The western honeybee (Apis mellifera) is indispensable for maintaining agricultural productivity as well as the abundance and diversity of wild flowering plants. However, bees suffer from environmental pollution, parasites, and pathogens, including viruses.
KEYWORDS: RNA, capsid opening, genome release, iflavirus
ABSTRACT
Infections with Kashmir bee virus (KBV) are lethal for honeybees and have been associated with colony collapse disorder. KBV and closely related viruses contribute to the ongoing decline in the number of honeybee colonies in North America, Europe, Australia, and other parts of the world. Despite the economic and ecological impact of KBV, its structure and infection process remain unknown. Here, we present the structure of the virion of KBV determined to a resolution of 2.8 Å. We show that the exposure of KBV to acidic pH induces a reduction in interpentamer contacts within capsids and the reorganization of its RNA genome from a uniform distribution to regions of high and low density. Capsids of KBV crack into pieces at acidic pH, resulting in the formation of open particles lacking pentamers of capsid proteins. The large openings of capsids enable the rapid release of genomes and thus limit the probability of their degradation by RNases. The opening of capsids may be a shared mechanism for the genome release of viruses from the family Dicistroviridae.
IMPORTANCE The western honeybee (Apis mellifera) is indispensable for maintaining agricultural productivity as well as the abundance and diversity of wild flowering plants. However, bees suffer from environmental pollution, parasites, and pathogens, including viruses. Outbreaks of virus infections cause the deaths of individual honeybees as well as collapses of whole colonies. Kashmir bee virus has been associated with colony collapse disorder in the United States, and no cure for the disease is currently available. Here, we report the structure of an infectious particle of Kashmir bee virus and show how its protein capsid opens to release the genome. Our structural characterization of the infection process determined that therapeutic compounds stabilizing contacts between pentamers of capsid proteins could prevent the genome release of the virus.
INTRODUCTION
Viruses that have the greatest impact on honeybees and other pollinators from the order Hymenoptera belong to the families Dicistroviridae and Iflaviridae (1–3). Kashmir bee virus (KBV) is an Aparavirus from the family Dicistroviridae. KBV, together with Israeli acute paralysis virus, and acute bee paralysis virus constitute a complex of closely related viruses that are distributed worldwide, with different members predominating in distinct geographical regions (4). The viruses ordinarily cause subclinical infections that are interspersed with sudden outbreaks marked by high virus titers and virulent pathology. KBV and the related viruses cause honeybee colony collapses, especially when the hives are infested with the parasitic mite Varroa destructor (4).
The structures of several viruses from the family Dicistroviridae have been determined: Israeli acute bee paralysis virus from the genus Aparavirus, triatoma virus and black queen cell virus from the genus Triatovirus, and cricket paralysis virus from the genus Cripavirus (5–8). Viruses from the family Dicistroviridae have nonenveloped capsids with icosahedral symmetry and diameters of about 30 nm. Capsids of dicistroviruses protect linear single-stranded positive-sense RNA genomes that are 9,000 to 10,500 nucleotides long (9). The genomes contain two open reading frames. The first one encodes nonstructural proteins that ensure the replication of dicistroviruses in infected cells. The second open reading frame contains structural (capsid-forming) proteins. The polyproteins are cotranslationally and posttranslationally cleaved by virus proteases to produce functional subunits. The capsid proteins originating from a single polyprotein form a protomer—the basic building block of the dicistrovirus capsid. The major capsid proteins VP1, VP2, and VP3 have β-sandwich jelly roll folds and form capsids with pseudo-T=3 icosahedral symmetry (5–8, 10). VP4s are peptides that are 70 to 90 residues long and attach to the inner faces of capsids of aparaviruses and cripaviruses. In contrast, VP4s are present in the virions of triatoviruses but are not attached to the shells formed by the other capsid proteins (5, 6, 8). Capsids protect dicistrovirus genomes from degradation by RNases present in the extracellular environment. To initiate infection, dicistroviruses bind to as-yet-unknown receptors at the cell surface. After endocytosis, the capsids release genomes and ensure their delivery into the cytoplasm. It has been shown that acidic pH induces the genome release of Israeli acute bee paralysis virus in vitro (11). Virions of Israeli acute bee paralysis virus convert to activated particles, which are characterized by the externalization of VP4 and the N termini of VP1 from a capsid and redistribution of their genomic RNA (11). Holes in the capsids of activated particles positioned on fivefold and twofold axes of picorna(-like) viruses were speculated to serve as channels for genome release (12–17). However, the dimensions of these holes are too small to enable the passage of single-stranded RNA. The dissociation of the capsid into pentamers was described as the mechanism of genome release of foot-and-mouth disease virus (18). It has been recently indicated that enterovirus particles expel pentamers of capsid protein protomers to release their genomes. The resulting openings, which are more than 120 Å in diameter, are sufficient to release the RNA genomes even if they contain double-stranded segments (19). It has been speculated that triatoma virus may release its genome by particle opening (20, 21).
Here, we present the virion structure of KBV. In addition, we show that the exposure of KBV to acidic pH induces the reorganization of its RNA genome and a reduction in interpentamer contacts within capsids. Capsids of KBV crack into pieces, resulting in the formation of open particles lacking pentamers of capsid proteins. The large opening of a capsid enables the rapid release of the genome and thus limits the probability of its degradation by RNases during the process.
RESULTS AND DISCUSSION
Virion structure of KBV.
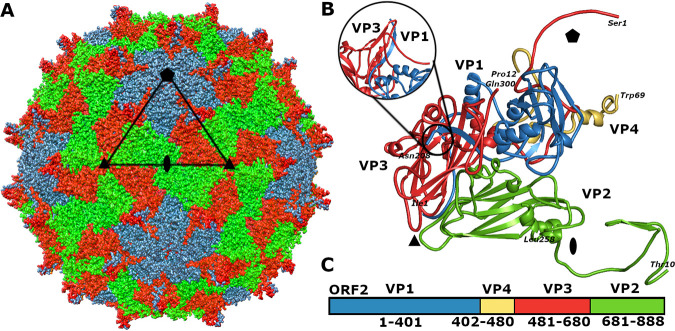
The structure of the KBV virion was determined to a resolution of 2.8 Å (Fig. 1; see also Fig. S1, S2, and S3 and Table S1 in the supplemental material). The capsid of KBV is built from VP1 subunits, which form pentamers around fivefold symmetry axes of the capsid, and VP2 and VP3 subunits that assemble into heterohexamers around the threefold symmetry axes (Fig. 1A and B). Sixty copies of the minor capsid protein VP4 are attached to the inner face of the capsid (Fig. 1B). The cryo-electron microscopy (cryo-EM) density map enabled building of the complete structure of 208 residues of VP1, residues 10 to 259 out of 259 of VP2, the complete structure of 300 residues of VP3, and residues 12 to 69 out of 69 of VP4 (Fig. 1B and C). The cores of major capsid proteins VP1 to VP3 have jelly roll β-sandwich folds with β-strands named B to I according to a convention (Fig. 1B) (10, 22, 23). VP4 of KBV forms two short α-helices at the C terminus (Fig. 1B and C). The surface of the KBV virion is decorated with 60 spiky protrusions located between fivefold and threefold axes of icosahedral symmetry of the capsid (Fig. 1A and B). The spikes, which protrude 23 Å above the capsid surface, are formed by two antiparallel β-strands from the CD loop of VP3 and the C-terminal β-strand of VP1 (Fig. 1B). Similar spikes decorate the particles of Israeli acute bee paralysis virus, black queen cell virus, and triatoma virus (5, 6, 8). The spikes of Israeli acute bee paralysis virus and black queen cell virus were speculated to play a role in receptor recognition (5, 6). Being the most prominent surface features, the spikes of KBV may have a similar function.
FIG 1.
Structure of KBV virion. (A) Molecular surface representation of KBV virion with subunits VP1 shown in blue, VP2 in green, and VP3 in red. The borders of one icosahedral asymmetric unit are outlined with a black triangle. The positions of selected symmetry axes are indicated with a pentagon for fivefold, triangle for threefold, and oval for twofold. Bar, 10 nm. (B) Cartoon representation of icosahedral asymmetric unit of KBV. VP1 subunit is shown in blue, VP2 in green, VP3 in red, and VP4 in yellow. The positions of icosahedral symmetry axes are indicated as in panel A. The inset shows a side view of a spike formed by a CD loop of VP3 and C-terminal β-strand VP1. (C) Organization of open reading frame 2 of KBV. Capsid proteins are colored as in panel B.
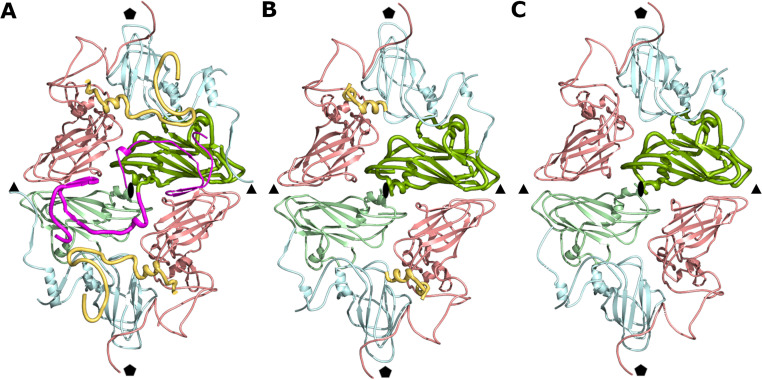
The capsid of KBV is built from 12 pentamers of capsid protein protomers VP1 to VP4. Interfaces between neighboring pentamers are predominantly formed by VP2 and VP3 subunits (Fig. 2A). Two β-strands from the N terminus of VP2 extend the β-sheet CHEF of the VP3 subunit from a neighboring pentamer, which is an example of domain swapping that is common in virus capsids (Fig. 2A) (24). The contacts between two pentamers mediated by the N termini of VP2 subunits account for 54% of the interface, which has a buried surface area of 4,900 Å2.
FIG 2.
Reduction of interpentamer contacts in KBV particles exposed to acidic pH. (A) Cartoon representation of subunits forming interface between two pentamers of capsid proteins in KBV virion. VP1 subunits are shown in blue, VP2 in green, VP3 in red, and VP4 in yellow. N termini of VP2 subunits mediating contacts between pentamers are highlighted in magenta. (B and C) Interpentamer interactions in activated particle (B) and empty particle (C). Coloring is the same as in panel A.
Maturation cleavage of VP0.
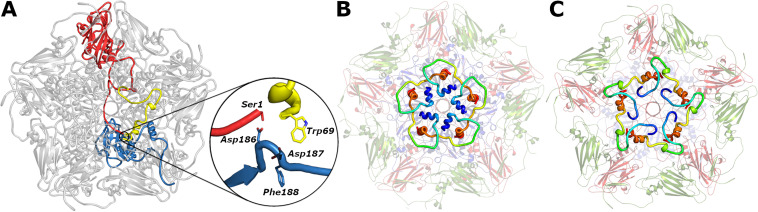
Immature particles of picorna-like viruses assemble from protomers containing a precursor protein, VP0, which in dicistroviruses is cleaved into N-terminal VP4 and C-terminal VP3 (Fig. 1C) (25). The cleavage occurs after the particles are filled with RNA genomes (26–28). It has been speculated that the Asp-Asp-Phe motif from the VP1 subunit cleaves the VP0 subunit of cricket paralysis virus and the capsid protein of black beetle virus from the family Nodaviridae (7, 27–29). Homologous residues Asp186, Asp187, and Phe188 from VP1 of KBV are positioned in the vicinity of the site of cleavage of VP0 into VP4 and VP3 (Fig. 3A).
FIG 3.
Structure and putative cleavage mechanism of VP0 subunits. (A) Cartoon representation of pentamer of capsid protein protomers of KBV viewed from inside the virion. Selected VP1 subunit is shown in blue, VP3 in red, and VP4 in yellow. The inset shows detail of Asp-Asp-Phe residues of VP1, constituting a putative proteolytic site, positioned close to the N terminus of VP3 and C terminus of VP4. (B and C) Pentamers of capsid protein protomers of KBV (B) and cricket paralysis virus (C) viewed from inside a particle. VP1 subunits are shown in pale blue, VP2 in pale green, and VP3 in pale red, and VP4s are highlighted with bright rainbow coloring from N terminus in blue to C terminus in red.
When viewed from inside the capsid, the VP4 of KBV resembles the Greek letter lambda (Fig. 3B). In contrast, VP4 of cricket paralysis virus has the shape of the letter sigma (Fig. 3C). VP4s of KBV, Israeli acute bee paralysis virus, and cricket paralysis virus interact with the other capsid proteins through interfaces with buried surface areas of 1,350, 1,600, and 1,400 Å2, respectively. The buried surface area of the interface between two neighboring copies of VP4 is 800 Å2 in KBV and Israeli acute bee paralysis virus and 1,000 Å2 in cricket paralysis virus. These extensive contacts between adjacent copies of VP4 may enable their synchronized release from particles together with the genome, as discussed below.
Intersubunit contacts in capsids of dicistroviruses.
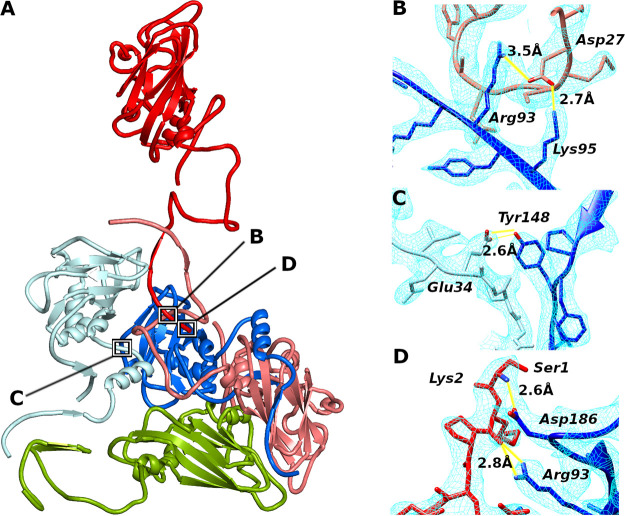
The availability of atomic structures enabled the comparison of intersubunit contacts in capsids of KBV, Israeli acute bee paralysis virus, cricket paralysis virus, black queen cell virus, and triatoma virus (5–8). Although the overall sequence conservation among the viruses is less than 41%, they share five interactions between capsid proteins that are formed by conserved residues. The interface between subunits VP1 and VP3 from one protomer contains two hydrogen bonds, which connect Arg93 and Lys95 from β-strand D of VP1 to Asp27 from the N terminus of VP3 (KBV residue numbering) (Fig. 4A and B). Another common interaction is that of Tyr148 from the GH-loop of VP1, which is linked by a hydrogen bond to Glu34 from the N terminus of the neighboring VP1 subunit (Fig. 4A and C). These interprotomer interactions may play a role in establishing the proper positioning of protomers during capsid assembly. In addition, Ser1 and Lys2 from the N terminus of VP3 form hydrogen bonds with Arg93 from β-strand D and Asp186 from the C terminus of the VP1 subunit from another protomer within the same pentamer (Fig. 4A and D). These contacts position the N terminus of the precursor subunit VP0 for cleavage by the putative proteolytic site formed by Asp186, Asp187, and Phe188 from VP1, as discussed above.
FIG 4.
Intersubunit contacts in capsids of dicistroviruses. (A) Cartoon representation of selected subunits interacting within a pentamer of KBV. Positions of interactions that are shared by structurally characterized dicistroviruses are indicated with black squares. VP1 subunits are shown in blue, VP2 in green, and VP3 in red. Please note that VP4 was omitted from the figure because it obscures the interaction sites. Subunits from different protomers are differentiated by bright and dark colors. (B to D) Details of interactions which are formed in the capsids of all dicistroviruses studied to date, as observed in the KBV structure. (B) Hydrogen bonds between Arg93 and Lys95 from β-strand D of subunit VP1 and Asp27 from N terminus of VP3. (C) Tyr148 from VP1 forms a hydrogen bond to Glu34 from neighboring VP1 subunit. (D) Ser1 and Lys2 of VP3 form hydrogen bonds with Arg93 and Asp186 of VP1, respectively. Distances indicated in panels B to D are in angstroms. The cryo-EM density map is shown as a light blue mesh.
Exposure of KBV to acidic buffer generates various kinds of particles.
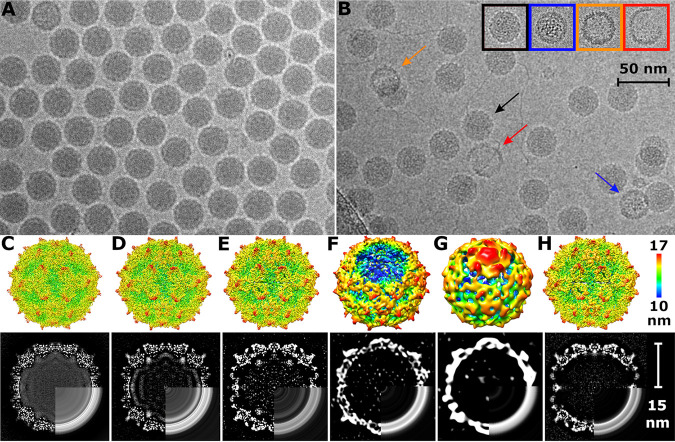
At neutral pH, the RNA genomes uniformly fill the capsid cavities of KBV virions (Fig. 5A and C). Empty or broken particles constituted less than 1% of the purified virus (Fig. 5A). In contrast, after exposure to acidic pH, the population of particles became structurally heterogeneous (Fig. 5B). Three-dimensional classification differentiated five types of particles: (i) particles that have a uniform distribution of RNA genomes and the same capsid structure as a native virus at neutral pH, except for missing resolved electron density for nine C-terminal residues of VP3 (Fig. 5B and D; Fig. S1, S2, and S3; Table S1); the residues which became disordered are located at the particle surface and probably became mobile at the acidic pH; (ii) activated particles with reduced interpentamer contacts and RNA reorganized into regions of high and low density (Fig. 2B and Fig. 5B and E; Fig. S1, S2, and S3; Table S1); VP4 remained attached to the shells formed by the other capsid proteins of these particles (Fig. 2B); (iii) open particles missing genomes and pentamers of capsid protein protomers (Fig. 5B and F; Fig. 6A and B; Fig. S1, S2, and S3; Table S1); (iv) empty particles in which one pentamer bulges out from the capsid (Fig. 5B and G; Fig. 6C; Fig. S1, S2, and S3; Table S1); (v) empty particles with intact capsids, which lack VP4 (Fig. 2C; Fig. 5B and H; Fig. S1, S2, and S3; Table S1). The formation of the different particles, after exposure to acidic pH, may be related to differences in the folds of the genomic RNA. The structures of packaged RNA genomes probably differ from particle to particle (30), and their reactions to exposure to acidic pH may be different. Furthermore, genomes of single-stranded RNA viruses are packaged in capsids together with positively charged polyamines, which shield the negative charge of the RNA (31–33). Individual particles might also differ in their polyamine content, which may cause differences in the reactions of KBV particles to acidic pH.
FIG 5.
Exposure of KBV to acidic buffer generates various kinds of particles. (A and B) Cryo-electron micrographs of native virions of KBV at neutral pH (A) and particles of KBV exposed to pH 6.0 (B). The black arrow indicates a particle with native-like structure, the blue arrow indicates an activated particle with nonuniform genome distribution, the red arrow indicates broken capsid, and the orange arrow indicates an empty particle. The inset shows additional examples of various types of particles with color coding the same as that for arrows in the main panel. Bar, 50 nm. (C to H) (Upper row) Surface representations of cryo-EM reconstructions. (Bottom row) Central slices of cryo-EM densities. Bottom-right quadrants of maps in panels C to H are replaced with central slices of spherically averaged maps. White color indicates high values of cryo-EM density. Native virion displayed at 4.0 σ (C), particle with native conformation at acidic pH at 4.0 σ (D), activated particle at acidic pH at 4.0 σ (E), open particle at 3.3 σ (F), empty particle with a pentamer of capsid proteins bulging out from its capsid at 2.5 σ (G), and empty capsid at 4.2 σ (H). Surfaces of the cryo-EM density maps are rainbow colored based on their distance from the particle center. Bar, 10 nm.
FIG 6.
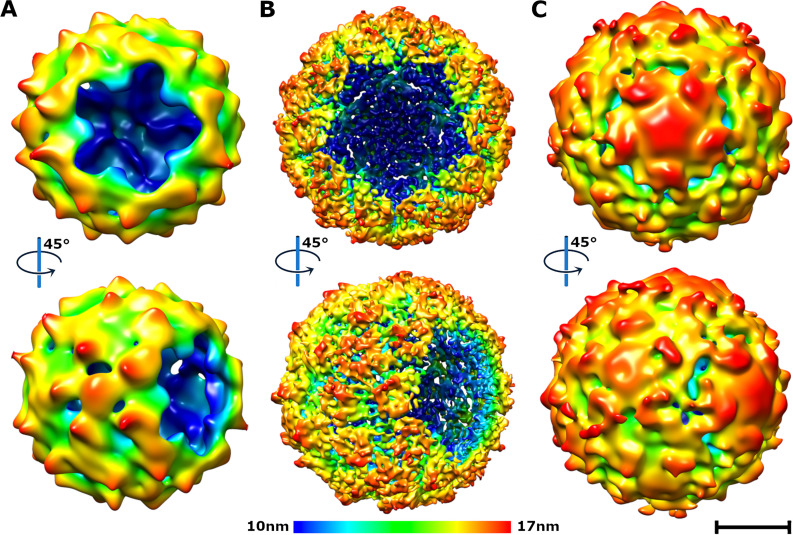
Particles of KBV that are missing a pentamer of capsid proteins (A and B) or contain a pentamer bulging from the capsid (C). (A and B) Surface representations of asymmetric (A) and fivefold symmetrized (B) cryo-EM reconstructions of KBV particles missing one pentamer of capsid protein protomers. (C) Particle with a pentamer of capsid protein protomers bulging out from its capsid. Surfaces of cryo-EM density maps are rainbow colored based on their distance from the particle center. Bar, 10 nm.
Capsids of KBV open to release genomes at acidic pH.
Capsids protect virus genomes from degradation in the extracellular environment, but viruses have to be able to release their RNAs to initiate infection. The genome release of KBV was induced by incubation at pH 6 at 34°C for 30 min, mimicking the conditions in endosomes (Fig. 5B). The acidic pH or the changes in the structure of the capsid induced by the acidic pH trigger the reorganization of RNA genomes in activated particles of KBV (Fig. 5E), which is a characteristic feature of activated particles of the well-studied enteroviruses from the family Picornaviridae (11, 19, 34–37). Changes in the capsid-genome interactions were also observed in particles of triatoma virus which were induced to release their genomes by exposure to alkaline pH (21). Furthermore, particles of KBV with nonuniform genome distribution lack the density resolved in the native virion for 53 residues from the N terminus of VP2, which causes a reduction in interpentamer contacts from 4,600 to 1,450 Å2 (Fig. 2B). This reduction in interpentamer contacts, which was also observed in Israeli acute bee paralysis virus and triatoma virus (11, 20), is another characteristic feature of activated particles. In contrast to the activated particles of enteroviruses, those of KBV are not expanded relative to virions at neutral pH (Fig. 5C and E) and do not contain pores along twofold axes of icosahedral symmetry of their capsids (Fig. 5E), and their VP4s remain attached to the shells formed by the other capsid proteins (Fig. 2B).
The asymmetric reconstruction of an open particle of KBV missing one pentamer of capsid protein protomers was determined to a resolution of 18 Å (Fig. S1 and S2; Table S1). The application of fivefold symmetry averaging enabled the determination of the structure of the open particle to a resolution of 11 Å (Fig. 6B; Fig. S1 and S2; Table S1). Furthermore, the micrographs of KBV exposed to acidic pH contain fragments of capsids (Fig. 5B). The observation of capsid fragments and open particles lacking a pentamer indicates that, at least in vitro, the genome release of KBV is enabled by capsid opening. It is likely that the reduction in interpentamer contacts, which is caused by the loss of structure of the N termini of VP2 subunits, destabilizes the KBV capsid. At the same time, the genome reorganization may induce an increase in the pressure exerted by the genome on the inner face of the capsid. The combination of capsid weakening and/or increased internal pressure enables capsid opening—either the cracking of the whole capsid into several pieces or expulsion of a pentamer of capsid proteins (19).
The genome release mechanism of dicistroviruses.
Activated and empty particles of dicistroviruses are not expanded relative to native virions and do not contain holes which may serve as channels for genome release (20, 38). It has been proposed that genomes of dicistroviruses are released from particles as single-stranded RNAs through transiently formed pores along twofold or fivefold axes of the icosahedral symmetry of their capsids (11). The observation of numerous empty particles in the sample of KBV exposed to acidic pH indicates that this mechanism of genome release is a possibility. However, extrusion of the 10,000-nucleotide-long RNA through a narrow pore would be a slow process, which would expose the genome to degradation by RNases (39, 40). The speculations about single-stranded RNA (ssRNA) release through a narrow pore in a capsid were based on the comparisons of icosahedrally symmetrized structures of capsids of native virions and those of empty particles resulting from the genome release (11). The reliance on icosahedrally symmetrized structures in the previous studies prevented the identification of asymmetric features that are essential for the genome release. In contrast, it has been indicated that the genome release of triatoma virus, which can be induced by alkaline pH, depends on capsid disruption or the lid-like opening of pentamers of capsid proteins (20, 21).
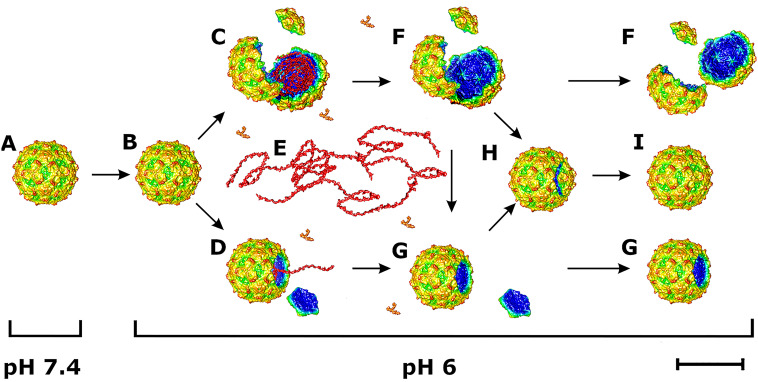
Here, we show that acidic pH induces the formation of activated particles of KBV, which are characterized by a reduction in interpentamer contacts and a genome organized into regions with low and high density (Fig. 7). The activated particles crack open or lose pentamers to release their genomes (Fig. 7). VP4s are probably released from KBV particles together with genomes. It has been shown that VP4 of triatoma virus forms toroidal pores in membranes and thus enables the delivery of the virus RNA from endosomes into the cytoplasm (41). It is likely that VP4 of KBV has the same function. Fragments of KBV capsids may reassemble into empty particles (Fig. 7). A subpopulation of the empty particles contains a pentamer of capsid proteins bulging out from their capsid (Fig. 7). Since these particles lack genomes, it is possible that they represent incompletely reassembled capsids, which have released their RNAs.
FIG 7.
Scheme of dicistrovirus genome release. (A) Native virions are stable at neutral pH. (B) Acidic pH induces formation of activated particles, which have reduced interpentamer contacts but are not expanded relative to virions at neutral pH. (C to E) Some activated particles crack open (C) or expel pentamers of capsid proteins (D), to release their genomes (red ribbon) and VP4 (orange Y shapes) (E). (F and G) Some capsids remain broken into pieces (F) or open (G). (H and I) Other capsid fragments reassemble into empty particles, some of which contain a pentamer bulging out from the capsid (H) or form complete capsids (I). Bar, 10 nm.
We analyzed disassembly intermediates of triatoma virus exposed to denaturing conditions. Particles of triatoma virus fragment into pentamers when exposed to urea at a concentration higher than 2.5 M (Fig. S4A). In contrast, holes with various shapes open in capsids of triatoma virus when treated with chaotropic salts (Fig. S4B). It has been shown previously that capsid proteins of triatoma virus reassemble into empty particles after the genome release (21).
The observation of open particles of both KBV and triatoma virus indicates that capsid opening may be a shared genome release mechanism of dicistroviruses. Previous in silico simulations of particles with properties similar to those of KBV demonstrated that disruption of a capsid enables genome release in less than a microsecond, which limits the probability of its degradation by RNases during the process (19). The dependence of dicistrovirus genome release on capsid opening indicates that compounds overstabilizing interpentamer interactions may block the infection process.
MATERIALS AND METHODS
KBV propagation in honeybee pupae.
The propagation of KBV was carried out as described previously (5, 6). In brief, brood areas with white-eyed pupae of Apis mellifera were identified by the color and structural features of the cell caps. White-eyed pupae were carefully extracted from the brood combs to prevent their injury. The pupae were placed on paper furrows with the ventral side up. The virus inoculum (1 μl) was injected into each pupa using a Hamilton micropipette with a 30-gauge 22-mm-long needle through the intersegmental cuticle between the 4th and 5th sternite. Pupae that leaked hemolymph after the injection were discarded. The optimal concentration of the virus in the inoculum for virus production was determined experimentally, by comparing virus yields when using different virus concentrations in the injection inoculum. Inoculated pupae were placed into petri dishes and incubated at 30°C and 75% humidity for 5 days. After incubation, the pupae were frozen at −20°C. For long-term storage, the pupae were kept at −80°C.
KBV purification.
Fifty experimentally infected honeybee pupae were homogenized with a Dounce homogenizer in 30 ml of phosphate-buffered saline (PBS), pH 7.5 (Sigma-Aldrich). Nonionic detergent NP-40 was added to a final concentration of 0.5%, and the homogenate was incubated for 1 h at room temperature. The homogenate was centrifuged at 8,000 × g for 30 min. The pellet was discarded, and the supernatant was centrifuged at 150,000 × g for 3 h in a Ti50.2 fixed-angle rotor (Beckman-Coulter). The resulting pellet was resuspended in PBS to a final volume of 5 ml. MgCl2 was added to a final concentration of 5 mM as well as 20 μg/ml DNase I and 20 μg/ml RNase. The solution was incubated at room temperature for 30 min and centrifuged at 4,000 × g for 15 min. The resulting supernatant was loaded onto a CsCl (0.6 g/ml) solution prepared in PBS. The ultracentrifugation proceeded for 16 h at 30,000 rpm in a Beckman SW-41 Ti rotor (Beckman Coulter Optima XPN-80 ultracentrifuge) at 10°C to establish the CsCl gradient. Virus bands were collected by gently piercing the ultracentrifuge tubes with an 18-gauge needle. The viruses were transferred to PBS by several rounds of concentration and dilution using centrifuge filter units with a 100-kDa-molecular-weight cutoff. This procedure yielded about 700 μg of the virus with sufficient purity for cryo-EM analysis.
Preparation of capsid protein pentamers and open particles of triatoma virus.
Native virions of triatoma virus were prepared as described previously (26). Purified virions at a concentration of 0.2 mg/ml were dialyzed against 8 M urea or 1 M guanidinium thiocyanate in NMT buffer (10 mM NaCl, 1 mM MgCl2, 50 mM Tris-HCl, pH 7.5) for 24 h at room temperature. The treated particles were applied to the nitrocellulose-covered copper grids and stained with 2.5% aqueous uranyl acetate for observation under an electron microscope.
Induction of genome release of KBV.
Purified KBV was diluted in 50 mM morpholineethanesulfonic acid (MES), pH 6.0, to a final virus concentration of 2.5 mg/ml and incubated at 34°C in a heat block for 30 min. The temperature was selected to mimic conditions in a beehive. After incubation, samples were immediately vitrified for cryo-EM analysis.
Preparation of samples for cryo-EM.
A solution of freshly purified KBV at a concentration of 2.5 mg/ml was applied onto holey carbon-coated copper grids (Quantifoil R2/1, mesh 300; Quantifoil Micro Tools), blotted, and plunge-frozen in liquid ethane using an FEI Vitrobot Mark IV, set to a 2-s blotting time and 0 blot force. The Vitrobot sample application chamber was held at 5°C and 100% humidity throughout the whole vitrification process. The samples were stored under liquid nitrogen until use.
Cryo-EM data collection.
Grids with the vitrified samples of KBV and KBV incubated at acidic pH were transferred to an FEI Titan Krios electron microscope operated at 300 kV and aligned for parallel illumination in nanoprobe mode. Images were recorded with an FEI Falcon II (native KBV) and FEI Falcon III (KBV incubated at acidic pH) direct electron detection camera under low-dose conditions (20 e−/Å2). Images were recorded using underfocus values ranging from 1.0 to 2.5 μm at a nominal magnification of ×75,000, resulting in a pixel size of 1.063 Å/px for native KBV. For the KBV sample treated with acidic pH, the underfocus ranged from 0.2 to 2.0 μm at a magnification of ×75,000, resulting in a pixel size of 1.07 Å/px. Each image of native KBV and KBV at acidic pH was recorded in movie mode with 0.5 s (7 fractions) and 1 s (39 fractions) of total acquisition time, respectively, and saved as separate movie frames.
Data processing.
The program MotionCor2 was used to align frames from each exposure to compensate for drift and beam-induced motion (42). Contrast transfer function (CTF) parameters were determined using the program Gctf (43). In total, 4,032 micrographs were acquired for the native virion and 9,545 micrographs for the virus incubated at acidic pH. Native KBV particles (total 530,217) were automatically picked using the software Gautomatch (K. Zhang, MRC-LMB, Cambridge, United Kingdom), whereas particles from the sample incubated at acidic pH (total 282,692) were automatically picked using the software crYOLO (44). The contrast transfer function parameters of each micrograph were automatically estimated using the program Gctf (43). The images were processed using the software package RELION 2.1 (native KBV) and RELION 3.0 (KBV incubated at acidic pH) (45). The particles were subjected to multiple rounds of two-dimensional classification, resulting in a near-homogeneous set of particles in the native virion sample and three major classes in the sample of the virus incubated at acidic pH. Particles from classes exhibiting high-resolution features were used for further calculations. A low-pass-filtered (50-Å) electron density map of the native virion of Israeli acute bee paralysis virus (EMD-4114) was used as an initial model for three-dimensional (3D) classification and for subsequent refinements of all the KBV reconstructions with imposed icosahedral symmetry, which was performed using the RELION 3dautorefine procedure (45). To further homogenize the data set, after initial refinement the particles were subjected to another round of 3D classification omitting the alignment step and using the particle shifts and orientations estimated in the previous refinement step. Particles belonging to the best class were reextracted and recentered. Reextracted particles were subjected to another round of autorefinement in RELION 3.1. The electrostatic potential map of the KBV particle featuring a missing pentamer, obtained from the initial asymmetric 3D classification, was subsequently used as an initial model for further 3D classification and subsequent refinement of the open particle. Fivefold symmetry was applied in all classifications and refinements of the open particle.
Final reconstructions, according to the gold standard (46), were calculated using RELION 3dautorefine. The resulting maps were masked with a threshold mask and B-factor sharpened using the RELION postprocess procedure.
The final resolution was estimated using the 0.143 Fourier shell correlation (FSC) cutoff.
Building of molecular structures.
The PDB structure of Israeli acute bee paralysis virus (PDB: 5CDC) was rigid-body fitted into the B-factor-sharpened electrostatic potential map of the KBV virion, using the program Phenix and manually rebuilt using the program Coot (5, 47, 48). The structure was refined by alternating rounds of real-space refinement in Phenix (47) and reciprocal space refinement in Refmac5 (49). After each refinement step, the structure was carefully inspected in Coot and irregularities were fixed manually. The structure quality was monitored throughout the model building and refinement using the MolProbity tool (50). The structure of the native KBV virion was used as the initial model for building the structures of particles treated with acidic pH.
Analysis of intersubunit contacts.
For the analysis of the intersubunit contacts, the following PDB models and their corresponding sequences were used: Israeli acute bee paralysis virus (PDB: 5CDC), cricket paralysis virus (PDB: 1B35), black queen cell virus (PDB: 5MQC), and triatoma virus (PDB: 3NAP) (5–8). Pairwise MUSCLE alignments of biological protomers were performed using the software Geneious Prime 2019. Structures of icosahedral asymmetric units of the five viruses were symmetry expanded to obtain complete capsids. Contacts representing all possible protein-protein interactions within the capsid were analyzed using the EMBL-EBI PDBePISA tool (51).
Preparation of figures.
The figures presented in this article were generated using the programs PyMOL (The PyMOL Molecular Graphics System, version 1.2r3pre; Schrödinger, LLC), Chimera (52), and Gimp (The GIMP Development Team, 2019, GIMP, https://www.gimp.org). Tables and graphs were generated using Microsoft Excel and Gnuplot (http://gnuplot.info), respectively.
Data deposition.
Cryo-EM maps and coordinates of the following structures were deposited in the Protein Data Bank (PDB) and Electron Microscopy Data Bank (EMDB; https://www.ebi.ac.uk/pdbe/emdb/) as follows: KBV virion at neutral pH, EMD-12182 and PDB 7BGK; particle with uniform genome distribution at acidic pH, EMD-12137 and PDB 7BC3; activated particle at acidic pH, EMD-12173 and PDB 7BG8; empty particle at acidic pH, EMD-12154 and PDB 7BE9; asymmetric reconstruction of open particle at acidic pH, EMD-12155; fivefold-symmetrized reconstruction of open particle at acidic pH, EMD-12167; and asymmetric reconstruction of particle with bulged pentamer of capsid protein protomers at acidic pH, EMD-12168.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jon Agirre for the preparation of triatoma virus and the Advanced Research Facilities (SGIker, UPV/EHU) for imaging of the virus. We gratefully acknowledge the Cryo-electron Microscopy and Tomography core facility of CEITEC supported by MEYS CR (LM2018127). We gratefully acknowledge the support of NVIDIA Corporation for the donation of the Titan Xp GPU used for this research.
This research was carried out under the project CEITEC 2020 (LQ1601), with financial support from the MEYS of the Czech Republic under National Sustainability Program II. This work was supported by IT4I project (CZ.1.05/1.1.00/02.0070), funded by the European Regional Development Fund and the national budget of the Czech Republic via the RDI-OP, as well as the MEYS via the grant (LM2011033). The research of G.A.M. was supported by the grants CONICET (PIP 2015-0288), 247 Agencia Nacional de Promoción Científica y Técnica, Argentina (PICT no. 2015-248 0665, PICT No. 2018-1545), and Universidad Nacional de La Plata, Argentina. The research of D.M.A.G. was supported by a Grupos Consolidados grant from the University of the Basque Country, Spain (GIU18/172). The research leading to these results received funding from the Grant Agency of the Czech Republic grant GX19-25982X to P.P.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.de Miranda JR, Gauthier L, Ribiere M, Chen YP. 2012. Honey bee viruses and their effect on bee and colony health, p 71–102. In Sammataro DYJ (ed), Honey bee colony health: challenges and sustainable solutions. CRC Press, Boca Raton, FL. [Google Scholar]
- 2.Chen YP, Siede R. 2007. Honey bee viruses. Adv Virus Res 70:33–80. 10.1016/S0065-3527(07)70002-7. [DOI] [PubMed] [Google Scholar]
- 3.Bailey L. 1976. Viruses attacking the honey bee. Adv Virus Res 20:271–304. 10.1016/s0065-3527(08)60507-2. [DOI] [PubMed] [Google Scholar]
- 4.de Miranda JR, Cordoni G, Budge G. 2010. The Acute bee paralysis virus-Kashmir bee virus-Israeli acute paralysis virus complex. J Invertebr Pathol 103(Suppl 1):S30–S47. 10.1016/j.jip.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 5.Mullapudi E, Přidal A, Pálková L, de Miranda JR, Plevka P. 2016. Virion structure of Israeli acute bee paralysis virus. J Virol 90:8150–8159. 10.1128/JVI.00854-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spurny R, Přidal A, Pálková L, Kiem HKT, de Miranda JR, Plevka P. 2017. Virion structure of black queen cell virus, a common honeybee pathogen. J Virol 91:e02100-16. 10.1128/JVI.02100-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tate J, Liljas L, Scotti P, Christian P, Lin T, Johnson JE. 1999. The crystal structure of cricket paralysis virus: the first view of a new virus family. Nat Struct Biol 6:765–774. 10.1038/11543. [DOI] [PubMed] [Google Scholar]
- 8.Squires G, Pous J, Agirre J, Rozas-Dennis GS, Costabel MD, Marti GA, Navaza J, Bressanelli S, Guerin DM, Rey FA. 2013. Structure of the Triatoma virus capsid. Acta Crystallogr D Biol Crystallogr 69:1026–1037. 10.1107/S0907444913004617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonning BC, Miller WA. 2010. Dicistroviruses. Annu Rev Entomol 55:129–150. 10.1146/annurev-ento-112408-085457. [DOI] [PubMed] [Google Scholar]
- 10.Procházková M, Škubník K, Füzik T, Mukhamedova L, Přidal A, Plevka P. 2020. Virion structures and genome delivery of honeybee viruses. Curr Opin Virol 45:17–24. 10.1016/j.coviro.2020.06.007. [DOI] [PubMed] [Google Scholar]
- 11.Mullapudi E, Füzik T, Přidal A, Plevka P. 2017. Cryo-electron microscopy study of the genome release of the dicistrovirus Israeli acute bee paralysis virus. J Virol 91:e02060-16. 10.1128/JVI.02060-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ren J, Wang X, Hu Z, Gao Q, Sun Y, Li X, Porta C, Walter TS, Gilbert RJ, Zhao Y, Axford D, Williams M, McAuley K, Rowlands DJ, Yin W, Wang J, Stuart DI, Rao Z, Fry EE. 2013. Picornavirus uncoating intermediate captured in atomic detail. Nat Commun 4:1929. 10.1038/ncomms2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garriga D, Pickl-Herk A, Luque D, Wruss J, Caston JR, Blaas D, Verdaguer N. 2012. Insights into minor group rhinovirus uncoating: the X-ray structure of the HRV2 empty capsid. PLoS Pathog 8:e1002473. 10.1371/journal.ppat.1002473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shingler KL, Yoder JL, Carnegie MS, Ashley RE, Makhov AM, Conway JF, Hafenstein S. 2013. The enterovirus 71 A-particle forms a gateway to allow genome release: a cryoEM study of picornavirus uncoating. PLoS Pathog 9:e1003240. 10.1371/journal.ppat.1003240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bostina M, Levy H, Filman DJ, Hogle JM. 2011. Poliovirus RNA is released from the capsid near a twofold symmetry axis. J Virol 85:776–783. 10.1128/JVI.00531-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, Peng W, Ren J, Hu Z, Xu J, Lou Z, Li X, Yin W, Shen X, Porta C, Walter TS, Evans G, Axford D, Owen R, Rowlands DJ, Wang J, Stuart DI, Fry EE, Rao Z. 2012. A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71. Nat Struct Mol Biol 19:424–429. 10.1038/nsmb.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seitsonen JJ, Shakeel S, Susi P, Pandurangan AP, Sinkovits RS, Hyvonen H, Laurinmaki P, Yla-Pelto J, Topf M, Hyypia T, Butcher SJ. 2012. Structural analysis of coxsackievirus A7 reveals conformational changes associated with uncoating. J Virol 86:7207–7215. 10.1128/JVI.06425-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan H, Li P, Ma X, Lu Z, Sun P, Bai X, Zhang J, Bao H, Cao Y, Li D, Fu Y, Chen Y, Bai Q, Zhang J, Liu Z. 2017. The pH stability of foot-and-mouth disease virus. Virol J 14:233. 10.1186/s12985-017-0897-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buchta D, Fuzik T, Hrebik D, Levdansky Y, Sukenik L, Mukhamedova L, Moravcova J, Vacha R, Plevka P. 2019. Enterovirus particles expel capsid pentamers to enable genome release. Nat Commun 10:1138. 10.1038/s41467-019-09132-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanchez-Eugenia R, Durana A, Lopez-Marijuan I, Marti GA, Guerin DMA. 2016. X-ray structure of Triatoma virus empty capsid: insights into the mechanism of uncoating and RNA release in dicistroviruses. J Gen Virol 97:2769–2779. 10.1099/jgv.0.000580. [DOI] [PubMed] [Google Scholar]
- 21.Snijder J, Uetrecht C, Rose RJ, Sanchez-Eugenia R, Marti GA, Agirre J, Guerin DM, Wuite GJ, Heck AJ, Roos WH. 2013. Probing the biophysical interplay between a viral genome and its capsid. Nat Chem 5:502–509. 10.1038/nchem.1627. [DOI] [PubMed] [Google Scholar]
- 22.Rossmann MG, Arnold E, Erickson JW, Frankenberger EA, Griffith JP, Hecht HJ, Johnson JE, Kamer G, Luo M, Mosser AG. 1985. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature 317:145–153. 10.1038/317145a0. [DOI] [PubMed] [Google Scholar]
- 23.Harrison SC, Olson AJ, Schutt CE, Winkler FK, Bricogne G. 1978. Tomato bushy stunt virus at 2.9 A resolution. Nature 276:368–373. 10.1038/276368a0. [DOI] [PubMed] [Google Scholar]
- 24.Bennett MJ, Schlunegger MP, Eisenberg D. 1995. 3D domain swapping: a mechanism for oligomer assembly. Protein Sci 4:2455–2468. 10.1002/pro.5560041202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le Gall O, Christian P, Fauquet CM, King AM, Knowles NJ, Nakashima N, Stanway G, Gorbalenya AE. 2008. Picornavirales, a proposed order of positive-sense single-stranded RNA viruses with a pseudo-T = 3 virion architecture. Arch Virol 153:715–727. 10.1007/s00705-008-0041-x. [DOI] [PubMed] [Google Scholar]
- 26.Agirre J, Aloria K, Arizmendi JM, Iloro I, Elortza F, Sanchez-Eugenia R, Marti GA, Neumann E, Rey FA, Guerin DM. 2011. Capsid protein identification and analysis of mature Triatoma virus (TrV) virions and naturally occurring empty particles. Virology 409:91–101. 10.1016/j.virol.2010.09.034. [DOI] [PubMed] [Google Scholar]
- 27.Fisher AJ, Johnson JE. 1993. Ordered duplex RNA controls capsid architecture in an icosahedral animal virus. Nature 361:176–179. 10.1038/361176a0. [DOI] [PubMed] [Google Scholar]
- 28.Munshi S, Liljas L, Cavarelli J, Bomu W, McKinney B, Reddy V, Johnson JE. 1996. The 2.8 A structure of a T = 4 animal virus and its implications for membrane translocation of RNA. J Mol Biol 261:1–10. 10.1006/jmbi.1996.0437. [DOI] [PubMed] [Google Scholar]
- 29.Zlotnick A, Reddy VS, Dasgupta R, Schneemann A, Ray WJ, Jr, Rueckert RR, Johnson JE. 1994. Capsid assembly in a family of animal viruses primes an autoproteolytic maturation that depends on a single aspartic acid residue. J Biol Chem 269:13680–13684. 10.1016/S0021-9258(17)36883-7. [DOI] [PubMed] [Google Scholar]
- 30.Gopal A, Zhou ZH, Knobler CM, Gelbart WM. 2012. Visualizing large RNA molecules in solution. RNA 18:284–299. 10.1261/rna.027557.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang P, Liu Y, Ma HC, Paul AV, Wimmer E. 2014. Picornavirus morphogenesis. Microbiol Mol Biol Rev 78:418–437. 10.1128/MMBR.00012-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fout GS, Medappa KC, Mapoles JE, Rueckert RR. 1984. Radiochemical determination of polyamines in poliovirus and human rhinovirus 14. J Biol Chem 259:3639–3643. 10.1016/S0021-9258(17)43142-5. [DOI] [PubMed] [Google Scholar]
- 33.Mounce BC, Olsen ME, Vignuzzi M, Connor JH. 2017. Polyamines and their role in virus infection. Microbiol Mol Biol Rev 81:e00029-17. 10.1128/MMBR.00029-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harutyunyan S, Kumar M, Sedivy A, Subirats X, Kowalski H, Kohler G, Blaas D. 2013. Viral uncoating is directional: exit of the genomic RNA in a common cold virus starts with the poly-(A) tail at the 3'-end. PLoS Pathog 9:e1003270. 10.1371/journal.ppat.1003270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levy HC, Bostina M, Filman DJ, Hogle JM. 2010. Catching a virus in the act of RNA release: a novel poliovirus uncoating intermediate characterized by cryo-electron microscopy. J Virol 84:4426–4441. 10.1128/JVI.02393-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pickl-Herk A, Luque D, Vives-Adrian L, Querol-Audi J, Garriga D, Trus BL, Verdaguer N, Blaas D, Caston JR. 2013. Uncoating of common cold virus is preceded by RNA switching as determined by X-ray and cryo-EM analyses of the subviral A-particle. Proc Natl Acad Sci U S A 110:20063–20068. 10.1073/pnas.1312128110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kalynych S, Füzik T, Přidal A, de Miranda J, Plevka P. 2017. Cryo-EM study of slow bee paralysis virus at low pH reveals iflavirus genome release mechanism. Proc Natl Acad Sci U S A 114:598–603. 10.1073/pnas.1616562114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Agirre J, Goret G, LeGoff M, Sanchez-Eugenia R, Marti GA, Navaza J, Guerin DM, Neumann E. 2013. Cryo-electron microscopy reconstructions of triatoma virus particles: a clue to unravel genome delivery and capsid disassembly. J Gen Virol 94:1058–1068. 10.1099/vir.0.048553-0. [DOI] [PubMed] [Google Scholar]
- 39.Groppelli E, Levy HC, Sun E, Strauss M, Nicol C, Gold S, Zhuang X, Tuthill TJ, Hogle JM, Rowlands DJ. 2017. Picornavirus RNA is protected from cleavage by ribonuclease during virion uncoating and transfer across cellular and model membranes. PLoS Pathog 13:e1006197. 10.1371/journal.ppat.1006197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Belnap DM, Filman DJ, Trus BL, Cheng N, Booy FP, Conway JF, Curry S, Hiremath CN, Tsang SK, Steven AC, Hogle JM. 2000. Molecular tectonic model of virus structural transitions: the putative cell entry states of poliovirus. J Virol 74:1342–1354. 10.1128/jvi.74.3.1342-1354.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanchez-Eugenia R, Goikolea J, Gil-Carton D, Sanchez-Magraner L, Guerin DM. 2015. Triatoma virus recombinant VP4 protein induces membrane permeability through dynamic pores. J Virol 89:4645–4654. 10.1128/JVI.00011-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA. 2017. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14:331–332. 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang K. 2016. Gctf: real-time CTF determination and correction. J Struct Biol 193:1–12. 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wagner T, Merino F, Stabrin M, Moriya T, Antoni C, Apelbaum A, Hagel P, Sitsel O, Raisch T, Prumbaum D, Quentin D, Roderer D, Tacke S, Siebolds B, Schubert E, Shaikh TR, Lill P, Gatsogiannis C, Raunser S. 2019. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun Biol 2:218. 10.1038/s42003-019-0437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scheres SH. 2012. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180:519–530. 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scheres SH, Chen S. 2012. Prevention of overfitting in cryo-EM structure determination. Nat Methods 9:853–854. 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. 2011. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr 67:355–367. 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66:12–21. 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krissinel E, Henrick K. 2007. Inference of macromolecular assemblies from crystalline state. J Mol Biol 372:774–797. 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 52.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.