Abstract
Reconstitution is an experimental strategy that seeks to recapitulate biological events outside their natural contexts using a reduced set of components. Classically, biochemical reconstitution has been extensively applied to identify the minimal set of molecules sufficient for recreating the basic chemistry of life. By analogy, reconstitution approaches to developmental biology recapitulate aspects of developmental events outside an embryo, with the goal of revealing the basic genetic circuits or physical cues sufficient for recreating developmental decisions. The rapidly growing repertoire of genetic, molecular, microscopic, and bioengineering tools is expanding the complexity and precision of reconstitution experiments. We review the emerging field of synthetic developmental biology, with a focus on the ways in which reconstitution strategies and new biological tools have enhanced our modern understanding of fundamental questions in developmental biology.
Keywords: reconstitution, synthetic biology, genetic circuits, embryo development, spatiotemporal dynamics
INTRODUCTION
In a single cell, thousands of genes act in an orchestrated way at any given time, and simplification of a large molecular network into the interactions among a few of its core components can be useful for understanding complex cellular processes. Biochemical reconstitution has proven to be a successful approach to simplify and isolate such core interactions. In biochemical reconstitution, a small set of molecular components are assembled outside the cell to recapitulate cellular processes that occur inside the cell (Fletcher 2016). To date, reconstitution using purified molecular components has recapitulated most of the central dogma including DNA replication, RNA transcription, and protein synthesis (Benne & Hershey 1978, Sayre et al. 1992, Yeeles et al. 2017). Although these reconstituted systems do not fully recapitulate all the features of a cellular process, they help define the minimal components needed for critical cellular processes.
Compared with that of single cells, the development of multicellular tissues involves additional levels of biological complexity, such as spatial patterning of different cell types, control of population sizes among the cell types, and changes in tissue shapes and mechanics. To accomplish these multicellular tasks, individual cells act as the basic unit that sends, perceives, and acts on signals to coordinate with one another. Combining these multicellular tasks in specific spatial and temporal orders allows cells to self-organize into tissues of certain cellular compositions, arrangements, shapes, and sizes. In the past several decades, research based on a handful of organisms has revealed amazing details about the genes involved in multicellular interactions. On the basis of this rich information, we can start putting together molecular models to explain developmental processes, but the sufficiency of the molecular models remains largely untested.
Sufficiency tests require taking cells outside their normal tissue contexts, or genes outside their normal cellular contexts, and testing their capability of reconstituting the behavior in a new environment. This way of thinking and experimenting has provided key insights into embryo development. For example, by transplanting cells from one part of an embryo to another, Spemann & Mangold (1924) famously demonstrated that a tissue fragment taken from the dorsal lip in a developing newt is sufficient to induce the formation of a secondary neural plate on the ventral side of the embryo. Going beyond these early transplantation experiments, it is possible to remove different parts of early Xenopus gastrula embryos, culture them in vitro either separately or in contact, and watch whether the reconstituted embryonic tissues can autonomously undergo central developmental processes such as gastrulation (Keller & Danilchik 1988). Ex situ gastrulation experiments revealed that tissues from different regions within the embryo have different degrees of convergence or extension capabilities and that their capability depends on the explant size and the presence of other tissues.
These early efforts demonstrated that reconstituting developmental processes outside embryos, as a complementary approach to using intact embryos, could offer a valuable opportunity for testing developmental models. In such reconstituted systems, just as in an embryo, individual cells are the basic building blocks. But unlike embryos, these cells do not have all the chemical or physical information needed to form tissues. By supplying the missing information using biological and/or engineering tools, one can discover the minimal set of components sufficient for programming multicellular events. Importantly, reconstituted systems provide the possibility of testing interactions between arbitrary cell types, the isolation of specific multicellular behaviors from parallel developmental processes, the flexibility of rewiring genetic pathways, and the precision to quantitatively manipulate key parameters in the system. Together, these technical advantages allow us to ask a broad range of questions: What is the minimal set of components sufficient for programming multicellular phenomena? What are the key parameters that determine the dynamics and precision of cell–cell interaction in space and time? How do modifications to the system during evolution lead to quantitative or qualitative changes in phenotype? Can different pathway designs enable similar multicellular outputs, and if so, what are the trade-offs? Answering these questions could help synthesize the general principles of embryo development and evolution. Building the system from the bottom up provides the knowledge and tools needed for rationally programming cells to form complex tissues, which is an engineering goal for regenerative medicine and embodies the emerging field of synthetic developmental biology.
Due to space limitations, we cannot comprehensively list all the examples that reconstitute different aspects of developmental biology, especially given the rapid progress at the interface between synthetic biology and developmental biology (Davies 2017, Ebrahimkhani & Ebisuya 2019, Santorelli et al. 2019, Toda et al. 2019). Instead, we focus on a few recent studies that apply modern molecular and engineering tools to reconstituted systems in order to answer fundamental questions in developmental biology. We start by discussing how cells determine what cell types to become. We then discuss examples of reconstituting several multicellular processes, including the maintenance of appropriate proportion among different cell types, the formation of spatial patterns due to chemical or physical cues, and the usage of temporal dynamics to organize cell fates.
CELL FATE DECISIONS
Perhaps no process is more fundamental to understanding development than cell fate determination. Although cell fate decisions are not unique to multicellular systems, they are among the earliest examples for which developmental functions have been reconstituted using molecular tools. Since realizing the importance of transcription factors in cell fate decisions, the field of developmental biology has been seeking the essential codes of transcription factor expression that can drive the differentiation of distinct lineages. A prominent approach to understanding cell fate specification has been to perform mutagenesis screens for mutants in which cell fate is perturbed. This strategy has identified many extracellular cell signaling pathways that coordinate developmental cell fate decisions, as well as the transcription factors that ultimately control a cell’s transcriptional response (Wieschaus & Nüsslein-Volhard 2016).
An alternative to the loss-of-function approach is to express the candidate fate-regulating genes in cells and investigate whether they can convert the cells to a different fate. If the cells can change their identity, this gain-of-function test would demonstrate the sufficiency of the genes for specifying cell identity in a particular context. Early experiments suggested that this approach might be feasible by using cell–cell fusion to show that cell identity can be converted (Blau et al. 1983, Gurdon et al. 1958). By applying this reasoning and molecular tools, investigators successfully reprogrammed the mouse fibroblast cell line C3H10T1/2 into myogenic cells, which led to the discovery of MyoD as a master regulator of muscle differentiation (Davis et al. 1987, Lassar et al. 1986). Subsequently, a series of studies demonstrated that converting cell identities by reconstituting the core transcriptional circuits can be applied to initiate many different cell fates (Figure 1a) (Morris & Daley 2013). For example, fibroblasts can be converted into neurons with a combination of three transcription factors (Ascl1, Brn2, and Myt1l) (Vierbuchen et al. 2010), and mouse and human fibroblast cells can be reprogrammed into induced pluripotent stem cells (iPSCs) by adding a few transcription factors (Oct3/4, Sox2, c-Myc, and Klf4) (Takahashi & Yamanaka 2006, Takahashi et al. 2007). The success of iPSC induction vividly demonstrated that a small set of genes encoding transcription factors can impart pluripotency to cells. Much of the excitement around iPSCs came from the possibility of deriving any cell type from fibroblasts through an iPSC intermediate, which would open many doors to regenerative medicine. Such applications require a second category of cell identity conversion that starts from pluripotent stem cells and forces their differentiation down a particular path. The efforts to induce pluripotent cell differentiation outside an embryo have led to key discoveries about embryo development, such as the identification of mesoderm-inducing signaling molecules using the animal cap from amphibian blastulas (Nieuwkoop 1969, Smith 1989). More recently, sophisticated protocols have been developed to differentiate mammalian embryonic stem cells (ESCs) or iPSCs into numerous cell types. For example, mouse ESCs can be induced to become motor neurons by supplying signaling molecules at a defined time in a stepwise manner that mimics embryonic development, or by synthetically expressing three motor neuron transcription factors to directly stimulate a lineage-specific transcriptional program (Velasco et al. 2017, Wichterle et al. 2002).
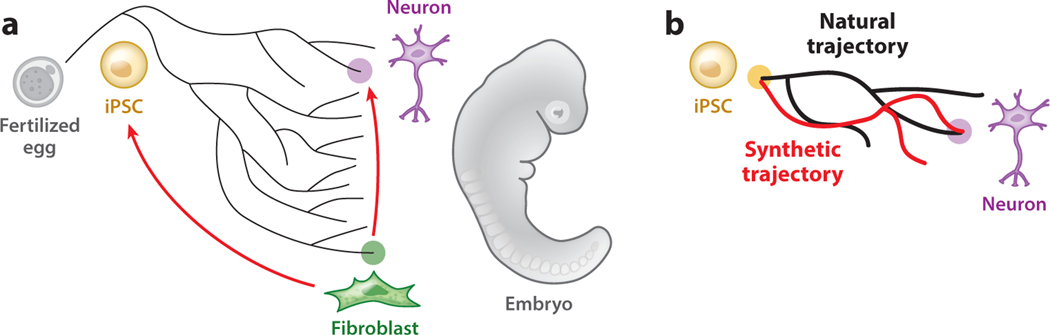
Figure 1.
Cell fate decisions can be programmed or reprogrammed. (a) From a fertilized egg to an embryo that is made of different cell types, cells have to make sequential fate decisions (branched tree). However, cell fates can be synthetically engineered by overexpressing a few transcription factors (red arrows). For example, fibroblasts can be converted into induced pluripotent stem cells (iPSCs) or neurons by different sets of transcription factors. (b) iPSCs can follow different trajectories, natural differentiation (black lines) versus synthetic engineering (red lines), both of which can give rise to the same neuronal fate, but transit through distinct intermediate cell identities.
Clearly, some examples of transcription factor–driven cell fate switches do not occur naturally, especially in the case of reprogramming differentiated cells into iPSCs or converting one type of terminally differentiated cell into another type of differentiated cell. Do these direct programming or reprogramming experiments, then, teach us anything about the principles of cell identity determination, maintenance, and switching, or are they interesting mainly for their potential biomedical applications? An interesting comparison between the two strategies of motor neuron differentiation revealed that whereas the stepwise differentiation protocol included a progenitor cell state that expressed oligodendrocyte markers, the direct differentiation strategy by synthetic expression of transcription factors entirely bypassed any intermediate progenitor-like cell state (Figure 1b) (Briggs et al. 2017). This surprising finding implies not only that cell identity is plastic but also that cells can take multiple paths to reach the same identity (Aydin & Mazzoni 2019, Briggs et al. 2017). These gain-of-function or reconstitution studies also demonstrated that synthetically expressed transcription factors are sufficient to instruct cell identity in the absence of differentiation cues from external sources. Even though these synthetic experiments might not fully recapitulate any known developmental process, they inspire new thoughts and questions about natural development. The possible alternative paths of cell differentiation make one wonder whether and how such plasticity can be tinkered with during evolution to produce different developmental dynamics and new cell types.
POPULATION SIZE CONTROL
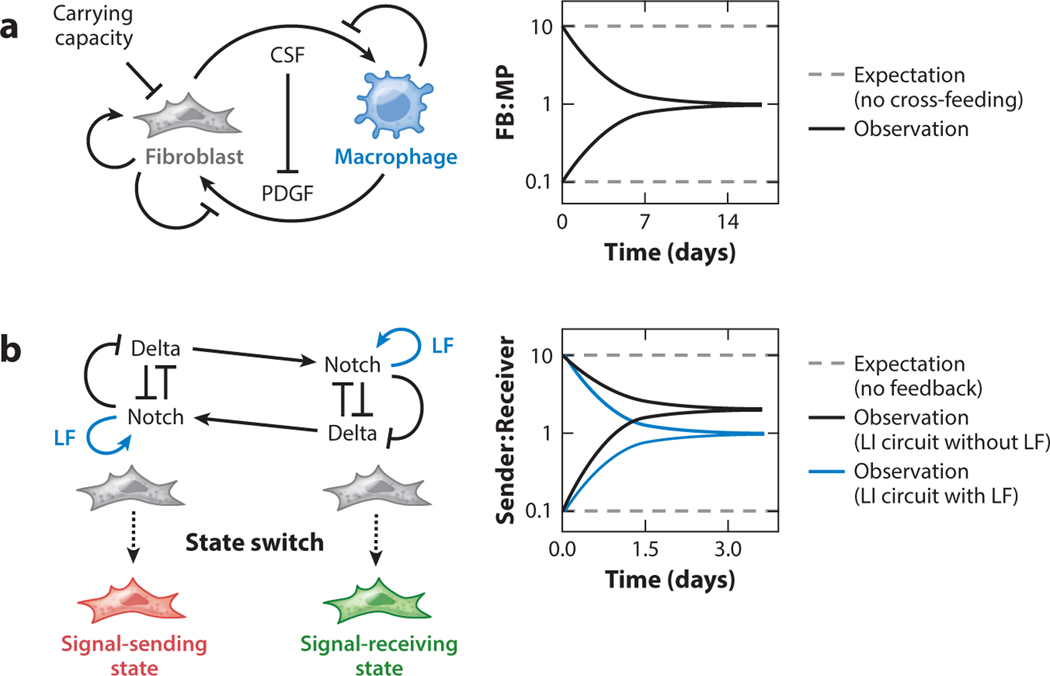
Tissue function depends not only on cells having the appropriate identities but also on the correct ratio of cell types that exist within the tissue. Loss of those properties can result in tumorigenesis. In ecology and microbiology, it is axiomatic that a cell will divide until the carrying capacity of its environment is reached; however, in healthy metazoan tissue, cells can be below the apparent carrying capacity of their niche. We can interpret the observation that cell abundances are at levels below the apparent carrying capacity in two ways: Either we do not fully understand the limiting resources in a cell’s environment, or biological processes like cell communication enforce a defined ratio of cell types. In one clearly articulated example of the latter idea, macrophage and fibroblast cocultures consistently converged to a stable ratio of ~1:1 after 2 weeks of cell culture (Zhou et al. 2018). Mathematical models based on previously characterized properties of macrophage and fibroblast growth factor requirements (Andrae et al. 2008, Chitu & Stanley 2006) describe how macrophages and fibroblasts in coculture can target a stable cell ratio by completing a cross-feeding circuit in which reciprocal growth factor exchange and negative feedback between two growth factor pathways can result in a stable population mixture (Figure 2a). Interestingly, in competitive coculture experiments that grew macrophages with two types of fibroblast simultaneously, including fibroblasts that were capable of cross-feeding with macrophages as well as fibroblasts that were not, fibroblasts that were capable of cross-feeding outcompeted non-cross-feeding fibroblasts, suggesting that maintaining a defined cell ratio by local cross-feeding enhances the survival of both cell types in the tissue (Zhou et al. 2018).
Figure 2.
Tissues coordinate proliferation or state transition of multiple cell lineages to achieve the correct ratio of cell types. (a) Natural communication circuits between fibroblasts (FB) and macrophages (MP) ensure a fixed ratio of the two population sizes, regardless of the initial ratio between the two. The circuits involve positive and negative feedback loops, as well as a fixed carrying capacity imposed by the cell culture environment. (b) Synthetically engineered circuits in Chinese hamster ovary cells instruct cell state bifurcation and a fixed ratio between the two states. Two cells with identical lateral inhibition (LI) circuit, in which cells expressing both Notch and Delta induce a negative feedback loop to repress Delta upon Notch activation, eventually adopt either the signal-sending (“sender”) or signal-receiving (“receiver”) state. Within a population of cells, the ratio between the two states depends on the LI circuit and a positive feedback loop involving Lunatic Fringe (LF). Abbreviations: CSF, colony-stimulating factor; PDGF, platelet-derived growth factor.
Whereas macrophages and fibroblasts coordinate their growth by using two distinct signaling pathways, the positive and negative feedback features that allow cells to achieve a defined population density can operate through a single extracellular signal. In CD4+ T cells, the autocrine cytokine interleukin-2 can activate both proliferation and cell death with different kinetics, such that cells converge to a target growth density—below the apparent carrying capacity—at which proliferation is exactly offset by cell death (Hart et al. 2014).
In addition to regulation of cell growth and death, cell-state switching provides another mechanism to maintain a desired ratio between two cell types. Notch signaling pathway has been observed in numerous developmental contexts to instruct binary fate switching between adjacent cells (reviewed in Mumm & Kopan 2000, Sjöqvist & Andersson 2017). In the Notch signaling pathway, the membrane-bound ligand Delta binds a Notch receptor, causing the intracellular domain of Notch to be cleaved and enter the nucleus, where it acts as a transcription factor to control cell fate decisions. Notch signaling is either positively activated by trans-presented Delta ligand expressed on adjacent cells or negatively regulated with cis-presented Delta ligand on the same cell (Sprinzak et al. 2010, 2011). Cells continuously monitor the amount of endogenous and exogenous Delta ligand, and on the basis of their observations each cell commits to either the signal-sending or signal-receiving state, a binary switch that has been synthetically recapitulated in cultured Chinese hamster ovary cells (Sprinzak et al. 2010). By further engineering Notch and Delta into a lateral inhibition circuit in which Notch activity downregulates Delta expression in the same cell, a stable ratio of the two cell states within the population can be achieved regardless of the initial ratio (Figure 2b) (Matsuda et al. 2015). In addition, the cis and trans interaction between Notch and Delta can be modified by Lunatic Fringe, a glycosylase that potentiates Notch signaling, to specify cells’ signaling states (Moloney et al. 2000, LeBon et al. 2014). Adding another feedback loop mediated by Lunatic Fringe could tune the ratio of cell types observed in culture (Matsuda et al. 2015). Through reconstitution of ligand, receptor, modifier, and feedback circuits, a picture has emerged of how a simple contact-dependent signaling circuit can specify the relative abundance of alternative cell types.
SPATIAL PATTERN FORMATION
Embryo development requires not only that the right number of the right cell types be made but also that they be present in the right place, a process resulting in pattern formation. Early embryonic patterns are established over multiple length scales and with varied geometry, and can arise through chemical-based cell–cell communication and/or in response to physical stimuli (Salazar-Ciudad 2003). In the following sections, we discuss reconstitution approaches to understand chemical or physical signals that influence developmental patterns.
Chemical Cues Instructing Spatial Patterns
Cells communicate with one another by producing ligands and receptors. To communicate with cells that are not immediately adjacent, developing animals rely on secreted molecules called morphogens, a name coined by Alan Turing (Turing 1952). Morphogens form concentration gradients outside cells and trigger graded intracellular signals that decay continuously as a function of distance (Figure 3a) (reviewed in Rogers & Schier 2011). Subsequently, it was suggested that each cell can autonomously calculate its position by interpreting its quantitative exposure to a diffusible signaling molecule, and that this process can instruct the development of discrete tissue boundaries (Stumpf 1966, Wolpert 1969). Despite the long history of this theory, the identity of morphogens was not revealed until decades later, when molecular genetics became available. These genetic studies in embryos have identified the key components and their interactions in the signaling pathways that mediate the effect of morphogens. However, pathway diagrams alone are not sufficient to explain key aspects of tissue patterning, such as its speed, size, robustness to perturbations, and changes during evolution. Spatiotemporal dynamics become increasingly relevant for the understanding tissue patterning because accumulating evidence suggests that correct tissue patterning depends on both the concentrations and the temporal dynamics of morphogen signals (Briscoe & Small 2015).
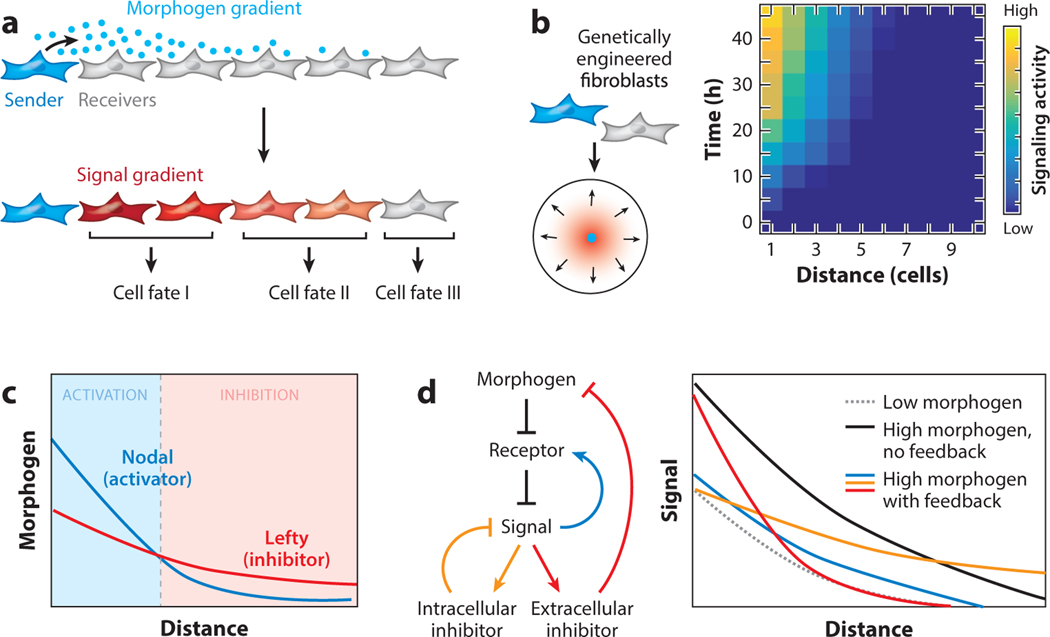
Figure 3.
Morphogen pathway architectures regulate patterning behavior. (a) Morphogens secreted by sender cells form spatial gradients, which trigger graded signaling responses in receiver cells and determine cell fates. (b) Morphogen signaling gradients can be reconstituted using genetically engineered fibroblasts in cell culture, and the gradients are dynamic in both time and space. (c) Nodal and Lefty morphogens secreted from the same sender cells form short-range activation and long-range inhibition zones, due to the differential diffusion rates of the two ligands. Nodal is an activator and Lefty is an inhibitor of the pathway. (d) The robustness of Sonic Hedgehog (SHH) signaling gradients to fluctuations in the morphogen production rate depends on the feedback loops in the pathway. Receiver cells with different types of negative feedback loops, which can be genetically engineered, show different capabilities to buffer the changes in the morphogen production level. Feedback that induces only an intracellular inhibitor (orange) or an extracellular inhibitor (red) buffer the gradient’s amplitude or length scale, respectively, against an increase in the morphogen production level. The evolutionarily conserved feedback loop (blue), in which receptor expression is upregulated upon signal activation, can provide both capabilities in the SHH pathway. Abbreviations: FB, feedback.
Constructing dynamic models of tissue patterning requires precise perturbation and quantitative measurements to determine the key biochemical and biophysical parameters. Furthermore, to understand not only how but also why the underlying genetic circuits are wired in a specific way, one would need to systematically rewire the circuit architecture while quantitatively monitoring the pattern dynamics in space and time. All of these goals can be achieved more easily in reconstituted systems than inside an embryo. Recent studies have shown that gradients of morphogens, such as Sonic Hedgehog (SHH) and Nodal/Lefty, can be reconstituted in confluently cultured cells, enabling quantitative analyses of gradient dynamics in space and time (Figure 3b) (Li et al. 2018, Sekine et al. 2018). An important criterion for this approach to be useful is that reconstituted morphogen gradients should behave in analogous ways to their embryonic counterparts. A critical feature of morphogen gradient formation in embryos is the appropriate movement of signaling molecules through the developing tissue, and it is not obvious that the two-dimensional cell culture extracellular matrix (ECM) and growth medium environment would be similarly permissive of morphogen diffusion compared with tissue in developing animals. In the case of SHH gradients reconstituted with mouse fibroblast cells, turbulent mixing of the cell culture medium—achieved by culturing cells on a rocking table or by frequently changing the media—did not affect morphogen signaling gradients, suggesting that the SHH ligands responsible for gradient formation stayed associated with the cell layer (Li et al. 2018). Additionally, SHH morphogen gradients could not cross gaps where there are no cells or ECM, consistent with the model in which a continuous layer of cells or ECM is required for gradient formation (Müller et al. 2013). In the case of reconstituted mouse Nodal/Lefty gradients in cultured human HEK293 cells, these ligands exhibit similar diffusion rates compared with their respective homologs in the zebrafish embryo (Müller et al. 2012, Sekine et al. 2018). With the inhibitory ligand Lefty diffusing faster than the activating ligand Nodal, the two ligands create two signaling zones: a local activation zone and a distal inhibition zone (Figure 3c). Together, these results suggest that the reconstituted system behaves in a similar way as embryonic tissues in supporting morphogen gradient formation.
Working with developmental pathways in cell culture makes available a wide range of genetic tools that can be used to test the causal relationship between genetic circuits and patterning behavior. For example, morphogen pathways often employ negative or positive feedback loops, some of which act only inside or outside the cell while others can influence both extracellular morphogen distribution and intracellular signal transduction (Figure 3d). What patterning capabilities can different feedback loops provide, and why do certain feedback designs act ubiquitously in response to their respective morphogen signals while others operate only in a tissue-specific manner? Using the reconstituted SHH gradient system, the SHH pathway architecture in the signal-receiving cells has been genetically rewired into different designs to test how different types of negative feedback loops affect the dynamic establishment and precision of the SHH signaling gradient. To switch the feedback loops on or off and to quantitatively tune the strength of the feedback, investigators implemented a synthetic circuit design that can be modulated by both SHH signal and an exogenously added drug. This manipulation revealed that the evolutionarily conserved negative feedback loop in the SHH pathway outperformed alternative designs by improving the robustness of signaling gradients to perturbations in the rate of morphogen secretion (Figure 3d) (Li et al. 2018). In this conserved feedback loop, HH signal transcriptionally upregulates its own receptor, Patched (PTCH), in signal-receiving cells. PTCH sequesters HH outside cells to regulate the size of the gradient and dampens the response inside cells to control the signal amplitude. Both the gradient size and amplitude provide crucial information for tissue patterning. While feedback loops with either intracellular or extracellular function exist and can improve the accuracy of the gradient, coupling these two control mechanisms in PTCH further protects the gradient from fluctuations in sender cells and therefore improves the accuracy of tissue patterning. An analogous negative feedback loop whereby an activator (Nodal) induces expression of its own repressor (Lefty) makes developmental processes more tolerant of perturbations to the level of Nodal signaling in zebrafish embryos (Rogers et al. 2017). Both reconstitution and embryo studies suggest that negative feedback is a central element of spatial patterning by morphogens. Although the bottom-up reconstitution approach to studying patterning is in its infancy, it has shown early promise in its ability to reconstruct, control, and compare genetic circuits and, importantly, to reveal their design principles for regulating developmental patterning.
Physical Cues Instructing Spatial Patterns
From cell cleavage in early embryos to gastrulation and organogenesis, embryo development is a manifestation of changes in physical properties. In particular, recent research has identified many examples in which physical forces within tissues can act as bona fide developmental signals (Mammoto & Ingber 2010). However, the molecular bases underlying generation of physical forces are often unclear, and there is much more to learn about how physical forces control tissue patterns and shapes. Reconstituted systems are well suited for addressing these questions because of their simplicity and manipulability.
A classic example of using reconstitution to study tissue mechanics and patterning is adhesion-based cell sorting. For more than 100 years, embryologists have been dissociating embryos or even adult tissues into cell suspension and reaggregating the cells to see whether the cells adhere to one another and whether they reform patterns and structures (Grunwald 1991, Kirillova et al. 2018). Cells from different tissue origins within the same species can often sort into distinct clusters; a classic example is the use of Hydra cells in sorting endoderm from ectoderm (Technau & Holstein 1992). With certain combinations of cell types, the arrangements of cells within the aggregates resemble the tissues they came from (Townes & Holtfreter 1955). This observation led to the suggestion that cell sorting was due to directed movement, meaning that cells move unidirectionally toward their destination possibly because of attractive signals. An alternative model was later proposed in which differential adhesion together with nondirectional cell movement could lead to cell sorting, with less cohesive cells surrounding a core of more cohesive cells achieving the lowest energy state (Steinberg 1963). Thirty years later, with better molecular tools, it became possible to test the alternative model directly by expressing P-cadherin in L cells at high or low levels and aggregating the two cell populations (Steinberg & Takeichi 1994); in these experiments, the two cell populations sorted into separate clusters. These reconstitution experiments elegantly demonstrated that the differential adhesion model is sufficient to explain cell sorting and the creation of spatial patterns. Furthermore, by using synthetic Notch signaling to induce differential cadherin expression properties between adjacent cells, cell aggregates initially composed of two cell types that are randomly mixed together can spontaneously form organized structures of three layers (Toda et al. 2018). In fact, cadherin-mediated cell sorting has been observed in the formation of sharp cell fate boundaries in the zebrafish neural tube, as well as in aggregates made of zebrafish cells from different tissue layers, suggesting that similar mechanisms can operate in both embryos and cultured cells (Krieg et al. 2008, Tsai et al. 2019, Xiong et al. 2013). Interestingly, in addition to cadherin-dependent differential adherence of cells within distinct tissue layers, generic actinomyosin cortex tension controls cell sorting by causing cells to favor distinct topologies within an aggregate (Krieg et al. 2008). These reconstitution experiments demonstrate that multiple physical processes act collectively to determine the cell sorting outcome.
Although processes that affect cell–cell interactions have been well described, the factors that control tension at the tissue scale remain unclear. To understand how developing tissues build tension, Hughes et al. (2018) reconstituted tissue folding behavior by plating fibroblasts on a collagen substrate. They found that on a collagen substrate, fibroblasts formed dense local cell aggregates and that aggregation required nonmuscular activity of myosin II. When cells were plated with defined spacing, for example, in a two-dimensional grid with unequal spacing in the x and y directions, cells selectively contracted toward their nearest neighbors, resulting in linear bundles of dense collagen and cords of tense cells. By manipulating the geometry of patterned cells, one can recreate complex tissue morphologies including helices, tubes, spheres, cubes, and sinusoidal wrinkles (Hughes et al. 2018). This study demonstrated that spatial patterns of tension can control three-dimensional morphogenesis of the tissue. The researchers took advantage of the manipulability in the reconstituted system to precisely control key parameters, which otherwise would be difficult to manipulate inside an embryo.
Dissecting Chemical Versus Physical Influences
In an embryo, the developmental cues mentioned above, including long-range morphogen signals, cell contact–mediated signals, and cell–ECM interaction, are all likely to coexist. In addition, these developmental cues can influence one another, forming convoluted feedback circuits (Howard et al. 2011). Therefore, the necessity and sufficiency of chemical or physical signals in a patterning process are often not easy to tease apart. Reconstituting patterning processes outside an embryo can help isolate and control each factor and test their direct consequences on patterning.
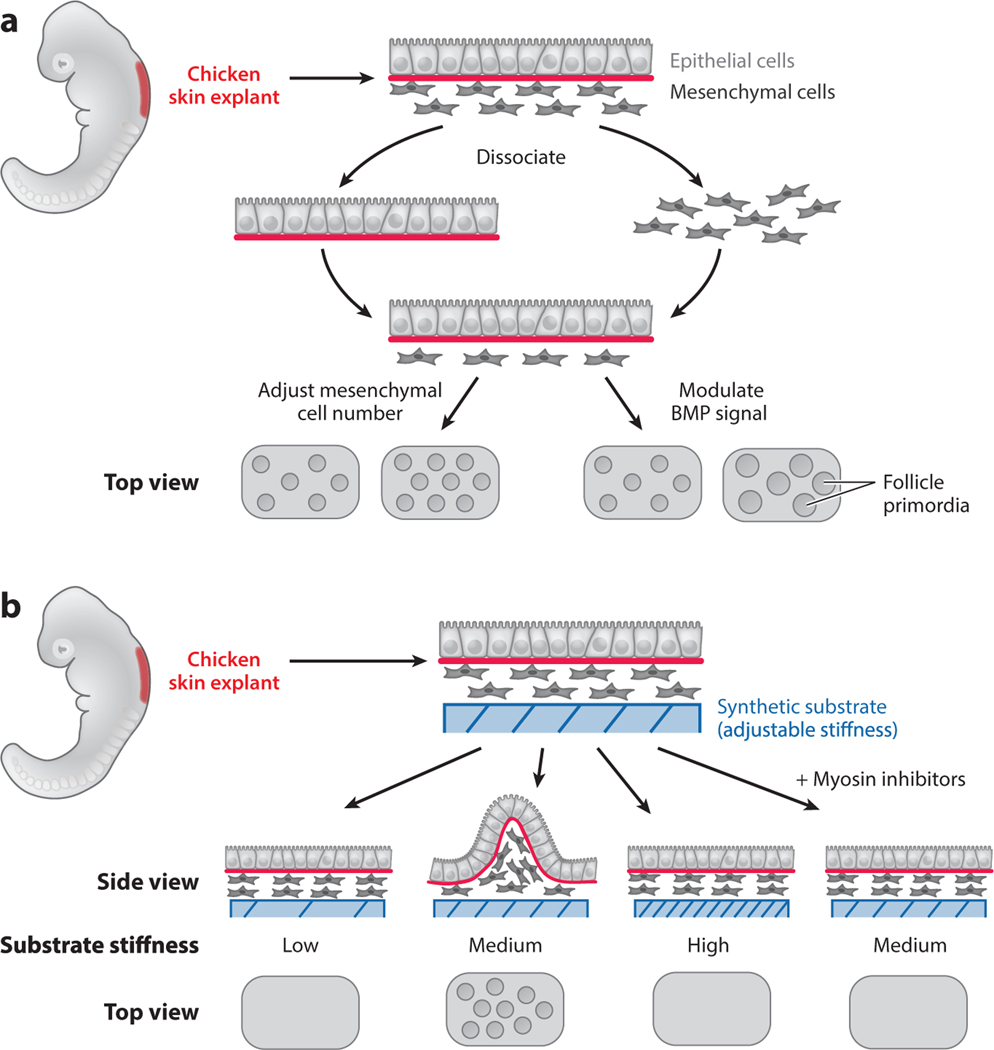
The involvement of both chemical and physical cues in tissue patterning is exemplified in the formation of the periodic patterns of hair follicles that give rise to feathers in birds. Starting as a uniform sheet of epithelium attached to the underlying mesenchyme, hair follicle primordia emerge as regularly spaced multicellular aggregates in the mesenchyme (Davidson 1983). A reconstitution system has been developed to capture the initial events of patterning and study how pattern emerges (Jiang et al. 1999, Widelitz et al. 1999). In order to harvest the right cell types, dermal mesenchymal cells were dissected out of early-stage embryos, dissociated into single cells, and plated on a tissue culture dish at high density. Intact epidermal epithelial tissue was then layered on top of the mesenchyme. The authors of these studies found that cells in the mesenchyme spontaneously formed periodic patterns of cell aggregates, a process that requires the presence of the epithelial tissue. While the spacing between aggregates depends on the number of mesenchymal cells initially seeded, the size of the aggregates can be modulated by bone morphogenetic protein signaling activities (Figure 4a) (Jiang et al. 1999). Importantly, the reconstituted system allowed the authors to test specific hypotheses. For example, one hypothesis suggests that two different cell populations exist in the mesenchymal compartment, one with the capability of becoming primordia and the other without, and patterns subsequently arise through differential adhesion-mediated sorting. By labeling subsets of cells and putting them through the dissociation–aggregation procedure, the authors could randomly mix the labeled cells with unlabeled cells. They elegantly demonstrated that all mesenchymal cells have equivalent capability of becoming primordia (Jiang et al. 1999). On the basis of these observations, they proposed that cells can randomly collide and adhere to one another to form unstable microaggregates, which can be refined and stabilized through physical and chemical feedback mechanisms.
Figure 4.
Chemical or physical cues can act on the same patterning process. Chicken skin explants were used to study the mechanism of feather follicle primordia formation. (a) Mesenchymal cells from the skin explant were dissociated from the epithelial cells and plated in vitro. An intact epithelial layer was then placed on top of the mesenchymal cells. Primordia formed spontaneously in the reconstituted system. The spacing between the primordia could be modulated by adjusting the number of mesenchymal cells seeded, and the size of the primordia could be modulated by the activity of the bone morphogenetic protein (BMP) signaling. (b) To test the role of physical cues in primordia formation, skin explants composed of both epithelial and mesenchymal cells were placed on top of engineered gels with defined stiffness. Primordia can form only on the substrate with intermediate stiffness (Medium). Primordia formation also depends on mesenchymal cell contraction, which can be blocked by myosin inhibitors.
A recent study in chicken skin explants demonstrated that physical tension in the dermal mesenchyme is sufficient for initiating pattern formation (Shyer et al. 2017). By culturing chicken skin samples on gels of different stiffness and chemically perturbing cellular contractility, the authors were able to control feather progenitor pattern establishment (Figure 4b). Cellular contractility acted as a local activator and tissue-level stiffness acted as a long-range inhibitor for forming cell aggregates. Both studies (Jiang et al. 1999, Shyer et al. 2017) suggested that physical cues can initiate pattern formation, but the two studies perturbed different physical parameters. A better understanding of the molecular/genetic nature of the mechanical properties will be the key to connecting different observations. In addition, in the reconstituted system, periodic patterns appear simultaneously across the entire tissue, whereas in chicken embryos, feather primordia in adjacent regions emerge with a stereotypical temporal order (Davidson 1983, Jiang et al. 1999). Reconstitution experiments demonstrated the sufficiency of certain factors and also suggested the existence of missing factors that await further investigation.
RECONSTITUTING TEMPORAL DYNAMICS
Accumulating evidence suggests that spatial patterns crucially depend on the temporal dynamics of developmental events, from the spatiotemporal onset of transcription of different Hox genes to the sequential patterning of the ventral neural tube (Briscoe & Small 2015, Kmita & Duboule 2003). Temporal oscillation in somite development is a beautiful example of the close relationship between temporal and spatial patterns. According to the classic clock and wavefront model, presomitic mesoderm (PSM) cells oscillate between a somite-permissive and a somite-nonpermissive state, while a somite-inducing signal moves as a wave from anterior to posterior through an embryo. Where a somite-inducing signal meets in space and time with somite-permissive cells, it induces the cells to differentiate into somites (reviewed in Dequéant & Pourquié 2008). This model depends on cell-autonomous oscillations that are coordinated between adjacent cells, an assumption difficult to assess inside an embryo.
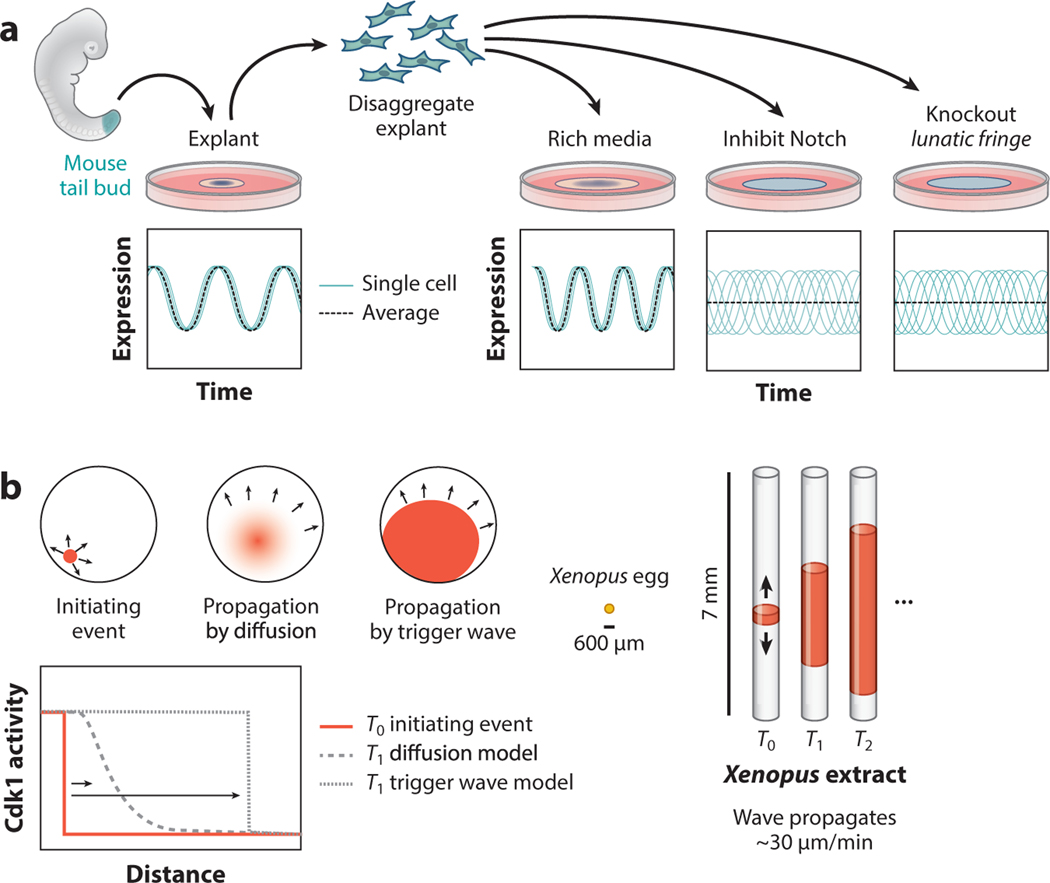
Recently, a reconstitution approach has helped with understanding the molecular basis of coordinated PSM oscillations by demonstrating that PSM cells surgically removed from a mouse tail bud continue to oscillate cell-autonomously after they have been dissociated into single cells. Furthermore, if they are reaggregated in cell culture they can spontaneously entrain their oscillations to recover the locally coordinated oscillation pattern observed in the PSM (Figure 5a) (Tsiairis & Aulehla 2016). Although the oscillations can appear to be oriented geometrically around an organizing point, PSM cells that oscillate in culture continue to oscillate even if cells at the center of the oscillating pattern are ablated, suggesting that the oscillations do not depend on a specific geometry to achieve synchronous oscillations (Hubaud et al. 2017)
Figure 5.
Mechanisms for generating temporal coordination. (a) Explant experiments using mouse tail buds demonstrated that presomitic mesoderm (PSM) cells achieve coordinated oscillations during somitogenesis by entraining locally with their neighbors via Notch signaling. When a mouse tail bud is explanted and cultured ex situ, PSM cells from the tail bud show coordinated oscillations in gene expression that reflect oscillations that occur during somitogenesis. Disaggregated tail bud cells spontaneously entrain with their neighbors if cells are disaggregated and then reaggregated, and neighbor entrainment requires Notch signaling between adjacent cells. Interfering with Notch signaling or Notch feedback (by interfering with Lunatic Fringe) prevents PSM cells from entraining with their neighbors. (b) Some signals in biology are propagated more quickly than could be achieved by diffusion of signaling molecules. Cell division in the Xenopus zygote is coordinated by a self-propagating wave of Cdk1 activity that does not slow down or change shape as it spreads from its source. The chemistry required to achieve self-propagation of Cdk1 waves can be reconstituted in tubes of zygotic extract, such that waves of activity that ordinarily travel ~300–600 μm can be made to travel 3–7 mm. Even over ~7-mm length scales, waves of Cdk1 activity were propagated at a constant rate without a decrease in amplitude, suggesting that early Xenopus cell cycles are coordinated by a Cdk1 trigger wave that travels with a uniform shape and speed to initiate cell division simultaneously throughout the zygote.
How, then, is oscillation entrained? One clue came from the fact that the oscillations are structured over very small spatial scales (i.e., cells in contact with one another have coordinated oscillations). This observation motivated experiments that tested whether contact-dependent Notch signaling is required for oscillation entrainment, and led to the discovery that PSM cells do not show entrained oscillations in the absence of Notch signaling (Tsiairis & Aulehla 2016). Coculturing explanted PSM cells with PSM cells mutated for Lunatic Fringe, a Notch modifier, revealed that Notch feedback through Lunatic Fringe affects the rate of entrainment (Yoshioka-Kobayashi et al. 2020). In addition, the entrainment depended on the precise density of the reaggregated cells, consistent with our understanding that Notch/Delta signaling requires direct physical contact between interacting cells.
When considering the temporal dynamics of development, it is useful to keep in mind the relative rates of different biological events. Oscillations during somite development occur with a ~90-min period in mice, which is sufficiently long for classically understood signal transduction and gene regulation (Lauschke et al. 2013). However, some developmentally critical events occur on much shorter timescales, such that signals cannot physically be transmitted by classically understood physical processes like diffusion (Howard et al. 2011). For example, a Xenopus egg has a 600-μm radius, and if mitosis were coordinated by a classical diffusion mechanism it would take ~2 h for an initiating signal at the center of the egg to reach the periphery of the egg. However, observations demonstrated that a fertilized egg proceeds through the cell cycle within only a few minutes (Chang & Ferrell 2013). One hypothesis that could explain this observation is that a mitotic signal propagates not through diffusion but instead as a trigger wave, which has the property that its speed and amplitude do not decrease as a function of distance from the source, and can theoretically arise from a signaling molecule that has fast positive autoregulation and slow negative autoregulation (Figure 5b) (Chang & Ferrell 2013). In one proof-of-principle experiment, Xenopus egg extract was mixed with sperm nuclei, which can divide mitotically in the egg extract and thus serve as a reporter for promitotic signals. The mixture was loaded into thin tubes, such that any wave that normally occurs in three dimensions over ~600 μm in a frog embryo can be observed in one dimension over ~7 mm. Remarkably, sperm nuclei showed coordinated mitosis, wherein a random initiating mitotic event results in a propagating wave of mitosis with a speed that decreased only slightly as it moved away from the source (Chang & Ferrell 2013). Sha et al. (2003) postulated and demonstrated that Cdk1, which is capable of rapid positive feedback and slow negative feedback, is involved in the trigger wave. Interestingly, the same framework in which Cdk1 coordinates mitosis in a developing Xenopus embryo translates directly into the corresponding process in early Drosophila embryos, suggesting that it is an ancient signaling circuit that can rapidly coordinate cell behavior throughout a tissue (Deneke et al. 2016, Vergassola et al. 2018).
Trigger wave propagation by Cdk1 enables an entire Xenopus egg to enter mitosis in a span of a few minutes, compared with diffusion-limited signaling, which would take hours to coordinate activity throughout the egg. In addition to rapid signal propagation during mitosis, Xenopus eggs show faster-than-diffusion synchrony as eggs undergo apoptosis. Using a similar reconstitution setup as had been used to model mitotic trigger waves, investigators observed apoptosis by monitoring the integrity of sperm nuclei suspended in thin tubes of egg extract, and found that the apoptotic signal propagated rapidly through thin tubes of Xenopus egg extract (Cheng & Ferrell 2018). Furthermore, by increasing the abundance of mitochondria (and therefore the positive feedback of proapoptotic signals) in the cell extract, the authors sped up apoptotic wave propagation. The measured propagation rate of the apoptotic wave—30 μm/min—closely matched the observed rate of apoptotic wave propagation when cytochrome c was injected into natural Xenopus oocytes, suggesting that the mechanism of apoptotic wave propagation observed in vitro is consistent with the process that occurs in vivo (Cheng & Ferrell 2018).
Although much progress has been made toward understanding entrainment of genetic oscillators and rapid propagation of certain signals in development, efforts to understand the basic temporal dynamics of embryogenesis have been hampered by the difficulty of quantitatively following dynamic processes in animals. Reconstituted systems are amenable to time-lapse imaging and can be studied to extract quantitative information about key parameters during embryo development. At the same time, it is crucial to evaluate whether the parameter values used in reconstituted experiments make sense in the context of developing embryos.
CHALLENGES AND OUTLOOK
As a cornerstone of synthetic developmental biology, reconstitution is not the goal but rather an approach to address questions that would be difficult to study inside an embryo, to distinguish between different models, or to discover novel biological features that can eventually be tied to actual embryo development (Davies 2017, Elowitz & Lim 2010). The examples discussed above demonstrate that reconstitution provides a unique angle for understanding development. Reconstituted systems enable isolation of concurrent developmental cues, quantitative observation of the spatiotemporal dynamics, fine control of individual parameters using synthetic tools, and even systematic rewiring of genetic circuits. For reconstitution of (aspects of) developmental processes, cells can come from many different sources, such as immortalized cell lines, primary cells isolated from an organism, or pluripotent stem cells. Different cells naturally have different self-organizing capabilities. In recent years, stem cells that can self-organize into three-dimensional tissues (e.g., organoids/embryoids/gastruloids) have been broadly used to recapitulate developmental processes outside an organism. The amount of literature on this exciting topic alone merits a separate review. We refer readers to recent excellent reviews on this topic (Huch et al. 2017, Rossi et al. 2018, Shahbazi et al. 2019, Siggia & Warmflash 2018, Simunovic & Brivanlou 2017). As the complexity of multicellular behavior that can be recapitulated increases, our control over the genetic programs decrease. Therefore, the right level of reconstitution should be carefully chosen depending on the question under study.
Synthetic developmental biology is still at a very early stage. In addition to reconstitution outside an embryo, synthetic biology tools have enabled numerous applications in live embryos, such as precise spatiotemporal control of protein activities (Johnson & Toettcher 2019) and tracking of developmental cell lineages (reviewed in McKenna & Gagnon 2019). However, to develop synthetic biology as a powerful approach for studying developmental questions, and to reconstitute basic building blocks for tissue engineering, several technical challenges must be solved. One challenge is to engineer the physical aspects of multicellular systems: to learn how to spatially arrange multiple cell types and how to supply missing boundary conditions in cell culture, such as ECM composition and tissue geometry. Miniaturized engineering tools, such as micropatterning and microfluidic techniques, have been incorporated into reconstituted systems to provide more precise control over cellular systems (Warmflash et al. 2014, Zheng et al. 2019), but new tools are still needed for dealing with submillimeter length scales. A second challenge is to genetically engineer cellular responses to environmental stimuli. Multicellular behavior depends on constant cell–cell communication and feedback, which often trigger changes in transcriptional programs. The ability to rationally engineer these transcriptional programs is crucial for building genetic circuits that enable complex multicellular behavior. Whereas analytical approaches have been effective in determining what function is computed by an enhancer, attempts to build enhancers from the bottom up using discrete regulatory elements have had limited success in recapitulating the behavior of natural enhancers, demonstrating how much we have left to discover about the design principles of enhancers (Crocker et al. 2017, Fakhouri et al. 2010, Ramos & Barolo 2013, Vincent et al. 2016). A third challenge is to introduce genetic perturbations that are precisely targeted in space and time. Natural developmental signals depend on precise spatial and temporal coordination, and we need equivalently precise experimental control to study such events. The rapidly progressing area of mammalian synthetic biology has ushered in new engineering tools that could introduce more precise perturbations in space and time (Lienert et al. 2014, Mathur et al. 2017). Once tested in reconstituted systems, these new tools can be adapted for genetic manipulation in animals. Finally, it is important to be able to efficiently integrate genes and large circuits into the genome, a shared goal in the field of synthetic biology. Together, reconstituting developmental processes will provide not only insights into developmental biology but also new tools for probing natural systems and programming multicellular behavior that will have key implications for tissue engineering and cell-based therapy.
ACKNOWLEDGMENTS
We thank Peter Reddien, Alex Schier, Fengzhu Xiong, Nagarajan Nandagopal, Christian Mosimann, Michael Pineda, Julia Kim, and Diep Nguyen for critical reading and thoughtful comments on the manuscript. P.L. acknowledges support from the Eunice Kennedy Shriver National Institute of Child Health and Human Development and the G. Harold & Leila Y. Mathers Foundation.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Contributor Information
Gavin Schlissel, Whitehead Institute for Biomedical Research, Cambridge, MA 02142, USA.
Pulin Li, Whitehead Institute for Biomedical Research, Cambridge, MA 02142, USA; Department of Biology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.
REFRENCES
- Andrae J, Gallini R, Betsholtz C. 2008. Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 22(10):1276–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydin B, Mazzoni EO. 2019. Cell reprogramming: the many roads to success. Annu. Rev. Cell Dev. Biol 35:433–52 [DOI] [PubMed] [Google Scholar]
- Benne R, Hershey JW. 1978. Mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. J. Biol. Chem 253(9):3078–87 [PubMed] [Google Scholar]
- Blau HM, Chiu C-P, Webster C. 1983. Cytoplasmic activation of human nuclear genes in stable heterocaryons. Cell 32(4):1171–80 [DOI] [PubMed] [Google Scholar]
- Briggs JA, Li VC, Lee S, Woolf CJ, Klein A, Kirschner MW. 2017. Mouse embryonic stem cells can differentiate via multiple paths to the same state. eLife 6:e26945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J, Small S. 2015. Morphogen rules: design principles of gradient-mediated embryo patterning. Development 142(23):3996–4009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JB, Ferrell JE Jr. 2013. Mitotic trigger waves and the spatial coordination of the Xenopus cell cycle. Nature 500(7464):603–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Ferrell JE Jr. 2018. Apoptosis propagates through the cytoplasm as trigger waves. Science 361(6402):607–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitu V, Stanley ER. 2006. Colony-stimulating factor 1 in immunity and inflammation. Curr. Opin. Immunol 18(1):39–48 [DOI] [PubMed] [Google Scholar]
- Crocker J, Tsai A, Stern DL. 2017. A fully synthetic transcriptional platform for a multicellular eukaryote. Cell Rep. 18(1):287–96 [DOI] [PubMed] [Google Scholar]
- Davidson D 1983. The mechanism of feather pattern development in the chick. 1. The time of determination of feather position. J. Embryol. Exp. Morphol 74:245–59 [PubMed] [Google Scholar]
- Davies J 2017. Using synthetic biology to explore principles of development. Development 144(7):1146–58 [DOI] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, Lassar AB. 1987. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51(6):987–1000 [DOI] [PubMed] [Google Scholar]
- Deneke VE, Melbinger A, Vergassola M, Di Talia S. 2016. Waves of Cdk1 activity in S phase synchronize the cell cycle in Drosophila embryos. Dev. Cell 38(4):399–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dequéant M-L, Pourquié O. 2008. Segmental patterning of the vertebrate embryonic axis. Nat. Rev. Genet 9(5):370–82 [DOI] [PubMed] [Google Scholar]
- Ebrahimkhani MR, Ebisuya M. 2019. Synthetic developmental biology: build and control multicellular systems. Curr. Opin. Chem. Biol 52:9–15 [DOI] [PubMed] [Google Scholar]
- Elowitz MB, Lim WA. 2010. Build life to understand it. Nature 468(7326):889–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakhouri WD, Ay A, Sayal R, Dresch J, Dayringer E, Arnosti DN. 2010. Deciphering a transcriptional regulatory code: modeling short‐range repression in the Drosophila embryo. Mol. Syst. Biol 6(1):341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher DA. 2016. Bottom-up biology: harnessing engineering to understand nature. Dev. Cell 38(6):587–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunwald GB. 1991. The conceptual and experimental foundations of vertebrate embryonic cell adhesion research. Dev. Biol 7:129–58 [DOI] [PubMed] [Google Scholar]
- Gurdon JB, Elsdale TR, Fischberg M. 1958. Sexually mature individuals of Xenopus laevis from the transplantation of single somatic nuclei. Nature 182(4627):64–65 [DOI] [PubMed] [Google Scholar]
- Hart Y, Reich-Zeliger S, Antebi YE, Zaretsky I, Mayo AE, et al. 2014. Paradoxical signaling by a secreted molecule leads to homeostasis of cell levels. Cell 158(5):1022–32 [DOI] [PubMed] [Google Scholar]
- Howard J, Grill SW, Bois JS. 2011. Turing’s next steps: the mechanochemical basis of morphogenesis. Nat. Rev. Mol. Cell Biol 12(6):392–98 [DOI] [PubMed] [Google Scholar]
- Hubaud A, Regev I, Mahadevan L, Pourquié O. 2017. Excitable dynamics and Yap-dependent mechanical cues drive the segmentation clock. Cell 171(3):668–82.e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch M, Knoblich JA, Lutolf MP, Martinez-Arias A. 2017. The hope and the hype of organoid research. Development 144(6):938–41 [DOI] [PubMed] [Google Scholar]
- Hughes AJ, Miyazaki H, Coyle MC, Zhang J, Laurie MT, et al. 2018. Engineered tissue folding by mechanical compaction of the mesenchyme. Dev. Cell 44(2):165–78.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang TX, Jung HS, Widelitz RB, Chuong CM. 1999. Self-organization of periodic patterns by dissociated feather mesenchymal cells and the regulation of size, number and spacing of primordia. Development 126(22):4997–5009 [DOI] [PubMed] [Google Scholar]
- Johnson HE, Toettcher JE. 2019. Signaling dynamics control cell fate in the early Drosophila embryo. Dev. Cell 48(3):361–70.e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller R, Danilchik M. 1988. Regional expression, pattern and timing of convergence and extension during gastrulation of Xenopus laevis. Development 103(1):193–209 [DOI] [PubMed] [Google Scholar]
- Kirillova AO, Kraus YA, Markov AV. 2018. Dissociation–reaggregation experiments in cnidarians as a model system for the study of the regulative capacity of metazoan development. Biol. Bull. Rev 8:1–11 [PubMed] [Google Scholar]
- Kmita M, Duboule D. 2003. Organizing axes in time and space: 25 years of colinear tinkering. Science 301(5631):331–33 [DOI] [PubMed] [Google Scholar]
- Krieg M, Arboleda-Estudillo Y, Puech PH, Käfer J, Graner F, et al. 2008. Tensile forces govern germ-layer organization in zebrafish. Nat. Cell Biol 10(4):429–36 [DOI] [PubMed] [Google Scholar]
- Lassar AB, Paterson BM, Weintraub H. 1986. Transfection of a DNA locus that mediates the conversion of 10T12 fibroblasts to myoblasts. Cell 47(5):649–56 [DOI] [PubMed] [Google Scholar]
- Lauschke VM, Tsiairis CD, François P, Aulehla A. 2013. Scaling of embryonic patterning based on phase-gradient encoding. Nature 493(7430):101–5 [DOI] [PubMed] [Google Scholar]
- LeBon L, Lee TV, Sprinzak D, Jafar-Nejad H, Elowitz MB. 2014. Fringe proteins modulate Notch-ligand cis and trans interactions to specify signaling states. eLife 3:e02950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Markson JS, Wang S, Chen S, Vachharajani V, Elowitz MB. 2018. Morphogen gradient reconstitution reveals Hedgehog pathway design principles. Science 360(6388):543–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienert F, Lohmueller JJ, Garg A, Silver PA. 2014. Synthetic biology in mammalian cells: next generation research tools and therapeutics. Nat. Rev. Mol. Cell Biol 15(2):95–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammoto T, Ingber DE. 2010. Mechanical control of tissue and organ development. Development 137(9):1407–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur M, Xiang JS, Smolke CD. 2017. Mammalian synthetic biology for studying the cell. J. Cell Biol 216(1):73–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M, Koga M, Woltjen K, Nishida E, Ebisuya M. 2015. Synthetic lateral inhibition governs cell-type bifurcation with robust ratios. Nat. Commun 6:6195. [DOI] [PubMed] [Google Scholar]
- McKenna A, Gagnon JA. 2019. Recording development with single cell dynamic lineage tracing. Development 146(12):dev169730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moloney DJ, Panin VM, Johnston SH, Chen J, Shao L, et al. 2000. Fringe is a glycosyltransferase that modifies Notch. Nature 406(6794): 369–75. [DOI] [PubMed] [Google Scholar]
- Morris SA, Daley GQ. 2013. A blueprint for engineering cell fate: current technologies to reprogram cell identity. Cell Res. 23(1):33–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller P, Rogers KW, Jordan BM, Lee JS, Robson D, et al. 2012. Differential diffusivity of Nodal and Lefty underlies a reaction-diffusion patterning system. Science 336(6082):721–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller P, Rogers KW, Yu SR, Brand M, Schier AF. 2013. Morphogen transport. Development 140(8):1621–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumm JS, Kopan R. 2000. Notch signaling: from the outside in. Dev. Biol 228(2):151–65 [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD. 1969. The formation of the mesoderm in urodelean amphibians. I. Induction by the endoderm. Wilhelm Roux Arch. Entwickl. Mech. Org 162(4):341–73 [DOI] [PubMed] [Google Scholar]
- Ramos AI, Barolo S. 2013. Low-affinity transcription factor binding sites shape morphogen responses and enhancer evolution. Philos. Trans. R. Soc. B 368(1632):20130018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers KW, Lord ND, Gagnon JA, Pauli A, Zimmerman S, et al. 2017. Nodal patterning without Lefty inhibitory feedback is functional but fragile. eLife 6:e28785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers KW, Schier AF. 2011. Morphogen gradients: from generation to interpretation. Annu. Rev. Cell Dev. Biol 27:377–407 [DOI] [PubMed] [Google Scholar]
- Rossi G, Manfrin A, Lutolf MP. 2018. Progress and potential in organoid research. Nat. Rev. Genet 19(11):671–87 [DOI] [PubMed] [Google Scholar]
- Salazar-Ciudad I 2003. Mechanisms of pattern formation in development and evolution. Development 130(10):2027–37 [DOI] [PubMed] [Google Scholar]
- Santorelli M, Lam C, Morsut L. 2019. Synthetic development: building mammalian multicellular structures with artificial genetic programs. Curr. Opin. Biotechnol 59:130–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayre MH, Tschochner H, Kornberg RD. 1992. Reconstitution of transcription with five purified initiation factors and RNA polymerase II from Saccharomyces cerevisiae. J. Biol. Chem 267(32):23376–82 [PubMed] [Google Scholar]
- Sekine R, Shibata T, Ebisuya M. 2018. Synthetic mammalian pattern formation driven by differential diffusivity of Nodal and Lefty. Nat. Commun 9:5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazi MN, Siggia ED, Zernicka-Goetz M. 2019. Self-organization of stem cells into embryos: a window on early mammalian development. Science 364(6444):948–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha W, Moore J, Chen K, Lassaletta AD, Yi C-S, et al. 2003. Hysteresis drives cell-cycle transitions in Xenopus laevis egg extracts. PNAS 100(3):975–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyer AE, Rodrigues AR, Schroeder GG, Kassianidou E, Kumar S, Harland RM. 2017. Emergent cellular self-organization and mechanosensation initiate follicle pattern in the avian skin. Science 357(6353):811–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siggia ED, Warmflash A. 2018. Modeling mammalian gastrulation with embryonic stem cells. Curr. Top. Dev. Biol 129:1–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simunovic M, Brivanlou AH. 2017. Embryoids, organoids and gastruloids: new approaches to understanding embryogenesis. Development 144(6):976–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöqvist M, Andersson ER. 2017. Do as I Say, Not(ch) as I Do: Lateral Control of Cell Fate. Dev. Biol 447(1):58–70. [DOI] [PubMed] [Google Scholar]
- Smith JC. 1989. Mesoderm induction and mesoderm-inducing factors in early amphibian development. Development 105(4):665–77 [DOI] [PubMed] [Google Scholar]
- Spemann H, Mangold H. 1924. Über Induktion von Embryonalanlagen durch Implantation artfremder Organisatoren. Arch. Mikrosk. Anat. Entwickl 101(1–3):458–638 [Google Scholar]
- Sprinzak D, Lakhanpal A, LeBon L, Garcia-Ojalvo J, Elowitz MB. 2011. Mutual inactivation of Notch receptors and ligands facilitates developmental patterning. PLOS Comput. Biol 7(6):e1002069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzak D, Lakhanpal A, LeBon L, Santat LA, Fontes ME, et al. 2010. Cis-interactions between Notch and Delta generate mutually exclusive signalling states. Nature 465(7294):86–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg MS. 1963. Reconstruction of tissues by dissociated cells. Science 141(3579):401–8 [DOI] [PubMed] [Google Scholar]
- Steinberg MS, Takeichi M. 1994. Experimental specification of cell sorting, tissue spreading, and specific spatial patterning by quantitative differences in cadherin expression. PNAS 91(1):206–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf HF. 1966. Mechanism by which cells estimate their location within the body. Nature 212(5060):430–31 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, et al. 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131(5):861–72 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126(4):663–76 [DOI] [PubMed] [Google Scholar]
- Technau U, Holstein TW. 1992. Cell sorting during the regeneration of Hydra from reaggregated cells. Dev. Biol 151(1):117–27 [DOI] [PubMed] [Google Scholar]
- Toda S, Blauch LR, Tang SKY, Morsut L, Lim WA. 2018. Programming self-organizing multicellular structures with synthetic cell–cell signaling. Science 361(6398):156–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda S, Brunger JM, Lim WA. 2019. Synthetic development: learning to program multicellular self-organization. Curr. Opin. Syst. Biol 14:41–49 [Google Scholar]
- Townes PL, Holtfreter J. 1955. Directed movements and selective adhesion of embryonic amphibian cells. J. Exp. Zool 128(1):53–120 [DOI] [PubMed] [Google Scholar]
- Tsai TY-C, Sikora M, Xia P, Colak-Champollion T, Knaut H, et al. 2019. An adhesion code ensures robust pattern formation during tissue morphogenesis. bioRxiv 803635. 10.1101/803635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiairis CD, Aulehla A. 2016. Self-organization of embryonic genetic oscillators into spatiotemporal wave patterns. Cell 164(4):656–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turing A 1952. The chemical basis of morphogenesis. Philos. Trans. R. Soc. B 237(641):37–72 [Google Scholar]
- Velasco S, Ibrahim MM, Kakumanu A, Garipler G, Aydin B, et al. 2017. A multi-step transcriptional and chromatin state cascade underlies motor neuron programming from embryonic stem cells. Cell Stem Cell 20(2):205–17.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergassola M, Deneke VE, Di Talia S. 2018. Mitotic waves in the early embryogenesis of Drosophila: bistability traded for speed. PNAS 115(10):E2165–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Südhof TC, Wernig M. 2010. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463(7284):1035–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent BJ, Estrada J, DePace AH. 2016. The appeasement of Doug: a synthetic approach to enhancer biology. Integr. Biol 8(4):475–84 [DOI] [PubMed] [Google Scholar]
- Warmflash A, Sorre B, Etoc F, Siggia ED, Brivanlou AH. 2014. A method to recapitulate early embryonic spatial patterning in human embryonic stem cells. Nat. Methods 11(8):847–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichterle H, Lieberam I, Porter JA, Jessell TM. 2002. Directed differentiation of embryonic stem cells into motor neurons. Cell 110(3):385–97 [DOI] [PubMed] [Google Scholar]
- Widelitz RB, Jiang TX, Chen CW, Stott NS, Jung HS, Chuong CM. 1999. Wnt-7a in feather morphogenesis: involvement of anterior–posterior asymmetry and proximal–distal elongation demonstrated with an in vitro reconstitution model. Development 126(12):2577–87 [DOI] [PubMed] [Google Scholar]
- Wieschaus E, Nüsslein-Volhard C. 2016. The Heidelberg screen for pattern mutants of Drosophila: a personal account. Annu. Rev. Cell Dev. Biol 32:1–46 [DOI] [PubMed] [Google Scholar]
- Wolpert L 1969. Positional information and the spatial pattern of cellular differentiation. J. Theor. Biol 25(1):1–47 [DOI] [PubMed] [Google Scholar]
- Xiong F, Tentner AR, Huang P, Gelas A, Mosaliganti KR, et al. 2013. Specified neural progenitors sort to form sharp domains after noisy Shh signaling. Cell 153(3):550–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeeles JTP, Janska A, Early A, Diffley JFX. 2017. How the eukaryotic replisome achieves rapid and efficient DNA replication. Mol. Cell 65(1):105–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka-Kobayashi K, Matsumiya M, Niino Y, Isomura A, Kori H, et al. 2020. Coupling delay controls synchronized oscillation in the segmentation clock. Nature 580(7801):119–23 [DOI] [PubMed] [Google Scholar]
- Zheng Y, Xue X, Shao Y, Wang S, Esfahani SN, et al. 2019. Controlled modelling of human epiblast and amnion development using stem cells. Nature 573(7774):421–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Franklin RA, Adler M, Jacox JB, Bailis W, et al. 2018. Circuit design features of a stable two-cell system. Cell 172(4):744–57.e17 [DOI] [PMC free article] [PubMed] [Google Scholar]