Abstract
Introduction
Ontology-based annotation of evidence, using disease-specific ontologies, can accelerate analysis and interpretation of the knowledge domain of diseases. Although many domain-specific disease ontologies have been developed so far, in the area of cardiovascular diseases, there is a lack of ontological representation of the disease knowledge domain of stroke.
Methods
The stroke ontology (STO) was created on the basis of the ontology development life cycle and was built using Protégé ontology editor in the ontology web language format. The ontology was evaluated in terms of structural and functional features, expert evaluation, and competency questions.
Results
The stroke ontology covers a broad range of major biomedical and risk factor concepts. The majority of concepts are enriched by synonyms, definitions, and references. The ontology attempts to incorporate different users’ views on the stroke domain such as neuroscientists, molecular biologists, and clinicians. Evaluation of the ontology based on natural language processing showed a high precision (0.94), recall (0.80), and F-score (0.78) values, indicating that STO has an acceptable coverage of the stroke knowledge domain. Performance evaluation using competency questions designed by a clinician showed that the ontology can be used to answer expert questions in light of published evidence.
Conclusions
The stroke ontology is the first, multiple-view ontology in the domain of brain stroke that can be used as a tool for representation, formalization, and standardization of the heterogeneous data related to the stroke domain. Since this is a draft version of the ontology, the contribution of the stroke scientific community can help to improve the usability of the current version.
Keywords: Knowledge representation, Ontology, Semantic web, Stroke, Stroke ontology, Translational research
Key Summary Points
| Why carry out this study? |
| Interpretation of the knowledge domain of diseases needs a semantic tool to retrieve information from evidence. |
| There is a lack of ontological representation of the stroke knowledge domain. |
| In this study, we developed the stroke ontology (STO), to capture and organize the stroke knowledge domain in a standard way. |
| What was learned from the study? |
| The STO enables researchers of the stroke domain to capture the knowledge stored in the high-dimensional biomedical repositories and databases. |
Digital Features
This article is published with digital features, including a summary slide, to facilitate understanding of the article. To view digital features for this article go to https://doi.org/10.6084/m9.figshare.14316173.
Introduction
Stroke, a non-communicable disease with debilitating and chronic nature, accounts for 5.5 million (95% UI 5.3–5.7) deaths and 116.4 million (111.4–121.4) DALYs (disability adjusted life years) worldwide [1]. Notably, the majority (80%) of modifiable risk factors for stroke are preventable [2]. Causes and effects of stroke have been thoroughly investigated in the past decades and consequently the knowledge domain of stroke has grown dramatically in different dimensions both preclinically and clinically. For instance, quantitative analysis of stroke literature within the 12-year interval between 1996 and 2008 showed continued growth in the number of publications and stroke research productivity worldwide [3]. Since 2008, this trend has continued to grow further [4]. Despite this growing volume of knowledge and data, translation of laboratory and animal model findings into efficacious clinical interventions for patients with stroke has not seen major success since the introduction of thrombolytic therapy [5]. A common translational challenge is to manage and integrate heterogeneous, multidimensional data types across basic, preclinical, and clinical phases of research so that all the knowledge and data generated along the continuum of the translational research can be consolidated into actionable, predictive, patient-centric models [6]. A prerequisite for data and knowledge integration and modeling is their interoperability across different stages of research and scientific groups, which is facilitated by standard knowledge representation of ontologies.
In 1998, Studer et al. defined ontology as a formal, explicit specification of a shared conceptualization [7]. Ontologies are a set of formal concepts defining a specific domain of knowledge, which indicate properties of those concepts (entities) and relationships between them [8]. Several initiatives have already undertaken the effort to develop disease taxonomies and standard ontologies specific to brain and neurodegenerative diseases including Alzheimer’s disease ontology [9], Parkinson’s disease ontology [10], and multiple sclerosis ontology [11]. In the realm of stroke, multiple classification systems and ontologies such as Harvard Stroke Registry classification [12], Oxfordshire Community Stroke Project Subtype Classification [13], Trial of ORG 10172 in Acute Stroke Treatment Subtype Classification [14], Stroke Subtype Classification [15], the NEUROWEB Reference Ontology [16], and National Institutes of Health Stroke Scale Ontology (https://bioportal.bioontology.org/ontologies/NIHSS) capture the granularity of phenotype representation in the clinic. NeuMORE ontology covers the domain of the recovery after stroke [17] and DStrokeOnto represents the knowledge domain of stroke diagnosis and patient management [18]. However, almost all of these resources focus on the clinical end of the translational continuum and none of them is sufficient to be used for the representation of the entire translational continuum, including preclinical aspects of the stroke research.
To address this shortcoming, we have undertaken the effort to develop the stroke ontology (STO). STO aims to capture and organize the knowledge domain of stroke in a standard way with more views than clinical only. One view, which positions itself at the opposite side of the clinical end in the translation continuum, is the prevention view because the study of lifestyle and risk factors facilitates the design of effective prevention strategies and early-life interventions [19]. The ontology attempts to cover intermediate views between the two extreme views of the translation continuum including the knowledge of risk factors, prevention, disease etiology, pathophysiology, biomarkers, preclinical models, and intervention options. The ontology is then evaluated for its functional coverage and also for its power to answer an expert’s competency questions.
Methods
STO was constructed under the ontology development life cycle [20]. We applied the construction principles based on the set of activities defined in the ontology development life cycle to ensure formal ontology construction.
Specifications of Requirement
The purpose of STO was to collect, organize, and represent the stroke knowledge domain in the standardized and computer-readable format so that it can be used by different expert users in the field of stroke. A set of competency questions was used to define and evaluate the scope and domain coverage of the STO. These competency questions were defined by a clinical expert during the development of the ontology and were focused on the stroke risk factors and prevention, preclinical models, and clinical diagnosis. Out of those questions, we selected the most relevant three questions for evaluation of the STO. To select the questions, various features were considered, such as the type of question (the elements mentioned in the question should be present in the ontology and the question should be formulated correctly), modifiers in the text of the question, and domain-independent elements.
Knowledge Acquisition
A collection of STO-related terms and concepts was generated by scanning different knowledge sources. A list of sources was compiled through a recommendation of the stroke experts, including medical textbooks such as Bradley’s Neurology in Clinical Practice [21] and Textbook of Stroke Medicine [22], encyclopedias like Encyclopedia of the Neurological Sciences [23] and The Gale Encyclopedia of Neurological Disorders [24], scientific articles (original or review), informative online sources, e-books, and websites. Also, we used some biomedical top-level ontologies (including Medical Subject Headings (MESH) (http://bioportal.bioontology.org/ontologies/MESH), Systematized Nomenclature of Medicine Clinical Terms (SNOMEDCT) (http://bioportal.bioontology.org/ontologies/SNOMEDCT), Online Mendelian Inheritance in Man (OMIM) (http://bioportal.bioontology.org/ontologies/OMIM), and Pathway Ontology (PW) (http://bioportal.bioontology.org/ontologies/PW) to capture stroke-related terms and definitions.
Conceptualization
We used the combined top-down and bottom-up strategy to identify the concepts and relations in the STO. It means that we identified the core basic terms and then specified and generalized concepts as required [20]. We have reviewed the existing ontologies to reuse their relevant concepts to facilitate interoperability. The National Institutes of Health Stroke Scale Ontology (NIHSS) is a suitable ontology for reuse in STO, as it is a domain ontology intended to evaluate the neurologic outcome and degree of recovery for patients with stroke (http://bioportal.bioontology.org/ontologies/NIHSS). The Stroke Diagnostic Ontology (DStrokeOnto ontology) includes concepts related to stroke diagnosis and management of patients with stroke [18]. Stroke subtype concepts (hemorrhagic stroke and ischemic stroke) were reused from the neurological disease ontology (ND) which has been designed for annotation and analysis of huge data sets and patient health records in Alzheimer’s disease, multiple sclerosis, and stroke [25]. In addition to these ontologies, we used some top-level ontologies to define general concepts and terms used in STO, including MESH, SNOMEDCT, OMIM, and PW, as mentioned in the “Knowledge Acquisition” section.
Integration and Formalization
To construct the hierarchical structure, first, any available hierarchical organization (structure) of the concepts was extracted and reused along with the concepts themselves from existing ontology resources. Then, we got help from clinical experts to confirm its integration into the STO overall structure. Moreover, we used some object properties to define semantic relation types between concepts, such as “is a”, “part of”, “causes”, “has-participant”, “contained-in”, “associated-with”, and “describes”. The Protégé ontology editor (version 5.1.0 Desktop System; http://protege.stanford.edu) was used to build the STO in Ontology Web Language (OWL) format [26].
Terminology Analysis and Concept Enrichment
For terminology analysis and enrichment, we incorporated the STO ontology into text mining tools. First, the ontology OWL format was transformed into Extensible Markup Language (XML) format, then the concepts’ labels and corresponding synonyms were extracted, and eventually transformed into a dictionary file. This dictionary file was incorporated into the text processing module of the KNIME analytical platform [27]. KNIME is the open solution for data-driven analysis and provides many nodes of data visualization. In the next step, the super-class concepts were used as keywords to build a search strategy in PubMed. After that, a corpus of 500 PubMed abstracts with informative contents about stroke was created. A set of 100 abstracts of the corpus was selected randomly. To optimize the ontology for the sake of the concept enrichment, these abstracts were automatically annotated for true-positive concepts of STO using the KNIME software so that false-negative entities were manually identified. Finally, these entities were added to the STO ontology after reviewing by an expert in the field.
Evaluation of STO
STO was evaluated for its functional features using the Natural Language Processing (NLP) assessment method in KNIME. To evaluate the quality of STO in terms of the boundary of the knowledge domain that it captures, the precision, recall, and F-score values were calculated using the following formulas [28]:
where true positives (TP) are the number of STO entities that were highlighted by KNIME; false positives (FP) are the number of entities that were annotated by KNIME but were not an STO entity, and false negatives (FN) are the number of non-STO entities that were not found by KNIME.
This study was approved by the ethical committee and the institutional review boards of the Golestan University of Medical Sciences (code IR.GOUMS.REC.1395.205).
Results
STO Structure and Contents
STO is a semantic framework that aims to standardize and integrate the stroke knowledge domain and covers a wide range of key terms and concepts specific to stroke. Furthermore, it can support information retrieval and knowledge extraction from any source (i.e., electronic medical records and bibliographic databases) by integration with data- and text-mining tools. The STO encompasses eight root classes describing the stroke knowledge domain including “comorbidity”, “complication”, “diagnosis”, “model of stroke”, “risk factor”, “stroke prevention”, “stroke type”, and “treatment” (Fig. 1).
Fig. 1.
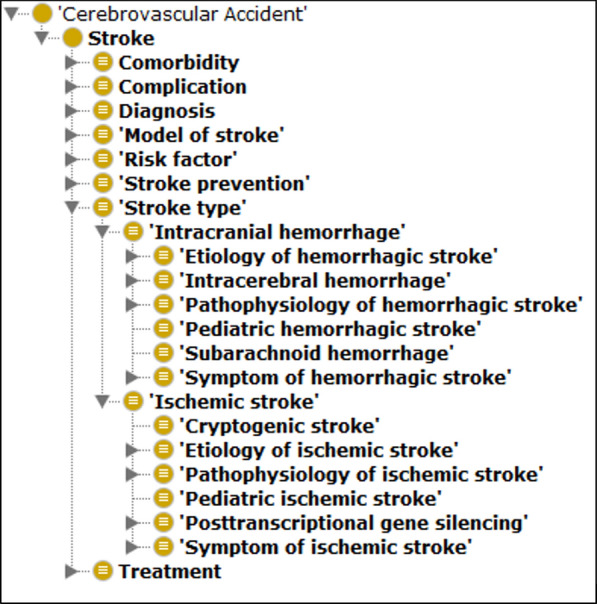
Root classes (super-classes) of STO as represented in the Protégé ontology editor software (a free, open-source ontology editor and framework for building intelligent systems, available from https://protege.stanford.edu/). Eight root classes describing the stroke knowledge domain suggested by experts. Also, subclasses of “Stroke type” are presented, which are categorized as “Intracranial hemorrhage” and “Ischemic stroke”. Musen and Protégé [26]
The super-class “comorbidity” contains “measures of comorbidity” and post- and pre-stroke diseases co-occurring with strokes such as “hypertension” and “diabetes”. The super-class “diagnosis” reflects those concepts related to OMICS-based, molecular and non-molecular biomarkers used for diagnosis of stroke including “differential diagnosis”, “evaluation of stroke”, “pathway”, and “brain anatomy”. Different concepts related to the animal and cell culture model of stroke were classified under the subclasses “model of stroke”. The super-class “risk factor” has been divided into two subclasses including modifiable and non-modifiable risk factors covering a broad range of conditions and attributes that may increase the risk of stroke. The super-class “stroke type” describes two common types of stroke (ischemic and hemorrhagic) and covers concepts related to etiology, pathophysiology, and symptoms of each subtype. The other three super-classes, namely “treatment”, “complication”, and “stroke prevention”, represent three separate aspects of the stroke knowledge domain.
In STO, each concept has been annotated by scientific definition from scientific publications, valid references, and synonyms. Some definitions have been reused from top-level ontologies such as SNOMED CT and MeSH with their database cross-reference as indicated by the “hasDBXref” annotation property. It should be noted that the field of stroke research is very dynamic and the current structure of STO is subject to change; thus, experts are invited to revise and update the draft ontology presented here.
STO Evaluation
Structural Evaluation
STO was evaluated for its structural properties including the number of classes, number of synonyms, and depth of the ontology, the last of these being indicative of the level of concept specificity in the stroke knowledge domain. Analysis of structural properties of STO was done using Protégé software. Values of these structural properties are summarized in Table 1.
Table 1.
Measures of the structural dimension of STO
| No. of roots | No. of concepts | No. of properties | No. of synonyms | Average no. of synonyms per concept | No. of leaves | Depth of ontology |
|---|---|---|---|---|---|---|
| 8 | 1712 | 29 | 4121 | 2.4 | 1263 | 14 |
Functional Evaluation
The functional dimension of the ontology was evaluated using the NLP approach, as explained in the “Methods”. A total of 538 STO entities (TP) were highlighted by KNIME, 34 entities (FP) were annotated by KNIME but were not an STO entity, and 133 entities (FN) were non-STO entities and were not found by KNIME. A high precision (0.94), recall (0.80), and an F-score of 0.78 indicate that STO has an acceptable coverage of the stroke knowledge domain (Table 2).
Table 2.
Measures of the functional dimension of STO using NLP-based evaluation approach
| Precision | Recall | F-score | |
|---|---|---|---|
| Independent test corpus of 100 PubMed abstracts | 0.94 | 0.80 | 0.78 |
Expert Evaluation
The ontology was reviewed and curated manually by a clinical neurologist against its relevancy of structure and contents. To evaluate the performance of the STO, the expert was asked to pose complex questions known as competency questions (CQs). We defined a search strategy for each of three selected CQs and evaluated the performance of the STO by screening abstracts that contained answers for these questions. Table 3 provides an overview of the number of relevant documents that were manually verified to contain a hypothetical answer to the corresponding question (accessed on 24 January 2017):
- CQ 1
- Return references that demonstrate the link between dehydration and cerebral venous sinus thrombosis (CVST) in stroke.((((((((stroke [STO Terms]) OR intracranial hemorrhage [STO Terms]) OR intracerebral hemorrhage [STO Terms]) OR hemorrhagic stroke [STO Terms]) OR ischemic stroke [STO Terms]))) AND dehydration [STO Terms]) AND ((((((((((((cerebral venous thrombosis [STO Terms]) OR cerebral venous sinus thrombosis [STO Terms]) OR deep venous thrombosis [STO Terms]) OR intracranial sinus thrombosis [STO Terms]) OR cranial sinus thrombosis [STO Terms]) OR sinus thrombosis [STO Terms]) OR cavernous sinus thrombosis [STO Terms]) OR venous thrombosis [STO Terms]) OR cerebral vein thrombosis [STO Terms])))
- CQ 2
- Return references associated with age and gender in intracerebral hemorrhage (ICH).((intracerebral hemorrhage [STO Terms]) AND age factor [STO Terms]) AND gender [STO Terms]
- CQ 3
- Return references that indicate the relation between stroke and cerebral vasculitis.(Cerebral vasculitis [STO Terms]) AND stroke [STO Terms]
Table 3.
Results of competency questions using the STO annotations within KNIME text processing platform
| Competency question | Number of relevant documents | Evidences (PubMed ID) | Answers |
|---|---|---|---|
| CQ 1 | 5 | 11510928, 18629575, 15100423, 18184942, 25900411 | Dehydration increases the risk of cerebral venous sinus thrombosis and causes stroke |
| CQ 2 | 12 | 26793409, 26088409, 25544173, 24483215, 24312335, 22012694, 20948201, 19659816, 15739042, 11728145, 10726330, 2063776 | In ICH case, men are younger age than women; age less than 60 and male gender has better survival of surgery; hypertension is a common risk factor of young patients; younger and female patients needed more examination; female sex at age more than 60 has a worst outcome |
| CQ 3 | 81 | 27113444, 26778046, 25778384, 25661835, 25316727, 25294561, 25095904, 24854370, 24508360, 24407026, 23710607, 23608691, 23329377, 23243263, 22980610, 22978371, 22735255, 22627088, 22418754, 21956650, 21940968, 21863270, 21773670, 21325772, 21122000, 20959356, 20884244, 20822714, 20661069, 20434741, 20425011, 20400168, 20370600, 20123232, 19646345, 19838657, 19593228, 19592058, 19235861, 19129348, 19011505, 18810458, 18334077, 18285353, 18021377, 17966442, 17630482, 17443285, 17352352, 17015122, 17009265, 16970859, 16428221, 16276375, 16113702, 15675128, 15026881, 15003300, 14616305, 12908757, 12401459, 12064698, 12040987, 11890855, 11784360, 11296969, 11196527, 11107576, 10948767, 10948755, 10909156, 8183760, 2543474, 10768635, 9742880, 8282839, 8478494, 2361038, 2792145, 3757384, 26773119 |
Vasculitis causes stroke, minocycline, methylphenidate, hepatitis C virus, neurosarcoidosis, West Nile virus, Cryptococcus, neuroborreliosis, Toxocara canis, Varicella-zoster virus, familial Mediterranean fever, Lyme disease, Behçet’s disease, elevation of circulating endothelial cells, HIV vasculopathy, and neuropsychiatric systemic lupus erythematosus (SLE) may induce, be complicated by, or associated with vasculitis Also, vasculitis associated with headache, schistosomiasis, and blindness CADASIL or Fabry’s disease can mimic clinical features of cerebral vasculitis |
Visualization of STO-Annotated Text
To visualize the named entities embedded in the ontology, a text file (i.e., dictionary) containing STO concepts and synonyms was integrated into the KNIME analytics platform, and PubMed abstracts were marked up with STO entities (Fig. 2). This facilitates rapid identification of stroke-related concepts within the text and assists the curator to spot relevant answers to competency questions.
Fig. 2.
The screenshot of the PubMed abstract visualization with STO entities using KNIME analytical platform (a free, open-
source software for creating data science, available from https://www.knime.com/downloads; KNIME automatically grabs free PubMed abstracts using the Document Grabber node). Red entities are STO terms. Zhu et al. [36]
Application Scenario
Application of STO to Hospital Electronic Medical Records
Electronic medical records (EMRs) capture and integrate patient’s health data in two forms, namely structured (e.g., diagnosis, laboratory tests, and drugs name) and unstructured (e.g., physician notes and any plain text in natural language) [29] Since EMRs are the primary source of phenotype information, they can be mined to find disease phenotype–genotype relations [30], disease comorbidity relations [9], or adverse drug events relations [31]. Accordingly, to demonstrate the utility of STO in capturing the stroke knowledge beyond publications, we analyzed risk factors for incidence of stroke among the patient population in Sayad Shirazi hospital, Gorgan, Northeastern Iran. We first retrieved 415 stroke EMRs from the hospital information system database according to International Classification of Diseases 10 revision (ICD10) codes (I60, I61, I62, I63, and I64). Afterward, these records were subjected to concept annotation by STO. Table 4 summarizes these annotations and their distribution based on the stroke subtype.
Table 4.
Risk factors of stroke in medical records and based on STO concept in risk factor class
| Hemorrhagic stroke (N, %) | Ischemic stroke (N, %) | Unspecified stroke (N, %) | |||
|---|---|---|---|---|---|
| Subarachnoid hemorrhage | Intracerebral hemorrhage | Total | |||
| Hypertension | 1 (7.7) | 76 (31.8) | 77 (30.6) | 174 (26.3) | 29 (24.6) |
| Diabetes mellitus | 1 (7.7) | 26 (10.9) | 27 (10.7) | 108 (16.3) | 26 (22) |
| Heart failure | 0 | 20 (8.4) | 20 (7.9) | 57 (8.6) | 12 (10.2) |
| Dyslipidemia | 1 (7.7) | 21 (8.8) | 22 (8.7) | 102 (15.4) | 18 (15) |
| Transient ischemic attack | 0 | 4 (1.7) | 4 (1.6) | 6 (0.9) | 0 |
| Anemia | 1 (7.7) | 0 | 1 (0.4) | 3 (0.5) | 1 (0.8) |
| Chronic kidney insufficiency | 0 | 3 (1.3) | 3 (1.2) | 5 (0.8) | 3 (2.5) |
| Prior stroke | 2 (15.4) | 30 (12.6) | 32 (12.7) | 81 (12.2) | 11 (9.3) |
| Vascular parkinsonism | 0 | 1 (0.4) | 1 (0.4) | 1 (0.2) | 0 |
| History of surgery | 1 (7.7) | 3 (1.3) | 4 (1.6) | 11 (1.7) | 1 (0.8) |
| Seizure | 1 (7.7) | 0 | 1 (0.4) | 3 (0.5) | 1 (0.8) |
| History of trauma | 2 (15.4) | 1 (0.4) | 3 (1.2) | 1 (0.2) | 0 |
| Smoking | 1 (7.7) | 11 (4.6) | 12 (4.8) | 18 (2.7) | 1 (0.8) |
| Total | 11 (100) | 196 (100) | 207 (100) | 570 (100) | 103 (100) |
As evident in Table 4, the most prevalent risk factors in the population of patients with hemorrhagic stroke are hypertension, prior stroke, diabetes, dyslipidemia, heart failure, and smoking, respectively, whereas, in the population of patients with ischemic stroke, the ranking favors hypertension, diabetes, dyslipidemia, prior stroke, heart failure, and smoking, respectively. The observed distribution indicates that a high number of patients with stroke belong to the ischemic category with a marked incidence of diabetes and dyslipidemia, which probably highlights the importance of diet patterns in the prevention of ischemic stroke in this particular patient population.
The STO is publicly and freely available to the research community for browsing and commenting at https://bioportal.bioontology.org/ontologies/STO. The STO will be maintained by its developers, and updates will be released after the incorporation of feedback from the scientific community.
Discussion
The importance of disease-specific ontologies for standardization, integration, annotation, and representation of the specific knowledge domain has been reported in the literature [32, 33]. There are already some ontologies related to the stroke knowledge domain, such as NIHSS ontology, DstrokeOntology [18], NEUROWEB reference ontology [16], and ND [25], but to our knowledge, none offers good coverage of the stroke knowledge domain. In contrast to these ontologies, STO contains major aspects of the stroke knowledge domain and provides hierarchical classes in the structured and formalized form with more than 4600 concepts and their synonyms. An advantage of STO over existing ontologies is the inclusion of several views onto the stroke knowledge domain from the standpoints of molecular biologists, translational scientists, and clinicians. These views have been reflected in the number of ontology roots and the depth of specificity in each root. The aim of including these various expert views was to make STO a preferred reference for stroke translational research as STO covers a wide spectrum of stroke research activities and features, from basic molecular research to clinical features and measurements.
However, the structural complexity of an ontology does not reflect its functional value. An accepted approach to evaluating the functional aspect of an ontology and its coverage is to apply the ontology terms to the scientific body of evidence in the literature [34]. The high values that we obtained for the metrics of the functional evaluation show that STO properly covers the major part of the stroke knowledge domain. Since the domain of stroke research is subject to active research and development, STO is not complete and thus requires the contribution of the scientific community to remain up-to-date.
Although functional evaluation can assess quality attributes of the ontology based on metrics, some quality attributes such as clarity and completeness can be difficult to evaluate. Thus, one or more experts are asked to pose so-called competency questions to the ontology [35]. Our expert evaluation of STO with three selected competency questions aiming at totally different areas of the ontology showed that STO was satisfactorily able to answer those competency questions by returning publications containing information relevant to questions. Therefore, if integrated into text-mining tools, STO can be used for faceted search within stroke literature to find targeted answers to expert questions. We have highlighted this possibility by annotating concepts of STO in both paper abstracts and electronic medical records. Annotation of hospital records with STO risk factor concepts and their analysis provided a picture of stroke risk factor prevalence among the patient population under study, which otherwise could remain hidden within hospital records. Application of such an approach can prove valuable when, for example, the pattern of extracted information from medical records is going to be used for defining prevention strategies by healthcare decision-makers.
Knowledge extraction of heterogeneous scientific resources with automated knowledge extraction tools was the major limitation of this work. Although using a text-mining tool facilitated the ontology-based information retrieval, the Portable Document Format (PDF) file format of many resources reduced the information retrieval process. We solved this problem just by spending a lot of time on knowledge extraction.
Conclusions
Despite the massive growth of the stroke knowledge domain during past decades, representing this knowledge domain in a standard manner with acceptable coverage and depth of information has not been realized. The STO was built to address this challenge as a semantic reference for representation, formalization, and standardization of heterogeneous stroke data. It enables researchers to capture the knowledge stored in the high-dimensional biomedical repositories and databases, and data stored in electronic clinical data repositories such as EMRs. This is the first version of STO which is freely available and needs the contribution of the stroke research community for enrichment.
Acknowledgements
This work was conducted using the Protégé resource, which is supported by grant GM10331601 from the National Institute of General Medical Sciences of the United States National Institutes of Health.
Funding
No funding or sponsorship was received for this study or publication of this article.
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Author’ Contributions
All authors have read the manuscript and approved it for publication. MHK drafted the article and EY revised it critically for important intellectual content. EY designed the study methodology and contributed to the writing of the manuscript. MHK created the STO and LS, SRK, HG, and EY contributed to its development and evaluation.
Disclosures
Mahdi Habibi-koolaee, Leila Shahmoradi, Sharareh R. Niakan Kalhori, Hossein Ghannadan, and Erfan Younesi declare that they have nothing to disclose regarding the content of this article.
Compliance with Ethics Guidelines
This study was approved by the ethical committee and the institutional review boards of the Golestan University of Medical Sciences (code IR.GOUMS.REC.1395.205).
Data Availability
The datasets generated during and/or analyzed during the current study are available in the BioPortal repository, https://bioportal.bioontology.org/ontologies/STO.
References
- 1.Johnson CO, Nguyen M, Roth GA, et al. Global, regional, and national burden of stroke, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2019;18(5):439–458. doi: 10.1016/S1474-4422(19)30034-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sherzai A, Heim LT, Boothby C, Sherzai AD. Stroke, food groups, and dietary patterns: a systematic review. Nutr Rev. 2012;70(8):423–435. doi: 10.1111/j.1753-4887.2012.00490.x. [DOI] [PubMed] [Google Scholar]
- 3.Chow D, Hauptman J, Wong T, et al. Changes in stroke research productivity: a global perspective. Surg Neurol Int. 2012;3:27. doi: 10.4103/2152-7806.92941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lau G, Kamalski J. Brain research: mining emerging trends and top research concepts. Res Trends. 2014;(39). https://www.researchtrends.com/issue-39-december-2014/brain-research/.
- 5.Boltze J, Ayata C. Challenges and controversies in translational stroke research—an introduction. Transl Stroke Res. 2016;7(5):355–357. doi: 10.1007/s12975-016-0492-4. [DOI] [PubMed] [Google Scholar]
- 6.Younesi E, Hofmann-Apitius M. From integrative disease modeling to predictive, preventive, personalized and participatory (P4) medicine. EPMA J. 2013;4(1):23. doi: 10.1186/1878-5085-4-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Studer R, Benjamins VR, Fensel D. Knowledge engineering: principles and methods. Data Knowl Eng. 1998;25(1):161–197. doi: 10.1016/S0169-023X(97)00056-6. [DOI] [Google Scholar]
- 8.Currás E. 1-From classifications to ontologies. ontologies, taxonomies and thesauri in systems science and systematics. Cambridge: Chandos; 2010. pp. 1–34. [Google Scholar]
- 9.Malhotra A, Younesi E, Gündel M, Müller B, Heneka MT, Hofmann-Apitius M. ADO: a disease ontology representing the domain knowledge specific to Alzheimer's disease. Alzheimers Dement. 2014;10(2):238–246. doi: 10.1016/j.jalz.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 10.Younesi E, Malhotra A, Gündel M, et al. PDON: Parkinson’s disease ontology for representation and modeling of the Parkinson’s disease knowledge domain. Theor Biol Med Model. 2015;12(1):20. doi: 10.1186/s12976-015-0017-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malhotra A, Gündel M, Rajput AM, et al. Knowledge retrieval from pubmed abstracts and electronic medical records with the multiple sclerosis ontology. PLoS ONE. 2015;10(2):e0116718. doi: 10.1371/journal.pone.0116718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mohr J, Caplan LR, Melski JW, et al. The Harvard Cooperative Stroke Registry a prospective registry. Neurology. 1978;28(8):754. doi: 10.1212/WNL.28.8.754. [DOI] [PubMed] [Google Scholar]
- 13.Bamford J, Sandercock P, Dennis M, Warlow C, Burn J. Classification and natural history of clinically identifiable subtypes of cerebral infarction. Lancet. 1991;337(8756):1521–1526. doi: 10.1016/0140-6736(91)93206-O. [DOI] [PubMed] [Google Scholar]
- 14.Adams HP, Bendixen BH, Kappelle LJ, et al. Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial TOAST. Trial of Org 10172 in Acute Stroke Treatment. Stroke. 1993;24(1):35–41. doi: 10.1161/01.STR.24.1.35. [DOI] [PubMed] [Google Scholar]
- 15.Amarenco P, Bogousslavsky J, Caplan L, Donnan G, Hennerici M. Classification of stroke subtypes. Cerebrovasc Dis. 2009;27(5):493–501. doi: 10.1159/000210432. [DOI] [PubMed] [Google Scholar]
- 16.Colombo G, Merico D, Boncoraglio G, et al. An ontological modeling approach to cerebrovascular disease studies: the NEUROWEB case. J Biomed Inform. 2010;43(4):469–484. doi: 10.1016/j.jbi.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 17.Townsend C, Huang J, Dou D, et al., editors. NeuMORE: Ontology in stroke recovery. Bioinformatics and Biomedicine Workshops (BIBMW), 2010 IEEE International Conference on; 2010: IEEE.
- 18.Podsiadly-Marczykowska T, Ciszek B, Przelaskowski A. Development of diagnostic stroke ontology—preliminary results. In: Piętka E, Kawa J, Wieclawek W, editors. Information technologies in biomedicine. Cham: Springer; 2014. pp. 261–272. [Google Scholar]
- 19.Feigin VL, Norrving B, George MG, Foltz JL, Roth GA, Mensah GA. Prevention of stroke: a strategic global imperative. Nat Rev Neurol. 2016;12(9):501–512. doi: 10.1038/nrneurol.2016.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gómez-Pérez A, Fernández-López M, Corcho O. Ontological engineering: with examples from the areas of knowledge management, e-commerce and the Semantic Web/Asunción Gómez-Pérez, Mariano Fernández-López, and Oscar Corcho. Advanced information and knowledge processing. New York: Springer; 2004. [Google Scholar]
- 21.Daroff RB, Bradley WG. Bradley’s neurology in clinical practice. 6. Philadelphia: Elsevier/Saunders; 2012. [Google Scholar]
- 22.Brainin M, Heiss WD, Heiss S. Textbook of stroke medicine. New York: Cambridge University Press; 2010. [Google Scholar]
- 23.Aminoff MJ, Daroff RB. Encyclopedia of the neurological sciences. 1. Amsterdam: Academic; 2003. [Google Scholar]
- 24.Chamberlin SL, Narins B, Gale Group . The Gale encyclopedia of neurological disorders. Detroit: Thomson Gale; 2005. [Google Scholar]
- 25.Jensen M, Cox AP, Chaudhry N, et al. The neurological disease ontology. J Biomed Semant. 2013;4:42. doi: 10.1186/2041-1480-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Musen MA, The Protégé Team The Protégé project: a look back and a look forward. AI Matters. 2015;1(4):4–12. doi: 10.1145/2757001.2757003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berthold M, Cebron N, Dill F, et al. KNIME: The Konstanz Information Miner In: studies in classification, data analysis, and knowledge organization (GfKL 2007) Springer; 2007. pp. 319–326. [Google Scholar]
- 28.Morgan AA, Lu Z, Wang X, et al. Overview of BioCreative II gene normalization. Genome Biol. 2008;9(2):S3. doi: 10.1186/gb-2008-9-s2-s3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson SB, Bakken S, Dine D, et al. An electronic health record based on structured narrative. J Am Med Inform Assoc. 2008;15(1):54–64. doi: 10.1197/jamia.M2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jensen PB, Jensen LJ, Brunak S. Mining electronic health records: towards better research applications and clinical care. Nat Rev Genet. 2012;13(6):395–405. doi: 10.1038/nrg3208. [DOI] [PubMed] [Google Scholar]
- 31.Henriksson A, Kvist M, Dalianis H, Duneld M. Identifying adverse drug event information in clinical notes with distributional semantic representations of context. J Biomed Inform. 2015;57:333–349. doi: 10.1016/j.jbi.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 32.Katayama T, Wilkinson MD, Aoki-Kinoshita KF, et al. BioHackathon series in 2011 and 2012: penetration of ontology and linked data in life science domains. J Biomed Semant. 2014;5:5. doi: 10.1186/2041-1480-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bodenreider O. Biomedical ontologies in action: role in knowledge management, data integration and decision support. Yearbook Med Inform. 2008;17:67–79. doi: 10.1055/s-0038-1638585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gómez-Pérez A. Evaluation of ontologies. Int J Intell Syst. 2001;16(3):391–409. doi: 10.1002/1098-111X(200103)16:3<391::AID-INT1014>3.0.CO;2-2. [DOI] [Google Scholar]
- 35.Obrst L, Ceusters W, Mani I, Ray S, Smith B. The evaluation of ontologies. Semant Web. 2007 doi: 10.1007/978-0-387-48438-9_8. [DOI] [Google Scholar]
- 36.Zhu X, Cao Y, Wei L, et al. von Willebrand factor contributes to poor outcome in a mouse model of intracerebral haemorrhage. Sci Rep. 2016;6:35901. doi: 10.1038/srep35901. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available in the BioPortal repository, https://bioportal.bioontology.org/ontologies/STO.