Antimicrobial resistance is a large concern in the health care field. With more multidrug-resistant bacterial pathogens emerging, other techniques for eliminating bacterial infections are being explored.
KEYWORDS: antibiotic resistance, bacteriophage therapy, flagellar motility
ABSTRACT
Bacteriophages are the most abundant biological entities in the biosphere. Due to their host specificity and ability to kill bacteria rapidly, bacteriophages have many potential health care applications, including therapy against antibiotic-resistant bacteria. Infection by flagellotropic bacteriophages requires a properly rotating bacterial flagellar filament. The flagella-dependent phage χ (chi) infects serovars of the pathogenic enterobacterium Salmonella enterica. However, cell surface receptors and proteins involved in other stages of χ infection have not been discovered to date. We screened a multigene deletion library of S. enterica serovar Typhimurium by spotting mutants on soft agar plates seeded with bacteriophage χ and monitoring their ability to grow and form a swim ring, a characteristic of bacteriophage-resistant motile mutants. Those multigene deletion regions identified to be important for χ infectivity were further investigated by characterizing the phenotypes of corresponding single-gene deletion mutants. In this way, we identified motile mutants with various degrees of resistance to χ. Deletions in individual genes encoding the AcrABZ-TolC multidrug efflux system drastically reduced infection by bacteriophage χ. Furthermore, an acrABtolC triple deletion strain was fully resistant to χ. Infection was severely reduced but not entirely blocked by the deletion of the gene tig, encoding the molecular chaperone trigger factor. Finally, deletion in genes encoding enzymes involved in the synthesis of the antioxidants glutathione (GSH) and uric acid resulted in reduced infectivity. Our findings begin to elucidate poorly understood processes involved in later stages of flagellotropic bacteriophage infection and inform research aimed at the use of bacteriophages to combat antibiotic-resistant bacterial infections.
IMPORTANCE Antimicrobial resistance is a large concern in the health care field. With more multidrug-resistant bacterial pathogens emerging, other techniques for eliminating bacterial infections are being explored. Among these is phage therapy, where combinations of specific phages are used to treat infections. Generally, phages utilize cell appendages and surface receptors for the initial attachment to their host. Phages that are flagellotropic are of particular interest because flagella are often important in bacterial virulence, making resistance to attachment of these phages harder to achieve without reducing virulence. This study discovered the importance of a multidrug efflux pump for the infection of Salmonella enterica by a flagellotropic phage. In theory, if a bacterial pathogen develops phage resistance by altering expression of the efflux pump, then the pathogen would simultaneously become more susceptible to the antibiotic substrates of the pump. Thus, coadministering antibiotics and flagellotropic phage may be a particularly potent antibacterial therapy.
INTRODUCTION
Bacteriophages, also called phages, are the viruses of bacteria and are the most abundant biological entities in the biosphere (1). Phages generally have very specific host requirements and only infect a small range of bacterial strains and species (2–4). Due to their specificity and ability to kill bacteria efficiently, phages have many potential health care applications, including phage therapy (5–8). Despite their differences in morphology and genome structure, most phages initiate infection via a receptor on the bacterial cell surface. Receptors include lipopolysaccharides as well as proteins such as channels, porins, pili, or flagella. After attachment, the phage ejects its DNA or RNA genome into the bacterial cell, where the cell machinery is used to replicate phage particles. When a sufficient quantity of phage is produced, the bacterial cell lyses, and the phages diffuse out to infect more host cells (9, 10).
Flagellotropic phages represent a distinct category of phages that infect via adsorption to the bacterial flagellar filament (11–15). It is hypothesized that they attach to the flagellar filament and use its rotation to work their way down to the cell surface like a nut on a bolt with threads in the same orientation (16), although the process of translocation along the flagellum has not been visualized to date. Bacteriophage χ is a virulent flagellotropic phage that infects some serotypes of enteric, Gram-negative bacteria, most notably from the genera Escherichia, Salmonella, and Serratia (11, 17, 18). The bacteriophage is a double-stranded DNA virus from the Siphoviridae family with a genome length of 59 kb, containing an estimate of 75 genes (17). The icosahedral head of χ is 65 nm in diameter. Its noncontractile tail has a length of 220 nm and a width of 13 nm, and contains a 200-nm single tail fiber (17). The dimensions of the tail fiber suggest that it fits along the right-handed grooves of the helical, left-handed flagellar filament, and that the counterclockwise (CCW) rotation of the flagellum forces the phage to follow the grooves to the base of the flagellar filament (16).
Many bacterial motility and chemotaxis genes have been shown to be required for χ to infect cells (19). In particular, absence of the flagellin proteins FliC and FljB causes a χ-resistant phenotype. However, the lack of flagellar filaments is partially offset by the production of polyhooks (20). The deletion of the motility genes motA or motB also confers resistance to χ, underlining the importance of flagellar rotation (16, 19, 21). Additionally, any mutations in chemotaxis genes that lock the flagellar motor in the clockwise (CW) rotating state result in resistance to χ, because the phage is not drawn to the base of the filament (11, 16, 19). Once the phage reaches the cell surface, it likely interacts with a secondary receptor (11, 14, 16), but infection processes following phage translocation on a host flagellum are unknown. To date, no host gene products unrelated to motility or chemotaxis have been shown to be involved or required for χ infection.
One biomedically important bacterial species infected by χ is Salmonella enterica of the Enterobacteriaceae family of Gammaproteobacteria (22). S. enterica is a diverse species, containing over 2,500 serovars, many of which are clinically relevant (23). Most pathogenic serovars, including serovar Typhimurium, cause human gastroenteritis and are often collectively referred to as nontyphoidal Salmonella (NTS) (22, 24). An NTS infection typically presents itself with severe diarrhea, fever, abdominal pain, and vomiting (22). Some serovars can be invasive, with infections potentially leading to bacteremia, meningitis, and even death in otherwise healthy patients (25). Serovars Typhi and Paratyphi cause typhoid and paratyphoid fever, respectively, which are potentially fatal systemic infections that result in high fever, rash, lethargy, abdominal pain, and vomiting (22). These conditions are a major cause of death in developing countries. At least 100,000 infections per year in the United States are due to antibiotic-resistant Salmonella, including those that are resistant to clinically important drugs such as ceftriaxone and ciprofloxacin (26). S. enterica subsp. enterica serovar Typhimurium 14028s (STM 14028s) is a pathogenic NTS that is susceptible to χ and is often used in laboratory settings.
To decipher the infection process by χ beyond its adhesion to flagella and the importance of flagellar rotation, we screened a library of defined deletion mutants in STM 14028s to identify genes not related to motility that play a role in χ infection.
RESULTS
Strategy for the identification of motile S. enterica mutants with reduced susceptibility to χ.
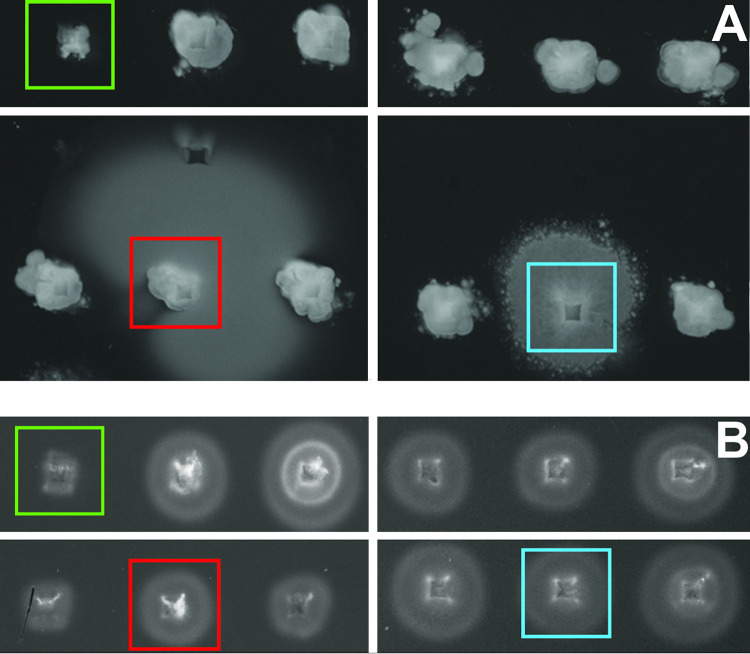
We hypothesized that χ infection required binding to secondary receptors on the cell surface following interactions with the flagella. We therefore screened a multigene deletion (MGD) mutant library of STM 14028s with 449 MGD mutants covering a total of 3,476 genes (27) for motile phage-resistant mutants by spotting mutants onto MSB swim agar containing 1 × 107 PFU of χ. We identified a total of 12 motile χ-resistant MGD library mutants, which informed a selection for further screening of single-gene deletion (SGD) mutants of the individual genes deleted in their respective MGD locus. This resulted in a second screen of 76 SGD mutants, carried out as described above. Phenotypes of χ-resistant SGD mutants were confirmed with a second independent mutant of the same gene disrupted by a different antibiotic cassette. Candidate mutants that retained motility were identified based on the size of their swim ring formed after 8 h of incubation. Phage-susceptible mutants formed very small, approximately 1- to 5-mm swim rings with inconsistent density throughout the ring. This is characteristic of the fact that χ does not lyse completely on solid medium, possibly due to downregulation of flagellar components under extreme selective pressure, resulting in the simultaneous survival and miniscule amount of swimming seen in these spots. Phage-resistant motile mutants formed larger, hazy swim rings of various sizes with more consistent density and shape, as seen in Fig. 1A. Motile mutants produced large swim rings of variable sizes on plates lacking phage (Fig. 1B) due to minor inconsistencies associated with the high-throughput 96-well pin replicator method of inoculation. Nonmotile mutants did not form rings regardless of the presence of phage (Fig. 1A and B).
FIG 1.
Multigene deletion (MGD) mutant library screening. (A) Screen of the STM 14028s MGD library on a rectangular plate containing MSB swim agar with approximately 1 × 107 PFU/ml χ phage after 8 h of incubation at 37°C. The colony highlighted in red is a ΔSTM14_0524-0529 mutant, which is motile and highly χ-resistant. The colony highlighted in blue is a ΔSTM14_0904-0923 mutant, which is motile and χ-resistant to a lesser degree than the red ΔSTM14_0524-0529, with an intermediate phenotype and irregular swim ring shape. The colony highlighted in green is a ΔSTM14_2324-2359 mutant, which includes a deletion in the essential flagellar motor gene motA and is nonmotile and χ-resistant. The remaining colonies consist of motile χ-susceptible mutants, which only exhibit small amounts of growth and swimming around the initial inoculation point compared to the nonmotile mutant. (B) Screen of the same STM 14028s MGD mutants on a rectangular plate containing MSB swim agar without added phage after a 3-h incubation at 37°C. Mutant colonies are highlighted as in Fig. 1A. The inconsistent swim ring size is characteristic of the inherent variability when inoculating swim agar using a 96-well pin replicator rather than applying a more controlled inoculation volume via pipet.
Next, we validated χ-resistant strains by analyzing newly constructed deletion mutants. We determined that disruption of eight genes negatively impacted susceptibility to χ while maintaining motility. These genes fell into three classes: (i) components of the multisubstrate efflux pump Acr/TolC; (ii) the molecular chaperone trigger factor; and (iii) enzymes involved in the synthesis of the cellular antioxidants glutathione (GSH) and uric acid (Table 1).
TABLE 1.
Candidate genes involved in infection of Salmonella by χ
| Gene | Gene no. | Function of gene product |
|---|---|---|
| acrB | STM14_0559 | AcrABZ-TolC multisubstrate efflux system, inner membrane component |
| acrA | STM14_0560 | AcrABZ-TolC, periplasmic adaptor protein |
| tolC | STM14_3859 | AcrABZ-TolC, outer membrane channel |
| acrZa | STM14_0906 | AcrABZ-TolC, AcrB-associated protein |
| acrF | STM14_4091 | AcrEF-TolC multisubstrate efflux system, inner membrane component |
| tig | STM14_0529 | Trigger factor molecular chaperone |
| gshA | STM14_3403 | Glutamate-cysteine ligase |
| gshB | STM14_3739 | Glutathione synthetase |
| ylbA | STM14_0616 | Putative (S)-ureidoglycine aminohydrolase |
The AcrZ homolog is encoded by the gene STM14_0906 in STM 14028s.
AcrABZ-TolC multidrug efflux system.
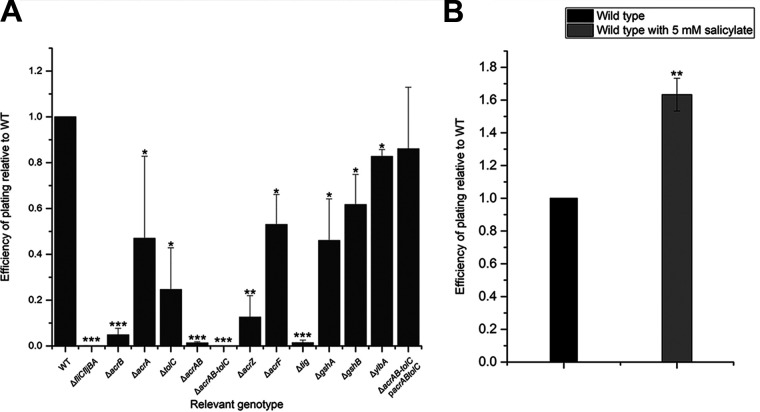
We found that deletions in genes coding for components of the AcrABZ-TolC multisubstrate efflux system confer high levels of resistance to bacteriophage χ. AcrABZ-TolC is part of the resistance-nodulation-division (RND) family of transporters that is well characterized in E. coli. RND efflux systems are tripartite pumps that extend fully across the cell envelope (28–30). They consist of an inner membrane pump, an outer membrane channel, and a periplasmic adaptor protein, and are powered by the proton motive force (PMF) (28, 29). In AcrABZ-TolC, TolC is the outer membrane channel, AcrB is the inner membrane pump, and AcrA is the adaptor protein, which connects AcrB and TolC in the periplasm (28, 29, 31). AcrZ is a small protein that assists AcrB in exporting certain substrates, including chloramphenicol and tetracycline (32). An AcrZ homolog is encoded by the gene STM14_0906 in STM 14028s. AcrABZ-TolC is involved in exporting numerous substrates, including antibiotics like penicillin G and nafcillin, detergents like sodium dodecyl sulfate, bile salts like deoxycholate, and dyes like crystal violet (28). Deletion of components of AcrABZ-TolC in E. coli and S. enterica increases sensitivity to these toxins (28, 33). To further characterize and quantify mutant phenotypes for their defects in phage infectivity, we performed efficiency-of-plating (EOP) assays (Fig. 2A). Deletion of acrB caused a reduction in EOP by 95%, whereas the effects of acrA, acrZ, or tolC deletions were more moderate (reductions between 35 and 87%). However, an acrAB double deletion strain exhibited a 99% reduction in EOP, and the additional deletion of tolC completely abolished infection, resulting in a phage-resistant phenotype.
FIG 2.
Efficiency of plating assays. (A) Efficiency of plating (EOP) of Salmonella enterica sv Typhimurium 14028s (STM 14028s) mutants compared to wild type. EOP results for all mutants were confirmed with a second deletion mutant constructed with a different antibiotic resistance cassette, with the exception of the ΔgshB, ΔacrAB, and ΔacrABtolC mutants. (B) EOP of wild-type STM 14028s grown in medium with and without 5 mM sodium salicylate. EOP is calculated using the formula EOP= (PFU per ml mutant)/(PFU per ml wild type). All data points are composed of three replicates. Error bars represent standard deviation and statistical significance was determined using Student’s t test.
To verify the involvement of AcrABZ-TolC in χ phage infection, we performed trans-complementation of the genes acrAacrB and tolC in the dual gene expression vector pETDuet-1. The genes acrA and acrB were cloned in MCS1 using the native chromosomal arrangement and tolC was cloned in MCS2, yielding plasmid pBS1250. When the ΔacrABtolC mutant was complemented with pBS1250 under induction with 1 mM IPTG (isopropyl-β-D-thiogalactopyranoside), the phage-resistant phenotype was restored to an EOP of 85% of wild type (Fig. 2A).
To provide further proof of the importance of AcrABZ-TolC in phage infection, we analyzed the effect of modulating its transcriptional regulation. AcrABZ-TolC is part of the complex multi-antibiotic-resistance (mar) regulon, which is conserved across many enteric bacteria. Overexpression of mar-regulated genes is associated with multidrug resistance in enteric pathogens (34–36). Expression of the mar regulon is directly controlled by the transcriptional activator MarA. The transcription of marA is negatively regulated by MarR through autorepression of the marRAB operon. MarR itself can be inactivated through binding of certain phenolic ligands, such as salicylate. Thus, marA transcription is derepressed in the presence of salicylate, which in turn causes increased AcrABZ-TolC expression (37, 38). We hypothesized that overexpression of AcrABZ-TolC would increase the susceptibility of S. enterica to χ. Indeed, when wild-type STM 14028s was grown in the presence of salicylate, cells exhibited an increased susceptibility to χ phage, showing an EOP of 156% compared to STM 14028s grown in medium lacking salicylate (Fig. 2B).
We also discovered that a deletion in acrF reduced S. enterica susceptibility to χ by 47%. AcrF is a component of the AcrEF-TolC efflux system, a functionally similar multisubstrate efflux pump. However, the STM 14028s SGD mutant in acrE did not show a significant χ-resistant phenotype (data not shown). The AcrEF-TolC system has not been studied in as much detail as AcrABZ-TolC, but it has been shown to export certain antibiotics, as well as indole compounds, from the E. coli cell (39, 40). Taking these findings together, we identified the AcrABZ-TolC complex as essential for χ infection and provide preliminary evidence for an involvement of other efflux systems.
Molecular chaperone trigger factor.
The deletion of tig, encoding the molecular chaperone trigger factor, confers 98% resistance to χ (Fig. 2). Trigger factor is a ribosome-associated chaperone that has peptidyl-prolyl cis/trans isomerase activity (41). It interacts with polypeptides exiting the ribosome and keeps them in an open conformation so they can be folded properly. It is also abundant as a free protein in the cytosol, where it promotes protein refolding (41, 42). The highly compromised infection phenotype of the tig mutant implies a major role of this chaperone in χ phage infection. Since the efflux system AcrABZ-TolC also had a chief impact on χ infection, we tested the possibility that the roles of trigger factor and AcrABZ-TolC are functionally linked. However, lack of trigger factor did not result in a reduction of the MIC of deoxycholate, a known substrate of AcrABZ-TolC (Table 2). Thus, trigger factor and AcrABZ-TolC play unrelated roles during χ infection.
TABLE 2.
MIC of sodium deoxycholate on selected deletion mutants
| Relevant genotype | MIC (% w/v deoxycholate)a |
|---|---|
| Wild type | 8 |
| ΔacrB | 2 |
| ΔacrABtolC | 2 |
| Δtig | 8 |
The assay was conducted in liquid medium with an increasing deoxycholate concentration in intervals of 1%. The assay was done in triplicate and resulted in a standard deviation of zero.
Enzymes involved in the synthesis of cellular antioxidants.
Deletion of gshA reduced χ infection of STM 14028s by 54% (Fig. 2), whereas a gshB deletion mutant showed an EOP of 61% compared to wild type. GshA and GshB are major components of the glutathione synthesis pathway and the lack of either one results in the absence of intracellular glutathione (43–46). Glutathione is an important antioxidant and responsible for maintaining a reducing environment in the cell to prevent damage from reactive oxygen species (ROS) such as superoxide and free radicals (47, 48).
Lack of the hypothetical protein-encoding gene ylbA confers a weak χ-resistant phenotype with an 84% EOP compared to wild type (Fig. 2). YlbA has no proven function in S. enterica and E. coli, but homologues have been shown to be involved in the ureide pathway that produces another reducing agent, uric acid (49), allowing for growth in low-nitrogen conditions (50, 51). In conclusion, the reduction of cellular levels of two antioxidants resulted in partial resistance to χ.
Evaluation of cell motility and growth in χ-resistant mutants.
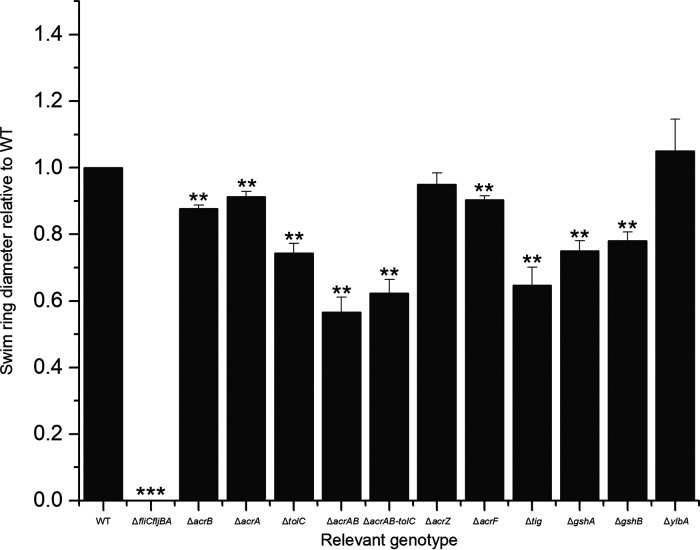
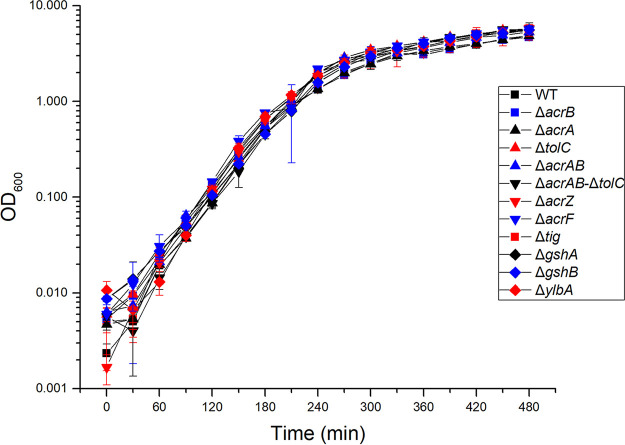
To verify that the observed resistance to χ was not due to reduced motility, we performed quantitative swim plate assays with the SGD mutants. Swim ring diameters after incubation for 8 h at 37°C were compared to those of the parental strain STM 14028s. All mutants were motile, as they formed swim rings at a size of at least 60% of wild type (Fig. 3). Motility was also confirmed by observation of cell cultures under a phase-contrast microscope, with no evident alteration in swimming behavior (data not shown). It should be noted that all investigated mutants, with the exception of ΔylbA and ΔacrZ, exhibited a statistically significant reduction in swim ring diameter. However, since there was no correlation between the degree of resistance to χ and the degree of the motility decrease, we concluded that a modest reduction in motility is not sufficient for a resistant phenotype. To confirm that the χ-resistant phenotype and the reduced motility of any of the investigated mutants was not caused by a growth defect, we assayed growth rates at 37°C using optical density at 600 nm (OD600) spectrophotometer readings over the course of 8 h. No statistically significant differences in growth rates between the deletion mutants and wild type were observed (Fig. 4).
FIG 3.
Swim ring diameter of STM 14028s mutants compared to wild type formed on 0.3% MSB agar after 5 h. All data points are composed of three replicates. Error bars represent standard deviation and statistical significance was determined using Student’s t test.
FIG 4.
Growth curves of select STM 14028s deletion mutants. All data points are the average of three replicates. Error bars represent standard deviation and statistical significance was determined using Student’s t test.
DISCUSSION
In this study, we employed swim plate assays of S. enterica serovar Typhimurium deletion mutants to identify genes that are involved in infection by phage χ, beyond those known to be required for flagellar motility. We found motile, phage-resistant mutants that were defective in genes coding for components of the AcrABZ-TolC and AcrEF-TolC efflux systems, the molecular chaperone trigger factor, and enzymes involved in the production of cellular antioxidants such as the glutathione synthesis proteins GshA and GshB, and a hypothetical protein YlbA.
The putative role of a multisubstrate efflux system in χ infection.
Multidrug resistance in E. coli and S. enterica is often associated with increased expression of AcrABZ-TolC (28, 33). The outer membrane channel TolC has been shown to be a phage receptor in other systems (52, 53), but this is the first report of a flagellotropic phage requiring the AcrABZ-TolC efflux system for infection. The exact role of AcrABZ-TolC in χ infection is not known. We hypothesize that the channel formed by AcrB and TolC is involved in the process of phage DNA entry into the host cell. Without production of AcrB, Salmonella cells were highly resistant to χ infection. Interestingly, absence of TolC had a lesser effect, showing approximately 75% resistance to χ (Fig. 2). It is possible that an outer membrane protein other than TolC can serve as a DNA channel into the periplasm, where AcrB then allows DNA entry into the cytoplasm. Since lack of the flagellar filament and hook or the absence of AcrABZ-TolC results in complete resistance to χ, we conclude that both of these structures are essential for the infection process. Bacteriophage χ uses the rotating flagellar filament to work its way down to the cell surface (11, 16), and we hypothesize that it then interacts with the AcrABZ-TolC complex to eject its DNA into the host cell. With the exception of the ΔacrZ mutant, which was fully motile, deletions of genes encoding components of the AcrABZ-TolC complex had various mild negative impacts on motility on swim plates. Reduced motility is perhaps due to a decrease in overall fitness caused by the inability of the cells to export toxic substrates (28, 29). It has been shown in other flagellotropic phage systems that even severe reductions in motility and flagellar rotational speed did not result in reductions in phage infection of the magnitude seen in the ΔacrABtolC mutant in this study (12). Therefore, we infer that these two behaviors are likely unrelated.
The role of the small protein AcrZ in χ infection is unknown. AcrZ directly associates with AcrB, and its absence results in a reduction in the ability of AcrB to export certain toxic substrates (29, 32). It has been suggested that AcrZ triggers conformational changes in the periplasmic domain of AcrB during the export of certain substrates (30, 32). This could explain why acrZ mutants exhibit an intermediate MIC phenotype (32). We hypothesize that the lack of AcrZ prevents conformational changes in AcrB that are necessary for the AcrABZ-TolC complex to efficiently serve as a phage DNA channel.
The putative role of a general chaperone in χ infection.
Trigger factor is an important ribosome-associated chaperone (41, 42, 54). Without production of trigger factor, there was a 98% reduction in plating efficiency of host bacteria when infected with χ (Fig. 2). Trigger factor has been shown to interact with T4 and T5 phage proteins (55, 56) and is implicated as a chaperone for the gp20 protein of T4 (56). We hypothesize that trigger factor is involved in the proper folding of crucial χ phage proteins. The absence of trigger factor would result in an aborted production of phage particles and, thus, a resistant phenotype. Alternatively, trigger factor could be responsible for the folding of one or more bacterial proteins required during the phage infection cycle, such as cell surface receptors. The absence of trigger factor also caused a reduction in swim ring diameter, suggesting that trigger factor possibly plays a role in the folding of certain motility and/or chemotaxis proteins. Finally, it is conceivable that the resistance of the tig mutant to χ is influenced by multiple other factors, because trigger factor is a chaperone for a wide array of proteins (54). However, it does not serve as a chaperone for proteins of the AcrABZ-TolC system, because the deoxycholate MIC of the tig mutant was equal to that observed in wild type, whereas acrB and acrABtolC deletion mutants exhibited drastic MIC reductions (Table 2). Therefore, the effect of trigger factor appears to be independent from that of the AcrABZ-TolC complex.
Involvement of host antioxidants in χ infection.
Glutathione (GSH) or l-γ-glutamyl-l-cysteinylglycine acts as a crucial antioxidant in prokaryotic as well as eukaryotic cells (46–48). This tripeptide maintains a reducing environment in the cell, neutralizing reactive oxygen species (ROS) such as superoxide, peroxides, and free radicals that would otherwise have the potential to damage cellular components (43, 46–48). GshA is a glutamate-cysteine ligase that conducts one branch of the GSH synthesis pathway, while the glutathione synthetase GshB conducts the final step of GSH production. Absence of either of these proteins results in abolished GSH production (44–46). The lack of GSH in a host cell caused by the deletion of gshA or gshB had a negative effect on the χ infection process (Fig. 2). A reducing environment may be just as important for the protection of phage proteins as it is for its bacterial host proteins. The lack of GSH seems to be more deleterious to the χ infection process than it is to the overall growth of its host under nonstress growth conditions. A reducing cellular environment has been shown to be beneficial in other phage systems, with high levels of oxidation being potentially inhibitory to infection (57). We hypothesize that this is also the case for χ. However, the reduced infectivity of the gshA and gshB mutants was more moderate than that of the two previously discussed classes, the efflux pump and the tig chaperone. Thus, reduction of ROS by GSH improves the ability of χ to infect and lyse its host, but it is not essential for phage propagation.
The role of YlbA in S. enterica is unknown, but a close homolog functions as (S)-ureidoglycine aminohydrolase in the ureide pathway of leguminous plants and prokaryotes, where it catabolizes purines to be used as an energy source in environments with low available nitrogen (50, 51). An (S)-ureidoglycine aminohydrolase enzyme is also present in Bacillus subtilis (58) and Klebsiella pneumoniae (59), and YlbA in S. enterica and E. coli likely performs the same function. The impact of a ylbA deletion on χ infection was relatively small (>80% EOP, Fig. 2) but sufficient to be detected in our screens. A link between the ureide pathway and phage infection has not been previously reported. This pathway ultimately produces allantoin from uric acid (50). Uric acid, similar to GSH, is an antioxidant (49). Absence of YlbA may therefore result in altered levels of uric acid in the cell, although this function has not been confirmed.
Potential applications and future studies.
The phage-seeded swim plate assay is a powerful method to screen mutant libraries and can be expanded to the studies of other flagellotropic phage systems. By screening only for mutants that resist the phage and remain motile, the high prevalence of motility-related gene disruptions can be ignored. Since swim ring formation also relies on chemotaxis in addition to motility (60), chemotaxis-related gene disruptions are also disregarded.
Bacteriophage χ has a broad host range that includes several clinically relevant human-pathogenic species of enterobacteria (11, 16, 18), including at least three genera in two different families, Enterobacteriaceae and Yersiniaceae. The necessity of an antibiotic efflux system in its host infection process makes χ an attractive potential future candidate for phage therapy. Wang-Kan et al. showed that Salmonella strains with an acrB deletion are unable to colonize the mouse intestine, thus significantly reducing virulence, because AcrABZ-TolC exports toxic bile salts such as cholate and deoxycholate (28, 33). The inclusion of χ in a phage cocktail against NTS would likely create selective pressure for pathogenic bacteria to reduce expression of a component of the AcrABZ-TolC system. The subset of resistance mutants that reduce expression would be more sensitive to bile salts, therefore lowering their virulence in the human intestine, as well as lowering resistance to certain antibiotics. A similar trade-off has been successfully exploited by Chan et al. in the Pseudomonas aeruginosa phage OMKO1, which also uses a multidrug efflux system, MexAB, as a cell-surface receptor (61). In this study, phage OMKO1 was coadministered with an antibiotic, forcing a trade-off by the cells to either express the mexAB operon, resulting in antibiotic resistance and phage susceptibility, or halt expression of the mexAB operon, resulting in phage resistance and antibiotic susceptibility (61). We hypothesize that the same phenomenon would occur in vivo with Salmonella in the mammalian intestine. Without the AcrABZ-TolC complex, Salmonella is resistant to χ, but highly susceptible to deoxycholate (62) (Table 2). However, its flagellotropic nature makes χ an even better additive to a phage cocktail against NTS. Nonflagellated Salmonella strains would be resistant to χ but have sacrificed flagellin as a virulence factor (22, 24, 63, 64). The virulence of Salmonella would be significantly reduced when downregulating flagellin or AcrABZ-TolC expression to develop resistance against χ.
The results obtained in this work allow for a myriad of opportunities for future studies. The role of the AcrABZ-TolC system in the infection process can be investigated by disrupting the function of this pump. The aspartate residue in position 408 in AcrB is crucial for proton translocation (33, 65, 66). A mutant strain with an Asp408Ala substitution has an antibiotic MIC profile equivalent to that of an acrB knockout strain (33, 66). An infectivity assay of an Asp408Ala mutant would therefore determine whether sensitivity to χ is affected by a functional AcrB pump.
While it is known that the single tail fiber of χ wraps around the flagellar filament of its host cell (11), phage protein(s) that interact with receptors on the cell surface have not been determined. With the identification of AcrABZ-TolC as putative cell surface receptor for χ, it is now possible to identify phage proteins that interact with this receptor.
One of the main features of phage therapy compared to conventional antibiotic administration is the specific customization of the treatment to the disease-causing species. Bacteriophage χ targets several prevalent pathogenic species, which makes it a good candidate for an antibacterial phage cocktail. However, certain serovars of Salmonella enterica subsp. enterica do not serve as a host for χ (67). Thus, our findings also open up opportunities to analyze whether variants in serovar antigens, flagella, and/or the AcrABZ-TolC system are determinants of host specificity of this bacteriophage.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Derivatives of S. enterica serovar Typhimurium 14028s (STM 14028s) used in this study are listed in Table 3.
TABLE 3.
Strains and plasmids used in this study
| Strain or plasmid name | Parent strain/plasmid | Relevant characteristics | Source |
|---|---|---|---|
| MZ1597 | STM 14028s | Wild type | ATCC |
| MZ1597 multigene deletion library | STM 14028s | Multigene deletion library, Kanr, Capr | (27) |
| TH2788 | Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 | FliY::Tn10dTc | Gift from Kelly Hughes |
| 14028s ΔacrA::tet | STM 14028s | ΔacrA, Tetr | This study |
| 14028s ΔacrA::kan | STM 14028s | ΔacrA, Kanr | (27) |
| 14028s ΔacrAB::tet | STM 14028s | ΔacrAB, Tetr | This study |
| 14028s ΔacrAB::tet ΔtolC::kan | STM 14028s | ΔacrAB ΔtolC, Tetr, Kanr | This study |
| 14028s ΔacrB::tet | STM 14028s | ΔacrB, Tetr | This study |
| 14028s ΔacrB::kan | STM 14028s | ΔacrB, Kanr | (27) |
| 14028s ΔacrF::kan | STM 14028s | ΔacrF, Kanr | (27) |
| 14028s ΔacrF::cap | STM 14028s | ΔacrF, Capr | (27) |
| 14028s ΔfliC::kan ΔfljBA::tet | STM 14028s | ΔfliC ΔfljBA, Kanr, Tetr | This study |
| 14028s ΔgshA::kan | STM 14028s | ΔgshA, Kanr | (27) |
| 14028s ΔgshA::tet | STM 14028s | ΔgshA, Tetr | This study |
| 14028s ΔgshB::tet | STM 14028s | ΔgshB, Tetr | This study |
| 14028s ΔSTM14_0524-0529::kan | STM 14028s | ΔSTM14_0524-0529, Kanr | (27) |
| 14028s ΔSTM14_0904-0923::kan | STM 14028s | ΔSTM14_0904-0923, Kanr | (27) |
| 14028s ΔSTM14_0906::kan | STM 14028s | ΔSTM14_0906, Kanr | (27) |
| 14028s ΔSTM14_0906::cap | STM 14028s | ΔSTM14_0906, Capr | (27) |
| 14028s ΔSTM14_2324-2359::kan | STM 14028s | ΔSTM14_2324-2359, Kanr | (27) |
| 14028s Δtig::tet | STM 14028s | Δtig, Tetr | This study |
| 14028s Δtig::cap | STM 14028s | Δtig, Capr | (27) |
| 14028s ΔtolC::kan | STM 14028s | ΔtolC, Kanr | (27) |
| 14028s ΔtolC::tet | STM 14028s | ΔtolC, Tetr | This study |
| 14028s ΔylbA::kan | STM 14028s | ΔylbA, Kanr | (27) |
| 14028s ΔylbA::tet | STM 14028s | ΔylbA, Kanr | This study |
| Bacteriophage χ | Wild type | Gift from Saeed Tavazoie | |
| pBS1250 | pETDuet-1 | plac STM 14028s acrAB (MCS1), tolC (MCS2), Ampr | This study |
| pETDuet-1 | plac MCS1, MCS2, Ampr | Novagen | |
| pKD46 | pINT-ts | pBAD gam bet exo, Ampr | Gift from Howard C. Berg |
Phage propagation and isolation.
To propagate and isolate χ phage particles, an overlay-based method was used. STM 14028s was grown in lysogeny broth (LB) (68) at 37°C to stationary phase. This stationary-phase culture was diluted 1:100 in LB and allowed to reach an optical density at 600 nm (OD600) of 1.0. Exactly 100 μl of culture was mixed with 100 μl of the appropriate serial dilution of χ phage to produce plates with confluent lysis. This mixture was combined with 4 ml of molten LB with 0.35% wt/vol agar and was poured onto an LB plate for a total of 15 plates. Plaques were allowed to form by incubating for approximately 8 h at 37°C, at which point 5 ml of TM buffer (50 mM Tris/HCl [pH 7.5], 10 mM MgSO4) was poured onto each plate. The plates were placed on a rocking platform overnight at 4°C, rotating at approximately 30 RPM. The TM buffer and soft agar from each plate was pooled in a centrifuge tube and chloroform was added to a final concentration of 2% vol/vol. The mixture was centrifuged at 10,000 × g for 30 min at 4°C. The supernatant was collected into a new tube, mixed with polyethylene glycol 8000 to a final concentration of 10% wt/vol and stirred overnight at 4°C. The mixture was centrifuged at 15,000 × g for 30 min at 4°C, and the pellet containing phage was suspended in TM buffer. This mixture was purified using an OptiPrep (Sigma) density gradient. Briefly, the gradient was made with 5 ml each of 50% and 10% OptiPrep in flexible polyallomer ultracentrifuge tubes (Beckman Coulter). The concentrated phage in TM buffer was layered on top of the gradient and centrifuged at 200,000 × g for 2 h at 15°C using an SW41 rotor (Beckman-Coulter). The white band containing phage was extracted with a syringe and 25-gauge needle and added to dialysis tubing in TM buffer. This was allowed to stir for 48 h at 4°C, changing the buffer approximately every 12 h. The TM buffer containing phage was extracted from the dialysis tubing, and the titer of the phage suspension was characterized for plaque forming units (PFU) by a plaque assay.
Library screening.
The STM 14028s multigene deletion (MGD) library (27) was screened via replica plating onto MSB swim plate medium. MSB medium is a modified lysogeny broth (LB) consisting of 1% Bacto tryptone, 0.5% yeast extract, 2 mM CaCl2, and 2 mM MgSO4. To make swim plates, agar was added to a final concentration of 0.3%, which allowed motile bacteria to form swim rings. After cooling to 50°C, bacteriophage χ in TM buffer was added to a final concentration of 1 × 107 PFU/ml. Approximately 40 ml of this molten agar was added to Thermo Scientific Nunc 128 × 86 mm 1-well rectangular plates and allowed to solidify. Libraries in 96-well plates were patched into the swim agar in the 1-well plates using a 96-well pin replicator tool (Boekel Scientific). After approximately 8 h of incubation at 37°C, plates were examined to determine swim ring formation. Colonies that formed swim rings in the presence of phage were investigated further with targeted gene deletions, which were generated by lambda red mutagenesis.
Generation of loss-of-function mutants.
We utilized established STM 14028s single-gene deletion (SGD) mutants of regions identified to be of interest in the MGD library screen (27). For acrAB and acrABtolC mutants, and select SGD mutants identified in Table 3, additional targeted mutagenesis was conducted using lambda red mutagenesis, modified from Datsenko and Wanner, 2000 (69). Briefly, plasmid pKD46 (pBAD λ-red) was introduced into wild type (or, in the case of acrABtolC, a tolC SGD) STM 14028s via electroporation, following a protocol modified from Binotto et al., 1991 (70). A 50-μl aliquot of electrocompetent STM 14028s cells was mixed with approximately 200 ng of plasmid pKD46, transferred to an electroporation cuvette with a 2-mm electrode gap (Bio Rad), and electroporated at 2.5 kV, 25μF, 2Ω, yielding a 5-ms exponentially decaying pulse. After recovery in Super Optimal broth with catabolite repression (SOC; 2% Bacto tryptone, 0.5% yeast extract, 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 10 mM MgSO4, and 20 mM glucose) at 30°C for 1 h, cells were plated onto LB with 100 μg/ml ampicillin and incubated overnight at 30°C. Cells containing plasmid pKD46 were subsequently made electrocompetent, adding 0.2% wt/vol l-arabinose to activate expression of the lambda red genes on pKD46.
To inactivate individual genes, the 2-kb tetracycline resistance cassette tetRA was inserted to replace the targeted region of the bacterial chromosome. Primers were designed with homology to the desired region and to the tetRA cassette. A PCR was performed with these custom primers and chromosomal DNA from S. enterica serovar Typhimurium TH2788 as the template, which contains a Tn10 insertion in fliY (71). This reaction yielded a fragment containing the tetRA cassette with ends homologous to the desired region for deletion in S. enterica 14028s. The PCR product was purified using a Promega gel and PCR cleanup kit, mixed with STM 14028s competent cells, and electroporated, as above. After the recovery period, cells were plated onto LB with 10 μg/ml tetracycline and incubated overnight at 37°C.
Construction of acrABtolC expression vector and complementation of the ΔacrABtolC mutant.
STM 14028s acrAB and tolC was ligated into pETDuet-1 MCS1 and MCS2 following standard restriction enzyme cloning procedures. Briefly, custom primers were designed with homology to the 5′ end of acrA and the 3′ end of acrB, flanked by restriction enzyme recognition sequences. These were used to amplify the entire acrAB locus via PCR. A restriction digest of this fragment and the pETDuet-1 vector was performed. This acrAB fragment was ligated into MCS1 of pETDuet-1 using T4 DNA ligase (New England BioLabs). The procedures were repeated for STM 14028s tolC, which was ligated into MCS2 of the plasmid constructed in the previous step. This construct was confirmed via Sanger sequencing and introduced into STM 14028s ΔacrABtolC via electroporation. Expression of acrABtolC was induced by the addition of 1 mM IPTG to solid and liquid media.
Quantitative motility assays.
Bacterial cultures were grown in LB at 37°C to an OD600 of 1.0. Next, 2.5 μl of culture was spotted in the center of a plate containing MSB swim plate medium, described above. These plates were incubated facing upward at 37°C. After approximately 5 h, the swim ring diameters were measured. Swim ring diameters relative to wild type were calculated by using the formula (diameter of swim ring formed by tested strain)/(diameter of swim ring formed by wild-type strain).
Assay of efficiency of plating.
Efficiency of plating (EOP) was determined via plaque assay. Briefly, LB medium was inoculated with a single bacterial colony and grown at 37°C. At an OD600 of 1.0, motility was verified via phase-contrast microscopy, and 100 μl of this culture was mixed with serial dilutions of bacteriophage χ stock (1.1 × 1011 PFU/ml) in 0.85% wt/vol NaCl. After an incubation time of 6 min to allow binding, this mixture was added to 4 ml of molten LB agar at 45°C and poured onto an LB agar plate. After 8 h of incubation, plaques were counted to determine PFU/ml. EOP was calculated using the formula (PFU/ml deletion mutant)/(PFU/ml wild type). To test the effect of salicylate, 5 mM sodium salicylate was added to all solid and liquid media before performing the EOP assay.
Determination of bacterial growth rate.
Bacterial cultures were grown in LB broth overnight at 37°C with shaking, then diluted 1:1,000 in 50 ml MSB and incubated under the same conditions. A 1-ml aliquot of culture was collected at 30 min intervals for 8 h and the OD600 was determined via spectrophotometer.
Assay of MIC.
MIC was determined in LB liquid medium. Sodium deoxycholate, a substrate of AcrABZ-TolC, was added to the medium to final concentrations of 1% to 10% (wt/vol). An overnight liquid culture was added at a dilution of 1:1,000 to LB supplemented with sodium deoxycholate and incubated in a 37°C roller drum for 8 h. MIC was recorded as the lowest concentration of sodium deoxycholate that resulted in no growth.
ACKNOWLEDGMENTS
We thank Saeed Tavazoie for bacteriophage χ, Kelly Hughes for S. enterica strain TH2788, and Howard Berg for plasmid pKD46. We thank Weiping Chu for technical assistance.
M.M. and S.P. were supported in part by grants R03 AI139557, USDA 2015-67017-23360, 2017-67015-26085, an NIFA Hatch grant (CA-d-PLS-2327-H), and an NIFA-BARD award (2017-67017-26180).
REFERENCES
- 1.Clokie MR, Millard AD, Letarov AV, Heaphy S. 2011. Phages in nature. Bacteriophage 1:31–45. 10.4161/bact.1.1.14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koskella B, Meaden S. 2013. Understanding bacteriophage specificity in natural microbial communities. Viruses 5:806–823. 10.3390/v5030806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Jonge PA, Nobrega FL, Brouns SJJ, Dutilh BE. 2019. Molecular and evolutionary determinants of bacteriophage host range. Trends Microbiol 27:51–63. 10.1016/j.tim.2018.08.006. [DOI] [PubMed] [Google Scholar]
- 4.Nobrega FL, Vlot M, de Jonge PA, Dreesens LL, Beaumont HJE, Lavigne R, Dutilh BE, Brouns SJJ. 2018. Targeting mechanisms of tailed bacteriophages. Nat Rev Microbiol 16:760–773. 10.1038/s41579-018-0070-8. [DOI] [PubMed] [Google Scholar]
- 5.Gordillo Altamirano FL, Barr JJ. 2019. Phage therapy in the postantibiotic era. Clin Microbiol Rev 32:e00066-18. 10.1128/CMR.00066-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chanishvili N. 2012. Phage therapy—history from Twort and d'Herelle through Soviet experience to current approaches. Adv Virus Res 83:3–40. 10.1016/B978-0-12-394438-2.00001-3. [DOI] [PubMed] [Google Scholar]
- 7.Romero-Calle D, Guimaraes Benevides R, Goes-Neto A, Billington C. 2019. Bacteriophages as alternatives to antibiotics in clinical care. Antibiotics (Basel) 8:138. 10.3390/antibiotics8030138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanlon GW. 2007. Bacteriophages: an appraisal of their role in the treatment of bacterial infections. Int J Antimicrob Agents 30:118–128. 10.1016/j.ijantimicag.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 9.Stone E, Campbell K, Grant I, McAuliffe O. 2019. Understanding and exploiting phage-host interactions. Viruses 11:567. 10.3390/v11060567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rakhuba DV, Kolomiets EI, Dey ES, Novik GI. 2010. Bacteriophage receptors, mechanisms of phage adsorption and penetration into host cell. Pol J Microbiol 59:145–155. 10.33073/pjm-2010-023. [DOI] [PubMed] [Google Scholar]
- 11.Schade SZ, Adler J, Ris H. 1967. How bacteriophage χ attacks motile bacteria. J Virol 1:599–609. 10.1128/JVI.1.3.599-609.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yen JY, Broadway KM, Scharf BE. 2012. Minimum requirements of flagellation and motility for infection of Agrobacterium sp. strain H13-3 by flagellotropic bacteriophage 7–7-1. Appl Environ Microbiol 78:7216–7222. 10.1128/AEM.01082-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raimondo LM, Lundh NP, Martinez RJ. 1968. Primary adsorption site of phage PBS1: the flagellum of Bacillus subtilis. J Virol 2:256–264. 10.1128/JVI.2.3.256-264.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guerrero-Ferreira RC, Viollier PH, Ely B, Poindexter JS, Georgieva M, Jensen GJ, Wright ER. 2011. Alternative mechanism for bacteriophage adsorption to the motile bacterium Caulobacter crescentus. Proc Natl Acad Sci U S A 108:9963–9968. 10.1073/pnas.1012388108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhilenkov EL, Popova VM, Popov DV, Zavalsky LY, Svetoch EA, Stern NJ, Seal BS. 2006. The ability of flagellum-specific Proteus vulgaris bacteriophage PV22 to interact with Campylobacter jejuni flagella in culture. Virol J 3:50. 10.1186/1743-422X-3-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Samuel AD, Pitta TP, Ryu WS, Danese PN, Leung EC, Berg HC. 1999. Flagellar determinants of bacterial sensitivity to chi-phage. Proc Natl Acad Sci U S A 96:9863–9866. 10.1073/pnas.96.17.9863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hendrix RW, Ko C-C, Jacobs-Sera D, Hatfull GF, Erhardt M, Hughes KT, Casjens SR. 2015. Genome sequence of Salmonella phage χ. Genome Announc 3:e01229-14. 10.1128/genomeA.01229-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iino T, Mitani M. 1967. Infection of Serratia marcescens by bacteriophage chi. J Virol 1:445–447. 10.1128/JVI.1.2.445-447.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Girgis HS, Liu Y, Ryu WS, Tavazoie S. 2007. A comprehensive genetic characterization of bacterial motility. PLoS Genet 3:e154. 10.1371/journal.pgen.0030154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kagawa H, Ono N, Enomoto M, Komeda Y. 1984. Bacteriophage chi sensitivity and motility of Escherichia coli K-12 and Salmonella typhimurium Fla- mutants possessing the hook structure. J Bacteriol 157:649–654. 10.1128/JB.157.2.649-654.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ravid S, Eisenbach M. 1983. Correlation between bacteriophage chi adsorption and mode of flagellar rotation of Escherichia coli chemotaxis mutants. J Bacteriol 154:604–611. 10.1128/JB.154.2.604-611.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jajere SM. 2019. A review of Salmonella enterica with particular focus on the pathogenicity and virulence factors, host specificity and antimicrobial resistance including multidrug resistance. Vet World 12:504–521. 10.14202/vetworld.2019.504-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grimont PA, Weill F-X. 2007. Antigenic formulae of the Salmonella serovars. Institut Pasteur, Paris, France. [Google Scholar]
- 24.Marcus SL, Brumell JH, Pfeifer CG, Finlay BB. 2000. Salmonella pathogenicity islands: big virulence in small packages. Microbes Infect 2:145–156. 10.1016/s1286-4579(00)00273-2. [DOI] [PubMed] [Google Scholar]
- 25.Huang K-Y, Wang Y-H, Chien K-Y, Janapatla RP, Chiu C-H. 2016. Hyperinvasiveness of Salmonella enterica serovar Choleraesuis linked to hyperexpression of type III secretion systems in vitro. Sci Rep 6:37642–37642. 10.1038/srep37642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.V T Nair D, Venkitanarayanan K, Kollanoor Johny A. 2018. Antibiotic-resistant Salmonella in the food supply and the potential role of antibiotic alternatives for control. Foods 7:167. 10.3390/foods7100167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Porwollik S, Santiviago CA, Cheng P, Long F, Desai P, Fredlund J, Srikumar S, Silva CA, Chu W, Chen X, Canals R, Reynolds MM, Bogomolnaya L, Shields C, Cui P, Guo J, Zheng Y, Endicott-Yazdani T, Yang HJ, Maple A, Ragoza Y, Blondel CJ, Valenzuela C, Andrews-Polymenis H, McClelland M. 2014. Defined single-gene and multi-gene deletion mutant collections in Salmonella enterica sv Typhimurium. PLoS One 9:e99820. 10.1371/journal.pone.0099820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobylka J, Kuth MS, Müller RT, Geertsma ER, Pos KM. 2020. AcrB: a mean, keen, drug efflux machine. Ann N Y Acad Sci 1459:38–68. 10.1111/nyas.14239. [DOI] [PubMed] [Google Scholar]
- 29.Anes J, McCusker MP, Fanning S, Martins M. 2015. The ins and outs of RND efflux pumps in Escherichia coli. Front Microbiol 6:587–587. 10.3389/fmicb.2015.00587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du D, Wang Z, James NR, Voss JE, Klimont E, Ohene-Agyei T, Venter H, Chiu W, Luisi BF. 2014. Structure of the AcrAB-TolC multidrug efflux pump. Nature 509:512–515. 10.1038/nature13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elkins CA, Nikaido H. 2003. 3D structure of AcrB: the archetypal multidrug efflux transporter of Escherichia coli likely captures substrates from periplasm. Drug Resist Updat 6:9–13. 10.1016/s1368-7646(03)00004-9. [DOI] [PubMed] [Google Scholar]
- 32.Hobbs EC, Yin X, Paul BJ, Astarita JL, Storz G. 2012. Conserved small protein associates with the multidrug efflux pump AcrB and differentially affects antibiotic resistance. Proc Natl Acad Sci U S A 109:16696–16701. 10.1073/pnas.1210093109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang-Kan X, Blair JMA, Chirullo B, Betts J, La Ragione RM, Ivens A, Ricci V, Opperman TJ, Piddock LJV. 2017. Lack of AcrB efflux function confers loss of virulence on Salmonella enterica serovar Typhimurium. mBio 8:e00968-17. 10.1128/mBio.00968-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alekshun MN, Levy SB. 1999. The mar regulon: multiple resistance to antibiotics and other toxic chemicals. Trends Microbiol 7:410–413. 10.1016/s0966-842x(99)01589-9. [DOI] [PubMed] [Google Scholar]
- 35.Thota SS, Chubiz LM. 2019. Multidrug resistance regulators MarA, SoxS, Rob, and RamA repress flagellar gene expression and motility in Salmonella enterica serovar Typhimurium. J Bacteriol 201:e00385-19. 10.1128/JB.00385-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferrari RG, Galiana A, Cremades R, Rodríguez JC, Magnani M, Tognim MC, Oliveira TC, Royo G. 2013. Expression of the marA, soxS, acrB and ramA genes related to the AcrAB/TolC efflux pump in Salmonella enterica strains with and without quinolone resistance-determining regions gyrA gene mutations. Braz J Infect Dis 17:125–130. 10.1016/j.bjid.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vila J, Soto SM. 2012. Salicylate increases the expression of marA and reduces in vitro biofilm formation in uropathogenic Escherichia coli by decreasing type 1 fimbriae expression. Virulence 3:280–285. 10.4161/viru.19205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang T, Kunze C, Dunlop MJ. 2019. Salicylate increases fitness cost associated with MarA-mediated antibiotic resistance. Biophys J 117:563–571. 10.1016/j.bpj.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang C-Z, Chang M-X, Yang L, Liu Y-Y, Chen P-X, Jiang H-X. 2018. Upregulation of AcrEF in quinolone resistance development in Escherichia coli when AcrAB-TolC function is impaired. Microb Drug Resist 24:18–23. 10.1089/mdr.2016.0207. [DOI] [PubMed] [Google Scholar]
- 40.Piñero-Fernandez S, Chimerel C, Keyser UF, Summers DK. 2011. Indole transport across Escherichia coli membranes. J Bacteriol 193:1793–1798. 10.1128/JB.01477-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haldar S, Tapia-Rojo R, Eckels EC, Valle-Orero J, Fernandez JM. 2017. Trigger factor chaperone acts as a mechanical foldase. Nat Commun 8:668. 10.1038/s41467-017-00771-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martinez-Hackert E, Hendrickson WA. 2009. Promiscuous substrate recognition in folding and assembly activities of the trigger factor chaperone. Cell 138:923–934. 10.1016/j.cell.2009.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malki L, Yanku M, Borovok I, Cohen G, Mevarech M, Aharonowitz Y. 2009. Identification and characterization of gshA, a gene encoding the glutamate-cysteine ligase in the halophilic archaeon Haloferax volcanii. J Bacteriol 191:5196–5204. 10.1128/JB.00297-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smirnova GV, Tyulenev AV, Bezmaternykh KV, Muzyka NG, Ushakov VY, Oktyabrsky ON. 2019. Cysteine homeostasis under inhibition of protein synthesis in Escherichia coli cells. Amino Acids 51:1577–1592. 10.1007/s00726-019-02795-2. [DOI] [PubMed] [Google Scholar]
- 45.Apontoweil P, Berends W. 1975. Mapping of gshA, a gene for the biosynthesis of glutathione in Eschericha coli K12. Mol Gen Genet 141:91–95. 10.1007/BF00267676. [DOI] [PubMed] [Google Scholar]
- 46.Watanabe K, Yamano Y, Murata K, Kimura A. 1986. The nucleotide sequence of the gene for gamma-glutamylcysteine synthetase of Escherichia coli. Nucleic Acids Res 14:4393–4400. 10.1093/nar/14.11.4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Forman HJ, Zhang H, Rinna A. 2009. Glutathione: overview of its protective roles, measurement, and biosynthesis. Mol Aspects Med 30:1–12. 10.1016/j.mam.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferguson GP, Booth IR. 1998. Importance of glutathione for growth and survival of Escherichia coli cells: detoxification of methylglyoxal and maintenance of intracellular K+. J Bacteriol 180:4314–4318. 10.1128/JB.180.16.4314-4318.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Glantzounis GK, Tsimoyiannis EC, Kappas AM, Galaris DA. 2005. Uric acid and oxidative stress. Curr Pharm Des 11:4145–4151. 10.2174/138161205774913255. [DOI] [PubMed] [Google Scholar]
- 50.Kim K, Park J, Rhee S. 2007. Structural and functional basis for (S)-allantoin formation in the ureide pathway. J Biol Chem 282:23457–23464. 10.1074/jbc.M703211200. [DOI] [PubMed] [Google Scholar]
- 51.Serventi F, Ramazzina I, Lamberto I, Puggioni V, Gatti R, Percudani R. 2010. Chemical basis of nitrogen recovery through the ureide pathway: formation and hydrolysis of S-ureidoglycine in plants and bacteria. ACS Chem Biol 5:203–214. 10.1021/cb900248n. [DOI] [PubMed] [Google Scholar]
- 52.German GJ, Misra R. 2001. The TolC protein of Escherichia coli serves as a cell-surface receptor for the newly characterized TLS bacteriophage. J Mol Biol 308:579–585. 10.1006/jmbi.2001.4578. [DOI] [PubMed] [Google Scholar]
- 53.Fan F, Li X, Pang B, Zhang C, Li Z, Zhang L, Li J, Zhang J, Yan M, Liang W, Kan B. 2018. The outer-membrane protein TolC of Vibrio cholerae serves as a second cell-surface receptor for the VP3 phage. J Biol Chem 293:4000–4013. 10.1074/jbc.M117.805689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ferbitz L, Maier T, Patzelt H, Bukau B, Deuerling E, Ban N. 2004. Trigger factor in complex with the ribosome forms a molecular cradle for nascent proteins. Nature 431:590–596. 10.1038/nature02899. [DOI] [PubMed] [Google Scholar]
- 55.Depping R, Lohaus C, Meyer HE, Rüger W. 2005. The mono-ADP-ribosyltransferases Alt and ModB of bacteriophage T4: target proteins identified. Biochem Biophys Res Commun 335:1217–1223. 10.1016/j.bbrc.2005.08.023. [DOI] [PubMed] [Google Scholar]
- 56.Quinten TA, Kuhn A. 2012. Membrane interaction of the portal protein gp20 of bacteriophage T4. J Virol 86:11107–11114. 10.1128/JVI.01284-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Loison P, Majou D, Gelhaye E, Boudaud N, Gantzer C. 2016. Impact of reducing and oxidizing agents on the infectivity of Qβ phage and the overall structure of its capsid. FEMS Microbiol Ecol 92:fiw153. 10.1093/femsec/fiw153. [DOI] [PubMed] [Google Scholar]
- 58.Ramazzina I, Costa R, Cendron L, Berni R, Peracchi A, Zanotti G, Percudani R. 2010. An aminotransferase branch point connects purine catabolism to amino acid recycling. Nat Chem Biol 6:801–806. 10.1038/nchembio.445. [DOI] [PubMed] [Google Scholar]
- 59.French JB, Ealick SE. 2010. Biochemical and structural characterization of a ureidoglycine aminotransferase in the Klebsiella pneumoniae uric acid catabolic pathway. Biochemistry 49:5975–5977. 10.1021/bi1006755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li X, Gonzalez F, Esteves N, Scharf BE, Chen J. 2020. Formation of phage lysis patterns and implications on co-propagation of phages and motile host bacteria. PLoS Comput Biol 16:e1007236. 10.1371/journal.pcbi.1007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chan BK, Sistrom M, Wertz JE, Kortright KE, Narayan D, Turner PE. 2016. Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Sci Rep 6:26717–26717. 10.1038/srep26717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oswald C, Tam H-K, Pos KM. 2016. Transport of lipophilic carboxylates is mediated by transmembrane helix 2 in multidrug transporter AcrB. Nat Commun 7:13819–13819. 10.1038/ncomms13819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dos Santos AMP, Ferrari RG, Conte-Junior CA. 2019. Virulence factors in Salmonella Typhimurium: the sagacity of a bacterium. Curr Microbiol 76:762–773. 10.1007/s00284-018-1510-4. [DOI] [PubMed] [Google Scholar]
- 64.Metcalfe HJ, Best A, Kanellos T, La Ragione RM, Werling D. 2010. Flagellin expression enhances Salmonella accumulation in TLR5-positive macrophages. Dev Comp Immunol 34:797–804. 10.1016/j.dci.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 65.Pos KM. 2009. Drug transport mechanism of the AcrB efflux pump. Biochim Biophys Acta 1794:782–793. 10.1016/j.bbapap.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 66.Seeger MA, von Ballmoos C, Verrey F, Pos KM. 2009. Crucial role of Asp408 in the proton translocation pathway of multidrug transporter AcrB: evidence from site-directed mutagenesis and carbodiimide labeling. Biochemistry 48:5801–5812. 10.1021/bi900446j. [DOI] [PubMed] [Google Scholar]
- 67.Meynell EW. 1961. A phage, øχ, which attacks motile bacteria. Microbiology 25:253–290. [DOI] [PubMed] [Google Scholar]
- 68.Bertani G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol 62:293–300. 10.1128/JB.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Binotto J, MacLachlan PR, Sanderson KE. 1991. Electrotransformation in Salmonella typhimurium LT2. Can J Microbiol 37:474–477. 10.1139/m91-078. [DOI] [PubMed] [Google Scholar]
- 71.Bonifield HR, Hughes KT. 2003. Flagellar phase variation in Salmonella enterica is mediated by a posttranscriptional control mechanism. J Bacteriol 185:3567–3574. 10.1128/jb.185.12.3567-3574.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]