Genome-wide association studies (GWAS) have identified associations between common genetic variants, single nucleotide polymorphisms (SNPs) and the risk of developing different cancers1–3. Proponents argue that polygenic risk score (PRS) testing, based on panels of risk SNPs, will revolutionize the prevention and early detection of cancer through individualised risk management strategies and streamlining of the current ‘one-size-fits all’ population screening programs4. Such a model is highly seductive for the rationalisation of healthcare provision. UK government enthusiasm for PRSs is well demonstrated within the recent Genome UK report and 2020 update to the Life Sciences Strategy5. Indeed, reflecting governmental endorsement of predictive genomics, the UK government’s Secretary of State for Health and Social Care, Matthew Hancock, rather questionably enthused that his recent PRS-derived lifetime prostate cancer risk estimate of 15% (compared to a prior of 13%) “may have saved his life”6,7. To establish the requisite governance and data infrastructure for population-level genomic profiling, national projects such as the 100,000 Genomes Project (>70,000 NHS patients) and the Accelerating Detection of Disease programme (up to 5 million volunteers) were initiated8,9. Following initial discontinuation by the U.S. Food and Drug Administration of the 23andMe PRS service10,11, there has been a resurgence within the direct-to-consumer genomics market of PRS predictions for many diseases. While the value of additional biomarkers to improve the targeting of measures for cancer prevention and early detection is indisputable, for PRS to be clinically useful, two assertions must be proven correct. The first assertion is that PRSs provides sufficient risk discrimination. The second is that this risk discrimination is meaningful in the context of absolute risk of that cancer and applicable in the context of respective tools available for prevention and early detection.
The risk discrimination for a given cancer afforded by PRS can be visualised most simply via PRS frequency distributions of those with the disease compared to those without the disease. (Fig. 1)12,13. Two commonly presented measures of discrimination derived from these distributions are: (i) comparison of the RR for those at the top and bottom tails of the PRS distribution (ii) comparison for specified PRS cut-offs of the proportion of affected individuals with ‘positive’ PRS (‘detection rate’) versus the proportion of unaffected individuals falling within the same PRS score range (the ‘false-positive rate’ (1–specificity)) (Supplementary Fig. 1). For the common cancers, PRS for prostate currently leads on discriminatory performance, with RR of 14.54 between the top 5% and bottom 5% of men (Fig. 1)14. This translates to a false-positive rate of 5% for a detection rate of 16%. Or, to achieve a detection rate of 50%, tolerance of a false-positive rate of 25%.
Fig. 1. Overlapping relative frequency distributions of polygenic risk score in prostate cancer cases and unaffected individuals.
The detection rate for a false-positive rate of 5% is 16%.
Quantitation of PRS performance tends to focus on the tails of the distribution, distracting from the fact that for 90% of individuals their PRS lies relatively close (<2 SD) to the mean. Up- or down-modification by these PRSs against baseline cancer risk results in minimal absolute difference in risk. Acknowledgement that the PRS for a chosen disease provides useful information for only a few percent of people is often countered by the argument that PRSs can be generated for dozens of cancer types, such that each individual is likely to be in an ‘extreme’ tail of PRS for at least one cancer type. However, in practice, clinical decision-making is driven by absolute risk, rather than relative risk per se. Hence, for all but the most common cancers, PRS reveals absolute increase in cancer risk against baseline that is miniscule even at the extreme upper tail of PRS. For example, for those in the respective top 5% of PRS, lifetime risk for breast cancer is elevated from 11.8 to 19.0% (1.6-fold), lifetime risk of prostate cancer is elevated from 12.7 to 22.2% (1.75-fold) and lifetime risk of colorectal cancer is elevated from 4.6 to 6.9% (1.5-fold). For less common cancers, the absolute risk increase is much lower: for women in the respective top 5% of ovarian cancer PRS lifetime risk is elevated 1.3-fold (from 1.6 to 2.1%)14,15.
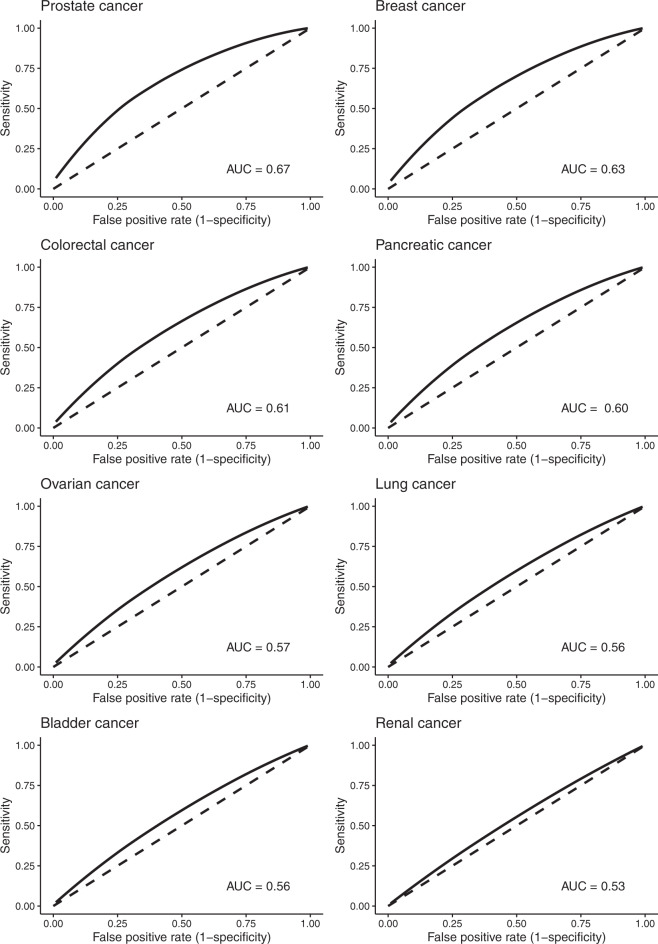
A presentation popular for capturing the discriminatory attributes of PRS is the Receiver Operating Characteristic (ROC) curve, with the probability that a randomly selected case having a higher PRS than a randomly selected control being quantified by the area under the ROC curve (AUC). An AUC of 0.5 reflects a testing tool with no discrimination. While the threshold for clinical discrimination clearly depends on the benefit-risk profile of the proposed intervention, broadly speaking AUC values of 0.7–0.8 are considered as acceptable and >0.8 as affording good discrimination16. As an example, digital mammography for breast cancer screening has an AUC of 0.7817. PRSs constructed from cancer SNP sets derived from GWAS yield only AUCs ranging from 0.53 (renal cancer) to 0.67 (prostate cancer) (Fig. 2).
Fig. 2. Receiver operator plots of polygenic risk scores for eight common cancers.
AUC area under the curve. The AUC provides an estimate of the probability a randomly selected subject with the condition has a test result indicating greater than that of a randomly chosen individual without the cancer. The solid line represents a receiver operator curve based on polygenic risk score from known risk SNPs based on reference13. An AUC of 0.5 (dashed line) indicates that the classifier does not provide any useful information in discriminating cases from controls.
The GWAS so far performed have only identified SNPs contributing 10–30% of common variant heritability for their respective cancers. It is widely asserted that discriminatory value of PRS will improve dramatically ‘in the future’, once additional rafts of new disease-associated SNPs are delivered. Imposing a threshold of 5 × 10−8 for declaring a SNP association guards against type 1 error but inevitably has limited the number of SNPs included in PRS testing panels. Methodologies such as LDpred apply a Bayesian genome-wide genetic risk prediction to incorporate SNP-correlations “tagged” by current arrays irrespective of association P-value18. However, to the extent that such methods have been explored, the discriminatory power for these ‘expanded’ PRSs is at best only very modestly improved19,20. The (likely) final generation of GWASs for common cancers such as breast and colorectal cancers is currently underway. Despite aggregating the’world-wide’ resource of available samples, power remains prohibitive and these experiments are unlikely to harvest more than 80% of the heritable risk for these cancers3. Even disregarding this and calculating PRS based on the full disease heritability, i.e. the hypothetical situation of identifying the full complement of risk SNPs, the AUCs for the common cancers remain disappointingly modest (0.64–0.73)3.
Another argument pertains to the boost in predictive value of PRS when applied in combination with non-genetic risk factors. Family history can be an important risk factor for cancer but is correlated with PRS21–23; some authors erroneously inflate their AUC by combining the two as if orthogonal24,25. For most cancer types, the AUC for non-genetic factors is modest26,27. When modelled, the well-established modifiable risk factors have additive effects with SNP associations and therefore only modestly improve the AUC27,28. Breast cancer has the best characterised set of non-genetic risk factors: the AUC for these risk factors is 0.637 while the AUC of the current PRS is 0.631; and in combination the AUC is 0.68329. Again, while aetiological epidemiological research continues to be a dynamic field, it would seem naïve to predict imminent discovery of robustly associated new non-genetic risk factors of sizeable effect.
There are many who acknowledge PRS to be a weak predictor of individual cancer risk but who suggest that it could still be useful to individualise population screening programmes, for example by excluding ‘low risk’ individuals from screening30,31. From modelled adaptation of the UK breast cancer screening programme such that screening of women age 35–79 would be offered on the basis of PRS rather than age alone, it has been predicted that screening could be reduced by 24% at the cost of reduction in screen-detectable cases of 14%32. Although this indicates potential for more efficient targeting of screening, there is concern that: (i) genomic profiling will do little to improve and could even reduce uptake of existing cancer screening programmes, which for breast cancer in the UK is currently only 69%33, and (ii) the incidence of breast cancers in those from whom breast screening has been ‘withheld’ (or in fact ‘withdrawn’) would be perceived by the public as too high. Another proposal is that PRSs could be combined as a Bayesian Prior to a population screening test, such that the more expensive and/or invasive confirmatory test is only triggered when a positive screening test (PSA, FIT) arises in individuals with high PRS. In practice, where the screening test itself has good performance characteristics, for example Faecal Immunochemical Test (FIT) for detection of colorectal cancer, the negative predictive value of PRSs will be insufficient to exclude disease in those with a positive screening test. Conversely, where the performance of the screening test is poorer (e.g. prostate specific antigen (PSA) for prostate cancer), the limited positive predictive value of the PRS adds little, resulting in a large group of PRS-false-positives receiving an invasive, confirmatory investigation. Hence, where screening is effective, inexpensive and safe, the limited additional boost in detection added by PRS stratification is arguably likely to be outweighed by the cost, complexity of delivery of population risk-profiling with the potential for reduced participation. Well-designed trials are therefore essential to avoid inadvertent disruption of existing screening programmes with documented evidence of benefit34. Where screening is not effective, inexpensive, or safe, the addition of a PRS is unlikely to make it so.
PRS testing has been suggested as a method of identifying individuals who may benefit from cancer chemopreventative agents35. While an attractive proposition, few agents are currently licenced for this purpose and clinical studies of new agents for chemoprevention are challenging. The risk-benefit profile of an agent dictates the level of disease risk at which administration is justified; thus the value of PRS stratification in determining administration would be very much contingent on the performance of PRS in discriminating those at sufficiently elevated risk. Administration of chemopreventative agents based on PRS-stratified groups has not yet been trialled for any agents and would require careful consideration36,37. While communicating DNA-based disease risk assessments may have a role in promoting risk-reducing behaviour38, evidence for this is currently lacking and there is a potential to introduce harm, particularly in the absence of counselling and clinical utility39. Furthermore, if PRS is to be used, it has to be universally applicable to all in the population regardless of ancestry to ensure equity in provision of healthcare resource. Presently the majority of PRSs are based on studies of European ancestry40 and their performance is poor in non-European populations41. This consideration is frequently offered as a minor technical ‘footnote’ to the value proposition of PRSs: given the limited predictive capabilities of existing PRS generated from decades of massive studies of available samples, it is unclear how this deficit will actually be addressed in the foreseeable future.
The clinical utility of identification of high-impact mutations in genes such as BRCA1 and MLH1 is not under dispute: such mutations provide effective risk discrimination and there are established clinical pathways for those in whom mutations are identified. However, the notion that PRSs will offer equivalent utility population-wide by providing informative risk stratification across multiple diseases is misleading. Raising unrealistic expectations and implementing programmes without careful evaluation risks compromising the application of PRSs for specific niches, and indeed, of genomic medicine as a whole.
Supplementary information
Acknowledgements
A.S., C.T. and R.S.H. researched, reviewed, drafted and edited the manuscript. A.S. generated the figures. R.S.H. acknowledges grant support from Cancer Research UK (C1298/A8362) and the Wellcome Trust (214388). C.T. acknowledges grant support from Cancer Research UK (C61296/A26688). A.S. is in receipt of a National Institute for Health Research (NIHR) Academic Clinical Lectureship and funding from the Royal Marsden Biomedical Research Centre. This is a summary of independent research supported by the NIHR Biomedical Research Centre at the Royal Marsden NHS Foundation Trust and the Institute of Cancer Research. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health. This research was funded in whole, or in part, by the Wellcome Trust [Grant number 214388]. For the purpose of open access, the author has applied a CC BY public copyright licence to any Author Accepted Manuscript version arising from this submission.
Data availability
No datasets were generated or analysed during the current study.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41698-021-00176-1.
References
- 1.Turnbull C, Sud A, Houlston RS. Cancer genetics, precision prevention and a call to action. Nat. Genet. 2018;50:1212–1218. doi: 10.1038/s41588-018-0202-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sud A, Kinnersley B, Houlston RS. Genome-wide association studies of cancer: current insights and future perspectives. Nat. Rev. Cancer. 2017;17:692–704. doi: 10.1038/nrc.2017.82. [DOI] [PubMed] [Google Scholar]
- 3.Zhang YD, et al. Assessment of polygenic architecture and risk prediction based on common variants across fourteen cancers. Nat. Commun. 2020;11:3353. doi: 10.1038/s41467-020-16483-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khera AV, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 2018;50:1219–1224. doi: 10.1038/s41588-018-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.UK Government. Genome UK: the future of healthcare. (Energy & Industrial Strategy Department for Business, 2020). https://www.gov.uk/government/publications/genome-uk-the-future-of-healthcare.
- 6.Nikolic, I. Health Secretary Matt Hancock discovers he is at a higher risk of developing prostate cancer after taking DNA test that could revolutionise NHS treatment. (Daily Mail, 2019). https://www.dailymail.co.uk/news/article-6828847/Health-Secretary-Matt-Hancock-discovers-higher-risk-developing-prostate-cancer.html.
- 7.Science Media Centre. Expert reaction to Matt Hancock’s speech on genetic testing.https://www.sciencemediacentre.org/expert-reaction-to-matt-hancocks-speech-on-genetic-testing/. (2019).
- 8.Turnbull C, et al. The 100 000 Genomes Project: bringing whole genome sequencing to the NHS. BMJ. 2018;361:k1687. doi: 10.1136/bmj.k1687. [DOI] [PubMed] [Google Scholar]
- 9.UK Government. UK to innovate new life-saving treatment and diagnosis technology. Accelerating Detection of Disease. https://www.gov.uk/government/news/uk-to-innovate-new-life-saving-treatment-and-diagnosistechnology#:~:text=The%20Accelerating%20Detection%20of%20Disease%20programme%20will%20put%20the%20UK,than%20treat%20it%20too%20late (2019).
- 10.U.S. Food & Drug Administration. FDA allows marketing of first direct-to-consumer tests that provide genetic risk information for certain conditions. https://www.fda.gov/news-events/press-announcements/fda-allows-marketing-first-direct-consumer-tests-provide-genetic-risk-information-certain-conditions (2018).
- 11.23andMe. 23andMe And The FDA. https://customercare.23andme.com/hc/en-us/articles/211831908-23andMe-and-the-FDA#:~:text=In%202013%2C%2023andMe%20received%20a,the%20agency's%20regulatory%20review%20process.&text=23andMe%20will%20continue%20to%20seek%20FDA%20authorization%20to%20offer%20new%20reports (2019).
- 12.Wald NJ, Old R. The illusion of polygenic disease risk prediction. Genet. Med. 2019;21:1705–1707. doi: 10.1038/s41436-018-0418-5. [DOI] [PubMed] [Google Scholar]
- 13.Wald NJ, Morris JK. Assessing risk factors as potential screening tests: a simple assessment tool. Arch. Intern. Med. 2011;171:286–291. doi: 10.1001/archinternmed.2010.378. [DOI] [PubMed] [Google Scholar]
- 14.Jia, G. et al. Evaluating the utility of polygenic risk scores in identifying high-risk individuals for eight common cancers. JNCI Cancer Spectrum. 10.1093/jncics/pkaa021 (2020). [DOI] [PMC free article] [PubMed]
- 15.Pal Choudhury P, et al. iCARE: An R package to build, validate and apply absolute risk models. PLoS ONE. 2020;15:e0228198. doi: 10.1371/journal.pone.0228198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mandrekar JN, et al. Receiver operating characteristic curve in diagnostic test assessment. J. Thorac. Oncol. 2010;5:1315–1316. doi: 10.1097/JTO.0b013e3181ec173d. [DOI] [PubMed] [Google Scholar]
- 17.Pisano ED, et al. Diagnostic performance of digital versus film mammography for breast-cancer screening. N. Engl. J. Med. 2005;353:1773–1783. doi: 10.1056/NEJMoa052911. [DOI] [PubMed] [Google Scholar]
- 18.Vilhjálmsson BJ, et al. Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am. J. Hum. Genet. 2015;97:576–592. doi: 10.1016/j.ajhg.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kulm, S., Mezey, J. & Elemento, O. Benchmarking the accuracy of polygenic risk scores and their generative methods. medRxiv. 10.1101/2020.04.06.20055574 (2020).
- 20.Thomas M, et al. Genome-wide modeling of polygenic risk score in colorectal cancer risk. Am. J. Hum. Genet. 2020;107:432–444. doi: 10.1016/j.ajhg.2020.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mavaddat N, et al. Prediction of breast cancer risk based on profiling with common genetic variants. J. Natl Cancer Inst. 2015;107:djv036. doi: 10.1093/jnci/djv036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yanes T, Young M-A, Meiser B, James PA. Clinical applications of polygenic breast cancer risk: a critical review and perspectives of an emerging field. Breast Cancer Res. 2020;22:21. doi: 10.1186/s13058-020-01260-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kachuri L, et al. Pan-cancer analysis demonstrates that integrating polygenic risk scores with modifiable risk factors improves risk prediction. Nat. Commun. 2020;11:6084. doi: 10.1038/s41467-020-19600-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shieh Y, et al. Breast cancer risk prediction using a clinical risk model and polygenic risk score. Breast Cancer Res. Treat. 2016;159:513–525. doi: 10.1007/s10549-016-3953-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conti DV, et al. Trans-ancestry genome-wide association meta-analysis of prostate cancer identifies new susceptibility loci and informs genetic risk prediction. Nat. Genet. 2021;53:65–75. doi: 10.1038/s41588-020-00748-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson IM, et al. Operating characteristics of prostate-specific antigen in men with an initial PSA level of 3.0 ng/mL or lower. JAMA. 2005;294:66–70. doi: 10.1001/jama.294.1.66. [DOI] [PubMed] [Google Scholar]
- 27.Frampton M, Houlston RS. Modeling the prevention of colorectal cancer from the combined impact of host and behavioral risk factors. Genet. Med. 2017;19:314–321. doi: 10.1038/gim.2016.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia-Closas, M., Gunsoy, N. B. & Chatterjee, N. Combined associations of genetic and environmental risk factors: implications for prevention of breast cancer. J. Natl Cancer Inst.10.1093/jnci/dju305 (2014). [DOI] [PMC free article] [PubMed]
- 29.Pal Choudhury P, et al. Comparative validation of breast cancer risk prediction models and projections for future risk stratification. J. Natl Cancer Inst. 2019;112:278–285. doi: 10.1093/jnci/djz113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Callender T, et al. Polygenic risk-tailored screening for prostate cancer: a benefit-harm and cost-effectiveness modelling study. PLoS Med. 2019;16:e1002998. doi: 10.1371/journal.pmed.1002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mavaddat N, et al. Polygenic risk scores for prediction of breast cancer and breast cancer subtypes. Am. J. Hum. Genet. 2019;104:21–34. doi: 10.1016/j.ajhg.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pashayan N, et al. Polygenic susceptibility to prostate and breast cancer: implications for personalised screening. Br. J. Cancer. 2011;104:1656–1663. doi: 10.1038/bjc.2011.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.NHS Digital. Breast Screening Programme. https://digital.nhs.uk/data-and-information/publications/statistical/breast-screening-programme/england---2019-20 (2021).
- 34.Eeles RA, ni Raghallaigh H. BARCODE 1: A pilot study investigating the use of genetic profiling to identify men in the general population with the highest risk of prostate cancer to invite for targeted screening. J. Clin. Oncol. 2020;38:1505–1505. doi: 10.1200/JCO.2020.38.15_suppl.1505. [DOI] [Google Scholar]
- 35.Kim JO, et al. Impact of a breast cancer (BC) polygenic risk score (PRS) on the decision to take preventive endocrine therapy (ET): The Genetic Risk Estimate (GENRE) trial. J. Clin. Oncol. 2019;37:1501–1501. doi: 10.1200/JCO.2019.37.15_suppl.1501. [DOI] [Google Scholar]
- 36.Smith SG, Sestak I, Howell A, Forbes J, Cuzick J. Participant-reported symptoms and their effect on long-term adherence in the International Breast Cancer Intervention Study I (IBIS I) J. Clin. Oncol. 2017;35:2666–2673. doi: 10.1200/JCO.2016.71.7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freedman AN, et al. Benefit/risk assessment for breast cancer chemoprevention with raloxifene or tamoxifen for women age 50 years or older. J. Clin. Oncol. 2011;29:2327–2333. doi: 10.1200/JCO.2010.33.0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hollands GJ, et al. The impact of communicating genetic risks of disease on risk-reducing health behaviour: systematic review with meta-analysis. BMJ. 2016;352:i1102. doi: 10.1136/bmj.i1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hudson K, Javitt G, Burke W, Byers P, Committee, A. S. I. ASHG statement on direct-to-consumer Genetic testing in the United States. Am. J. Hum. Genet. 2007;81:635–637. doi: 10.1086/521634. [DOI] [PubMed] [Google Scholar]
- 40.Park SL, Cheng I, Haiman CA. Genome-Wide Association Studies of cancer in diverse populations. Cancer Epidemiol. Biomark. Prev. 2018;27:405–417. doi: 10.1158/1055-9965.EPI-17-0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duncan L, et al. Analysis of polygenic risk score usage and performance in diverse human populations. Nat. Commun. 2019;10:3328. doi: 10.1038/s41467-019-11112-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.