The four-residue insert (PRRA) at the boundary between the S1 and S2 subunits of SARS-CoV-2 has been widely recognized since day 1 for its role in SARS-CoV-2 S protein processing and activation. As this PRRA insert is unique to SARS-CoV-2 among group b betacoronaviruses, it is thought to affect the tissue and species tropism of SARS-CoV-2.
KEYWORDS: SARS-CoV-2, RaTG13, pangolin GX, spike protein, furin cleavage site, coronavirus entry, ACE2, cross-species transmission
ABSTRACT
Biochemical and structural analyses suggest that severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) efficiently infects humans, and the presence of four residues (PRRA) at the S1/S2 site within the spike (S) protein may lead to unexpected tissue or host tropism. Here, we report that SARS-CoV-2 efficiently utilized angiotensin-converting enzyme 2 (ACE2) of 9 species to infect 293T cells. Similarly, pseudoviruses bearing S protein derived from either bat RaTG13 or pangolin GX, two closely related animal coronaviruses, utilized ACE2 of a diverse range of animal species to gain entry. The removal of PRRA from SARS-CoV-2 S protein displayed distinct effects on pseudoviral entry into different cell types. Unexpectedly, the insertion of PRRA into the RaTG13 S protein selectively abrogated the usage of horseshoe bat and pangolin ACE2 but enhanced the usage of mouse ACE2 by the relevant pseudovirus to enter cells. Together, our findings identified a previously unrecognized effect of the PRRA insert on SARS-CoV-2 and RaTG13 S proteins.
IMPORTANCE The four-residue insert (PRRA) at the boundary between the S1 and S2 subunits of SARS-CoV-2 has been widely recognized since day 1 for its role in SARS-CoV-2 S protein processing and activation. As this PRRA insert is unique to SARS-CoV-2 among group b betacoronaviruses, it is thought to affect the tissue and species tropism of SARS-CoV-2. We compared the usages of 10 ACE2 orthologs and found that the presence of PRRA not only affects the cellular tropism of SARS-CoV-2 but also modulates the usage of ACE2 orthologs by the closely related bat RaTG13 S protein. The binding of pseudovirions carrying RaTG13 S with a PRRA insert to mouse ACE2 was nearly 2-fold higher than that of pseudovirions carrying RaTG13 S.
INTRODUCTION
The outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (or 2019 novel coronavirus [2019-nCoV]) has quickly turned into a global pandemic. Multiple studies have suggested a zoonotic origin of SARS-CoV-2, highlighting the potential of coronaviruses (CoVs) to continuously evolve in wild-animal reservoirs and jump to new species (1, 2). Various bat species are believed to be reservoirs of progenitors of diverse CoVs, including severe acute respiratory syndrome CoV (SARS-CoV) and SARS-CoV-2. The closest virus to SARS-CoV-2 found to date is the bat RaTG13 CoV, with over 96% identity at the genomic level (3). A newly reported bat CoV, RmYN02, shares 93.3% nucleotide identity with SARS-CoV-2 genomewide and 97.2% identity in the 1ab gene (2). Besides bat CoVs, pangolin/GD/2019 CoV (pangolin-GD) and pangolin/GX/P5L/2017 CoV (pangolin-GX) share up to 90% and 85.2% sequence identities with SARS-CoV-2, respectively (4, 5). While the origin of SARS-CoV-2 appears to be linked to bat reservoirs, there is still no definitive evidence of an intermediate host that could have transmitted the virus to humans.
The coronavirus S protein plays a pivotal role in mediating coronavirus entry (6, 7). The ectodomain of SARS-CoV S protein is divided into two subunits, S1 and S2. SARS-CoV S1 contains a receptor-binding domain (RBD) that specifically binds to human angiotensin-converting enzyme 2 (ACE2), whereas the S2 subunit contains the fusion peptide (8, 9). Like SARS-CoV, the SARS-CoV-2 S protein in the prefusion conformation is a homotrimer, with three S1 heads sitting on top of a trimeric S2 stalk (10–12). It was initially proposed that at a given time, only one of three RBDs is in an upward position for receptor binding (12). However, a very recent structural study of a full-length, fully wild-type SARS-CoV-2 S protein indicated that the prefusion structure is in the “all-down” configuration (13). The interaction between the RBD and ACE2 is thought to lock the SARS-CoV-2 spike protein in a conformation that makes it susceptible to multiple cellular proteases (14), including cell surface transmembrane serine protease 2 (TMPRSS2) and the lysosomal proteases cathepsins (15–20). In addition to receptor binding, sequential proteolytic cleavages of SARS-CoV S protein at the S1/S2 site and the S2 site are also important for triggering conformational changes in the S2 subunit and the final exposure of the fusion peptide (16). All current evidence implies that SARS-CoV-2 may follow the footsteps of SARS-CoV to enter host cells.
The S protein of SARS-CoV-2 displays two distinct features compared to SARS-CoV and other betacoronaviruses: (i) the SARS-CoV-2 S protein appears to have been well adapted for binding to human ACE2 (hACE2) despite significant sequence divergence from SARS-CoV S protein in the RBD region (21, 22), and (ii) a distinct 4-amino-acid insert within the S protein (underlined, SPRRAR↓S) between the S1 receptor-binding subunit and the S2 fusion subunit presumably creates a functional polybasic (furin) cleavage site. This novel S1/S2 site within the SARS-CoV-2 S protein is hypothesized to affect the stability, transmission, and/or host range of the virus (23, 24).
In this study, we set out to determine (i) if SARS-CoV-2 infects cells via diverse animal ACE2 and (ii) how the PRRA insert at the S1/S2 boundary affects the cellular and host range of SARS-CoV-2 and the closely related CoV RaTG13. Our findings not only demonstrate the wide usage of ACE2 from diverse animal species by SARS-CoV-2, RaTG13, and pangolin GX but also reveal a striking effect of the PRRA insert on the RaTG13 spike protein regarding its usage of ACE2 derived from horseshoe bat, pangolin, and mouse.
RESULTS
Efficient infection of cells expressing ACE2 of diverse species by SARS-CoV-2.
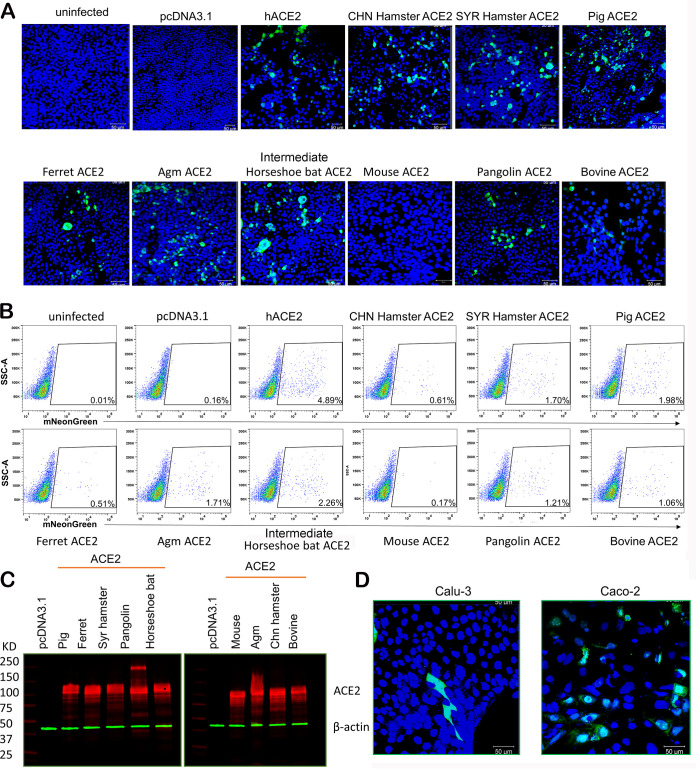
Although SARS-CoV-2 S differs significantly from SARS-CoV S in the receptor-binding motif (RBM) within the RBD, SARS-CoV-2 S protein appears to have evolved a mechanism to efficiently utilize human ACE2 to gain entry (1, 25). Besides human, SARS-CoV-2 reportedly infects domesticated pets such as cats and dogs (26) and wild animals such as mink (27), raising the concern of cross-species transmissions. To test whether SARS-CoV-2 may use ACE2 of different species to infect cells, we transfected 293T cells to express ACE2 from 10 different species, including human, mouse, intermediate horseshoe bat (Rhinolophus affinis), pangolin, Syrian (SYR) hamster, Chinese (CHN) hamster, African green monkey (Agm), bovine, pig, and ferret. Cells were subsequently infected by a SARS-CoV-2 reporter virus (icSARS-CoV-2-mNG) that expresses the monomeric neon green (mNeonGreen [mNG]) fluorescent protein (28). Except for mouse ACE2 (mACE2), the expressions of all ACE2 orthologs rendered cells susceptible to SARS-CoV-2 infection (Fig. 1A), and the percentages of cells positive for mNeonGreen were quantified by flow cytometry (Fig. 1B). Of note, although the expression of each ACE2 ortholog was verified by Western blotting (Fig. 1C), we did not quantify each ACE2 ortholog on the cell surface, nor do we know whether each ACE2 ortholog is equally compatible with the endocytic pathway, which is probably utilized in SARS-CoV-2 entry into 293T cells. Therefore, the percentages of positive cells in Fig. 1B may not directly reflect the efficiencies of different ACE2 orthologs in mediating SARS-CoV-2 entry. The icSARS-CoV-2-mNG reporter virus also readily infected the human airway cell line Calu-3 and the intestinal cell line Caco-2, although only small percentages of both cell lines are susceptible (Fig. 1D).
FIG 1.
ACE2 orthologs of diverse species mediate SARS-CoV-2 infection. (A) 293T cells transfected with plasmids expressing 10 ACE2 orthologs or an empty vector and then infected by icSARS-CoV-2-mNG at a multiplicity of infection of 1 for 48 h. Images were captured by using a Zeiss LSM 880 laser scanning microscope. (B) Quantification of infection by flow cytometry. Cells from panel A were fixed in 4% paraformaldehyde, and mNG-positive cells were then quantified by flow cytometric analysis. SSC, side scatter. (C) The expression of ACE2 ortholog plasmids in 293T cells was detected using a mouse anti-V5 tag monoclonal antibody targeting the C-terminal V5 tag (in red). Green, anti-β-actin antibody. (D) Representative confocal images of Caco-2 and Calu-3 cells infected with icSARS-CoV-2-mNG at a multiplicity of infection of 1 for 48 h.
PRRA-led proteolytic cleavage of SARS-CoV-2 spike protein.
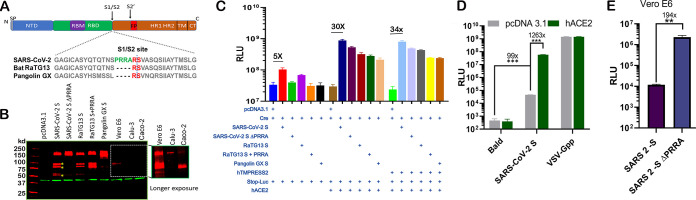
Next, we sought to investigate the role of the PRRA insert in SARS-CoV-2 entry. To explore the proteolytic cleavage mediated by the insert, we transfected 293T cells with constructs expressing wild-type SARS-CoV-2 spike (SARS-CoV-2 S), a mutant with PRRA removed (SARS-CoV-2 S ΔPRRA), bat RaTG13 spike (RaTG13 S), an insertion mutant containing the PRRA insert (RaTG13 S+PRRA), and the pangolin GX S protein (pangolin GX S) (Fig. 2A). We used a mouse monoclonal antibody that recognizes the S2 domains of both SARS-CoV spike and SARS-CoV-2 spike proteins for detection. Partial cleavage was noticed when SARS-CoV-2 S was expressed in 293T cells, and the deletion of PRRA abolished the cleaved products (Fig. 2B). Interestingly, RaTG13 S was partially cleaved even in the absence of PRRA. The insertion of PRRA into RaTG13 S further enhanced cleavage by 2- to 3-fold. Pangolin GX S protein did not show a similar cleavage pattern in transfected 293T cells. Altogether, these results suggest that CoV S proteins undergo intensive intracellular processing before being loaded onto virions. To confirm that cleavage occurs in the context of infection, Western blotting was performed using SARS-CoV-2-infected Vero E6 cell lysates. Again, a similar cleavage pattern was observed (Fig. 2B). Of note, the extent of the observed cleavage appears to vary significantly among susceptible cell lines. While the ratio of the cleaved over noncleaved SARS-CoV-2 S protein appears to be comparable between 293T and Vero E6 cells, the infected human lung epithelial cell line Calu-3 and the intestinal cell line Caco-2 yielded banding patterns that represent rather complete cleavage of the S protein in Caco-2 and less cleavage in Calu-3 cells (Fig. 2B).
FIG 2.
PRRA-led proteolytic cleavage of SARS-CoV-2 spike protein and effect on fusion. (A) Organization of SARS-CoV-2 spike protein and sequence alignment at the S1/S2 site with RaTG13 and pangolin GX S proteins. SP, signal peptide; NTD, N-terminal domain; FP, fusion peptide; TM, transmembrane; CT, C terminus. (B) 293T cells transfected with SARS-CoV-2 S, SARS-CoV-2 S ΔPRRA, bat RaTG13 spike (RaTG13 S), an insertion mutant containing PRRA (RaTG13 S+PRRA), and the pangolin GX spike protein (Pangolin GX S) (lanes 3 to 7) and SARS-CoV-2 S-infected Vero E6, Calu-3, and Caco-2 cells (lanes 8 to 10). A 5-min exposure of this part of the gel is included. Red, anti-S antibody; green, anti-β-actin antibody. (C) Cell-cell fusion mediated by CoV S proteins. 293T cells expressing Stop‐Luc, ACE2, and/or ACE2/TMPRSS2 (acceptor cells) were mixed at a 1:1 ratio with donor cells expressing Cre, CoV S, or both to initiate cell‐cell fusion. Data are presented as means ± standard errors of the means (SEM). (D) 293T cells or 293T-hACE2 cells were infected by pseudovirus bearing SARS-CoV-2 S. Bald viruses without any viral envelope or VSV-Gpp were included as negative and positive controls. (E) Entry of MLV pseudoviruses bearing SARS-CoV-2 S or SARS-CoV-2 S ΔPRRA in Vero cells. **, P < 0.001; ***, P < 0.0001. RLU, relative luciferase units.
A precleaved S protein at S1/S2 could confer an unexpected fusogenic ability on the plasma membrane and allow coronaviruses to undergo receptor-independent entry (virus-cell fusion) (7, 29). To assess S protein-mediated membrane fusion, we set up a quantitative cell-cell fusion assay in which 293T acceptor cells, containing a LoxP-flanked stop cassette that blocks the transcription of the downstream luciferase reporter gene, were cocultured with Cre-expressing donor cells. Fusion between donor and acceptor cell membranes removes the stop cassette and hence permits luciferase production. Various spike constructs were transfected into donor cells, and the hACE2 plasmid was then transfected into the acceptor cells. Notably, even in the absence of hACE2, SARS-CoV-2 spike induced fusion at a level about 5-fold over the background (Fig. 2C). The deletion of PRRA from SARS-CoV-2 S protein completely abolished this basal level of fusion activity. In the presence of hACE2, all S proteins induced cell-cell fusion to various extents (up to 30-fold above the background), with wild-type SARS-CoV-2 S being the most efficient one. Surprisingly, both the insertion of the novel PRRA into RaTG13 S and the deletion of PRRA from SARS-CoV-2 S reduced the fusogenic ability in the presence of ACE2, in comparison to their wild-type counterparts. Finally, the addition of TMPRSS2 marginally affected S protein-driven cell-cell fusion (Fig. 2C).
The PRRA insert at the S1/S2 cleavage site distinctly modulates cell susceptibility to SARS-CoV-2 pseudovirions.
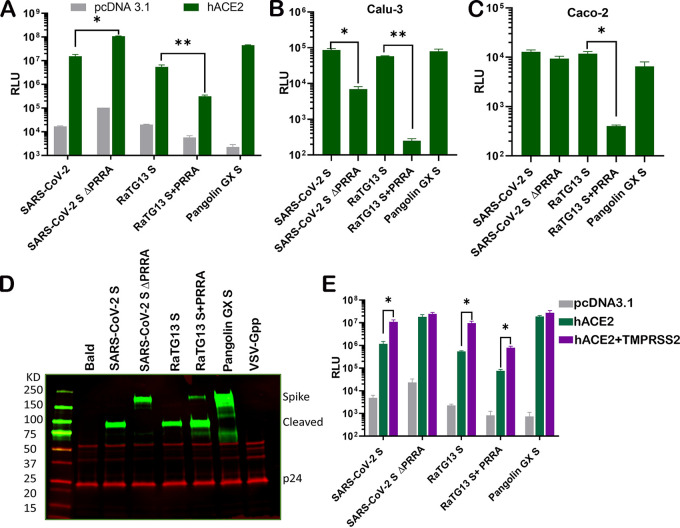
Next, we sought to investigate the impact of the PRRA insert on virus entry. To this end, we packaged pseudoviruses bearing SARS-CoV-2 S, SARS-CoV-2 S ΔPRRA, RaTG13 S, RaTG13 S+PRRA, and pangolin GX S. Again, we noticed a basal level of infection by pseudovirus bearing SARS-CoV-2 S in 293T cells even in the absence of ACE2 (Fig. 2D). This finding is consistent with the observed basal level of cell-cell fusion that is mediated by SARS-CoV-2 spike. Surprisingly, the infectivity of pseudovirus carrying SARS-CoV-2 S ΔPRRA on 293T-hACE2 and Vero E6 cells was about 1/2 to even 2 log10 units higher than that of the pseudovirus carrying wild-type SARS-CoV-2 S (Fig. 2D and Fig. 3A). In contrast, infections of the human lung epithelial cell line Calu-3 as well as the human intestinal cell line Caco-2 by the same pseudovirus were reduced by 2 and 1.5 logs, respectively, compared to pseudoviruses bearing wild-type SARS-CoV-2 S (Fig. 3B and C). Among the five pseudoviruses, bat RaTG13 spike appears to be the least efficient one in mediating pseudovirus entry through hACE2. The insertion of PRRA into RaTG13 S further reduced pseudovirus entry into all cell types that were tested (Fig. 3A to C). Pseudovirus bearing pangolin GX S is highly infectious on 293T-hACE2 cells (Fig. 3A). Interestingly, the S proteins displayed on pseudoviruses bearing SARS-CoV-2 S, RaTG13 S, and RaTG13 S+PRRA were almost completely cleaved but were not cleaved on pseudovirions carrying SARS-CoV-2 S ΔPRRA (Fig. 3D). Pangolin GX S on the pseudovirions appears to display an entirely different cleavage pattern (Fig. 3D). Finally, the expression of TMPRSS2 increased the infectivity of pseudoviruses bearing SARS-CoV-2 S, RaTG13 S, and RaTG13 S+PRRA by about 1 log but not those bearing SARS-CoV-2 S ΔPRRA and pangolin GX S (Fig. 3E).
FIG 3.
The PRRA insert at the S1/S2 site distinctly modulates cell susceptibility to SARS-CoV-2 pseudovirions. (A to C) Entry of HIV pseudotyped with SARS-CoV-2 S, SARS-CoV-2 S ΔPRRA, bat RaTG13 S, RaTG13 S+PRRA, and pangolin GX S into 293T cells transiently expressing ACE2 (A), Calu-3 cells (B), and Caco-2 cells (C). (D) Western blot analysis of pseudovirions bearing SARS-CoV-2 S, SARS-CoV-2 S ΔPRRA, bat RaTG13 S, RaTG13 S+PRRA, and pangolin GX S. Red, anti-p24 antibody; green, antispike antibody. (E) Infection by pseudovirions harboring the indicated viral glycoproteins in 293T cells transfected with ACE2 alone or in combination with TMPRSS2. Data are presented as means ± SEM. *, P < 0.01.
The novel S1/S2 site alters the dependence of RaTG13 pseudovirus on ACE2 proteins of different species.
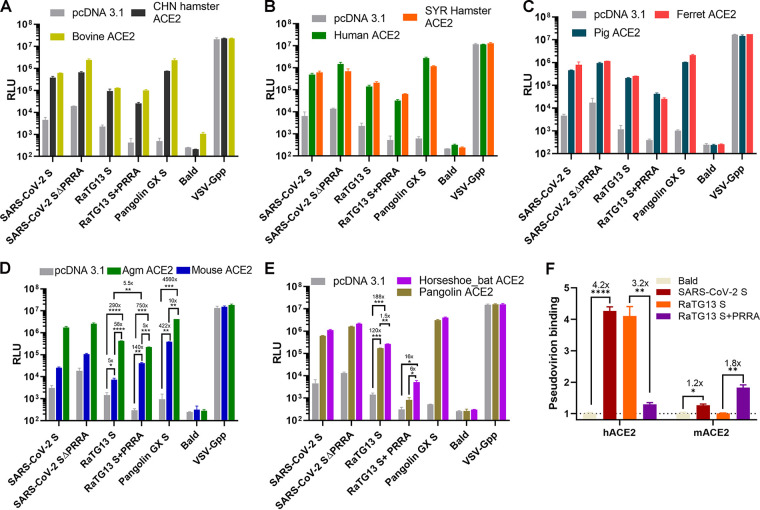
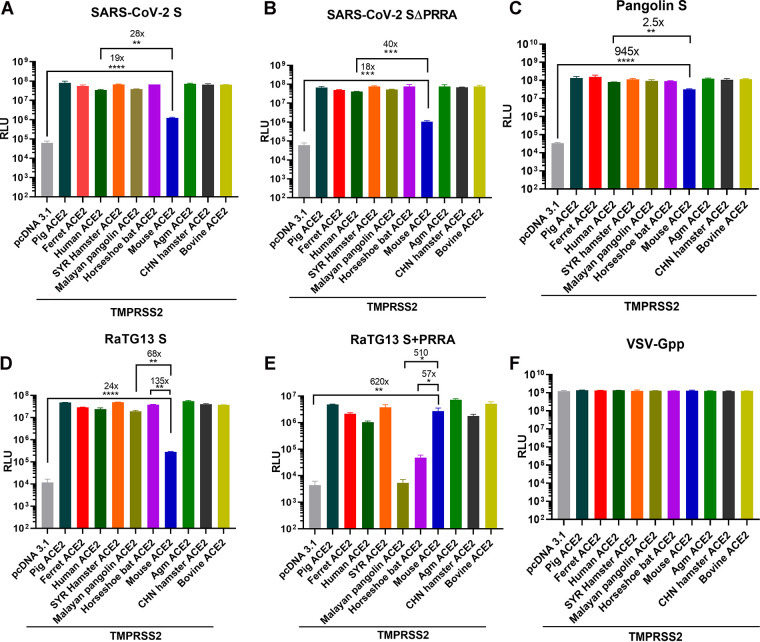
The PRRA insert at the S1/S2 boundary of SARS-CoV-2 S is suspected to alter the stability, transmission, and even host range of SARS-CoV-2. Furthermore, recent studies suggest that a bat virus, rather than a pangolin virus, remains the most probable origin of SARS-CoV-2 (30). Given the evolutionary gap, RaTG13 is unlikely to be the direct ancestor virus of SARS-CoV-2; rather, it branched out from the same ancestor (2, 31). To compare the usages of ACE2 proteins from different species by SARS-CoV-2 and RaTG13, we transfected 293T cells with ACE2 proteins of 10 different species and then infected them with pseudoviruses bearing SARS-CoV-2 S, SARS-CoV-2 S ΔPRRA, RaTG13 S, RaTG13 S+PRRA, and pangolin GX S. Bald virus (free of S protein) and pseudovirus bearing vesicular stomatitis virus glycoprotein G (VSV-Gpp) were included in the study as the negative and positive controls. Pseudovirions bearing SARS-CoV-2 S efficiently infected all but mouse ACE2-expressing cells. The same observation was made with pseudoviruses bearing SARS-CoV-2 S ΔPRRA and RaTG13 S. Strikingly, pseudovirus bearing RaTG13 S+PRRA lost most of its infectivity via pangolin and horseshoe bat ACE2 but gained infectivity by nearly 30-fold via mouse ACE2 (Fig. 4). Pseudovirions bearing pangolin GX S efficiently entered cells via all ACE2 proteins (including mouse ACE2 albeit with a 1-log reduction). The above-described experiment was repeated in the presence of TMPRSS2, and the same observations were made (Fig. 5).
FIG 4.
The novel S1/S2 site alters the dependence of RaTG13 pseudovirus on ACE2 orthologs. Shown are the infectivities of pseudovirions harboring the indicated viral spike proteins on 293T cells transiently transfected with Chinese hamster ACE2 and bovine ACE2 (A), Syrian hamster ACE2 and human ACE2 (B), ferret ACE2 and pig ACE2 (C), Agm ACE2 and mouse ACE2 (D), and horseshoe bat ACE2 and Malayan pangolin ACE2 (E). Data are presented as means ± SEM. *, P < 0.01; **, P < 0.001; ***, P < 0.0001; ****, P < 0.00001. Numbers above the asterisks denote the fold changes. (F) Binding of pseudovirions carrying SARS-CoV-2 S, RaTG13 S, and RaTG13 S+PRRA to full-length recombinant human and mouse ACE2 was quantified by qPCR (see Materials and Methods). Binding relative to that of bald virus is plotted.
FIG 5.
The novel S1/S2 site alters the dependence of RaTG13 pseudovirus on ACE2 orthologs in the presence of TMPRSS2. 293T cells expressing each ACE2 ortholog with TMPRSS2 were infected by pseudoviruses harboring SARS-CoV-2 S (A), SARS-CoV-2 S ΔPRRA (B), RaTG13 S (C), RaTG13 S+PRRA (D), pangolin S (E), and VSV-G (F). Luciferase activities in cell lysates were determined at 48 h postinfection. Data are presented as means ± SEM. *, P < 0.01; **, P < 0.001; ***, P < 0.0001; ****, P < 0.00001.
Although the insertion of PRRA into RaTG13 S does not appear to alter the incorporation of S protein into pseudovirions (Fig. 3D), it might alter ACE2-binding properties via allosteric effects. To this end, we measured the binding of pseudovirions to full-length recombinant ACE2 proteins. Pseudovirions carrying SARS-CoV-2 S efficiently bound to human ACE2 but displayed very little binding to mouse ACE2 (Fig. 4F), which is consistent with what has been reported previously (32). Most notably, the binding of pseudovirions carrying RaTG13 S+PRRA to mouse ACE2 was nearly 2-fold higher than that of pseudovirions carrying RaTG13 S (Fig. 4F). Overall, the profiles of binding of pseudovirions to human and mouse ACE2 correlate with the infectivity of each pseudovirion (Fig. 4B and D).
DISCUSSION
Here, we experimentally demonstrated that SARS-CoV-2 is capable of infecting cells via ACE2 of a diverse range of species. Consistent with recent studies (33, 34), pseudoviruses bearing SARS-CoV-2 S, bat RaTG13 S, and pangolin GX S entered cells via a wide range of ACE2 proteins. Pangolin GX S protein appears to use ACE2 from an even broader range of species to gain entry, at least in the case of mouse ACE2. Despite 87% identity to SARS-CoV-2 in the RBD region (10), the RBM of pangolin GX S protein contains both residues that are favorable for ACE2 recognition (L455 and the 482–485 loop) and those that are less favorable for ACE2 recognition (L486, E493, and T501). Therefore, it was predicted that pangolin GX may not recognize human ACE2 (10). Yet our results here showed otherwise. Overall, our findings support potential cross-species transmissions of bat or pangolin coronaviruses but also highlight the importance of putting animal coronaviruses under surveillance for potential spillovers into humans.
This study also revealed a rather convoluted role of the PRRA insert between the S1 and S2 subunits in SARS-CoV-2 entry. The presence of PRRA in SARS-CoV-2 S protein clearly resulted in one (or two) cleavage site. However, at least on 293T-hACE2 cells, pseudovirus bearing SARS-CoV-2 S ΔPRRA exhibited even greater infectivity, suggesting that PRRA is dispensable for spike-driven viral entry. The RaTG13 S protein, which does not contain the PRRA sequence, is readily cleavable under the same conditions (Fig. 3D). Hence, while the furin cleavage site is key in SARS-CoV-2 S processing, other factors outside the PRRA insertion play a role in the efficient cleavage of the RaTG13 S protein. The addition of PRRA to RaTG13 S further enhanced cleavage but decreased the relevant pseudovirus infectivity on 293T-hACE2 cells. It is worth mentioning that SARS-CoV-2 and RaTG13 S protein sequences are identical within 76 residues downstream and upstream of the PRRA S1/S2 site. Wrobel et al. reported that RaTG13 S adopts a structure similar to that of uncleaved SARS-CoV-2 S protein, although the furin cleavage site at the S1/S2 boundary of SARS-CoV-2 S is located in a surface-exposed and disordered loop (32). We envision that SARS-CoV-2 and RaTG13 spike proteins may still differ subtly near the S1/S2 boundary such that the exact region within SARS-CoV-2 is less accessible to proteases, as opposed to that in RaTG13. Notably, the PRRA insert appears to be essential for efficient SARS-CoV-2 S-dependent pseudovirus entry into the human lung epithelial cell line Calu-3. Two recent studies also reported similar findings (15, 20). In contrast, pseudovirions bearing intact full-length SARS-CoV-2 S without precleavage at S1/S2 displayed higher infectivity on 293T-hACE2 cells (15, 20). Such a discrepancy may be attributed to the relative abundance of proteases and different routes of entry in each cell type. Precleavage of SARS-CoV-2 S at the S1/S2 site by furin may be important only for entry into human lung cells, which make TMPRSS2 but little cathepsin L. On the contrary, entry into certain TMPRSS2-negative cell lines depends on cathepsin L and does not require precleavage of S protein at the S1/S2 site. From a structural biology standpoint, efficient protease cleavage increases the proportion of S protein on the virus surface that adopts an open conformation in its RBD while decreasing the overall stability of SARS-CoV-2 S (32). Thus, the pseudovirions without the furin cleavage site may be more stable and hence promote infection of 293T-hACE2 cells via the endosomal pathway.
The acquisition of the 4-amino-acid insert (PRRA) has been thought to broaden the activating protease repertoire of the SARS-CoV-2 S1/S2 cleavage site. Jaimes et al. reported that the S1/S2 site of SARS-CoV-2 S is efficiently cleaved by furin-like, trypsin-like, and cathepsin proteases (24). Depending on the availability of the activating protease in each cell type, PRRA-guided cleavage may be subjected to differential regulations. Bestle et al. reported that S can be cleaved by the proprotein convertase furin at the S1/S2 site and transmembrane serine protease 2 (TMPRSS2) at the S2′ site (19). RNA sequencing (RNA-Seq) analyses of SARS-CoV-2-infected Calu-3 and Caco-2 cells revealed different protease repertoires, with cathepsin A being expressed to a much higher level in Caco-2 cells (see Fig. S1 in the supplemental material). It is conceivable that the S protein is cleaved by different host proteases near the PRRA insert in 293T, Calu-3, and Caco-2 cells. Depending on the host protease profiles, human CoVs (hCoVs) enter the host cells via the endocytic pathway or from the plasma membrane (35). Clinical isolates of hCoVs reportedly become activated by trypsin-like serine proteases and use endosomal cathepsins in the absence of suitable trypsin-like proteases in cell culture (36, 37). Consequently, SARS-CoV-2 appears to enter human airway cells primarily from the plasma membrane in a TMPRSS2-dependent manner, whereas it enters 293T-hACE2 cells through an endosomal pathway, during which the S protein is cleaved by cathepsins (3, 17–20, 37).
To date, the species tropism of SARS-CoV-2 has largely been attributed to S protein-ACE2 interactions. The SARS-CoV-2 S RBD bound human, pangolin, and horseshoe bat ACE2 more efficiently than the RaTG13 S RBD, but the latter reportedly bound rodent ACE2 orthologs more efficiently (33, 38). The most striking finding of this study is that the insertion of PRRA into the RaTG13 S protein altered the ability of the relevant pseudovirus to utilize ACE2 of three species to gain entry. Specifically, pseudovirus bearing RaTG13 S+PRRA lost most of its infectivity via pangolin and horseshoe bat ACE2 but gained infectivity by nearly 30-fold via mouse ACE2. The latter part of this finding is particularly astounding given that the entry of all other pseudoviruses was somewhat impaired via mouse ACE2. Several possible explanations are as follows. First, the RBM within the RaTG13 RBD significantly differs from that of SARS-CoV-2. Therefore, it is not surprising that RaTG13 S exhibits distinct properties when interacting with ACE2 of different species. Second, the increased infectivity of RaTG13 S+PRRA in the presence of mouse ACE2 is at least partially explained by an increase in binding to mouse ACE2. In support of this, structural and biochemical analyses now suggest that cleavage at the furin cleavage site decreases the overall stability of SARS-CoV-2 S and facilitates the adoption of the open conformation in its RBD that is required for S to bind to the ACE2 receptor (32). Finally, the insertion of PRRA may have a previously unrecognized impact on spike-mediated virus entry. Based on the known structure of SARS-CoV-2 S protein (13), PRRA is part of a protruding loop that is rather distal to the RBD and hence is not expected to directly modulate the S protein-ACE2 interaction. Nonetheless, residues outside the RBD can affect SARS-CoV-2 infectivity. An example is the prevalent SARS-CoV-2 variant carrying the D614G mutation in its S protein (39). Cryo-electron microscopy (cryo-EM) studies of the SARS-CoV-2 S protein trimer found that D614G adopted a more open conformation in its RBD, representing an intermediate that is on its way to S-mediated membrane fusion (34). As the binding of the RBD to ACE2 triggers a conformational rearrangement that causes S1 shedding and the proteolytic cleavage of S2 and hence primes the fusion peptide for subsequent viral-cell fusion (40), an all-open conformation of D614G would reflect an intermediate that is on-pathway to S-mediated membrane fusion (32). It is possible that the PRRA insertion into RaTG13 S decreases the overall stability of the S protein, facilitates the adoption of the open conformation of the RBD that is more efficient in transitioning to the postfusion state after binding to mACE2, and hence increases viral-cell fusion. Future structural analyses will be needed to explore this possibility.
In summary, we showed that S proteins from all three viruses, SARS-CoV-2, bat RaTG13, and pangolin GX, have the potential to mediate entry using ACE2 from multiple animal species besides human. The PRRA insert enhances infection of the human lung cell line Calu-3 by SARS-CoV-2 but unexpectedly alters the dependence of RaTG13 S on ACE2 of three species.
MATERIALS AND METHODS
Viruses and cells.
The Vero E6 cell line (ATCC CRL-1586) and the Caco-2 cell line (ATCC HTB-37) were purchased from the American Type Culture Collection (ATCC) and cultured in Eagle’s minimal essential medium (EMEM) supplemented with 10% fetal bovine serum (FBS; Invitrogen), 1% penicillin-streptomycin, and l-glutamine. The Calu-3 cell line (ATCC HTB-55) was obtained from the ATCC and maintained in EMEM plus 20% FBS. The Huh7.5.1 cell line was a gift of F. Chisari and was maintained in Dulbecco’s modified Eagle’s medium (DMEM) plus 10% FBS (41).
The SARS-CoV-2 isolate USA-WA1/2020 was obtained from BEI Resources, NIAID, NIH, and had been passed three times on Vero cells and one time on Vero E6 cells prior to acquisition. It was further passaged once on Vero E6 cells in our laboratory. The virus has been sequence verified to contain no mutation in its original seed virus. A reporter virus (icSARS-CoV-2-mNG) that contains an insert of monomeric neon green (mNG) in the SARS-CoV-2 USA-WA1/2020 backbone was obtained from Pei-Yong Shi’s laboratory (University of Texas Medical Branch) at passage 2 and was further expanded once on Vero E6 cells (28) at the FDA.
Virus titration.
The titer of SARS-CoV-2 isolate USA-WA1/2020 was determined using the Reed and Muench 50% tissue culture infectious dose (TCID50) assay system (TCID50 per milliliter) (42). Vero cells were plated the day before infection into 96-well plates at 1.5 × 104 cells/well. On the day of the experiment, serial dilutions of virus were made in medium, and a total of 6 to 8 wells were infected with each serial dilution of the virus. After 48 h of incubation, cells were fixed in 4% paraformaldehyde followed by staining with 0.1% crystal violet. The TCID50 was then calculated using the formula log(TCID50) = log(do) + log(R)(f + 1), where do represents the dilution giving a positive well, f is a number derived from the number of positive wells calculated by a moving average, and R is the dilution factor.
Constructs.
Full-length SARS-CoV-2 spike (GenBank accession number YP_009724390.1), bat RaTG13 spike (accession number QHR63300.2), and pangolin Guangxi spike (accession number QIA48614) were human codon optimized and synthesized by GenScript. Human ACE2 (GenBank accession number NM_021804.2) was purchased from GenScript with a C-terminal Flag tag. Other ACE2 genes were synthesized (GenScript Biotech) and subcloned into the pcDNA3.1(+) vector between the BamHI and XhoI sites with a C-terminal V5 tag. The sequences of all spike proteins and ACE2 orthologs can be found in the methods in the supplemental material. TMPRSS2, psPAX2, pCMV-CREM, and pSV-STOP-luc were purchased from Addgene.
SARS-CoV-2 S ΔPRRA was generated using overlap PCR and cloned into the pcDNA3.1(+) vector between the BamHI and XhoI sites. The primers are SARS-2S-F (5′-GCTCGGATCCCCACCATGTT-3′), SARS-2S-PRRA R (5′-CTTGCCACAGACCGGGAGTTTGTCTGGGTCTGGTA-3′), SARS-2S-PRRA F (5′-AGACAAACTCCCGGTCTGTGGCAAGCCAGTC-3′), and SARS-2S-R (5′-TCTAGACTCGAGTTAGGTGT-3′). The RaTG13 S+PRRA mutation was created by overlap PCR. The mutant PCR fragment was digested by BamHI/XhoI and ligated into the BamHI/XhoI-digested pcDNA3.1 vector. The primers are as follows: RaTG13 S F (5′-GCGGATCCCCACCATGTTCGTGTTTCTGGTGCT-3′), RaTG13 S PRRA R (5′-GGCCACGCTCCTTGCTCTCCTTGGAGAGTTTGTCTGGGTCTGAT-3′), RaTG13 S PRRA F (5′-CAAACTCTCCAAGGAGAGCAAGGAGCGTGGCCTCCCAGTC-3′), and RaTG13 S R (5′-ATATAGCTCGAGTTAGGTATAGTGCAG-3′).
SARS-CoV-2 pseudovirus production.
Human codon-optimized cDNA encoding SARS-CoV-2 S glycoprotein (GenBank accession number NC_045512) was synthesized by GenScript and cloned into the eukaryotic cell expression vector pcDNA3.1 between the BamHI and XhoI sites. Pseudovirions were produced by the cotransfection of Lenti‐X 293T cells with psPAX2, pTRIP-luc, and a SARS-CoV-2 S-expressing plasmid using Lipofectamine 3000. The supernatants were harvested at 48 h and 72 h posttransfection and filtered through 0.45-mm membranes. To pellet down pseudovirions, the virus-like particles were centrifuged at 20,000 × g for 2 h at 4°C and then resuspended in SDS-PAGE sample buffer after removing all the supernatant. Alternatively, pseudovirions were produced by the cotransfection of Lenti‐X 293T cells with pMLV-gag-pol, pFBluc, and pcDNA3.1 SARS-CoV-2 S using Lipofectamine 3000. The supernatants were harvested at 48 h and 72 h posttransfection and filtered through 0.45-mm membranes. The murine leukemia virus (MLV)-based pseudovirus infects Vero E6 cells more efficiently than the HIV-based counterpart.
CoV spike-mediated cell-cell fusion assay.
In brief, for the CoV spike-mediated cell-cell fusion assay, 293T cells were transfected with hACE2 and a Stop-Luc construct, which contains a firefly luciferase reporter gene whose transcription is prevented by a stop cassette flanked by LoxP sites. At 24 h posttransfection, these recipient cells were mixed at a 1:1 ratio with 293T cells expressing Cre and CoV S (donor cells) to initiate cell-cell fusion. Luciferase activity was measured 48 h thereafter.
Pseudovirion-ACE2-binding assay.
To ensure that equal amounts of pseudovirions were used in the study, input pseudovirions were quantified by a reverse transcription-quantitative PCR (RT-qPCR) assay. To eliminate the contamination of packaging plasmids in the assay, pseudovirion stocks were pretreated with DNase I (catalog number M0303L; New England BioLabs) at 37°C for 20 min, followed by viral RNA isolation (QIAamp viral RNA minikit, catalog number 52906). RT-qPCR quantification was conducted using the following primers and probe: forward primer GGCTGAATACAAACCATCGG, reverse primer CGCTCGTTGTAGATGTCGTTAG, and probe FAM (6-carboxyfluorescein)-CCCTGTTCATCGGTGTGGCTGT-BHQ1 (black hole quencher 1).
A total of 1010 RNA copies of pseudovirions were incubated with 1.5 μg recombinant human ACE2 protein (catalog number 933-ZN-010; R&D Systems) or mouse ACE2 protein (catalog number 3437-ZN-010; R&D Systems) and 15 μl His tag Dynabeads (catalog number 10103D; Thermo Fisher) for 3 h at room temperature. After incubation, the resulting beads were washed three times with phosphate-buffered saline (PBS), followed by RNA isolation with TRIzol reagent (catalog number 15596026; Thermo Fisher). Bound pseudoviral RNA was quantified using RT-qPCR. The binding of each pseudovirion was normalized against the level of binding (“noise”) obtained when an equal amount of bald pseudovirions was used in the assay.
Immunofluorescence assay.
Cells were plated on collagen-coated glass coverslips the day before infection in 24-well plates at 5 × 104 cells/well. The cells were washed for 5 min 3 times with 1× phosphate-buffered saline. After washing, cells were fixed in 4% paraformaldehyde for 15 min at room temperature. Images were captured by using a Zeiss LSM 880 laser scanning confocal microscope.
Flow cytometry.
Infected cells were trypsinized and washed in 1× PBS. mNG-positive (mNG+) cells were quantified by using a BD FACSCanto II instrument (BD Biosciences). Data were analyzed by using FlowJo software.
Immunoblotting.
Cell lysates were prepared with radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 7.4], 1% NP-40, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA, protease inhibitor cocktail [Sigma], 1 mM sodium orthovanadate), and insoluble material was precipitated by a brief centrifugation. The protein concentration of lysates was determined by a bicinchoninic acid (BCA) protein assay (Thermo Scientific). Lysates containing equal amounts of protein were loaded onto 4 to 20% SDS-PAGE gels, transferred to a nitrocellulose membrane (Li-Cor, Lincoln, NE), blocked with 10% milk for 1 h, and incubated with the primary antibody overnight at 4°C. Membranes were blocked with Odyssey blocking buffer (Li-Cor, Lincoln, NE), followed by incubation with primary antibodies at 1:1,000 dilutions. Membranes were washed three times with 1× PBS containing 0.05% (vol/vol) Tween 20, incubated with IRDye secondary antibodies (Li-Cor, Lincoln, NE) for 1 h, and washed again to remove unbound antibody. Images were captured by using the Odyssey CLx infrared imaging system (Li-Cor Biosystems, Lincoln, NE).
mRNA-Seq by next-generation sequencing.
Total RNAs extracted from Calu-3 and Caco-2 cells were used for sequencing library preparation. RNA quality was assessed by using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA), and the RNA integration numbers (RINs) were all >9. Poly(A)-enriched stranded libraries were generated using a TruSeq stranded mRNA library preparation kit (Illumina, San Diego, CA, USA), according to Illumina’s protocol. In brief, 0.5 μg of total RNA was poly(A) enriched, fragmented, and reverse transcribed into cDNAs. Double-stranded cDNAs were adenylated at the 3′ ends and individually indexed, followed by 15-cycle PCR amplification. The quality and yield of the prepared libraries were evaluated using an Agilent 2100 Bioanalyzer and Qubit 4 (Thermo Fisher Scientific). Libraries from mRNA-Seq were sequenced on an Illumina NovaSeq 6000 instrument according to the manufacturer’s instruction. Data analysis was performed using the RNA-Seq tool of CLC Genomics Workbench V12 (Qiagen, Hilden, Germany), which aligned the paired-end reads to the human hg38 reference genome and calculated gene expression. For each sample, reads per kilobase per million mapped reads (RPKM) were calculated to show the expression level. The full data set can be found in Table S1 in the supplemental material.
Statistical analysis.
All experiments were independently performed at least twice, as indicated in the figure legends. Except where specified otherwise, bar graphs were plotted to show means ± standard deviations (SD). Statistical analyses were performed using Prism 8.
Supplementary Material
ACKNOWLEDGMENTS
The following reagent was deposited by the Centers for Diseases Control and Prevention and obtained through BEI Resources, NIAID, NIH: SARS-related coronavirus 2, isolate USA-WA1/2020, NR-52281. We are grateful to P. Shi (UTMB) for the generous gift of the mNeonGreen SARS-CoV-2.
S.L., P.S., C.Z.L., W.W.W., C.-K.C., and T.T.W. conducted the experiments; S.L., P.S., I.A.N., and T.T.W. analyzed the data; and T.T.W. designed the experiments and wrote the paper.
We declare no competing interests.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. 2020. The proximal origin of SARS-CoV-2. Nat Med 26:450–452. 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou H, Chen X, Hu T, Li J, Song H, Liu Y, Wang P, Liu D, Yang J, Holmes EC, Hughes AC, Bi Y, Shi W. 2020. A novel bat coronavirus closely related to SARS-CoV-2 contains natural insertions at the S1/S2 cleavage site of the spike protein. Curr Biol 30:2196–2203.e3. 10.1016/j.cub.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J, Jia W, Zhu J, Li B, Xing J, Liao M, Qi W. 2020. Insights into the cross-species evolution of 2019 novel coronavirus. J Infect 80:671–693. 10.1016/j.jinf.2020.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu P, Jiang J-Z, Wan X-F, Hua Y, Li L, Zhou J, Wang X, Hou F, Chen J, Zou J, Chen J. 2020. Are pangolins the intermediate host of the 2019 novel coronavirus (SARS-CoV-2)? PLoS Pathog 16:e1008421. 10.1371/journal.ppat.1008421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belouzard S, Millet JK, Licitra BN, Whittaker GR. 2012. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses 4:1011–1033. 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li F. 2016. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol 3:237–261. 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li F, Li W, Farzan M, Harrison SC. 2005. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 309:1864–1868. 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 9.Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, Choe H, Farzan M. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426:450–454. 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, Geng Q, Auerbach A, Li F. 2020. Structural basis of receptor recognition by SARS-CoV-2. Nature 581:221–224. 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. 2020. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181:281–292.e6. 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. 2020. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367:1260–1263. 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai Y, Zhang J, Xiao T, Peng H, Sterling SM, Walsh RM, Jr, Rawson S, Rits-Volloch S, Chen B. 2020. Distinct conformational states of SARS-CoV-2 spike protein. Science 369:1586–1592. 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. 2020. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367:1444–1448. 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffmann M, Kleine-Weber H, Pohlmann S. 2020. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol Cell 78:779–784.e5. 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Millet JK, Whittaker GR. 2015. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res 202:120–134. 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffmann M, Mosbauer K, Hofmann-Winkler H, Kaul A, Kleine-Weber H, Kruger N, Gassen NC, Muller MA, Drosten C, Pohlmann S. 2020. Chloroquine does not inhibit infection of human lung cells with SARS-CoV-2. Nature 585:588–590. 10.1038/s41586-020-2575-3. [DOI] [PubMed] [Google Scholar]
- 18.Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Muller MA, Drosten C, Pohlmann S. 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181:271–280.e8. 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bestle D, Heindl MR, Limburg H, Van Lam van T, Pilgram O, Moulton H, Stein DA, Hardes K, Eickmann M, Dolnik O, Rohde C, Klenk H-D, Garten W, Steinmetzer T, Bottcher-Friebertshauser E. 2020. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci Alliance 3:e202000786. 10.26508/lsa.202000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shang J, Wan Y, Luo C, Ye G, Geng Q, Auerbach A, Li F. 2020. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci U S A 117:11727–11734. 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu A, Peng Y, Huang B, Ding X, Wang X, Niu P, Meng J, Zhu Z, Zhang Z, Wang J, Sheng J, Quan L, Xia Z, Tan W, Cheng G, Jiang T. 2020. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe 27:325–328. 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, Wang X. 2020. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581:215–220. 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 23.Jaimes JA, Andre NM, Chappie JS, Millet JK, Whittaker GR. 2020. Phylogenetic analysis and structural modeling of SARS-CoV-2 spike protein reveals an evolutionary distinct and proteolytically sensitive activation loop. J Mol Biol 432:3309–3325. 10.1016/j.jmb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaimes JA, Millet JK, Whittaker GR. 2020. Proteolytic cleavage of the SARS-CoV-2 spike protein and the role of the novel S1/S2 site. iScience 23:101212. 10.1016/j.isci.2020.101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhan SH, Deverman BE, Chan YA. 2020. SARS-CoV-2 is well adapted for humans. What does this mean for re-emergence? bioRxiv 10.1101/2020.05.01.073262. [DOI]
- 26.Shi J, Wen Z, Zhong G, Yang H, Wang C, Huang B, Liu R, He X, Shuai L, Sun Z, Zhao Y, Liu P, Liang L, Cui P, Wang J, Zhang X, Guan Y, Tan W, Wu G, Chen H, Bu Z. 2020. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science 368:1016–1020. 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oreshkova N, Molenaar R-J, Vreman S, Harders F, Oude Munnink BB, Hakze R, Gerhards N, Tolsma P, Bouwstra R, Sikkema R, Tacken M, de Rooij MMT, Weesendorp E, Engelsma M, Bruschke C, Smit LAM, Koopmans M, van der Poel WHM, Stegeman A. 2020. SARS-CoV2 infection in farmed mink, Netherlands, April 2020. bioRxiv 10.1101/2020.05.18.101493. [DOI] [PMC free article] [PubMed]
- 28.Xie X, Muruato A, Lokugamage KG, Narayanan K, Zhang X, Zou J, Liu J, Schindewolf C, Bopp NE, Aguilar PV, Plante KS, Weaver SC, Makino S, LeDuc JW, Menachery VD, Shi PY. 2020. An infectious cDNA clone of SARS-CoV-2. Cell Host Microbe 27:841–848.e3. 10.1016/j.chom.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miura TA, Travanty EA, Oko L, Bielefeldt-Ohmann H, Weiss SR, Beauchemin N, Holmes KV. 2008. The spike glycoprotein of murine coronavirus MHV-JHM mediates receptor-independent infection and spread in the central nervous systems of Ceacam1a−/− mice. J Virol 82:755–763. 10.1128/JVI.01851-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lau SKP, Luk HKH, Wong ACP, Li KSM, Zhu L, He Z, Fung J, Chan TTY, Fung KSC, Woo PCY. 2020. Possible bat origin of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis 26:1542–1547. 10.3201/eid2607.200092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang X, Wu C, Li X, Song Y, Yao X, Wu X, Duan Y, Zhang H, Wang Y, Qian Z, Cui J, Lu J. 2020. On the origin and continuing evolution of SARS-CoV-2. Natl Sci Rev 7:1012–1023. 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wrobel AG, Benton DJ, Xu P, Roustan C, Martin SR, Rosenthal PB, Skehel JJ, Gamblin SJ. 2020. SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects. Nat Struct Mol Biol 27:763–767. 10.1038/s41594-020-0468-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, Wang H, Tang X, Fang S, Ma D, Du C, Wang Y, Pan H, Yao W, Zhang R, Zou X, Zheng J, Xu L, Farzan M, Zhong G. 2020. SARS-CoV-2 and three related coronaviruses utilize multiple ACE2 orthologs and are potently blocked by an improved ACE2-Ig. J Virol 94:e01283-20. 10.1128/JVI.01283-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yurkovetskiy L, Wang X, Pascal KE, Tomkins-Tinch C, Nyalile TP, Wang Y, Baum A, Diehl WE, Dauphin A, Carbone C, Veinotte K, Egri SB, Schaffner SF, Lemieux JE, Munro JB, Rafique A, Barve A, Sabeti PC, Kyratsous CA, Dudkina NV, Shen K, Luban J. 2020. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell 183:739–751.e8. 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zumla A, Chan JF, Azhar EI, Hui DS, Yuen KY. 2016. Coronaviruses—drug discovery and therapeutic options. Nat Rev Drug Discov 15:327–347. 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shirato K, Kanou K, Kawase M, Matsuyama S. 2017. Clinical isolates of human coronavirus 229E bypass the endosome for cell entry. J Virol 91:e01387-16. 10.1128/JVI.01387-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shirato K, Kawase M, Matsuyama S. 2018. Wild-type human coronaviruses prefer cell-surface TMPRSS2 to endosomal cathepsins for cell entry. Virology 517:9–15. 10.1016/j.virol.2017.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mou H, Quinlan BD, Peng H, Guo Y, Peng S, Zhang L, Davis-Gardner ME, Gardner MR, Crynen G, Voo ZX, Bailey CC, Alpert MD, Rader C, Choe H, Farzan M. 2020. Mutations from bat ACE2 orthologs markedly enhance ACE2-Fc neutralization of SARS-CoV-2. bioRxiv 10.1101/2020.06.29.178459. [DOI] [PMC free article] [PubMed]
- 39.Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, Bhattacharya T, Foley B, Hastie KM, Parker MD, Partridge DG, Evans CM, Freeman TM, de Silva TI, Sheffield COVID-19 Genomics Group, McDanal C, Perez LG, Tang H, Moon-Walker A, Whelan SP, LaBranche CC, Saphire EO, Montefiori DC. 2020. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 182:812–827.e19. 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walls AC, Xiong X, Park YJ, Tortorici MA, Snijder J, Quispe J, Cameroni E, Gopal R, Dai M, Lanzavecchia A, Zambon M, Rey FA, Corti D, Veesler D. 2019. Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell 176:1026–1039.e15. 10.1016/j.cell.2018.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. 2005. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci U S A 102:9294–9299. 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Epidemiol 27:493–497. 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.