Given the presumed zoonotic origin of SARS-CoV-2, the human-animal-environment interface of the COVID-19 pandemic is an area of great scientific and public and animal health interest. Identification of animal species that are susceptible to infection by SARS-CoV-2 may help to elucidate the potential origin of the virus, identify potential reservoirs or intermediate hosts, and define the mechanisms underlying cross-species transmission to humans.
KEYWORDS: ACE2, SARS-CoV-2, cervids, deer, host species, pathogenesis, host range
ABSTRACT
The origin of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus causing the global coronavirus disease 2019 (COVID-19) pandemic, remains a mystery. Current evidence suggests a likely spillover into humans from an animal reservoir. Understanding the host range and identifying animal species that are susceptible to SARS-CoV-2 infection may help to elucidate the origin of the virus and the mechanisms underlying cross-species transmission to humans. Here, we demonstrated that white-tailed deer (Odocoileus virginianus), an animal species in which the angiotensin-converting enzyme 2 (ACE2)—the SARS-CoV-2 receptor—shares a high degree of similarity to that of humans, are highly susceptible to infection. Intranasal inoculation of deer fawns with SARS-CoV-2 resulted in established subclinical viral infection and shedding of infectious virus in nasal secretions. Notably, infected animals transmitted the virus to noninoculated contact deer. Viral RNA was detected in multiple tissues 21 days postinoculation (p.i.). All inoculated and indirect contact animals seroconverted and developed neutralizing antibodies as early as day 7 p.i. The work provides important insights into the animal host range of SARS-CoV-2 and identifies white-tailed deer as a wild animal species susceptible to the virus.
IMPORTANCE Given the presumed zoonotic origin of SARS-CoV-2, the human-animal-environment interface of the COVID-19 pandemic is an area of great scientific and public and animal health interest. Identification of animal species that are susceptible to infection by SARS-CoV-2 may help to elucidate the potential origin of the virus, identify potential reservoirs or intermediate hosts, and define the mechanisms underlying cross-species transmission to humans. Additionally, it may also provide information and help to prevent potential reverse zoonosis that could lead to the establishment of new wildlife hosts. Our data shows that upon intranasal inoculation, white-tailed deer became subclinically infected and shed infectious SARS-CoV-2 in nasal secretions and feces. Importantly, indirect contact animals were infected and shed infectious virus, indicating efficient SARS-CoV-2 transmission from inoculated animals. These findings support the inclusion of wild cervid species in investigations conducted to assess potential reservoirs or sources of SARS-CoV-2 infection.
INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a novel coronavirus, within the genus Betacoronavirus (subgenus Sarbecovirus) of the family Coronaviridae, that causes coronavirus disease 2019 (COVID-19) in humans (1). COVID-19 was first reported in Wuhan, Hubei Province, China, in December 2019 (2). The early clusters of the disease in humans had an epidemiological link to the Huanan Seafood Wholesale market in Wuhan, where several live wild animal species were sold (2–4). Genome sequence analysis of SARS-CoV-2 revealed a high degree of similarity to coronaviruses circulating in bats (3, 5, 6), with current evidence pointing to horseshoe bats as the most likely source of the ancestral virus that crossed the species barrier to cause the global COVID-19 pandemic in humans (7, 8).
Other pathogenic zoonotic coronaviruses, including severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV), are also believed to have originated in bat reservoirs. However, there is no evidence of direct bat-to-human transmission, and current data indicate that human infections with SARS-CoV and MERS-CoV resulted from interactions with intermediate animal hosts, such as palm civet cats (Paguma larvata) and dromedary camels (Camelus dromedaries), respectively (9–11). The epidemiological link of the first reported human cases of COVID-19 with the Huanan animal market in Wuhan suggests that SARS-CoV-2 may have spilled over into humans from an animal host (3–5, 11–13). Early studies proposed that pangolins (Manis sp.) may have served as an intermediate host for SARS-CoV-2, as they are a natural reservoir for SARS-CoV-2-like coronaviruses (14). However, phylogenetic analyses and amino acid sequence analysis of the S gene of SARS-CoV-2 did not support the hypothesis of the virus arising directly from the closely related pangolin betacoronaviruses (15). A better understanding of the host range and species susceptibility of SARS-CoV-2 is critical to elucidate the origin of the virus and to identify potential animal reservoirs and routes of transmission to humans.
The tropism and host range of coronaviruses is largely determined by the spike (S) glycoprotein, which binds to host cell receptors, triggering fusion and virus entry into susceptible cells (16). The SARS-CoV-2 S protein binds host cells through the angiotensin-converting enzyme 2 (ACE2) protein receptor (17). Comparison of the human ACE2 protein to that of over 400 vertebrate species demonstrated that the ACE2 protein of several animal species presents a high degree of amino acid conservation to the human protein (18). Further analysis of the ACE2/S binding motif and predictions of the SARS-CoV-2 S binding propensity led to the identification of several animal species with an ACE2 protein with a high binding probability to the SARS-CoV-2 S protein (18). Not surprisingly, the majority of species with the highest S/ACE2 binding propensity are nonhuman primates (18). Notably, the ACE2/S protein binding motif of three species of deer, including Père David’s deer (Elaphurus davidianus), reindeer (Rangifer tarandus), and white-tailed deer (Odocoileus virginianus), share a high homology with the human ACE2 (18). These observations suggest a putative broad host range for SARS-CoV-2; however, the susceptibility of most of these animal species to SARS-CoV-2 infection remains unknown.
Natural SARS-CoV-2 infections have been reported in dogs, cats, mink, tigers, and lions in Hong Kong, Netherlands, China, and the United States (19–22). The increased interest in the virus-host range and in understanding the array of susceptible animal species, and the need to develop reliable animal models for SARS-CoV-2 infection, led to several experimental inoculations in domestic and wild animal species. Experimentally infected nonhuman primates, ferrets, minks, cats, dogs, raccoon dogs, golden Syrian hamsters, and deer mice have displayed mild to moderate clinical disease upon SARS-CoV-2 infection (23–28), whereas experimental inoculation of swine, cattle, poultry, and fruit bats have shown that these species are either not susceptible to SARS-CoV-2 or that inoculation on these studies did not result in productive infection and sustained viral replication (23, 29–31). Here, we assessed the susceptibility of white-tailed deer to SARS-CoV-2. Viral infection, clinical outcomes, shedding patterns, and tissue distribution were evaluated. Additionally, transmission of SARS-CoV-2 to indirect contact animals was also investigated.
RESULTS
Susceptibility of deer cells to SARS-CoV-2 infection and replication.
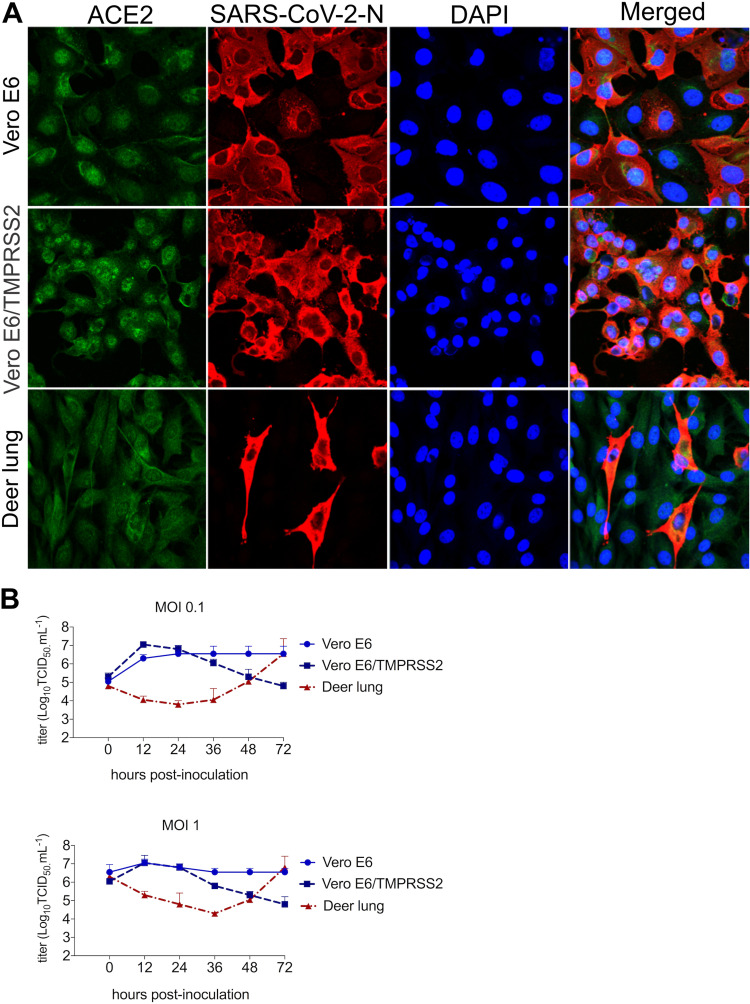
The susceptibility of deer cells to SARS-CoV-2 infection and replication was assessed in vitro. Deer lung (DL) cells were inoculated with SARS-CoV-2 isolate TGR/NY/20 (22), and the ability of SARS-CoV-2 to infect these cells was compared to Vero-E6 and Vero-E6/TMPRRS2 cells. Virus infection and replication were assessed by immunofluorescence (IFA) staining using a SARS-CoV-2 N-specific monoclonal antibody. As shown in Fig. 1A, SARS-CoV-2 N expression was detected in DL cells at 24 h postinoculation (hpi), indicating that these cells are susceptible to SARS-CoV-2 infection. Importantly, staining for ACE2 confirmed expression of the SARS-CoV-2 receptor in DL cells (Fig. 1A).
FIG 1.
Susceptibility and replication properties of the SARS-CoV-2 in deer cells in vitro. (A) Deer lung (DL), Vero E6, and Vero E6/TMPRSS2 cells were inoculated with SARS-CoV-2 at a multiplicity of infection (MOI) of 1. At 24 h postinoculation (hpi), cells were fixed and subjected to an immunofluorescence assay using a monoclonal antibody (MAb) anti-ACE2 (green) and with a MAb anti-SARS-CoV-2-nucleoprotein (N) (red). Nuclear counterstain was performed with DAPI (blue); ×40 magnification. (B) To assess the kinetics of replication of SARS-CoV-2, DL, Vero E6, and Vero E6/TMPRSS2 cells were inoculated with SARS-CoV-2 isolate TGR/NY/20 (MOI of 0.1 and 1) and harvested at various time points postinoculation (12, 24, 36, 48, and 72 hpi; time zero shown in the graphs represent the back titration of input virus). Virus titers were determined on each time point using endpoint dilutions and the Spearman and Karber's method and expressed as log10 . Results represent the average of three independent experiments.
The replication kinetics of SARS-CoV-2 was investigated in DL cells. For comparison, we included Vero-E6 and Vero-E6/TMPRSS2 cells, which are known to support efficient SARS-CoV-2 replication (32, 33). All cells were inoculated with SARS-CoV-2 at multiplicities of infection (MOI) of 0.1 and 1, and cells plus supernatant were harvested at 12, 24, 36, 48, and 72 hpi. Consistent with previous studies showing efficient replication of SARS-CoV-2 in Vero-E6 and Vero-E6/TMPRSS2 cells (32, 33), SARS-CoV-2 replicated well in these cells, reaching peak titers (∼106 to 107 50% tissue culture infectious dose per milliliter ) within 12 to 24 hpi (Fig. 1B). Interestingly, while SARS-CoV-2 also replicated to high titers in DL cells (∼106 to 107 ), replication kinetics was delayed with peak viral titers being reached by 72 hpi (Fig. 1B). These results show that DL cells are susceptible to SARS-CoV-2 infection and support productive virus replication in vitro.
Infection and transmission of SARS-CoV-2 in white-tailed deer.
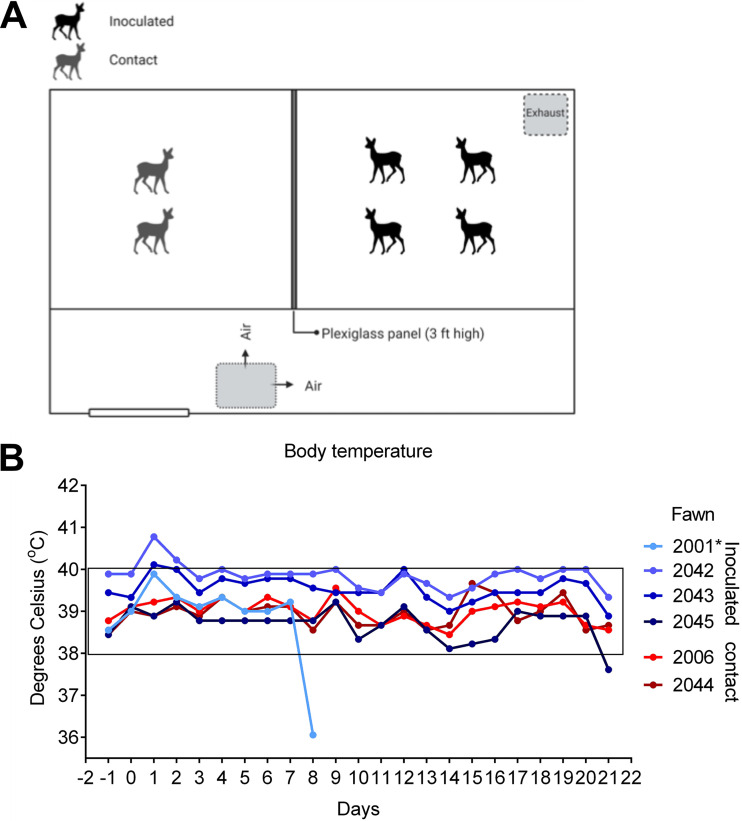
Given that white-tailed deer have been recently identified as one of the species with an ACE2 protein with a high binding probability to the SARS-CoV-2 S protein (18), we investigated the susceptibility of white-tailed deer fawns to SARS-CoV-2 infection. Six-week-old fawns (n = 4) were inoculated intranasally with 5 ml (2.5 ml per nostril) of a virus suspension containing 106.3 of SARS-CoV-2 TGR/NY/20, an animal SARS-CoV-2 (D614G) strain that is identical to human viral strains (22). To assess the potential transmission of SARS-CoV-2 between white-tailed deer, two fawns (n = 2) were maintained as noninoculated contacts in the same biosafety level 3 (agriculture) (BSL-3Ag) room. The inoculated and indirect contact animals were kept in separate pens divided by a plexiglass barrier to prevent direct nose-to-nose contact between inoculated and contact animals (Fig. 2A). Following inoculation, animals were monitored daily for clinical signs and body temperature. No clinical signs of overt respiratory distress were observed in any of the inoculated or contact animals during the 21-day experimental period. Interestingly, a slight and transient increase in body temperature was noted in 3/4 (no. 2001, 2042, and 2043) inoculated fawns between days 1 to 3 postinoculation (p.i.) (Fig. 2B). One fawn (no. 2001) died on day 8 p.i. of unrelated intestinal perforation and peritonitis. The body temperature in both indirect contact animals (no. 2006 and 2044) remained within physiological ranges throughout the experimental period (Fig. 2B). Postmortem examination of fawns at 8 days p.i. (animal no. 2001) or 21 days p.i. (animal no. 2042, 2043, 2045, 2006, and 2044) revealed no gross lesions. Microscopic changes observed were not associated with the presence of the virus, as no viral RNA (genomic and subgenomic RNA) was detected by PCR or in situ hybridization (ISH) (data not shown).
FIG 2.
Infection and transmission of SARS-CoV-2 in white-tailed deer. (A) Room setup of animal experiment. Fawns were kept in a room of a biosafety level 3 (agriculture) (BSL-3Ag) facility. Four fawns were inoculated intranasally with a virus suspension containing 5 × 106.3 TCID50 of SARS-CoV-2 isolate TGR/NY/20, and two fawns were maintained as noninoculated room contact animals. All fawns were maintained in a 3.7-m by 3.7-m room and inoculated, and room contact animals were kept in two pens separated by a plexiglass barrier approximately 0.9 m (∼3 ft) in height to prevent direct nose-to-nose contact. Airflow in the room was maintained at 10 to 11 air exchanges per hour and was directional from the contact pen toward the inoculated pen. (B) Fawns were microchipped subcutaneously for identification and monitored daily for clinical signs and body temperature starting on day 1 before inoculation or contact day (day −1). Body temperatures are expressed in degrees Celsius.
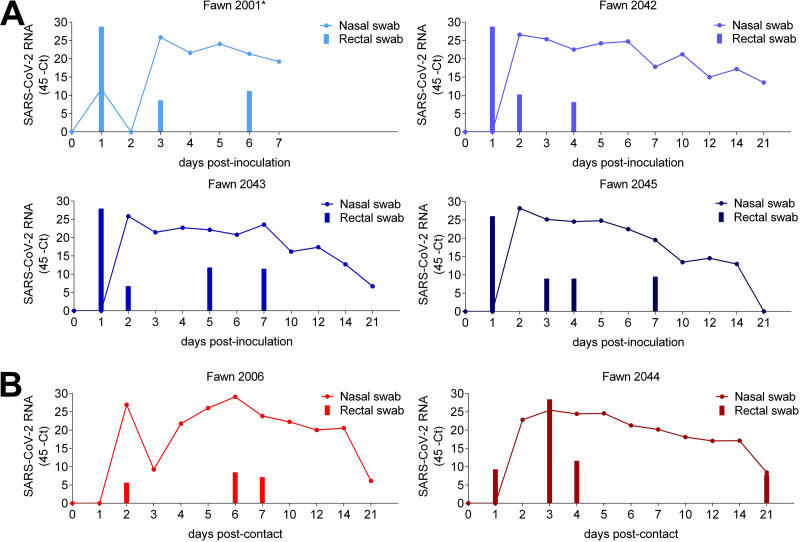
Replication of SARS-CoV-2 in the upper respiratory and gastrointestinal tracts and the dynamics and patterns of virus shedding and viremia were assessed in inoculated and indirect contact fawns. Nasal secretions and feces were collected by nasal and rectal swabs on days 0, 1, 2, 3, 4, 5, 6, 7, 10, 12, 14, and 21 p.i.; serum and buffy coat collected on days 0, 7, 14, and 21 p.i. were subjected to nucleic acid extraction and tested for the presence of SARS-CoV-2 RNA (genomic and subgenomic RNA) by real-time reverse transcriptase PCR (rRT-PCR). While viremia was not detected in serum and buffy coat samples, viral RNA was detected between days 2 and 21 p.i. in nasal secretions from inoculated animals (Fig. 3A), with higher viral RNA loads being detected between days 2 and 7 p.i. and decreasing thereafter through day 21 p.i. (Fig. 3A). Notably, high levels of viral RNA were also detected in nasal secretions from indirect contact animals throughout the experimental period (Fig. 3B). Viral RNA was detected in fecal samples in all inoculated and indirect contact animals (Fig. 3A and B); however, intermittent and short-duration fecal shedding was observed, with most animals (4/5) only transiently shedding detectable SARS-CoV-2 in feces through days 6 to 7 p.i. (Fig. 3A and B).
FIG 3.
Viral RNA in nasal secretion and feces. (A) Dynamics of virus shedding was assessed in nasal secretions (line) and feces (bars) of four inoculated fawns (no. 2001, 2042, 2043, and 2045). Nasal and rectal swabs collected on days 0 to 7, 10, 12, 14, and 21 postinoculation (p.i.) were tested for the presence of SARS-CoV-2 RNA by real-time reverse transcriptase PCR (rRT-PCR). (B) Dynamics of virus shedding of contact fawns (no. 2006 and 2044). Nasal and rectal swabs collected on the same time points as the inoculated animals were subjected to nucleic acid extraction and tested for the presence of SARS-CoV-2 RNA by rRT-PCR.
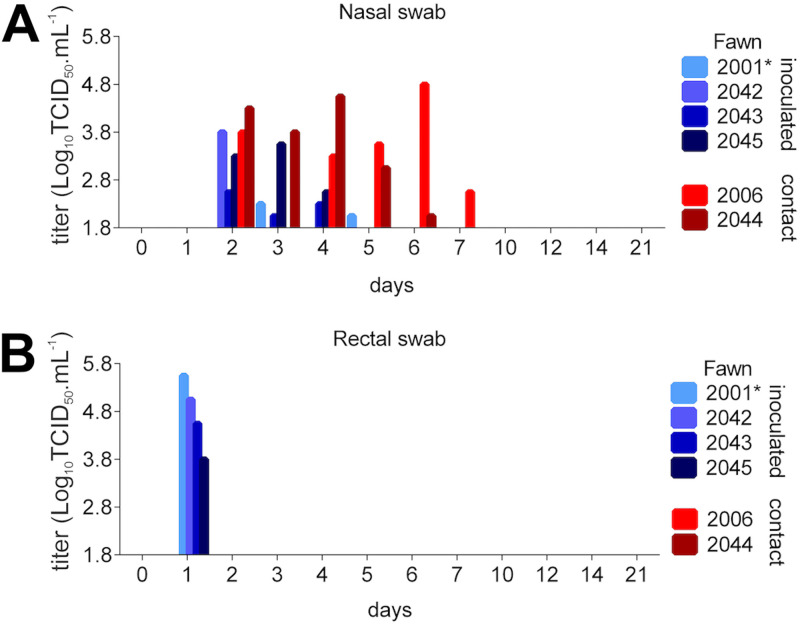
Notably, infectious SARS-CoV-2 was detected in nasal secretions of all inoculated and indirect contact animals between days 2 and 5 p.i. (Fig. 4A). Indirect contact animals shed infectious SARS-CoV-2 in nasal secretions through day 7 p.i. Viral titers shed in nasal secretions ranged from 2.0- to 4.8-log10 (Fig. 4A). Infectious SARS-CoV-2 shedding in feces was detected in inoculated animals on day 1 p.i. (Fig. 4B). Sequencing of SARS-CoV-2 from select nasal secretions from inoculated and indirect contact animals confirmed the identity of the virus, matching the sequence of the inoculated virus. Together, these results indicate that SARS-CoV-2 productively infected and replicated in the upper respiratory tract of white-tailed deer. In addition, the virus was efficiently transmitted to indirect contact animals.
FIG 4.
Shedding of infectious SARS-CoV-2 by inoculated and contact fawns. (A) Infectious virus was assessed by virus isolation in nasal secretions in rRT-PCR-positive samples. (B) Infectious SARS-CoV-2 shedding in feces in rRT-PCR-positive samples. Virus titers were determined using endpoint dilutions and the Spearman and Karber's method and expressed as log10 .
Viral load and tissue distribution.
Viral RNA load and tissue distribution of SARS-CoV-2 were assessed on day 8 p.i. (animal no. 2001, which died of an unrelated cause [intestinal perforation]) and day 21 p.i. (animal no. 2042, 2043, 2045, 2006, and 2044). Tissues were collected and processed for rRT-PCR, virus isolation, and ISH. On day 8 p.i. (fawn 2001), SARS-CoV-2 RNA was detected in nasal turbinates, palatine tonsil, spleen, ileocecal junction, and in submandibular, medial retropharyngeal, tracheobronchial, and mediastinal lymph nodes (Fig. 5). Among the tissues collected on day 21 p.i. from the remaining inoculated and indirect contact fawns, SARS-CoV-2 RNA was consistently detected in nasal turbinates, palatine tonsil, and retropharyngeal lymph node (Fig. 5). This subset of tissues presented higher viral RNA loads on both days 8 and 21 p.i. than the other tissues tested (Fig. 5).
FIG 5.
Tissue distribution of SARS-CoV-2 RNA. Tissues were collected and processed for rRT-PCR on day 8 postinoculation (p.i.) for animal no. 2001 (which died of an unrelated cause) and 21 p.i. of the remaining animals (animal no. 2042, 2043, 2045, 2006, and 2044).
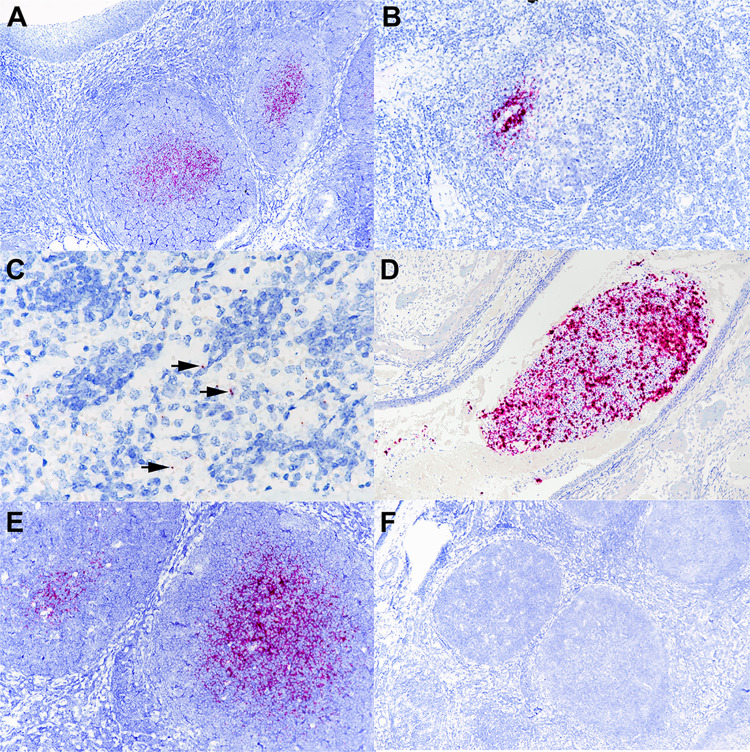
The presence of SARS-CoV-2 RNA was confirmed by ISH in the palatine tonsils and medial retropharyngeal lymph nodes of all inoculated and indirect contact fawns. Labeling of viral RNA was intense and limited to central regions of lymphoid follicles (Fig. 6A and B). Viral RNA was also noted in tracheobronchial and/or mediastinal lymph nodes of 5/6 deer but was less intense than that seen in tonsils and was limited to the medulla of the lymph node (Fig. 6C). Only the nasal turbinates of fawn 2001, collected on day 8 p.i., exhibited viral RNA labeling. Within the nasal lumen, there was strong labeling of aggregates of mucus and cell debris (Fig. 6D). No viral RNA was detected in sections of lung, kidney, brain, intestine, or mesenteric lymph nodes. Tissues were also analyzed using the negative sense-specific RNA probe (V-nCoV2019-S-sense probe) to detect virus replication. While intense labeling was noted with the probe detecting genomic and subgenomic positive-sense viral RNA (Fig. 6E), no labeling was observed with the negative sense-specific probe in any of the tissues (Fig. 6F), suggesting lack of virus replication. These findings are consistent with lack of virus isolation from rRT-PCR-positive tissues. The ability of the negative sense-specific probe to bind actively replicating virus was confirmed in Vero cells infected with SARS-CoV-2 isolate TGR/NY/20 (data not shown).
FIG 6.
Tissues from white-tailed deer fawn inoculated intranasally with SARS-CoV-2 and examined 21 days later. (A) Note intense labeling of viral RNA in the centers of lymphoid follicles located subjacent to tonsillar epithelium (upper left). (B and C) Note labeling for SARS-CoV-2 RNA within the medial retropharyngeal lymph node follicle (B) and mediastinal lymph node medulla (C). (D) Nasal turbinate lumen contains aggregate of mucus, cells, and debris with intense labeling for SARS-CoV-2 RNA. (E and F) Adjacent microscopic sections demonstrate intense labeling of lymphoid follicles with probe for SARS-CoV-2 RNA (E) but no labeling using the anti-genomic sense probe (F). ISH-RNAscope.
Antibody responses in white-tailed deer following SARS-CoV-2 infection.
The serological responses to SARS-CoV-2 were assessed using a Luminex and a virus neutralization (VN) assay. Serum samples collected on days 0, 7, 14, and 21 p.i. were used to assess nucleocapsid (N) and S receptor binding domain (RBD)-specific or neutralizing antibody (NA) levels following SARS-CoV-2 infection. All inoculated and indirect contact animals seroconverted to SARS-CoV-2, with antibodies against S-RBD and NA being detected as early as day 7 p.i. (Fig. 7A and B, respectively) and increasing thereafter on days 14 and 21 p.i. Overall, titers of antibodies against the N protein were lower and appeared at later time points (days 14 to 21) (Fig. 7A). Geometric mean titers (GMT) of NA ranged from 37 to 107, 85 to 214, and 85 to 256 on days 7, 14, and 21, respectively (Fig. 7C). These results confirm infection of all inoculated fawns and further confirm transmission to indirect contact animals.
FIG 7.
Antibody responses in white-tailed deer following SARS-CoV-2 infection. (A) Luminex assay to assess IgG anti-SARS-CoV-2 nucleocapsid (N) in serum samples collected on days 0, 7, 14, and 21 postinoculation (p.i.) (fawn no. 2001, 2042, 2043, and 2045), or contact (fawn no. 2006 and 2044). (B) Luminex assay to assess IgG anti-SARS-CoV-2-S-receptor binding domain (RBD) specificity at the same time point and fawns as above. (C) Virus neutralization (VN) assay. Neutralizing antibody (NA) titers were expressed as the reciprocal of the highest dilution of serum that completely inhibited SARS-CoV-2 infection/replication in serum at the same time point and fawns as above. Results represent the geometric mean titers (GMT) of three independent experiments.
DISCUSSION
Here, we demonstrated that white-tailed deer are susceptible to SARS-CoV-2 infection. Infection of deer cells in vitro resulted in productive virus replication. Most importantly, intranasal inoculation of white-tailed deer fawns led to subclinical infection and productive virus replication in the upper respiratory tract with shedding of infectious virus in nasal secretions of infected animals.
One of the most remarkable characteristics of SARS-CoV-2 is its highly efficient transmissibility (34, 35). Consistent with this, rapid SARS-CoV-2 transmission was observed in several experimentally infected animal species (27, 28, 31, 36–41). Similarly, results here show efficient SARS-CoV-2 transmission between white-tailed deer. Indirect contact fawns became infected and supported productive SARS-CoV-2 replication, as evidenced by virus shedding in nasal secretions and seroconversion. Evidence to date indicates that transmission of SARS-CoV-2 occurs mainly through direct, indirect, or close contact with infected individuals through secretions such as respiratory, salivary, and/or fecal droplets (42). Under experimental conditions, transmission via direct contact has been demonstrated in ferrets, minks, raccoon dogs, hamsters, cats, and deer mice (23, 28, 31, 36, 38, 39, 41), while indirect/aerosol transmission was observed in ferrets and hamsters (36, 38). The experimental setup in the present study prevented direct contact between animals, suggesting that transmission of SARS-CoV-2 from inoculated to room indirect contact fawns most likely occurred via aerosols or droplets generated by inoculated animals. Importantly, in a follow-up study conducted to assess the transmission dynamics of SARS-CoV-2 in white-tailed deer, we have confirmed the rapid transmission of the virus from inoculated to contact animals. Contact animals became positive within ∼24 to 48 h, when inoculated animals were placed in contact with naive animals on day 3 postinoculation (M. Martins et al., unpublished data). These findings are consistent with the present study and with the transmission dynamics of the virus in other animal species (23, 28, 31, 36, 38, 39, 41).
Despite the potential for severe illness and mortality due to SARS-CoV-2 infection in humans (43, 44), asymptomatic and mild cases represent approximately 80% of human COVID-19 cases, while severe and critical disease outcomes account for approximately 15 and 5% of the cases, respectively (45). Notably, experimental infections in several animal species, including nonhuman primates (26), cats (19, 23, 46), ferrets (23, 31, 37, 47), minks (27), deer mice (39), and raccoon dogs (28), have mostly resulted in subclinical infections or only mild respiratory distress. Consistent with these observations, white-tailed deer fawns inoculated here did not present overt clinical disease following SARS-CoV-2 infection. Only a transient and modest increase in body temperature was observed in 3 of 4 inoculated animals between days 1 to 3 p.i. It is important to note that most severe and critical cases of COVID-19 in humans have been associated with underlying clinical conditions (e.g., heart and pulmonary disease, cancer, and diabetes, among others), which may complicate full recapitulation of the clinical manifestations of the human disease in animal species. Differences in expression and/or tissue distribution of the SARS-CoV-2 receptor (ACE2), cellular proteases (transmembrane serine protease 2, TMPRRS2, cathepsins), or other cofactor(s) that might be required for efficient virus infection and replication in target tissues may help to explain the diverse clinical outcomes of infection in humans and animals.
While no viremia was detected in infected animals, active virus shedding was observed in nasal secretions of both inoculated and indirect contact animals. The patterns and dynamics of SARS-CoV-2 shedding observed in fawns here are consistent with what has been described for other susceptible animal species (23, 28, 31, 36, 39, 41, 46). Most importantly, the duration and magnitude of virus shedding observed in deer corroborate observations in humans in which prolonged viral RNA has been detected by rRT-PCR (21 to 35 days), with virus infectivity being usually limited to the first 7 to 10 days of infection (48, 49). This could be partially explained by the activity of effector host immune responses elicited against the virus, especially of neutralizing antibodies, which were detected as early as day 7 p.i. in all fawns here, paralleling decreased virus infectivity in nasal secretions.
Viral RNA load and tissue distribution were assessed on day 8 p.i. (animal no. 2001) following acute infection and on day 21 p.i. after termination of the experiment (remaining fawns). High viral RNA loads were detected in nasal turbinates, palatine tonsils, and medial retropharyngeal lymph nodes, suggesting that these tissues may potentially serve as sites of SARS-CoV-2 replication in white-tailed deer. Since no infectious virus was recovered from any of the tissues sampled on days 8 or 21 p.i., additional studies involving serial tissue collection over time are needed to determine specific sites of virus replication. Interestingly, no viral RNA was detected in the lung tissues from inoculated or indirect contact fawns, suggesting that these tissues may not be targeted by SARS-CoV-2 following infection of white-tailed deer. The possibility that the virus may have been cleared from these sites by the time of tissue collection, however, cannot be excluded.
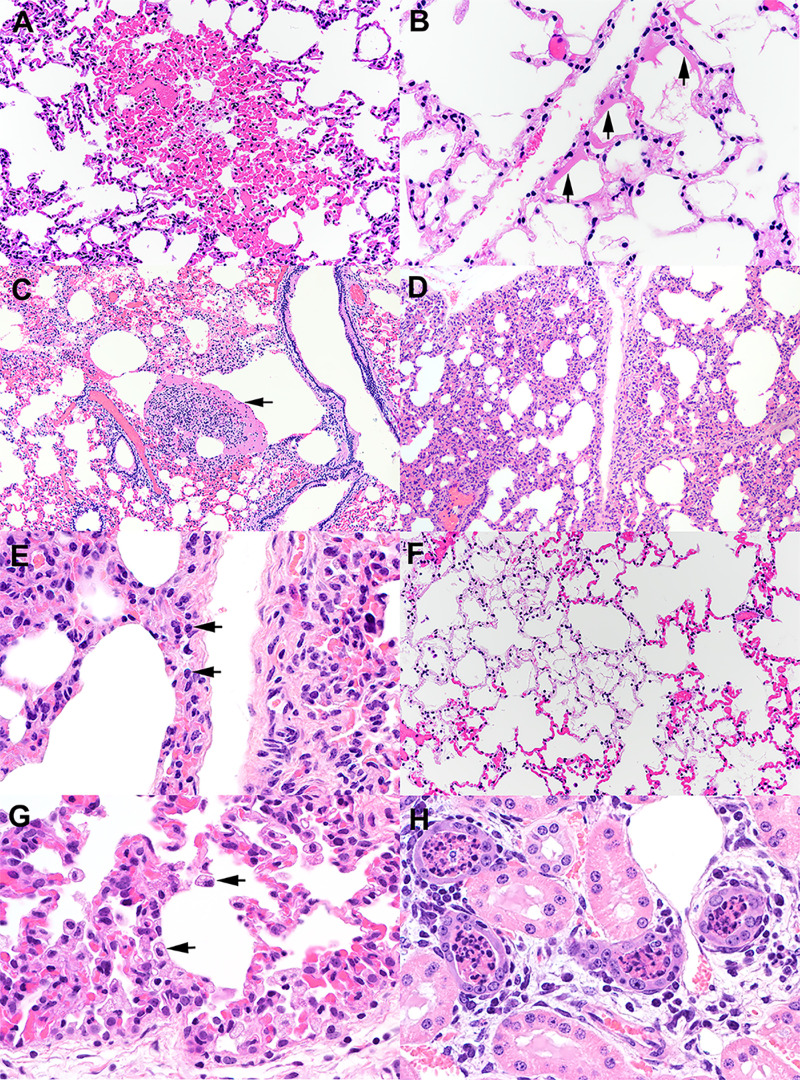
Microscopic changes were observed in the lungs of 2 of 4 inoculated fawns and the 1 indirect contact fawn examined 21 days p.i. These changes were characterized by congestion, lymphohistiocytic interstitial pneumonia, hyaline membranes, and intra-alveolar fibrin (Fig. 8), all of which are characteristic of the acute phase of diffuse alveolar damage, the predominant finding in the lungs of patients affected with COVID-19 (50, 51). Mild to moderate pulmonary changes have been described in other species following experimental SARS-CoV-2 infection, including domestic cats (40), rhesus macaques (25, 52, 53), hACE2 mice (54, 55), ferrets (36), and minks (27). It is also notable that no SARS-CoV-2 viral RNA was associated with lesions in white-tailed deer, which could be a result of earlier viral clearance from these sites due to potent immunological responses following SARS-CoV-2 infection.
FIG 8.
Histological examination of lung from white-tailed deer fawns intranasally inoculated with SARS-CoV-2. (A) Note the well-demarcated focus of congestion. Alveolar septal capillaries are engorged with blood surrounded by normal appearing alveolar septa. HE. (B) Multiple alveolar septa are lined by bands of eosinophilic hyalinized proteinaceous material (arrows) consistent with hyaline membranes. Multiple alveoli contain flocculent to fibrillar eosinophilic material consistent with fibrin. HE. (C) Expanded alveolus contains a large collection of fibrin, inflammatory cells, and cell debris (arrow). HE. (D) Alveolar septa are expanded by an inflammatory infiltrate (interstitial pneumonia) composed primarily of lymphocytes (arrows) and macrophages (E). HE. (F) Within a field of congested alveolar septa are irregular regions characterized by hypocellular septa containing few erythrocytes. Septal stroma is fibrillar and lightly eosinophilic. Multiple alveoli within these regions contain flocculent strands of fibrin. HE. (G) There is type II pneumocyte hyperplasia and an increase in alveolar macrophages (arrows). HE. (H) Lumens of cortical tubules in the kidney are filled with necrotic cellular debris. Renal tubules are variably lined by attenuated epithelium, occasionally have hypereosinophilic cytoplasm and pyknotic nuclei (degeneration and necrosis), and overall exhibit increased cytoplasmic basophilia (regeneration). Tubules are separated by interstitial edema and a cellular infiltrate composed of lymphocytes, plasma cells, and fewer macrophages. HE. Tissue sections were examined on day 21 postinoculation.
Given the susceptibility of white-tailed deer to SARS-CoV-2 and the fact that coronaviruses may adapt to host species through mutations that may enhance binding affinity of the S protein to the ACE2 receptor, it will be interesting to assess potential evolution of SARS-CoV-2 in this species in the future. Studies to investigate evolution as the virus is transmitted between animals (multiple rounds of transmission) are warranted. The best example of SARS-CoV-2 adaptation to a new species comes from mink, a species that is naturally susceptible to the virus and in which variant SARS-CoV-2 strains have been identified and shown to be transmitted back to humans (56). It is important to note, however, that the mink ACE2 protein is less conserved with the human protein than the white-tailed deer receptor (18). Whether these differences result in different evolutionary rates and differential adaptation of the virus remains to be demonstrated. In summary, our study shows that white-tailed deer are susceptible to SARS-CoV-2 infection and can transmit the virus to indirect contact animals. These results confirmed in silico predictions describing a high propensity of interaction between SARS-CoV-2 S protein and the cervid ACE2 receptor. Our findings indicate that deer and other cervids should be considered in investigations conducted to identify the origin and potential intermediate host species that may have served as the link host reservoir to humans.
MATERIALS AND METHODS
Cells and virus.
Vero cells (ATCC CCL-81), Vero E6 (ATCC CRL-1586), and Vero E6/TMPRSS2 (JCRB Cell Bank; JCRB1819) were cultured in Dulbecco’s modified eagle medium (DMEM), while deer lung (DL; ATCC CRL6195) cells were cultured in minimum essential medium (MEM). Both DMEM and MEM were supplemented with 10% fetal bovine serum (FBS), l-glutamine (2 mM), penicillin (100 U·ml−1), streptomycin (100 μg·ml−1), and gentamicin (50 μg·ml−1). The cell cultures were maintained at 37°C with 5% CO2. The SARS-CoV-2 isolate TGR/NY/20 obtained from a Malayan tiger naturally infected with SARS-CoV-2 and presenting with respiratory disease compatible with SARS-CoV-2 infection (22) was propagated in Vero CCL-81 cells. Low-passage virus stocks (passage 4) were prepared, cleared by centrifugation (1,966 × g for 10 min), and stored at −80°C. The endpoint titer was determined by limiting dilution following the Spearman and Karber method. A viral suspension containing 106.3 50% tissue culture infectious dose per milliliter () was used for all in vitro experiments and fawn inoculations.
Cell susceptibility and growth curves.
The susceptibility and kinetics of replication of the SARS-CoV-2 in DL cells was assessed in vitro and compared to virus replication in Vero E6 and Vero E6/TMPRSS2. For this, DL, Vero E6, and Vero E6/TMPRSS2 cells were inoculated with SARS-CoV-2 isolate TGR/NY/20 at a multiplicity of infection (MOI) of 1 and adsorbed for 1 h at 37°C with 5% CO2. After adsorption, cells were washed 3 times with MEM, and the cells were incubated for 24 h at 37°C with 5% CO2. At 24 h postinoculation, cells were fixed with 3.7% formaldehyde for 30 min at room temperature, permeabilized with 0.2% Triton X-100 (in phosphate-buffered saline [PBS]), and subjected to an immunofluorescence assay (IFA) using a monoclonal antibody (MAb) anti-ACE2 (Sigma-Aldrich), then incubated with a goat anti-rabbit IgG (Alexa Fluor 488) and using a MAb anti-SARS-CoV-2 nucleoprotein (N) (clone B6G11) produced and characterized in D. G. Diel’s laboratory, and then incubated with a goat anti-mouse IgG secondary antibody (Alexa Fluor 594). Nuclear counterstain was performed with 4′,6-diamidino-2-phenylindole (DAPI) and visualized under a fluorescence microscope. To assess the kinetics of replication of SARS-CoV-2 isolate TGR/NY/20, Vero E6, Vero E6/TMPRSS2, and DL cells were cultured in 12-well plates, inoculated with SARS-Cov-2 isolate TGR/NY/20 (MOI of 0.1 and 1) and adsorbed for 1 h at 37°C with 5% CO2. After adsorption, the inoculum was removed, and cells were washed 3 times with MEM. Inoculated cell cultures were maintained at 37°C with 5% CO2 and harvested at various time points postinoculation (12, 24, 36, 48, and 72 hpi). Cell monolayers and supernatants were collected and kept at −80°C until further processing. The input virus amount was confirmed by back titration of the inoculum virus (time zero). Virus titers were determined on each time point using endpoint dilutions and the Spearman and Karber's method and expressed as .
Animals, inoculation, and sampling.
All animals were handled in accordance with the Animal Welfare Act Amendments (7 U.S. Code §2131 to §2156), and all study procedures were reviewed and approved by the Institutional Animal Care and Use Committee at the National Animal Disease Center (NADC; IACUC approval number ARS-2020-861). White-tailed deer fawns (n = 6) were obtained from a breeding herd maintained at the National Animal Disease Center in Ames, Iowa. Within 24 h of birth, fawns were removed from the pasture, moved to indoor housing, and bottle-fed. Hand-raising fawns has been found to greatly decrease stress when animals are moved to containment housing (57). At approximately 4 weeks of age, fawns were moved to the biosafety level 3 (agriculture) (BSL-3Ag) facility at NADC and were microchipped (subcutaneously [s.c.]) for identification and body temperature monitoring. Fawns were fed white-tailed deer milk replacer, and hay was also available.
After a 2-week acclimation period, four fawns were inoculated intranasally with 5 ml (2.5 ml per nostril) of a virus suspension containing 106.3 of SARS-CoV-2, isolated from respiratory secretions from a tiger at the Bronx Zoo (TGR/NY/20) (22). Two fawns were maintained as noninoculated indirect contact animals to evaluate potential transmission of SARS-CoV-2 from inoculated to indirect contact animals. All fawns were maintained in a 3.7-m by 3.7-m room and inoculated, and indirect contact animals were kept in two pens separated by a plexiglass barrier approximately 0.9 m (∼3-feet) in height to prevent direct nose-to-nose contact (Fig. 2A). Airflow in the room was maintained at 10 to 11 air exchanges per hour at a standard exchange rate for BSL-3Ag housing of large animals. Body temperatures were recorded daily; nasal swabs (NS) and rectal swabs (RS) were collected on days 0, 1, 2, 3, 4, 5, 6, 7, 10, 12, 14, and 21 postinoculation (p.i.). Upon collection, swabs were placed individually in sterile tubes containing 2 ml of viral transport media (MEM with 1,000 U·ml−1 of penicillin, 1,000 μg·ml−1 of streptomycin, and 2.5 μg·ml−1 of amphotericin B) and stored at −80°C until analysis. Blood was collected through jugular venipuncture in EDTA and serum separator tubes on days 0, 7, 14, and 21 p.i. The tubes were centrifuged for 25 min at 1,200 × g, and buffy coat (BC) was collected from EDTA tubes and stored at −80°C. Serum from the serum separator tubes was aliquoted and stored at −80°C until analysis.
Fawns were humanely euthanized on day 21 p.i. Following necropsy, multiple specimens, including tracheal wash (TW), lung lavage (LL), and several tissues (nasal turbinates, palatine tonsil, thymus, trachea, lung, bronchi, kidney, liver, spleen, ileum, ileocecal junction, spiral colon, cerebellum, cerebrum, olfactory bulbs, and medial retropharyngeal, mandibular, tracheobronchial, mediastinal and mesenteric lymph nodes) were collected. Samples were processed for rRT-PCR and virus isolation (VI) and were individually bagged, placed on dry ice, and transferred to a −80°C freezer until testing. Additionally, tissue samples were collected and processed for standard microscopic examination and in situ hybridization (ISH). For this, tissue fragments of approximately ≤0.5 cm in width were fixed by immersion in 10% neutral-buffered formalin (≥20 volumes fixative to 1 volume tissue) for approximately 24 h and then transferred to 70% ethanol, followed by standard paraffin embedding techniques. Slides for standard microscopic examination were stained with hematoxylin and eosin (HE).
Nucleic acid extraction and rRT-PCR.
Nucleic acid was extracted from nasal secretions, feces, BC, serum tracheal wash, and lung lavage, and all the tissue samples were collected at necropsy. Before extraction, 0.5 g of tissues were minced with a sterile scalpel and resuspended in 5 ml DMEM (10% wt/vol) and homogenized using a stomacher (Stomacher 80 Biomaster; one speed cycle of 60 s). Homogenized tissue samples were cleared by centrifugation (1,966 × g for 10 min), and 200 μl of the homogenate supernatant was used for RNA extraction using the MagMax Core extraction kit (Thermo Fisher, Waltham, MA, USA) and the automated KingFisher Flex nucleic acid extractor (Thermo Fisher) following the manufacturer’s recommendations. The real-time reverse transcriptase PCR (rRT-PCR) was performed using the EZ-SARS-CoV-2 real-time RT-PCR assay (Tetracore Inc., Rockville, MD), which detects both genomic and subgenomic viral RNA for increased diagnostic sensitivity. An internal inhibition control was included in all reactions. Positive and negative amplification controls were run side by side with test samples. All RNA extractions and rRT-PCR were performed at the Cornell Animal Health Diagnostic Center (AHDC).
Virus isolation and titration.
Nasal and rectal swabs collected on days 0, 1, 2, 3, 4, 5, 6, 7, 10, 12, 14, and 21, and tissues collected during the necropsy that tested positive for SARS-CoV-2 by rRT-PCR were subjected to virus isolation under biosafety level 3 conditions at Cornell University. Twenty-four-well plates were seeded with ∼75,000 Vero E6/TMPRSS2 cells per well 24 h prior to sample inoculation. Cells were rinsed with PBS (Corning) and inoculated with 150 μl of each sample and inoculum adsorbed for 1 h at 37°C with 5% CO2. Mock-inoculated cells were used as negative controls. After adsorption, replacement cell culture medium supplemented as described above was added, and cells were incubated at 37°C with 5% CO2 and monitored daily for cytopathic effect (CPE) for 3 days. SARS-CoV-2 infection in CPE-positive cultures was confirmed with an immunofluorescence assay (IFA) as described above. Cell cultures with no CPE were frozen, thawed, and subjected to two additional blind passages/inoculations in Vero E6/TMPRSS2 cell cultures. At the end of the third passage, the cell cultures were subjected to IFA as above. Positive samples were subjected to endpoint titrations by limiting dilution using the Vero E6/TMPRSS2 cells, and virus titers were determined in the original swab sample using the Spearman and Karber’s method and expressed as .
ISH.
Paraffin-embedded tissues were sectioned at 5 μm and subjected to ISH using the RNAscope ZZ probe technology (Advanced Cell Diagnostics, Newark, CA). In situ hybridization was performed to assess tissue distribution of SARS-CoV-2 nucleic acid in palatine tonsil; medial retropharyngeal, tracheobronchial, mediastinal, and mesenteric lymph nodes; nasal turbinate; brain; lung; and kidney, using the RNAscope 2.5 HD reagents-Red kit (Advanced Cell Diagnostics) as previously described (58). Proprietary ZZ probes targeting SARS-CoV-2 RNA (V-nCoV2019-S probe; reference no. 8485561) or anti-sense RNA (V-nCoV2019-S-sense; reference no. 845701) designed and manufactured by Advance Cell Diagnostics were used for detection of viral RNA. The V-nCoV2019-S probe detects the subgenomic RNA encoding the S gene, while the V-nCoV2019-S-sense detects negative-sense genomic RNA intermediates of virus replication. A positive-control probe targeted the Bos taurus-specific cyclophilin B (PPIB; catalog no. 3194510) or ubiquitin (UBC; catalog no. 464851) housekeeping genes, while a probe targeting dapB of Bacillus subtilis (catalog no. 312038) was used as a negative control. Slides were counterstained with hematoxylin and examined by light microscopy using a Nikon Eclipse Ci microscope. Digital images were captured using a Nikon DS-Ri2 camera.
Serological analysis.
Antibody responses to SARS-CoV-2 were assessed by a Luminex assay, and virus neutralization assays were developed in-house. For the Luminex assay, SARS-CoV-2 antigens were expressed as interleukin 4 (IL-4) fusion proteins in mammalian cells as previously described (59). SARS-CoV-2 RNA (strain Hu-WA-1) was derived from Vero cells infected with the virus, and cDNA was synthesized using the SuperScript III reverse transcriptase (Life Technologies) and oligo(dT) and six hexamer random primers. The cDNA was used to amplify the receptor binding domain (RBD; amino acids 319 to 529) of the spike protein and the whole nucleocapsid protein (NP; amino acids 1 to 419) by PCR. The primer sequences for RBD were AAGGATCCAAGAGTCCAACCAACAGAATCTATTGTT (forward) and AAAGGTACCTTACTTTTTAGGTCCACAAACAGTTGCT (reverse). N primers were AAGGATCCAATGTCTGATAATGGACCCCAAAATC (forward) and AAAGGTACCTTAGGCCTGAGTTGAGTCAGCACTG (reverse). The restriction sites for expression cloning are underlined. Both PCR products were cloned into the multiple cloning site of the mammalian expression vector pcDNA3.1 (Thermo Fisher Scientific, Waltham, MA, USA) downstream of the equine IL-4 sequence as described previously (59). Nucleotide sequences of RBD and N were verified by Sanger sequencing and identical to respective sequences of the Hu-WA-1 strain (GenBank accession number NC_045512).
CHO-K1 cells were transiently transfected with the recombinant plasmid constructs. Expression and secretion of the recombinant SARS-CoV-2 antigen fusion proteins by CHO transfectants were confirmed using the IL-4 tag by flow cytometric analysis and enzyme-linked immunosorbent assay (ELISA) as previously described (59). After 24 to 30 h of incubation, the supernatants were harvested for fluorescent bead-based multiplex assays.
Fluorescent beads were coupled with the anti-equine IL-4 antibody, clone 25 (RRID AB_2737308) as previously described (60). The RBD/IL-4 and N/IL-4 fusion proteins were bound by incubating the anti-IL-4-coupled beads with the fusion protein supernatant solution for 30 min at room temperature, followed by a wash. The assay was performed with a few modifications from previously described procedures (60, 61). Briefly, beads were incubated with deer serum samples diluted 1:100. The assay was detected using a biotinylated mouse anti-goat IgG (H+L) (RRID AB_2339061; Jackson ImmunoResearch Laboratories, West Grove, PA) cross-reactive with deer immunoglobulin. Afterwards, streptavidin-phycoerythrin (Invitrogen, Carlsbad, CA) was added as a final detection step. All incubation steps were for 30 min at room temperature, and the assay was washed after each incubation step. The assay was measured in a Luminex 200 instrument (Luminex Corp., Austin, TX). Assay results were expressed as median fluorescence intensity (MFI).
Neutralizing antibody responses to SARS-CoV-2 were assessed by a virus neutralization (VN) assay performed under BSL-3 conditions at the Cornell AHDC. Twofold serial dilutions (1:8 to 1:4,096) of serum samples were incubated with 100 to 200 TCID50 of SARS-CoV-2 isolate TGR/NY/20 for 1 h at 37°C. Following incubation of serum and virus, 50 μl of a cell suspension of Vero cells was added to each well of a 96-well plate and incubated for 48 h at 37°C with 5% CO2. The cells were fixed and permeabilized as described above and subjected to IFA using a rabbit polyclonal antibody (pAb) specific for the SARS-CoV-2 nucleoprotein (N) (produced in D. G. Diel’s laboratory) followed by incubation with a goat anti-rabbit IgG (DyLight 594 conjugate, ImmunoReagents, Inc.). Unbound antibodies were washed from cell cultures by rinsing the cells in PBS, and virus infectivity was assessed under a fluorescence microscope. Neutralizing antibody titers were expressed as the reciprocal of the highest dilution of serum that completely inhibited SARS-CoV-2 infection/replication after three independent VN assays were performed. Fetal bovine serum (FBS) and convalescent human serum (kindly provided by Elizabeth Plocharczyk, Cayuga Medical Center [CMC], under CMC’s Institutional Review Board; protocol number 0420EP) were used as negative and positive controls, respectively.
ACKNOWLEDGMENTS
We thank clinical veterinarian, Rebecca Cox, and animal caretakers, Tiffany Williams, Kolby Stallman, Derek Vermeer, and Robin Zeisneiss, for excellent animal care and Patricia Federico for excellent technical assistance.
Antibody reagents to SARS-CoV-2 and the virus neutralization assay used to assess serological responses were developed with support from the Cornell Feline Health Center.
Mention of tradenames or commercial products is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
REFERENCES
- 1.Gorbalenya AE, Baker SC, Baric RS, de Groot RJ, Drosten C, Gulyaeva AA, Haagmans BL, Lauber C, Leontovich AM, Neuman BW, Penzar D, Perlman S, Poon LLM, Samborskiy DV, Sidorov IA, Sola I, Ziebuhr J. 2020. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol Nature Res 5:536–544. 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team . 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. 2020. A new coronavirus associated with human respiratory disease in China. Nature 579:265–269. 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. 2020. The proximal origin of SARS-CoV-2. Nat Med 26:450–454. 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Latinne A, Hu B, Olival KJ, Zhu G, Zhang L, Li H, Chmura AA, Field HE, Zambrana-Torrelio C, Epstein JH, Li B, Zhang W, Wang LF, Shi ZL, Daszak P. 2020. Origin and cross-species transmission of bat coronaviruses in China. Nat Commun 11:4235. 10.1038/s41467-020-17687-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murakami S, Kitamura T, Suzuki J, Sato R, Aoi T, Fujii M, Matsugo H, Kamiki H, Ishida H, Takenaka-Uema A, Shimojima M, Horimoto T. 2020. Detection and characterization of bat sarbecovirus phylogenetically related to SARS-CoV-2, Japan. Emerg Infect Dis 26:3025–3029. 10.3201/eid2612.203386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lau SKP, Luk HKH, Wong ACP, Li KSM, Zhu L, He Z, Fung J, Chan TTY, Fung KSC, Woo PCY. 2020. Possible bat origin of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis 26:1542–1547. 10.3201/eid2607.200092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu DKW, Poon LLM, Gomaa MM, Shehata MM, Perera RAPM, Zeid DA, El Rifay AS, Siu LY, Guan Y, Webby RJ, Ali MA, Peiris M, Kayali G. 2014. MERS coronaviruses in dromedary camels, Egypt. Emerg Infect Dis 20:1049–1053. 10.3201/eid2006.140299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alagaili AN, Briese T, Mishra N, Kapoor V, Sameroff SC, Burbelo PD, de Wit E, Munster VJ, Hensley LE, Zalmout IS, Kapoor A, Epstein JH, Karesh WB, Daszak P, Mohammed OB, Lipkin WI. 2014. Middle East respiratory syndrome coronavirus infection in dromedary camels in Saudi Arabia. mBio 5:e00884-14. 10.1128/mBio.00884-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Childs JE, Mackenzie JS, Richt JA. 2007. Current topics in microbiology and immunology. The biology, circumstances and consequences of cross-species transmission. Springer-Verlag, Berlin, Heidelberg, Germany. [Google Scholar]
- 12.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. 2020. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395:565–574. 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paraskevis D, Kostaki EG, Magiorkinis G, Panayiotakopoulos G, Sourvinos G, Tsiodras S. 2020. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect Genet Evol 79:104212. 10.1016/j.meegid.2020.104212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lam TTY, Jia N, Zhang YW, Shum MHH, Jiang JF, Zhu HC, Tong YG, Shi YX, Ni XB, Liao YS, Li WJ, Jiang BG, Wei W, Yuan TT, Zheng K, Cui XM, Li J, Pei GQ, Qiang X, Cheung WYM, Li LF, Sun FF, Qin S, Huang JC, Leung GM, Holmes EC, Hu YL, Guan Y, Cao WC. 2020. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature 583:282–285. 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- 15.Liu P, Jiang J-Z, Wan X-F, Hua Y, Li L, Zhou J, Wang X, Hou F, Chen J, Zou J, Chen J. 2020. Are pangolins the intermediate host of the 2019 novel coronavirus (SARS-CoV-2)? PLoS Pathog 16:e1008421. 10.1371/journal.ppat.1008421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Letko M, Marzi A, Munster V. 2020. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol 5:562–569. 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wan Y, Shang J, Graham R, Baric RS, Li F. 2020. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol 94:1–9. 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Damas J, Hughes GM, Keough KC, Painter CA, Persky NS, Corbo M, Hiller M, Koepfli KP, Pfenning AR, Zhao H, Genereux DP, Swofford R, Pollard KS, Ryder OA, Nweeia MT, Lindblad-Toh K, Teeling EC, Karlsson EK, Lewin HA. 2020. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc Natl Acad Sci U S A 117:22311–22322. 10.1073/pnas.2010146117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sit THC, Brackman CJ, Ip SM, Tam KWS, Law PYT, To EMW, Yu VYT, Sims LD, Tsang DNC, Chu DKW, Perera RAPM, Poon LLM, Peiris M. 2020. Infection of dogs with SARS-CoV-2. Nature 586:776–778. 10.1038/s41586-020-2334-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oreshkova N, Molenaar RJ, Vreman S, Harders F, Oude MBB, Van Der Honing RWH, Gerhards N, Tolsma P, Bouwstra R, Sikkema RS, Tacken MGJ, De RM, Weesendorp E, Engelsma MY, Bruschke CJM, Smit LAM, Koopmans M, Van Der Poel WHM, Stegeman A. 2020. SARS-CoV-2 infection in farmed minks, the Netherlands, April and May 2020. Eurosurveillance 25:2001005. 10.2807/1560-7917.ES.2020.25.23.2001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rockx B, Kuiken T, Herfst S, Bestebroer T, Lamers MM, Munnink BBO, De Meulder D, Van Amerongen G, Van Den Brand J, Okba NMA, Schipper D, Van Run P, Leijten L, Sikkema R, Verschoor E, Verstrepen B, Bogers W, Langermans J, Langermans J, Drosten C, Van Vlissingen MF, Fouchier R, De Swart R, Koopmans M, Haagmans BL. 2020. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science 368:1012–1015. 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McAloose D, Laverack M, Wang L, Killian ML, Caserta LC, Yuan F, Mitchell PK, Queen K, Mauldin MR, Cronk BD, Bartlett SL, Sykes JM, Zec S, Stokol T, Ingerman K, Delaney MA, Fredrickson R, Ivančić M, Jenkins-Moore M, Mozingo K, Franzen K, Bergeson NH, Goodman L, Wang H, Fang Y, Olmstead C, McCann C, Thomas P, Goodrich E, Elvinger F, Smith DC, Tong S, Slavinski S, Calle PP, Terio K, Torchetti MK, Diel DG. 2020. From people to Panthera: natural SARS-CoV-2 infection in tigers and lions at the Bronx Zoo. mBio 11:e02220-20. 10.1128/mBio.02220-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi J, Wen Z, Zhong G, Yang H, Wang C, Huang B, Liu R, He X, Shuai L, Sun Z, Zhao Y, Liu P, Liang L, Cui P, Wang J, Zhang X, Guan Y, Tan W, Wu G, Chen H, Bu Z. 2020. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science 368:1016–1020. 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chandrashekar A, Liu J, Martino AJ, McMahan K, Mercad NB, Peter L, Tostanosk LH, Yu J, Maliga Z, Nekorchuk M, Busman-Sahay K, Terry M, Wriji LM, Ducat S, Martine DR, Atyeo C, Fischinger S, Burk JS, Slei MD, Pessaint L, Van Ry A, Greenhouse J, Taylor T, Blade K, Cook A, Finneyfrock B, Brown R, Teow E, Velasco J, Zahn R, Wegmann F, Abbink P, Bondzi EA, Dagotto G, Gebr MS, He X, Jacob-Dolan C, Kordana N, Li Z, Lifto MA, Mahrokhia SH, Maxfiel LF, Nityanandam R, Nkolol JP, Schmid AG, Mille AD, Bari RS, Alter G, Sorge PK, Este JD, Andersen H, Lewi MG, Barou DH. 2020. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science 369:812–817. 10.1126/science.abc4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Munster VJ, Feldmann F, Williamson BN, van Doremalen N, Pérez-Pérez L, Schulz J, Meade-White K, Okumura A, Callison J, Brumbaugh B, Avanzato VA, Rosenke R, Hanley PW, Saturday G, Scott D, Fischer ER, de Wit E. 2020. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature 585:268–272. 10.1038/s41586-020-2324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hartman AL, Nambulli S, McMillen CM, White AG, Tilston-Lunel NL, Albe JR, Cottle E, Dunn MD, Frye LJ, Gilliland TH, Olsen EL, O'Malley KJ, Schwarz MM, Tomko JA, Walker RC, Xia M, Hartman MS, Klein E, Scanga CA, Flynn JL, Klimstra WB, McElroy AK, Reed DS, Duprex WP. 2020. SARS-CoV-2 infection of African green monkeys results in mild respiratory disease discernible by PET/CT imaging and shedding of infectious virus from both respiratory and gastrointestinal tracts. PLoS Pathog 16:e1008903. 10.1371/journal.ppat.1008903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Z, Wang L, Liu W, Yan Z, Zhu Y, Zhou S, Guan S. 2020. Replication, pathogenicity, and transmission of SARS-CoV-2 in minks. Natl Sci Rev 10.1093/nsr/nwaa291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freuling CM, Breithaupt A, Müller T, Sehl J, Balkema-Buschmann A, Rissmann M, Klein A, Wylezich C, Höper D, Wernike K, Aebischer A, Hoffmann D, Friedrichs V, Dorhoi A, Groschup MH, Beer M, Mettenleiter TC. 2020. Susceptibility of raccoon dogs for experimental SARS-CoV-2 infection. Emerg Infect Dis 26:2982–2985. 10.3201/eid2612.203733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meekins DA, Morozov I, Trujillo JD, Gaudreault NN, Bold D, Carossino M, Artiaga BL, Indran SV, Kwon T, Balaraman V, Madden DW, Feldmann H, Henningson J, Ma W, Balasuriya UBR, Richt JA. 2020. Susceptibility of swine cells and domestic pigs to SARS-CoV-2. Emerg Microbes Infect 9:2278–2288. 10.1080/22221751.2020.1831405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ulrich L, Wernike K, Hoffmann D, Mettenleiter TC, Beer M. 2020. Experimental infection of cattle with SARS-CoV-2. Emerg Infect Dis 26:2979–2981. 10.3201/eid2612.203799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlottau K, Rissmann M, Graaf A, Schön J, Sehl J, Wylezich C, Höper D, Mettenleiter TC, Balkema-Buschmann A, Harder T, Grund C, Hoffmann D, Breithaupt A, Beer M. 2020. SARS-CoV-2 in fruit bats, ferrets, pigs, and chickens: an experimental transmission study. Lancet Microbe 1:e218–e225. 10.1016/S2666-5247(20)30089-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsuyama S, Nao N, Shirato K, Kawase M, Saito S, Takayama I, Nagata N, Sekizuka T, Katoh H, Kato F, Sakata M, Tahara M, Kutsuna S, Ohmagari N, Kuroda M, Suzuki T, Kageyama T, Takeda M. 2020. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc Natl Acad Sci U S A 117:7001–7003. 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harcourt J, Tamin A, Lu X, Kamili S, Sakthivel SK, Murray J, Queen K, Tao Y, Paden CR, Zhang J, Li Y, Uehara A, Wang H, Goldsmith C, Bullock HA, Wang L, Whitaker B, Lynch B, Gautam R, Schindewolf C, Lokugamage KG, Scharton D, Plante JA, Mirchandani D, Widen SG, Narayanan K, Makino S, Ksiazek TG, Plante KS, Weaver SC, Lindstrom S, Tong S, Menachery VD, Thornburg NJ. 2020. Severe acute respiratory syndrome coronavirus 2 from patient with 2019 novel coronavirus disease, United States. Emerg Infect Dis 26:1266–1273. 10.3201/eid2606.200516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, Bhattacharya T, Foley B, Hastie KM, Parker MD, Partridge DG, Evans CM, Freeman TM, de Silva TI, Angyal A, Brown RL, Carrilero L, Green LR, Groves DC, Johnson KJ, Keeley AJ, Lindsey BB, Parsons PJ, Raza M, Rowland-Jones S, Smith N, Tucker RM, Wang D, Wyles MD, McDanal C, Perez LG, Tang H, Moon-Walker A, Whelan SP, LaBranche CC, Saphire EO, Montefiori DC. 2020. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 182:812–827.e19. 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arons MM, Hatfield KM, Reddy SC, Kimball A, James A, Jacobs JR, Taylor J, Spicer K, Bardossy AC, Oakley LP, Tanwar S, Dyal JW, Harney J, Chisty Z, Bell JM, Methner M, Paul P, Carlson CM, McLaughlin HP, Thornburg N, Tong S, Tamin A, Tao Y, Uehara A, Harcourt J, Clark S, Brostrom-Smith C, Page LC, Kay M, Lewis J, Montgomery P, Stone ND, Clark TA, Honein MA, Duchin JS, Jernigan JA. 2020. Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. N Engl J Med 382:2081–2090. 10.1056/NEJMoa2008457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim YI, Kim SG, Kim SM, Kim EH, Park SJ, Yu KM, Chang JH, Kim EJ, Lee S, Casel MAB, Um J, Song MS, Jeong HW, Lai VD, Kim Y, Chin BS, Park JS, Chung KH, Foo SS, Poo H, Mo IP, Lee OJ, Webby RJ, Jung JU, Choi YK. 2020. Infection and rapid transmission of SARS-CoV-2 in ferrets. Cell Host Microbe 27:704–709.e2. 10.1016/j.chom.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richard M, Kok A, de Meulder D, Bestebroer TM, Lamers MM, Okba NMA, Fentener van Vlissingen M, Rockx B, Haagmans BL, Koopmans MPG, Fouchier RAM, Herfst S. 2020. SARS-CoV-2 is transmitted via contact and via the air between ferrets. Nat Commun 11:3496. 10.1038/s41467-020-17367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sia SF, Yan LM, Chin AWH, Fung K, Choy KT, Wong AYL, Kaewpreedee P, Perera RAPM, Poon LLM, Nicholls JM, Peiris M, Yen HL. 2020. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature 583:834–838. 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fagre A, Lewis J, Eckley M, Zhan S, Rocha SM, Sexton NR, Burke B, Geiss B, Peersen O, Kading R, Rovnak J, Ebel GD, Tjalkens RB, Aboellail T, Schountz T. 2020. SARS-CoV-2 infection, neuropathogenesis and transmission among deer mice: implications for reverse zoonosis to New World rodents. BioRxiv 10.1101/2020.08.07.241810. [DOI] [PMC free article] [PubMed]
- 40.Bosco-Lauth AM, Hartwig AE, Porter SM, Gordy PW, Nehring M, Byas AD, VandeWoude S, Ragan IK, Maison RM, Bowen RA. 2020. Experimental infection of domestic dogs and cats with SARS-CoV-2: pathogenesis, transmission, and response to reexposure in cats. Proc Natl Acad Sci U S A 117:26382–26388. 10.1073/pnas.2013102117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Halfmann PJ, Hatta M, Chiba S, Maemura T, Fan S, Takeda M, Kinoshita N, Hattori S, Sakai-Tagawa Y, Iwatsuki-Horimoto K, Imai M, Kawaoka Y. 2020. Transmission of SARS-VoV-2 in domestic cats. N Engl J Med 383:592–594. 10.1056/NEJMc2013400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Honein MA, Christie A, Rose DA, Brooks JT, Meaney-Delman D, Cohn A, Sauber-Schatz EK, Walker A, McDonald LC, Liburd LC, Hall JE, Fry AM, Hall AJ, Gupta N, Kuhnert WL, Yoon PW, Gundlapalli AV, Beach MJ, Walke HT, CDC COVID-19 Response Team . 2020. Summary of guidance for public health strategies to address high levels of community transmission of SARS-CoV-2 and related deaths, December 2020. MMWR Morb Mortal Wkly Rep 69:1860–1867. 10.15585/mmwr.mm6949e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bialek S, Boundy E, Bowen V, Chow N, Cohn A, Dowling N, Ellington S, Gierke R, Hall A, MacNeil J, Patel P, Peacock G, Pilishvili T, Razzaghi H, Reed N, Ritchey M, Sauber-Schatz E. 2020. Severe outcomes among patients with coronavirus disease 2019 (COVID-19) — United States, February 12–March 16, 2020. MMWR Morb Mortal Wkly Rep 69:343–346. 10.15585/mmwr.mm6912e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nachtigall I, Lenga P, Jóźwiak K, Thürmann P, Meier-Hellmann A, Kuhlen R, Brederlau J, Bauer T, Tebbenjohanns J, Schwegmann K, Hauptmann M, Dengler J. 2020. Clinical course and factors associated with outcomes among 1904 patients hospitalized with COVID-19 in Germany: an observational study. Clin Microbiol Infect 26:1663–1669. 10.1016/j.cmi.2020.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu Z, McGoogan JM. 2020. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China. JAMA 323:1239. 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 46.Gaudreault NN, Trujillo JD, Carossino M, Meekins DA, Morozov I, Madden DW, Indran SV, Bold D, Balaraman V, Kwon T, Artiaga BL, Cool K, García-Sastre A, Ma W, Wilson WC, Henningson J, Balasuriya UBR, Richt JA. 2020. SARS-CoV-2 infection, disease and transmission in domestic cats. Emerg Microbes Infect 9:2322–2332. 10.1080/22221751.2020.1833687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oude Munnink BB, Sikkema RS, Nieuwenhuijse DF, Molenaar RJ, Munger E, Molenkamp R, van der Spek A, Tolsma P, Rietveld A, Brouwer M, Bouwmeester-Vincken N, Harders F, Hakze-van der Honing R, Wegdam-Blans MCA, Bouwstra RJ, GeurtsvanKessel C, van der Eijk AA, Velkers FC, Smit LAM, Stegeman A, van der Poel WHM, Koopmans MPG. 2021. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science 371:172–177. 10.1126/science.abe5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Widders A, Broom A, Broom J. 2020. SARS-CoV-2: the viral shedding vs infectivity dilemma. Infect Dis Health 25:210–215. 10.1016/j.idh.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perera RAPM, Tso E, Tsang OTY, Tsang DNC, Fung K, Leung YWY, Chin AWH, Chu DKW, Cheng SMS, Poon LLM, Chuang VWM, Peiris M. 2020. SARS-CoV-2 virus culture and subgenomic RNA for respiratory specimens from patients with mild coronavirus disease. Emerg Infect Dis 26:2701–2704. 10.3201/eid2611.203219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suess C, Hausmann R. 2020. Gross and histopathological pulmonary findings in a COVID-19 associated death during self-isolation. Int J Legal Med 134:1285–1290. 10.1007/s00414-020-02319-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arrossi AV, Farver C. 2020. The pulmonary pathology of COVID-19. Cleve Clin J Med 10.3949/ccjm.87a.ccc063. [DOI] [PubMed] [Google Scholar]
- 52.Yu P, Qi F, Xu Y, Li F, Liu P, Liu J, Bao L, Deng W, Gao H, Xiang Z, Xiao C, Lv Q, Gong S, Liu J, Song Z, Qu Y, Xue J, Wei Q, Liu M, Wang G, Wang S, Yu H, Liu X, Huang B, Wang W, Zhao L, Wang H, Ye F, Zhou W, Zhen W, Han J, Wu G, Jin Q, Wang J, Tan W, Qin C. 2020. Age‐related rhesus macaque models of COVID‐19. Anim Model Exp Med 3:93–97. 10.1002/ame2.12108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rockx B, Feldmann F, Brining D, Gardner D, LaCasse R, Kercher L, Long D, Rosenke R, Virtaneva K, Sturdevant DE, Porcella SF, Mattoon J, Parnell M, Baric RS, Feldmann H. 2011. Comparative pathogenesis of three human and zoonotic SARS-CoV strains in cynomolgus macaques. PLoS One 6:e18558. 10.1371/journal.pone.0018558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun SH, Chen Q, Gu HJ, Yang G, Wang YX, Huang XY, Liu SS, Zhang NN, Li XF, Xiong R, Guo Y, Deng YQ, Huang WJ, Liu Q, Liu QM, Shen YL, Zhou Y, Yang X, Zhao TY, Fan CF, Zhou YS, Qin CF, Wang YC. 2020. A mouse model of SARS-CoV-2 infection and pathogenesis. Cell Host Microbe 28:124–133.e4. 10.1016/j.chom.2020.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bao L, Deng W, Huang B, Gao H, Liu J, Ren L, Wei Q, Yu P, Xu Y, Qi F, Qu Y, Li F, Lv Q, Wang W, Xue J, Gong S, Liu M, Wang G, Wang S, Song Z, Zhao L, Liu P, Zhao L, Ye F, Wang H, Zhou W, Zhu N, Zhen W, Yu H, Zhang X, Guo L, Chen L, Wang C, Wang Y, Wang X, Xiao Y, Sun Q, Liu H, Zhu F, Ma C, Yan L, Yang M, Han J, Xu W, Tan W, Peng X, Jin Q, Wu G, Qin C. 2020. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature 583:830–833. 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- 56.Hammer AS, Quaade ML, Rasmussen TB, Fonager J, Rasmussen M, Mundbjerg K, Lohse L, Strandbygaard B, Jørgensen CS, Alfaro-Núñez A, Rosenstierne MW, Boklund A, Halasa T, Fomsgaard A, Belsham GJ, Bøtner A. 2021. SARS-CoV-2 transmission between mink (Neovison vison) and humans, Denmark. Emerg Infect Dis 27:547–551. 10.3201/eid2702.203794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Palmer MV, Cox RJ, Waters WR, Thacker TC, Whipple DL. 2017. Using white-tailed deer (Odocoileus virginianus) in infectious disease research. J Am Assoc Lab Anim Sci 56:350–360. [PMC free article] [PubMed] [Google Scholar]
- 58.Palmer MV, Wiarda J, Kanipe C, Thacker TC. 2019. Early pulmonary lesions in cattle infected via aerosolized Mycobacterium bovis. Vet Pathol 56:544–554. 10.1177/0300985819833454. [DOI] [PubMed] [Google Scholar]
- 59.Wagner B, Hillegas JM, Babasyan S. 2012. Monoclonal antibodies to equine CD23 identify the low-affinity receptor for IgE on subpopulations of IgM + and IgG1 + B-cells in horses. Vet Immunol Immunopathol 146:125–134. 10.1016/j.vetimm.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 60.Wimer CL, Schnabel CL, Perkins G, Babasyan S, Freer H, Stout AE, Rollins A, Osterrieder N, Goodman LB, Glaser A, Wagner B. 2018. The deletion of the ORF1 and ORF71 genes reduces virulence of the neuropathogenic EHV-1 strain Ab4 without compromising host immunity in horses. PLoS One 13:e0206679. 10.1371/journal.pone.0206679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wagner B, Freer H, Rollins A, Erb HN. 2011. A fluorescent bead-based multiplex assay for the simultaneous detection of antibodies to B. burgdorferi outer surface proteins in canine serum. Vet Immunol Immunopathol 140:190–198. 10.1016/j.vetimm.2010.12.003. [DOI] [PubMed] [Google Scholar]