and extremely challenging. The approach to characterizing IAVs reported here is designed to aid risk assessment efforts by generating rigorous and quantitative data on viral phenotypes relevant for emergence. Our data suggest that the ability to replicate to high titers and transmit efficiently irrespective of the initial dose are key characteristics distinguishing IAVs that have established sustained circulation in the human population from IAVs that circulate in nonhuman mammalian hosts.
INTRODUCTION
Diverse influenza A viruses (IAVs) circulate in multiple nonhuman hosts, such as wild waterfowl and several domestic animals to which humans have routine and close-range exposure (1). Owing to host species barriers, zoonotic transmission of IAV typically leads to isolated cases without onward transmission (2). Nevertheless, such zoonotic introductions create the potential for an influenza pandemic (3). For a pandemic to occur, an IAV introduced into humans must be antigenically distinct from seasonal IAVs in current or recent circulation and must overcome species barriers to transmit efficiently among humans. To better prepare for an IAV pandemic, much effort is spent assessing the risk posed by viruses present at the interface of humans and livestock, with a particular focus on viruses that have been transmitted to humans from their nonhuman hosts. Indeed, in attempts to standardize risk assessment and gauge the relative risk posed by different IAV strains, the U.S. Centers for Disease Control and Prevention developed the Influenza Risk Assessment Tool (IRAT), and the World Health Organization devised a similar approach with the Tool for Influenza Pandemic Risk Assessment (TIPRA) (4–6). Both of these algorithms rely heavily on laboratory data generated in animal models of infection. To ensure the utility and predictive power of these tools, it is therefore essential that well-powered and quantitative data be obtained from appropriate animal models.
Transmissibility in mammals, and in humans in particular, has been understood for many years to be a critical factor in determining whether an IAV adapted to nonhuman hosts will cause a pandemic (7). Significant effort has therefore been spent to develop animal models of IAV transmission. Most commonly, ferrets (Mustela putorious furo) are used in a system in which inoculated animals receive a high dose of virus intranasally and exposed animals are introduced 24 h later, either into the same cage as an inoculated ferret or into a neighboring cage that is open for exchange of air (8). These two methods of exposure are designed to allow transmission via the contact and respiratory droplet routes, respectively. Notably, however, the contact setup also allows for close-range transmission of respiratory droplets. The efficiency of transmission is then assessed by monitoring exposed ferrets for viral shedding and/or seroconversion. Owing to the costs of purchasing and maintaining ferrets, efforts to assess the transmissibility of IAV strains of interest in this model typically rely on a sample size of 3 to 4 transmission pairs per group (8–11).
Although ferrets are an excellent model for influenza, the use of multiple animal models can be extremely valuable in the study of infectious disease. This is because no model system recapitulates all aspects of human infection, and each model species bears different strengths and weaknesses. In the context of pandemic risk assessment, use of multiple animal models may be particularly important for differentiating viruses that replicate well in diverse mammals from those that have great potential to emerge in humans. When considering IAVs that circulate in nonhuman mammals, making this distinction can be challenging.
Several years ago, we reported the suitability of guinea pigs (Cavia porcellus) as an alternative model in which to assess IAV transmission (12). We found that, similar to ferrets, guinea pigs were highly susceptible to infection with a broad range of IAVs and that viruses of both the seasonal H3N2 and pandemic H1N1 lineages transmitted efficiently among guinea pigs housed together or in neighboring cages (12, 13). Importantly, typical low-pathogenicity avian IAVs did not transmit, and swine-adapted strains showed an intermediate phenotype (14, 15). H5N1, H7N9, and H9N2 subtype IAVs, representative of lineages with known zoonotic potential, have shown a range of transmission outcomes in guinea pigs, with some failing to transmit and others showing an intermediate phenotype, with partial transmission by a respiratory droplet route (15–20). These data suggested that the guinea pig is an ideal model host for assessing the risk of IAV emergence in humans. Studies to date have been limited in scope, however, and focused on transmission, with little consideration of other measures of fitness. From a practical point of view, the lower cost of guinea pigs compared to ferrets facilitates the use of larger group sizes and replicate experiments.
In the work reported here, we sought to capitalize on this last feature of the guinea pig model to develop a highly informative approach for evaluating the outbreak potential of IAVs. In particular, we had two goals: (i) to devise a transmission assay sensitive enough to differentiate highly transmissible strains (e.g., human seasonal viruses) from those that are likely to be moderately to poorly transmissible in humans (e.g., swine-adapted IAV) and (ii) to establish multiple metrics in addition to transmissibility that could inform assessments of fitness. Our approach includes the following features. Donor animals were infected at a range of relatively low inoculation doses, a larger number of replicates than are typically employed for transmission studies was used, and multiple measures of viral infectivity, replication, and transmission were analyzed to support a more comprehensive assessment of viral fitness compared to analysis of transmission efficiency alone. Our results reveal that the ability to replicate to high titers and transmit efficiently irrespective of the initial dose are key characteristics distinguishing human-adapted IAVs from those that circulate in nonhuman mammalian hosts.
RESULTS
We selected five IAV strains for in-depth analysis in guinea pigs: A/Netherlands/602/2009 (H1N1) (NL/09), A/Panama/2007/99 (H3N2) (Pan/99), A/swine/Spain/53207/2004 (H1N1) (sw/Sp/04), A/swine/KS/77778/2007 (H1N1) (sw/KS/07), and A/canine/IL/41915/2015 (H3N2) (can/IL/15). These viruses were chosen to represent lineages that are well adapted to human hosts and lineages that circulate in domestic mammals to which humans are routinely exposed. NL/09 virus is an early isolate of the 2009 pandemic, and Pan/99 virus is representative of the seasonal H3N2 lineage that was established in humans with the 1968 pandemic. The sw/Sp/04 strain is representative of the Eurasian avian-like swine lineage, which circulated widely in Europe and Asia prior to 2009 and caused a small number of documented human infections (21). sw/KS/07 virus is derived from the triple reassortant lineage prevalent in North American swine and is closely related to strains that were transmitted to humans in the context of a 2007 agricultural fair (22, 23). can/IL/15 is an early American isolate of an avian origin lineage that emerged in dogs ca. 2006 in South Korea and China (24). No human infections with canine IAVs have been reported to date (25). With this panel of viruses, we aimed to develop an experimental system that would allow phenotypic differentiation of IAVs that are successful in humans from those that have not become established in the human population despite exposure through domestic animals.
To evaluate viral infectivity, as well as replication and transmissibility from low inoculation doses, groups of guinea pigs were inoculated intranasally with 1 × 103, 1 × 102, or 1 × 101 PFU of each virus. For a subset of the viruses, doses of 1 × 104 and 1 × 105 PFU were also included. At 24 h postinoculation, a naive guinea pig was introduced into the same cage with each inoculated animal. Nasal washes were collected on days 2, 4, 6, and 8 postinoculation from both inoculated and exposed guinea pigs. Infectious virus present in nasal washings was then quantified by plaque assay. Results obtained with all five strains at all doses tested are plotted against time in Fig. 1. These data were analyzed further to derive the specific metrics of viral fitness detailed below.
FIG 1.
Replication and transmission of mammalian-adapted IAVs in a guinea pig model. A total of 12 guinea pigs were inoculated intranasally with the indicated doses of the indicated IAV strains. One naive guinea pig was introduced into the same cage with each inoculated animal at 24 h postinoculation. Viral titers detected in nasal washes are plotted against time (days). The limit of detection is 1.70 log10 PFU/ml, and data below the limit of detection are plotted at 0 log10 PFU/ml. Dashed lines represent inoculated animals, and solid lines show contact animals. N.D., not done. Results obtained with can/IL/15 virus were reported previously (38).
Infectivity.
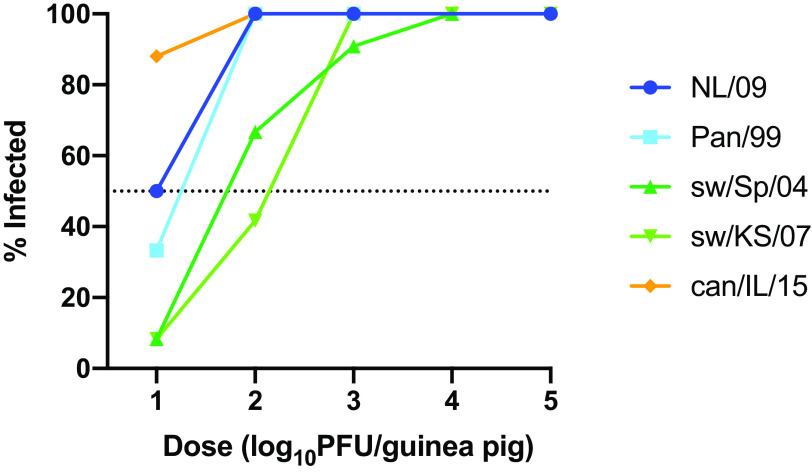
The use of multiple inoculation doses allowed comparison of infectivities across the panel of IAVs tested (Fig. 2). At the lowest dose used, 10 PFU, none of the IAVs examined initiated a productive infection in 100% of guinea pigs. The proportion of animals infected at this low dose ranged from 8.3% for the two swine isolates to 88% for can/IL/15 virus. With a 100-PFU inoculum, can/IL/15 and both of the human isolates initiated robust infection in 100% of guinea pigs, while sw/Sp/04 and sw/KS/07 infected 66.7 and 41.7% of animals, respectively. With a 1,000-PFU inoculum, all viruses infected efficiently, with 90 to 100% of animals showing sustained viral shedding. Thus, can/IL/15 was the most infectious virus, followed by the two human viruses and then the two swine viruses.
FIG 2.
Human and canine IAVs show higher infectivity than swine IAVs in a guinea pig model. A total of 12 guinea pigs were inoculated intranasally with each dose of virus. The percentage of guinea pigs found to be infected following inoculation is plotted for each dose of each virus. An animal was scored as infected if nasal washes collected on day 2, 4, 6, or 8 postinoculation contained ≥1 × 103 PFU/ml.
Viral load in nasal washes.
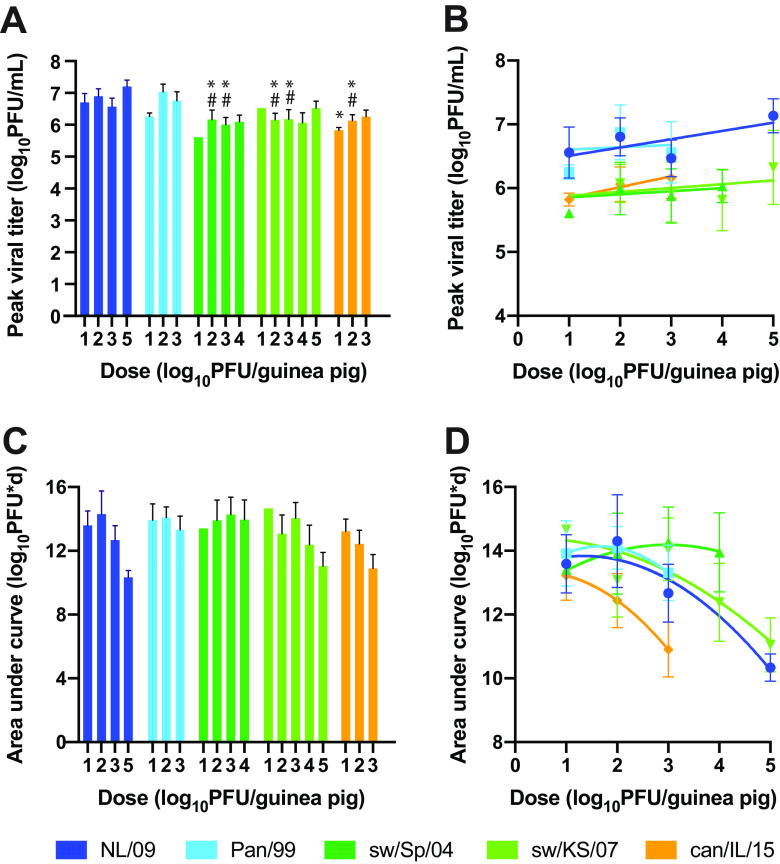
To assess the extent of viral propagation within the infected guinea pigs, we analyzed (i) peak titers attained and (ii) the amount of virus shed throughout the course of infection, calculated as the area under the curve (Fig. 3). Peak titers were determined by taking the average of the highest titers detected for each guinea pig in a group, regardless of sampling day. This metric revealed improved replication for Pan/99 and NL/09 viruses compared to sw/Sp/04 and sw/KS/07 viruses, particularly in the groups with doses of 1 × 102 and 1 × 103 PFU, which had a larger sample size of productively infected animals than the group with a dose of 1 × 101 PFU. Peak titers were also significantly higher for Pan/99 and NL/09 viruses compared to can/IL/15 virus, but only in the group with the dose 1 × 102 PFU (Fig. 3A). Analysis of the relationship between inoculation dose and peak viral titers using linear regression revealed a weak positive effect for NL/09 and can/IL/15 viruses. For the other three viral strains, peak viral titers did not change significantly with inoculation dose (Fig. 3B).
FIG 3.
Human IAVs achieve higher peak titers relative to swine and canine IAVs but comparable total viral loads. A total of 12 guinea pigs were inoculated intranasally with each dose of virus. The mean with standard error is shown, with n = 1 to 12, as some animals were not productively infected. (A) Viral titers at the time of peak shedding. Data from doses of 1 × 101, 1 × 102, and 1 × 103 PFU were compared across groups using a two-way ANOVA with Tukey’s correction for multiple comparisons to evaluate significance. NL/09 and Pan/99 were not significantly different at any dose. sw/Sp/04 and sw/KS/07 were not significantly different at any dose. * indicates significant difference relative to NL/09 (P < 0.01); # indicates significant difference relative to Pan/99 (P < 0.01). (B) All doses tested were analyzed by linear regression. Slopes were significantly nonzero only for NL/09 and can/IL/15 viruses (P < 0.05). (C) The area under the curve was calculated for each animal individually to quantify total virus shed into nasal washes over the time course. Two-way ANOVA with Tukey’s correction for multiple comparisons revealed no significant difference among virus strains. (D) Relationship between inoculation dose and area under the curve. Curve fits use a quadratic model.
Conversely, total virus shed over the course of infection, estimated by calculating the area under the replication curve for each animal, did not show significant differences among viral strains at any of the doses examined (Fig. 3C). The relationship between inoculation dose and total virus shed was not well described by a linear model, but rather a quadratic model, with higher inoculation doses tending to result in lower area under the curve values (Fig. 3D). This finding that overall viral load declines at high inoculation doses is likely accounted for by more rapid viral clearance under these conditions (Fig. 1).
Kinetics of viral replication.
To evaluate differences in kinetics of replication among the IAVs examined, we compared the time at which peak viral titers were observed in nasal wash samples. Specifically, the average time to peak shedding was evaluated for each strain-dose combination (Fig. 4A). Comparing among strains, the most rapid replication observed was with can/IL/15 virus. The two human isolates were similar to each other and reached peak titers somewhat more slowly than can/IL/15 virus. The two swine IAVs were also comparable to each other and replicated more slowly than the two human isolates and can/IL/15 virus. Thus, significant differences among the IAVs in time to peak shedding were detected, and the rank order of the viruses was the same as that seen for infectivity: can/IL/15 shows the fastest replication, followed by the two human IAVs and then the two swine IAVs.
FIG 4.
Time to reach peak viral titers is lower for human and canine versus swine IAVs and decreases with inoculation dose. The time at which peak viral shedding was detected is plotted for each virus and each dose. The mean and standard error for each strain-dose pairing is shown (n = 1 to 12). Animals that were not productively infected were excluded from the analysis, leaving n = 1 for the 10-PFU doses of the two swine IAVs. (A) Data from doses of 1 × 101, 1 × 102, and 1 × 103 PFU were compared across groups using a two-way ANOVA with Tukey’s correction for multiple comparisons to evaluate significance. NL/09 and Pan/99 were not significantly different at any dose. sw/Sp/04 and sw/KS/07 were not significantly different at any dose. * indicates significant difference relative to NL/09 (P < 0.001, except versus can/IL/15 at 10 PFU/guinea pig, where P = 0.020); # indicates significant difference relative to Pan/99 (P < 0.001). (B) All doses tested were analyzed by linear regression, revealing significant negative relationships between time to peak shedding and log10 inoculation dose for all strains (P < 0.0001 for NL/09, sw/KS/07, and sw/Sp/04, P = 0.027 for Pan/99, and P = 0.0085 for can/IL/15).
We also analyzed the relationship between inoculation dose and time to peak shedding for each of the viral strains tested (Fig. 4B). Linear regression revealed an inverse relationship between the log10 inoculation dose and time needed to reach peak titers, which was statistically significant for all strains tested (P < 0.05).
Transmissibility.
The efficiency of transmission to contact animals was assessed by tabulating the proportion of transmission pairs in which the exposed animal contracted infection, as detected by the presence of infectious virus in nasal washes. For this analysis, pairs of guinea pigs in which the inoculated animal was not productively infected were excluded. The results reveal that both human IAVs transmitted to 100% of contacts, irrespective of inoculation dose (Fig. 5). can/IL/15 and sw/KS/07 viruses showed intermediate transmission efficiency, with between 50 and 100% of contacts infected across the dosing groups. Among the viruses tested, sw/Sp/04 virus showed the lowest efficiency of transmission, with 0 to 70% of contacts contracting infection.
FIG 5.
Human IAVs exhibit more efficient transmission among guinea pigs compared to swine and canine IAVs. The percentage of contact guinea pigs that contracted infection following cohousing with an infected donor guinea pig is plotted for each virus strain and each dose. Ratios above each bar indicate the number of transmission events in the numerator and the number of productively infected donor animals in the denominator.
Kinetics of transmission.
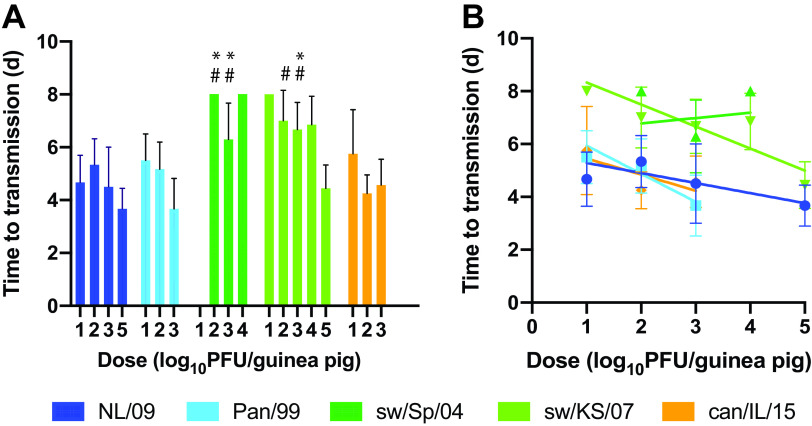
The kinetics of transmission were evaluated by calculating the average time after inoculation of the donor animal at which virus was initially detected in the nasal washings of contact animals. Transmission pairs in which no transmission was observed were excluded from this analysis. For all IAV strains and doses tested, the mean times to transmission ranged from 3.7 to 8.0 days postinoculation. Transmission tended to take longer for the two swine IAVs compared to the two human-adapted strains. The times to transmission were, however, similar between the human viruses and the can/IL/15 strain (Fig. 6A). Regression of time to transmission against dose revealed that increasing inoculation dose typically resulted in a shorter time to transmission. This relationship was significant for the two human strains and sw/KS/07 virus (Fig. 6B).
FIG 6.
Time to transmission is lower for human and canine versus swine IAVs and typically decreases with inoculation dose. The average time to first detection of infectious virus in nasal washes of exposed guinea pigs is plotted, with standard error. (A) Two-way ANOVA with Tukey’s correction for multiple comparisons was used to evaluate significance of differences among virus strains. Only data from doses of 1 × 102 and 1 × 103 PFU were included in this analysis. Differences among NL/09, Pan/99, and can/IL/15 were not significant. Differences between sw/Sp/04 and sw/KS/07 were not significant. * indicates significant difference relative to NL/09 (P < 0.01); # indicates significant difference relative to Pan/99 (P < 0.01). (B) All doses tested were analyzed by linear regression. Slopes were significantly nonzero for NL/09 (P = 0.0040), Pan/99 (P = 0.0014), and sw/KS/07 (P < 0.0002) viruses.
Relationships among the metrics evaluated.
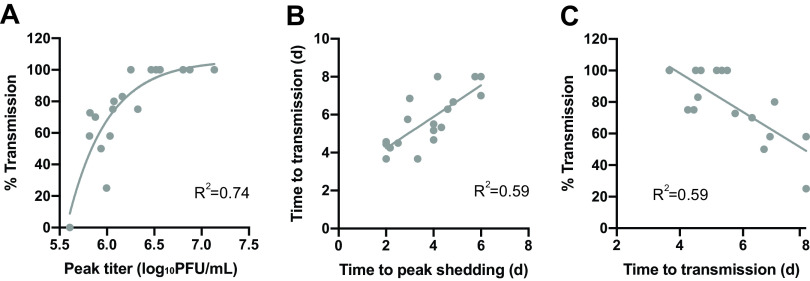
Finally, with the goal of identifying relationships among metrics, additional analyses that combined data from all five IAV strains were performed. Of note, this analysis revealed a clear correspondence between peak viral load and transmission, suggesting that transmission success is dependent on the peak magnitude of viral replication within the donor host (Fig. 7A). This relationship became saturated at higher values of peak viral titers, with transmission at 100%. A reliance of transmission on high peak titers is further supported by a correlation between time to peak shedding and time to transmission (Fig. 7B). This concordance of kinetics suggests that transmission proceeds only once the viral load is sufficiently high. We also noted that the efficiency of transmission was negatively correlated with the time to transmission (Fig. 7C), indicating that dose-strain pairings that are more favorable for transmission tend to have a shorter time to transmission. Conversely, time to peak shedding and percentage of transmission did not correlate (data not shown).
FIG 7.
Relationships among metrics. Results obtained with all viral strains and inoculation doses were pooled to evaluate relationships among measures of fitness. (A) The relationship between percentage of transmission and log10 peak viral titers is well described using an exponential growth model. (B) Linear regression revealed a positive correlation between time to peak shedding and time to transmission (P = 0.0002). (C) Linear regression revealed a negative correlation between time to transmission and percentage of transmission (P = 0.0003). One data point was excluded from this last analysis—that of sw/KS/07 at a 10-PFU inoculation dose, which showed 100% transmission with a sample size of one transmission pair.
DISCUSSION
The careful examination presented here of multiple viral phenotypes in a guinea pig model was undertaken with the goal of establishing metrics to better assess the risk of emergence in humans posed by IAVs adapted to nonhuman hosts. Such risk assessment is particularly challenging for IAVs that circulate in nonhuman mammals owing to subtle phenotypic differences from seasonal IAVs. We therefore focused our analysis on swine and canine IAVs and elected to compare outcomes to those seen with human IAVs representative of both lineages currently circulating in humans. Our findings indicate that certain measures of fitness, such as total viral load, do not vary greatly among the mammalian-adapted strains tested and are therefore unlikely to be predictive of outbreak potential. Conversely, the higher peak viral replication and higher transmission efficiency observed for both human IAVs compared to swine and canine strains suggest that these fitness indicators are likely to be informative for risk assessment purposes.
Use of multiple inoculation doses in donor animals allowed evaluation of whether this parameter modulates the phenotypes measured. In particular, we sought to determine whether replication and transmission of swine/canine IAV are more sensitive to reductions in inoculation dose than human IAV. The data reveal subtle effects of dose on peak viral titers for some strains and no clear relationship between dose and the likelihood of transmission to recipients. However, the kinetics of both peak viral load and transmission were clearly impacted by dose, with more rapid viral growth and onward transmission seen with higher inoculation doses. The effect of dose on time to transmission is likely a consequence of the effect on peak viral loads. We note that the typical dose of IAV mediating infection in natural settings is thought to be low (26), arguing for routine use of lower doses in experimental models.
In the experimental design used here, inoculated and exposed guinea pigs were placed together in the same cage. In this context, transmission is possible by all modes: direct contact, indirect contact, droplet spray, and aerosol inhalation. When inoculated and exposed animals are instead placed in separate but adjacent cages—often referred to as a respiratory droplet transmission model—viral spread must occur through transfer of aerosol particles or potentially droplet spray (27, 28). These two methods of modeling transmission in the lab are complementary in that placing animals in direct contact is more sensitive, while placing animals in separate cages is more stringent (29, 30). Because a complete lack of transmission is difficult to interpret in quantitative terms, and because a number of the metrics evaluated herein cannot be assessed in the absence of transmission, the more sensitive, contact model is well suited for the strategy pursued here. However, it would be of interest to evaluate the relationships of the various metrics analyzed here with transmissibility in a respiratory droplet model. If transmission within the cage and between cages occurs through distinct modes, the correlates of transmission may differ between the two experimental setups. For example, one might expect virion shape or extent of aggregation to have a greater impact on aerosol transmission compared to transmission via direct or indirect contact. This comparison may also be important for further refinement of risk assessment algorithms, given the likely importance of transmission via aerosols or droplets in human-to-human spread (31, 32).
The limitations of the study are important to consider in interpreting the results. As outlined above, one limitation is uncertainty regarding the mode(s) of transmission between animals housed in the same cage. An additional limitation stems from the timing of viral sample collection. This aspect of the experimental design defines the resolution with which the timing of peak viral replication and of transmission can be determined. Because nasal wash samples were collected every other day, kinetic differences of less than 48 h between virus strains or inoculation doses may be missed. Finally, while the viral strains examined herein were chosen with the goal of better differentiating the phenotypes of mammalian-adapted IAVs, a similar analysis of avian IAV would give important context to the results obtained. It is therefore noteworthy that a number of prior studies have examined avian IAV replication and transmission in guinea pigs. This prior work showed that low-pathogenicity strains of the H1N1, H3N8, and H7N1 subtypes replicated in but did not transmit among cohoused guinea pigs (15). Strains of the H9N2, H7N9, and H5N1 subtypes showed mixed outcomes, ranging from no transmission to full transmission in a contact model and partial transmission in a respiratory droplet model (15–20). Although direct comparisons between human and avian IAVs in guinea pigs are rare, a global analysis of the available data strongly suggests that viral loads are higher for human compared to avian IAV in this model (15–20). Of note, our prior work with variants of the highly pathogenic A/VN/1203/2004 (H5N1) virus implicated infectivity in recipient animals, rather than viral load in donors, was an important determinant of transmission (16). This finding highlights the complexity of transmission and underlines the fact that no single fitness metric is likely to be sufficient for predicting transmissibility and outbreak potential of IAVs.
The IRAT and TIPRA tools were developed in response to a need for evidence-based prioritization of preparedness efforts, such that resources are focused on viruses that pose the greatest pandemic risk (4, 5). Our analyses point to measures of viral fitness that are important to consider in these risk assessment tools. Namely, we find that peak viral load and efficiency of transmission clearly distinguish IAVs well adapted to humans from swine and canine strains. Although infectivity, kinetics of viral growth, and kinetics of transmission did not separate can/IL/15 virus from the human strains tested, these parameters differentiated swine and human IAVs well and are therefore also likely to be valuable metrics in risk assessment. We note that, within epidemiological models, transmission efficiency is expected to relate to the basic reproduction number, R0, and transmission kinetics are expected to correlate with generation time, both of which are critical to determining the outbreak potential of a pathogen (33).
In addition to informing methods of IAV risk assessment, our data provide primary insight into the impact of within-host viral dynamics on transmission between hosts. Namely, integrated analysis of the metrics examined herein suggests a model in which the likelihood of transmission is determined by peak viral load within the donor host and the kinetics of transmission is determined by the timing with which the peak viral load is reached.
MATERIALS AND METHODS
Cells.
Madin-Darby canine kidney (MDCK) cells (a kind gift from Peter Palese, Icahn School of Medicine at Mount Sinai) were maintained in minimal essential medium (Gibco) supplemented with 10% fetal bovine serum and penicillin-streptomycin.
Viruses.
All viruses used in this work except A/canine/IL/41915/2015 (H3N2) (can/IL/15) were generated using reverse-genetics techniques (34). The can/IL/15 strain was obtained from Colin Parrish (Cornell University) as an MDCK passage 2 sample and propagated once in 10- to 11-day-old embryonated chicken’s eggs incubated at 37°C. The reverse-genetics system for A/Panama/2007/99 (H3N2) (Pan/99) virus was initially described in reference 16, and that for A/swine/Spain/53207/2004 (H1N1) (sw/Sp/04) was initially described in reference 14. The reverse-genetics system for A/Netherlands/602/2009 (H1N1) (NL/09) virus was a gift from Ron Fouchier (Erasmus Medical Center) (35), and the reverse-genetics system for A/swine/KS/77778/2007 (H1N1) (sw/KS/07) was a gift from Juergen Richt and Wenjun Ma (Kansas State University) (36). The NL/09 virus was propagated from a low multiplicity of infection (MOI) in MDCK cells for two passages. All other virus stocks were generated in 10- to 11-day-old embryonated chicken’s eggs incubated at 37°C.
Viral growth and transmission in guinea pigs.
All animal experiments were conducted in accordance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. Guinea pig studies were conducted under animal biosafety level 2 (ABSL2) containment and approved by the IACUC of Emory University (protocol PROTO201700595). Animals were humanely euthanized following guidelines approved by the American Veterinary Medical Association (AVMA).
Female Hartley strain guinea pigs weighing 300 to 350 g were obtained from Charles River Laboratories. Prior to inoculation or nasal lavage, animals were anesthetized with a mixture of ketamine (30 mg/kg) and xylazine (4 mg/kg). Virus used for inoculation was diluted in phosphate-buffered saline (PBS) to allow intranasal inoculation of guinea pigs with 1 × 101 to 1 × 105 PFU in a 300-μl volume. One contact guinea pig was introduced into the same cage with each of the inoculated animals at 24 h postinoculation. Nasal wash samples were collected as described previously (12), with PBS as the collection fluid. Samples were aliquoted and stored at −80°C prior to analysis. Animals were housed in a Caron 6040 environmental chamber set to 10°C and 20% rH throughout the 7-day exposure period, and lids were left off the cages during this time to ensure environmental control within the cages (37). Four cages, each containing two animals, were placed in each chamber. Three replicate experiments were performed for each strain-dose combination; data were pooled for analysis. Results obtained with can/IL/15 virus were reported previously (38).
Titration of nasal lavage samples.
Nasal lavage samples were aliquoted at the time of collection and stored at −80°C. Standard plaque assays on MDCK cells were used to determine viral titers. Briefly, one aliquot was thawed, mixed well, and subjected to 10-fold serial dilution in PBS. Dilutions of 10−1 to 10−6 were then used to inoculate confluent MDCK cells in 6-well plates. Following a 1-h attachment period, inoculum was removed, monolayers were washed with PBS, and the cells were overlaid with serum-free culture medium containing 0.6% Oxoid agar. Cultures were incubated for 2 days at 37°C, at which time plaques were counted and titer determined by taking into account the initial dilution of the sample. The limit of detection of the plaque assay is 50 PFU/ml, equivalent to one plaque from the 10−1 dilution.
Data analysis.
Statistical analyses were performed in GraphPad Prism version 8.4. All analyses were performed with log-transformed values of viral load and inoculation dose. The rationale for analysis of log-transformed data is that viral replication is an exponential process, and thus differences perceptible only on a linear scale are unlikely to be biologically significant.
Pairwise comparisons between virus strains of peak viral titers, area under the curve, time to peak shedding, and time to transmission were carried out for each dose separately, using two-way analysis of variance (ANOVA) with Tukey’s correction for multiple comparisons. The rationale for this analysis approach is as follows. A two-way ANOVA was used to allow the effect of two factors (viral strain and dose) to be evaluated. Multiple pairwise comparisons were made using Tukey’s correction because differences between any two viral strains are of interest: in other words, we did not consider any one strain to be the control. In addition, when multiple comparisons are made, a correction must be applied to adjust P values accordingly. As required when performing an ANOVA, any doses for which data were not available for all five strains were excluded from these analyses: these exclusions are indicated in the corresponding figure legends.
Where error bars are displayed in figures, they represent standard error of the mean. Standard error, rather than standard deviation, was used because this measure is more appropriate for comparisons between means.
To test whether each metric (peak viral titer, area under the curve, time to peak shedding, and time to transmission) trends linearly with inoculation dose, linear regression was used. Unlike ANOVA, this analysis takes into account the fact that dose is a quantitative factor. Regression can therefore test for a trend, as opposed to only differences among the dose groups. In the case of area under the curve, a quadratic model was found to give a better fit to the data than a linear model for all five strains (as indicated by R2 values) and was therefore employed in Fig. 3D.
Similarly, regression was used to test for relationships among the different metrics examined. Trends supported by R2 values greater than 0.5 were included in Fig. 7.
ACKNOWLEDGMENTS
We thank Colin Parrish for the A/canine/IL/41915/2015 virus, Wenjun Ma for the A/swine/KS/77778/2007 virus reverse-genetics plasmids, and Ron Fouchier for the A/NL/602/2009 reverse-genetics plasmids.
This work was funded by the National Institute of Allergy and Infectious Diseases through Centers of Excellence for Influenza Research and Surveillance (CEIRS) contract no. HHSN272201400004C.
REFERENCES
- 1.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. 1992. Evolution and ecology of influenza A viruses. Microbiol Rev 56:152–179. 10.1128/MR.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Subbarao K. 2019. The critical interspecies transmission barrier at the animal-human interface. Trop Med Infect Dis 4:72. 10.3390/tropicalmed4020072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lipsitch M, Barclay W, Raman R, Russell CJ, Belser JA, Cobey S, Kasson PM, Lloyd-Smith JO, Maurer-Stroh S, Riley S, Beauchemin CA, Bedford T, Friedrich TC, Handel A, Herfst S, Murcia PR, Roche B, Wilke CO, Russell CA. 2016. Viral factors in influenza pandemic risk assessment. eLife 5:e18491. 10.7554/eLife.18491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cox NJ, Trock SC, Burke SA. 2014. Pandemic preparedness and the Influenza Risk Assessment Tool (IRAT). Curr Top Microbiol Immunol 385:119–136. 10.1007/82_2014_419. [DOI] [PubMed] [Google Scholar]
- 5.World Health Organization. 2016. Tool for Influenza Pandemic Risk Assessment (TIPRA). https://www.who.int/influenza/publications/TIPRA_manual_v1/en/.
- 6.Burke SA, Trock SC. 2018. Use of Influenza Risk Assessment Tool for prepandemic preparedness. Emerg Infect Dis 24:471–477. 10.3201/eid2403.171852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox NJ, Subbarao K. 2000. Global epidemiology of influenza: past and present. Annu Rev Med 51:407–421. 10.1146/annurev.med.51.1.407. [DOI] [PubMed] [Google Scholar]
- 8.Belser JA, Barclay W, Barr I, Fouchier RAM, Matsuyama R, Nishiura H, Peiris M, Russell CJ, Subbarao K, Zhu H, Yen HL. 2018. Ferrets as models for influenza virus transmission studies and pandemic risk assessments. Emerg Infect Dis 24:965–971. 10.3201/eid2406.172114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richard M, Schrauwen EJ, de Graaf M, Bestebroer TM, Spronken MI, van Boheemen S, de Meulder D, Lexmond P, Linster M, Herfst S, Smith DJ, van den Brand JM, Burke DF, Kuiken T, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. 2013. Limited airborne transmission of H7N9 influenza A virus between ferrets. Nature 501:560–563. 10.1038/nature12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Munster VJ, de Wit E, van den Brand JM, Herfst S, Schrauwen EJ, Bestebroer TM, van de Vijver D, Boucher CA, Koopmans M, Rimmelzwaan GF, Kuiken T, Osterhaus AD, Fouchier RA. 2009. Pathogenesis and transmission of swine-origin 2009 A(H1N1) influenza virus in ferrets. Science 325:481–483. 10.1126/science.1177127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Itoh Y, Shinya K, Kiso M, Watanabe T, Sakoda Y, Hatta M, Muramoto Y, Tamura D, Sakai-Tagawa Y, Noda T, Sakabe S, Imai M, Hatta Y, Watanabe S, Li C, Yamada S, Fujii K, Murakami S, Imai H, Kakugawa S, Ito M, Takano R, Iwatsuki-Horimoto K, Shimojima M, Horimoto T, Goto H, Takahashi K, Makino A, Ishigaki H, Nakayama M, Okamatsu M, Takahashi K, Warshauer D, Shult PA, Saito R, Suzuki H, Furuta Y, Yamashita M, Mitamura K, Nakano K, Nakamura M, Brockman-Schneider R, Mitamura H, Yamazaki M, Sugaya N, Suresh M, Ozawa M, Neumann G, Gern J, Kida H, et al. 2009. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature 460:1021–1025. 10.1038/nature08260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lowen AC, Mubareka S, Tumpey TM, Garcia-Sastre A, Palese P. 2006. The guinea pig as a transmission model for human influenza viruses. Proc Natl Acad Sci U S A 103:9988–9992. 10.1073/pnas.0604157103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steel J, Staeheli P, Mubareka S, Garcia-Sastre A, Palese P, Lowen AC. 2010. Transmission of pandemic H1N1 influenza virus and impact of prior exposure to seasonal strains or interferon treatment. J Virol 84:21–26. 10.1128/JVI.01732-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Campbell PJ, Kyriakis CS, Marshall N, Suppiah S, Seladi-Schulman J, Danzy S, Lowen AC, Steel J. 2014. Residue 41 of the Eurasian avian-like swine influenza A virus matrix protein modulates virion filament length and efficiency of contact transmission. J Virol 88:7569–7577. 10.1128/JVI.00119-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gabbard JD, Dlugolenski D, Van Riel D, Marshall N, Galloway SE, Howerth EW, Campbell PJ, Jones C, Johnson S, Byrd-Leotis L, Steinhauer DA, Kuiken T, Tompkins SM, Tripp R, Lowen AC, Steel J. 2014. Novel H7N9 influenza virus shows low infectious dose, high growth rate, and efficient contact transmission in the guinea pig model. J Virol 88:1502–1512. 10.1128/JVI.02959-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steel J, Lowen AC, Mubareka S, Palese P. 2009. Transmission of influenza virus in a mammalian host is increased by PB2 amino acids 627K or 627E/701N. PLoS Pathog 5:e1000252. 10.1371/journal.ppat.1000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hai R, Schmolke M, Leyva-Grado VH, Thangavel RR, Margine I, Jaffe EL, Krammer F, Solorzano A, Garcia-Sastre A, Palese P, Bouvier NM. 2013. Influenza A(H7N9) virus gains neuraminidase inhibitor resistance without loss of in vivo virulence or transmissibility. Nat Commun 4:2854. 10.1038/ncomms3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao Y, Zhang Y, Shinya K, Deng G, Jiang Y, Li Z, Guan Y, Tian G, Li Y, Shi J, Liu L, Zeng X, Bu Z, Xia X, Kawaoka Y, Chen H. 2009. Identification of amino acids in HA and PB2 critical for the transmission of H5N1 avian influenza viruses in a mammalian host. PLoS Pathog 5:e1000709. 10.1371/journal.ppat.1000709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun Y, Bi Y, Pu J, Hu Y, Wang J, Gao H, Liu L, Xu Q, Tan Y, Liu M, Guo X, Yang H, Liu J. 2010. Guinea pig model for evaluating the potential public health risk of swine and avian influenza viruses. PLoS One 5:e15537. 10.1371/journal.pone.0015537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lv J, Wei B, Yang Y, Yao M, Cai Y, Gao Y, Xia X, Zhao X, Liu Z, Li X, Wang H, Yang H, Roesler U, Miao Z, Chai T. 2012. Experimental transmission in guinea pigs of H9N2 avian influenza viruses from indoor air of chicken houses. Virus Res 170:102–108. 10.1016/j.virusres.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 21.Van Reeth K. 2007. Avian and swine influenza viruses: our current understanding of the zoonotic risk. Vet Res 38:243–260. 10.1051/vetres:2006062. [DOI] [PubMed] [Google Scholar]
- 22.Ma W, Vincent AL, Lager KM, Janke BH, Henry SC, Rowland RR, Hesse RA, Richt JA. 2010. Identification and characterization of a highly virulent triple reassortant H1N1 swine influenza virus in the United States. Virus Genes 40:28–36. 10.1007/s11262-009-0413-7. [DOI] [PubMed] [Google Scholar]
- 23.Vincent AL, Swenson SL, Lager KM, Gauger PC, Loiacono C, Zhang Y. 2009. Characterization of an influenza A virus isolated from pigs during an outbreak of respiratory disease in swine and people during a county fair in the United States. Vet Microbiol 137:51–59. 10.1016/j.vetmic.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 24.Song D, Kang B, Lee C, Jung K, Ha G, Kang D, Park S, Park B, Oh J. 2008. Transmission of avian influenza virus (H3N2) to dogs. Emerg Infect Dis 14:741–746. 10.3201/eid1405.071471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pulit-Penaloza JA, Simpson N, Yang H, Creager HM, Jones J, Carney P, Belser JA, Yang G, Chang J, Zeng H, Thor S, Jang Y, Killian ML, Jenkins-Moore M, Janas-Martindale A, Dubovi E, Wentworth DE, Stevens J, Tumpey TM, Davis CT, Maines TR. 2017. Assessment of molecular, antigenic, and pathological features of canine influenza A(H3N2) viruses that emerged in the United States. J Infect Dis 216:S499–S507. 10.1093/infdis/jiw620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCrone JT, Woods RJ, Martin ET, Malosh RE, Monto AS, Lauring AS. 2018. Stochastic processes constrain the within and between host evolution of influenza virus. eLife 7:e35962. 10.7554/eLife.35962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frise R, Bradley K, van Doremalen N, Galiano M, Elderfield RA, Stilwell P, Ashcroft JW, Fernandez-Alonso M, Miah S, Lackenby A, Roberts KL, Donnelly CA, Barclay WS. 2016. Contact transmission of influenza virus between ferrets imposes a looser bottleneck than respiratory droplet transmission allowing propagation of antiviral resistance. Sci Rep 6:29793. 10.1038/srep29793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lowen AC, Bouvier NM, Steel J. 2014. Transmission in the guinea pig model. Curr Top Microbiol Immunol 385:157–183. 10.1007/82_2014_390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fobian K, Fabrizio TP, Yoon SW, Hansen MS, Webby RJ, Larsen LE. 2015. New reassortant and enzootic European swine influenza viruses transmit efficiently through direct contact in the ferret model. J Gen Virol 96:1603–1612. 10.1099/vir.0.000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun H, Xiao Y, Liu J, Wang D, Li F, Wang C, Li C, Zhu J, Song J, Sun H, Jiang Z, Liu L, Zhang X, Wei K, Hou D, Pu J, Sun Y, Tong Q, Bi Y, Chang KC, Liu S, Gao GF, Liu J. 2020. Prevalent Eurasian avian-like H1N1 swine influenza virus with 2009 pandemic viral genes facilitating human infection. Proc Natl Acad Sci U S A 117:17204–17210. 10.1073/pnas.1921186117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tellier R. 2009. Aerosol transmission of influenza A virus: a review of new studies. J R Soc Interface 6(Suppl 6):S783–S790. 10.1098/rsif.2009.0302.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan J, Grantham M, Pantelic J, Bueno de Mesquita PJ, Albert B, Liu F, Ehrman S, Milton DK, EMIT Consortium. 2018. Infectious virus in exhaled breath of symptomatic seasonal influenza cases from a college community. Proc Natl Acad Sci U S A 115:1081–1086. 10.1073/pnas.1716561115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boelle PY, Ansart S, Cori A, Valleron AJ. 2011. Transmission parameters of the A/H1N1 (2009) influenza virus pandemic: a review. Influenza Other Respir Viruses 5:306–316. 10.1111/j.1750-2659.2011.00234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fodor E, Devenish L, Engelhardt OG, Palese P, Brownlee GG, Garcia-Sastre A. 1999. Rescue of influenza A virus from recombinant DNA. J Virol 73:9679–9682. 10.1128/JVI.73.11.9679-9682.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chutinimitkul S, Herfst S, Steel J, Lowen AC, Ye J, van Riel D, Schrauwen EJA, Bestebroer TM, Koel B, Burke DF, Sutherland-Cash KH, Whittleston CS, Russell CA, Wales DJ, Smith DJ, Jonges M, Meijer A, Koopmans M, Rimmelzwaan GF, Kuiken T, Osterhaus ADME, García-Sastre A, Perez DR, Fouchier RAM. 2010. Virulence-associated substitution D222G in the hemagglutinin of 2009 pandemic influenza A(H1N1) virus affects receptor binding. J Virol 84:11802–11813. 10.1128/JVI.01136-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee J, Henningson J, Ma J, Duff M, Lang Y, Li Y, Li Y, Nagy A, Sunwoo S, Bawa B, Yang J, Bai D, Richt JA, Ma W. 2017. Effects of PB1-F2 on the pathogenicity of H1N1 swine influenza virus in mice and pigs. J Gen Virol 98:31–42. 10.1099/jgv.0.000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lowen AC, Mubareka S, Steel J, Palese P. 2007. Influenza virus transmission is dependent on relative humidity and temperature. PLoS Pathog 3:1470–1476. 10.1371/journal.ppat.0030151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martinez-Sobrido L, Blanco-Lobo P, Rodriguez L, Fitzgerald T, Zhang H, Nguyen P, Anderson CS, Holden-Wiltse J, Bandyopadhyay S, Nogales A, DeDiego ML, Wasik BR, Miller BL, Henry C, Wilson PC, Sangster MY, Treanor JJ, Topham DJ, Byrd-Leotis L, Steinhauer DA, Cummings RD, Luczo JM, Tompkins SM, Sakamoto K, Jones CA, Steel J, Lowen AC, Danzy S, Tao H, Fink AL, Klein SL, Wohlgemuth N, Fenstermacher KJ, El Najjar F, Pekosz A, Sauer L, Lewis MK, Shaw-Saliba K, Rothman RE, Liu ZY, Chen KF, Parrish CR, Voorhees IEH, Kawaoka Y, Neumann G, Chiba S, Fan S, Hatta M, Kong H, Zhong G, et al. 2020. Characterizing emerging canine H3 influenza viruses. PLoS Pathog 16:e1008409. 10.1371/journal.ppat.1008409. [DOI] [PMC free article] [PubMed] [Google Scholar]