Abstract
BACKGROUND AND PURPOSE: Functional MR imaging has been used to study patterns of hippocampal activation that distinguish pathologic from normal memory loss in the elderly population. Our objective was to assess whether hippocampal atrophy confounds measurements of hippocampal activation in subjects with mild cognitive impairment (MCI).
METHODS: Twenty subjects with MCI and 20 elderly control subjects with objectively normal memory were studied at 4T during a face-name paradigm designed to activate the hippocampus. Hippocampal activation was measured using 2 separate approaches: applying a preset region of interest (ROI) in standardized template space and applying a manually drawn ROI in native subject space. Pearson correlation coefficients were calculated to compare group-dependent relationships between hippocampal volume and activation. Analysis of covariance (ANCOVA) was performed to assess group differences in hippocampal activation during encoding and retrieval. Age and hippocampal volume were included as covariates, as was a term for the interaction between hippocampal volume and group.
RESULTS: When hippocampal activation was measured by the template-based method, the correlation coefficient in the right hippocampus of subjects with MCI but not control subjects during retrieval differed significantly from zero. There was a significant (P < .05) group-by-volume interaction in the ANCOVA model. No significant correlations or interactions were demonstrated when activation was measured in native subject space with manually drawn ROIs.
CONCLUSION: Our findings suggest a potential confounding relationship between hippocampal volume and activation for subjects with MCI in template-based analyses. Template-based measures of hippocampal activation that do not adequately account for hippocampal atrophy should be used with caution in patients with MCI.
Isolated memory complaints characterize a subset of the elderly population with mild cognitive impairment (MCI), a condition that is likely to represent a transitional state between normal aging and Alzheimer disease (AD).1 Previous studies have demonstrated subtle anatomic changes in the medial temporal lobes in MCI compared with healthy aging.2 Various functional imaging modalities, including functional MR imaging (fMRI) and positron-emission tomography, have been applied in the population with MCI in an attempt to identify areas of early cortical dysfunction, before the appearance of anatomic changes.3,4 Unfortunately, measures of functional change can be complicated by the presence of cortical and subcortical atrophy, which occur to various degrees and in different patterns in normal aging5 and various types of dementia.6,7 Accounting for such atrophy and increased CSF volume during analysis of functional data may be important to prevent reporting spurious results. Previous PET studies indicate that a correction for partial volume averaging is necessary in the AD population.8 Because fMRI has higher spatial resolution, partial volume averaging is not as significant a concern; however, other methods of standardization are applied in fMRI to correct for individual differences in brain volume and morphology. Such methods may not adequately compensate for local tissue loss and thus may confound measurements of cortical function in these regions.
Standardizing fMRI data allows patterns of activation to be assessed among subjects and groups. In one of the more common standardizing procedures used in fMRI, interindividual structural differences, including those produced by atrophy, are accounted for by an algorithm that preserves tissue concentration but warps the brain to a standardized template.9 The whole-brain template may be based on a single brain, such as the Talairach model,10 or on a probabilistic map developed across a group of subjects, as in the Montreal Neurologic Institute (MNI) template.11 Such template-based methods allow fast and reproducible regional or whole-brain analysis within and across groups. The assumption is that such techniques remove the confounding variability in different cortical structures and thus allow direct comparison of activation in a single area across subjects and across studies.
This assumption, however, may be problematic, especially for structures affected by local pathology, such as the medial temporal lobes (MTL) in AD. Structural volumetric studies of the MTL region, particularly the hippocampus, have found a significant degree of hippocampal atrophy in subjects with MCI compared with healthy elderly control subjects.12–16 Such disproportionate atrophy could complicate the detection of functional activation in the hippocampus when using global standardized template methods based upon a whole-brain stereotactic space.17 A previous fMRI study that used a template-based analysis technique demonstrated a correlation between atrophy and activation in the inferior frontal gyrus of patients with AD but not control subjects during a semantic decision task.18 Recent studies have instead used a manually drawn region-of-interest (ROI) approach,3,4 which intrinsically takes into account individual differences in hippocampal volume and probably yields the most accurate measurement of hippocampal activation independent of volume. This technique, however, is time-consuming, labor-intensive, and has uncertain reproducibility across individual operators and institutions.
The purpose of this study was to assess whether hippocampal atrophy in the MCI population confounds fMRI measures of hippocampal activation that use template-based ROIs. To answer this question, we studied the relationship between hippocampal volume and activation using both template-based and manually drawn ROI methods in a group of subjects with MCI and elderly control subjects. We hypothesize that template-based ROI analysis methods of assessing fMRI activation will demonstrate a correlation with brain atrophy in the MCI group, suggesting that atrophy is a confounding factor in measurements of hippocampal activation. Further, we hypothesize that ROI-based analysis, which accounts for changes in hippocampal volume, will not be significantly affected by hippocampal atrophy.
Subjects and Methods
Subjects
The study received approval from our Institutional Review Board. Subjects satisfying entry criteria were recruited from the local community via advertisements and referrals. Twenty subjects with MCI and 20 cognitively normal elderly subjects were studied.
Entry Criteria
Requirements for entry included fluency in English, completion of at least 8 years of formal education, and willingness to participate in a functional MR imaging scan. All subjects provided written informed consent before any testing or neuropsychologic evaluation.
MCI.
Inclusion criteria were as follows for the MCI group: (1) recent history of symptomatic worsening in memory supported by informant, (2) objective memory impairment (>1 SD below normal) as evidenced by performance on the California Verbal Learning Test (CVLT) II and logical memory and visual reproduction tests from the Wechsler Memory Scale III, (3) normal/near-normal performance on global cognitive test, defined as a Mini-Mental State Examination (MMSE) score >24, (4) global rating on the Clinical Dementia Rating (CDR) scale of 0.5 (questionable dementia) with at least 0.5 on the memory score, (5) does not meet National Institute of Neurological and Communicative Diseases and Stroke/Alzheimer’s Disease and Related Disorders Association (NINCDS-ADRDA) or Diagnostic and Statistical Manual of Mental Disorders, 4th edition (DSM-IV) criteria for dementia, (6) normal/near-normal independent function, and (7) absence of other factors that might better explain memory loss (eg, current major depression).19,20
Control Subjects.
Criteria were as follows for inclusion in the control group: (1) MMSE score >28, (2) does not meet NINCDS-ADRDA or DSM-IV criteria for dementia, (3) normal/near-normal independent function, (4) normal memory, and (5) CDR global score of 0.
Exclusion Criteria
Subjects were excluded on the basis of the following: (1) uncontrolled depression or other psychiatric illness, (2) taking psychoactive medications known to significantly affect memory, (3) standard contraindications to MR imaging, including size incompatible with scanner, metal implants, or cardiac pacemakers, (4) technical difficulties that prevented the completion of successful anatomic imaging and all functional MR imaging runs, and (5) excessive motion during the functional MR imaging examination in excess of 5 mm determined by center of mass plots.
fMRI Stimuli and Paradigm
Our study used a variation of a face-name associative memory encoding task developed by Sperling et al21 in young adults and later applied to elderly subjects and patients with AD.22 In addition to the encoding task, our paradigm also included a retrieval task, for which responses were monitored within the scanner. Subjects were required to encode and later retrieve face-name associations of 2 conditions, novel face-name pairs and familiar face-name pairs, which were presented to the subject within a blocked experimental design. Sixty novel and 2 familiar face-name pairs drawn from the AR Face Database23 were presented within a block design over a period of 6 minutes 50 seconds per run for a total of 3 runs.
Anatomic and Functional Whole-Brain Imaging
Imaging was performed on a 4T MR scanner. Axial T2-weighted spin-echo images (matrix, 256 × 256; 3.75-mm sections with no intersection gap; field of view (FOV), 240 mm) were obtained through the brain for diagnostic purposes to assess for intracranial pathology. Anatomic scans, consisting of 44 contiguous 3.75-mm coronal sections, were acquired for normalization of the functional scans using high-resolution T1-weighted images (IR prep3D SPGR, echo time [TE], 5.4 ms; repetition time [TR], 12.2 ms; inversion time [TI], 500 ms; flip angle [FA], 20°; matrix, 256 × 256; FOV, 240 mm). Functional scans, consisting of a time series of 164 T2*-weighted isotropic image volumes (inverse spiral echo-planar imaging; TE, 31 ms; TR, 2500 ms; FA, 60°; matrix, 64 × 64; FOV, 240 mm), were acquired during each of the 3 functional runs per subject from the same 44 continuous anatomic series coronal section locations.
All functional images were initially screened for quality control purposes using center-of-mass plots to detect excessive motion or section acquisition errors. Anatomic images were screened for intracranial pathology by a board-certified neuroradiologist (J.R.P.).
Hippocampal Volume Measurements
For hippocampal tracing, T1-weighted images were viewed using ITK-SnAP 1.0 software,24 a computer program that displays structural images simultaneously in 3 planes and allows for mouse-driven manual segmentation of images. The left and right hippocampi were traced independently by a single experimenter (S.K.) who was blinded to clinical classification. Tracings were similar to those reported in a previous volumetric study.25 The hippocampus was first identified at the most posterior portion, where the hippocampal tail became visible under the fornix, and proceeded on contiguous sections to the most anterior section on which the hippocampus could be delineated from the amygdala. The selected area was then reviewed for consistency on the axial and sagittal representations. The traced region encompassed the subiculum, CA1–4, and the internal and external digitations. Each hippocampus spanned approximately 10–11 image sections. The traced region became the manually drawn hippocampal ROI later used to detect fMRI activation within the hippocampus in native subject space.
After manual tracing was completed, the volume within the encompassed hippocampal region was calculated for right (RHV) and left (LHV) sides. Hippocampal volumes were then adjusted to account for individual differences in intracranial volume. As an index of total intracranial volume, single-section intracranial volume was reported by Laakso et al26 for the adjustment of hippocampal volume. Applying this method, intracranial volume (ICA) was measured on the coronal section at the level of the anterior commissure. Right and left hippocampal volumes were divided by this measure of intracranial volume resulting in RHV/ICA and LHV/ICA, respectively.
Hippocampal ROI Analysis in Template-Based Space
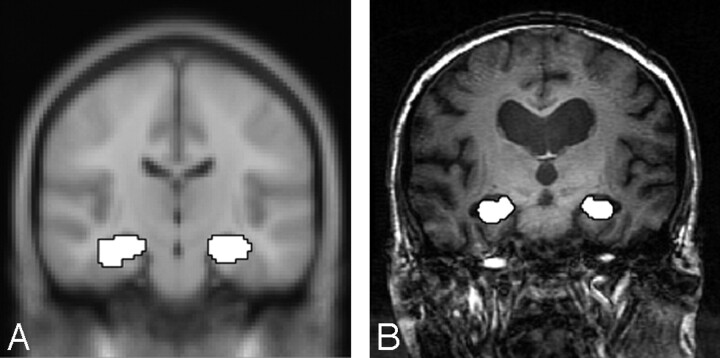
Processing of images was performed using Statistical Parametric Mapping 2 (SPM2) software.27 For the template-based method, preprocessing of each subject’s data consisted of section timing and motion correction, normalization to the MNI template, and spatial smoothing with an 8-mm Gaussian kernel. Two contrast maps of the novel versus familiar condition were created for each subject using the general linear model approach in SPM2, one for encoding and one for retrieval. Right and left hippocampi were analyzed independently for each part of the memory task (encoding and retrieval). Statistical t-maps were created for each subject using a first-level analysis with a voxel-wise significance level of P ≤ .01 (uncorrected) for all maps.28,29 This threshold was selected for group comparisons consistent with previously published fMRI studies in similar populations.4,22,30 The number of activated voxels was measured within the standardized hippocampal ROI (Fig 1A) defined by WFU PickAtlas.31–33 The proportion of activated voxels was calculated for each subject by dividing the number of activated voxels by the number of voxels in the standardized hippocampal ROI (922 and 946 voxels for left and right hippocampi, respectively).
Fig 1.
A, Hippocampal ROI (from WFU PickAtlas31–33) on the MNI template.11
B, Seventy-five-year-old man with MCI, with hippocampal ROI drawn manually on image.
Hippocampal ROI Analysis in Native Subject Space
For the analysis within native subject space, processing of each subject’s data was identical to that of the template-based analysis with the exception of normalization to the MNI template. Before spatial smoothing, the data were resampled to match the voxel size of the standard MNI space (2 × 2 × 2 mm). The manually drawn ROIs, which were obtained from anatomic MR images in the same orientation as those of the functional data (Fig 1B), were then applied to each subject’s contrast map to produce the activated voxel count within each ROI. The voxel count was then adjusted for hippocampal volume by dividing activated voxels by voxel count within each ROI. This yielded the proportion of activated voxels within the subject’s manually drawn ROIs.
Statistical Analysis
The 2 groups were compared for differences in demographic and clinical variables using 2-tailed Student t tests (for age, education, MMSE, CVLT, and hippocampal volume) or χ2 analysis (for sex). Pearson correlation coefficients were calculated to directly compare the group-dependent relationships between hippocampal volume and activation. Likewise, Pearson correlation coefficients were used to examine correlations between hippocampal activation and clinical data of MMSE and CVLT. Analysis of covariance (ANCOVA) was performed with hippocampal activation during encoding or retrieval as the independent variable, group as the main effect, and age and hippocampal volume as covariates. The model also included an interaction term between hippocampal volume and group.
Results
Sex and education did not differ significantly between the 20 subjects with MCI and the 20 control subjects (Table 1). The MCI group was marginally older than the control group (P = .056), so age was accounted for in the statistical model. As expected, CVLT and MMSE scores were significantly lower in subjects with MCI (P < .001). There was no significant correlation between hippocampal activation detected with a manual ROI and clinical data, including CVLT or MMSE. After adjusting for differences in intracranial volume, RHV and LHV were significantly smaller in subjects with MCI, with a greater difference in mean volume between the groups noted on the left.
Table 1:
Demographic and clinical characteristics
| Characteristics | Mild Cognitive Impairment (N = 20) | Control (N = 20) |
|---|---|---|
| Age, y (SD) | 75.0 (7.6) | 71.2 (4.5) |
| Age range, y | 55.5–83.3 | 63.3–80.5 |
| Men/women | 12/8 | 9/11 |
| Education, y | 15.0 (2.2) | 15.9 (2.9) |
| CVLT (SD) | 5.0 (2.5) | 11.0 2.6)* |
| MMSE (SD) | 26.7 (1.5) | 28.4 (1.4)* |
| LHV (as % of ICA) (SD) | 6.19 (1.16) | 7.29 (1.01)* |
| RHV (as % of ICA) (SD) | 6.40 (1.00) | 7.16 (0.91)* |
Note:—Values shown are means (SD) unless otherwise noted. CVLT indicates delayed recall score on the California Verbal Learning Test-II; MMSE, Mini Mental State Examination; LHV, left hippocampal volume; RHV, right hippocampal volume; ICA, intracranial area.
P < .05.
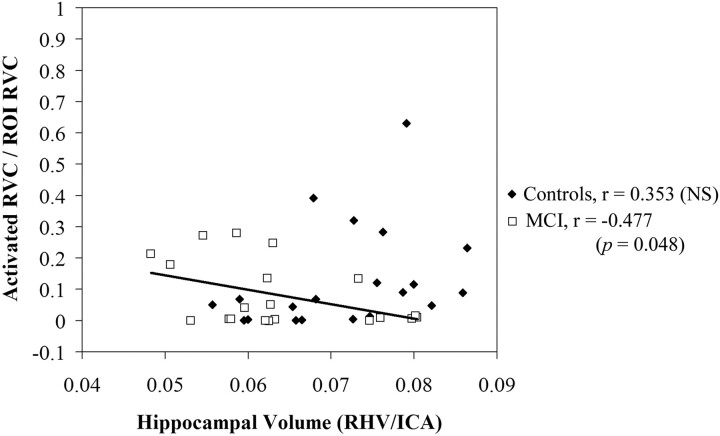
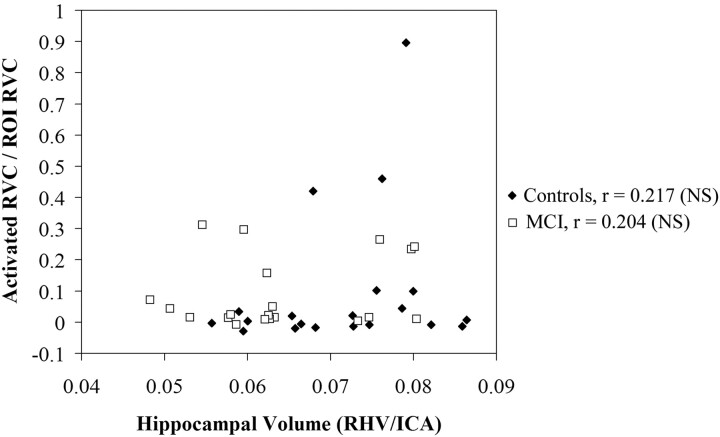
In subjects with MCI, a significant negative correlation between hippocampal volume and activation (r = −0.477, P < .05) was noted in the right hippocampus during retrieval; this correlation was detected only in template-based analysis. Native space analysis with manually drawn ROIs failed to reveal a significant relationship (r = 0.204) in subjects with MCI. Conversely, correlations in control subjects did not differ significantly from zero or from each other, regardless of the method of analysis used for hippocampal activation. These group-specific relationships in the right hippocampus during retrieval are shown in Fig 2 (template-based analysis with automated hippocampal ROI) and Fig 3 (native subject space with manual ROI). The relationship between volume and activation was not statistically significant (P > .05) in the left hippocampus during retrieval and in either side of the hippocampus during encoding for either group by using either ROI approach.
Fig 2.
Correlation of right hippocampal activation in a template-space-based ROI with right hippocampal volume in subjects with MCI and control subjects during retrieval. Hippocampal volumes have been normalized by single-section intracranial area and hippocampal activation has been adjusted for subject age. Units of activation are represented as proportion of ROI activated. The line demonstrates a significant negative correlation between hippocampal volume and activation in the subjects with MCI.
Fig 3.
Correlation of right hippocampal activation in a native-space-based manual ROI with right hippocampal volume in subjects with MCI and control subjects during retrieval. Hippocampal volumes have been normalized by single-section intracranial area and hippocampal activation has been adjusted for subject age. Units of activation are represented as proportion of ROI activated.
In the ANCOVA model, there was a significant effect of both group (P = .030) and the interaction term (P = .020) only on right hippocampal activation during retrieval using template-based analysis (Table 2). Native space analysis failed to show a significant effect of group or of the interaction term. Thus, when the effects of atrophy were accounted for by manually drawing the hippocampal ROI and performing analysis in native space, no significant difference was identified in the amount of activation between control subjects and those with MCI, as evidenced by lack of a statistically significant group effect or interaction term.
Table 2:
Results from ANCOVA (with P values for independent variables included in the model)
| ANCOVA Model | Template-Based Analysis | Native Subject Space with Manual ROI |
|---|---|---|
| LH, encoding | ||
| Group | .699 | .617 |
| Age | .616 | .017 |
| LHV/ICA | .428 | .490 |
| Group X LHV/ICA | .789 | .700 |
| LH, retrieval | ||
| Group | .591 | .542 |
| Age | .271 | .181 |
| LHV/ICA | .496 | .619 |
| Group X LHV/ICA | .620 | .593 |
| RH, encoding | ||
| Group | .909 | .724 |
| Age | .148 | .070 |
| RHV/ICA | .731 | .943 |
| Group X RHV/ICA | .108 | .897 |
| RH, retrieval | ||
| Group | .030* | .694 |
| Age | .707 | .828 |
| RHV/ICA | .812 | .121 |
| Group X RHV/ICA | .020 | .712 |
Note:—Dependent variable is proportion of activated voxels in hippocampal region of interest.
LH indicates left hippocampal activation; LHV/ICA, left hippocampal volume divided by single-section intracranial volume; RH, right hippocampal activation; RHV/ICA, right hippocampal volume divided by single-section intracranial volume; Group X LHV/ICA and Group X RHV/ICA, interaction term between group and hippocampal volume. Significant values of P are indicated in boldface type.
Group term becomes nonsignificant (P = .314) when the interaction term, Group X RHV/ICA, is removed from the ANCOVA model.
Discussion
Template-based analyses offer speed, reproducibility, and operator independence and thus have wide appeal, particularly for use in whole-brain comparisons of functional activation. Whole-brain, template-based analyses allow detection of patterns of cortical function and dysfunction throughout the entire brain in an exploratory analysis or within one or multiple standardized ROIs in a hypothesis-driven analysis. The operating assumption is that spatial normalization corrects for differences in size of various cortical structures. However, this might not be true for localized areas of atrophy; in fact, the use of standardized ROI analysis in the MTL of memory-impaired subjects has been questioned in the past.17 When localized atrophy is a concern, such as in subjects with MCI, manually drawn ROIs in native subject are frequently used, because they directly account for individual differences in hippocampal volume. We report a significant negative correlation between hippocampal volume and hippocampal activation in a sample of subjects with MCI using a standardized ROI approach in template space, but not a manually drawn ROI approach in native subject space. This suggests that atrophy in a memory-impaired population may act as a confounder in template-based approaches, despite their obvious logistic advantages.
The implication is that reports of areas of significant activation and nonactivation might be spurious when failing to account for differences in volume in structurally and functionally complex regions, such as the hippocampus. A negative correlation between hippocampal volume and hippocampal activation was only evident in subjects with MCI during template-based analysis. This would suggest that subjects with MCI with high hippocampal volumes tend to have much lower levels of activation during retrieval, whereas subjects with MCI with small RHVs have increased levels of hippocampal activation, indicating compensatory right hippocampal activation. Although this hypothesis has been suggested previously in the pathologic progression of MCI,1,4,30,34 we propose that in template-based studies, this is, at least in part, a confounding rather than real effect. For example, with the current study, a group-specific relationship is no longer evident when using the manually drawn ROI analysis in native subject space, which takes into account individual differences in volume when detecting hippocampal activation. When a standardized template is used to define activation within the hippocampus in subjects with localized hippocampal atrophy, the resulting ROI may spuriously include surrounding tissues, such as the parahippocampal gyrus, and activation within these tissues may account for the apparent increase. In fact, it has been suggested that it may be these extrahippocampal MTL tissues that display compensatory activation to overcome dysfunctional activation within the hippocampus.4 Unfortunately, our study did not assess activation in these extra-hippocampal regions, and we therefore cannot verify this explanation. Nevertheless, it is important to keep in mind that when comparing healthy control subjects with lesser degrees of hippocampal atrophy to memory-impaired subjects with significant hippocampal volume loss, the spurious inclusion of nonhippocampal tissue in template-based ROIs might lead to the false conclusion of increased hippocampal activation in the group with more profound hippocampal atrophy. A similar inverse relationship between local volume loss and activation has been reported in the left inferior frontal gyrus of patients with AD, but not in healthy control subjects, during a semantic decision task.18 Although the measure of activation and voxel-wise approach differed slightly from our methods, this study also used a template-based approach. Template-based analyses comparing different clinical groups, therefore, could report significant results that might be reflective of morphologic changes rather than effects of neuronal dysfunction in the targeted region.
Manual tracing of hippocampal ROIs provides a direct measure of volume that can be applied to analyses of hippocampal activation to account for atrophy. In this study, we analyzed hippocampal activation within a manually drawn ROI in native subject space that encompassed the full volume of the hippocampus. This approach probably yields the most accurate measurement of hippocampal activation independent of volume but is time-consuming, labor-intensive, and unlikely to be suited to a clinical environment. Other less time-consuming methods of accounting for hippocampal atrophy might be equally valid in the context of template-based analyses. For example, a visual rating scale of hippocampal atrophy (as a gross assessment of CSF accumulation in the perihippocampal fissures) has been used effectively in volumetric studies.35 Alternatively, Gao et al36 measured the thinnest width of the MTL at the level of the intercollicular sulcus to differentiate between patients with mild AD and control subjects. Indices of hippocampal volume such as these might be acquired quickly and yet still adequately account for atrophy in template-based fMRI analyses.
It is noteworthy that we were unable to demonstrate a significant correlation between clinical measures of disease and activation, even for the native subject space approach. Other groups have found group differences in activation between subjects with MCI and control subjects with the use of the native space approach.3,30 However, the direction of these changes has conflicted, possibly reflecting high- versus low-functioning samples within the MCI population. We were unable to demonstrate a group difference in activation, perhaps reflecting the heterogeneity of our sample of the MCI population and the use of very high field strength at 4T, which may decrease signal intensity-to-noise in areas near the skull base secondary to susceptibility artifacts.37
A limitation of this study is that we studied only a single standardized whole-brain template, though the one chosen is commonly used in studies of this nature.22 We did not specifically investigate the validity of the standardization process itself. It is noteworthy that its validity in a memory-impaired population has been previously addressed.17,38 Analysis using probabilistic atlases in disease populations may avoid such confounders, though this complicates comparisons between diseased and nondiseased populations. A second limitation is that different volumes of hippocampal tissue were labeled in the native and template space. A manually drawn ROI was applied to data in native subject space, whereas a predefined anatomic label (WFU PickAtlas Tool) was applied to identify the hippocampus in template space. We cannot comment on which label more accurately identifies hippocampal tissue and excludes surrounding tissue. Nevertheless, we acknowledge that the use of automated spatial normalization and preconfigured anatomic labels has substantial appeal in allowing for standardization across studies. A third limitation is that our findings apply only in the hippocampus. Although there is evidence that template-based measurements of activation may be confounded by atrophy in other areas of the brain in a memory impaired population,18 comparisons of template-based and manual ROI approaches were not performed for other cortical structures; thus, we cannot speculate on the effect of atrophy outside the hippocampus.
In conclusion, we have shown, using a template-based ROI analysis approach, that hippocampal atrophy in subjects with MCI confounds measures of hippocampal activation. When comparing functional activation in the hippocampus across different groups of subjects with MCI, such as in a trial of drug efficacy, template-based analyses might be adequate. However, when using a template-based analysis to compare a memory-impaired population and a nondiseased control population, our findings suggest that some index of atrophy should take into account the interaction between local morphologic changes and fMRI measures of activation in the hippocampus.
Acknowledgments
We thank the subjects who participated in this study.
Footnotes
This work was supported by National Institute on Aging grant 1RO1-AG019728-03.
References
- 1.Bookheimer SY, Strojwas MH, Cohen MS, et al. Patterns of brain activation in people at risk for Alzheimer’s disease. N Engl J Med 2000;343:450–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jack CR, Dickson DW, Parisi JE, et al. Antemortem MRI findings correlate with hippocampal neuropathology in typical aging and dementia. Neurology 2002;58:750–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Machulda MM, Ward HA, Borowski B, et al. Comparison of memory fMRI response among normal, MCI, and Alzheimer’s patients. Neurology 2003;61:500–06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dickerson BC, Salat DH, Bates JF, et al. Medial temporal lobe function and structure in mild cognitive impairment. Ann Neurol 2004;56:27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyer JS, Takashima S, Terayama Y, et al. CT changes associated with normal aging of the human brain. J Neurol Sci 1994;123:200–08 [DOI] [PubMed] [Google Scholar]
- 6.Karas GB, Scheltens P, Rombouts SA, et al. Global and local gray matter loss in mild cognitive impairment and Alzheimer’s disease. Neuroimage 2004;23:708–16 [DOI] [PubMed] [Google Scholar]
- 7.Galton CJ, Patterson K, Graham K, et al. Differing patterns of temporal atrophy in Alzheimer’s disease and semantic dementia. Neurology 2001;57:216–25 [DOI] [PubMed] [Google Scholar]
- 8.Meltzer CC, Zubieta JK, Brandt J, et al. Regional hypometabolism in Alzheimer’s disease as measured by positron emission tomography after correction for effects of partial volume averaging. Neurology 1996;47:454–61 [DOI] [PubMed] [Google Scholar]
- 9.Veltman DJ, Hutton C. SPM99 Manual. London, UK: Wellcome;2001
- 10.Talairach J, Tournoux P. Co-planar stereotaxic atlas of the human brain: 3-dimensional proportional system. New York: Thieme Medical Publishers;1988
- 11.Evans AC, Collins DL, Mills SR, et al. 3D statistical neuroanatomical models from 305 MRI volumes. In: IEEE-Nuclear Science Symposium and Medical Imaging Conference;1993
- 12.Convit A, De Leon MJ, Tarshish C, et al. Specific hippocampal volume reductions in individuals at risk for Alzheimer’s disease. Neurobiol Aging 1997;18:131–38 [DOI] [PubMed] [Google Scholar]
- 13.Killiany RJ, Gomez-Isla T, Moss M, et al. Use of structural magnetic resonance imaging to predict who will get Alzheimer’s disease. Ann Neurol 2000;47:430–39 [PubMed] [Google Scholar]
- 14.Dickerson BC, Goncharova I, Sullivan MP, et al. MRI-derived entorhinal and hippocampal atrophy in incipient and very mild Alzheimer’s disease. Neurobiol Aging 2001;22:747–54 [DOI] [PubMed] [Google Scholar]
- 15.Pennanen C, Testa C, Laakso MP, et al. A voxel based morphometry study on mild cognitive impairment. J Neurol Neurosurg Psychiatry 2005;76:11–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolf H, Grunwald M, Kruggel F, et al. Hippocampal volume discriminates between normal cognition, questionable and mild dementia in the elderly. Neurobiol Aging 2001;22:177–86 [DOI] [PubMed] [Google Scholar]
- 17.Vandenbroucke MW, Goekoop R, Duschek EJ, et al. Interindividual differences of medial temporal lobe activation during encoding in an elderly population studied by fMRI. Neuroimage 2004;21:173–80 [DOI] [PubMed] [Google Scholar]
- 18.Johnson SC, Saykin AJ, Baxter LC, et al. The relationship between fMRI activation and cerebral atrophy: comparison of normal aging and Alzheimer disease. Neuroimage 2000;11:179–87 [DOI] [PubMed] [Google Scholar]
- 19.Petersen RC. Mild cognitive impairment clinical trials. Nat Rev Drug Discov 2003;2:646–53 [DOI] [PubMed] [Google Scholar]
- 20.Petersen RC, Smith GE, Waring SC, et al. Mild cognitive impairment: clinical characterization and outcome. Arch Neurol 1999;56:303–08 [DOI] [PubMed] [Google Scholar]
- 21.Sperling RA, Bates JF, Cocchiarella AJ, et al. Encoding novel face-name associations: a functional MRI study. Hum Brain Mapp 2001;14:129–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sperling RA, Bates JF, Chua EF, et al. fMRI studies of associative encoding in young and elderly controls and mild Alzheimer’s disease. J Neurol Neurosurg Psychiatry 2003;74:44–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinez AM, Benavente R. The AR face database. CVC Technical Report 24,1998
- 24.Gerig G, Yushkevich P, Ho S. ITK-SnAP [computer program]. Version 1.0. Chapel Hill, NC: Cognitica Corporation;2004
- 25.Jack CR, Petersen RC, Xu YC, et al. Prediction of AD with MRI-based hippocampal volume in mild cognitive impairment. Neurology 1999;52:1397–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laakso MP, Soininen H, Partanen K, et al. MRI of the hippocampus in Alzheimer’s disease: sensitivity, specificity, and analysis of the incorrectly classified subjects. Neurobiol Aging 1998;19:23–31 [DOI] [PubMed] [Google Scholar]
- 27.Frackowiak RSJ, Friston KJ, Frith CD, et al. Human Brain Function. San Diego, Calif: Academic Press;1997
- 28.Saykin AJ, Wishart HA, Rabin LA, et al. Cholinergic enhancement of frontal lobe activity in mild cognitive impairment. Brain 2004;127:1574–83 [DOI] [PubMed] [Google Scholar]
- 29.Goekoop R, Rombouts SARB, Jonker C, et al. Challenging the cholinergic system in mild cognitive impairment: a pharmacological fMRI study. Neuroimage 2004;23:1450–59 [DOI] [PubMed] [Google Scholar]
- 30.Dickerson BC, Salat DH, Greve DN, et al. Increased hippocampal activation in mild cognitive impairment compared to normal aging and AD. Neurology 2005;65:404–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maldjian JA, Laurienti PJ, Burdette JH. Precentral gyrus discrepancy in electronic versions of the Talairach Atlas. Neuroimage 2004;21:450–55 [DOI] [PubMed] [Google Scholar]
- 32.Maldjian JA, Laurienti PJ, Burdette JH, et al. An automated method for neuroanatomic and cytoarchitectonic atlas-based interrogation of fMRI data sets. Neuroimage 2003;19:1233–39 [DOI] [PubMed] [Google Scholar]
- 33.Tzourio-Mazoyer N, Landeau B, Papathanassiou D, et al. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 2002;15:273–89 [DOI] [PubMed] [Google Scholar]
- 34.Bondi MW, Houston WS, Eyler LT, et al. fMRI evidence of compensatory mechanisms in older adults at genetic risk for Alzheimer disease. Neurology 2005;64:501–08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Leon MJ, George AE, Golomb J, et al. Frequency of hippocampal formation atrophy in normal aging and Alzheimer’s disease. Neurobiol Aging 1997;18:1–11 [DOI] [PubMed] [Google Scholar]
- 36.Gao FQ, Black SE, Leibovitch FS, et al. Linear width of the medial temporal lobe can discriminate Alzheimer’s disease from normal aging: the Sunnybrook dementia study. Neurobiol Aging 2004;25:441–48 [DOI] [PubMed] [Google Scholar]
- 37.Krasnow B, Tamm L, Greicius M, et al. Comparison of fMRI activation at 3 and 1.5 T during perceptual, cognitive, and affective processing. Neuroimage 2003;18:813–26 [DOI] [PubMed] [Google Scholar]
- 38.Krishnan S, Slavin MJ, Tran TT, et al. Accuracy of spatial normalization of the hippocampus: Implications for fMRI research in memory disorders. NeuroImage 2006;31:560–71 [DOI] [PubMed] [Google Scholar]