Several of the mechanisms that function during the infection of target cells by HPV particles have been previously described. However, many aspects of this process remain unknown.
KEYWORDS: HPV, infection, CCT complex
ABSTRACT
Human papillomavirus (HPV) infection is a multistep process that implies complex interactions of the viral particles with cellular proteins. The HPV capsid includes the two structural proteins L1 and L2, which play crucial roles in infectious viral entry. L2 is particularly relevant for the intracellular trafficking of the viral DNA toward the nucleus. Here, using proteomic studies we identified CCT proteins as novel interaction partners of HPV-16 L2. The CCT multimeric complex is an essential chaperonin which interacts with a large number of protein targets. We analyzed the binding of different components of the CCT complex to L2. We confirmed the interaction of this structural viral protein with the CCT subunit 3 (CCT3), and we found that this interaction requires the N-terminal region of L2. Defects in HPV-16 pseudoviral particle (PsV) infection were revealed by small interfering RNA-mediated knockdown of some CCT subunits. While a substantial drop in the viral infection was associated with the ablation of CCT component 2, even more pronounced effects on infectivity were observed upon depletion of CCT component 3. Using confocal immunofluorescence assays, CCT3 colocalized with HPV PsVs at early times after infection, with L2 being required for this to occur. Further analysis showed the colocalization of several other subunits of CCT with the PsVs. Moreover, we observed a defect in capsid uncoating and a change in PsV intracellular normal processing when ablating CCT3. Taken together, these studies demonstrate the importance of CCT chaperonin during HPV infectious entry.
IMPORTANCE Several of the mechanisms that function during the infection of target cells by HPV particles have been previously described. However, many aspects of this process remain unknown. In particular, the role of cellular proteins functioning as molecular chaperones during HPV infections has been only partially investigated. To the best of our knowledge, we describe here for the first time a requirement of the CCT chaperonin for HPV infection. The role of this cellular complex seems to be determined by the binding of its component 3 to the viral structural protein L2. However, CCT’s effect on HPV infection most probably comprises the whole chaperonin complex. Altogether, these studies define an important role for the CCT chaperonin in the processing and intracellular trafficking of HPV particles and in subsequent viral infectious entry.
INTRODUCTION
Human papillomaviruses (HPVs) are part of a numerous group of small DNA viruses ubiquitously infecting human epithelia and that are transmitted by direct contact. More than 200 different HPV types have been identified, each one being evolutionarily adapted to infect different human epithelial tissues. Therefore, these viruses are classified into cutaneous or mucosal types, based on their specific tropism. Although HPV infections are mainly asymptomatic or associated with benign epithelial lesions, the normal infectious cycle can be perturbed and result in tumor development. Thus, these viral infections cause practically all cases of cervical cancer and a large proportion of other anogenital and oropharyngeal tumors (1). Despite HPVs’ relevance as human pathogens, the cellular mechanisms required for effective viral infections are not completely understood.
All HPVs have a nonenveloped icosahedral capsid that is 50 to 60 nm in diameter. Housed within the virion is a histone-associated circular viral DNA genome of ∼8 kbp. The virus particle structure comprises two proteins, the major and the minor structural proteins L1 and L2. The outer shell of the virion contains 360 units of the major structural protein L1 arranged into 72 pentameric capsomeres. The interactions between these capsomeric units are achieved through the C-terminal section of the L1 protein, and mature viral particles present a more rigid conformation that is further stabilized by disulfide bonds (2, 3). L1 constitutes the major virion component; however, the viral structure also includes the capsid protein L2, which is present in variable but smaller amounts. L2 provides minor but significant contributions to the physical features of the particle. Only some of its regions are typically exposed on the surface of the mature virion, while most of the protein remains hidden inside (4–6).
Both structural proteins are known to perform essential functions during viral infectious entry. Initial virion structural remodeling takes place at the level of the extracellular matrix and the cell surface, where L1 engages with heparan sulfate proteoglycans (7, 8). While L2’s contribution to viral uptake is still uncertain (5, 9–11), L1’s interaction with this and other cellular attachment factors triggers specific conformational changes in both structural proteins (11–13). Further structural alterations are caused by the cleavage of both L1 and L2 by specific cell surface proteases (14, 15). This complex matrix of events partially alters the conformation of the virion, which is thus prepared for recognition by a still-debated uptake receptor and for subsequent endocytosis through a process related to macropinocytosis (16–19). Afterward, endocytic vesicles containing viral particles fuse with early endosomes, and further virion processing depends on the usual endocytic maturation toward late endosomes (20–23), in which capsid disassembly takes place, at least in part, facilitated by its acidic environment. These additional conformational changes allow L2 to emerge from the inner part of the virion. Moreover, cellular cyclophilins aid in segregating L1 from L2, the majority of L1 being then targeted for lysosomal degradation (24). Through specific sequences on L2, as well as by the action of the host factor γ-secretase, L2 protein inserts into the endosomal membrane, thereby becoming partially translocated to the cytoplasm (25–30). In this way, L2 can interact with the cellular cargo-sorting/recycling machinery (21, 22, 31–36), thus allowing complexes of L2 and viral DNA (L2/vDNA) to be transported toward the trans-Golgi network (TGN) in a process involving endosome tubulation and the endoplasmic reticulum (ER)-anchored protein VAP (37). This retrograde transport seems to involve transient contact sites with the ER (23, 37–39) and leads to the progressive accumulation of L2/vDNA in the TGN prior to nuclear entry during mitosis (40–42). This process is most likely mediated by microtubular traffic of L2/vDNA-positive vesicles toward the condensed chromosomes once the mitotic nuclear envelope breaks down (16, 43). Altogether, both L1 and L2 play essential roles in viral entry by interacting with a large number of cellular proteins. Although much effort has been made to unravel the processes that take place during the establishment of HPV infection, given the complexity and multiplicity of these sequential events, many of the cellular mechanisms involved still need to be elucidated.
Although host chaperones have been widely reported to be coopted by several viruses to complete their life cycle (44), there is only limited information available for their roles in the case of HPV. The relevance of chaperones has been mostly examined in regards of the late steps of genome replication and virus assembly (45–48), while only the cyclophilin chaperones have been proposed to facilitate the release of L1 from the L2/vDNA complex (24). For the early stages of the infection, comprising the initial conformational changes in the viral capsid, there has been no compelling information concerning the requirement for chaperones (13, 15, 27).
Among functional chaperones that are known to be important for diverse viruses, it is worth mentioning the eukaryotic cytosolic containing TCP1 (CCT), also named TCP1 ring complex (TRiC), which is required for the folding of multiple newly made proteins, as well as for the control of aggregation (49). A member of the group II of chaperonins, CCT complex presents a hetero-oligomeric barrel-like structure that can alternate between open and closed conformations in an ATP-dependent manner and forms a structure with two optomeric back-to-back stacked rings, each composed of eight different paralogous subunits of about 60 kDa (50). The mammalian subunits of CCT are commonly designated using the Greek alphabet (α to θ), while Arabic numerals (1 to 8) are usually applied to CCT in yeast (51). However, as a matter of simplicity, the latter system is used here to refer to the mammalian cell components.
The exact arrangement of the complex has been a matter of controversy, but recent findings (52–54) have led to a better comprehension of CCT’s interaction with its substrates. Different cellular targets are recognized by distinct subsets of CCT subunits through specific motifs (55–57). Most likely, this complex at least partially encapsulates its target proteins to assist in their folding, presenting a broad range of cellular substrates, many with intricate topologies, including the well-reported cytoskeletal targets tubulin and actin, as well as relevant tumor suppressors and cell cycle regulators (56, 58).
As already mentioned, CCT has been reported to be subverted by several viruses to achieve a successful infection, including influenza A virus (59), reovirus (60), dengue virus (61), Zika virus (62), and hepatitis C virus (63). Overall, many reports have established the relevance of CCT to the correct folding of newly synthesized viral proteins required for viral replication (59, 61–66) or for the assembly and release of new viral particles (60, 67, 68). Here, we report an interaction of the HPV L2 protein with components of the CCT complex and we further demonstrate that this complex, and in particular CCT subunit 3, plays an essential role in HPV infection.
RESULTS
Identification of CCT component 3 as a novel interacting partner of L2.
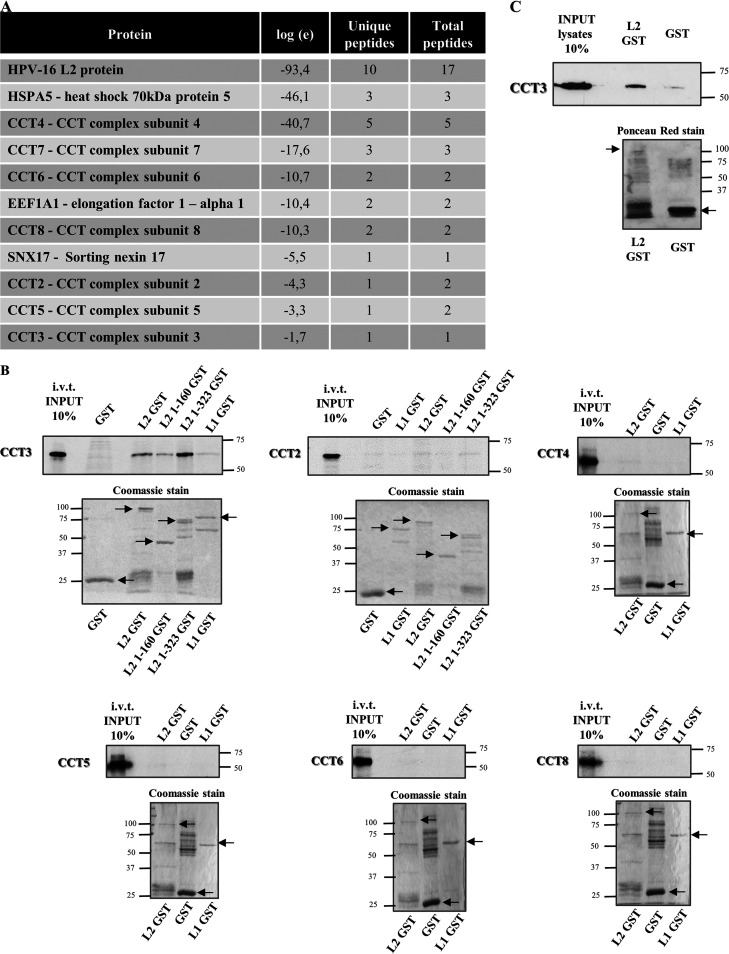
Although it is well known that the human papillomavirus L2 capsid protein plays a crucial role in viral infectious entry, the exact cellular mechanisms implicated have been only partially described (4). Therefore, to identify novel interacting partners of the viral structural protein L2, we performed a proteomic analysis. HEK293TT cells were transfected with a plasmid expressing Flag-HA-tagged HPV-16 L2. The cellular extracts were subjected to immunoprecipitation using anti-HA antibody-conjugated agarose beads, and the resulting immunoconjugates were analyzed by mass spectrometry. Mock-transfected cells were used as a control to exclude nonspecifically interacting proteins. As presented in Fig. 1A, among the potential interacting proteins identified, most of the components of the CCT complex were observed, constituting a group of interesting novel putative interacting candidates. While the number of peptides identified was quite low, previously reported interactors such as SNX17, a well-known binding partner of HPV-L2 (34, 35), was also found, providing confidence in the performance of the assay. In addition, these results confirmed our previous findings, in which CCT component 2 had also been identified in the mass spectrometric analysis of HPV-16 L2 cellular partners (35).
FIG 1.
CCT subunit 3 interacts with HPV-16 L2. (A) Extracts from HEK293 cells expressing FLAG-HA tagged HPV-16 L2 were immunoprecipitated using anti-HA antibody-conjugated agarose beads, and the immunoconjugates were subjected to mass spectrometric analysis. The table presents a selection of L2-specific hits (protein), their corresponding E value (representing the log of the expectation that the assignment is stochastic), the number of peptide sequences assigned, and the total number of tandem mass spectra that can be assigned to each protein. (B) The purified GST fusion proteins L1 and L2 were incubated with several in vitro translated (i.v.t.) and radiolabeled CCT components. GST alone was used as control. Two different GST-fusion truncated forms of L2 were also incubated with i.v.t. and radiolabeled CCT subunits 2 and 3. After an extensive washing, the bound proteins were eluted and analyzed by SDS-PAGE and autoradiography (upper panels for each subunit analysis). The lower panels show the corresponding Coomassie blue-stained gels and the purified proteins are indicated. (C) Endogenous CCT3 was extracted from HEK293 cells and subjected to pulldown assays with purified GST HPV-16 L2 fusion protein (indicated by the arrow) and empty GST as a control. Bound proteins were analyzed by Western blotting assays with anti-CCT3 antibody (upper panel). The corresponding Ponceau red staining of the nitrocellulose membrane is shown (lower panel).
Consequently, we proceeded to further examine the putative interaction of the HPV L2 protein with the CCT complex subunits. We incubated different in vitro-translated (i.v.t.) radiolabeled CCT components with glutathione S-transferase (GST) alone or with GST-L1 or GST-L2, in each case bound to glutathione agarose. After several washes, bound proteins were analyzed by SDS-PAGE and autoradiography. As shown in Fig. 1B (upper left panel), i.v.t. CCT subunit 3 (CCT3) clearly binds to L2 and only interacts marginally with L1. We further analyzed whether L2 truncated mutants, terminated after amino acid 160 or 323, could interact with CCT3. Both bound to CCT3, as well as full-length L2. These results indicate that L2 binding to CCT3 involves the amino-terminal region of the viral protein and that the sequences in the carboxy terminus of L2 are not apparently required for this interaction. In contrast, we saw no binding of HPV structural proteins to the other CCT subunits assayed (2, 4, 5, 6 and 8; Fig. 1B), despite the fact that they were prominent in the proteomic analysis. These results indicate that CCT3 is most likely being bound directly by HPV-16 L2, which in turn is in complex with the other CCT subunits. Since this proteomic screen is not quantitative, it is difficult to comment upon the reasons underlying differences in the number of peptides of each subunit that were detected.
In view of the results with i.v.t. CCT proteins, we next studied the binding of HPV-16 L2 to CCT3 by pulldown assays using extracts from HEK293 cells. The bound endogenously expressed CCT3 was detected by Western blotting with specific antibodies. The results shown in Fig. 1C confirm that L2 protein interacts specifically with endogenous CCT3 from mammalian cells.
CCT is required for efficient infection by HPV-16 pseudovirions.
Taking into account the important role that CCT complex plays in the life cycle of different viruses (59–68), we next wanted to address the effect of CCT in HPV infection. Having shown the interaction between HPV16-L2 and CCT3, we aimed to further investigate the possible role of this and other subunits of the CCT chaperonin complex in HPV infectious entry. When produced in diverse recombinant expression systems, L1 self-assembles to form synthetic virus-like particles (VLPs) and, when coexpressed with L1, L2 can be incorporated into these VLPs. We here employed a well-established and widely used HPV-pseudovirion (PsV) system (69, 70), expressing and further assembling L1 and L2 in HEK293TT cells, encapsidating a luciferase reporter plasmid as the pseudogenome (71).
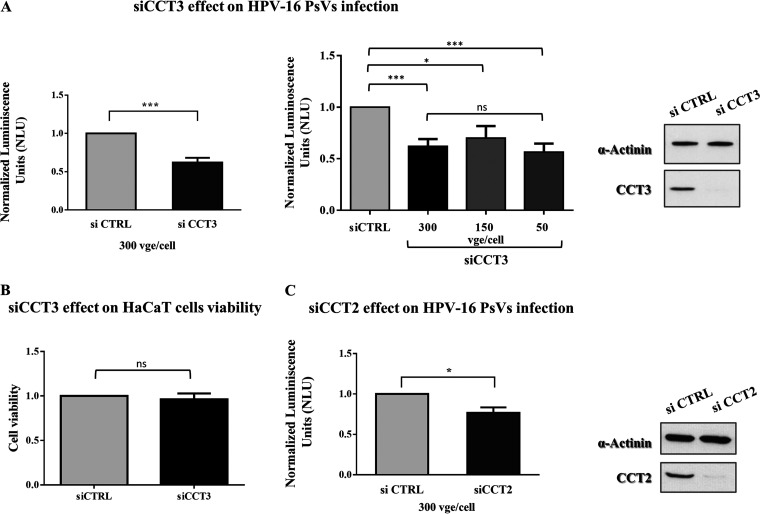
These particles were used to infect HaCaT cells previously transfected either with scrambled small interfering RNA (siRNA) as a control (siCTRL) or with siRNA against CCT3 or CCT2. After 24 h, we monitored the infection efficiency by detecting luciferase activity in the cell lysates. The results obtained demonstrate that a loss of CCT3 expression leads to a reduction of around 40% in the level of infection attained by HPV-16 PsVs (Fig. 2A, left panel), an effect which was obtained across a range of viral genome equivalents (vge) per cell (Fig. 2A, middle panel). This decrease in the infection levels when CCT3 is knocked down is comparable to the decrease previously observed when other cellular components required for efficient HPV infection are removed (21, 33–35). No adverse effects on cell viability (Fig. 2B) or proliferation (data not shown) were observed following siCCT3 transfection. Interestingly, the ablation of CCT subunit 2 also caused a clear defect in HPV infectious entry (Fig. 2C), although the reduction was of around 25% and thus milder than the one observed when depleting CCT3. Altogether, these results strongly point toward a function for the complete CCT complex in HPV infection, while also suggest a key role of CCT3 in the interplay between the chaperonin and the viral particles under study.
FIG 2.
Loss of CCT3 or CCT2 reduces HPV16 infection. HaCaT cells were transfected with scrambled siRNA as a control (siCTRL) or siRNA targeting CCT3 (A) or CCT2 (C). At 48 h posttransfection, the cells were infected with indicated amounts of HPV16 PsVs carrying a luciferase reporter plasmid. After further 24 h, the cells were harvested, and the luciferase activity was measured. The values were normalized to those of siCTRL-transfected cells. The data are expressed as the mean luminescence from at least three independent experiments in each case, the bars indicate the SE (***, P < 0.001; *, P < 0.05). The efficiency of CCT3 or CCT2 knockdown was corroborated by Western blotting of cell lysates; α-actinin was used as loading control (A and C, right panels). (B) Viability of HaCaT cells transfected with scrambled siRNA as a control (siCTRL) or siRNA targeting CCT3 was assessed by using a trypan blue exclusion assay. At 72 h after transfection, the cells were trypsinized, collected, and stained with 0.2% trypan blue. The number of viable cells in siCCT-treated samples was normalized to the number of viable cells obtained for siCTRL-treated samples. The data are expressed as the mean viability from three independent experiments in each case, and the bars indicate the SE. ns, nonsignificant.
HPV-16 PsVs colocalize with CCT at early times after infection.
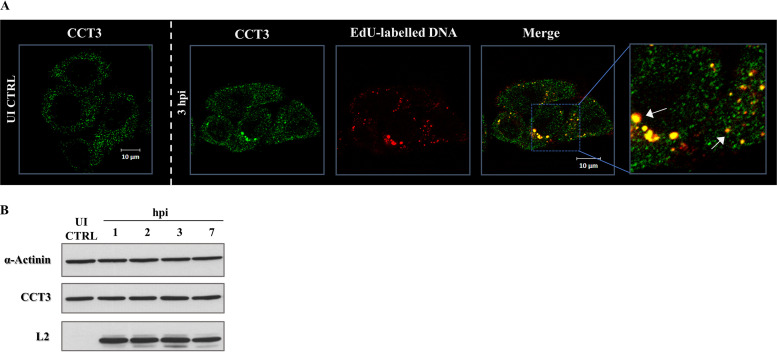
To further explore the role of CCT in HPV infection, we performed a series of immunofluorescence assays to determine the localization of HPV-16 capsids and CCT3 protein. HeLa cells were infected with HPV-16 PsVs encapsidating the reporter DNA labeled with 5-ethynyl-2′-deoxyuridine (EdU). The cells were fixed at 3 h postinfection (hpi), together with uninfected cells used as a control. The samples were subsequently subjected to specific EdU staining and immunodetection of endogenous CCT3. The specificity of the anti-CCT3 antibody was confirmed in siCCT3-silenced HeLa and HaCaT cells (data not shown). As presented in Fig. 3A, endogenous CCT3 protein was observed predominantly in the cytoplasm of uninfected HeLa cells, as expected for a protein reported to be cytosolic. On the other hand, its distribution was altered in cells infected with HPV-16 PsVs, where bright-intensity CCT3 spots were observed. A great number of these structures were also positive for EdU staining, indicating a certain degree of colocalization between CCT3 and the PsVs. Since the signal from EdU-labeled particles can be used as a surrogate marker for the corresponding L2/vDNA complexes (34, 37), these results indicate that CCT3 is in close proximity with the L2/vDNA complex. No variation in the level of expression of CCT3 was observed in response to PsV infection (Fig. 3B), confirming a redistribution event rather than a change in CCT3 levels.
FIG 3.
CCT3 colocalizes with HPV-16 PsVs during infection while maintaining its expression levels. (A) HeLa cells were infected with EdU-labeled HPV-16 PsVs using 500 vge/cell. At 3 h postinfection (3 hpi), the cells were fixed and stained for endogenous CCT3 with specific anti-CCT3 antibody (green) and for EdU (red) to detect reporter DNA encapsidated in PsVs. As a control, CCT3 staining in uninfected (UI) cells was also assayed. Micrographs representative of the results from at least three independent experiments are shown. White arrows indicate the observed colocalization of EdU-labeled pseudogenome and endogenous CCT3 in infected cells. (B) Western blot showing the expression of CCT3 in HeLa cells infected with HPV-16 PsVs at 500 vge/cell. Cell extracts were obtained at 1, 2, 3, and 7 hpi, while uninfected cells (UI CTRL) were used as reference sample. α-Actinin was used as loading control. L2 viral protein was detected as an indicator of cell infection.
To further examine this interaction, we performed a series of immunofluorescence assays with infected cells over time. HeLa cells infected with EdU-labeled PsVs were fixed at different times postinfection. We monitored the distribution of CCT3, together with the EdU-labeled DNA at 1, 3, and 7 h postinfection. Cells only temporarily exposed to PsVs for potential attachment were washed and immediately fixed (0 hpi) as a control (CTRL). As observed previously in uninfected cells (Fig. 3), endogenous CCT3 shows a diffuse and predominantly cytosolic expression in the control samples. Conversely, as shown in Fig. 4, cells infected with pseudovirions consistently showed intense CCT3 positive structures. This clustering was evident even at early times after infection, and a clear colocalization of these CCT3 spots with the internalized pseudovirion particles was detected.
FIG 4.
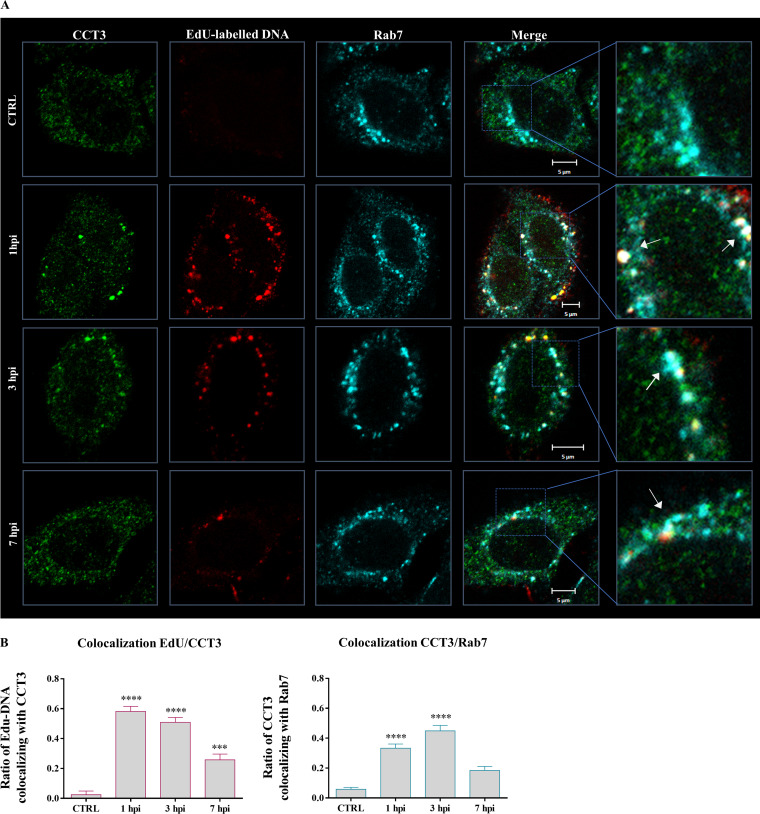
HPV-16 PsVs colocalize with CCT3 at early times after infection in Rab7-positive structures. (A) HeLa cells were exposed to infection with EdU-labeled HPV-16 PsVs using 500 vge/cell. After initial incubation to allow virus binding, the cells were washed and further incubated during 1, 3, or 7 h before being fixed (1, 3, and 7 hpi). Cells immediately fixed after washing were used as a control (CTRL). Representative micrographs from two independent experiments of the corresponding staining for EdU (red), endogenous CCT3 (green), and the GTPase Rab7 (cyan) are shown. White arrows indicate the observed colocalization of the EdU-labeled PsV pseudogenome, CCT3, and the late endosomes marker Rab7 in infected cells. (B) Ratios of CCT3 colocalizing with PsVs (left panel) and CCT3 colocalizing with Rab7 (right panel) were determined for at least 25 cells for each time point. Mander’s colocalization coefficients (MCCs) were determined using JACoP plug-in in Fiji software. MCCs could range from 0 to 1, where MCC = 0 indicates that no colocalization is present and MCC = 1 indicates perfect colocalization between the signals. Statistically significant variations of colocalization over time are shown (***, P < 0.001; ****, P < 0.0001).
Based on our results indicating that CCT3 plays an essential role in efficient HPV infection (Fig. 2), we addressed its probable involvement in the initial steps of viral infection. Simultaneously, we analyzed the distribution of the Rab7 GTPase, a broadly used marker for late endosomes (72). As presented in Fig. 4A, we observed a clear colocalization of the dual CCT3/Edu-positive structures together also with Rab7, particularly at early times after infection, suggesting that CCT3 might be involved in HPV intracellular trafficking. Fewer structures concomitantly stained for EdU, Rab7, and CCT3 were observed at 7 hpi, although the colocalization was still noticeable. Fewer CCT3 clusters were also evident at this later time after infection.
To quantitatively assess this codistribution of CCT3, HPV PsVs and late endosomes, Mander’s colocalization coefficients (MCCs) were calculated, which measure the fractional overlap of the corresponding fluorescent signals (73, 74). In Fig. 4B, the quantified fractional overlapping of EdU-labeled PsVs and CCT3 (left panel), as well as the ratio of CCT3 and Rab7 (right panel), are shown. We found that the signals corresponding to EdU-labeled DNA and CCT3 show statistically significant colocalization at 1 and 3 hpi, with overlapping ratios of 0.58 ± 0.03 and 0.51 ± 0.03, respectively, then dropping to a value of 0.26 ± 0.04 (mean ± the standard errors [SE]) at 7 hpi. In addition, endogenous CCT3 also colocalized with Rab7 to a significantly greater extent at early times after infection, with overlapping ratios at 1 and 3 hpi of 0.33 ± 0.03 and 0.45 ± 0.04, respectively. On the other hand, their remaining colocalization at 7 hpi was not significant (MCC = 0.18 ± 0.03), while endogenous CCT3 and Rab7 showed no basal cooccurrence in control cells (MCC = 0.06 ± 0.01).
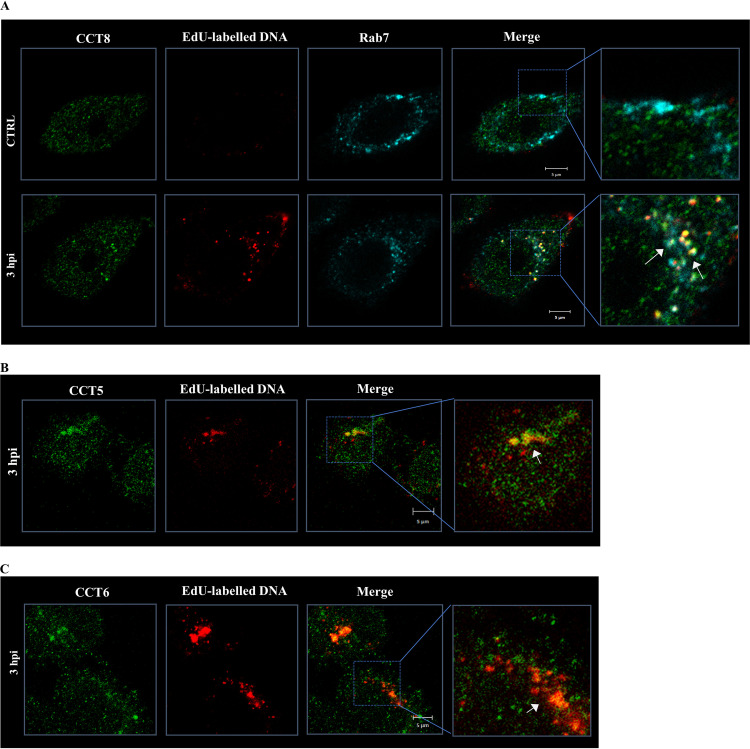
Since other CCT subunits were found in the proteomic analysis, we also proceeded to investigate whether they were also closely associated with the HPV-16 PsVs. As can be seen from Fig. 5, CCT5, CTT6, and CCT8 also colocalized with HPV-16 PsVs, while CCT8-PsV structures were also at least partially positive for Rab7, pointing toward a joint function for the complete CCT complex.
FIG 5.
Diverse CCT subunits colocalize with HPV particles at early times after infection. HeLa cells were exposed to infection with EdU-labeled HPV-16 PsVs using 500 vge/cell. After initial incubation to allow the binding of the particles, the cells were washed and further incubated for 3 h. Representative micrographs of the staining for EdU (red) and different CCT subunits (green) are shown. (A) CCT8; (B) CCT5; (C) CCT6. The Rab7 GTPase (cyan) was also detected when analyzing CCT8 cellular distribution, using as a control cells immediately fixed after washing (CTRL) (A). In every case, white arrows indicate the observed colocalization of EdU-labeled pseudogenome and the indicated cellular proteins.
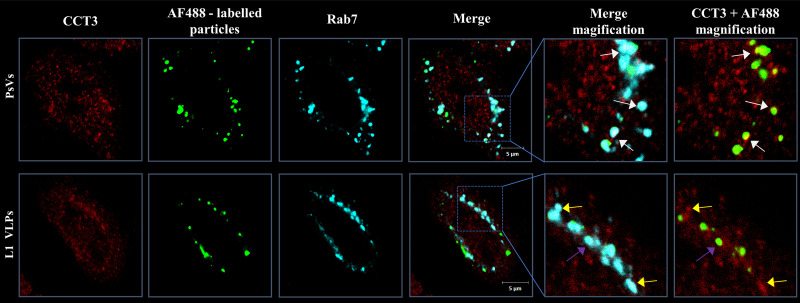
In order to determine whether the colocalization of CCT components was influenced by L2, HeLa cells were infected either with Alexa Fluor 488 (AF488)-labeled PsVs or with L1-only VLPs, which lack the minor capsid protein L2. The fixed samples were subjected to immunodetection of endogenous CCT3 and Rab7. As shown in Fig. 6, the AF488-PsVs are readily detected in structures positive for CCT3 and Rab7, similar to what was seen with EdU-labeled particles. However, when L1-only VLPs were used, minimal clustering with CCT3 spots was observed, further supporting a role for L2 in this process.
FIG 6.
L2 plays a role in colocalization with CCT3. HeLa cells were infected with equal amounts of AF488-labeled PsVs (comprising both L1 and L2) or VLPs (L1 only). After initial incubation to allow virus binding, the cells were washed and further incubated for 1 h before fixation. The micrographs show the fluorescent particles (green) and the staining for CCT3 (red) and Rab7 (cyan). White arrows indicate the observed colocalization of CCT3, AF488-labeled PsVs, and Rab7. Fewer partially colocalizing spots were observed for AF488-labeled VLPs (purple arrows). Yellow arrows indicate the colocalization of CCT3 and Rab7.
Taken together, these results demonstrate that the CCT complex accumulates early after infection with the internalized particles, most likely at the level of L2/vDNA complexes, and as a result of an L2-CCT3 interaction. This colocalization occurs within or near Rab7-positive structures and therefore probably during PsV intracellular trafficking involving late endosomes.
CCT3 plays a role in the intracellular processing of HPV particles.
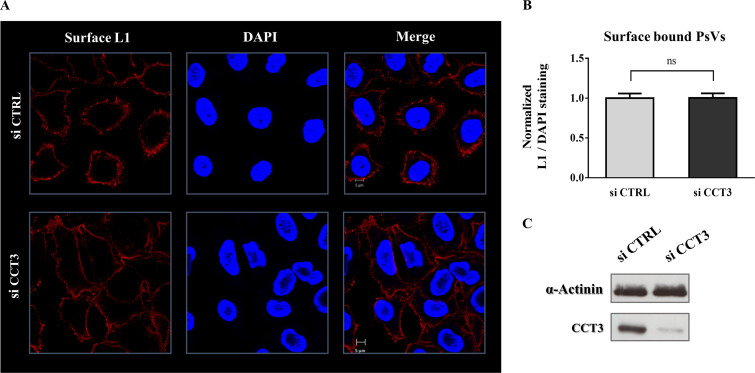
To ascertain which stages in HPV infection are affected by CCT3, we next sought to determine whether CCT3 is required for virion attachment to the cellular surface. HeLa cells were initially transfected either with scrambled siRNA as a control (siCTRL) or with siRNA specific against CCT3. After allowing proper silencing, the cells were briefly incubated with HPV-16 PsVs, permitting particles to bind. Immunofluorescence assays using an L1-specific antibody were then performed on nonpermeabilized cells. As shown in Fig. 7, similar levels of L1 protein were detected on the cellular surface of mock-silenced and CCT3-depleted cells. These results indicate that, regardless of the level of CCT3, comparable amounts of PsVs can bind to the cellular surface of the target cells, demonstrating that CCT3 does not have a role in initial HPV-16 attachment.
FIG 7.
CCT3 depletion has no effect on virus binding to the cell surface. (A) HeLa cells were transfected with siRNA targeting CCT3 (siCCT3) or scrambled siRNA as a control (siCTRL). At 72 h posttransfection, cells were exposed to HPV16 PsVs at 500 vge/cell. After incubation for 1 h at 4°C, the cells were washed and immediately fixed. Total L1 in the cellular surface was consequently assayed by immunofluorescence in nonpermeabilized samples. Representative micrographs of the corresponding staining for surface L1 using a specific anti-L1 antibody (red) and nuclear marker DAPI used as control (blue) are shown. (B) L1 signal relative to DAPI staining was quantified and normalized to the control treatment (siCTRL). At least 100 infected cells from two independent experiments were analyzed. Corresponding means are shown, with bars representing the SE. ns, nonsignificant. (C) CCT3 knockdown was corroborated by Western blotting of cell lysates. α-Actinin was used as loading control.
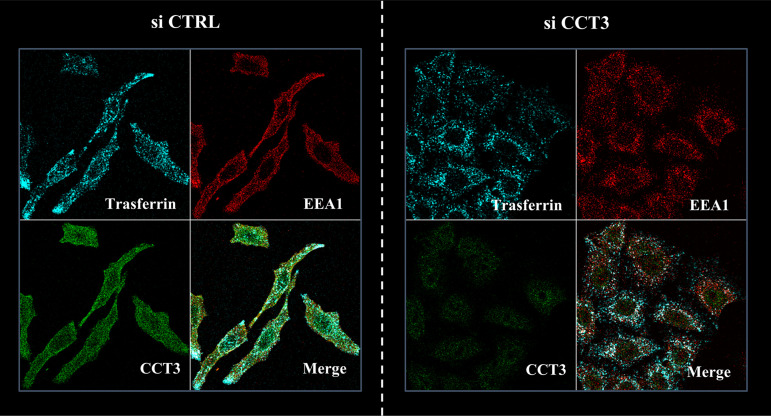
In order to determine whether CCT3 ablation was causing a general defect in endocytic uptake, we analyzed the uptake of the well-studied cellular cargo transferrin (75). CCT3- or control-silenced HeLa cells were incubated with Alexa Fluor 647 (AF647)-labeled transferrin for 30 min before cell fixation. As shown in Fig. 8, CCT3-depleted cells do not show any changes in the endocytic uptake of transferrin.
FIG 8.
CCT3 knockdown does not influence transferrin uptake. HeLa cells were transfected with siRNA targeting CCT3 (siCCT3) or with scrambled siRNA as a control (siCTRL). At 72 h posttransfection, the cells were incubated with AF647-labeled transferrin for 30 min before cell fixation. Representative micrographs of the corresponding staining for the internalized transferrin (cyan) and the endogenous early endosome marker EEA1 (red) are shown, with the colocalization being clear in both conditions. Note that transferrin uptake and distribution is similar in the presence or absence of CCT3 (green).
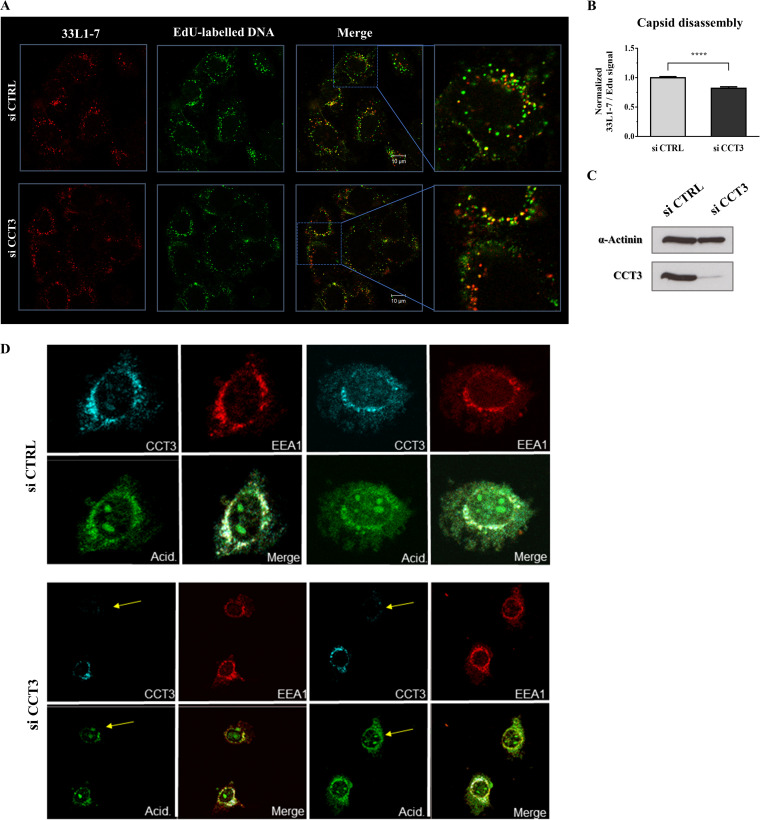
Having shown that CCT3 depletion is detrimental to infectious viral entry and given that we did not find any associated general effect in endocytic uptake, we next analyzed whether this defect in viral infection occurs before or after capsid uncoating. We used the 33L1-7 antibody, which specifically recognizes an epitope on L1 that is only exposed after the capsid has initiated its disassembly during the process of endocytic acidification (76). HeLa cells were transfected with siCCT3 or a control siRNA, and the cells were later exposed to infection with EdU-labeled PsVs. Subsequently, we performed a series of immunofluorescence assays, examining 33L1-7-reactive signal and EdU staining. Thus, we simultaneously monitored the degree of disassembly and the corresponding quantity of infecting particles. As presented in Fig. 9A and B, CCT3 depletion resulted in a significant reduction (20%) in the number of disassembled virions at 7 hpi, with a normalized 33L1-7/EdU ratio of 0.82 ± 0.03 (mean ± SE). Therefore, CCT3 seems to play a role in capsid disassembly during HPV intracellular processing. In order to determine whether loss of CCT3 resulted in a general defect in endocytic trafficking we analyzed endosomal acidification following CCT3 knockdown. As can be seen from Fig. 9D, loss of CCT3 has no major impact on endosomal acidification, indicating that the effects on viral trafficking are specific and not due to a generalized defect in endocytic transport processes.
FIG 9.
Loss of CCT3 leads to defects in PsV capsid disassembly but it does not affect endosomal acidification. (A) HeLa cells were transfected with siRNA targeting CCT3 (siCCT3) or scrambled siRNA as a control (siCTRL). At 72 h posttransfection, the cells were infected with EdU-labeled HPV-16 PsVs at 500 vge/cell and fixed at 7 hpi. Representative micrographs of the corresponding staining for L1 in disassembled capsids using an uncoating-specific anti-L1 antibody (33L1-7, green) and EdU-stained pseudoviral particles (red) are shown. (B) 33L1-7 staining relative to EdU signal was quantified in at least 150 infected cells from each condition corresponding to three independent experiments. The data are expressed as mean ratios normalized to the siCTRL-treated cells, with bars indicating the SE (****, P < 0.0001). (C) CCT3 knockdown was corroborated by Western blotting of cell lysates. α-Actinin was used as loading control. (D) Endosome acidification of HeLa cells transfected with siRNA targeting CCT3 (siCCT3) or scrambled siRNA as a control (siCTRL) was assessed 72 h after transfection using pHrodo Green AM intracellular pH indicator (Thermo Fisher Scientific), together with EEA1 (red) and CCT3 (cyan) staining. Yellow arrows indicate the unchanged endosomal acidification in cells effectively silenced for CCT3.
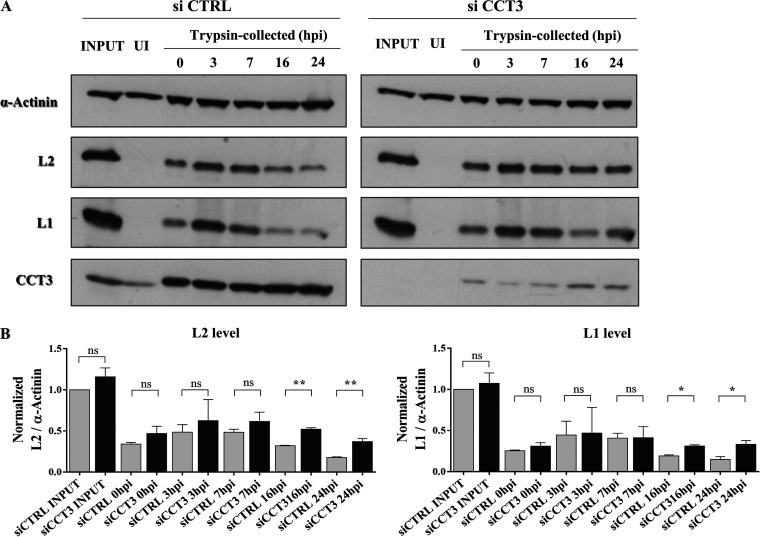
These data were strengthened when we analyzed particle internalization by Western blotting assays. To perform these experiments, HeLa cells were first transfected with either a control siRNA (siCTRL) or with siCCT3. Subsequently, mock-silenced or CCT3-depleted cells were exposed to infection with HPV-16 PsVs. At various times postinfection, the cells were treated with trypsin to remove noninternalized particles and then harvested. The amount of L1 and L2 proteins were then determined in these lysates. As can be seen in Fig. 10A (and as quantified in Fig. 10B), similar amounts of PsVs were internalized regardless of the level of cellular CCT3 (trypsin-collected samples, 0 hpi). These results further complement the information presented above, pointing to an unaltered uptake of diverse cargos in the absence of CCT3, including HPV particles. Likewise, no major differences were observed in L1 and L2 levels at early times after infection. Interestingly, significantly higher intensities of both L1 and L2 were detected at later times (16 and 24 hpi) in CCT3-depleted cells, indicating a slower processing of PsVs.
FIG 10.
CCT3 is necessary for PsV intracellular processing (A) HeLa cells silenced for CCT3 or a control scrambled siRNA (siCTRL) were infected with HPV16 PsVs at 500 vge/cell. Uninfected (UI) and total infected samples (INPUT) were obtained to use as control. At different times after infection cells were washed, treated with trypsin in order to remove noninternalized viruses, and then harvested. PsV internalization and subsequent digestion were assessed by Western blotting with anti-L2 anti anti-L1 specific antibodies. α-Actinin was used as a loading control and CCT3 was monitored to confirmed silencing when corresponding. (B) Relative levels over time after infection of L2 (left panel) or L1 (right panel) were quantified and normalized to the initial amount of the corresponding viral structural protein (INPUT). Data from three independent experiments is expressed as mean ratios, bars representing the SE (*, P < 0,05; **, P < 0,01; ns, nonsignificant).
DISCUSSION
The early steps of HPV infections are a series of consecutive and interdependent processes comprising the initial attachment to the target cell and the virion internalization, followed by the progressive disassembly of viral capsids. However, many of the mechanisms associated with HPV infectious entry are still unclear (4, 16, 23, 77). This study was orientated toward the identification and characterization of novel cellular factors relevant for infectious entry.
Chaperones, which tightly control cellular protein quality, are frequently exploited by viruses to support the completion of their life cycles. However, the role of these cellular proteins during the HPV cycle had only partially been described: two heat shock proteins, Hsp70 and Hsp40, are implicated in the genome replication of these viruses (78); the interaction of HPV structural proteins with NPM1 and Hsc70 have important functions in viral assembly (46, 47); and karyopherins were proposed to play a role both in capsid assembly and in viral genome nuclear entry (48, 79). All of these events occur in the late stages of an HPV infection, but less information is currently available concerning chaperone function in the corresponding early or middle steps. In this regard, while the effect of the host cyclophilins in the initial conformational changes of L2 has been demonstrated (13), the importance of this event in L2 cleavage and thus its relevance for infection are still controversial (4, 15). It is noteworthy that cyclophilins have also been shown to mediate the dissociation of L1 and L2 structural proteins following the acidification of endosomes (24). Also, the in vitro disassembly of bovine papillomavirus particles has been associated with the Hsc70/40 chaperones (80). Altogether, the analysis of the association of host chaperones to the initial stages of HPV infection is a highly relevant and only partially comprehended topic.
The specific requirement for CCT chaperonin in several viral processes has been previously identified, but chiefly concerning replication or assembly, both late phases of the viral life cycles (59–68). Moreover, no CCT involvement in any step of the HPV viral cycle had previously been described. In this study, we have identified the involvement of the CCT chaperonin complex in the HPV life cycle. We present here compelling evidence supporting CCT, and particularly its subunit 3, as a critical component of the HPV-16 entry pathway.
We considered that further exploration of the nonstructural functions of L2 and its interactions with host cell proteins was particularly relevant. The results from our proteomic screen indicated the CCT complex as a novel putative cellular partner of L2. Moreover, we further reinforced these data by showing the interaction between L2 and the component 3 of the chaperonin complex, both in vitro and ex vivo, while confirming that L1 has no relevant association with CCT3. Interestingly, we were unable to demonstrate any significant degree of association between HPV-16 L2 and several other CCT subunits, despite them being present in the proteomic analysis. Overall, L2 is most likely interacting only with CCT3, which in turn would be responsible for the cointeraction of the other CCT subunits. Indeed, the immunofluorescence analyses also support this notion, where there is a clear colocalization between incoming PsVs and several subunits of the CCT complex.
We found that the L2/CCT3 interaction involves the N-terminal region of L2, as a truncated form of L2 comprising only the first 160 amino acid residues binds to CCT3 equally well. This was surprising, given that the CCT complex has been extensively reported to localize in the cytosol (81) and considering that the HPV-L2 C-terminal region mediates contact with diverse cytosolic cellular partners, including a variety of cell-sorting/recycling factors (21, 22, 33, 35, 82). However, the exact topology and stoichiometric distribution of L2 is still a matter of controversy (4, 40). A large portion of L2 is thought to be exposed to the cytosol, most probably extending from the region C-terminal to the transmembrane-like domain, which is situated at residues 45 to 67 in HPV16-L2 (15, 26). Therefore, although more data are needed to determine the precise sites of contact of L2 with CCT3, the results presented here clearly point toward a region of L2 within the first 160 residues.
Moreover, we assessed the effect that abrogating CCT might have upon viral infection and we found a clear decrease in the levels of infection with HPV-16 PsVs when CCT3 expression levels were knocked down. Significantly, under the conditions used for CCT3 ablation, no effect on the cellular viability was observed. A milder but still significant defect in HPV infection was observed by the ablation of CCT2. Although CCT-independent mechanisms cannot be ruled out, these observations point toward a role for the complete CCT complex in HPV infection, while they also highlight the main relevance of its subunit 3, probably by interacting directly with L2.
We obtained immunofluorescence data indicating the proximity of HPV particles, and most likely the L2/vDNA complexes, with CCT3 in characteristic spots that were mainly present at early times after infection. This colocalization is clearly disturbed when using L1-only VLPs, further pointing toward an L2-dependent mechanism. Furthermore, we showed that this clustering is only due to a redistribution of CCT3, while its total expression levels were unchanged during HPV infection. We examined the colocalization of CCT3 with vDNA over time and found that it reaches its peak early after infection and occurs in structures positive for Rab7. This GTPase is present in late endosomes and its different forms play important roles in trafficking both to lysosomes and the TGN (72). Moreover, its importance in HPV intracellular transport has been demonstrated (17, 31, 83–87). Thus, the aforementioned results initially point toward the participation of CCT in the viral trafficking within the infected cell. While it is not possible to estimate the percentage of viral particles which associate with the CCT complex, the overall levels of colocalization and alteration in CCT distribution upon infection are indicative of a substantial proportion of PsVs following this route. No general defects in endocytic transport pathways were observed when silencing CCT3, since neither transferrin uptake nor endosomal acidification were affected. In contrast, CCT3 depletion resulted in a defect in the intracellular processing of the viral capsid, with a significant reduction in the exposure of disassembly-specific L1 epitopes in the CCT3-depleted cells and a corresponding decrease in the degradation of the structural proteins.
In addition, CCT has previously been shown to take part in the dissociation of oligomeric structures (88). Thus, in the processing of HPV capsids during the maturation of the endosomal compartments, the evidence here clearly suggests a role for this chaperonin in the natural disassembly of the virions. CCT appears to have an impact on this process by directly interacting with the cytosolic region of L2 and probably by modulating its interplay with other effectors important for viral traffic.
Regardless of CCT’s well-studied effect on the structure of its substrates, it is also interesting to highlight that CCT proteins have also been shown to directly bind and regulate some targets without affecting their folding (89). Several reports have shown the individual action of specific subunits of CCT (89–94), many of which act in a complex-independent manner. Therefore, in addition to the more obvious possible action of this cellular chaperonin, other possibilities might also be considered. The question arises whether the effect of CCT on HPV infective entry shown here concerns the complete chaperonin complex or the individual subunit 2 and, particularly, the component 3. However, we identified other CCT components colocalizing with HPV virions, similarly to CCT3, and, in addition, our proteomic screen also identified several CCT subunits in the L2 pull downs. These observations once again suggest that the interaction between L2 and CCT3 results in a recruitment of the whole CCT complex to sites of viral infection, indicating that it is not just the monomeric CCT3 which is required for infectious entry but also the integrity of the CCT complex.
The CCT chaperonin has a wide range of substrates comprising several viral proteins (59–65, 67, 68), although it is principally known to be an important cytoskeletal regulator center, actin and tubulin being its most studied and well-reported substrates (95–97). Therefore, the modulation of CCT may have relevant effects on HPV infection by affecting any of these cytoskeletal proteins. It is particularly noteworthy that the intracellular transport of HPV L2/vDNA complexes depends on the microtubule network both to reach the TGN and for their migration toward the condensed chromosomes (4, 17, 41, 87, 98). Nevertheless, the precise mechanisms by which CCT influences the viral infectious entry remains to be elucidated.
Altogether, this study sheds light onto the previously unreported participation of the CCT chaperonins in the HPV life cycle, demonstrating a role for these cellular proteins at early steps during infection. Further studies will be focused on whether the observed effect in HPV infection is due solely to the identified direct interaction with L2 or whether, as previously discussed, it might also be a consequence of affecting the viral endosome trafficking, probably via alteration of important cytoskeletal players. Determining the underlying mechanisms by which CCT modulates the HPV infection will enhance our understanding of the cellular basis of HPV-host interaction.
MATERIALS AND METHODS
Cell culture and transfections.
HeLa cervical cancer cells, HEK293 and HEK293TT (71) human embryonic kidney cells, and HaCaT human skin keratinocytes were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum, penicillin-streptomycin (100 U/ml), and glutamine (300 mg/ml).
HEK293 and HEK293TT cells were transfected by the calcium phosphate precipitation method (99). For transient siRNA experiments, HeLa or HaCaT cells were transfected by using Lipofectamine RNAiMAX (Invitrogen) with a pool of siRNAs specific against CCT3 (5′-CAGACUGACAUUGAGAUUACA, 5′-AGCGGCCAAGUCCAUGAUCGA, 5′-AUCCACGUAUGCGGCGCUAUA, and 5′-CUUGCGUGGAGUCAUGAUUAA) (Sigma; as previously reported [60]) or by using an ON-TARGETplus SMART pool against CCT2 (Dharmacon). Concomitantly, a scrambled siSTABLE nontargeting siRNA (Dharmacon) was used as a control.
Plasmids.
Plasmids expressing FLAG-HA-tagged HPV-16 L2 were described previously (100). The pUC-derived CCT-expressing plasmids were described previously (101) and were kindly provided by Hiroaki Imataka (University of Hyogo, Hyogo, Japan). The expression vectors for HPV-16 L1 and HPV-16 L2 full-length GST fusion proteins were already described (35). The plasmids for the expression of truncated forms of GST-tagged HPV-16 L2 were generated by the GENEART site-directed mutagenesis system (Invitrogen) and further confirmed by DNA sequencing. In-house designed oligonucleotides synthesized by Eurofins MWG were used. Plasmid p16shell L2-3×FLAG-HA expresses HPV-16 L1 and L2 in a codon-optimized bicistronic manner, where L2 is FLAG and hemagglutinin (HA) tagged (28). The pXull L1 plasmid expresses HPV-16 L1 only and was generated by deleting the L2 sequence from WT pXull (37). pGL3 Luci construct, carrying the firefly luciferase gene, was purchased from Promega.
Antibodies.
In this study, the following primary antibodies were used: mouse anti-CCT3 (F-3; Santa Cruz), mouse anti-CCT2 (D-8; Santa Cruz), mouse anti-CCT8 (E-1; Santa Cruz), mouse anti-CCT5 (D-5; Santa Cruz), mouse anti-CCT6 (F-12; Santa Cruz), mouse anti-α-actinin (H-2; Santa Cruz) for loading control in Western blotting experiments and rabbit anti-Rab7 (D95F2; Cell Signaling), goat anti-EEA1 (N19; Santa Cruz), mouse anti-HPV-16 L1 (CAMVIR-1; Santa Cruz), mouse anti-HPV-16 L2 (16.D4 64–81; kindly provided by Martin Müller), and mouse anti-33L1-7 (kindly provided by Martin Sapp). The secondary antibodies AF488 donkey anti-mouse, AF647 donkey anti-rabbit, AF647 donkey anti-mouse, rhodamine goat anti-mouse, and AF546 donkey anti-goat (Invitrogen) were used for the immunofluorescence assays, while horseradish peroxidase-conjugated swine anti-rabbit and rabbit anti-mouse secondary antibodies (Dako) were used in Western blotting experiments.
Mass spectrometry analysis.
HEK293 cells were transfected with FLAG-HA-tagged HPV-16 L2 expression plasmid or the empty plasmid as control. After 48 h, the corresponding extracts were obtained with mass spectrometry lysis buffer (50 mM HEPES [pH 7.4], 150 mM NaCl, 50 mM NaF, 1 mM EDTA, 0.5% NP-40) and consequently incubated with EZview Red Anti-HA affinity gel beads (Sigma) for 2 h on a rotating wheel at 4°C. The beads were rinsed first with wash buffer (25 mM HEPES [pH 7.0], 0.1% NP-40, 150 mM NaCl) and then with phosphate-buffered saline (PBS) and subsequently subjected to proteomic analysis as previously described (35).
Briefly, proteins were eluted from the affinity beads using 50 ng of sequencing-grade trypsin in 20 mM triethanolamine bicarbonate (pH 8.5) for 12 h at room temperature. After digestion, the supernatant was removed from the beads, and the beads were briefly incubated with an additional aliquot of 20 mM triethanolamine bicarbonate (pH 8.5), which was removed and pooled with the first supernatant. The reactions were stopped by the addition of formic acid to 0.1%, and the resulting mixture was desalted using C18 STAGE (STop And Go Extraction) tips and lyophilized to dryness.
Nanobore columns were constructed using in-house constructed columns packed with 15 cm of 3-μm Ascentis RPA particles (Sigma) using a high-pressure column loader. The desalted samples were resuspended in 0.1% formic acid and injected onto the column in 0.1% formic acid. The column was developed with a discontinuous gradient of 0% to 80% acetonitrile in 0.1% formic acid for 60 min and sprayed directly into the orifice of an Amazon ETD ion trap mass spectrometer (Bruker). A cycle of one full scan (375 to 1,700 m/z), followed by eight data-dependent tandem mass spectrometry (MS/MS) scans, was performed throughout the liquid chromatography (LC) separation. Raw data files from the Amazon ETD instrument were converted to mgf files by a data analysis software package (Bruker) and searched against the Ensembl human protein database and the NCBInr Viral database using the Global Proteome Machine interfaced with X!Tandem, and the resulting matches were filtered at a 1% false discovery rate.
Fusion protein purification and binding assays.
GST alone or GST-tagged fusion proteins were expressed and purified as described previously (102) and immobilized on glutathione agarose (Sigma). CCT2 and CCT3 proteins were in vitro transcribed and translated using a TNT T7 coupled reticulocyte lysate system (Promega) and radiolabeled with [35S]methionine (Perkin-Elmer). Equal amounts of in vitro translated (i.v.t.) proteins were added to GST alone or GST-tagged fusion proteins, followed by incubation on a rotating wheel for 2 h at room temperature in E1A buffer (25 mM HEPES [pH 7.0], 0.5% NP-40, 150 mM NaCl) plus protease inhibitors. After extensive washing in the same buffer, retained proteins were eluted and separated by SDS-PAGE. The gel was stained with Coomassie blue, dried, and analyzed by autoradiography.
For the pulldown analysis, HEK293 cell were extracted with radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 5 mM EDTA, 50 mM Tris [pH 8.0], 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS]) plus protease inhibitors, followed by incubation with GST or HPV-16 L2 GST-purified proteins for 1 h at 4°C on a rotating wheel. After extensive washing in E1A buffer, the retained proteins were separated by SDS-PAGE and analyzed by Western blotting. The nitrocellulose membrane was stained with Ponceau red to detect the GST or GST-tagged proteins.
Synthetic HPV particle production and labeling.
HPV-16 PsVs containing a luciferase reporter plasmid (pGL3 Luci) were generated in HEK293TT cells as described previously (71). The purity and the capsid protein content of the PsV preparations were analyzed by SDS-PAGE and Coomassie brilliant blue staining, using solutions of bovine serum albumin (BSA) at different concentrations as standards. For EdU labeling, the growth medium was supplemented with 25 μM EdU at 12 h posttransfection during PsV production. The encapsidated DNA was analyzed by real-time PCR, using a standard curve of the reported plasmid DNA to quantify the copy number of each PsV preparation. HPV-16 L1-only VLPs were generated using a similar protocol as for PsVs. Labeling of both types of synthetic particles, i.e., PsVs and VLPs, with the AF488 fluorophore was performed according to the manufacturer’s protocol (Molecular Probes).
Infectivity assays.
To check the infectivity in HaCaT cells, silencing using siRNA specifically targeting CCT3, CCT2, or a control siRNA (siCTRL) was performed for 48 h. These cells were further challenged with HPV-16 PsVs at a concentration of 300, 150, or 50 vge/cell, as indicated in each case. Firefly luciferase activity was monitored after 24 h as a measure of infection using a luciferase assay system (Promega). Total cell protein extracts were quantified to ensure the use of equal protein inputs in the luciferase measurements. The change in luciferase activity in siCCT-treated cells was normalized to the luciferase readings obtained for siCTRL-treated cells in each experiment, and statistical significance was analyzed by performing a one-tailed t test. The cellular lysates were also used to confirm the efficiency of CCT3 or CCT2 knockdown by Western blotting.
Cell viability assay.
To measure the number of viable cells, a trypan blue exclusion assay was used. HaCaT cells were seeded in a 12-well plate, incubated for 24 h, and treated with siRNA specifically targeting CCT3 or a control siRNA (siCTRL). After 72 h, the cells were trypsinized, collected, and stained with 0.2% trypan blue. The numbers of viable cells were determined by direct counting of cells under the microscope by using a standard hemocytometer. The number of viable cells in siCCT-treated samples was normalized to the number of viable cells obtained for siCTRL-treated samples in each experiment, and statistical significance was analyzed by performing a one-tailed t test. The cellular lysates were also used to confirm the efficiency of CCT3 by Western blotting.
HPV tracking assays.
HeLa cells previously seeded on sterile glass coverslips were challenged with the synthetic HPV particles and incubated at 4°C on an orbital shaker for 1 h to allow its attachment to the cells. Cells were then washed, supplemented with new medium, and incubated at 37°C. EdU-labeled PsVs were used at a concentration of 500 vge/cell. For AF488-labeled PsVs and VLPs, the total protein content was measured, and the preparations were subsequently used in equivalent amounts. Uninfected samples, or cells immediately fixed after this incubation time, were used as control, as indicated in each case. Cells were fixed at one or more time points postinfection (1, 3, or 7 h) with 3.7% paraformaldehyde for 15 min at room temperature. After being washed in PBS, the cells were permeabilized with 0.5% Triton in PBS at room temperature for 5 min, washed in PBS, briefly incubated with 0.1 M glycine in PBS to reduce background staining, and further analyzed by immunofluorescence. The samples were incubated with primary and secondary antibodies for 1 h each at 37°C, with intermediate washes in PBS.
For the detection of EdU-labeled PsVs, cells were then washed in 3% BSA in PBS and incubated with freshly prepared Click-iT reaction solution (Molecular Probes) for 40 min at room temperature. The cells were then washed with 3% BSA in PBS and finally with water before being mounted on glass slides. Samples were visualized using an Axiovert 100 M microscope (Zeiss) supported by an LSM 510 confocal unit. Colocalization analysis was performed using Fiji software to determine the corresponding Mander’s colocalization coefficients for two channels. Statistical significance was ascertained by one-way analysis of variance, followed by a Bonferroni post hoc test.
In parallel, to assess CCT3 protein levels during PsV infection, cells were challenged as described with PsVs at an approximate concentration of 500 vge/cell, collected in RIPA buffer with protease inhibitors at different time points after infection (1, 2, 3, or 7 h), and subsequently analyzed by Western blotting. Samples from uninfected cells were used as a control.
PsV cell attachment assays.
To analyze HPV PsV binding to the cell surface, HeLa cells, previously seeded on sterile glass coverslips, were treated with siRNA specifically targeting CCT3 or a control siRNA (siCTRL). After 72 h, the cells were further challenged with PsVs at a concentration of 500 vge/cell and incubated at 4°C on an orbital shaker for 1 h. The cells were then washed and fixed with 3.7% paraformaldehyde for 15 min at room temperature. These nonpermeabilized cells were then subjected to quantitative immunofluorescence to detect L1 signal on the cellular surface and counterstained with DAPI (4′,6′-diamidino-2-phenylindole) as a reference signal before being washed with water and mounted on glass slides. Samples were visualized using an Axiovert 100 M microscope (Zeiss) supported by an LSM 510 confocal unit. Statistical significance was analyzed by performing a one-tailed t test. In parallel, to confirm the efficiency of CCT3 knockdown, the cells were treated as described with the corresponding siRNAs and, after 72 h, cellular lysates were collected in RIPA buffer with protease inhibitors and subsequently analyzed by Western blotting.
Transferrin uptake.
In order to analyze transferrin internalization, HeLa cells previously attached to sterile glass coverslips were treated with siRNA specifically targeting CCT3 or a control siRNA (siCTRL). At 72 h after transfection, cells were incubated for 30 min at 37°C in serum-free medium with the addition of 50 μg/ml AF647-labeled transferrin (Molecular Probes). After incubation, the cells were washed several times with PBS and fixed in 3.7% paraformaldehyde for 15 min at room temperature. The cells were incubated for 1 h at room temperature with 10% goat serum and later stained for EEA1. Slides were visualized using a Zeiss Axiovert 100M microscope attached to an LSM 510 confocal unit.
PsV capsid disassembly analysis.
HeLa cells previously attached to sterile glass coverslips were treated with siRNA specifically targeting CCT3 or a control siRNA (siCTRL). After 72 h, the cells were exposed to EdU-labeled PsVs at a concentration of 500 vge/cell and incubated at 4°C on orbital oscillation for 1 h. The cells were then washed, supplemented with new medium, and incubated at 37°C for further 7 h. Subsequently, the samples were fixed with 3.7% paraformaldehyde for 15 min at room temperature, washed, and permeabilized with 0.5% Triton in PBS for 5 min at room temperature. After a brief wash with 0.1 M glycine in PBS, the cells were further subjected to quantitative immunofluorescence to detect the uncoating-specific L1 epitopes using the primary antibody 33L1-7 and an AF488-tagged secondary antibody. EdU-labeled PsVs were detected as described above, according to the manufacturer’s indication (Molecular Probes). After final washes with 3% BSA in PBS and water, the cells were mounted on glass slides and visualized using an Axiovert 100M microscope (Zeiss) with an LSM 510 confocal unit. Statistical significance was analyzed by performing a one-tailed t test. At the same time, cells were treated with CCT3 or CTRL siRNA and, after 72 h, lysed in RIPA buffer containing protease inhibitors and then analyzed by Western blotting to confirm the corresponding silencing.
Acidification assay.
To assess endosome acidification, HeLa cells previously attached to sterile glass coverslips were treated with siRNA specifically targeting CCT3 or a control siRNA (siCTRL). After 72 h, the cells were incubated with pHrodo Green AM intracellular pH indicator (Thermo Fisher Scientific) at a final concentration of 1 mg/ml for ∼4 h. The cells were then washed with PBS, fixed with 3.7% paraformaldehyde, and processed for immunofluorescence as previously described. To localize early endosomes (EEA1) and CCT3, immunofluorescence was carried out with specific primary antibodies and AF546- and AF647-tagged secondary antibodies, respectively. A Zeiss Axiovert 100 M microscope attached to an LSM 510 confocal unit was used for the analysis.
PsV internalization and intracellular digestion assays.
HeLa cells were initially treated with siRNA specifically targeting CCT3 or a control siRNA (siCTRL). After 72 h, the cells were challenged with HPV-16 PsVs at a concentration of 500 vge/cell, followed by incubation at 4°C on an orbital shaker for 1 h. The cells were then washed, supplemented with new medium, and further incubated at 37°C for 0, 3, 7, 16, or 24 h before being collected by trypsin treatment and harvested in RIPA buffer plus protease inhibitors, thus collecting only internalized viruses. Uninfected (UI) and total infected samples (INPUT) were immediately lysed with SDS sample buffer (100 mM Tris-HCl [pH 7], 200 mM dithiothreitol, 4% SDS, 0.2% bromophenol, 20% glycerol) to monitor the total input virus. PsV internalization levels and subsequent digestion were assessed by Western blotting using anti-L2- and anti-L1-specific antibodies, while CCT3 silencing was also analyzed. Statistical significance in the levels of structural viral proteins were analyzed for each time point by performing one-tailed t tests.
ACKNOWLEDGMENTS
We are very grateful to Miranda Thomas for valuable comments on the manuscript.
The research leading to these results has received funding from the Associazione Italiana per la Ricerca sul Cancro under IG2019 (ID 23572 project; principal investigator, Lawrence Banks). M.B.V. received an ICGEB Arturo Falaschi postdoctoral fellowship and is now supported by a postdoctoral fellowship from CONICET. J.B. gratefully acknowledges support from the Umberto Veronesi Foundation (postdoctoral fellowship, years 2018 and 2020).
REFERENCES
- 1.Schiffman M, Doorbar J, Wentzensen N, de Sanjosé S, Fakhry C, Monk BJ, Stanley MA, Franceschi S. 2016. Carcinogenic human papillomavirus infection. Nat Rev Dis Primers 2:16086. 10.1038/nrdp.2016.86. [DOI] [PubMed] [Google Scholar]
- 2.Doorbar J, Egawa N, Griffin H, Kranjec C, Murakami I. 2015. Human papillomavirus molecular biology and disease association. Rev Med Virol 25:2–23. 10.1002/rmv.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buck CB, Day PM, Trus BL. 2013. The papillomavirus major capsid protein L1. Virology 445:169–174. 10.1016/j.virol.2013.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campos SK. 2017. Subcellular trafficking of the papillomavirus genome during initial infection: the remarkable abilities of minor capsid protein l2. Viruses 9:370. 10.3390/v9120370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guan J, Bywaters SM, Brendle SA, Ashley RE, Makhov AM, Conway JF, Christensen ND, Hafenstein S. 2017. Cryoelectron microscopy maps of human papillomavirus 16 reveal L2 densities and heparin binding site. Structure 25:253–263. 10.1016/j.str.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 6.Bywaters SM, Brendle SA, Tossi KP, Biryukov J, Meyers C, Christensen ND. 2017. Antibody competition reveals surface location of HPV L2 minor capsid protein residues 17–36. Viruses 9:336–321. 10.3390/v9110336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giroglou T, Florin L, Schäfer F, Streeck RE, Sapp M. 2001. Human papillomavirus infection requires cell surface heparan sulfate. J Virol 75:1565–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Selinka H-C, Florin L, Patel HD, Freitag K, Schmidtke M, Makarov VA, Sapp M. 2007. Inhibition of transfer to secondary receptors by heparan sulfate-binding drug or antibody induces noninfectious uptake of human papillomavirus. J Virol 81:10970–10980. 10.1128/JVI.00998-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Day PM, Lowy DR, Schiller JT. 2008. Heparan sulfate-independent cell binding and infection with furin-precleaved papillomavirus capsids. J Virol 82:12565–12568. 10.1128/JVI.01631-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang JW, Roden RBS. 2013. L2, the minor capsid protein of papillomavirus. Virology 445:175–186. 10.1016/j.virol.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang R, Day PM, Yutzy WH, Lin K-Y, Hung C-F, Roden RBS. 2003. Cell surface-binding motifs of L2 that facilitate papillomavirus infection. J Virol 77:3531–3541. 10.1128/jvi.77.6.3531-3541.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richards KF, Bienkowska-Haba M, Dasgupta J, Chen XS, Sapp M. 2013. Multiple heparan sulfate binding site engagements are required for the infectious entry of human papillomavirus type 16. J Virol 87:11426–11437. 10.1128/JVI.01721-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bienkowska-Haba M, Patel HD, Sapp M. 2009. Target cell cyclophilins facilitate human papillomavirus type 16 infection. PLoS Pathog 5:e1000524. 10.1371/journal.ppat.1000524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richards RM, Lowy DR, Schiller JT, Day PM. 2006. Cleavage of the papillomavirus minor capsid protein, L2, at a furin consensus site is necessary for infection. Proc Natl Acad Sci U S A 103:1522–1527. 10.1073/pnas.0508815103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bronnimann MP, Calton CM, Chiquette SF, Li S, Lu M, Chapman JA, Bratton KN, Schlegel AM, Campos SK. 2016. Furin cleavage of L2 during papillomavirus infection: minimal dependence on cyclophilins. J Virol 90:6224–6234. 10.1128/JVI.00038-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DiGiuseppe S, Bienkowska-Haba M, Guion LG, Sapp M. 2017. Cruising the cellular highways: how human papillomavirus travels from the surface to the nucleus. Virus Res 231:1–9. 10.1016/j.virusres.2016.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schelhaas M, Shah B, Holzer M, Blattmann P, Kühling L, Day PM, Schiller JT, Helenius A. 2012. Entry of human papillomavirus type 16 by actin-dependent, clathrin- and lipid raft-independent endocytosis. PLoS Pathog 8:e1002657. 10.1371/journal.ppat.1002657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spoden G, Kuhling L, Cordes N, Frenzel B, Sapp M, Boller K, Florin L, Schelhaas M. 2013. Human papillomavirus types 16, 18, and 31 share similar endocytic requirements for entry. J Virol 87:7765–7773. 10.1128/JVI.00370-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bannach C, Brinkert P, Kühling L, Greune L, Schmidt MA, Schelhaas M. 2020. Epidermal growth factor receptor and Abl2 kinase regulate distinct steps of human papillomavirus 16 endocytosis. J Virol 94:1–15. 10.1128/JVI.02143-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grässel L, Fast LA, Scheffer KD, Boukhallouk F, Spoden GA, Tenzer S, Boller K, Bago R, Rajesh S, Overduin M, Berditchevski F, Florin L. 2016. The CD63-syntenin-1 complex controls post-endocytic trafficking of oncogenic human papillomaviruses. Sci Rep 6:32337–32318. 10.1038/srep32337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Broniarczyk J, Bergant M, Goździcka-Józefiak A, Banks L. 2014. Human papillomavirus infection requires the TSG101 component of the ESCRT machinery. Virology 460–461:83–90. 10.1016/j.virol.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 22.Broniarczyk J, Pim D, Massimi P, Bergant M, Goździcka-Józefiak A, Crump C, Banks L. 2017. The VPS4 component of the ESCRT machinery plays an essential role in HPV infectious entry and capsid disassembly. Sci Rep 7:45159–45111. 10.1038/srep45159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siddiqa A, Broniarczyk J, Banks L. 2018. Papillomaviruses and endocytic trafficking. Int J Mol Sci 19:2619. 10.3390/ijms19092619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bienkowska-Haba M, Williams C, Kim SM, Garcea RL, Sapp M. 2012. Cyclophilins facilitate dissociation of the human papillomavirus type 16 capsid protein L1 from the L2/DNA complex following virus entry. J Virol 86:9875–9887. 10.1128/JVI.00980-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bronnimann MP, Chapman JA, Park CK, Campos SK. 2013. A transmembrane domain and GxxxG motifs within L2 are essential for papillomavirus infection. J Virol 87:464–473. 10.1128/JVI.01539-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DiGiuseppe S, Keiffer TR, Bienkowska-Haba M, Luszczek W, Guion LGM, Müller M, Sapp M. 2015. Topography of the human papillomavirus minor capsid protein L2 during vesicular trafficking of infectious entry. J Virol 89:10442–10452. 10.1128/JVI.01588-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inoue T, Zhang P, Zhang W, Goodner-Bingham K, Dupzyk A, DiMaio D, Tsai B. 2018. γ-Secretase promotes membrane insertion of the human papillomavirus L2 capsid protein during virus infection. J Cell Biol 217:3545–3559. 10.1083/jcb.201804171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang W, Kazakov T, Popa A, DiMaio D. 2014. Vesicular trafficking of incoming human papillomavirus 16 to the Golgi apparatus and endoplasmic reticulum requires γ-secretase activity. mBio 5:e01777-14. 10.1128/mBio.01777-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang P, Monteiro da Silva G, Deatherage C, Burd C, DiMaio D. 2018. Cell-penetrating peptide mediates intracellular membrane passage of human papillomavirus L2 protein to trigger retrograde trafficking. Cell 174:1465–1476.e13. 10.1016/j.cell.2018.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harwood MC, Dupzyk AJ, Inoue T, DiMaio D, Tsai B. 2020. p120 catenin recruits HPV to γ-secretase to promote virus infection. PLoS Pathog 16:e1008946–20. 10.1371/journal.ppat.1008946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lipovsky A, Popa A, Pimienta G, Wyler M, Bhan A, Kuruvilla L, Guie MA, Poffenberger AC, Nelson CDS, Atwood WJ, DiMaio D. 2013. Genome-wide siRNA screen identifies the retromer as a cellular entry factor for human papillomavirus. Proc Natl Acad Sci U S A 110:7452–7457. 10.1073/pnas.1302164110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Popa A, Zhang W, Harrison MS, Goodner K, Kazakov T, Goodwin EC, Lipovsky A, Burd CG, DiMaio D. 2015. Direct binding of retromer to human papillomavirus type 16 minor capsid protein L2 mediates endosome exit during viral infection. PLoS Pathog 11:e1004699. 10.1371/journal.ppat.1004699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pim D, Broniarczyk J, Bergant M, Playford MP, Banks L. 2015. A novel PDZ domain interaction mediates the binding between human papillomavirus 16 L2 and sorting nexin 27 and modulates virion trafficking. J Virol 89:10145–10155. 10.1128/JVI.01499-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bergant M, Peternel Š, Pim D, Broniarczyk J, Banks L. 2017. Characterizing the spatio-temporal role of sorting nexin 17 in human papillomavirus trafficking. J Gen Virol 98:715–725. 10.1099/jgv.0.000734. [DOI] [PubMed] [Google Scholar]
- 35.Bergant Marušič M, Ozbun MA, Campos SK, Myers MP, Banks L. 2012. Human papillomavirus L2 facilitates viral escape from late endosomes via sorting nexin 17. Traffic 13:455–467. 10.1111/j.1600-0854.2011.01320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang P, Moreno R, Lambert PF, DiMaio D. 2020. Cell-penetrating peptide inhibits retromer-mediated human papillomavirus trafficking during virus entry. Proc Natl Acad Sci U S A 117:6121–6128. 10.1073/pnas.1917748117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siddiqa A, Massimi P, Pim D, Broniarczyk J, Banks L. 2018. Human papillomavirus 16 infection induces VAP-dependent endosomal tubulation. J Virol 92:1–14. 10.1128/JVI.01514-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siddiqa A, Massimi P, Pim D, Banks L. 2019. Diverse papillomavirus types induce endosomal tubulation. Front Cell Infect Microbiol 9:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dong R, Saheki Y, Swarup S, Lucast L, Harper JW, De Camilli P. 2016. Endosome-ER contacts control actin nucleation and retromer function through VAP-dependent regulation of PI4P. Cell 166:408–423. 10.1016/j.cell.2016.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Calton CM, Bronnimann MP, Manson AR, Li S, Chapman JA, Suarez-Berumen M, Williamson TR, Molugu SK, Bernal RA, Campos SK. 2017. Translocation of the papillomavirus L2/vDNA complex across the limiting membrane requires the onset of mitosis. PLoS Pathog 13:e1006200. 10.1371/journal.ppat.1006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aydin I, Villalonga-Planells R, Greune L, Bronnimann MP, Calton CM, Becker M, Lai KY, Campos SK, Schmidt MA, Schelhaas M. 2017. A central region in the minor capsid protein of papillomaviruses facilitates viral genome tethering and membrane penetration for mitotic nuclear entry. PLoS Pathog 13:e1006308. 10.1371/journal.ppat.1006308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Day PM, Weisberg AS, Thompson CD, Hughes MM, Pang YY, Lowy DR, Schiller JT. 2019. Human papillomavirus 16 capsids mediate nuclear entry during infection. J Virol 93:1–18. 10.1128/JVI.00454-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DiGiuseppe S, Bienkowska-Haba M, Guion LGM, Keiffer TR, Sapp M. 2017. Human papillomavirus major capsid protein L1 remains associated with the incoming viral genome throughout the entry process. J Virol 91:1–17. 10.1128/JVI.00537-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sullivan CS, Pipas JM. 2001. The virus-chaperone connection. Virology 287:1–8. 10.1006/viro.2001.1038. [DOI] [PubMed] [Google Scholar]
- 45.Liu X, Qu L, Ye X, Yi C, Zheng X, Hao M, Su W, Yao Z, Chen P, Zhang S, Feng Y, Wang Q, Yan Q, Li P, Li H, Li F, Pan W, Niu X, Xu R, Feng L, Chen L. 2018. Incorporation of NS1 and prM/M are important to confer effective protection of adenovirus-vectored Zika virus vaccine carrying E protein. NPJ Vaccines 3:1–8. 10.1038/s41541-018-0072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Florin L, Becker KA, Sapp C, Lambert C, Sirma H, Müller M, Streeck RE, Sapp M. 2004. Nuclear translocation of papillomavirus minor capsid protein L2 requires Hsc70. J Virol 78:5546–5553. 10.1128/JVI.78.11.5546-5553.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Day PM, Thompson CD, Pang YY, Lowy DR, Schiller JT. 2015. Involvement of nucleophosmin (NPM1/B23) in assembly of infectious HPV16 capsids. Papillomavirus Res 1:74–89. 10.1016/j.pvr.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bird G, O’Donnell M, Moroianu J, Garcea RL. 2008. Possible role for cellular karyopherins in regulating polyomavirus and papillomavirus capsid assembly. J Virol 82:9848–9857. 10.1128/JVI.01221-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grantham J. 2020. The molecular chaperone CCT/TRiC: an essential component of proteostasis and a potential modulator of protein aggregation. Front Genet 11:172. 10.3389/fgene.2020.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yébenes H, Mesa P, Muñoz IG, Montoya G, Valpuesta JM. 2011. Chaperonins: two rings for folding. Trends Biochem Sci 36:424–432. 10.1016/j.tibs.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 51.Kabir MA, Uddin W, Narayanan A, Reddy PK, Jairajpuri MA, Sherman F, Ahmad Z. 2011. Functional subunits of eukaryotic chaperonin CCT/TRiC in protein folding. J Amino Acids 2011:843206–843216. 10.4061/2011/843206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leitner A, Joachimiak LA, Bracher A, Mönkemeyer L, Walzthoeni T, Chen B, Pechmann S, Holmes S, Cong Y, Ma B, Ludtke S, Chiu W, Hartl FU, Aebersold R, Frydman J. 2012. The molecular architecture of the eukaryotic chaperonin TRiC/CCT. Structure 20:814–825. 10.1016/j.str.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jin M, Han W, Liu C, Zang Y, Li J, Wang F, Wang Y, Cong Y. 2019. An ensemble of cryo-EM structures of TRiC reveal its conformational landscape and subunit specificity. Proc Natl Acad Sci U S A 116:19513–19522. 10.1073/pnas.1903976116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Balchin D, Miličić G, Strauss M, Hayer-Hartl M, Hartl FU. 2018. Pathway of actin folding directed by the eukaryotic chaperonin TRiC. Cell 174:1507–1521.e16. 10.1016/j.cell.2018.07.006. [DOI] [PubMed] [Google Scholar]
- 55.Yam AY, Xia Y, Lin H-TJ, Burlingame A, Gerstein M, Frydman J. 2008. Defining the TRiC/CCT interactome links chaperonin function to stabilization of newly made proteins with complex topologies. Nat Struct Mol Biol 15:1255–1262. 10.1038/nsmb.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Willison KR. 2018. The substrate specificity of eukaryotic cytosolic chaperonin CCT. Philos Trans R Soc Lond B Biol Sci 373:20170192. 10.1098/rstb.2017.0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gestaut D, Limatola A, Joachimiak L, Frydman J. 2019. The ATP-powered gymnastics of TRiC/CCT: an asymmetric protein folding machine with a symmetric origin story. Curr Opin Struct Biol 55:50–58. 10.1016/j.sbi.2019.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lopez T, Dalton K, Frydman J. 2015. The mechanism and function of group II chaperonins. J Mol Biol 427:2919–2930. 10.1016/j.jmb.2015.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fislová T, Thomas B, Graef KM, Fodor E. 2010. Association of the influenza virus RNA polymerase subunit PB2 with the host chaperonin CCT. J Virol 84:8691–8699. 10.1128/JVI.00813-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Knowlton JJ, Fernández De Castro I, Ashbrook AW, Gestaut DR, Zamora PF, Bauer JA, Forrest JC, Frydman J, Risco C, Dermody TS. 2018. The TRiC chaperonin controls reovirus replication through outer-capsid folding. Nat Microbiol 3:481–493. 10.1038/s41564-018-0122-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hafirassou ML, Meertens L, Umaña-Diaz C, Labeau A, Dejarnac O, Bonnet-Madin L, Kümmerer BM, Delaugerre C, Roingeard P, Vidalain PO, Amara A. 2017. A global interactome map of the dengue virus NS1 identifies virus restriction and dependency host factors. Cell Rep 21:3900–3913. 10.1016/j.celrep.2017.11.094. [DOI] [PubMed] [Google Scholar]
- 62.Wang Y, Uraki R, Hwang J, Fikrig E. 2020. TRiC/CCT complex, a binding partner of NS1 protein, supports the replication of Zika virus in both mammalians and mosquitoes. Viruses 12:519. 10.3390/v12050519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Inoue Y, Aizaki H, Hara H, Matsuda M, Ando T, Shimoji T, Murakami K, Masaki T, Shoji I, Homma S, Matsuura Y, Miyamura T, Wakita T, Suzuki T. 2011. Chaperonin TRiC/CCT participates in replication of hepatitis C virus genome via interaction with the viral NS5B protein. Virology 410:38–47. 10.1016/j.virol.2010.10.026. [DOI] [PubMed] [Google Scholar]
- 64.Gopal S, Perez E, Xia AY, Knowlton JJ, Cerqueira F, Dermody TS, Upton JW. 2018. Murine cytomegalovirus M72 promotes acute virus replication in vivo and is a substrate of the TRiC/CCT complex. Virology 522:92–105. 10.1016/j.virol.2018.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang J, Wu X, Zan J, Wu Y, Ye C, Ruan X, Zhou J. 2013. Cellular chaperonin CCT contributes to rabies virus replication during infection. J Virol 87:7608–7621. 10.1128/JVI.03186-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang Q, Huang WR, Chih WY, Chuang KP, Chang CD, Wu Y, Huang Y, Liu HJ. 2019. Cdc20 and molecular chaperone CCT2 and CCT5 are required for the Muscovy duck reovirus p10.8-induced cell cycle arrest and apoptosis. Vet Microbiol 235:151–163. 10.1016/j.vetmic.2019.06.017. [DOI] [PubMed] [Google Scholar]
- 67.Lingappa JR, Martin RL, Wong ML, Ganem D, Welch WJ, Lingappa VR. 1994. A eukaryotic cytosolic chaperonin is associated with a high molecular weight intermediate in the assembly of hepatitis B virus capsid, a multimeric particle. J Cell Biol 125:99–112. 10.1083/jcb.125.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hong S, Choi G, Park S, Chung A-S, Hunter E, Rhee SS. 2001. Type D retrovirus Gag polyprotein interacts with the cytosolic chaperonin TRiC. J Virol 75:2526–2534. 10.1128/JVI.75.6.2526-2534.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Buck CB, Pastrana DV, Lowy DR, Schiller JT. 2004. Efficient intracellular assembly of papillomaviral vectors. J Virol 78:751–757. 10.1128/jvi.78.2.751-757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Conway MJ, Meyers C. 2009. Replication and assembly of human papillomaviruses. J Dent Res 88:307–317. 10.1177/0022034509333446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Buck CB, Pastrana DV, Lowy DR, Schiller JT. 2005. Generation of HPV pseudovirions using transfection and their use in neutralization assays. Methods Mol Med 119:445–462. 10.1385/1-59259-982-6:445. [DOI] [PubMed] [Google Scholar]
- 72.Guerra F, Bucci C. 2016. Multiple roles of the small GTPase Rab7. Cells 5:34. 10.3390/cells5030034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Manders EMM, Verbeek FJ, Aten JA. 1993. Measurement of colocalization of objects in dual-colour confocal images. J Microsc 169:375–382. 10.1111/j.1365-2818.1993.tb03313.x. [DOI] [PubMed] [Google Scholar]
- 74.Pike JA, Styles IB, Rappoport JZ, Heath JK. 2017. Quantifying receptor trafficking and colocalization with confocal microscopy. Methods 115:42–54. 10.1016/j.ymeth.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 75.Mayle KM, Le AM, Kamei DT. 2012. The intracellular trafficking pathway of transferrin. Biochim Biophys Acta 1820:264–281. 10.1016/j.bbagen.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sapp M, Kraus U, Volpers C, Snijders PJF, Walboomers JMM, Streeck RE. 1994. Analysis of type-restricted and cross-reactive epitopes on virus-like particles of human papillomavirus type 33 and in infected tissues using monoclonal antibodies to the major capsid protein. J Gen Virol 75:3375–3383. 10.1099/0022-1317-75-12-3375. [DOI] [PubMed] [Google Scholar]
- 77.Aksoy P, Gottschalk EY, Meneses PI. 2017. HPV entry into cells. Mutat Res Rev Mutat Res 772:13–22. 10.1016/j.mrrev.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu JS, Kuo SR, Makhov AM, Cyr DM, Griffith JD, Broker TR, Chow LT. 1998. Human Hsp70 and Hsp40 chaperone proteins facilitate human papillomavirus-11 E1 protein binding to the origin and stimulate cell-free DNA replication. J Biol Chem 273:30704–30712. 10.1074/jbc.273.46.30704. [DOI] [PubMed] [Google Scholar]
- 79.Bordeaux J, Forte S, Harding E, Darshan MS, Klucevsek K, Moroianu J. 2006. The L2 minor Capsid protein of low-risk human papillomavirus type 11 interacts with host nuclear import receptors and viral DNA. J Virol 80:8259–8262. 10.1128/JVI.00776-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chromy LR, Oltman A, Estes PA, Garcea RL. 2006. Chaperone-mediated in vitro disassembly of polyoma- and papillomaviruses. J Virol 80:5086–5091. 10.1128/JVI.80.10.5086-5091.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Young JC, Barral JM, Hartl FU. 2003. More than folding: localized functions of cytosolic chaperones. Trends Biochem Sci 28:541–547. 10.1016/j.tibs.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 82.McNally KE, Faulkner R, Steinberg F, Gallon M, Ghai R, Pim D, Langton P, Pearson N, Danson CM, Nägele H, Morris LL, Singla A, Overlee BL, Heesom KJ, Sessions R, Banks L, Collins BM, Berger I, Billadeau DD, Burstein E, Cullen PJ. 2017. Retriever is a multiprotein complex for retromer-independent endosomal cargo recycling. Nat Cell Biol 19:1214–1225. 10.1038/ncb3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Smith JL, Campos SK, Wandinger-Ness A, Ozbun MA. 2008. Caveolin-1-dependent infectious entry of human papillomavirus type 31 in human keratinocytes proceeds to the endosomal pathway for pH-dependent uncoating. J Virol 82:9505–9512. 10.1128/JVI.01014-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Young JM, Zine El Abidine A, Gómez-Martinez RA, Ozbun MA. 2019. The known and potential intersections of Rab-GTPases in human papillomavirus infections. Front Cell Dev Biol 7:139. 10.3389/fcell.2019.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xie J, Heim EN, Crite M, DiMaio D. 2020. TBC1D5-catalyzed cycling of Rab7 is required for retromer-mediated human papillomavirus trafficking during virus entry. Cell Rep 31:107750. 10.1016/j.celrep.2020.107750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Salvo MA, Kingstad-Bakke B, Salas-Quinchucua C, Camacho E, Osorio JE. 2018. Zika virus like particles elicit protective antibodies in mice. PLoS Negl Trop Dis 12:e0006210. 10.1371/journal.pntd.0006210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Day PM, Thompson CD, Schowalter RM, Lowy DR, Schiller JT. 2013. Identification of a role for the trans-Golgi network in human papillomavirus 16 pseudovirus infection. J Virol 87:3862–3870. 10.1128/JVI.03222-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kaisari S, Sitry-Shevah D, Miniowitz-Shemtov S, Teichner A, Hershko A. 2017. Role of CCT chaperonin in the disassembly of mitotic checkpoint complexes. Proc Natl Acad Sci U S A 114:956–961. 10.1073/pnas.1620451114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brackley KI, Grantham J. 2010. Subunits of the chaperonin CCT interact with F-actin and influence cell shape and cytoskeletal assembly. Exp Cell Res 316:543–553. 10.1016/j.yexcr.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 90.Chen XQ, Fang F, Florio JB, Rockenstein E, Masliah E, Mobley WC, Rissman RA, Wu C. 2018. T-complex protein 1-ring complex enhances retrograde axonal transport by modulating tau phosphorylation. Traffic 19:840–853. 10.1111/tra.12610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Echbarthi M, Vallin J, Grantham J. 2018. Interactions between monomeric CCTδ and p150Glued: a novel function for CCTδ at the cell periphery distinct from the protein folding activity of the molecular chaperone CCT. Exp Cell Res 370:137–149. 10.1016/j.yexcr.2018.06.018. [DOI] [PubMed] [Google Scholar]
- 92.Spiess M, Echbarthi M, Svanström A, Karlsson R, Grantham J. 2015. Overexpression analysis of all eight subunits of the molecular chaperone CCT in mammalian cells reveals a novel function for CCTδ. J Mol Biol 427:2757–2764. 10.1016/j.jmb.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 93.Roobol A, Sahyoun ZP, Carden MJ. 1999. Selected subunits of the cytosolic chaperonin associate with microtubules assembled in vitro. J Biol Chem 274:2408–2415. 10.1074/jbc.274.4.2408. [DOI] [PubMed] [Google Scholar]
- 94.Elliott KL, Svanström A, Spiess M, Karlsson R, Grantham J. 2015. A novel function of the monomeric CCTε subunit connects the serum response factor pathway to chaperone-mediated actin folding. Mol Biol Cell 26:2801–2809. 10.1091/mbc.E15-01-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vallin J, Grantham J. 2019. The role of the molecular chaperone CCT in protein folding and mediation of cytoskeleton-associated processes: implications for cancer cell biology. Cell Stress Chaperones 24:17–27. 10.1007/s12192-018-0949-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Grantham J, Ruddock LW, Roobol A, Carden MJ. 2002. Eukaryotic chaperonin containing T-complex polypeptide 1 interacts with filamentous actin and reduces the initial rate of actin polymerization in vitro. Cell Stress Chaperones 7:235–242. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Llorca O, Martín-Benito J, Ritco-Vonsovici M, Grantham J, Hynes GM, Willison KR, Carrascosa JL, Valpuesta JM. 2000. Eukaryotic chaperonin CCT stabilizes actin and tubulin folding intermediates in open quasi-native conformations. EMBO J 19:5971–5979. 10.1093/emboj/19.22.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schneider MA, Spoden GA, Florin L, Lambert C. 2011. Identification of the dynein light chains required for human papillomavirus infection. Cell Microbiol 13:32–46. 10.1111/j.1462-5822.2010.01515.x. [DOI] [PubMed] [Google Scholar]
- 99.Graham FL, Van der Eb A. 1973. A new technique for the assay of infectivity of adenovirus 5 DNA. Virology 52:456–467. 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 100.Marušič MB, Mencin N, Ličen M, Banks L, Grm HŠ. 2010. Modification of human papillomavirus minor capsid protein L2 by sumoylation. J Virol 84:11585–11589. 10.1128/JVI.01269-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.MacHida K, Masutani M, Kobayashi T, Mikami S, Nishino Y, Miyazawa A, Imataka H. 2012. Reconstitution of the human chaperonin CCT by coexpression of the eight distinct subunits in mammalian cells. Protein Expr Purif 82:61–69. 10.1016/j.pep.2011.11.010. [DOI] [PubMed] [Google Scholar]
- 102.Thomas M, Massimi P, Jenkins J, Banks L. 1995. HPV-18 E6 mediated inhibition of p53 DNA binding activity is independent of E6 induced degradation. Oncogene 10:261–268. [PubMed] [Google Scholar]