Abstract
Mucin 5AC (MUC5AC) is a secreted gel-forming mucin expressed by several epithelia. In the colon, MUC5AC is expressed in scattered normal epithelial cells but can be abundant in colorectal cancers. To clarify the relationship of MUC5AC expression with parameters of tumor aggressiveness and mismatch repair deficiency (dMMR) in colorectal cancer, a tissue microarray containing 1812 colorectal cancers was analyzed by immunohistochemistry. MUC5AC expression was found in 261 (15.7%) of 1,667 analyzable colorectal cancers. MUC5AC expression strongly depended on the tumor location and gradually decreased from proximal (27.4% of cecum cancers) to distal (10.6% of rectal cancers; p < 0.0001). MUC5AC expression was also strongly linked to dMMR. dMMR was found in 21.3% of 169 cancers with MUC5AC positivity but in only 4.6% of 1051 cancers without detectable MUC5AC expression (p < 0.0001). A multivariate analysis showed that dMMR status and tumor localization predicted MUC5AC expression independently (p < 0.0001 each). MUC5AC expression was unrelated to pT and pN status. This also applied to the subgroups of 1136 proficient MMR (pMMR) and of 84 dMMR cancers. The results of our study show a strong association of MUC5AC expression with proximal and dMMR colorectal cancers. However, MUC5AC expression is unrelated to colon cancer aggressiveness.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00795-020-00274-2.
Keywords: MUC5AC, Mismatch repair deficiency, Colon cancer, Progression, Tissue microarray
Introduction
Colorectal cancer was the third most common cancer worldwide in 2018 and the second most common cause for cancer related death [1]. Standard treatment of colorectal cancer consists of surgical removal. In high-risk cancers, adjuvant chemotherapy is also given in order to destroy micrometastasis and to reduce the risk of local recurrence. Possible chemotherapies include conventional cytotoxic chemotherapy and several antiangiogenic substances. In case of BRAF, KRAS, and NRAS wild-type cancers, anti-EGFR therapy antibodies can also be applied (summarized in [2]). Immune checkpoint inhibitors can be administered in cancers harboring microsatellite instability (MSI) or mismatch repair deficiency (dMMR) (summarized in [3]). Established prognostic factors of colorectal carcinomas include pT, pN, M status and histologic tumor features [4, 5]. They are statistically powerful but cannot reliably predict disease course in individual patients.
Mucin 5AC (MUC5AC) is a biomarker of potential interest in colorectal cancer. MUC5AC is a secreted gel-forming mucin [6, 7] expressed in normal mucus-producing cells of the stomach, the lung, and the uterine cervix [8–10] as well as in cancer cells of the ovarian, the pancreas, and the gastrointestinal tract [11–14]. MUC5AC expression was earlier described to occur in 0–95% of colorectal cancers in studies analyzing 22–649 cancers [11, 12, 15–35], and it was shown to be linked to MSI or dMMR in studies analyzing 35–649 cancers [16–18, 20, 22, 26, 28, 30, 33, 34]. Since MSI is linked to favorable prognosis in colorectal cancer [36], a favorable disease course could be expected in MUC5AC-positive cancers. However, multiple studies have provided evidence that MUC5AC expression may drive cancer aggressiveness in colorectal cancer cell lines and xenograft models [37–39]. The 11 studies analyzing the prognostic relevance of MUC5AC expression in colorectal cancer in cohorts of 35–649 patients have found divergent results [11, 12, 17, 18, 20, 21, 23, 25, 31, 34, 35].
None of the earlier studies on the putative clinical role of MUC5AC expression in colon cancer have considered the association between MUC5AC expression and clinic-pathological parameters in the subgroups of mismatch repair proficient (pMMR) and dMMR cancers in their analyses. We thus analyzed the relationship between MUC5AC expression and features of tumor aggressiveness (pT and pN) in all cancers and in the subgroup of pMMR and dMMR cancers in a cohort of 1812 colorectal cancers by immunohistochemistry (IHC) in a tissue microarray (TMA) format.
Material and methods
Tissue microarray (TMA)
Our colon cancer TMA consisted of 1,812 colon cancers diagnosed at the Institutes of Pathology of the University Medical Center Hamburg-Eppendorf (Hamburg, Germany) and the Department of Pathology of the Academic Hospital Fuerth (Fuerth, Germany) between 2009 and 2019. TMA construction was done as previously described [40]. Clinical, pathological and molecular parameters were obtained from patient records (Table 1). The use of archived remnants of diagnostic tissues for manufacturing of tissue microarrays and their analysis for research purpose as well as patient data analysis has been approved by local laws (HmbKHG, §12) and by the local ethics committee (Ethics Commission Hamburg, WF-049/09). All work has been carried out in compliance with the Helsinki Declaration.
Table 1.
Patient cohort
| All tumors (n = 1812) | |
|---|---|
| Age | |
| Median | 73.2 |
| Mean | 72.1 |
| Tumor localization | |
| Caecum | 172 (9.6%) |
| c. ascendens | 200 (11.2%) |
| c. transversum | 110 (6.2%) |
| c. descendens | 115 (6.5%) |
| c. sigmoideum | 725 (40.7%) |
| Rectum | 461 (25.9%) |
| Colon side | |
| Left | 1311 (73.1%) |
| Right | 483 (26.9%) |
| Tumor stage | |
| pT1 | 76 (4.3%) |
| pT2 | 354 (19.8%) |
| pT3 | 989 (55.4%) |
| pT4 | 365 (20.5%) |
| Lymph node status | |
| pN− | 926 (52.4%) |
| pN + | 841 (47.6%) |
| Mismatch repair status | |
| Deficient | 94 (7.2%) |
| Proficient | 1203 (92.3%) |
c. colon
Immunohistochemistry (IHC)
Freshly prepared TMA sections were immunostained on one day in one experiment. Slides were deparaffinized and exposed to heat-induced antigen retrieval for 5 min in an autoclave at 121 °C in pH 7.8 Dako Target Retrieval Solution buffer (Dako, Glostrup, Denmark). Primary antibody specific against MUC5AC protein (mouse monoclonal, MSVA-109, MS Validated Antibodies, Hamburg, Germany) was applied at 37 °C for 60 min at a dilution of 1:200. Bound antibody was then visualized using the EnVision Kit (Dako, Glostrup, Denmark) according to the manufacturer’s instructions. For immunostaining of the MMR proteins, primary antibodies (ready to use) specific for MLH1 (clone ES05, mouse), PMS2 (clone EP51, rabbit), MSH2 (clone FE11, mouse), and MSH6 (clone EP49, rabbit) from Dako (Glostrup, Denmark) were applied for 20 min (MLH1, MSH2, MSH) or 30 min (PMS2) in an automated immunostainer (Dako/Agilent Autostainer Link 48 (Santa Clara, USA). MUC5AC staining was seen in the membrane and cytoplasm of the colon cancer cells and immunostaining was interpreted as follows: Negative: no staining at all tumor cells, weak staining: staining intensity of 1 + in ≤ 70% of the tumor cells or staining intensity of 2 + in ≤ 30% of the tumor cells, moderate staining: staining intensity of 1 + in > 70% of the tumor cells, staining intensity of 2 + in > 30% but in ≤ 70% of the tumor cells or staining intensity of 3 + in ≤ 30% of the tumor cells, strong staining: staining intensity of 2 + in > 70% of the tumor cells or staining intensity of 3 + in > 30% of the tumor cells. Nuclear staining of the MMR proteins was interpreted as negative (no staining) and positive (at least weak staining).
Statistics
Statistical calculations were performed with JMP® software (SAS Institute Inc., NC, USA). Contingency tables and the Chi-square test were performed to search for associations between MUC5AC expression, clinical-pathological parameters, and MSI. Multinominal logistic regression was performed to test the impact of MUC5AC and tumor localization (right/left, proximal to distal) on dMMR status and test the impact of dMMR and tumor localization (right/left, proximal to distal) on MUC5AC status. A p value ≤ 0.05 was regarded as statistically significant.
Results
Technical issues
MUC5AC expression was informative in 1667 (92%) of the 1,812 arrayed cancers in our IHC analysis. Reasons for non-informative cases (n = 145; 8%) included lack of tissue samples or the absence of unequivocal cancer cells in the TMA spot.
MUC5AC expression and tumor phenotype
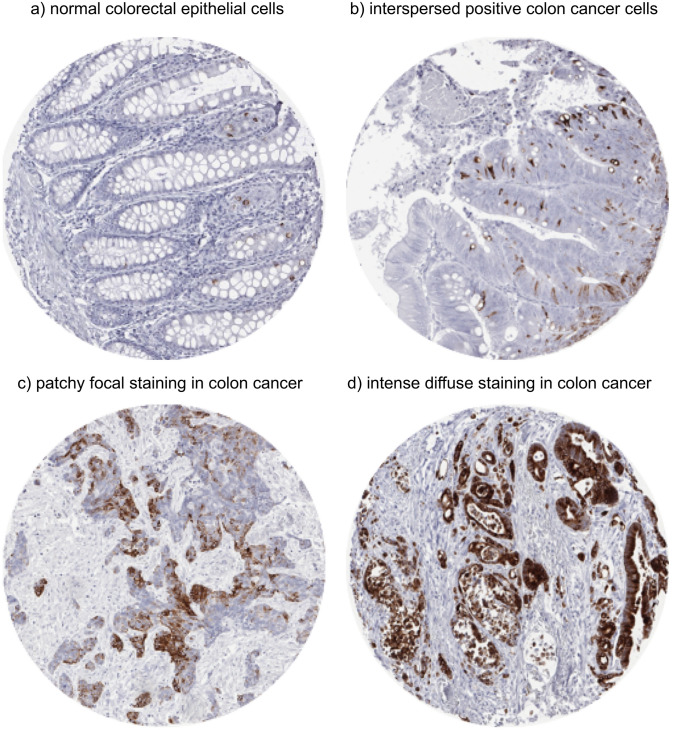
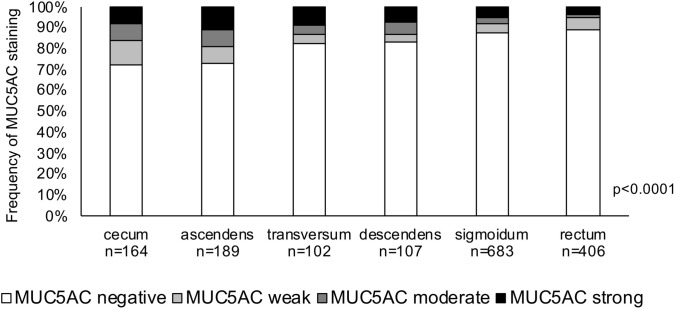
In normal colorectal epithelial cells, MUC5AC expression was only seen in few scattered epithelial cells (Fig. 1a). In colorectal cancer, positive MUC5AC staining was seen in 261 (15.7%) of 1667 analyzable tumor spots. The staining patterns included variable numbers of interspersed positive cells, patchy focal staining, and intense diffuse positivity (Fig. 1b–d). According to our classification, positive cases included 97 (5.8%) cancers with weak, 63 (3.8%) with moderate, and 101 (6.0%) with strong MUC5AC staining. MUC5AC positivity was significantly associated with colorectal cancer localization. The positivity rate gradually decreased from proximal (27.4% of 164 cecum cancers) to distal (10.6% of 406 rectal cancers; p < 0.0001; Fig. 2). MUC5AC expression was unrelated to pT and pN (Table 2).
Fig. 1.
MUC5AC staining in normal and cancerous tissue (magnification: 10×)
Fig. 2.
MUC5AC expression and colorectal cancer localization
Table 2.
MUC5AC expression and tumor phenotype
| N | Negative (%) | Weak (%) | Moderate (%) | Strong (%) | p | ||
|---|---|---|---|---|---|---|---|
| MUC5AC staining in all cancers | |||||||
| Tumor stage | 0.3387 | ||||||
| pT1 | 70 | 87.1 | 8.6 | 1.4 | 2.9 | ||
| pT2 | 336 | 83.6 | 5.1 | 5.1 | 6.3 | ||
| pT3 | 907 | 85.7 | 5.8 | 3.0 | 5.5 | ||
| pT4 | 338 | 81.7 | 5.9 | 5.0 | 7.4 | ||
| Lymph node status | 0.4523 | ||||||
| pN− | 852 | 84.0 | 5.5 | 3.6 | 6.8 | ||
| pN + | 782 | 85.0 | 6.0 | 4.0 | 5.0 | ||
| Side | < 0.0001 | ||||||
| Left | 1204 | 88.0 | 4.7 | 2.5 | 4.8 | ||
| Right | 457 | 74.8 | 8.8 | 7.2 | 9.2 | ||
| Mismatch repair status | < 0.0001 | ||||||
| Deficient | 84 | 57.1 | 7.1 | 9.5 | 26.2 | ||
| Proficient | 1136 | 88.3 | 5.7 | 2.5 | 3.5 | ||
| MUC5AC in mismatch repair deficient cancers | |||||||
| Tumor stage | 0.0793 | ||||||
| pT1 | 6 | 83.3 | 0 | 0 | 16.7 | ||
| pT2 | 19 | 36.8 | 10.5 | 26.3 | 26.3 | ||
| pT3 | 40 | 52.5 | 7.5 | 7.5 | 32.5 | ||
| pT4 | 19 | 78.9 | 5.3 | 0 | 15.8 | ||
| Lymph node status | 0.0581 | ||||||
| pN− | 55 | 52.7 | 5.4 | 14.5 | 27.7 | ||
| pN + | 27 | 63.0 | 11.1 | 0 | 25.9 | ||
| MUC5AC in mismatch repair proficient cancers | |||||||
| Tumor stage | 0.0688 | ||||||
| pT1 | 45 | 88.9 | 11.1 | 0 | 0 | ||
| pT2 | 240 | 91.3 | 5.0 | 2.5 | 1.3 | ||
| pT3 | 618 | 88.7 | 5.5 | 2.1 | 3.7 | ||
| pT4 | 222 | 85.1 | 5.8 | 3.6 | 5.4 | ||
| Lymph node status | 0.7912 | ||||||
| pN− | 574 | 89.4 | 5.2 | 2.1 | 3.3 | ||
| pN + | 541 | 87.6 | 6.1 | 2.7 | 3.5 | ||
MUC5AC expression and mismatch repair deficiency (dMMR)
dMMR as a surrogate for MSI was detected by MLH1, MSH2, PMS2, and MSH6 IHC. In brief, MLH1 was negative in 12.3% and positive in 87.7% of 1379 analyzable cases, MSH2 was negative in 2.3% and positive in 97.7% of 1356 analyzable cases, MSH6 was negative in 3.9% and positive in 96.1% of 1416 analyzable cases, and PMS2 was negative in 9.9% and positive in 90.1% of 1401 analyzable cases (Supplementary Table 1 and supplementary Fig. 1). Overall, dMMR was found in 94 (7.2%) and pMMR in 1203 (92.8%) of 1297 cases analyzable for all four proteins. Of these cases, 1220 cancers were analyzable for both MMR and MUC5AC. MUC5AC expression was significantly associated with dMMR. dMMR was found in 21.3% of 169 cancers with MUC5AC positivity but in only 4.6% of 1051 cancers without detectable MUC5AC expression (p < 0.0001, Table 2). This association was independent of the tumor localization, as the frequency of dMMR cancers is higher in proximal and distal colorectal tumors with MUC5AC positive in comparison with MUC5AC-negative cancers. Lack of significance in some subgroups may be due to low case numbers (Table 3). In addition, our multivariate analyses showed that firstly MUC5AC expression and tumor localization independently predicted dMMR status and secondly dMMR status and tumor localization independently predicted MUC5AC expression status (Table 4). In the subgroups of dMMR and pMMR cancers, MUC5AC expression was unrelated to pT and pN status (Table 2).
Table 3.
MUC5AC expression, tumor localization, and mismatch repair status
| MUC5AC | N | Mismatch repair status | P | |
|---|---|---|---|---|
| Deficient (%) | Proficient (%) | |||
| Caecum | 0.0004 | |||
| Negative | 74 | 8.1 | 91.9 | |
| Weak | 11 | 36.4 | 63.6 | |
| Moderate | 9 | 22.2 | 77.8 | |
| Strong | 9 | 66.7 | 33.3 | |
| C. ascendens | < 0.0001 | |||
| Negative | 98 | 9.2 | 90.8 | |
| Weak | 13 | 15.4 | 84.6 | |
| Moderate | 8 | 62.5 | 37.5 | |
| Strong | 11 | 63.4 | 36.4 | |
| C. transversum | 0.0004 | |||
| Negative | 56 | 1.8 | 98.2 | |
| Weak | 3 | 0 | 100 | |
| Moderate | 3 | 33.3 | 66.7 | |
| Strong | 5 | 40.0 | 60.0 | |
| C. descendens | 0.7063 | |||
| Negative | 65 | 15.4 | 84.6 | |
| Weak | 3 | 0 | 100 | |
| Moderate | 1 | 0 | 100 | |
| Strong | 5 | 20.0 | 80.0 | |
| C. sigmoideum | 0.0064 | |||
| Negative | 453 | 3.5 | 96.5 | |
| Weak | 22 | 0 | 100 | |
| Moderate | 12 | 0 | 100 | |
| Strong | 22 | 22.7 | 77.3 | |
| Rectum | 0.4502 | |||
| Negative | 297 | 2.0 | 98.0 | |
| Weak | 18 | 0 | 100 | |
| Moderate | 2 | 0 | 100 | |
| Strong | 9 | 11.1 | 88.9 | |
c. colon
Table 4.
Multivariate analysis
| Parameter vs mismatch repair status (MMR) | Chi-square | P | Parameter vs MUC5AC | Chi-square | p |
|---|---|---|---|---|---|
| Model 1 | |||||
| MUC5AC (negative, weak, moderate, strong) | 47.8 | < 0.0001 | MMR (proficient, deficient) | 47.8 | < 0.0001 |
| Localization (6 loci) | 43.6 | < 0.0001 | Localization (6 loci) | 33.1 | 0.0045 |
| Model 2 | |||||
| MUC5AC (negative, weak, moderate, strong) | 49.6 | < 0.0001 | MMR (proficient, deficient) | 49.6 | < 0.0001 |
| Left vs right | 22.6 | < 0.0001 | Localization (left vs right) | 24.6 | < 0.0001 |
MMR mismatch repair, 6 loci: Caecum, caecum ascendens, caecum transversum, caecum descendens, caecum sigmoideum, rectum
Discussion
MUC5AC expression was found in 16% of colorectal cancers in our study. This is in the lower range of data from previous studies. A total of 23 studies have earlier analyzed MUC5AC expression in colorectal cancer (Table 5) and described MUC5AC expression to occur in 0–95% analyzing 22–649 cancers [11, 12, 15–35]. Likely reasons for discrepant results include the use of different antibodies, IHC protocols, and cut-off levels or scores to define MUC5AC positivity as well as the composition of the tumor cohorts. Notably higher rates of MUC5AC expression were, for example, found in studies analyzing mucinous colorectal cancers only (MUC5AC expression in 23–90% of 32–194 tumors) [19 , 26, 30–32].
Table 5.
Summary of previous MUC5AC studies
| Author and year | Analyzable tumors | Antibody | Cut-off/score | Expression frequency (%) | Prognosis | Phenotype |
|---|---|---|---|---|---|---|
| Li et al. (2019) [12] | Meta-anaylsis | Different | Different | 15–60 | Favorable | No association |
| Mesa et al. (2020) [28] | 88 | 292-M94 | Score: percentage and intensity | 37–59 | – | – |
| Hiromoto et al. (2018) [19] | – | NCL-MUC-5AC | Score: percentage and intensity | 26–53 | – | – |
| Al-Khayal et al. (2016) [15] | 22 | sc-33667 | Score: percentage and intensity | 0 | – | – |
| Betge et al. (2016) [17] | 381 | 45M1 | > 50% positive tumor cells | 8–42 | Favorable | – |
| Krishn et al. (2016) [24] | – | 45M1 | Score: percentage and intensity | 38 | – | – |
| Kesari et al. (2015) [11] | 50 | CLH2 | > 30% positive tumor cells | 25 | – | No association |
| Kim et al. (2015) [22] | 274 | NCL-MUC-5AC | ≥ 10% positive tumor cells | 35 | – | – |
| Raghoebir et al. (2014) [32] | 32 | 45M1 | ≥ 5% tumor cells with high staining intensity | 27–90 | – | – |
| Tsai et al. (2015) [33] | 123 | CLH2 | Score: percentage and intensity | 55 | – | – |
| Nishida et al. (2014) [29] | 265 | CLH2 | ≥ 20% positive tumor cells | 17 | – | – |
| Imai et al. (2013) [20] | 250 | CLH2 | Score: percentage and intensity | 30–95 | Favorable | Inverse |
| Walsh et al. (2013) [34] | 649 | 45M1 | > 0% positive tumor cells | 49 | – | Associated |
| Khanh et al. (2013) [21] | 206 | CLH2 | score: percentage and intensity | 34 | Favorable | No association |
| Matsuda et al. (2010) [27] | 569 | CLH2 | / | 15 | – | – |
| Arai et al. (2007) [16] | 35 | CLH2 | ≥ 25% positive tumor cells | 63 | – | – |
| Park et al. (2006) [30] | 194 | CLH2 | ≥ 10% positive tumor cells | 47 | – | – |
| Losi et al. (2004) [26] | 136 | 45M1 | Percentage of positive cells | 20–54 | – | – |
| Kocer et al. (2002) [23] | 41 | 45M1 | Score: percentage and intensity | 34 | Favorable | Inverse |
| Biemer-Hüttmann et al. (2000) [18] | 93 | Neomarkers | Score: percentage and intensity | 41 | – | No association |
| Lennerz (2016) [25] | 33 | – | – | 45 | Unfavorable | – |
| Perez et al. (2008) [31] | 35 | CLH2 | Score: percentage and intensity | 23 | Favorable | No association |
| Wang et al. (2017) [35] | 139 | NCL-MUC5-AC | > 20% positive tumor cells | 28 | Favorable | Associated |
Our data demonstrate a striking link of MUC5AC expression with right colon tumor location and dMMR. Both findings are consistent with data from previous studies. A total of 13 studies have earlier analyzed the relationship of MUC5AC expression and tumor location in colon cancer, and almost all of them (11/13) have found a significant association of MUC5AC expression with proximal cancers (Supplementary Table 2). For example, MUC5AC expression was found by Betge et al. in 66% of 107 right-sided cancers and in 47% of 107 left-sided cancers [17], Nishida et al. in 27% of 89 right-sided cancers and in 12% of 27 left-sided cancers [29], Imai et al. in 63% of 114 right-sided cancers and in 31% of 121 left-sided cancers [20], Walsh et al. in 60% of 220 right-sided cancers and in 43% of 417 left-sided cancers [34], Park et al. in 53% of 76 right-sided cancers and in 17% of 118 left-sided cancers [30], and Biemer-Hüttmann et al. in 55% of 40 proximal and in 22% of 23 rectal cancers [18]. A gradual change of biomarker expression from proximal to distal colon could, for example, be explained by continuing changes in the density and the composition of the stool, and the exposure time to different carcinogenic factors during the colon passage or be related to the embryonal development of the colon (summarized in [41]). Other examples of biomarkers that were described to vary dependent on the localization within the colon, for example, include AMACR [42], p53 [30] as well as amplification of EGFR and HER2 [43].
At least 9 studies have analyzed MUC5AC expression and dMMR/MSI in colon cancer, and all of them have described significant associations (Supplementary Table 3). For example, MUC5AC expression was found by Betge et al. in 7% of 350 pMMR and in 17% of 23 dMMR cancers [17], Imai et al. in 43% of 72 pMMR and in 84% of 19 dMMR cancers [20], Arai et al. in 45% of 20 MSS and in 87% of 15 MSI cancers [16], Losi et al. in 47% of 23 pMMR and in 67% of 27 dMMR cancers [26], and Biemer-Hüttmann et al. in 28% of 47 MSS and 77% of 22 MSI tumors [18]. Given the well-known associations of MSI with right-sided colon cancer location (reviewed in [44]), it was expected that MUC5AC (as any biomarker) would be linked to both or none. However, our multivariate analysis revealed that tumor location and dMMR independently enhanced the likelihood for detectable MUC5AC expression in colorectal cancer. The underlying molecular mechanism is not known. However, one study hypothesizes an increased likelihood of MUC5AC promotor hypomethylation in MSI cancers. MUC5AC promotor hypomethylation results in MUC5AC upregulation and was shown to be associated with a “mutator” phenotype in—especially MSI—colon cancers [45].
In a thorough experimental study, Pothuraju et al. have recently shown that differential MUC5AC expression drives tumorigenesis and promotes aggressiveness of colorectal cancer cells in vitro and in vivo. The authors examined the impact of reduced (shRNA-mediated) or absent (CRISPR/Cas9-mediated) MUC5AC expression on cell proliferation, anchorage independent cell growth, cell migration, and cell invasion in two endogenous MUC5AC expressing colorectal cancer cell lines. In addition, they prepared a MUC5AC knockout xenograft model to investigate the impact of MUC5AC on tumorigenesis in vivo. Overall, the authors showed that MUC5AC expression enhanced cell growth, invasion and migration and decreased apoptosis in vitro and increased tumorigenesis in vivo [38]. However, the absence of associations between MUC5AC expression and pT as well as pN argues against a clinically relevant role of MUC5AC expression for driving aggressiveness of human colorectal cancer cells. A lack of clinical relevant prognostic impact of MUC5AC expression is in line with the conflicting results of 11 earlier studies analyzing the clinical relevance of MUC5AC expression in colorectal cancer (summarized in Table 5). Of these, seven studies with 35–381 patients described an association of high MUC5AC expression with favorable phenotype and/or prognosis [12, 17, 20, 21, 23, 31, 35], three studies with 33–250 patients reported a link between high MUC5AC expression and poor phenotype and/or prognosis [25, 34, 35], and five studies involving 35–206 patients could not find any relationship between MUC5AC expression and clinic-pathological features [11, 12, 18, 21, 31]. The fact that MUC5AC expression was also unrelated to aggressive cancer phenotype in our 1051 pMMR cancers demonstrates that adverse prognostic effects of MUC5AC are not obscured by the favorable prognostic influence of dMMR.
It is of note that MUC5AC may also represent a suitable drug target. Ensituximab (Neo-102), a novel chimeric monoclonal antibody, binds to an aberrantly glycosylated cancer-associated MUC5AC variant and is able to activate the immune system to exert a cytotoxic T-lymphocyte response [47]. In a phase I study of pancreatic cancer patients preselected for MUC5AC expression, a favorable toxicity profile was found for Ensituximab [47]. In a recent phase II study, Ensituximab resulted in stable disease in 21% of 56 patients with heavily pretreated refractory colorectal cancers and was well tolerated [48]. Given the high numbers of inflammatory cells occurring in MSI/dMMR-positive colorectal cancers, one might speculate that Ensituximab treatment might be potentially promising in these carcinomas.
In summary, the results of our study show that elevated MUC5AC expression is independently linked to proximal location and dMMR in colorectal cancers. However, both in dMMR and in pMMR cancers, MUC5AC expression is unrelated to aggressive cancer phenotype.
Competing interest
The Institute of Pathology of the UKE receives royalties on the sales of MUC5AC clone MSVA-109 from MS Validated Antibodies GmbH (owned by a family member of GS).
Ethical approval
The usage of archived diagnostic left-over tissues for manufacturing of tissue microarrays, their analysis for research purposes and patient data analysis has been approved by local laws (HmbKHG, §12,1) and by the local ethics committee (Ethics commission Hamburg, WF-049/09). All work has been carried out in compliance with the Helsinki Declaration.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file1 Supplementary figure 1. MMR protein immunostaining in colorectal cancer. Supplementary table 1. Frequency of mismatch repair protein staining in colorectal cancer. Supplementary table 2. MUC5AC immunostaining and tumor location in previous studies. Supplementary table 3. MUC5AC immunostaining and mismatch repair deficiency/microsatellite instability in previous studies (DOCX 14617 KB)
Acknowledgments
We are grateful to Melanie Witt, Inge Brandt, Maren Eisenberg, and Sünje Seekamp for excellent technical assistance.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Afrasanie VA, Marinca MV, Alexa-Stratulat T, et al. KRAS, NRAS, BRAF, HER2 and microsatellite instability in metastatic colorectal cancer—practical implications for the clinician. Radiol Oncol. 2019;53:265–274. doi: 10.2478/raon-2019-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sahin IH, Akce M, Alese O, et al. Immune checkpoint inhibitors for the treatment of MSI-H/MMR-D colorectal cancer and a perspective on resistance mechanisms. Br J Cancer. 2019;121:809–818. doi: 10.1038/s41416-019-0599-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amin MB, Edge S, Greene F, et al. AJCC Cancer Staging Manual. New York: Springer; 2017. [Google Scholar]
- 5.Fleming M, Ravula S, Tatishchev SF, Wang HL. Colorectal carcinoma: pathologic aspects. J Gastrointest Oncol. 2012;3:153–173. doi: 10.3978/j.issn.2078-6891.2012.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verma M, Davidson EA. Mucin genes: structure, expression and regulation. Glycoconj J. 1994;11:172–179. doi: 10.1007/BF00731215. [DOI] [PubMed] [Google Scholar]
- 7.Kim YS, Gum JR., Jr Diversity of mucin genes, structure, function, and expression. Gastroenterology. 1995;109:999–1001. doi: 10.1016/0016-5085(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 8.Adler KB, Tuvim MJ, Dickey BF. Regulated mucin secretion from airway epithelial cells. Front Endocrinol (Lausanne) 2013;4:129. doi: 10.3389/fendo.2013.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riethdorf L, O'Connell JT, Riethdorf S, Cviko A, Crum CP. Differential expression of MUC2 and MUC5AC in benign and malignant glandular lesions of the cervix uteri. Virchows Arch. 2000;437:365–371. doi: 10.1007/s004280000273. [DOI] [PubMed] [Google Scholar]
- 10.Van de Bovenkamp JH, Mahdavi J, Korteland-Van Male AM, et al. The MUC5AC glycoprotein is the primary receptor for Helicobacter pylori in the human stomach. Helicobacter. 2003;8:521–532. doi: 10.1046/j.1523-5378.2003.00173.x. [DOI] [PubMed] [Google Scholar]
- 11.Kesari MV, Gaopande VL, Joshi AR, Babanagare SV, Gogate BP, Khadilkar AV. Immunohistochemical study of MUC1, MUC2 and MUC5AC in colorectal carcinoma and review of literature. Indian J Gastroenterol. 2015;34:63–67. doi: 10.1007/s12664-015-0534-y. [DOI] [PubMed] [Google Scholar]
- 12.Li C, Zuo D, Liu T, Yin L, Li C, Wang L. Prognostic and clinicopathological significance of MUC family members in colorectal cancer: a systematic review and meta-analysis. Gastroenterol Res Pract. 2019;2019:2391670. doi: 10.1155/2019/2391670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Albarracin CT, Jafri J, Montag AG, Hart J, Kuan SF. Differential expression of MUC2 and MUC5AC mucin genes in primary ovarian and metastatic colonic carcinoma. Hum Pathol. 2000;31:672–677. doi: 10.1053/hupa.2000.6799. [DOI] [PubMed] [Google Scholar]
- 14.Sierzega M, Mlynarski D, Tomaszewska R, Kulig J. Semiquantitative immunohistochemistry for mucin (MUC1, MUC2, MUC3, MUC4, MUC5AC, and MUC6) profiling of pancreatic ductal cell adenocarcinoma improves diagnostic and prognostic performance. Histopathology. 2016;69:582–591. doi: 10.1111/his.12994. [DOI] [PubMed] [Google Scholar]
- 15.Al-Khayal K, Abdulla M, Al-Obaid O, et al. Differential expression of mucins in Middle Eastern patients with colorectal cancer. Oncol Lett. 2016;12:393–400. doi: 10.3892/ol.2016.4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arai T, Kasahara I, Sawabe M, et al. Microsatellite-unstable mucinous colorectal carcinoma occurring in the elderly: comparison with medullary type poorly differentiated adenocarcinoma. Pathol Int. 2007;57:205–212. doi: 10.1111/j.1440-1827.2007.02082.x. [DOI] [PubMed] [Google Scholar]
- 17.Betge J, Schneider NI, Harbaum L, et al. MUC1, MUC2, MUC5AC, and MUC6 in colorectal cancer: expression profiles and clinical significance. Virchows Arch. 2016;469:255–265. doi: 10.1007/s00428-016-1970-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Biemer-Huttmann AE, Walsh MD, McGuckin MA, et al. Mucin core protein expression in colorectal cancers with high levels of microsatellite instability indicates a novel pathway of morphogenesis. Clin Cancer Res. 2000;6:1909–1916. [PubMed] [Google Scholar]
- 19.Hiromoto T, Murakami T, Akazawa Y, et al. Immunohistochemical and genetic characteristics of a colorectal mucin-rich variant of traditional serrated adenoma. Histopathology. 2018;73:444–453. doi: 10.1111/his.13643. [DOI] [PubMed] [Google Scholar]
- 20.Imai Y, Yamagishi H, Fukuda K, Ono Y, Inoue T, Ueda Y. Differential mucin phenotypes and their significance in a variation of colorectal carcinoma. World J Gastroenterol. 2013;19:3957–3968. doi: 10.3748/wjg.v19.i25.3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khanh DT, Mekata E, Mukaisho K, et al. Transmembrane mucin MUC1 overexpression and its association with CD10(+) myeloid cells, transforming growth factor-beta1 expression, and tumor budding grade in colorectal cancer. Cancer Sci. 2013;104:958–964. doi: 10.1111/cas.12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim JH, Kim KJ, Rhee YY, et al. Gastric-type expression signature in serrated pathway-associated colorectal tumors. Hum Pathol. 2015;46:643–656. doi: 10.1016/j.humpath.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 23.Kocer B, Soran A, Erdogan S, et al. Expression of MUC5AC in colorectal carcinoma and relationship with prognosis. Pathol Int. 2002;52:470–477. doi: 10.1046/j.1440-1827.2002.01369.x. [DOI] [PubMed] [Google Scholar]
- 24.Krishn SR, Kaur S, Smith LM, et al. Mucins and associated glycan signatures in colon adenoma-carcinoma sequence: prospective pathological implication(s) for early diagnosis of colon cancer. Cancer Lett. 2016;374:304–314. doi: 10.1016/j.canlet.2016.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lennerz JK, van der Sloot KWJ, Le LP, et al. Colorectal cancer in Crohn's colitis is comparable to sporadic colorectal cancer. Int J Colorectal Dis. 2016;31:973–982. doi: 10.1007/s00384-016-2574-x. [DOI] [PubMed] [Google Scholar]
- 26.Losi L, Scarselli A, Benatti P, et al. Relationship between MUC5AC and altered expression of MLH1 protein in mucinous and non-mucinous colorectal carcinomas. Pathol Res Pract. 2004;200:371–377. doi: 10.1016/j.prp.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 27.Matsuda M, Sentani K, Noguchi T, et al. Immunohistochemical analysis of colorectal cancer with gastric phenotype: claudin-18 is associated with poor prognosis. Pathol Int. 2010;60:673–680. doi: 10.1111/j.1440-1827.2010.02587.x. [DOI] [PubMed] [Google Scholar]
- 28.Mesa H, Manivel JC, Larson WS, Dachel SK, Reinink AR, Jessurun J. Immunophenotypic comparison of neoplasms of the appendix, right colon, and left colon in search of a site-specific phenotypic signature. Int J Surg Pathol. 2020;28:20–30. doi: 10.1177/1066896919859096. [DOI] [PubMed] [Google Scholar]
- 29.Nishida T, Egashira Y, Akutagawa H, et al. Predictors of lymph node metastasis in T1 colorectal carcinoma: an immunophenotypic analysis of 265 patients. Dis Colon Rectum. 2014;57:905–915. doi: 10.1097/DCR.0000000000000168. [DOI] [PubMed] [Google Scholar]
- 30.Park SY, Lee HS, Choe G, Chung JH, Kim WH. Clinicopathological characteristics, microsatellite instability, and expression of mucin core proteins and p53 in colorectal mucinous adenocarcinomas in relation to location. Virchows Arch. 2006;449:40–47. doi: 10.1007/s00428-006-0212-7. [DOI] [PubMed] [Google Scholar]
- 31.Perez RO, Bresciani BH, Bresciani C, et al. Mucinous colorectal adenocarcinoma: influence of mucin expression (Muc1, 2 and 5) on clinico-pathological features and prognosis. Int J Colorectal Dis. 2008;23:757–765. doi: 10.1007/s00384-008-0486-0. [DOI] [PubMed] [Google Scholar]
- 32.Raghoebir L, Biermann K, Kempen MB, et al. Aberrant SOX2 expression in colorectal cancers does not correlate with mucinous differentiation and gastric mucin MUC5AC expression. Virchows Arch. 2014;465:395–400. doi: 10.1007/s00428-014-1638-y. [DOI] [PubMed] [Google Scholar]
- 33.Tsai JH, Lin YL, Cheng YC, et al. Aberrant expression of annexin A10 is closely related to gastric phenotype in serrated pathway to colorectal carcinoma. Mod Pathol. 2015;28:268–278. doi: 10.1038/modpathol.2014.96. [DOI] [PubMed] [Google Scholar]
- 34.Walsh MD, Clendenning M, Williamson E, et al. Expression of MUC2, MUC5AC, MUC5B, and MUC6 mucins in colorectal cancers and their association with the CpG island methylator phenotype. Mod Pathol. 2013;26:1642–1656. doi: 10.1038/modpathol.2013.101. [DOI] [PubMed] [Google Scholar]
- 35.Wang H, Jin S, Lu H, et al. Expression of survivin, MUC2 and MUC5 in colorectal cancer and their association with clinicopathological characteristics. Oncol Lett. 2017;14:1011–1016. doi: 10.3892/ol.2017.6218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Battaglin F, Naseem M, Lenz HJ, Salem ME. Microsatellite instability in colorectal cancer: overview of its clinical significance and novel perspectives. Clin Adv Hematol Oncol. 2018;16:735–745. [PMC free article] [PubMed] [Google Scholar]
- 37.Reynolds IS, Fichtner M, McNamara DA, Kay EW, Prehn JHM, Burke JP. Mucin glycoproteins block apoptosis; promote invasion, proliferation, and migration; and cause chemoresistance through diverse pathways in epithelial cancers. Cancer Metastasis Rev. 2019;38:237–257. doi: 10.1007/s10555-019-09781-w. [DOI] [PubMed] [Google Scholar]
- 38.Pothuraju R, Rachagani S, Krishn SR, et al. Molecular implications of MUC5AC-CD44 axis in colorectal cancer progression and chemoresistance. Mol Cancer. 2020;19:37. doi: 10.1186/s12943-020-01156-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu X, Long X, Luo X, Song Z, Li S, Wang H. Abrogation of MUC5AC expression contributes to the apoptosis and cell cycle arrest of colon cancer cells. Cancer Biother Radiopharm. 2016;31:261–267. doi: 10.1089/cbr.2016.2054. [DOI] [PubMed] [Google Scholar]
- 40.Kononen J, Bubendorf L, Kallioniemi A, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4:844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 41.Baran B, Mert Ozupek N, Yerli Tetik N, Acar E, Bekcioglu O, Baskin Y. Difference between left-sided and right-sided colorectal cancer: a focused review of literature. Gastroenterol Res. 2018;11:264–273. doi: 10.14740/gr1062w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marx A, Simon P, Simon R, et al. AMACR expression in colorectal cancer is associated with left-sided tumor localization. Virchows Arch. 2008;453:243–248. doi: 10.1007/s00428-008-0646-1. [DOI] [PubMed] [Google Scholar]
- 43.Missiaglia E, Jacobs B, D'Ario G, et al. Distal and proximal colon cancers differ in terms of molecular, pathological, and clinical features. Ann Oncol. 2014;25:1995–2001. doi: 10.1093/annonc/mdu275. [DOI] [PubMed] [Google Scholar]
- 44.Gelsomino F, Barbolini M, Spallanzani A, Pugliese G, Cascinu S. The evolving role of microsatellite instability in colorectal cancer: a review. Cancer Treat Rev. 2016;51:19–26. doi: 10.1016/j.ctrv.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 45.Renaud F, Vincent A, Mariette C, et al. MUC5AC hypomethylation is a predictor of microsatellite instability independently of clinical factors associated with colorectal cancer. Int J Cancer. 2015;136:2811–2821. doi: 10.1002/ijc.29342. [DOI] [PubMed] [Google Scholar]
- 46.Renaud F, Mariette C, Vincent A, et al. The serrated neoplasia pathway of colorectal tumors: identification of MUC5AC hypomethylation as an early marker of polyps with malignant potential. Int J Cancer. 2016;138:1472–1481. doi: 10.1002/ijc.29891. [DOI] [PubMed] [Google Scholar]
- 47.Beg MS, Azad NS, Patel SP, et al. A phase 1 dose-escalation study of NEO-102 in patients with refractory colon and pancreatic cancer. Cancer Chemother Pharmacol. 2016;78:577–584. doi: 10.1007/s00280-016-3108-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim RD, Azad NS, Morse MA, et al. Phase II study of Ensituximab, a novel chimeric monoclonal antibody, in adults with unresectable, metastatic colorectal cancer. Clin Cancer Res. 2020;26(14):3557–3564. doi: 10.1158/1078-0432.CCR-20-0426. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file1 Supplementary figure 1. MMR protein immunostaining in colorectal cancer. Supplementary table 1. Frequency of mismatch repair protein staining in colorectal cancer. Supplementary table 2. MUC5AC immunostaining and tumor location in previous studies. Supplementary table 3. MUC5AC immunostaining and mismatch repair deficiency/microsatellite instability in previous studies (DOCX 14617 KB)