Avian H9N2 influenza virus, although primarily restricted to chicken populations, is a major threat to human public health by acting as a donor of variant viral genes through reassortment to cocirculating influenza viruses. We established that the high prevalence of evolving H9N2 virus in vaccinated flocks played a key role as a donor of new subclade PB2 and PA genes in the generation of a dominant H7N9 virus genotype (genotype 72 [G72]) with enhanced infectivity in humans during the 2016–2017 N7N9 virus epidemic.
KEYWORDS: avian influenza virus, H9N2, H7N9, PB2, PA, genetic evolution, reassortment, zoonosis, interspecies transmission
ABSTRACT
H9N2 avian influenza virus (AIV) is regarded as a principal donor of viral genes through reassortment to cocirculating influenza viruses that can result in zoonotic reassortants. Whether H9N2 virus can maintain a sustained evolutionary impact on such reassortants is unclear. Since 2013, avian H7N9 virus had caused five sequential human epidemics in China; the fifth wave in 2016 to 2017 was by far the largest, but the mechanistic explanation behind the scale of infection is not clear. Here, we found that just prior to the fifth H7N9 virus epidemic, H9N2 viruses had phylogenetically mutated into new subclades, changed antigenicity, and increased their prevalence in chickens vaccinated with existing H9N2 vaccines. In turn, the new H9N2 virus subclades of PB2 and PA genes, housing mammalian adaptive mutations, were reassorted into cocirculating H7N9 virus to create a novel dominant H7N9 virus genotype that was responsible for the fifth H7N9 virus epidemic. H9N2-derived PB2 and PA genes in H7N9 virus conferred enhanced polymerase activity in human cells at 33°C and 37°C and increased viral replication in the upper and lower respiratory tracts of infected mice, which could account for the sharp increase in human cases of H7N9 virus infection in the 2016–2017 epidemic. The role of H9N2 virus in the continual mutation of H7N9 virus highlights the public health significance of H9N2 virus in the generation of variant reassortants of increasing zoonotic potential.
IMPORTANCE Avian H9N2 influenza virus, although primarily restricted to chicken populations, is a major threat to human public health by acting as a donor of variant viral genes through reassortment to cocirculating influenza viruses. We established that the high prevalence of evolving H9N2 virus in vaccinated flocks played a key role as a donor of new subclade PB2 and PA genes in the generation of a dominant H7N9 virus genotype (genotype 72 [G72]) with enhanced infectivity in humans during the 2016–2017 H7N9 virus epidemic. Our findings emphasize that the ongoing evolution of prevalent H9N2 virus in chickens is an important source, via reassortment, of mammalian adaptive genes for other influenza virus subtypes. Thus, close monitoring of the prevalence and variants of H9N2 virus in chicken flocks is necessary for the detection of zoonotic mutations.
INTRODUCTION
Since the first human outbreak in 2013 of avian H7N9 influenza virus, there had been a total of five waves of the virus in China (1, 2). The fifth wave of 2016 to 2017 was by far the largest epidemic to date, where the number of human cases (n = 758) was almost the same as the sum of the cases from all four previous outbreaks (2). In the fifth wave, the affected regions had expanded from eastern and southern China to the central and western provinces (2). Highly pathogenic avian influenza (HPAI) H7N9 viruses were first identified in this epidemic to cause clinical infections in both chickens and humans (3–5); however, most human cases continued to be from low-pathogenicity avian influenza (LPAI) H7N9 viruses (6). Although human-to-human H7N9 virus transmission has not been conclusively documented, the risk still exists (5, 7).
H9N2 avian influenza virus (AIV) has been in circulation in chickens in China for over 20 years (8, 9). To reduce the impact of H9N2 virus infection, flocks have been vaccinated since 1998 with commercial inactivated vaccines (10–12); however, the virus continues to circulate in vaccinated chickens (10, 11, 13). We previously showed that a particular genotype (genotype 57 [G57]) of H9N2 virus was responsible for outbreaks in chicken flocks across the country between 2010 and 2013 and that through reassortment, it provided six internal genes to generate the novel H7N9 virus of 2013 (8). Subsequently, the H7N9 virus crossed the human barrier to cause the first epidemic wave (14). The cocirculation of H9N2 and H7N9 viruses in chickens has continued to perpetuate H7N9 virus evolution, resulting in increased viral diversity in subsequent waves of infection (14–19). Several studies have described molecular characteristics of the evolving H7N9 virus of the fifth wave (4–6, 20). However, it remains unclear whether H9N2 influenza viruses can maintain a sustained evolutionary impact on the largest H7N9 epidemic in humans so far.
Here, we found that the evolution of H9N2 virus in chickens has continued to impact H7N9 virus genetic changes and contributes to the largest epidemic of human infection in the fifth wave. Through systematic analysis of the evolution of H9N2 influenza viruses in chickens in China in the period from 2014 to 2017, we discovered that H9N2 viruses isolated since 2015 have undergone significant genetic and antigenic changes, with a corresponding increased prevalence in vaccinated chicken flocks. The evolving H9N2 virus reassorted its PB2 and PA genes containing mammalian adaptive mutations with cocirculating H7N9 virus, thus creating a novel dominant H7N9 genotype that led to the fifth H7N9 virus epidemic. These findings highlight the ongoing public health threat posed by avian H9N2 viruses in generating novel or variant reassortants of H7N9 virus with epidemic/pandemic potential.
RESULTS
New subclades of H9N2 AIVs in surface and internal genes emerged before the fifth wave of H7N9 virus infection in humans.
To ascertain the possible role of H9N2 virus in the fifth H7N9 virus epidemic (2016 to 2017), we examined the evolutionary changes of chicken H9N2 viruses before and during the human H7N9 virus outbreak. The genomes of 143 chicken-derived H9N2 viruses from chicken flocks and live-bird markets collected between 2014 and 2017 in China were sequenced for phylogenetic analysis of all eight gene segments, supplemented by publicly available H9N2 sequences from the 2010–2017 period.
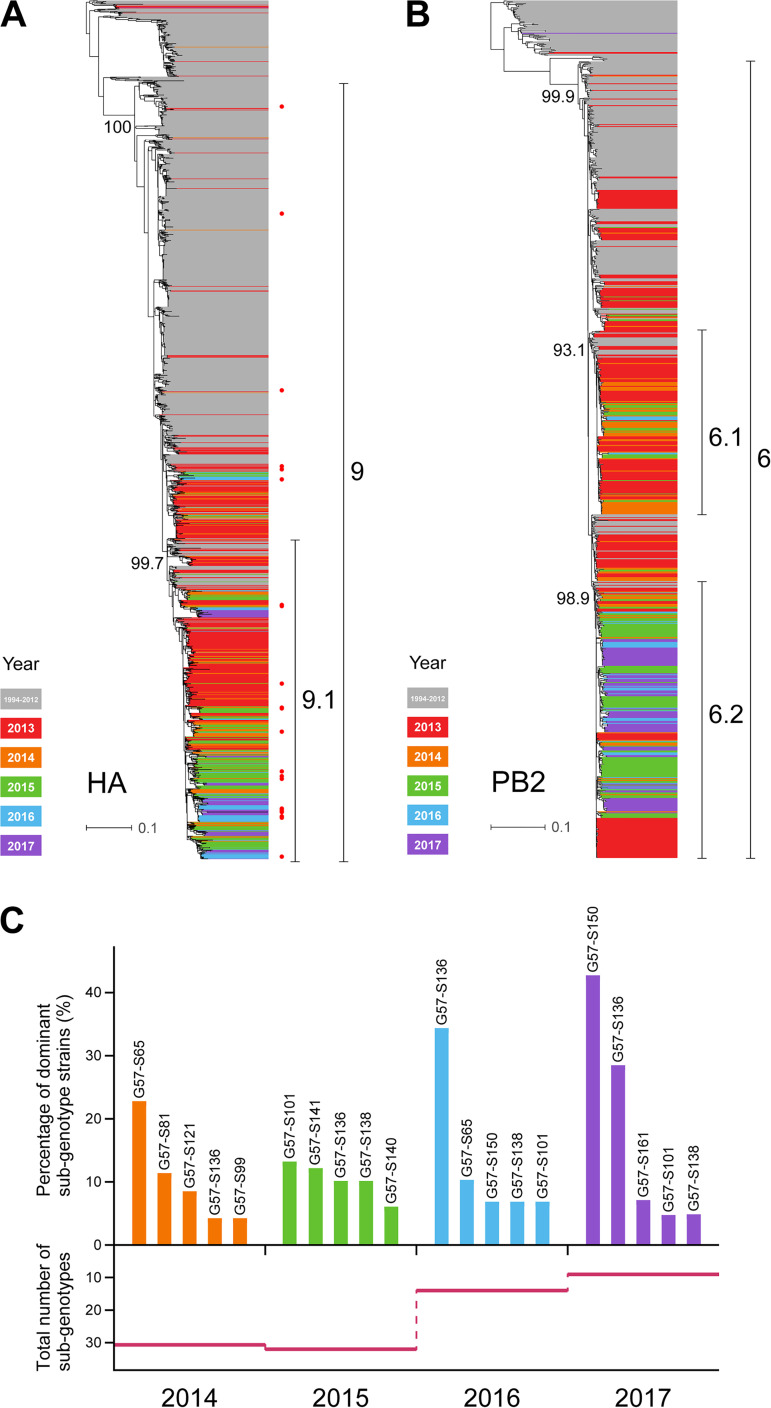
We found that H9N2 virus strains from the 2014–2017 period were all of the previously identified G57-like clades (Fig. 1A and B; see also Fig. S1 in the supplemental material) (8) but had formed relatively independent branches (identified as subclades) in the eight segments. In the hemagglutinin (HA) phylogenetic tree, a new subclade, 9.1, was evident in G57-like clade 9; most of the 2014–2017 strains formed this 9.1 subclade. In the phylogenetic tree of PB2, PB1, or PA, most isolates from 2014 onward were different from the earlier isolates and formed relatively independent subclades (PB2-6.2, PB1-3.2, and PA-3.1).
FIG 1.
Phylogenetic analysis of H9N2 influenza viruses isolated from chickens. (A) HA phylogenetic tree of H9N2 influenza viruses from 2010 to 2017 denoted by different colors. Viruses labeled with a red dot were selected for HI assays. (B) PB2 phylogenetic tree of H9N2 influenza viruses. (C) Abundance of the subgenotypes of H9N2 influenza virus during the period from 2014 to 2017. The top graph depicts the percentages of strains of the five most dominant subgenotypes in each year; the bottom graph depicts the total numbers of subgenotypes in each year from 2014 to 2017. The total numbers of H9N2 viruses in each year are 72, 101, 29, and 41.
According to their subclades, the viruses analyzed could be categorized into multiple subgenotypes (G57-S1 to -S162) (Table S1). Notably, this diversity was reduced over time. Viruses from 2016 to 2017 had the lowest diversity, with two major dominant subgenotypes (Fig. 1C), which had the same evolved genes of PB2, PB1, and PA, indicating that H9N2 viruses with such a gene constellation were prevalent in chickens during this period.
Rising rates of isolation of H9N2 virus in vaccinated chickens preceded and overlapped the fifth human outbreak of H7N9 virus infection.
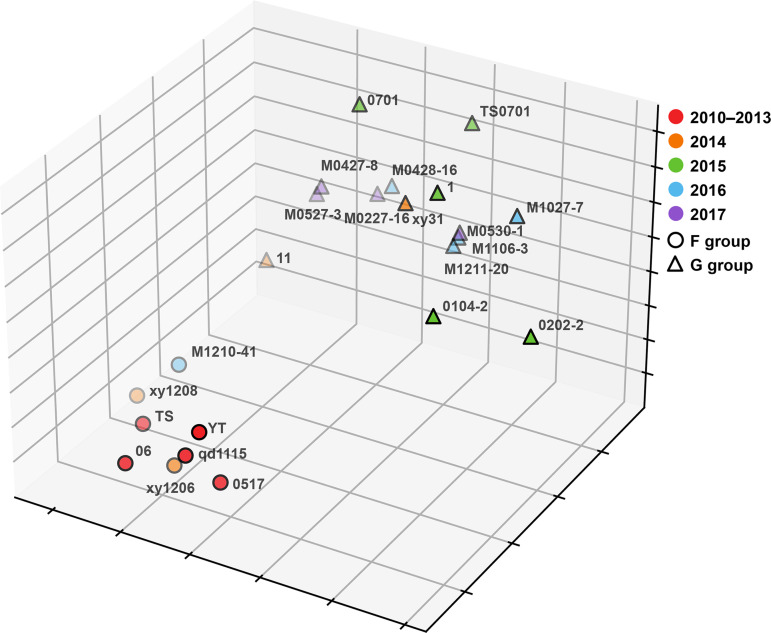
As most H9N2 viruses isolated between 2014 and 2017 belonged to a new HA subclade, the associated HA gene changes would amount to viral antigenic drift (21–24). To determine the extent of antigenic drift, hemagglutination inhibition (HI) assays, using 7 selected sera, were performed on 23 representative H9N2 viruses from the 2010–2017 period (Fig. 2 and Table S2). Three of the seven sera were previously identified as being specific for viral antigenic group F (8), and the remaining four were derived from the viruses from 2015 to 2017. H9N2 isolates from 2010 to 2013 belonged to antigenic group F, as previously found (8). However, some strains in 2014 showed distinct antigenic changes. By 2015 to 2017, significant antigenic drift was commonly found; most isolates had HI titers that were 4 to 32 times lower than those of the F group viruses. Thus, these new variants were classified as belonging to novel antigenic group G. Out of the 23 representative H9N2 viruses tested, 5 from HA clade 9 belonged to antigenic group F; the remaining 18 viruses were from HA subclade 9.1, of which 15 belonged to antigenic group G (Fig. 2 and Table S2).
FIG 2.
Antigenic cartography representation of the HI data generated by a panel of chicken antisera. The map was drawn by performing multidimensional scaling (MDS) with downscaling of the HI data to three-dimensional space after log2 transformation, using K-means clustering. Circles and triangles represent the locations of strains classified as belonging to the F and G antigen groups, respectively. Different colors represent different years of isolation. Virus names are indicated by using their abbreviations, and details of the HI data are shown in Table S2 in the supplemental material.
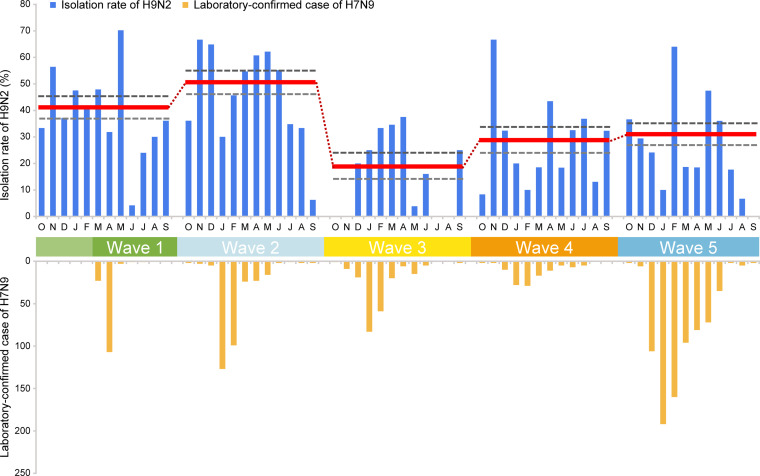
To determine if the antigenic drift of the H9N2 virus was connected to a rising prevalence of H9N2 virus infection in chicken flocks, we retrospectively examined the rates of isolation of H9N2 virus in vaccinated chickens across 21 provinces in China from January 2014 to December 2017 (Table S3). Out of a total of 1,455 flocks, 465 were positive for H9N2 virus infection, with a virus isolation rate of 31.96% (Table S3). Mean H9N2 isolation rates varied annually. Compared with our previously determined isolation rate (47.08%) in 2013 (8), the isolation rate declined in 2014 (41.13%), followed by a further reduction in 2015 (25.20%). However, in the 2016–2017 period, isolation rates began to rise again and reached 31.15% in 2017. Notably, the rising H9N2 virus prevalence in chickens over the 2016–2017 period preceded and overlapped the fifth H7N9 virus epidemic (Fig. 3), which suggests that these recent H9N2 viruses could have been sources of genetic transmission via reassortment to H7N9 viruses.
FIG 3.
Rate of isolation (percent) of H9N2 influenza viruses in chicken flocks reporting illness and number of human H7N9 cases in the five epidemic waves. Red horizontal lines (with connecting dots) indicate mean annual isolation rates, and gray horizontal dashed lines indicate 95% confidence intervals (CIs). The 2013 H9N2 isolation rate data obtained during the first epidemic wave are from our previous study (8). The total numbers of human cases from wave 1 to wave 5 were 135, 320, 226, 119, and 758, respectively.
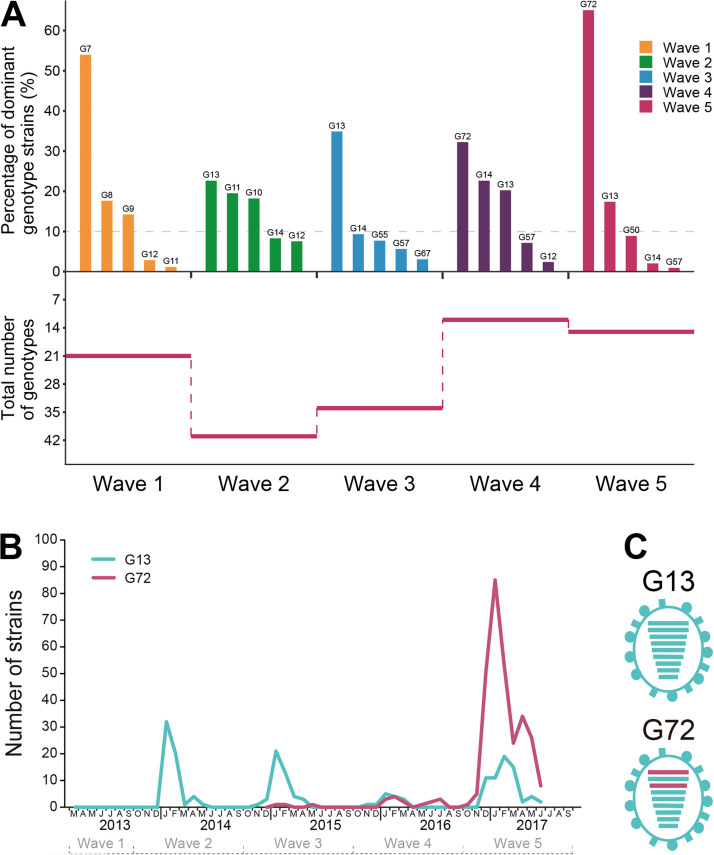
The dominant H7N9 genotype (G72) identified in the fifth wave of human infection comprised PB2 and PA genes from chicken H9N2 virus.
Using chicken H7N9 virus sequences generated in this study, along with all available public H7N9 sequences from humans and chickens, we constructed phylogenetic trees of the six internal segments to examine genotype evolution (Fig. S2) as previously described (6, 18). We found that the internal genes of H7N9 virus formed diversified clades as reported previously (5, 6, 20, 25). Based on the genomic diversity of the internal genes, the five waves of H7N9 virus could be classified into 78 genotypes (Table S1); each genotype with 10% or more of the total number of strains in each epidemic was regarded as a major genotype (Fig. 4A). Genotype diversity showed a sharp increase from the first to the second wave and was followed by successive reductions, with the lowest diversity being found during the fourth and fifth waves (Fig. 4A). H7N9 genotype 13 (G13) was the most dominant genotype in the third wave of human infection (Fig. 4A). In this third wave, G72 was first detected in January 2015 (Fig. 4B and Table S1); its rate of isolation subsequently rose going into the fourth and fifth waves to replace the dominance of G13 (Fig. 4A) in frequency and geographical distribution (Fig. S3). The number of G72 viruses isolated from humans (n = 15) in the fourth wave rose sharply (n = 287) into the fifth wave (Fig. 4B). It is worth noting that the number of G72 viruses from humans showed an abrupt increase after November 2016 (Fig. 4B) and was preceded a year earlier by an increased prevalence of avian H9N2 virus with newly evolved genes in chicken flocks (Fig. 3).
FIG 4.
Genetic evolution of H7N9 influenza viruses from epidemic wave 1 through wave 5. (A) Abundance of different genotypes of H7N9 influenza virus. The top graph shows the rate of prevalence of the five most abundant genotypes, and the bottom graph illustrates the total number of genotypes in each wave. (B) Number of G13 and G72 H7N9 strains in each month from 2013 to 2017 in humans. The first isolation of G72 H7N9 virus was in January 2015 during the third wave. The total numbers of G72 viruses isolated from humans in the third, fourth, and fifth waves were 3, 15, and 286, respectively. (C) Genomic constitution of G13 and G72 H7N9 viruses. Virus particles are represented by ovals. The eight gene segments are horizontal bars (from the top, PB2, PB1, PA, HA, NP, NA, MP, and NS). Scarlet bars represent the internal segments in the G72 genotype that are different from the G13 H7N9 viruses.
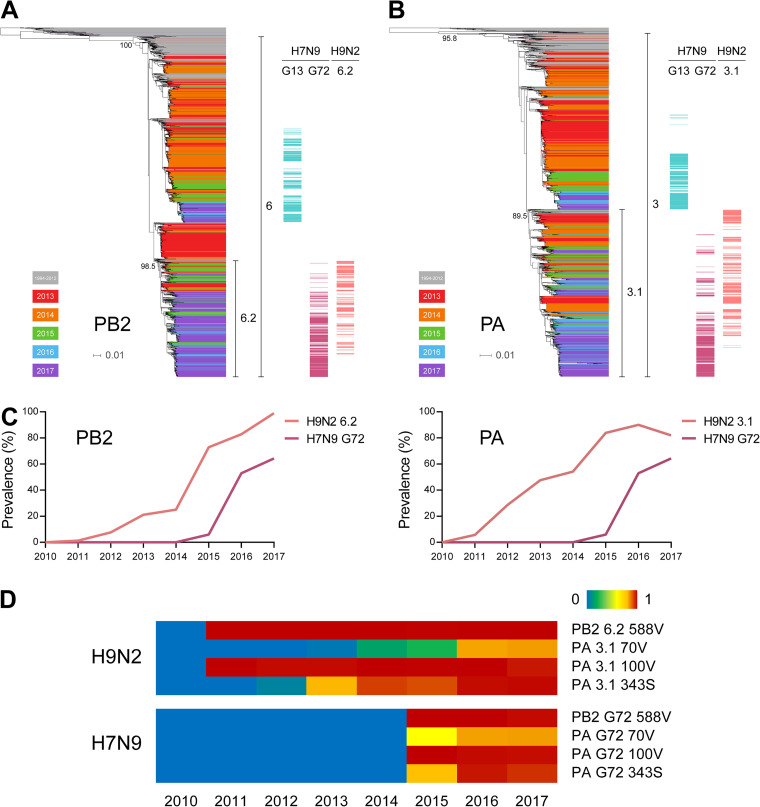
We compared the genetic makeups of G72 and G13 genotypes and found distinct PB2 and PA gene combinations (Fig. 4C and Table S1), where the two genes in G72, distinct from G13, could contribute to the prevalence of G72 in the fifth wave. As changes in PB2 and PA segments were found in both recent H9N2 and the dominant (G72) H7N9 viruses, these genes could have been donated from cocirculating H9N2 to H7N9 viruses. We constructed PB2 and PA phylogenetic trees using all available H9N2 sequences from 2010 to 2017 and H7N9 strain sequences from 2013 to 2017, which clearly showed that the PB2 and PA genes of the H7N9 G72 strains, but not the H7N9 G13 strains, were derived from H9N2 viruses (Fig. 5A and B and Fig. S4). The PB2 and PA genes of the G72 genotype were from the H9N2 PB2-6.2 and PA-3.1 subclades, respectively.
FIG 5.
Genetic relatedness of H7N9 and H9N2 influenza viruses in PB2 and PA genes. (A) PB2 phylogenetic tree of H7N9 and H9N2 influenza viruses. On the right side of the tree, PB2 genes of G13 H7N9, G72 H7N9, and 6.2 subclade H9N2 viruses are denoted by different-colored bars. (B) PA phylogenetic tree of H7N9 and H9N2 influenza viruses. On the right side of the tree, PA genes of G13 H7N9, G72 H7N9, and 3.1 subclade H9N2 viruses are denoted by different-colored bars. (C) Prevalence of PB2-6.2 and PA-3.1 in H7N9 and H9N2 viruses over time. (D) Prevalence of critical amino acid residues encoded by the PB2 and PA genes of the indicated H7N9 and H9N2 viruses. Red indicates high-prevalence (up to 100%) substitutions, and blue indicates no mutation or no virus isolated in the given year.
To confirm the delivery of PB2-6.2 and PA-3.1 from H9N2 virus to the G72 genotype of H7N9, we compared the temporal prevalences of PB2 and PA genes in both subtypes. As shown in Fig. 5C, PB2-6.2 and PA-3.1 genes first appeared in the 2011 H9N2 virus strains, while those from G72 H7N9 virus were first found in 2015. The increasing prevalence of PB2-6.2 and PA-3.1 in H9N2 viruses preceded a similar rise in their prevalence in H7N9 viruses. Thus, H9N2 viruses were likely to be responsible for the provision of PB2 and PA genes to the H7N9 viruses to form the G72 genotype. Furthermore, comparisons of PB2 and PA amino acid sequences revealed the delivery of 12 possible critical mammalian adaptive residues from H9N2 virus to the H7N9 virus, including PB2-588V (26), PA-70V (27), PA-100V (28), and PA-343S (29, 30) (Fig. 5D and Table S4). Thus, the earliest PB2 and PA reassortments between H9N2 and H7N9 viruses had likely taken place in or before 2015, which led to the generation of the predominant G72 genotype responsible for the fifth H7N9 virus outbreak in humans.
H9N2-derived PB2 and PA genes increased polymerase activity in the dominant G72 genotype H7N9 virus in human cells.
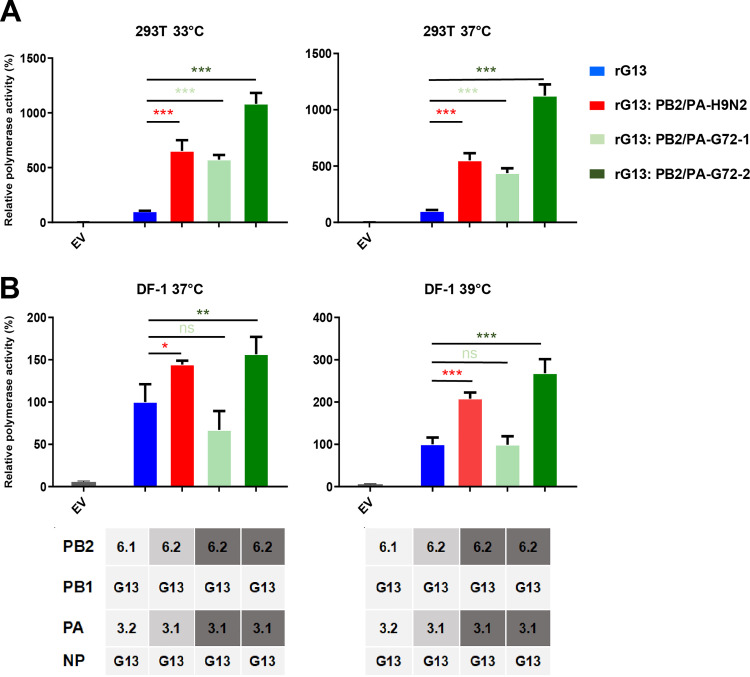
Adaptation of viral polymerase is necessary for efficient virus replication in new host species (31). To determine the effect of the H9N2 PB2 and PA gene substitutions on H7N9 viral polymerase function, viral minigenome polymerase assays were performed in human 293T cells at 33°C and 37°C, respectively. We used the PB2, PB1, PA, and NP genes from an LPAI H7N9 G13-like virus as the backbone of the polymerase complex and generated a series of recombinant polymerase complexes by replacing G13-like PB2 and PA genes with PB2-6.2 and PA-3.1 genes derived from different H9N2 viruses and G72 H7N9 viruses (Fig. 6 and Table S5). PB2-6.2 and PA-3.1 genes from the test H9N2 and G72 H7N9 viruses carry 16 specific amino acid residues reported to confer mammalian adaptive functions (Table S6). Viral minigenome polymerase assays showed that replacement with PB2-6.2 and PA-3.1 genes of H9N2 or G72 H7N9 virus significantly enhanced the polymerase activity of the LPAI H7N9 polymerase complex at both 33°C and 37°C by 4.4- to 11.2-fold (P < 0.001) (Fig. 6A). Thus, the recent H9N2-derived PB2 and PA genes conferred increased polymerase activity in H7N9 viruses, possibly making the G72 genotype replicate better in mammals. The new subclade of PB2 and PA genes also increased the viral polymerase activity of H7N9 viruses in chicken DF-1 cells at 37°C and 39°C, respectively, by 1.4- to 2.7-fold (P < 0.05), with the exception of genes from one G72 H7N9 virus (Fig. 6B).
FIG 6.
Contribution of H9N2-derived PB2 and PA genes to polymerase activity in H7N9 virus. Viral polymerase activities in 293T cells (A) were determined by minigenome replication assays at 33°C and 37°C, and those in DF-1 cells (B) were determined at 37°C and 39°C (expressed as mean percentages ± standard deviations, with the activity of the corresponding wild-type G13 H7N9 virus being set to 100%, from three independent experiments). Test RNP complexes were variants of a G13 H7N9 virus with the indicated substitutions of PB2 and PA genes. rG13:PB2/PA-H9N2 refers to the RNP complex with the PB2 and PA genes from an H9N2 virus; rG13:PB2/PA-G72-1 and rG13:PB2/PA-G72-2 refer to RNP complexes with the PB2 and PA genes from different G72 H7N9 viruses. All substituted genes were from the H9N2-derived PB2-6.2 and PA-3.1 subclades. Details of segment and viral information for these reassortants are shown in Table S5 in the supplemental material. Statistical significance was based on one-way ANOVA (ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001). EV, empty vector.
H9N2-derived PB2 and PA genes increased the infectivity of G72 genotype H7N9 virus in mice.
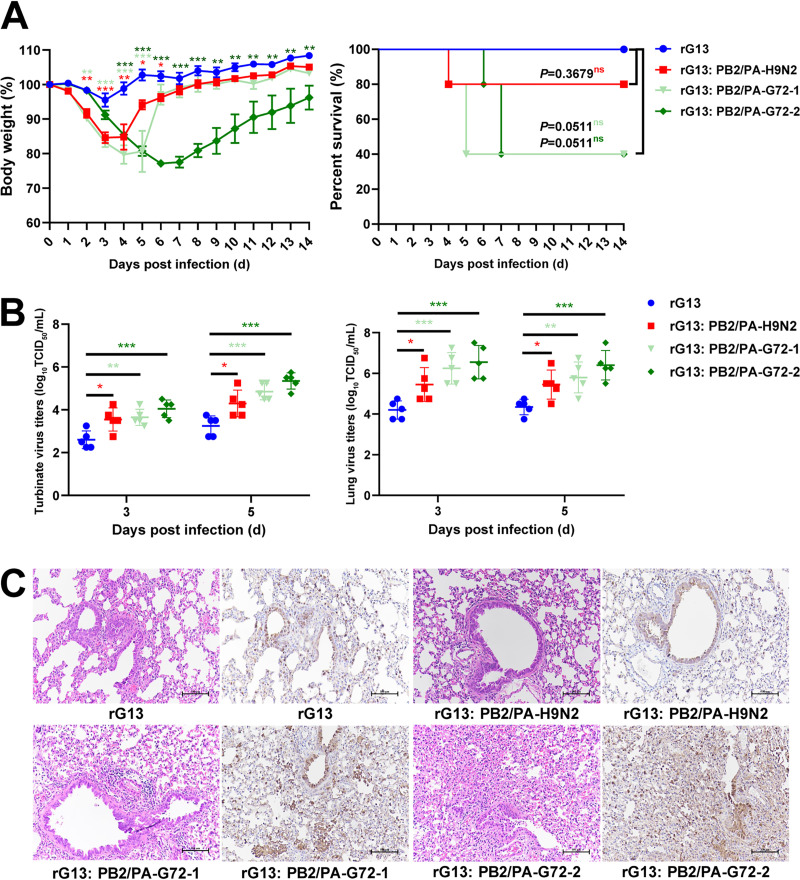
To further assess the contribution of the H9N2-derived PB2-6.2 and PA-3.1 genes to the pathogenicity and replication of H7N9 virus in mammals (Fig. 7), we used LPAI G13 H7N9 virus as a backbone control virus (rG13) (Table S5 and Fig. S5) to generate three reassortants (rG13:PB2/PA-H9N2, rG13:PB2/PA-H7N9-1, and rG13:PB2/PA-H7N9-2). These reassortants had PB2-6.2 and PA-3.1 genes from H9N2 and G72 H7N9 viruses (Table S5). The resulting 50% mouse infective dose (MID50) values of rG13, rG13:PB2/PA-H9N2, rG13:PB2/PA-H7N9-1, and rG13:PB2/PA-H7N9-2 were 4.5, 3.5, 3.5, and 1.5 log10 50% tissue culture infective doses (TCID50), respectively, demonstrating that PB2-6.2 and PA-3.1 genes conferred increased infectivity by between 10- and 1,000-fold (Table S7).
FIG 7.
Contribution of PB2 and PA genes of H9N2 virus origin to infectivity of H7N9 virus in mice. (A) Weight loss and percent survival of mice (n = 5) inoculated with 106 TCID50 of each virus. Mice that lost >25% of their baseline weight were euthanized. (B) Virus production from nasal turbinates and lungs. Five mice from each group were euthanized at 3 and 5 dpi to determine viral titers in nasal turbinates and lung tissues. Statistical significance was based on one-way ANOVA (*, P < 0.05; **, P < 0.01; ***, P < 0.001). (C) Representative histopathological changes in lung sections at 3 dpi by hematoxylin and eosin (H&E) staining (left) and immunodetection of influenza viral NP antigen (right). Mice infected with rG13:PB2/PA-H9N2, rG13:PB2/PA-H7N9-1, and rG13:PB2/PA-H7N9-2 viruses presented with more severe histopathology and a greater abundance of viral antigen-positive cells in the lung fields.
Next, the four viruses were inoculated into mice to determine their pathogenicity and viral load in the respiratory tract. Each mouse was intranasally inoculated with the indicated virus at a dose of 106 TCID50. Nasal turbinate and lung samples from five mice per group were collected for viral titration at 3 and 5 days postinfection (dpi). rG13-infected mice showed no obvious weight loss; the other three reassortants caused a range of weight losses, with a maximum loss of 15.4% to 22.86% (Fig. 7A). All mice infected with the rG13 virus recovered; those infected with the other three reassortants showed 1 to 3 deaths from each group, although they were not statistically different from the rG13 group (Fig. 7A). For viral loads in the nasal turbinates and lungs, the three reassortants produced significantly higher viral titers than those of rG13 virus at 3 and 5 dpi (P < 0.05) (Fig. 7B). Among the three reassortants, rG13:PB2/PA-H7N9-2 virus produced the highest titers in the turbinates (>100-fold) and lungs (>200-fold) relative to rG13 virus. Histopathology findings of virus infection in mice were consistent with the clinical response. Lungs from rG13 virus infection showed mild bronchitis (Fig. 7C), while the 3 reassortant viruses caused extensive and severe peribronchiolitis and bronchopneumonia; interstitial pneumonia was detected (Fig. 7C). Viral-antigen-positive cells in the lung were also more abundant with the 3 reassortant viruses (Fig. 7C). In summary, recent substitutions of H9N2-derived PB2 and PA genes with those in H7N9 virus conferred increased infectivity in mice.
DISCUSSION
We demonstrated here that continually evolving H9N2 virus not only resulted in its increased prevalence in vaccinated chickens but also led to the transfer of its PB2 and PA gene variants to cocirculating H7N9 viruses that in turn caused the largest H7N9 virus epidemic to date between 2016 and 2017. Our findings highlight chicken H9N2 virus as a key donor in virus reassortment and thus an ongoing threat to public health.
In recent years, the H9N2 virus has undergone rapid mutational changes, leading to repeated outbreaks in vaccinated chicken flocks. In 1998, H9N2 virus caused the first epizootic in chickens along the eastern coast of China (11). Despite an existing nationwide poultry vaccination program, during 2010 to 2013, it caused a second large-scale outbreak in chickens across most provinces in China (8). Subsequently, updated vaccines were able to reduce the virus prevalence in 2014 to 2015, according to our findings. However, a year later, between 2016 and 2017, the prevalence of H9N2 virus in vaccinated flocks had again risen. Although this increase was not as high as that in the 2010–2013 outbreak in chickens, it produced new H9N2 subclades of zoonotic significance.
The formation of the H7N9 virus G72 genotype, responsible for the fifth epidemic wave, was the result of newly acquired subclade PB2 and PA gene segments from cocirculating H9N2 virus. Previous studies have also analyzed the internal genes of the dominant H7N9 virus genotype of the fifth wave (4–6, 20); most identified H7N9 PB2 and PA genes were from the cocirculating H9N2 virus, but the functional and genetic details of these genes are hitherto unclear. Here, we found that the PB2 and PA genes of 2016–2017 H7N9 virus were different from those isolated in 2013. The PB2 and PA genes from the dominant H9N2 virus subclades had undergone further mutations to carry more mammalian adaptive variations. Experimentally, we demonstrated that the PB2 and PA genes donated by H9N2 virus significantly enhanced the polymerase activity of the recipient H7N9 virus in human cells at 37°C and 33°C. The latter temperature could favor G72 H7N9 virus replication along the nasal passage, thus promoting initial upper respiratory tract infection in humans. A challenge study in mice confirmed that the 2016–2017 H7N9 virus with the recently acquired H9N2-originating PB2 and PA genes produced significantly higher viral titers in nasal turbinates and lungs at 3 and 5 dpi. Notably, the H9N2 virus-derived PB2 and PA genes carried a number of known mammalian adaptive residues, PB2-588V, PA-70V, PA-100V, and PA-343S, in PB2 and PA (26–28). We previously reported additional mammalian adapted amino acid residues, such as PA-356R and PB2-292V, in recent chicken H9N2 isolates (32–34). Thus, H9N2 virus is highly adept at producing mammalian adaptive mutations in its natural chicken host and contributes to viral transmissibility to humans.
There are at least three major factors behind the largest H7N9 virus epidemic to date (2, 35). First, over the four previous waves, the H7N9 virus had steadily spread through chickens from the original center of infection (the Yangtze River Delta) to across China (2, 25, 35). With the growing cocirculation of H7N9 and H9N2 viruses in most of China, it is not surprising that reassortment events took place. Second, the 2016–2017 H7N9 virus was more infectious. We found that the new subclade of PB2 and PA genes increased the viral polymerase activity of H7N9 viruses in chicken cells, which would facilitate viral spread among chicken populations. Moreover, the H7N9 viruses were found to replicate better than their predecessor in the upper and lower respiratory tracts of mice, which suggests more effective transmission of G72 H7N9 virus from chickens to humans. Third, other gene segments, in addition to PB2 and PA genes, with mammalian mutations could have contributed to increased human infection by H7N9 virus (2, 6).
The clinical severity of human cases in the fifth H7N9 virus wave was not significantly different from those in previous waves (36). Our mouse challenge results also found no significant difference in survival rates between G13 and its reassortants with H9N2-derived PB2 and PA genes. Thus, the evolved PB2 and PA genes of H9N2 virus could have conferred higher infectivity of H7N9 virus in humans without significantly enhancing its pathogenicity. In addition to H7N9 virus, H5N6 and other viruses with H9N2-original genes are also widely prevalent in poultry populations in China (37). The impact of H9N2 virus on these reassortants should also be given more attention.
In summary, the dominant G72 genotype H7N9 virus, responsible for the largest 2016–2017 H7N9 virus epidemic to date, had arisen from the reassortment of new subclade PB2 and PA genes from continually evolving H9N2 virus. At present, H9N2 virus is the most dominant AIV in China and shows little seasonality of transmission in chicken populations (7, 8). Although the number of H7N9 virus isolates in chickens has recently declined (4), H9N2 virus with mammalian adaptive mutations remains widespread among chickens. Therefore, it is necessary to continue to gather epidemiological data on H9N2 virus and its reassortants in chickens throughout the country to chart their evolutionary progress and better predict zoonotic transmission to humans.
MATERIALS AND METHODS
Ethics statement.
All animal research was approved by the Beijing Association for Science and Technology and performed in compliance with Beijing Laboratory Animal Welfare and Ethics guidelines, as issued by the Beijing Administration Committee of Laboratory Animals, and in accordance with China Agricultural University (CAU) Institutional Animal Care and Use Committee guidelines (SKLAB-B-2010-003).
Virus isolation, identification, and genomic sequencing.
In our retrospective survey for the period from January 2014 to December 2017, samples (chicken carcasses and lung samples) from 1,455 chicken flocks located across 21 provinces in China (Anhui, Beijing, Guangxi, Guansu, Fujuan, Hebei, Heilongjiang, Henan, Hubei, Jiangsu, Jiangxi, Jilin, Liaoning, Neimenggu, Ningxia, Shanxi, Shaanxi, Shandong, Sichuan, Yunnan, and Zhejiang) were received by our laboratory for diagnosis (see Table S1 in the supplemental material). Samples were from H9N2 virus-vaccinated chicken flocks that showed respiratory signs and/or a 5 to 20% drop in egg production. During 2014 to 2017, H9N2 virus vaccines used in chickens in China were mostly derived from strains belonging to antigen group F (38). Virus isolation using 10-day-old specific-pathogen-free (SPF) embryonated chicken eggs and virus identification by a hemagglutination inhibition (HI) assay were carried out as previously described (8). Typically, several chickens or lung samples from each flock were sent to the laboratory each time. If H9N2 virus was isolated from one or more samples, the flock was considered H9N2 virus positive. The virus isolation rate was determined by dividing the number of H9N2-positive flocks by the total number of flocks. Oropharyngeal and cloacal swabs from healthy chickens from live poultry markets were also taken in four provinces (Beijing, Gansu, Hebei, and Shandong) for virus isolation. H9N2 viruses (n = 143) isolated from 2014 through 2017 and H7N9 viruses (n = 17) isolated from 2013 through 2017 were sequenced as previously described (8). All isolated H7N9 viruses were LPAI viruses.
Human cases of H7N9 virus infection in China.
The number of confirmed human cases of H7N9 virus infection cited in this study were derived from data from the World Health Organization (www.who.int/influenza/human_animal_interface/avian_influenza/archive/en/) and the National Health Commission of the People’s Republic of China (www.nhc.gov.cn).
Sequence collection and alignment.
All previously published sequences of H9N2 chicken influenza A virus isolated in China (2010 to 2017) and related sequences were obtained from the Global Initiative on Sharing Avian Influenza Data (www.gisaid.org) and the Influenza Virus Resource at the National Center for Biotechnology Information (NCBI) (www.ncbi.nlm.nih.gov/genomes/FLU) (Tables S8 and S9). All sequences of H7N9 virus published before September 2017 were acquired from the above-mentioned databases. H9N2 and H7N9 virus sequences, combined with sequences generated in this study (Table S9), were used for further analysis. All duplicate submissions were removed by identifying sets of isolates with identical viral names and sequences in submitted segments. The resulting sequences of each gene segment were aligned using MUSCLE v3.7 (39) via the CIPRES Science Gateway (40), manually adjusted to correct frameshift errors, and subsequently translated. Downstream phylogenetic analyses were performed on regions of the alignments containing few gaps across sequences. These regions consist of the following intervals (nucleotide numbers are from the start of the untranslated region): PB2 nucleotides 28 to 2307, PB1 nucleotides 25 to 2298, PA nucleotides 25 to 2175, HA nucleotides 34 to 1716, NP nucleotides 46 to 1542, NA nucleotides 20 to 1420, MP nucleotides 26 to 1007, and NS nucleotides 27 to 864.
Phylogenetic analysis and clade classification.
IQ-TREE was used to construct maximum likelihood phylogenies for each segment (41). A generalized time-reversible substitution model with a gamma distribution (GTR+G) was used, and branch supports were assessed through the Shimodaira-Hasegawa approximate likelihood-ratio test (SH-aLRT) (42) with 1,000 replicated tests. Clades with SH-aLRT support of ≥90% in the tree were selected as clusters. Clades were then manually merged, if necessary, based on the branch length (>0.01) and reported classification (6, 8). In the identified clades, clusters that formed relatively independent branches (SH-aLRT support of >90% and branch length of >0.001) were identified as subclades. Each clade/subclade was assigned a unique clade/subclade identification number. Figures were generated using iTOL (43).
Genotypic analysis.
Isolates were genotyped/subgenotyped if sequences and clade/subclade assignments were available for all eight segments of H9N2 viruses (8) or for all six internal segments of H7N9 viruses (6, 18, 20). Virus subgenotypes were analyzed in chicken H9N2 viruses isolated in China from 2010 to 2017. Virus genotypes were analyzed in H7N9 viruses isolated from humans and other hosts in China during wave 1 to wave 5. Genotype/subgenotype identifications were assigned according to the isolation time of the initial founder isolate.
Antigenic analysis.
HI assays, performed as previously described (44), were used to antigenically characterize H9N2 viruses isolated in China in this study. The HI titer was expressed as the reciprocal of the highest serum dilution in which hemagglutination was inhibited. For analysis of antigenic variation, the HI data were log2 transformed, and the transformed data were downscaled to three-dimensional space by multidimensional scaling (MDS) (45) with an explained variance of 96.78%. Antigenic clustering of virulent strains was performed using K-means clustering (46).
Plasmids and cells.
Protein expression plasmids, pcDNA-PB2, pcDNA-PB1, pcDNA-PA, and pcDNA-NP, were generated by subcloning the corresponding coding segments into the pcDNA3.1 vector. Human embryonic kidney (293T) cells, human lung adenocarcinoma epithelial (A549) cells, and chicken embryo fibroblast (DF-1) cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) supplemented with 10% fetal bovine serum (FBS; Gibco), 100 U/ml of penicillin, and 100 μg/ml of streptomycin at 37°C in a 5% CO2 atmosphere.
Polymerase activity assay.
A dual-luciferase reporter assay system (Promega, Madison, WI, USA) was used to compare the polymerase activities of different viral RNP complexes (32). PB2, PB1, PA, and NP gene segments of the indicated viruses were separately cloned into the pCDNA3.1 expression plasmid. PB2, PB1, PA, and NP plasmids (125 ng each plasmid) along with the pLuci luciferase reporter plasmid (10 ng) and the renilla internal control plasmid (2.5 ng) were used to transfect 293T cells or DF-1 cells. 293T cell cultures were incubated at 33°C or 37°C; DF-1 cell cultures were incubated at 37°C or 39°C. Cell lysates were analyzed at 24 h posttransfection for firefly and renilla luciferase activities using a GloMax 96 microplate luminometer (Promega). The PB2, PB1, PA, and NP genes from an LPAI G13 genotype H7N9 virus (A/chicken/Shandong/M0303-10/2017) were used as a reference viral RNP complex in polymerase activity assays. Series of recombinant viral RNP complexes were generated by replacing the reference complex with specific PB2 and PA genes from H9N2 and G72 genotype H7N9 viruses, whose PB2 and PA genes were from the PB2-6.2 and PA-3.1 subclades, respectively. The H9N2 viruses used were A/chicken/Shandong/217/2017 and A/chicken/Hebei/M1211-20/2016, and the G72 genotype H7N9 viruses used were A/chicken/Shandong/F0513-60/2017 and A/chicken/Beijing/F0606-1/2017. Further details of virus and segment information are shown in Table S5 in the supplemental material.
Generation of reassortant viruses by reverse genetics.
A recombinant reference vG13 genotype (rG13) virus was generated by reverse genetics (47). All six internal gene segments were amplified by reverse transcription-PCR (RT-PCR) from the LPAI G13 genotype H7N9 virus (A/chicken/Shandong/M0303-10/2017), and two surface genes were from the LPAI G13 virus (A/chicken/Jiangsu/BD145/2014). Each gene was individually cloned into a dual-promoter plasmid, pHW2000. Based on the rG13 virus, three additional reassortants (rG13:PB2/PA-H9N2, rG13:PB2/PA-H7N9-1, and rG13:PB2/PA-H7N9-2) were generated by substituting the PB2 and PA genes (from the PB2-6.2 and PA-3.1 subclades) of H9N2 and G72 H7N9 viruses. Rescued viruses were generated in 293T cells as previously described (47). Further details of segment and viral information for these reassortants are shown in Table S5.
Mouse challenge study.
Fifteen mice (6-week-old female BALB/c mice; Vital River Laboratory, Beijing, China) per group were anesthetized with tiletamine-zolazepam (Zoletil; Virbac SA, Carros, France) (20 mg/g), and each mouse was inoculated intranasally with 106 TCID50 of the indicated test virus diluted to 50 μl with phosphate-buffered saline (PBS). Five mice from each group were monitored daily for 14 days, and mice that lost >25% of their original body weight were humanely euthanized. Five mice from each group were euthanized at 3 and 5 days postinfection (dpi) for the determination of virus titers and histopathology. Nasal turbinates and lungs were collected and homogenized in 1 ml of cold PBS. Virus titers were determined by TCID50 assays. A portion of the lung from each euthanized mouse at 3 dpi was fixed in 10% phosphate-buffered formalin for histopathological examination, which was performed as described previously (48). The MID50 was determined according to a previous study (49).
Statistical analysis.
Experimental groups were statistically compared by analysis of variance (ANOVA). A P value of <0.05 was considered to indicate a statistically significant difference.
Data availability.
The sequences generated in this study have been deposited in the GenBank database, and the accession numbers are listed in Table S9 in the supplemental material.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jixun Zhao (China Agricultural University, China) and Robert G. Webster (St. Jude Children’s Research Hospital, USA) for helpful suggestions on data analysis.
This work was supported by the National Natural Science Foundation of China (81961128002), the National Key Research and Development Program (2016YFD0500204, 2018YFD0501400, and 2018YFD0501404), the National Natural Science Foundation of China (31672568 and 31761133003), and the Project of Poultry Industry Innovation Team of Shandong Provincial Modern Agricultural Industry Technology System (SDAIT-11-03).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, Xu X, Lu H, Zhu W, Gao Z, Xiang N, Shen Y, He Z, Gu Y, Zhang Z, Yang Y, Zhao X, Zhou L, Li X, Zou S, Zhang Y, Li X, Yang L, Guo J, Dong J, Li Q, Dong L, Zhu Y, Bai T, Wang S, Hao P, Yang W, Zhang Y, Han J, Yu H, Li D, Gao GF, Wu G, Wang Y, Yuan Z, Shu Y. 2013. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med 368:1888–1897. 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 2.Su S, Gu M, Liu D, Cui J, Gao GF, Zhou J, Liu X. 2017. Epidemiology, evolution, and pathogenesis of H7N9 influenza viruses in five epidemic waves since 2013 in China. Trends Microbiol 25:713–728. 10.1016/j.tim.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 3.Qi W, Jia W, Liu D, Li J, Bi Y, Xie S, Li B, Hu T, Du Y, Xing L, Zhang J, Zhang F, Wei X, Eden J-S, Li H, Tian H, Li W, Su G, Lao G, Xu C, Xu B, Liu W, Zhang G, Ren T, Holmes EC, Cui J, Shi W, Gao GF, Liao M. 2018. Emergence and adaptation of a novel highly pathogenic H7N9 influenza virus in birds and humans from a 2013 human-infecting low-pathogenic ancestor. J Virol 92:e00921-17. 10.1128/JVI.00921-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi J, Deng G, Ma S, Zeng X, Yin X, Li M, Zhang B, Cui P, Chen Y, Yang H, Wan X, Liu L, Chen P, Jiang Y, Guan Y, Liu J, Gu W, Han S, Song Y, Liang L, Qu Z, Hou Y, Wang X, Bao H, Tian G, Li Y, Jiang L, Li C, Chen H. 2018. Rapid evolution of H7N9 highly pathogenic viruses that emerged in China in 2017. Cell Host Microbe 24:558–568.e7. 10.1016/j.chom.2018.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi J, Deng G, Kong H, Gu C, Ma S, Yin X, Zeng X, Cui P, Chen Y, Yang H, Wan X, Wang X, Liu L, Chen P, Jiang Y, Liu J, Guan Y, Suzuki Y, Li M, Qu Z, Guan L, Zang J, Gu W, Han S, Song Y, Hu Y, Wang Z, Gu L, Yang W, Liang L, Bao H, Tian G, Li Y, Qiao C, Jiang L, Li C, Bu Z, Chen H. 2017. H7N9 virulent mutants detected in chickens in China pose an increased threat to humans. Cell Res 27:1409–1421. 10.1038/cr.2017.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu W, Dong J, Zhang Y, Yang L, Li X, Chen T, Zhao X, Wei H, Bo H, Zeng X, Huang W, Li Z, Tang J, Zhou J, Gao R, Xin L, Yang J, Zou S, Chen W, Liu J, Shu Y, Wang D. 2018. A gene constellation in avian influenza A (H7N9) viruses may have facilitated the fifth wave outbreak in China. Cell Rep 23:909–917. 10.1016/j.celrep.2018.03.081. [DOI] [PubMed] [Google Scholar]
- 7.Bi Y, Li J, Li S, Fu G, Jin T, Zhang C, Yang Y, Ma Z, Tian W, Li J, Xiao S, Li L, Yin R, Zhang Y, Wang L, Qin Y, Yao Z, Meng F, Hu D, Li D, Wong G, Liu F, Lv N, Wang L, Fu L, Yang Y, Peng Y, Ma J, Sharshov K, Shestopalov A, Gulyaeva M, Gao GF, Chen J, Shi Y, Liu WJ, Chu D, Huang Y, Liu Y, Liu L, Liu W, Chen Q, Shi W. 2020. Dominant subtype switch in avian influenza viruses during 2016-2019 in China. Nat Commun 11:5909. 10.1038/s41467-020-19671-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pu J, Wang S, Yin Y, Zhang G, Carter RA, Wang J, Xu G, Sun H, Wang M, Wen C, Wei Y, Wang D, Zhu B, Lemmon G, Jiao Y, Duan S, Wang Q, Du Q, Sun M, Bao J, Sun Y, Zhao J, Zhang H, Wu G, Liu J, Webster RG. 2015. Evolution of the H9N2 influenza genotype that facilitated the genesis of the novel H7N9 virus. Proc Natl Acad Sci U S A 112:548–553. 10.1073/pnas.1422456112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gu M, Xu L, Wang X, Liu X. 2017. Current situation of H9N2 subtype avian influenza in China. Vet Res 48:49. 10.1186/s13567-017-0453-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li C, Yu K, Tian G, Yu D, Liu L, Jing B, Ping J, Chen H. 2005. Evolution of H9N2 influenza viruses from domestic poultry in Mainland China. Virology 340:70–83. 10.1016/j.virol.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 11.Zhang P, Tang Y, Liu X, Peng D, Liu W, Liu H, Lu S, Liu X. 2008. Characterization of H9N2 influenza viruses isolated from vaccinated flocks in an integrated broiler chicken operation in eastern China during a 5 year period (1998-2002). J Gen Virol 89:3102–3112. 10.1099/vir.0.2008/005652-0. [DOI] [PubMed] [Google Scholar]
- 12.Wu ZQ, Ji J, Zuo KJ, Xie QM, Li HM, Liu J, Chen F, Xue CY, Ma JY, Bi YZ. 2010. Cloning and phylogenetic analysis of hemagglutinin gene of H9N2 subtype avian influenza virus from different isolates in China during 2002 to 2009. Poult Sci 89:1136–1143. 10.3382/ps.2010-00695. [DOI] [PubMed] [Google Scholar]
- 13.Sun Y, Pu J, Jiang Z, Guan T, Xia Y, Xu Q, Liu L, Ma B, Tian F, Brown EG, Liu J. 2010. Genotypic evolution and antigenic drift of H9N2 influenza viruses in China from 1994 to 2008. Vet Microbiol 146:215–225. 10.1016/j.vetmic.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 14.Lam TT-Y, Wang J, Shen Y, Zhou B, Duan L, Cheung C-L, Ma C, Lycett SJ, Leung CY-H, Chen X, Li L, Hong W, Chai Y, Zhou L, Liang H, Ou Z, Liu Y, Farooqui A, Kelvin DJ, Poon LLM, Smith DK, Pybus OG, Leung GM, Shu Y, Webster RG, Webby RJ, Peiris JSM, Rambaut A, Zhu H, Guan Y. 2013. The genesis and source of the H7N9 influenza viruses causing human infections in China. Nature 502:241–244. 10.1038/nature12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu X, Jin T, Cui Y, Pu X, Li J, Xu J, Liu G, Jia H, Liu D, Song S, Yu Y, Xie L, Huang R, Ding H, Kou Y, Zhou Y, Wang Y, Xu X, Yin Y, Wang J, Guo C, Yang X, Hu L, Wu X, Wang H, Liu J, Zhao G, Zhou J, Pan J, Gao GF, Yang R, Wang J. 2014. Influenza H7N9 and H9N2 viruses: coexistence in poultry linked to human H7N9 infection and genome characteristics. J Virol 88:3423–3431. 10.1128/JVI.02059-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lam TT-Y, Zhou B, Wang J, Chai Y, Shen Y, Chen X, Ma C, Hong W, Chen Y, Zhang Y, Duan L, Chen P, Jiang J, Zhang Y, Li L, Poon LLM, Webby RJ, Smith DK, Leung GM, Peiris JSM, Holmes EC, Guan Y, Zhu H. 2015. Dissemination, divergence and establishment of H7N9 influenza viruses in China. Nature 522:102–105. 10.1038/nature14348. [DOI] [PubMed] [Google Scholar]
- 17.Wu A, Su C, Wang D, Peng Y, Liu M, Hua S, Li T, Gao GF, Tang H, Chen J, Liu X, Shu Y, Peng D, Jiang T. 2013. Sequential reassortments underlie diverse influenza H7N9 genotypes in China. Cell Host Microbe 14:446–452. 10.1016/j.chom.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 18.Cui L, Liu D, Shi W, Pan J, Qi X, Li X, Guo X, Zhou M, Li W, Li J, Haywood J, Xiao H, Yu X, Pu X, Wu Y, Yu H, Zhao K, Zhu Y, Wu B, Jin T, Shi Z, Tang F, Zhu F, Sun Q, Wu L, Yang R, Yan J, Lei F, Zhu B, Liu W, Ma J, Wang H, Gao GF. 2014. Dynamic reassortments and genetic heterogeneity of the human-infecting influenza A (H7N9) virus. Nat Commun 5:3142. 10.1038/ncomms4142. [DOI] [PubMed] [Google Scholar]
- 19.Wang D, Yang L, Gao R, Zhang X, Tan Y, Wu A, Zhu W, Zhou J, Zou S, Li X, Sun Y, Zhang Y, Liu Y, Liu T, Xiong Y, Xu J, Chen L, Weng Y, Qi X, Guo J, Li X, Dong J, Huang W, Zhang Y, Dong L, Zhao X, Liu L, Lu J, Lan Y, Wei H, Xin L, Chen Y, Xu C, Chen T, Zhu Y, Jiang T, Feng Z, Yang W, Wang Y, Zhu H, Guan Y, Gao GF, Li D, Han J, Wang S, Wu G, Shu Y. 2014. Genetic tuning of the novel avian influenza A(H7N9) virus during interspecies transmission, China, 2013. Euro Surveill 19:20836. 10.2807/1560-7917.es2014.19.25.20836. [DOI] [PubMed] [Google Scholar]
- 20.Quan C, Shi W, Yang Y, Yang Y, Liu X, Xu W, Li H, Li J, Wang Q, Tong Z, Wong G, Zhang C, Ma S, Ma Z, Fu G, Zhang Z, Huang Y, Song H, Yang L, Liu WJ, Liu Y, Liu W, Gao GF, Bi Y. 2018. New threats from H7N9 influenza virus: spread and evolution of high- and low-pathogenicity variants with high genomic diversity in wave five. J Virol 92:e00301-18. 10.1128/JVI.00301-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rambaut A, Pybus OG, Nelson MI, Viboud C, Taubenberger JK, Holmes EC. 2008. The genomic and epidemiological dynamics of human influenza A virus. Nature 453:615–619. 10.1038/nature06945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McHardy AC, Adams B. 2009. The role of genomics in tracking the evolution of influenza A virus. PLoS Pathog 5:e1000566. 10.1371/journal.ppat.1000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boni MF. 2008. Vaccination and antigenic drift in influenza. Vaccine 26(Suppl 3):C8–C14. 10.1016/j.vaccine.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. 1992. Evolution and ecology of influenza A viruses. Microbiol Rev 56:152–179. 10.1128/MR.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang D, Yang L, Zhu W, Zhang Y, Zou S, Bo H, Gao R, Dong J, Huang W, Guo J, Li Z, Zhao X, Li X, Xin L, Zhou J, Chen T, Dong L, Wei H, Li X, Liu L, Tang J, Lan Y, Yang J, Shu Y. 2016. Two outbreak sources of influenza A (H7N9) viruses have been established in China. J Virol 90:5561–5573. 10.1128/JVI.03173-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao C, Ma W, Sun N, Huang L, Li Y, Zeng Z, Wen Y, Zhang Z, Li H, Li Q, Yu Y, Zheng Y, Liu S, Hu P, Zhang X, Ning Z, Qi W, Liao M. 2016. PB2-588 V promotes the mammalian adaptation of H10N8, H7N9 and H9N2 avian influenza viruses. Sci Rep 6:19474. 10.1038/srep19474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun Y, Xu Q, Shen Y, Liu L, Wei K, Sun H, Pu J, Chang K-C, Liu J. 2014. Naturally occurring mutations in the PA gene are key contributors to increased virulence of pandemic H1N1/09 influenza virus in mice. J Virol 88:4600–4604. 10.1128/JVI.03158-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamayoshi S, Yamada S, Fukuyama S, Murakami S, Zhao D, Uraki R, Watanabe T, Tomita Y, Macken C, Neumann G, Kawaoka Y. 2014. Virulence-affecting amino acid changes in the PA protein of H7N9 influenza A viruses. J Virol 88:3127–3134. 10.1128/JVI.03155-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu L, Bao L, Zhou J, Wang D, Deng W, Lv Q, Ma Y, Li F, Sun H, Zhan L, Zhu H, Ma C, Shu Y, Qin C. 2011. Genomic polymorphism of the pandemic A (H1N1) influenza viruses correlates with viral replication, virulence, and pathogenicity in vitro and in vivo. PLoS One 6:e20698. 10.1371/journal.pone.0020698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng X, Wu H, Peng X, Wu X, Cheng L, Liu F, Ji S, Wu N. 2016. Amino acid substitutions occurring during adaptation of an emergent H5N6 avian influenza virus to mammals. Arch Virol 161:1665–1670. 10.1007/s00705-016-2826-7. [DOI] [PubMed] [Google Scholar]
- 31.Naffakh N, Tomoiu A, Rameix-Welti M-A, van der Werf S. 2008. Host restriction of avian influenza viruses at the level of the ribonucleoproteins. Annu Rev Microbiol 62:403–424. 10.1146/annurev.micro.62.081307.162746. [DOI] [PubMed] [Google Scholar]
- 32.Xu G, Zhang X, Gao W, Wang C, Wang J, Sun H, Sun Y, Guo L, Zhang R, Chang K-C, Liu J, Pu J. 2016. Prevailing PA mutation K356R in avian influenza H9N2 virus increases mammalian replication and pathogenicity. J Virol 90:8105–8114. 10.1128/JVI.00883-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao W, Zu Z, Liu J, Song J, Wang X, Wang C, Liu L, Tong Q, Wang M, Sun H, Sun Y, Liu J, Chang K-C, Pu J. 2019. Prevailing I292V PB2 mutation in avian influenza H9N2 virus increases viral polymerase function and attenuates IFN-β induction in human cells. J Gen Virol 100:1273–1281. 10.1099/jgv.0.001294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun Y, Tan Y, Wei K, Sun H, Shi Y, Pu J, Yang H, Gao GF, Yin Y, Feng W, Perez DR, Liu J. 2013. Amino acid 316 of hemagglutinin and the neuraminidase stalk length influence virulence of H9N2 influenza virus in chickens and mice. J Virol 87:2963–2968. 10.1128/JVI.02688-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X, Jiang H, Wu P, Uyeki TM, Feng L, Lai S, Wang L, Huo X, Xu K, Chen E, Wang X, He J, Kang M, Zhang R, Zhang J, Wu J, Hu S, Zhang H, Liu X, Fu W, Ou J, Wu S, Qin Y, Zhang Z, Shi Y, Zhang J, Artois J, Fang VJ, Zhu H, Guan Y, Gilbert M, Horby PW, Leung GM, Gao GF, Cowling BJ, Yu H. 2017. Epidemiology of avian influenza A H7N9 virus in human beings across five epidemics in mainland China, 2013–17: an epidemiological study of laboratory-confirmed case series. Lancet Infect Dis 17:822–832. 10.1016/S1473-3099(17)30323-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X, Wu P, Pei Y, Tsang TK, Gu D, Wang W, Zhang J, Horby PW, Uyeki TM, Cowling BJ, Yu H. 2019. Assessment of human-to-human transmissibility of avian influenza A(H7N9) virus across 5 waves by analyzing clusters of case patients in Mainland China, 2013–2017. Clin Infect Dis 68:623–631. 10.1093/cid/ciy541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bi Y, Chen Q, Wang Q, Chen J, Jin T, Wong G, Quan C, Liu J, Wu J, Yin R, Zhao L, Li M, Ding Z, Zou R, Xu W, Li H, Wang H, Tian K, Fu G, Huang Y, Shestopalov A, Li S, Xu B, Yu H, Luo T, Lu L, Xu X, Luo Y, Liu Y, Shi W, Liu D, Gao GF. 2016. Genesis, evolution and prevalence of H5N6 avian influenza viruses in China. Cell Host Microbe 20:810–821. 10.1016/j.chom.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 38.Wei Y, Xu G, Zhang G, Wen C, Anwar F, Wang S, Lemmon G, Wang J, Carter R, Wang M, Sun H, Sun Y, Zhao J, Wu G, Webster RG, Liu J, Pu J. 2016. Antigenic evolution of H9N2 chicken influenza viruses isolated in China during 2009-2013 and selection of a candidate vaccine strain with broad cross-reactivity. Vet Microbiol 182:1–7. 10.1016/j.vetmic.2015.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees, p 1–8. In Proceedings of the 2010 Grid Computing Environments Workshop. IEEE, Piscataway, NJ. [Google Scholar]
- 41.Trifinopoulos J, Nguyen L-T, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res 44:W232–W235. 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 43.Letunic I, Bork P. 2019. Interactive Tree of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47:W256–W259. 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Edwards S. 2006. OIE laboratory standards for avian influenza. Dev Biol (Basel) 124:159–162. [PubMed] [Google Scholar]
- 45.Borg I, Groenen P. 2003. Modern multidimensional scaling: theory and applications. J Educ Meas 40:277–280. 10.1111/j.1745-3984.2003.tb01108.x. [DOI] [Google Scholar]
- 46.Lapedes A, Farber R. 2001. The geometry of shape space: application to influenza. J Theor Biol 212:57–69. 10.1006/jtbi.2001.2347. [DOI] [PubMed] [Google Scholar]
- 47.Sun Y, Qin K, Wang J, Pu J, Tang Q, Hu Y, Bi Y, Zhao X, Yang H, Shu Y, Liu J. 2011. High genetic compatibility and increased pathogenicity of reassortants derived from avian H9N2 and pandemic H1N1/2009 influenza viruses. Proc Natl Acad Sci U S A 108:4164–4169. 10.1073/pnas.1019109108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y, Sun Y, Sun H, Pu J, Bi Y, Shi Y, Lu X, Li J, Zhu Q, Gao GF, Yang H, Liu J. 2012. A single amino acid at the hemagglutinin cleavage site contributes to the pathogenicity and neurovirulence of H5N1 influenza virus in mice. J Virol 86:6924–6931. 10.1128/JVI.07142-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramesh AK, Parreño V, Schmidt PJ, Lei S, Zhong W, Jiang X, Emelko MB, Yuan L. 2020. Evaluation of the 50% infectious dose of human norovirus Cin-2 in gnotobiotic pigs: a comparison of classical and contemporary methods for endpoint estimation. Viruses 12:955. 10.3390/v12090955. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequences generated in this study have been deposited in the GenBank database, and the accession numbers are listed in Table S9 in the supplemental material.