RSV is an important respiratory pathogen. This study describes a prefusion F-specific VHH that primarily binds to antigenic site I of RSV F. This is the first time that a prefusion F-specific antibody that binds this site has been reported. In general, antibodies that bind to site I are poorly neutralizing, whereas the VHH described here neutralizes RSV A at subnanomolar concentrations. Our findings contribute to insights into the RSV F antigenic map.
KEYWORDS: fusion protein, RSV, single-domain antibody
ABSTRACT
Human respiratory syncytial virus (RSV) is a major cause of lower respiratory tract disease, especially in young children and the elderly. The fusion protein (F) exists in a pre- and postfusion conformation and is the main target of RSV-neutralizing antibodies. Highly potent RSV-neutralizing antibodies typically bind sites that are unique to the prefusion conformation of F. In this study, we screened a single-domain antibody (VHH) library derived from a llama immunized with prefusion-stabilized F and identified a prefusion F-specific VHH that can neutralize RSV A at subnanomolar concentrations. Structural analysis revealed that this VHH primarily binds to antigenic site I while also making contacts with residues in antigenic sites III and IV. This new VHH reveals a previously underappreciated membrane-proximal region sensitive to neutralization.
IMPORTANCE RSV is an important respiratory pathogen. This study describes a prefusion F-specific VHH that primarily binds to antigenic site I of RSV F. This is the first time that a prefusion F-specific antibody that binds this site has been reported. In general, antibodies that bind to site I are poorly neutralizing, whereas the VHH described here neutralizes RSV A at subnanomolar concentrations. Our findings contribute to insights into the RSV F antigenic map.
INTRODUCTION
Human respiratory syncytial virus (RSV) is a major respiratory pathogen that is responsible for approximately 33 million cases of acute lower respiratory infection in young children worldwide (1). Despite many years of research, still no active vaccination strategy has been licensed to prevent disease caused by RSV, and there are no antiviral drugs available to treat RSV patients. The monoclonal antibody (MAb) palivizumab (Synagis) is used for prophylaxis in high-risk children. Monthly intramuscular injections of this biologic during the RSV season reduce the chance of RSV-related hospitalization by 55% (2). Clinical trials are currently ongoing with the more potent and half-life-extended MAb nirsevimab, which preferentially binds to the prefusion conformation of the RSV fusion protein (F) at the so-called “site Ø.” A single intramuscular injection of preterm infants with this antibody resulted in a 78.4% reduction in RSV-associated hospitalization (3).
Next to some antibodies targeting the RSV attachment protein G, most of the antiviral antibodies that are in development to prevent or treat RSV target F, a trimeric type I membrane protein, which is well conserved and exists in a metastable prefusion or an energetically more favorable postfusion conformation (3–8). Based on secondary structure elements and the analysis of RSV antibody repertoires of human memory B cells, the surface of RSV F has been divided into six antigenic sites: Ø, I, II, III, IV, and V (9, 10). Most of the RSV-neutralizing activity in human serum is specifically directed against prefusion F (9, 11, 12). Antigenic sites Ø and V, present in the apex of prefusion F, are unique to the prefusion conformation, and monoclonal antibodies directed to these sites have potent RSV-neutralizing activity (50% infective concentration [IC50] of ≤50 ng/ml) (9). Site III recognition is also primarily associated with prefusion F-specific antibodies, even though the conformations of this site are almost identical in pre- and postfusion F (9, 13). The three-dimensional structure of antigenic sites I, II, and IV is largely conserved between the pre- and postfusion F conformations (13). While the neutralizing activity of antibodies directed at site II, III, or IV ranges from low to very high, site I-specific antibodies typically have poor RSV-neutralizing activity or even lack such activity (9, 14).
Previously, we described two RSV A- and B-neutralizing single-domain antibodies (VHHs) from a llama that had been immunized with recombinant, prefusion-stabilized F (7). These VHHs, F-VHH-4 and F-VHH-L66, can neutralize RSV in vitro at picomolar concentrations. F-VHH-4 and F-VHH-L66 are prefusion F specific and bind a highly conserved cavity between two F protomers and thereby partly cover antigenic sites II, III, and V on a first protomer and IV on a neighboring protomer.
We were interested in the identification of new RSV-neutralizing VHHs that are prefusion F specific and target an epitope that does not overlap that of F-VHH-4. Here, we report on the isolation, functional, and structural characterization of a new prefusion F-specific RSV-neutralizing VHH that binds to a unique epitope that is located in the lower part of prefusion F.
RESULTS
Isolation of a potent RSV-neutralizing VHH.
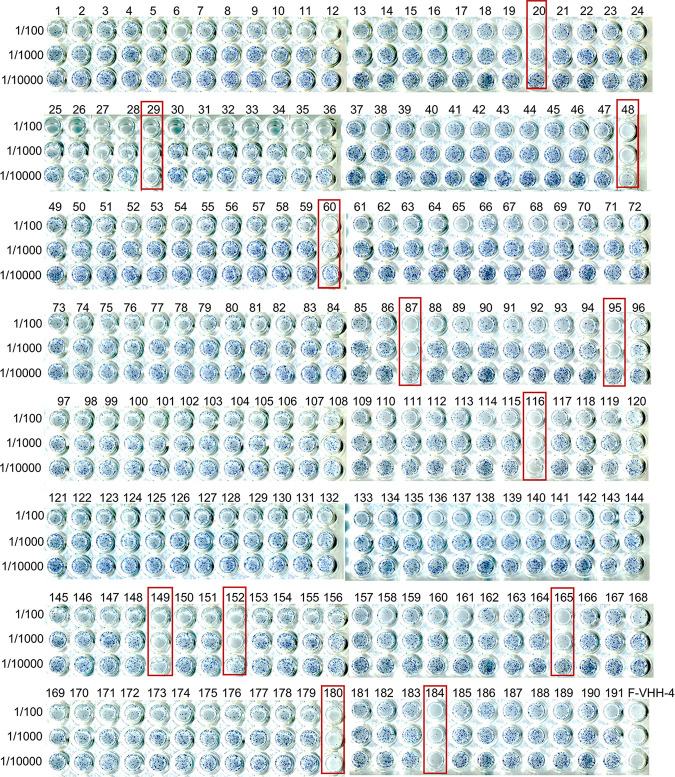
To investigate which sites on prefusion F, besides the F-VHH-4 epitope, are vulnerable for potent neutralizing VHHs, we aimed to identify new RSV-neutralizing VHHs that do not compete with F-VHH-4 for binding to prefusion F (7). Therefore, we screened a VHH-phage display library derived from a llama that had been immunized with recombinant F protein stabilized in the prefusion conformation (DS-Cav1) by panning on immobilized DS-Cav1 that had been presaturated with F-VHH-4 (15). We note that F-VHH-4 has an extremely low dissociation rate once bound to DS-Cav1 (7). After two rounds of panning on F-VHH-4-saturated DS-Cav1, we obtained a 242-fold enrichment of VHH-displaying phages. The enriched VHH cDNA inserts were subsequently cloned as a pool into a Pichia pastoris expression vector and the resulting library was used to transform P. pastoris cells. Next, the RSV-neutralizing activity of crude yeast culture medium of 191 individual P. pastoris transformants was determined in a 96-well-format plaque reduction assay. Several VHH candidates with modest to high RSV A2-neutralizing activity were identified this way (Fig. 1). Based on the apparent neutralizing activity, we selected 12 P. pastoris transformants that were expanded, and the respective VHHs were purified from the culture medium and then tested again in an RSV A2-neutralization assay (Fig. 2A). Six of the 12 candidates did not exhibit neutralizing activity in this assay. This could be explained by high expression of these clones in the initial screen, where there was no normalization for the amount of VHH present, and the low starting concentration used in this assay with purified VHHs. The most potent candidate, F-VHH-Cl184, was selected for further characterization. Although F-VHH-Cl184 does not reach the very high neutralizing activity of F-VHH-4, it is much more potent against RSV A2 than palivizumab (Fig. 2B and C). F-VHH-Cl184 can neutralize RSV A2, with an IC50 of 0.4 nM (5.0 ng/ml), but has only moderate neutralization activity against RSV B subtype strains, such as RSV B49, with an IC50 of 21.6 nM (305.3 ng/ml) (Fig. 2C). Unlike F-VHH-4, F-VHH-Cl184 lacks a cysteine residue in its complementarity-determining region 3 (CDR3), a feature often present in VHHs to help stabilize the extended CDR3 loop (Fig. 2D) (16).
FIG 1.
RSV-neutralizing activity in Pichia pastoris culture supernatants. VHHs were produced in 400-μl P. pastoris cultures in 96-deep-well plates. Ten-fold serial dilutions (from 1/100 to 1/10,000) of the cleared culture supernatants were tested for RSV A2-neutralizing activity in a 96-well-formatted plaque-reduction assay. Boxes indicate P. pastoris clones that secrete VHHs that may have neutralizing activity. Supernatants of F-VHH-4-expressing P. pastoris cells were included as a positive control.
FIG 2.
F-VHH-Cl184 can neutralize RSV A and B. (A) RSV A2 was incubated with different concentrations of VHH before infection of Vero cells. Three days later, the viral plaques were visualized with a polyclonal anti-RSV serum. A curve was fitted using a log versus response variable-slope 4-parameter equation. (B) RSV A2 or RSV B49 was preincubated with different concentrations of F-VHH-4, F-VHH-Cl184, or palivizumab before infection of Vero cells. Viral plaques were visualized 3 days later. Assays were performed twice; mean values with standard deviation (SD) (n = 2) based on the pooled data from 2 independently performed experiments are shown. A curve was fitted using a log versus response variable-slope 4-parameter equation. (C) IC50 values of F-VHHs and palivizumab against RSV A2 and RSV B49 as determined by plaque reduction assay. (D) Amino acid sequences of F-VHH-Cl184 and F-VHH-4 with Kabat numbering. The complementarity-determining regions (CDRs) are boxed. F-VHH-Cl184 CDRs were predicted by abysis.org. The amino acid sequence of F-VHH-4 was aligned to the sequence of F-VHH-Cl184.
F-VHH-Cl184 is prefusion specific.
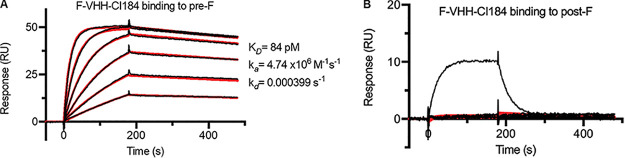
To determine the affinity as well as the preferred F conformation specificity of F-VHH-Cl184, we performed a comparative surface plasmon resonance (SPR) analysis with immobilized recombinant RSV A2-derived pre- and postfusion F (Fig. 3). This analysis showed that F-VHH-Cl184 binds tightly to prefusion F, with an equilibrium dissociation constant (KD) of 84 pM, an association rate constant (ka) of 4.74 × 106 M−1 s−1, and a dissociation rate constant (kd) of 3.99 × 10−4 s−1 (Fig. 3A). Binding to postfusion F could not be detected, indicating that F-VHH-Cl184 is prefusion F specific (Fig. 3B).
FIG 3.
F-VHH-Cl184 is specific for prefusion F. Shown are surface plasmon resonance (SPR) sensorgrams for the binding of F-VHH-Cl184 to immobilized prefusion F (A) and postfusion F (B). Two-fold serial dilutions of F-VHH-Cl184 ranging from 12.5 nM to 0.39 nM in HBS-P+ buffer were injected over the prefusion F and reference control cells. For binding experiments to postfusion F, 2-fold serial dilutions of F-VHH-Cl184 ranging from 100 nM to 3.13 nM were used. ADI-14359 (100 nM), a postfusion F-specific Fab, was included as a positive control for binding to postfusion F (black line). Binding data are shown as black lines, and the best fit to a 1:1 binding model is shown as red lines.
F-VHH-Cl184 binds to a prefusion-specific, membrane-proximal epitope in F.
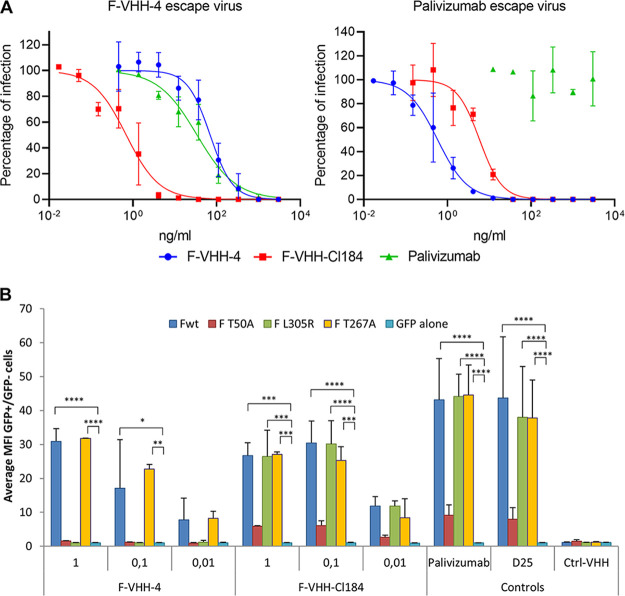
To confirm that F-VHH-Cl184 binds to an epitope that is different from that of F-VHH-4, we performed a set of experiments using various F variants harboring single amino acid substitutions within the F-VHH-4 epitope. We previously described the isolation of an F-VHH-4 RSV A in vitro escape variant, which was attenuated and carried a Thr-to-Asn substitution at position 50 in F (7). F-VHH-Cl184 could neutralize this F-VHH-4 escape virus as efficiently as wild-type (WT) RSV A2 (Fig. 2B and Fig. 4A). Also, a palivizumab escape virus with a Lys272Asn substitution in antigenic site II was neutralized equally well as WT RSV A2 by F-VHH-Cl184 (Fig. 2B and Fig. 4A). These results suggest that neither Thr50 nor Lys272 is involved in the binding of F-VHH-Cl184 to F. We next determined the binding of F-VHH-Cl184 to a set of F variants that were expressed on the surface of transfected cells by flow cytometry. F variants Leu305Arg and Thr267Ala were selected because these are located in the epitope of F-VHH-4. The site II-binding palivizumab and the site Ø-binding D25 were included as positive controls. F-VHH-Cl184 could efficiently bind to the three F variants (Thr50Ala, Leu305Arg, and Thr267Ala), while F-VHH-4 failed to bind to the Thr50Ala and Leu305Arg variants (Fig. 4B). Together, these results show that F-VHH-Cl184 can neutralize in vitro selected RSV A2 variants that are poorly neutralized by palivizumab and F-VHH-4, as efficiently as WT RSV A2.
FIG 4.
F-VHH-Cl184 can neutralize RSV viruses that are resistant to palivizumab or partially resistant to F-VHH-4. (A) F-VHH-4 escape virus (left panel) or palivizumab escape virus (right panel) was preincubated with the indicated concentrations of VHH or palivizumab before infection of Vero cells. Three days later, the viral plaques were stained with polyclonal anti-RSV serum. Assays were performed twice; mean values with standard deviation (n = 2) based on the pooled data from 2 independently performed experiments are shown. A curve was fitted using a log versus response variable-slope 4-parameter equation. (B) HEK293T cells were transfected with the wild type (Fwt) or T50A, L305R, or T267A mutant RSV F expression plasmid in combination with a green fluorescent protein-nuclear localization signal expression vector (peGFP-NLS) or with the peGFP-NLS expression vector combined with an empty expression vector. Cells were subsequently stained with a dilution series of F-VHH-4 or F-VHH-Cl184 or the indicated control antibodies and analyzed by flow cytometry. The graphs show for each VHH the average ± SD of the ratio of the median fluorescence intensity (MFI) of the AF-633-labeled VHH detection antibody bound to GFP+ cells over the mean fluorescent intensity (MFI) of GFP− cells that were stained with the indicated VHH concentration in μg/ml (or at 1 μg/ml for the controls) (n = 2). *, P < 0.05, **, P < 0.01, ***, P < 0.001, and ****, P < 0.0001, by two-way ANOVA.
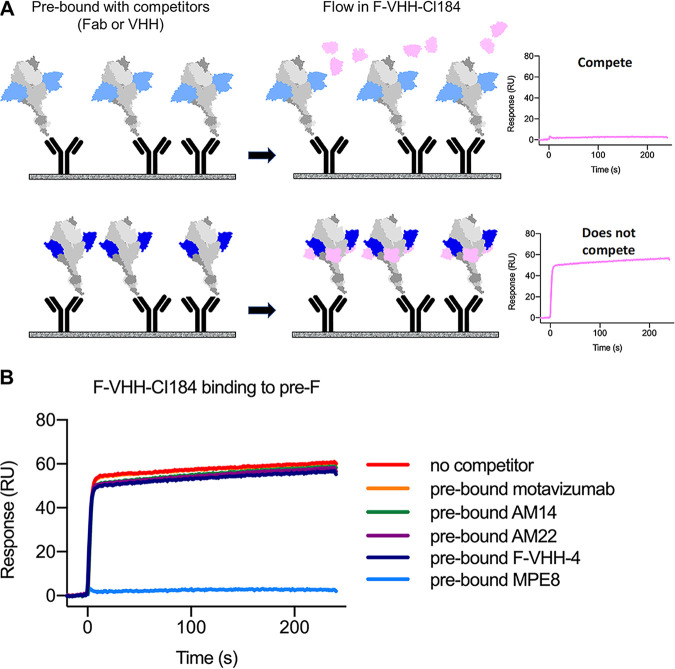
Our neutralization and F-binding experiments are in line with the assumption that F-VHH-Cl184 and F-VHH-4 bind to different epitopes in prefusion F and validate our panning approach. We next performed SPR-based competition experiments to try to confirm this assumption. In this experiment, immobilized prefusion F was first saturated with a competitor, and then responses resulting from F-VHH-Cl184 binding were recorded by SPR (Fig. 5A). This revealed that presaturation of immobilized prefusion F with F-VHH-4 did not interfere with the binding of F-VHH-Cl184 to prefusion F, indicating that F-VHH-Cl184 and F-VHH-4 bind nonoverlapping epitopes (Fig. 5B). A variety of Fabs, including Fabs derived from motavizumab (site II), AM14 (sites IV and V), AM22 (site Ø), and MPE8 (sites II, III, IV, and V), as well as F-VHH-4 (sites II, III, IV, and V), were also used to evaluate potential competition with F-VHH-CI184 for binding to prefusion F (7, 17–21). Interestingly, only Fabs derived from MPE8, a human monoclonal antibody that can neutralize RSV as well as human metapneumovirus, prevented binding of F-VHH-Cl184 (Fig. 5B) (21). This suggests that F-VHH-CI184 might bind to a site that overlaps or is in proximity to antigenic site III, the binding site of MPE8.
FIG 5.
F-VHH-Cl184 competes with MPE8 Fab. (A) Schematic diagram of the SPR-based competition assay. Prefusion F (gray) is first saturated with a competitor Fab or VHH (blue), and then additional responses from F-VHH-Cl184 (pink) binding are measured through SPR. (B) SPR sensorgrams for the binding of F-VHH-Cl184 to prefusion F in the presence of the indicated Fabs or F-VHH-4.
To narrow down the likely epitope of F-VHH-Cl184, we selected for in vitro escape viruses. After four rounds of selection of RSV A Long in the presence of the F-VHH-Cl184, four escape viruses were selected. Remarkably, these viruses all contained mutations that led to a change of the asparagine residue at position 380 (Asn380Ser, Asn380Ile, Asn380Thr, and Asn380Tyr), which is part of antigenic site I. These mutant viruses remained susceptible to neutralization by F-VHH-4 and palivizumab but completely (Asn380Ile, Asn380Thr, and Asn380Tyr) or partially (Asn380Ser) resist F-VHH-Cl184 at a concentration of 3 μg/ml (Fig. 6).
FIG 6.

Neutralization of F-VHH-Cl184 escape RSV strains. RSV A2 or the different escape viruses were preincubated with different concentrations of the indicated VHH or palivizumab before addition to a monolayer of Vero cells. Three days later, cells were fixed and stained with polyclonal anti-RSV serum to visualize the plaques. A curve was fitted using a log versus response variable-slope 4-parameter equation.
To define the F-VHH-Cl184 epitope, we determined the crystal structures of F-VHH-Cl184 alone and in complex with prefusion F. The crystal structures of F-VHH-Cl184 alone was determined to 1.9-Å resolution, whereas the structure of the complex was determined to 2.1-Å resolution (Fig. 7A and Table 1). F-VHH-Cl184 buries approximately 904 Å2 of surface area on the membrane-proximal region of the globular prefusion F ectodomain. The epitope of the VHH is confined to one protomer and largely overlaps antigenic site I, the conformation of which is conserved in pre- and postfusion F (13). While all three CDRs mainly contact residues on antigenic site I, CDR1 also reaches to the edge of antigenic site III (Fig. 7B). This region partially overlaps the MPE8 epitope, which explains why MPE8 competes with F-VHH-Cl184 binding to prefusion F (Fig. 7C). The residues in CDR1 and CDR3 contribute to the majority of the polar interactions with F. For example, Tyr32 from CDR1 interacts with Asp344 via a hydrogen bond. On the other hand, Trp52a from CDR2 forms hydrophobic interactions with Leu381, Val384, Pro389, and Lys390 from antigenic site I. CDR3 also contacts the β22 strand that is part of the neighboring antigenic site IV (Fig. 7D). Two hydrophobic residues of F-VHH-Cl184 CDR3, Val100, and Trp100e are neatly packed in a small cavity between antigenic sites I and IV, and Glu100d forms a salt bridge with Lys465 at β22 (Fig. 7D). The prefusion F specificity of F-VHH-Cl184 most likely originates from this contact with β22, as this strand changes conformation considerably during the transition of F from the prefusion to the postfusion state. Our structural analysis also revealed that the epitope of F-VHH-Cl184 is poorly conserved between RSV A and B, explaining the reduced neutralizing activity of the VHH against RSV B. Indeed, position 380 in F of RSV B viruses is a serine instead of an asparagine, which leads to the loss of the hydrogen bonds with CDR1 Gly31 and CDR3 Gly97. The importance of this residue for the binding of F-VHH-Cl184 to F is in line with those of the selected escape mutants, which all had an amino acid substitution at Asn380. Finally, superimposition of F-VHH-4 on F-VHH-Cl184-bound prefusion F structure showed that the epitopes of both VHHs do not overlap and that simultaneous binding is possible without any clashes, indicating that prefusion F can be bound by both VHHs simultaneously (Fig. 7E).
FIG 7.
The epitope of F-VHH-Cl184 is mainly located in antigenic site I, contacts sites III and IV, and does not overlap F-VHH-4. (A) F-VHH-Cl184 (blue) bound to prefusion RSV F as viewed from the side. (B) Close-up of the VHH-binding site as viewed from the side (left) and from the bottom (right). The key hydrophobic VHH residues on the CDRs are highlighted. (C) The epitope of F-VHH-Cl184 partially overlaps the epitope of MPE8. Shown is the structure of prefusion F bound to F-VHH-Cl184 along with the footprints of the Fabs of AM22 (red), AM14 (orange), motavizumab (yellow), MPE8 (green), and F-VHH-Cl184 (blue). (D) F-VHH-Cl184 also contacts antigenic site IV. A close-up of the VHH-binding site is shown; residues of the RSV F protein contacted by the VHH are labeled. The key residues of VHH CDRs are labeled in blue. The β22 strand is shown in dark green. (E) Prefusion RSV F in complex with F-VHH-4 (magenta) and F-VHH-Cl184 (blue).
TABLE 1.
Crystallographic data collection and refinement statistics
| Parameter | Value fora: |
|
|---|---|---|
| F-VHH-CI184 | F-VHH-CI184-prefusion F | |
| PDB ID | 7LVU | 7LVW |
| Data collection statistics | ||
| Space group | P4322 | P21 |
| Cell constants | ||
| a, b, c (Å) | 73.6, 73.6, 222.0 | 104.5, 200.5, 134.7 |
| α, β, γ (°) | 90, 90, 90 | 90, 105.9, 90 |
| Wavelength (Å) | 0.9774 | 1.0332 |
| Resolution (Å) | 73.9–1.9 (1.99–1.94) | 89.9–2.1 (2.14–2.10) |
| No. of reflections: | ||
| Total | 504,205 (19,524) | 993,485 (50,385) |
| Unique | 46,193 (2,987) | 301,087 (15,113) |
| Rmerge | 0.098 (0.802) | 0.083 (0.696) |
| Rpim | 0.031 (0.331) | 0.054 (0.447) |
| I/σ〈I〉 | 17.1 (2.4) | 8.4 (1.5) |
| CC1/2 | 0.998 (0.814) | 0.989 (0.568) |
| Completeness (%) | 99.9 (98.2) | 97.4 (98.3) |
| Redundancy | 10.9 (6.5) | 3.3 (3.3) |
| Refinement statistics | ||
| Resolution (Å) | 73.6–1.94 (2.01–1.94) | 50.2–2.1 (2.12–2.10) |
| No. of unique reflections | 46,099 | 301,032 |
| Rwork/Rfree (%) | 20.8/23.2 | 20.2/23.1 |
| No. of atoms | ||
| Protein | 3,730 | 28,340 |
| Ligand/ion | 0 | 86 |
| Water | 354 | 2,254 |
| B-factors | ||
| Protein | 29.6 | 43.9 |
| Ligand/ion | 56.4 | |
| Water | 36.9 | 43.8 |
| RMSDb | ||
| Bond length (Å) | 0.007 | 0.006 |
| Bond angle (°) | 0.92 | 0.85 |
| Ramachandran plot (%) | ||
| Favored | 99.2 | 96.5 |
| Allowed | 0.8 | 3.4 |
| Outliers | 0.0 | 0.2 |
Values in parentheses are for the highest-resolution shell.
RMSD, root mean square deviation.
DISCUSSION
In this study, we describe the isolation and characterization of an RSV-neutralizing VHH that binds to a unique prefusion F-specific epitope. This VHH was isolated from a previously described VHH library (7). Our panning strategy on immobilized prefusion F that was presaturated with F-VHH-4 was successful for the isolation of F-VHH-Cl184, which binds to an epitope that is distinct from that of F-VHH-4. F-VHH-Cl184 displays potent RSV A-neutralizing activity and binds specifically and with high affinity to prefusion F. The epitope of F-VHH-Cl184 has not been described before and mainly overlaps with antigenic site I, with additional interactions with antigenic sites III and IV. In the screens described here, no VHHs with a neutralizing potency as high as that of F-VHH-4 or F-VHH-L66 were identified. F-VHH-Cl184 is approximately 10 times less potent than F-VHH-4, yet it still reaches subnanomolar RSV-neutralizing activity against RSV A2 and, in line with the elucidated epitope, shows a 540-fold-lower neutralizing activity (IC50, 21.6 nM) for RSV B.
The specificity of F-VHH-Cl184 for prefusion F is surprising because its epitope largely overlaps antigenic site I, which is often considered to be a postfusion F-specific site, although the site is largely conserved during the transformation from prefusion to postfusion F (10, 13, 22). The membrane-proximal position of this site in prefusion F probably results in limited accessibility for conventional antibodies, while the site is readily accessible at the tip of postfusion F. Previously described antibodies that bind to antigenic site I, such as monoclonal antibodies 131-2a and 2F, preferentially bind postfusion F and are poorly neutralizing (10, 23, 24). The (poor) neutralization of RSV by these antibodies could be brought about by steric hindrance, limiting the accessibility of the virus to the host cell (24). F-VHH-Cl184, however, can potently neutralize RSV A2 and is specific for prefusion F. Residues in CDR3 make critical polar and hydrophobic contacts with the β22 strand located in antigenic site IV, which undergoes a considerable transformation upon refolding of the F protein. Therefore, we speculate that F-VHH-Cl184 neutralizes RSV by locking β22 in a prefusion conformation and thus prevents the transformation to postfusion F.
Despite several variations between RSV A and B in the epitope of F-VHH-Cl184, the VHH can still neutralize RSV B, although less potently. This reduced neutralizing activity can be traced back to one amino acid residue difference between RSV A and B, Asn380, which is a serine in RSV B. Four different RSV A2 escape mutants contain a mutation leading to a substitution of Asn380, including the Ser corresponding to RSV B. Remarkably, RSV B is still much more susceptible to neutralization by F-VHH-Cl184 than the RSV A mutant that has the single Asn380Ser change. Therefore, other variations in RSV B, either inside or outside the F-VHH-Cl184 epitope, may compensate for this serine. In addition, bivalent F-VHH-Cl184 constructs—for example, by fusion with an antibody Fc domain—could potentially have increased RSV B neutralizing potency.
The neutralizing epitope of F-VHH-Cl184 is relatively novel since it does not overlap the epitopes of F-VHH-4 or several neutralizing antibodies that recognize site Ø (AM22), site V (AM14), or site II (motavizumab). The epitope of F-VHH-Cl184 partially overlaps that of MPE8. Antigenic site I has not yet been associated with prefusion F-specific antibodies, although such antibodies are detected in human serum in low levels (9). Yet, in serum no highly neutralizing antibodies that bind to this site could be detected. Possibly, neutralizing antibodies directed to sites located at the membrane-proximal region of prefusion F can be more readily elicited by immunization with prefusion F than by RSV infection. This could mean that a more diverse anti-F antibody response could be evoked by immunization with F outside its viral context, such as with the DS-Cav1 prefusion F subunit vaccine candidate, which has recently been evaluated in a phase I clinical trial (25). It still needs to be assessed if an increased antibody response against the membrane-proximal region of RSV F would also result in better protection.
The reduced neutralizing activity against RSV B is a limitation for the possible development of F-VHH-Cl184 as a therapeutic. F-VHH-4 and -L66 escape viruses, which are attenuated in vitro, have been isolated. A combination of F-VHH-Cl184 with, e.g., F-VHH-4, or even merged in a single molecule could potentially lower the risk even further that escape viruses emerge upon the usage of F-VHH-4 for the treatment of RSV-associated disease. In line with this, it has been shown for rabies virus, Zika virus, simian HIV, and, more recently, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), that a cocktail of two antibodies that bind to nonoverlapping epitopes can mitigate such a risk (26–29). Further research is necessary to verify if such a biparatopic construct is indeed advantageous compared to the already very potent F-VHH-4 alone.
MATERIALS AND METHODS
Generation of prefusion F-specific VHH.
A library of VHH-displaying phages was synthesized from the lymphocytes of a llama immunized with prefusion F protein (DS-Cav1) as described in reference 7. This library was subjected to two rounds of panning on prefusion F protein, captured with antifoldon antibody (100 ng of MF4 [30]), which was presaturated with equimolar amounts of F-VHH-4 in one well of a 96-well microtiter plate (Maxisorp; Nunc). Panning was performed in the same way as described before (7). Sea Block blocking buffer (Thermo Scientific) was used during the first panning round, Pierce protein-free blocking buffer (Thermo Scientific) was used during the second panning round.
Expression of VHHs in Pichia pastoris.
The complete VHH library after the second panning round was cloned as a single pool into a yeast expression vector. The VHH coding sequences were PCR amplified from the pHEN4 plasmid library using the respective forward and reverse primers 5′-GGGCTCTTCAAGGCAGGTGCAGCTGCAGGAGTC-3′ and 5′-GGGCTCTTCAAGTCTAGTGATGGTGATGGTGGTGGCTGGAGACGGTGACCTGGG-3′. The resulting PCR product was mixed with pKai61 plasmid (described in reference 7, modified to include SapI restriction sites), SapI, and T4 DNA ligase, and a thermocycled restriction-ligation reaction was performed. The resulting library was linearized with PmeI before transformation of P. pastoris strain GS115 using the lithium acetate-dithiothreitol (DTT) method described by Wu and Letchworth (31).
Purification of VHHs produced by Pichia pastoris.
Expression of VHHs by single P. pastoris transformants for the RSV neutralization screening was performed in 400-μl cultures in 96-deep-well plates. Production was performed as described before, adapted to the reduced culture volume (7). P. pastoris transformants that produced RSV-neutralizing VHHs were selected for scaling up using 4-, 25-, or 100-ml cultures. Purification was performed as described before (7).
Cells and viruses.
Vero cells (ATCC CCL-81) and HEp-2 cells (ATCC CCL-23) were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum (FCS), 2 mM l-glutamine, nonessential amino acids (Invitrogen, Carlsbad, CA, USA), and 1 mM sodium pyruvate at 37°C in the presence of 5% carbon dioxide. RSV A2, an A subtype of RSV (ATCC VR-1540; ATCC, Gaithersburg, MD), RSV B49, a B subtype of RSV (BE/5649/08 [described in reference 32]), RSV A Long (ATCC VR-26 [a kind gift from Rik De Swart, Erasmus MC, Rotterdam, The Netherlands]), and palivizumab-escape and F-VHH-4-escape RSV mutants (described in reference 7) were propagated on HEp-2 cells and quantified on Vero cells by plaque assay using goat anti-RSV serum (AB1128; Chemicon International).
Plaque reduction assay.
The plaque reduction assay was performed as described previously (7). In short, a dilution series of the VHHs, palivizumab, or raw yeast supernatant was prepared in Opti-MEM (Gibco), incubated with RSV for 30 min at 37°C, and used to infect confluent Vero cells. After 3 h, an equal volume of 1.2% Avicel RC-591 and growth medium was added to each well, and the infection was allowed to continue for 3 days. Viral infection was tested by immunostaining of the viral plaques with goat anti-RSV serum (AB1128; Chemicon International) and horseradish peroxidase-conjugated anti-goat IgG (SC2020; Santa Cruz). The plaques were visualized by applying TrueBlue peroxidase substrate (KPL, Gaithersburg, MD).
VHH binding to cells expressing F.
HEK293T cells were transfected with pCAGGS containing a codon-optimized WT, T50A, L305R, or T267A mutant RSV F cDNA with polyethyleneimine (PEI) transfection reagent (Polysciences, Inc.). To trace transfected cells, transfections were performed in the presence of peGFP-NLS. Control transfections were performed with peGFP-NLS and an empty pCAGGS vector. Twenty-four hours after transfection, the cells were detached, washed, and blocked. Subsequently, the cells were incubated with 1, 0.1, or 0.01 μg/ml of VHH or 1 μg/ml control VHH (specific for an irrelevant target) or MAb in phosphate-buffered saline (PBS) with 0.5% bovine serum albumin (BSA). One hour later, the cells were washed and stained with mouse anti-histidine tag antibody (MCA1396; Abd Serotec) followed by anti-mouse IgG conjugated with Alexa 633 (Invitrogen) for the VHH samples or anti-human IgG conjugated with Alexa 633 (Invitrogen) for the MAb samples. The stained cells were analyzed using a BD LSRII flow cytometer. All procedures were performed on ice or at 4°C.
Surface plasmon resonance.
The surface plasmon resonance (SPR) experiment was performed as described previously, with some modifications using a Biacore X100 (GE Healthcare) (7). In each cycle, either purified prefusion F (DS-Cav1) or postfusion F (FΔTM) was captured to ∼400 response units (RU) by a CM5 sensor chip that had been immobilized with an anti-Strep·TagII Fab. The sensor chip was regenerated between cycles using 0.1% SDS followed by 10 mM glycine at pH 2. Serial 2-fold dilutions of F-VHH-CI184 from 12.5 nM to 0.39 nM were injected through reference flow cells and then prefusion F-coated cells in HBS-P+ buffer (0.01 M HEPES, pH 7.4, 0.15 M NaCl, 0.05 [vol/vol] surfactant P20). The buffer-only control was also injected through both flow cells as a reference. For the binding experiment to postfusion F, 2-fold dilutions of F-VHH-CI184 from 100 nM to 3.13 nM were used. ADI-14359, a postfusion F-specific Fab, was also included as a positive control (14). The data were double-reference subtracted and fit to a 1:1 binding model using BIAevaluation software.
Competition assay with SPR.
Competition experiments were conducted by saturating captured DS-Cav1 with a 100 nM (each) set of Fabs (AM14, AM22, motavizumab, and MPE8) or F-VHH-4 for 300 s followed by 100 nM F-VHH-CI184 for 240 s. The additional resonance response (RU) generated by F-VHH-CI184 binding was measured by a Biacore X100 (GE Healthcare).
RSV escape mutant analysis.
RSV A Long (10,000 PFU) was mixed with a dose of palivizumab or F-VHH-Cl184 corresponding to 10× and 100× the IC50 and added to confluent HEp-2 cells in a 96-well plate. This was performed in triplicate. After 7 days of infection, an aliquot of the supernatant was transferred to infect a subsequent series of HEp-2 cells in the presence of 10× and 100× the IC50 of palivizumab or F-VHH-Cl184 to continue the selection. After every round, a separate aliquot of supernatant was transferred to fresh HEp-2 cells and mixed with a dose of 500× IC50 of palivizumab or F-VHH-Cl184. These cells were stained after 3 days of infection. After four rounds of selection, clonal RSV isolates were obtained by serial dilution and then amplified on Vero cells. Total RNA was isolated from the resulting virus stocks, cDNA was prepared, and the F coding information was amplified and sequenced.
Expression and purification of F-VHH-CI184 and prefusion F.
To express F-VHH-Cl184 on a large scale, the pαH vector-based construct containing F-VHH-Cl184 with a C-terminal His tag was transfected into 1 liter of Freestyle 293-F cells at 37°C for 6 days. The protein was then purified from cell supernatants using Ni-nitrilotriacetic acid (NTA) resin, and the concentrated eluate was further applied to a Superdex 200 column in 2 mM Tris buffer at pH 8 with 200 mM NaCl and 0.02% NaN3. To prepare F-VHH-Cl184-F complex, two plasmids, F-VHH-Cl184 without tags, and PR-DM (prefusion-stabilized F) with a C-terminal 3C cleavage site and Strep tag, were cotransfected to Freestyle 293-F cells (33). After 6 days of transfection, the supernatant containing the protein complex was purified with a Strep-Tactin resin (IBA Lifesciences). The coeluted complex was then treated with human rhinovirus (HRV) 3C protease prior to further purification using a Superdex 200 column in 2 mM Tris buffer at pH 8 with 200 mM NaCl and 0.02% NaN3. All proteins were concentrated and stored at −80°C.
Crystallization and data collection.
F-VHH-CI184 crystals were produced by sitting-drop vapor diffusion by mixing 100 nl of F-VHH-CI184 (22 mg/ml) with 100 nl of reservoir solution containing 0.1 M HEPES (pH 7.5) and 2.0 M ammonium formate. Crystals were soaked in reservoir supplemented with 20% glycerol and frozen in liquid nitrogen. Diffraction data were collected to 1.94 Å at SBC beamline 5.0.2 (Advanced Light Source, Lawrence Berkeley National Laboratory). Initial crystals of F-VHH-CI184 in complex with PR-DM were grown by sitting-drop vapor diffusion by mixing 100 nl of the complex (4.4 mg/ml) with 100 nl of reservoir solution containing 0.2 M ammonium citrate (pH 4.5), 12.5% (vol/vol) isopropanol, and 21% (wt/vol) polyethylene glycol 3350 (PEG 3350). A single crystal that diffracted to 2.1 Å was obtained by seeding microcrystals from the initial condition into a reservoir solution containing 0.1 M HEPES (pH 7.5) and 25% PEG MME2K. Diffraction data were collected at the SBC beamline 19ID (Advanced Photon Source, Argonne National Laboratory).
Structure determination.
All data were indexed and integrated in iMOSFLM and scaled and merged using AIMLESS. F-VHH-CI184 crystals formed in space group P4322. A homologous VHH structure (PDB ID 5VAK) was used as a search model for molecular replacement solution using PHASER. Each asymmetric unit contains four molecules of VHH. After manual model building in Coot, the structure was refined in PHENIX to Rwork/Rfree values of 20.8%/23.2%.
The F-VHH-CI184 in complex with PR-DM formed crystals in P21 space groups. The unbound PR-DM structure (PDB ID 5C69) was initially used as a search model for molecular replacement solution, and VHH-CI184 was included as another assembly to search for the complex in the second run of molecular replacement using PHASER. Each asymmetric unit contained two molecules of the complex. After iterative model building in Coot and refinement in PHENIX, the structure was refined to Rwork/Rfree values of 20.2%/23.1%. Complete data collection and refinement statistics are presented in Table 1.
Data availability.
The structures of RSV F-directed VHH Cl184 and RSV F in complex with VHH Cl184 are available in the Protein Data Bank under PDB IDs 7LVU and 7LVW, respectively.
ACKNOWLEDGMENTS
This study was supported by the Industrial Research Fund of Ghent University (F2016/IOF-Advanced/393). I.R. is a postdoctoral fellow supported by FWO-Vlaanderen, B.S. was supported by FWO-EOS project VIREOS, and K.S. was supported by research project BOF17-GOA-018 from Ghent University.
We thank the staff of the VIB Flow Core for providing access to flow cytometry equipment. We thank Vicente Mas for kindly providing us with the MF4 antifoldon antibody. We acknowledge the team of the VIB Nanobody Service Facility for their services. We also thank Morgan Gilman for providing thoughtful advice and feedback. Results shown in this article are derived in part from work performed at Argonne National Laboratory (ANL), the Structural Biology Center (SBC) at the Advanced Photon Source (APS), under U.S. Department of Energy, Office of Biological and Environmental Research, contract DE-AC02-06CH11357. In addition, the Advanced Light Source is a Department of Energy Office of Science User Facility under contract no. DE-AC02-05CH11231.
We declare that no competing interests exist.
REFERENCES
- 1.Shi T, McAllister DA, O'Brien KL, Simoes EAF, Madhi SA, Gessner BD, Polack FP, Balsells E, Acacio S, Aguayo C, Alassani I, Ali A, Antonio M, Awasthi S, Awori JO, Azziz-Baumgartner E, Baggett HC, Baillie VL, Balmaseda A, Barahona A, Basnet S, Bassat Q, Basualdo W, Bigogo G, Bont L, Breiman RF, Brooks WA, Broor S, Bruce N, Bruden D, Buchy P, Campbell S, Carosone-Link P, Chadha M, Chipeta J, Chou M, Clara W, Cohen C, de Cuellar E, Dang DA, Dash-Yandag B, Deloria-Knoll M, Dherani M, Eap T, Ebruke BE, Echavarria M, de Freitas Lazaro Emediato CC, Fasce RA, Feikin DR, Feng L, RSV Global Epidemiology Network, et al. 2017. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: a systematic review and modelling study. Lancet 390:946–958. 10.1016/S0140-6736(17)30938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The IMPACT-RSV Study Group. 1998. Palivizumab, a humanized respiratory syncytial virus monoclonal antibody, reduces hospitalization from respiratory syncytial virus infection in high-risk infants. Pediatrics 102:531–537. 10.1542/peds.102.3.531. [DOI] [PubMed] [Google Scholar]
- 3.Griffin MP, Yuan Y, Takas T, Domachowske JB, Madhi SA, Manzoni P, Simoes EAF, Esser MT, Khan AA, Dubovsky F, Villafana T, DeVincenzo JP, Nirsevimab Study Group. 2020. Single-dose nirsevimab for prevention of RSV in preterm infants. N Engl J Med 383:415–425. 10.1056/NEJMoa1913556. [DOI] [PubMed] [Google Scholar]
- 4.Collarini EJ, Lee FE, Foord O, Park M, Sperinde G, Wu H, Harriman WD, Carroll SF, Ellsworth SL, Anderson LJ, Tripp RA, Walsh EE, Keyt BA, Kauvar LM. 2009. Potent high-affinity antibodies for treatment and prophylaxis of respiratory syncytial virus derived from B cells of infected patients. J Immunol 183:6338–6345. 10.4049/jimmunol.0901373. [DOI] [PubMed] [Google Scholar]
- 5.Han J, Takeda K, Wang M, Zeng W, Jia Y, Shiraishi Y, Okamoto M, Dakhama A, Gelfand EW. 2014. Effects of anti-G and anti-F antibodies on airway function after respiratory syncytial virus infection. Am J Respir Cell Mol Biol 51:143–154. 10.1165/rcmb.2013-0360OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O'Brien KL, Chandran A, Weatherholtz R, Jafri HS, Griffin MP, Bellamy T, Millar EV, Jensen KM, Harris BS, Reid R, Moulton LH, Losonsky GA, Karron RA, Santosham M, Respiratory Syncytial Virus Prevention Study Group. 2015. Efficacy of motavizumab for the prevention of respiratory syncytial virus disease in healthy Native American infants: a phase 3 randomised double-blind placebo-controlled trial. Lancet Infect Dis 15:1398–1408. 10.1016/S1473-3099(15)00247-9. [DOI] [PubMed] [Google Scholar]
- 7.Rossey I, Gilman MS, Kabeche SC, Sedeyn K, Wrapp D, Kanekiyo M, Chen M, Mas V, Spitaels J, Melero JA, Graham BS, Schepens B, McLellan JS, Saelens X. 2017. Potent single-domain antibodies that arrest respiratory syncytial virus fusion protein in its prefusion state. Nat Commun 8:14158. 10.1038/ncomms14158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang A, Chen Z, Cox KS, Su HP, Callahan C, Fridman A, Zhang L, Patel SB, Cejas PJ, Swoyer R, Touch S, Citron MP, Govindarajan D, Luo B, Eddins M, Reid JC, Soisson SM, Galli J, Wang D, Wen Z, Heidecker GJ, Casimiro DR, DiStefano DJ, Vora KA. 2019. A potent broadly neutralizing human RSV antibody targets conserved site IV of the fusion glycoprotein. Nat Commun 10:4153. 10.1038/s41467-019-12137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilman MS, Castellanos CA, Chen M, Ngwuta JO, Goodwin E, Moin SM, Mas V, Melero JA, Wright PF, Graham BS, McLellan JS, Walker LM. 2016. Rapid profiling of RSV antibody repertoires from the memory B cells of naturally infected adult donors. Sci Immunol 1:eaaj1879. 10.1126/sciimmunol.aaj1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lopez JA, Bustos R, Orvell C, Berois M, Arbiza J, Garcia-Barreno B, Melero JA. 1998. Antigenic structure of human respiratory syncytial virus fusion glycoprotein. J Virol 72:6922–6928. 10.1128/JVI.72.8.6922-6928.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Magro M, Mas V, Chappell K, Vazquez M, Cano O, Luque D, Terron MC, Melero JA, Palomo C. 2012. Neutralizing antibodies against the preactive form of respiratory syncytial virus fusion protein offer unique possibilities for clinical intervention. Proc Natl Acad Sci U S A 109:3089–3094. 10.1073/pnas.1115941109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ngwuta JO, Chen M, Modjarrad K, Joyce MG, Kanekiyo M, Kumar A, Yassine HM, Moin SM, Killikelly AM, Chuang GY, Druz A, Georgiev IS, Rundlet EJ, Sastry M, Stewart-Jones GB, Yang Y, Zhang B, Nason MC, Capella C, Peeples ME, Ledgerwood JE, McLellan JS, Kwong PD, Graham BS. 2015. Prefusion F-specific antibodies determine the magnitude of RSV neutralizing activity in human sera. Sci Transl Med 7:309ra162. 10.1126/scitranslmed.aac4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rossey I, McLellan JS, Saelens X, Schepens B. 2018. Clinical potential of prefusion RSV F-specific antibodies. Trends Microbiol 26:209–219. 10.1016/j.tim.2017.09.009. [DOI] [PubMed] [Google Scholar]
- 14.Goodwin E, Gilman MSA, Wrapp D, Chen M, Ngwuta JO, Moin SM, Bai P, Sivasubramanian A, Connor RI, Wright PF, Graham BS, McLellan JS, Walker LM. 2018. Infants infected with respiratory syncytial virus generate potent neutralizing antibodies that lack somatic hypermutation. Immunity 48:339–349.e5. 10.1016/j.immuni.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McLellan JS, Chen M, Joyce MG, Sastry M, Stewart-Jones GB, Yang Y, Zhang B, Chen L, Srivatsan S, Zheng A, Zhou T, Graepel KW, Kumar A, Moin S, Boyington JC, Chuang GY, Soto C, Baxa U, Bakker AQ, Spits H, Beaumont T, Zheng Z, Xia N, Ko SY, Todd JP, Rao S, Graham BS, Kwong PD. 2013. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science 342:592–598. 10.1126/science.1243283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vu KB, Ghahroudi MA, Wyns L, Muyldermans S. 1997. Comparison of llama VH sequences from conventional and heavy chain antibodies. Mol Immunol 34:1121–1131. 10.1016/s0161-5890(97)00146-6. [DOI] [PubMed] [Google Scholar]
- 17.Wu H, Pfarr DS, Johnson S, Brewah YA, Woods RM, Patel NK, White WI, Young JF, Kiener PA. 2007. Development of motavizumab, an ultra-potent antibody for the prevention of respiratory syncytial virus infection in the upper and lower respiratory tract. J Mol Biol 368:652–665. 10.1016/j.jmb.2007.02.024. [DOI] [PubMed] [Google Scholar]
- 18.Gilman MS, Moin SM, Mas V, Chen M, Patel NK, Kramer K, Zhu Q, Kabeche SC, Kumar A, Palomo C, Beaumont T, Baxa U, Ulbrandt ND, Melero JA, Graham BS, McLellan JS. 2015. Characterization of a prefusion-specific antibody that recognizes a quaternary, cleavage-dependent epitope on the RSV fusion glycoprotein. PLoS Pathog 11:e1005035. 10.1371/journal.ppat.1005035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwakkenbos MJ, Diehl SA, Yasuda E, Bakker AQ, van Geelen CM, Lukens MV, van Bleek GM, Widjojoatmodjo MN, Bogers WM, Mei H, Radbruch A, Scheeren FA, Spits H, Beaumont T. 2010. Generation of stable monoclonal antibody-producing B cell receptor-positive human memory B cells by genetic programming. Nat Med 16:123–128. 10.1038/nm.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McLellan JS, Chen M, Leung S, Graepel KW, Du X, Yang Y, Zhou T, Baxa U, Yasuda E, Beaumont T, Kumar A, Modjarrad K, Zheng Z, Zhao M, Xia N, Kwong PD, Graham BS. 2013. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science 340:1113–1117. 10.1126/science.1234914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wen X, Mousa JJ, Bates JT, Lamb RA, Crowe JE, Jr, Jardetzky TS. 2017. Structural basis for antibody cross-neutralization of respiratory syncytial virus and human metapneumovirus. Nat Microbiol 2:16272. 10.1038/nmicrobiol.2016.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson LJ, Hierholzer JC, Stone YO, Tsou C, Fernie BF. 1986. Identification of epitopes on respiratory syncytial virus proteins by competitive binding immunoassay. J Clin Microbiol 23:475–480. 10.1128/JCM.23.3.475-480.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Widjaja I, Wicht O, Luytjes W, Leenhouts K, Rottier PJM, van Kuppeveld FJM, Haijema BJ, de Haan CAM. 2016. Characterization of epitope-specific anti-respiratory syncytial virus (anti-RSV) antibody responses after natural infection and after vaccination with formalin-inactivated RSV. J Virol 90:5965–5977. 10.1128/JVI.00235-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Magro M, Andreu D, Gomez-Puertas P, Melero JA, Palomo C. 2010. Neutralization of human respiratory syncytial virus infectivity by antibodies and low-molecular-weight compounds targeted against the fusion glycoprotein. J Virol 84:7970–7982. 10.1128/JVI.00447-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crank MC, Ruckwardt TJ, Chen M, Morabito KM, Phung E, Costner PJ, Holman LA, Hickman SP, Berkowitz NM, Gordon IJ, Yamshchikov GV, Gaudinski MR, Kumar A, Chang LA, Moin SM, Hill JP, DiPiazza AT, Schwartz RM, Kueltzo L, Cooper JW, Chen P, Stein JA, Carlton K, Gall JG, Nason MC, Kwong PD, Chen GL, Mascola JR, McLellan JS, Ledgerwood JE, Graham BS, VRC 317 Study Team. 2019. A proof of concept for structure-based vaccine design targeting RSV in humans. Science 365:505–509. 10.1126/science.aav9033. [DOI] [PubMed] [Google Scholar]
- 26.Bakker AB, Marissen WE, Kramer RA, Rice AB, Weldon WC, Niezgoda M, Hanlon CA, Thijsse S, Backus HH, de Kruif J, Dietzschold B, Rupprecht CE, Goudsmit J. 2005. Novel human monoclonal antibody combination effectively neutralizing natural rabies virus variants and individual in vitro escape mutants. J Virol 79:9062–9068. 10.1128/JVI.79.14.9062-9068.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keeffe JR, Van Rompay KKA, Olsen PC, Wang Q, Gazumyan A, Azzopardi SA, Schaefer-Babajew D, Lee YE, Stuart JB, Singapuri A, Watanabe J, Usachenko J, Ardeshir A, Saeed M, Agudelo M, Eisenreich T, Bournazos S, Oliveira TY, Rice CM, Coffey LL, MacDonald MR, Bjorkman PJ, Nussenzweig MC, Robbiani DF. 2018. A combination of two human monoclonal antibodies prevents Zika virus escape mutations in non-human primates. Cell Rep 25:1385–1394.e7. 10.1016/j.celrep.2018.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shingai M, Nishimura Y, Klein F, Mouquet H, Donau OK, Plishka R, Buckler-White A, Seaman M, Piatak M, Jr, Lifson JD, Dimitrov DS, Nussenzweig MC, Martin MA. 2013. Antibody-mediated immunotherapy of macaques chronically infected with SHIV suppresses viraemia. Nature 503:277–280. 10.1038/nature12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baum A, Fulton BO, Wloga E, Copin R, Pascal KE, Russo V, Giordano S, Lanza K, Negron N, Ni M, Wei Y, Atwal GS, Murphy AJ, Stahl N, Yancopoulos GD, Kyratsous CA. 2020. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science 369:1014–1018. 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Battles MB, Mas V, Olmedillas E, Cano O, Vazquez M, Rodriguez L, Melero JA, McLellan JS. 2017. Structure and immunogenicity of pre-fusion-stabilized human metapneumovirus F glycoprotein. Nat Commun 8:1528. 10.1038/s41467-017-01708-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu S, Letchworth GJ. 2004. High efficiency transformation by electroporation of Pichia pastoris pretreated with lithium acetate and dithiothreitol. Biotechniques 36:152–154. 10.2144/04361DD02. [DOI] [PubMed] [Google Scholar]
- 32.Tan L, Coenjaerts FE, Houspie L, Viveen MC, van Bleek GM, Wiertz EJ, Martin DP, Lemey P. 2013. The comparative genomics of human respiratory syncytial virus subgroups A and B: genetic variability and molecular evolutionary dynamics. J Virol 87:8213–8226. 10.1128/JVI.03278-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krarup A, Truan D, Furmanova-Hollenstein P, Bogaert L, Bouchier P, Bisschop IJM, Widjojoatmodjo MN, Zahn R, Schuitemaker H, McLellan JS, Langedijk JPM. 2015. A highly stable prefusion RSV F vaccine derived from structural analysis of the fusion mechanism. Nat Commun 6:8143. 10.1038/ncomms9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The structures of RSV F-directed VHH Cl184 and RSV F in complex with VHH Cl184 are available in the Protein Data Bank under PDB IDs 7LVU and 7LVW, respectively.