Abstract
Deregulated immune response and raised inflammation are the cardinal laboratory features in COVID-19 infection reflecting severity of condition. Detection of the markers will help in early diagnosis with timely therapeutic implementation and effective outcome. Observational studies have suggested alteration in these parameters with severity of the condition. This systematic review and meta-analysis was conducted to assess the relevance of the fact. Observational studies from databases were scrutinised and 3669 articles were identified. Further screening, based on the inclusion criteria a total of 19 articles with 3115 participants, were reviewed for meta-analysis using random effects model. Any data in median and interquartile range were converted to mean ± SD. There was a significant rise in total leukocyte count, C-reactive protein, ferritin, IL-6, IL-10, procalcitonin in severe cases but absolute lymphocyte count, CD4+ and CD8+ registered a fall in severe cases in comparison to non-severe group. Immune and inflammatory markers are significantly altered and related to severity of manifestation in COVID-19 infection.
Keywords: Cytokine storm, immunological, immune response, risk factor, SARS CoV2
Introduction
With the advent of a pandemic with COVID-19, that has spread to 213 countries, a lot of varied characteristics of the disease has evolved, both in clinical and laboratory. The most common manifestations are typical symptoms of viral pneumonia, such as fever, fatigue, dry cough, anosmia, and headache, that may evolve into respiratory failure.[1] The pathogen, however, displays a wide range of severity causing difficulty in determining infection outcome. Inflammatory response along with immune deregulation plays a critical role in this infection as evidenced by the development of cytokine storm in severe acute respiratory syndrome corona virus-2 (SARS-CoV-2) and the Middle East respiratory syndrome (MERS-CoV).[2,3]
As of 20 October 2020, India has crossed the 7.6 million mark for COVID-19 cases, with a case fatality rate of 1.9%.[4] According to the COVID-19 Situation report by WHO, 80% of infections are mild or asymptomatic and 15% are severe infection requiring oxygen and nearly 5% are critical infections, requiring ventilation.[5] The chance of transmission being more during pre-symptomatic period. Diagnosis of these cases at early stage with biomarkers may help in timely detection, implementation of therapy as delay may lead to its progression to severe stage of the disease. In India because of COVID-19 care centres at all district hospitals and sub-divisional hospitals, this information will bring awareness among health care workers.
Deregulated host immune response is the hallmark of COVID-19 infection, that may lead to acute respiratory distress syndrome (ARDS). Autopsy analyses as well as critically ill COVID-19 cases have revealed involvement of both cellular and humoral host immune response pathway resulting in fatal outcome of the patients. Pathogenic role of inflammation with release of pro-inflammatory cytokines like IL-6, procalcitonin, ferritin, C-reactive protein, etc., cause lung injury along with other manifestation of COVID-19 infection. Resultant systemic inflammation also affects circulating blood cells like neutrophils, lymphocytes along with neutrophil to lymphocyte ratio.
COVID-19 infection although associated with various laboratory changes the role of immunologic and inflammatory indicators have not been comprehensively studied. The knowledge regarding the changes in cytokines and other parameters of inflammation and immune deregulation reported in various population studies can help in identifying key molecular predictors that can be further explored for a deeper understanding of the molecular pathogenesis of COVID-19.
Aims and Objective
Therefore, the objective of this study is to summarize the alteration in the immunological and inflammatory markers of COVID-19 cases reported in currently available observational studies and their associations with the severity of disease process.
Methods
Eligibility criteria
Observational studies that reported laboratory characteristics and risk factors were included for quantitative synthesis, i.e., meta-analysis. Articles that were included in the study with the following inclusion criteria;
Population: All individuals ≥19 years of age with confirmed COVID-19 infection by RT-PCR
-
Exposure: Varied severity of COVID-19 infection
Severe COVID-19 was defined as patients who had any of the following features at the time of, or after, admission: (1) respiratory rate ≥30 breaths per min, (2) xygen saturation ≤93% (at rest), (3) ratio of partial pressure of arterial oxygen to fractional concentration of oxygen inspired air (PaO2 to fiO2 ratio) ≤300 mmHg, or (4) specific complications, such as septic shock, respiratory failure, and or multiple organ dysfunction[6,7]
Study design: Case series with more than 10 patients and cross sectional studies reporting laboratory parameters of patients based on level of severity of disease presentation
Outcome: Evaluation of inflammatory and immunological parameters [total leukocyte count (TLC), absolute lymphocyte count (ALC), erythrocyte sedimentation rate (ESR), C-reactive protein (CRP), ferritin, IL-6, IL-10, CD4+, CD8+ and procalcitonin (PCT)] in patients with clinical manifestation of COVID-19 clinical infection.
Studies with the following conditions were excluded;
Opinion pieces, systematic reviews, review articles, clinical trials, consensus documents and clinical guidelines, original articles with samples below 10 or case reports and non-English articles
Studies relating to other corona serotypes of severe acute respiratory syndrome and other related illness like Middle East Respiratory syndrome (MERS) were excluded from study
Studies involving pregnant women
Studies where data could not be extracted.
Search strategy and study selection criteria
We searched Pubmed/Medline and Google scholar databases for studies published between 24th January 2020 to 8th May 2020 using the following search terms in all possible combinations: “Corona Virus Disease-2019 “[Mesh] OR “2019 novel coronavirus”[Mesh] OR” SARS-CoV-2 “[Mesh] OR “COVID-19 “[Mesh] OR“2019-nCoV “[Mesh] AND” inflammatory markers” AND “immune markers” OR “Total leukocyte count” OR “Erythrocyte Sedimentation rate” OR “C-Reactive Protein” OR “Absolute lymphocyte count” OR “Ferritin” OR “CD4” OR “CD8” OR “IL-6” OR “IL-10” OR “TNF-α”. The search was limited to human subjects and to English language only. Two authors (RN and SP) independently screened the results from the initial search by titles and abstracts for relevance and the full texts were reviewed for the eligibility criteria. Third (PKT) and fourth (MM) researcher checked the article list and data extraction with MM ensuring no duplication of articles and resolved any dispute in the study inclusion. After detailed evaluation, a total of twenty three articles of which nineteen were used in meta-analysis and four for qualitative studies.[8,9,10,11]
The protocol follows the recommendations established by Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement.
Data extraction
Data extraction was done independently by two authors (SP and RN) using standardized format comprising of author, year, type of study, total number of study subjects, age, sex, laboratory variables in the severe and non-severe groups. Some of the included studies classified the patients based on the severity into mild, moderate, severe and critically ill. In such studies-mild and moderate patients were grouped as non-severe whereas severe and critically ill patients were considered in the severe group. Levels of systemic inflammation and immunological markers like TLC, absolute lymphocyte count, ESR, CRP, ferritin, procalcitonin, IL-6, IL-10, TNF-α, CD4+ and CD8+ were also retrieved from the articles.
Quality assessment
Quality of the study was assessed by using the critical Appraisal tool for Cross sectional Studies (AXIS). Quality assessment was based on patient selection, reporting of laboratory tests, method of defining and applying outcome, and timing of laboratory tests. Four investigators conducted the quality assessment independently and disagreements were mutually sorted out. Each study was given a score, which was used in the sensitivity analysis of the meta-analysis. The score ranged from 0 to 20, where 20 was considered excellent.
Statistical analysis
The meta-analysis of studies was performed using Review Manager 5.4 (Cochrane Collaboration). Any discrepancy in units were checked and converted to same units. To pool continuous variables, we used an inverse variance method to obtain mean differences (MDs) and its standard deviations (SDs). Maentel-Haenszel formula was used to calculate odds ratio along with its 95% confidence intervals (CIs). We used random-effects models for pooled analysis regardless of heterogeneity. All P values were two-tailed, and statistical significance was set at ≤0.05. Heterogeneity among studies was evaluated using Q test and I2 statistic. Graphically, we displayed data using forest plots. Heterogeneity was identified using I2 that describes the percentage of variation across the included studies caused by heterogeneity rather than chance. The heterogeneity between studies was assessed by the Chi-squared and I-squared tests, with values of 0–25, 25.1–75, and 75.1–100% indicating a low, moderate, and high degree of heterogeneity, respectively.[12] For studies which reported median and interquartile ranges, mean and standard deviation were estimated using method described by Wan et al.[13]
Results
Baseline characteristics and study selection
A total of 3669 articles were identified after search of the databases. After the removal of duplicates (1158) and screening through title and abstracts, 260 articles were assessed for eligibility as per criteria already defined. Sixty-eight full text articles were assessed for eligibility and 45 were excluded, not being able to extract data as proper classification and required investigations were not available. Thereby, 23 studies remained for qualitative studies and meta-analysis (n = 19) [Figure 1]. All the studies were conducted in China and were observational in nature. The total study population consisted of 3386 patients who were categorised into non-severe (n = 2067, 61.05%) and severe (n = 1319, 38.95%) groups.
Figure 1.
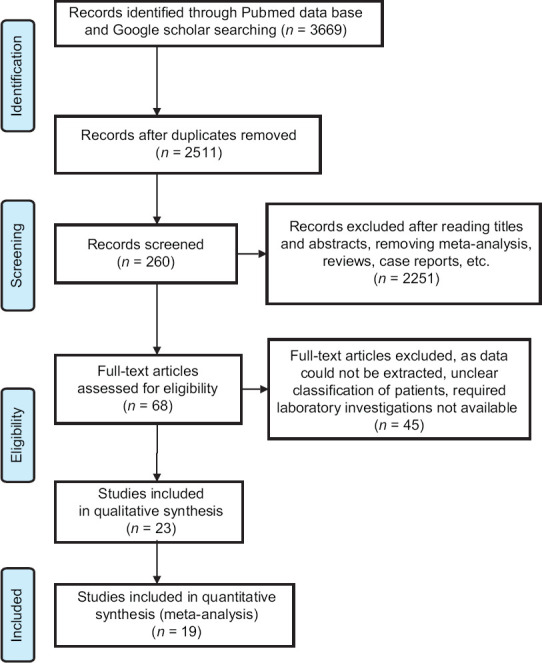
Flow diagram of study selection
The demographic characteristics [Table 1] revealed that the reported age (years) in mean ± SD of the severe and non-severe categories were, 61.89 ± 18.66 and 50.29 ± 21.40, respectively. About 48.2% of non-severe patients were males whereas in severe category 58.9% were males. All studies were of good quality with AXIS score of 13 or more.
Table 1.
Characteristics of included studied of patients with COVID-19
| Author, Study date, Country/region | Journal | Study type | Total number of patients | Outcomes | Severe patients | Non severe patients | Parameters | Quality Score# | References | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n(%) | Age | M/F | n(%) | Age | M/F | ||||||||
| Huang et al., 1/24, Wuhan, China | The Lancet | Retrospective cohort study | 41 | ARDS ICU admission | 13 (31.7) | 49 (41-61) | 11/2 | 28 (68.3) | 49 (41-57.5) | 19/9 | TLC, PCT, ALC | 17 | 9 |
| Chaomin Wu et al., 3/13, Wuhan, China | JAMA Internal Medicine | Retrospective cohort study | 201 | ARDS Hospitalisation Death | 84 (41.8) | 58.5 (50-69) | 60/24 | 117 (68.2) | 48 (40-54) | 68/49 | TLC, ALC, ESR, CRP, Ferritin, IL-6, CD4+, CD8+ | 16 | 17 |
| Yong Gao et al., 3/13, Fuyang, China | Journal of Medical Virology | Retrospective cohort study | 43 | ARDS | 15 (34.8) | 45.20±7.68 | 9/6 | 28 (65.2) | 42.96±14.00 | 17/11 | TLC, ALC, CRP, IL-6, PCT | 19 | 18 |
| Lo LL et al., 3/15, Macau, China | International J. BiolSci | Retrospective case series | 10 | Increased duration of hospital stay | 4 (40) | 61±5.0 | 1/3 | 6 (60) | 37±19 | 2/4 | TLC, ALC, CRP | 18 | 35 |
| Wang R et al., 3/24, Fuyang, China | International Journal of Infectious Diseases | Retrospective study | 125 | Pneumonia ICU admission | 25 (20) | 49.4±13.64 | 16/9 | 100 (80) | 39.47±14.84 | 55/45 | TLC, IL-6, PCT, ALC, ESR | 18 | 50 |
| Wang L et al., 3/31, KailiGuizhou, China | Médecine et Maladies Infectieuses | Retrospective analysis | 27 | Pneumonia | 4 (14.8) | 40.50±4.95, 41.00±25.46 | 3/1 | 23 (85.2) | 30.55±10.46, 33.29±14.97 | 10/13 | CRP | 17 | 10 |
| He R et al., 4/5, Wuhan, China | Journal of Clinical Virology | Retrospective data analysis | 204 | ICU, Invasive mechanical ventilator | 69 (33.8) | 61 (52-74) | 37/32 | 135 (66.2) | 43 (31-53) | 42/93 | TLC, ALC, CRP, IL-6, IL-10, TNF-α, CD4+, CD8+, PCT | 14 | 33 |
| Zhang G, et al., 4/9, Wuhan, China | Journal of Clinical Virology | Retrospective case series study | 221 | ARDS | 55 (24.9) | 62 (52-74) | 35/20 | 166 (75.1) | 51 (36-64.3) | 73/93 | TLC, ALC, PCT | 15 | 16 |
| Bo XU, 4/13 , Wuhan, China | Journal of Infection | Retrospective single centred study | 187 | ARDS ICU admission | 107 (57.2) | 60 (46-67) 70 (60.24-76.75) | 73/34 | 80 (42.8) | 56.00 [44.00-67.00] | 30/50 | CRP, IL-6, CD4+, CD8+PCT | 14 | 31 |
| Zhu Z et al., 4/17, Ningbo Zhejiang, China | International Journal of Infectious Diseases, | Retrospective data analysis | 127 | Respiratory failure, mechanical ventilation, shock | 16 (12.6) | 57.5±11.7 | 9/7 | 111 (87.4) | 49.95±15.52 | 73/38 | TLC, ALC, ESR, CRP, IL-6, IL-10, TNF-α | 17 | 30 |
| Sun Y, 4/17, Beijing, China | Journal of Autoimmunity | Retrospective single centred study | 63 | Alveolar damage and progressive respiratory failure | 19 (30.1) | Severe 59 (33-85) cri-63 (34-79) | 11/8 | 44 (69.9) | 23 (3-48) 47 (13-78) | 26/18 | TLC, ALC, ESR, CRP, Ferritin, IL-6, CD4+, CD8+ | 20 | 29 |
| Liu J et al., 4/18, Wuhan, China | EBio Medicin e | Retrospective data analysis | 40 | ARDS, multiple organ failure, death | 13 (32.5) | 59.7±10.1 | 7/6 | 27 (67.5) | 43.2±12.3 | 8/19 | TLC, ALC, CRP, Ferritin | 17 | 34 |
| Wei X et al., 4/22, Wuhan, China | Journal of Clinical Lipidology | Retrospective data analysis | 597 | Pneumonia ARDS | 203 (34) | 69 (64-77), 69 (61-83) | 116/87 | 394 (66) | 64 (53-69) | 189/215 | TLC, ALC, CRP, IL-6, CD4+, CD8+ | 20 | 36 |
| Fu J et al., 5/4, Sozhou, China | Thrombosis research | Retrospective cohort | 75 | ARDS | 16 (21.3) | 51.8±12.8 | 10/6 | 59 (78.7) | 45.1±14 | 35/24 | TLC, ALC, CRP, PCT | 17 | 32 |
| Liu R et al., 5/8, Wuhan, China | Clinica Chimica Acta | Cross-sectional | 154 | ARDS Pneumonia | 105 (68.2) | 63±14, 65±15 | 58/47 | 49 (31.8) | 63±13 | 26/23 | TLC, ALC, CD4+, CD8+ | 16 | 19 |
| Liu Y et al., 2/8, Shenzhen, China | Science China Life Sciences | Retrospective | 12 | ARDS Pneumonia | 6 (50) | 64 (62.75-65.25) | 3/3 | 6 (50) | 43.5 (28.75-60) | 5/1 | TLC, ALC, CRP, CD4+, CD8+, PCT | 16 | 8 |
| Qin C et al., 3/12, Tongji, China | Clin Infectious disease | Retrospective | 452 | ARDS Death | 286 (63.3) | 61 (51-69) | 155/131 | 166 (36.7) | 53 (41.25-62) | 80/86 | TLC, ESR, CRP, Ferritin, TNF-α, IL-6, IL-10, PCT | 14 | 28 |
| Zhou F et al., 3/9, Wuhan, China | The Lancet | Retrospective | 191 | Death | 54 (28.3) | 69.0 (63.0-76.0) | 38/16 | 137 (71.7) | 52.0 (45.0-58.0) | 81/56 | TLC, ALC, Ferritin, IL-6, PCT | 13 | 11 |
| Zhang J et al., 2/18, Wuhan, China | Epidemiology and Genetics | Retrospective | 140 | Pneumonia | 58 (41.4) | 64 (25-87) | 33/25 | 82 (58.6) | 51.5 (26-78) | 38/44 | TLC, CRP, PCT | 16 | 15 |
| Mo P et al., 3/16, Wuhan, China | Clin Infectious disease | Retrospective | 155 | ARDS | 85 (64.8) | 61 (51-70) | 55/30 | 70 (45.2) | 46 (35-56) | 31/39 | TLC, ALC, ESR, CRP, IL-6, PCT | 17 | 26 |
| Jiang Mei, et al., 5/1, Nanchang, Jiangxi, China | Journal of Infectious Disease | Prospective | 103 | Pneumonia ARDS | 17 (16.5) | 64 (41-88) | 11/6 | 86 (83.5) | 44 (17-83) | 47/39 | CD4, CD8 | 16 | 46 |
| Wan S et al., 3/18, Chongqing, China | J Med Virol | Retrospective | 135 | ARDS | 40 (29.6) | 56 (52-73) | 21/19 | 95 (70.4) | 44 (33-49) | 52/43 | TLC, ALC, CRP, PCT | 16 | 14 |
| Li Kunhua et al., 2/24, China. | Investigative Radiology | Retrospective | 83 | ARDS | 25 (30.1) | 53.7±12.3 | 15/10 | 58 (69.9) | 41.9±10.6 | 29/29 | TLC, ALC CRP, PCT | 16 | 16 |
All patients who were positive by real time PCR were recruited for study. Parameters studied: Total lymphocyte count (TLC), Absolute lymphocyte count (ALC), Erythrocyte sedimentation rate (ESR), C-reactive protein (CRP), Procalcitonin (PCT), IL-6, IL-10, Ferritin, TNF-α, CD4+, CD8+
Table 2 depicts the inflammatory and immunological markers in severe and non-severe disease. Amongst the analysed parameters total leukocyte count, and CRP were the most common parameters studied. Next to them were absolute lymphocyte count and procalcitonin, respectively. IL-6 has been analysed in ten studies followed by CD4 and CD8 count, while TNF-α was the least studied parameter.
Table 2.
Laboratory parameters in mean±SD in the severe and non-severe COVID- 19 patients
| Author, Study | TLC (×109/L) | Absolute lymphocyte count (×109/L) | ESR (mm/hr) | CRP (mg/L) | Ferritin (ng/ml) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sev | Non sev | Sev | Non Sev | Sev | Non sev | Sev | Non sev | Sev | Non sev | |||
| Huang et al. | 9.73±5.23 | 5.46±3.51 | 0.46±0.49 | 0.93±0.31 | ||||||||
| Chaomin Wu et al. | 8.19±4.62 | 5.19±2.85 | 0.71±0.37 | 1.08±0.54 | 54.46±23.38 | 50.66±18.23 | 91.6±85.19 | 29.28±38.39 | 1191.84±1096.56 | 461.34±359.48 | ||
| Yong Gao et al. | 4.26±1.64 | 4.96±1.85 | 1.20±0.42 | 1.07±0.40 | 39.37±27.68 | 18.76±22.20 | ||||||
| Lo LL et al. | 5.57±1.99 | 4.51±1.18 | 2.63±3.02 | 1.65±3.16 | ||||||||
| Wang R et al. | 5.11±2.32 | 4.93±1.70 | 0.79±0.33 | 1.10±0.40 | ||||||||
| Wang L et al. | 79.57±42.16 | 9.47±3.24 | ||||||||||
| R He, et al. | 5.5±3.04 | 4.84±1.28 | 0.74±0.28 | 1.47±0.56 | 42.23±46.18 | 7.01±12.4 | ||||||
| Zhang G, et al. | 6.56±4.03 | 4.3±2.01 | 0.66±0.38 | 0.9±0.44 | ||||||||
| Bo XU, et al. | 0.68±0.37 | 1.24±0.52 | 70.07±56.10 | 12.04±16.70 | ||||||||
| Zhu Z et al. | 5.67±2.76 | 5.26±1.65 | 85.08±36.78 | 66.66±40.56 | 40.63±44.38 | 9.15±10.06 | ||||||
| Sun Y et al. | 5.87±2.84 | 1.32 | 48.83±29.7 | 22.5±20.79 | 40.4±55.8 | 10.60±16 | ||||||
| Liu J et al. | 6.6±3.4 | 3.9±1.5 | 0.66±0.16 | 1.1±0.46 | 22.66±45.01 | 63.9±34.59 | 354.83±265.2 | 664.3±94.1 | ||||
| Wei X et al. | 6.23±3.2 | 5.5±1.48 | 1.25±0.69 | 1.46±0.52 | 16.27±35.48 | 3.63±3.34 | ||||||
| Fu J et al. | 7.14±3.61 | 4.87±1.70 | 0.97±0.33 | 1.42±0.66 | 80.5±146.8 | 23.5±45.13 | ||||||
| Liu R et al. | 0.81±0.31 | 0.98±0.54 | 0.34±0.44 | 0.98±0.54 | ||||||||
| Liu Y et al. | 5.46±1.22 | 6.46±3.58 | 53.48±19.81 | 28.8±30.17 | ||||||||
| Qin C et al. | 6.1±3.05 | 4.9±1.78 | 0.83±0.37 | 1.0±0.44 | 37.6±30.5 | 30.6±26.9 | 60.6±61.3 | 33.7±38.5 | 901.6±744.1 | 554.4±404.8 | ||
| Zhou F et al. | 10.2±5.28 | 5.73±2.54 | 562.9±492.6 | 1388.4±967.3 | ||||||||
| Zhang J et al. | 6.1±3.8 | 4.63±1.81 | 51.7±60.5 | 40.1±32.14 | ||||||||
| Mo P et al. | 4.87±2,78 | 4.19±1.33 | 0.8±0.36 | 1.01±0.37 | 31.66±26.3 | 25.6±21.19 | 58±63.3 | 26.6±28.0 | ||||
| Jiang Mei et al. | ||||||||||||
| Wan S et al. | 5.66±1.53 | 5.83±3.0 | 0.8±0.30 | 1.2±0.60 | 93.32±64.28 | 13.5±21.98 | ||||||
| Li K et al. | 5.76±2.83 | 5.25±1.59 | 1.19±0.37 | 0.69±0.40 | 90.57±68.2 | 13.85±21.14 | ||||||
| Author, Study | IL6 (pg/ml) | IL-10 (pg/ml)± | TNF-α (pg/ml) | CD4 (counts/µL) | CD8 (counts/µL) | Procalcitonin (ng/ml) | ||||||
| Sev | Non sev | Sev | Non sev | Sev | Non sev | Sev | Non sev | Sev | Non sev | Sev | Non sev | |
| Huang et al. | 0.2±0.24 | 0.1±0 | ||||||||||
| Chaomin Wu et al. | 7.97±3.96 | 6.49±1.85 | 256.25±197.06 | 408.66±216.92 | 174.16±161.04 | 241±123.09 | ||||||
| Yong Gao et al. | 39.43±29.6 | 13.30±14.88 | 0.05±0.05 | 0.02±0.02 | ||||||||
| Lo LL et al. | ||||||||||||
| Wang R et al. | 0.06±0.09 | 0.03±0.03 | ||||||||||
| Wang L et al. | ||||||||||||
| R He, et al. | 11.23±2.87 | 12.16±6.06 | 7.16±1.40 | 6.25±0.71 | 2.90±0.36 | 2.55±0.21 | 193.3±143.86 | 633.33±249.53 | 124±108.27 | 374±155.86 | 0.140±0.181 | 0.031±0.022 |
| Zhang G, et al. | ||||||||||||
| Bo XU, et al. | 20.64±28.1 | 13.7±13.65 | 9.79±7.73 | 5.68±1.70 | 247.29±148.93 | 590.41±259.89 | 142.37±84.40 | 335±163.07 | 0.35±0.05 | 0.02±0.01 | ||
| Zhu Z et al. | 26.54±43.26 | 5.29±5.76 | 6.89±6.32 | 3.28±1.81 | 1.53±0.28 | 1.4±0.45 | ||||||
| Sun Y et al. | 33.63±31.04 | 12.55±9.67 | 263.66±92.1 | 357.38±244.42 | 203.7±171.28 | 374.88±168.237 | ||||||
| Liu J et al. | ||||||||||||
| Wei X et al. | 38.43±53.13 | 17.0±20.68 | 45.04±15.63 | 49±8.92 | 15.63±9.94 | 24.6±7.46 | ||||||
| Fu J et al. | 1.6±3.62 | 0.312±0.67 | ||||||||||
| Liu R et al. | 271.11±144.68 | 322.8±254.7 | 151.62±115.41 | 1.92±1.06 | ||||||||
| Liu Y et al. | 36.47±10.73 | 47.97±4.31 | 15.5±4.39 | 9.88±11.19 | 1.58±3.72 | 0.09±0.11 | ||||||
| Qin C et al. | 29.7±33.5 | 19.4±27.8 | 7.63±4.69 | 5.6±1.49 | 9.13±3.35 | 8.56±2.61 | 285.1±168.0 | 420.5±207.8 | 154.7±116.5 | 201.9±107.1 | 0.1±0.14 | 0.05±0.04 |
| Zhou F et al. | 6.4±2.17 | 10.9±5.25 | 0.1±0 | 0.23±0.3 | ||||||||
| Zhang J et al. | 0.15±0.18 | 0.06±0.05 | ||||||||||
| Mo P et al. | 86.6±101.05 | 26.3±28.76 | 0.05±0 | 0.096±0.1 | ||||||||
| Jiang Mei et al. | 130.3±67.87 | 227.58±181.9 | 65.66±49.31 | 227.58±181 | ||||||||
| Wan S et al. | 0.11±0.06 | 0.04±0.02 | ||||||||||
| Li K et al. | 0.1±0.08 | 0.04±0.03 | ||||||||||
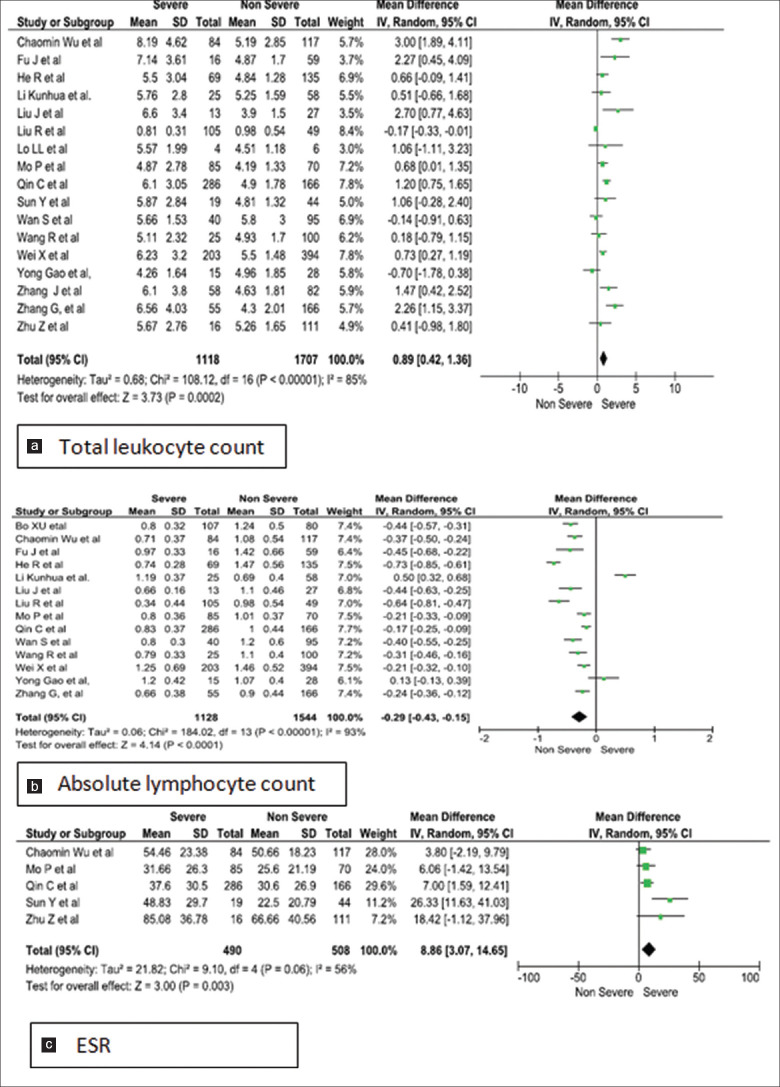
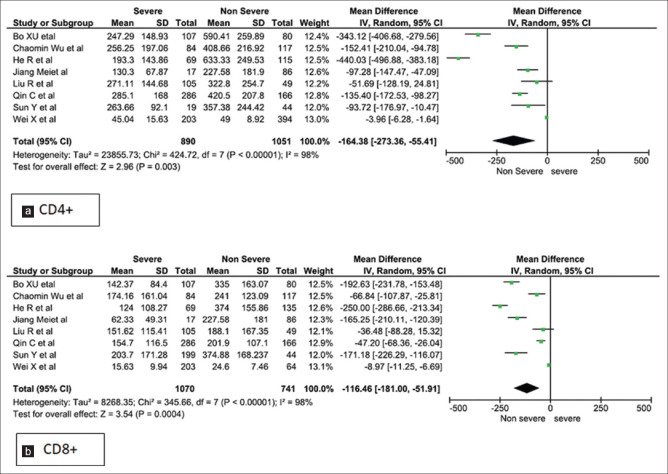
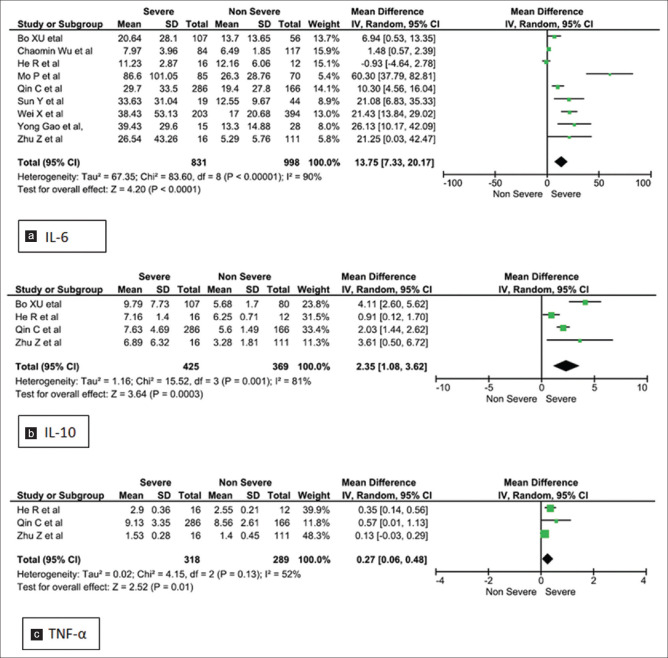
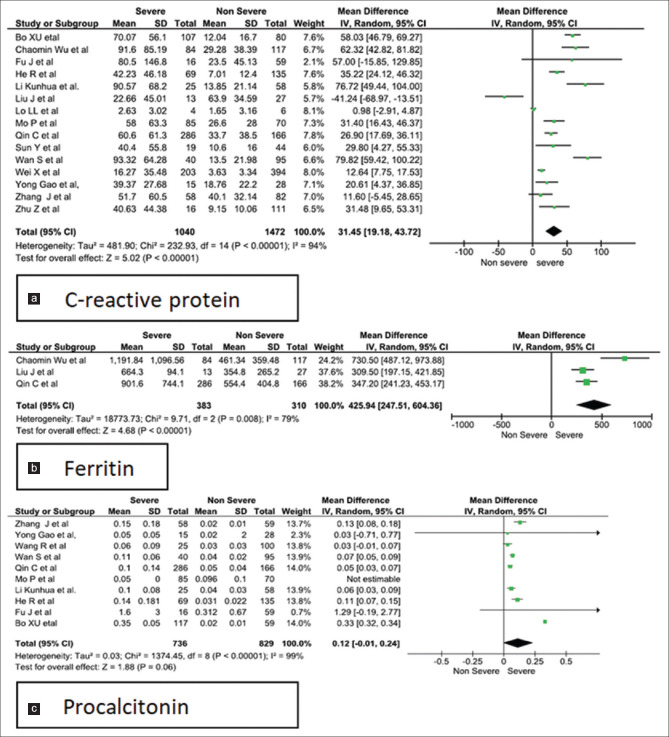
Forest plots [Figures 2-5] demonstrates the inflammatory and the immunological parameters in severe and non-severe patients suffering from COVID-19 infection. Random effect results demonstrated that patients in the severe group had higher levels for TLC (WMD 0.89, 95%CI; 0.42-1.36, P = 0.0002), lower ALC (WMD -0.29, 95%CI; -0.44,-0.15, p=<0.0001) higher ESR (WMD 8.86, 95%CI; 3.07-14.65, P = 0.003), raised CRP (WMD 31.45, 95%CI; 19.18-43.72, P < 0.00001), elevated procalcitonin (WMD 0.11, 95%CI; -0.01, 0.23, p=<0.07) and ferritin (WMD 425.94, 95%CI; 247.51-604.36, P < 0.00001). The same patients when stratified by severity, demonstrated that in comparison to patients of severe group, non-severe groups had low levels of IL-6 (WMD 13.75, 95%CI; 7.33, 20.17, p=<0.0001), IL-10 (WMD 2.35, 95%CI; 1.08, 3.62, p=<0.0003), TNF-α (WMD 0.27, 95%CI; 0.06, 0.48, p=<0.01), CD4+ (WMD -164.38, 95%CI; -273.36, -55.41, p=<0.003) and CD8+ (WMD -116.46, 95%CI; -181.00, -51.91, p=<0.0004) respectively.
Figure 2.
Meta-analysis of (a). total leukocyte count, (b). absolute lymphocyte count and (c). ESR levels with severe vs non-severe patients
Figure 5.
Meta-analysis of (a). CD4+ and (b). CD8+ levels with severe and non-severe patients
Figure 4.
Meta-analysis of (a). IL-6, (b). IL-10 and (c). TNF- α level with severe vs non-severe patients
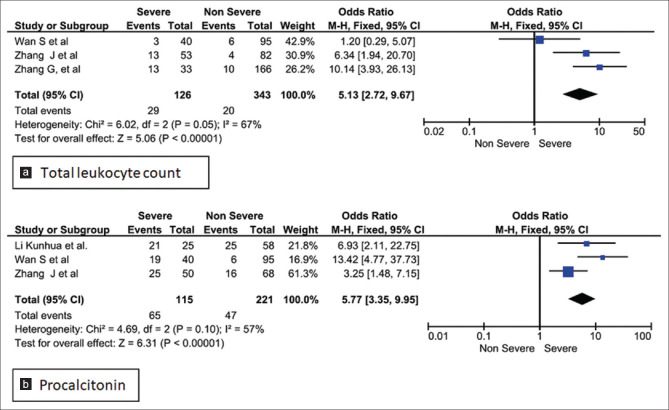
Three studies were homogenous and considered by fixed effect model for the calculation of odds ratio for TLC in the study group. Our analysis revealed that, patients with raised total leukocyte count (>9.5 × 109/L) had a 5-fold higher risk of developing into the severe form of COVID-19 infection (Odds ratio 5.13; 95% CI: .72–9.67, P < 0.00001). However, the studies showed significant heterogeneity as they used different cut-off values [Figure 6a].[14,15,16] Conversely on assessment of all studies for TLC, leukopenia was revealed in the severe cases [Figure 2a].[14,17,18,19] Procalcitonin assay conducted in 10 studies on 736 non severe and 852 severe patients registered a raised value in severe group [Figure 3a], with fixed effect results arriving at a similar conclusion. Three homogenous studies reporting procalcitonin documented a highly significant value (p < 0.00001) with a I2 value of 57% and odd ratio of 5.77 (95%CI: 3.35, 9.95) beyond the cut off value of 0.1 ng/mL [Figure 6b].[14,15,20]
Figure 6.
Forest plot of odds ratio for association of elevated (a). TLC and (b). procalcitonin with disease severity
Figure 3.
Meta-analysis of (a). C-reactive protein (b). ferritin and (c). procalcitonin levels with severe vs non-severe patients
Investigation of heterogeneity
Heterogeneity was found in all the comparisons with I2 value ranging from 56% to 99% [Table 3]. Moderate heterogeneity was observed in ESR and TNF-α. Pooled analysis of studies using fixed effect model revealed a moderate heterogeneity for TLC (I2 = 67%, P = 0.05) and PCT (I2 = 57%, P = 0.10). Therefore, COVID-19 patients who present with TLC > 9.5 × 109/L and PCT > 0.1 ng/mL should be monitored earnestly for implementation of therapy.
Table 3.
The association between laboratory parameters and disease severity in patients with COVID-19
| Parameters | No of studies | Participants | Mean difference (95% CI) | P | Effect model - REM (random effect model) | Heterogeneity | |
|---|---|---|---|---|---|---|---|
| I2(%) | P | ||||||
| Total leukocyte count | 17 | 2825 | 0.89 (0.42, 1.36) | 0.0002 | REM | 85 | <0.00001 |
| Absolute lymphocyte count | 14 | 2672 | -0.29 (-0.43, -0.15) | <0.0001 | REM | 93 | <0.00001 |
| ESR | 5 | 998 | 8.86 (3.07, 14.65) | 0.003 | REM | 56 | 0.06 |
| CRP | 13 | 2512 | 31.45 (19.18, 43.72) | <0.00001 | REM | 94 | <0.00001 |
| Ferritin | 4 | 693 | 425.94 (247.51, 604.36) | <0.00001 | REM | 79 | 0.008 |
| IL-6 | 9 | 1829 | 13.75 (7.33, 20.17) | <0.0001 | REM | 90 | <0.00001 |
| IL-10 | 4 | 794 | 2.35 (1.08, 3.62) | 0.0003 | REM | 81 | <0.001 |
| TNF-α | 3 | 601 | 1.01 (0.08, 1.94) | 0.003 | REM | 98 | <0.00001 |
| CD4+ | 7 | 1941 | -164.38 (-273.36, -55.41) | 0.003 | REM | 98.33 | <0.0001 |
| CD8+ | 6 | 1811 | -116.46 (-181.0, -51.91) | 0.0004 | REM | 98 | <0.0001 |
| Procalcitonin | 8 | 1565 | 0.12 (-0.01, 0.24) | 0.06 | REM | 99 | <0.00001 |
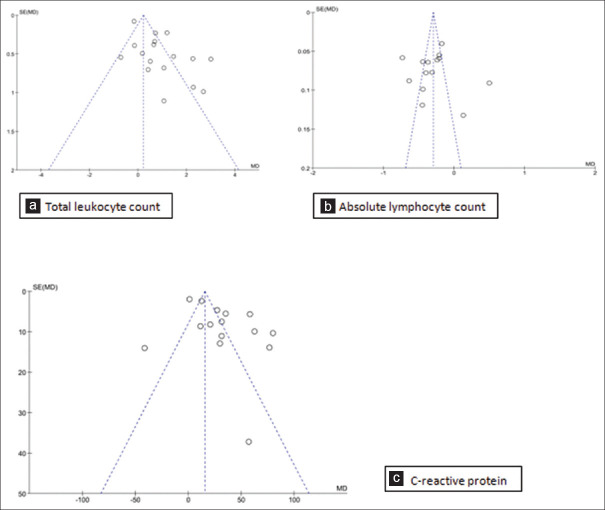
Publication bias
Publication bias was evaluated using a funnel plot for parameters with ten or more studies by visual observation. A fixed effect model was used in RevMan. The vertical line of the funnel gives the summary effect and the guidelines represent the 95% CI. Substantial asymmetry was detected for the funnel plots as the studies were clustered towards the top of the funnel [Figure 7]. This can be attributed to their cross sectional in nature, clinical difference due to statistical heterogeneity due to clinical difference between the studies such as setting of the study, variance in the included participants and the type of intervention.[21]
Figure 7.
Funnel plots for association of (a). Total leukocyte count, (b). Absolute lymphocyte count, (c). C-reactive protein with disease severity
Discussion
COVID-19 infection has become pandemic affecting the entire world. However, the management of patients still remains a challenge. Just like any infection, here too the innate immunity is evoked in the early stage. Later there is activation of adaptive immunity for a specific response. Like HIV infection, there is a defective adaptive immunity characterised by a decline in lymphocyte subsets of CD4+ and CD8+.[22] Although many patients remain asymptomatic, some of them develop pneumonia leading to acute respiratory distress syndrome requiring mechanical ventilation and care in the intensive care unit (ICU). These patients have higher levels of immune-active molecules like proinflammatory cytokines. Therefore, cytokine storm is induced by external stimuli i.e., the virus resulting in severe deterioration of patients. Early identification of this virus induced inflammation and immune deregulation can provide valuable guidance for early diagnosis and clinical treatment at the first point of contact of patient with health care worker.
The majority of COVID-19 patients have relatively mild symptoms, but a considerable number of patients progress to severe pneumonia and even eventually develop acute respiratory distress syndrome (ARDS), septic shock, and/or multiple organ failure. Therefore, assessment of inflammatory and immunological characteristics of peripheral blood is essential in severe patients for timely diagnosis, precise treatment, for retarding the progression of the disease. Many systematic reviews and meta-analysis have been conducted so far on the immunological and inflammatory parameters. Zeng et al., reported the association of inflammatory parameters such as IL-6, PCT, ESR in severe and non-severe COVID-19.[23] Our study includes the commonly tested parameters TLC, ALC, ESR, CRP and many other immunological markers like IL-6, IL-10, TNF-α, ferritin, procalcitonin, CD4+ and CD8+. We also found similar positive association of the immunological and inflammatory parameters in COVID-19 severe patients in line with the meta analysis conducted by Zeng et al.[23] Various other observational studies have been conducted, based on clinical features, CT imaging and few specific laboratory findings.[24,25]
We found that TLC of severe patients was higher than that of non-severe patients. The pooled results statistically supported the conclusions that elevated TLC was strongly associated with the deterioration of COVID-19 patients. Several studies have addressed the difference of baseline TLC between the clinical stages where refractory patients had higher TLC than non-severe patients.[26] Since this is one of the most basic investigations that is done at the primary care setting the importance of this parameter cannot be overlooked.
Excessive pro-inflammatory cytokines responsible for the lymphopenia associated with COVID-19 also promotes expression of proteins like programmed cell death protein and mucin domain 3 and other markers of T cell exhaustion in CD4+ and CD8+ cells.[27] Certain genes related to T cell activation are down regulated in severe COVID-19 infection which return to normal on recovery.[27] Since lymphocytes carry ACE2 receptors they can also get infected by the virus leading to death.[27] Understanding the mechanism of lymphopenia in COVID-19 infection can provide an effective strategy for treatment.
ESR, a non-specific inflammatory marker is still being used as a routine parameter of investigation. Five studies were included in this analysis which comprised of 490 severe and 508 non-severe COVID-19 patients.[17,26,28,29,30] There was a statistical difference between severe and non-severe COVID-19 cases reflecting the presence of intense inflammatory response in the acute stage. However, a heterogeneity of 56% (p = 0.003) was observed.
C-reactive protein (CRP) was high in severe and critically ill patients.[14,15,17,18,20,26,28,29,30,31,32,33,34,35,36] The mean difference in CRP ranged from 19.18 to 43.72 with I2 of 94%. Raised CRP is mostly indicative of bacterial infections, unlike a study, showing evidence of increased CRP with respiratory virus infection and also a predictive factor for hospitalisation in ICU and for mechanical ventilation.[37] Further raised CRP levels have also been found in individuals infected with virulent influenza A virus being associated with its severity and mortality.[38] Therefore, the observations of CRP from our meta-analysis can be used as a biomarker to categorise COVID-19 infections as per the severity.
Cytokines and chemokines are critical mediators of the immune system and play a pivotal role in anti-viral immunity. Studies have demonstrated that the primary cause of ARDS and multi-organ failure in COVID-19 is its association with cytokine storm. Multiple logistic regression demonstrate cytokines, as an independent risk factor to predict the disease severity.[34] The exaggerated, excessive cytokines lead to an acute, severe systemic inflammatory response called 'cytokine storm' increasing disease severity. Our analysis showed that severe category of patients had a significantly higher levels of IL-6, IL-10, TNF-α. However, there was a lower expression of CD4+ and CD8+ counts in severe cases.
IL-6 is involved in the acute phase response of the infection, and induces Th17 cells which further induces production of IL-6. Like the SARS-CoV infection, the SARS-CoV-2 is also associated with elevated IL-6 whose pleotropic role, due to activation of numerous genes, may play a key factor in the immune response.[39] IL-6 as a promising marker of prognosis has been investigated in sepsis as well as in acute organ injuries. They have also been studied in plasma and broncho alveolar lavage in lung injury serving as predictive factors of mechanical ventilation.[40] IL-6 values were higher in the severe patients as compared to the non-severe groups.[17,18,26,28,29,30,31,33,36] In the retrospective cohort study baseline IL-6 positively correlated with bilateral pulmonary involvement (r = 0.45, P = 0.001), and maximum body temperature (r = 0.52, P = 0.001).[8] IL-6 is an independent risk factor for severity of COVID-19 infection and a value of >24.3 pg/mL could predict the severity of COVID-19 with a sensitivity and specificity of 73.3% and 89.3% respectively.[18] A linear mixed model, also strengthens the role of IL-6 as an inflammatory parameter with highest AUROC related to severity of the disease.[30] This evidence has recommended the usage of monoclonal antibody against IL-6 receptor. IL-10, inhibits the pro-inflammatory response from innate to adaptive immunity and suppresses the inflammation, but viral infections evade the immune regulatory role of IL-10 and uphold their survival.[41] In the context of cytokine release syndrome (CRS), the IL-10 released downregulates the neutrophil and monocyte function inducing a protective phenomenon of immunoparalysis. However, the persistent depression leads to higher mortality, so a recovery from immunoparalysis is critical for patient survival.[42] Thus these cytokines may help to predict the worsening of mild patients and can be useful as predictors of the pathology. Only one of the nine studies of IL-6 in the forest plot had a lowered value, however the mean difference between severe and non-severe was -0.93.[29] This could be because of the small sample size of the study with 16 in severe and 12 in non-severe group. In contrast, IL-10 has been analysed in four studies, with heterogeneity of 81% was achieved with P value of 0.0003.
TNF-α is a pro-inflammatory cytokine that was found to be elevated in all the three research papers in our analysis. The very high heterogeneity of 98% (p = 0.03) could be because of the different sampling times in the studies as well as the short half-life of TNF-α.[43] At the start of infection the spike protein of SARS-CoV-2 induces a TNF- α converting enzyme-dependent shedding of ACE2 ectodomain that is coupled to viral entry into cells. Later TNF-α through its NF-kB activation pathway induces production of IL-6, that causes extensive vascular leakage and lung injury, acting as a central cytokine for activation and maintenance of CRS.[44]
The T cell response (CD4+ and CD8+) in patients of COVID-19 revealed that both were significantly reduced in severe patients when compared with non-severe patients.[17,19,28,29,31,33,36,45] Studies suggest that cytokine storm may promote apoptosis of T cells and a decline in CD4+ and CD8+ counts as shown by dynamic changes in T lymphocyte subset confirmed their correlation with the severity of disease.[33]
The mechanism of the association of ferritin with COVID-19 is not well delineated but it is suggested to increase due to the influence of pro-inflammatory cytokines.[46] Three studies revealed a high level of ferritin at the time of admission, with the severe group of patients having more comorbidities.[17,28,34] The various suggested mechanisms of raised ferritin level are the pro-inflammatory cytokines like IL-16 and TNF-α promoting synthesis of ferritin, leakage of intracellular ferritin by cellular damage.[47] Raised ferritin leads to thymosuppression with lowering of absolute lymphocyte count affecting CD4+ and CD8+ count adding to the state of inflammation.[48] Thus hyperferritinemia is also associated with systemic inflammation due to co-morbidities and can correlate with the severity of the disease.
Procalcitonin (PCT), serving as a biomarker in sepsis due to bacterial infection. is also released by many extrathyroidal tissues. Cytokines IL-6 and TNF-α released during sepsis, mediate release of PCT and is associated with disease severity.[49] In this meta-analysis patients in severe group had a higher level of procalcitonin in comparison with the non-severe group.[14,15,18,20,26,28,31,32,33,50] In severe groups there may be an associated secondary bacterial infection which must be responsible for heterogeneity and non-significance in the forest plot (I2 = 99%, P = 0.06). Although a mean difference of 0.12 ng/mL with CI; -0.01, 0.24 was observed the pooled analysis of three studies revealed more than five fold rise of procalcitonin in severe COVID-19 patients [Figure 3c, 6b].[10,11,16] The rising procalcitonin is another indicator of increased severity of the disease.
The pandemic COVID-19 infection has affected millions of people around the world. Being prevalent for more than one year, COVID-19 has reached a stage where there has been a large inflow of patients requiring critical care, putting immense pressure on the available health care facility. The identification of biomarkers not only serve to detect the severity of the disease but are also essential for therapeutic interventions and future prognosis. The present study could identify two crucial biomarkers, i.e., TLC and PCT that can detect the severity of the condition at an early stage for timely implementation of therapy to retard the morbidity and mortality in these COVID-19 cases at primary care setting.
Limitations of study
This meta-analysis involved retrospective studies that were primarily from China where the primary infection occurred. Since the outbreak in China, there is little information about what has changed in the COVID-19 virus infection in India. Studies from India are required to reach at a conclusion about the nature of the immunological and inflammatory markers as genetic and environmental factors can also play a role in the outcome. The cross-sectional retrospective nature of the articles also were able to show association rather than causality. Prospective studies if designed would be better for stronger evidence. In few studies comprehensive information about patients and their detailed clinical outcomes were not available. Further there were varied definitions on the gradation of severity making it difficult for placement of participants in the correct group. There was also minimal information on the analytical methods used and the different units of measurements which was responsible for gross heterogeneity across the studies.
Conclusion
Severe COVID-19 infection is associated with increased levels of TLC, ESR, CRP, Ferritin, IL 6, IL 10, TNFα and procalcitonin with a fall in absolute lymphocyte count as well as CD4+ and CD8+ count. The most promising markers for assessing the risk of severity are total leukocyte count and procalcitonin as evidenced from the meta-analysis. Thus, it supports the opinion to evaluate these parameters for early detection of severe COVID-19 cases in a primary care setup.
Ethical approval
Not required.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome [published correction appears in Lancet Respir Med.2020 Feb 25;]? Lancet Respir Med. 2020;8:420–2. doi: 10.1016/S2213-2600(20)30076-X. doi: 10.1016/S2213-2600 (20) 30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Channappanavar R, Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin Immunopathol. 2017;39:529–39. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Min CK, Cheon S, Ha NY, Sohn KM, Kim Y, Aigerim A, et al. Comparative and kinetic analysis of viral shedding and immunological responses in MERS patients representinga broad spectrum of disease severity? Sci Rep. 2016;6:25359. doi: 10.1038/srep25359. doi: 10.1038/srep25359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Max R, Ritchie, Ortiz-Ospina E, Hasell J. Coronavirus Pandemic (COVID-19). 2020. Published online at OurWorldInData.org. Retrieved from 'https://ourworldindata.org/covid-cases' . Online resource.
- 5.Coronavirus disease 2019 (COVID-19). Situation Report-46. Published online at http“//www.who.int/docs/defaultsource/coronaviruse/situation reports . Online resource.
- 6.Guideline on the management of COVID-19, version 6, National Health Commission of the People's Republic of China. [Last accessed on 2020 Feb 22]. Available from: http://www.gov.cn/zhengce/zhengceku/2020-02/19/content . published online 2020 Feb 18.
- 7.World Health Organization. [Last accessed on 2020 Mar 05];Clinical management of severe acute respiratory infection when novel coronavirus (nCoV) infection in suspected: interim guidance. January 28, 2020. [Google Scholar]
- 8.Liu Y, Yang Y, Zhang C, Huang F, Wang F, Yuan J, et al. Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and ling injury. Sci China Life Sci. 2020;63:364–74. doi: 10.1007/s11427-020-1643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang L. C-reactive protein levels in the early stage of COVID-19. Med Mal Infect. 2020;50:332–4. doi: 10.1016/j.medmal.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical course and risk factors for mortality of adult in patients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet. 2020;395:1054–62. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wan X, Wang W, Liu J, Tong T. Estimating the sample mean and standard deviation from the sample size, median, range and/or interquartile range? BMC Med Res Methodol. 2014;14:135. doi: 10.1186/1471-2288-14-135. doi: 10.1186/1471-2288-14-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wan S, Xiang Y, Fang W, Zheng Y, Li B, Hu Y, et al. Clinical features and treatment of COVID-19 patients in northeast Chongqing. J Med Virol. 2020;92:797–806. doi: 10.1002/jmv.25783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J, Dong X, Cao Y, Yuan Y, Yang Y, Yan Y, et al. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy. 2020;75:1730–41. doi: 10.1111/all.14238. [DOI] [PubMed] [Google Scholar]
- 16.Zhang G, Hu C, Luo L, Fang F, Chen Y, Li J, et al. Clinical features and short-term outcomes of 221 patients with COVID-19 in Wuhan China? J Clin Virol. 2020;127:104364. doi: 10.1016/j.jcv.2020.104364. doi: 10.1016/j.jcv. 2020.104364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu C, Chen X, Cai Y, Xia J, Zhou X, Xu S, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med. 2020;180:934–43. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao Y, Li T, Han M, Li X, Wu D, Xu Y, et al. Diagnostic utility of clinical laboratory data determinations for patients with severe COVID-19? J Med Virol. 2020:1–6. doi: 10.1002/jmv.25770. doi: 10.1002/jmv.25770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu R, Wang Y, Li J, Han H, Xia Z, Liu F, et al. Decreased T cell populations contribute to the increased severity of OCVID-19. Clin Chim Acta. 2020;508:110–4. doi: 10.1016/j.cca.2020.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li K, Wu J, Wu F, Guo D, Chen L, Fang Z, et al. The clinical and chest CT features associated with severe and critical COVID-19 pneumonia. Invest Radiol. 2020;55:327–31. doi: 10.1097/RLI.0000000000000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dekkers OM, Vandenbroucke JP, Cevallos M, Renehan AG, Altman DG, Egger M. COSMOS-E: Guidance on conducting systematic reviews and meta-analyses of observational studies of etiology. PLoS Med. 2019;16:e1002742. doi: 10.1371/journal.pmed.1002742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kagan JM, Sanchez AM, Landay A, Denny TN. A brief chronicle of CD4 as a biomarker for HIV/AIDS: A tribute to the memory of John L Fahey. For Immunopathol Dis Therap. 2015;6:55–64. doi: 10.1615/ForumImmunDisTher.2016014169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeng F, Huang Y, Guo Y, Yin M, Chen X, Xiao L, et al. Association of inflammatory markers with the severity of COVID-19: A meta-analysis. Int J Infect Dis. 2020;96:467–74. doi: 10.1016/j.ijid.2020.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng Z, Lu Y, Cao Q, Qin L, Pan Z, Yan F, et al. Clinical features and chest CT manifestations of coronavirus disease 2019 (COVID-19) in a Single-Center Study in Shanghai, China. AJR Am J Roentgenol. 2020;215:121–6. doi: 10.2214/AJR.20.22959. [DOI] [PubMed] [Google Scholar]
- 25.Xiong Y, Sun D, Liu Y, Fan Y, Zhao L, Li X, et al. Clinical and high-resolution CT features of the COVID-19 Infection: Comparison of the initial and follow-up changes. Invest Radiol. 2020;55:332–9. doi: 10.1097/RLI.0000000000000674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mo P, Xing Y, Xiao Y, Deng L, Zhao L, Wang H, et al. Clinical characteristics of refractory COVID-19 pneumonia in Wuhan, China? Clin Infect Dis. 2020:ciaa270. doi: 10.1093/cid/ciaa270. doi: 10.1093/cid/ciaa270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tavakolpour S, Rakhshandehroo T, Wei EX, Rashidian M. Lymphopenia during the COVID-19 infection: What it shows and what can be learned. Immunol Lett. 2020;225:31–2. doi: 10.1016/j.imlet.2020.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qin C, Zhou L, Hu Z, Zhang S, Tao Y, Xie C, et al. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin Infect Dis. 2020;71:762–8. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun Y, Dong Y, Wang L, Xie H, Li B, Chang C, et al. Characteristics and prognostic factors of disease severity in patients with COVID-19: The Beijing experience? Beijing experience. J Autoimmun. 2020;112:102473. doi: 10.1016/j.jaut.2020.102473. doi: 10.1016/j.jaut. 2020.102473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu Z, Cai T, Fan L, Lou K, Hua X, Huang Z, et al. Clinical value of immune-inflammatory parameters to assess the severity of coronavirus disease 2019. Int J Infect Dis. 2020;95:332–9. doi: 10.1016/j.ijid.2020.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu B, Fan C, Wang A, Zou Y, Yu Y, He C, et al. Suppressed T cell-mediated immunity in patients with COVID-19: A clinical retrospective study in Wuhan, China. J Infect. 2020;81:e51–60. doi: 10.1016/j.jinf.2020.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu J, Kong J, Wang W, Wu M, Yao L, Wang Z, et al. The clinical implication of dynamic neutrophil to lymphatic ratio and D-dimer in COVID-19: A retrospective study in Suzhou, China. Thromb Res. 2020;192:3–8. doi: 10.1016/j.thromres.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He R, Lu Z, Zhang L, Fan T, Xiong R, Shen X, et al. The clinical course and its correlated immune status in COVID-19? J Clin Virol. 2020;127:104361. doi: 10.1016/j.jcv.2020.104361. doi: 10.1016/j.jcv. 2020.104361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu J, Li S, Liu J, Liang B, Wang X, Wang X, et al. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients? EBioMedicine. 2020;55:102763. doi: 10.1016/j.ebiom.2020.102763. doi: 10.1016/j.ebiom. 2020.102763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lo LL, Lio CF, Cheong HH, Lei CI, Cheong TH, Zhong X, et al. Evaluation of SARS-CoV-2 RNA shedding in clinical specimens and characteristics of 10 patients with COVID-19 in Macau. Int J Biol Sci. 2020;16:1698–707. doi: 10.7150/ijbs.45357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei X, Zeng W, Su J, Wan H, Yu X, Cao X, et al. Hypolipidemia is associated with the severity of COVID-19. J Clin Lipidol. 2020;14:297–304. doi: 10.1016/j.jacl.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeon JS, Rheem I, Kim JK. C-reactive protein and respiratory viral infection. Korean J Clin Sci. 2017;489:15–21. [Google Scholar]
- 38.Perez L. Acute phase proteins response to viral infection and vaccination. Arch Biochem Biophys. 2019;671:196–202. doi: 10.1016/j.abb.2019.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y, van Boxel-Dezaire AH, Cheon H, Yang J, Stark GR. STAT3 activation in response to IL-6 is prolonged by the binding of IL-6 receptor to EGF receptor. Proc Natl Acad Sci USA. 2013;1110:16975–80. doi: 10.1073/pnas.1315862110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stuber F, Wrigge H, Schroeder S, Wetegrove S, Zinserling J, Hoeft A, et al. Kinetic and reversibility of mechanical ventilation-associated pulmonary and systemic inflammatory response in patients with acute lung injury. Intensive Care Med. 2002;28:834–41. doi: 10.1007/s00134-002-1321-7. [DOI] [PubMed] [Google Scholar]
- 41.Rojas JM, Avia M, Martin V, Sevilla N. IL-10: A multifunctional cytokine in viral infections? J Immunol Res. 2017;2017:6104054. doi: 10.1155/2017/6104054. doi: 10.1155/2017/6104054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Diamanti AP, Rosado MM, Pioli C, Sesti G, Lagana B. Cytokine release syndrome in COVID-19 patients, a new scenario for an old concern: The fragile balance between infections and autoimmunity. Int J Mol Sci. 2020;21:3330. doi: 10.3390/ijms21093330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oliver JC, Bland LA, Oettinger CW, Arduino MJ, McAllister SK, Aguero MS, et al. Cytokine kinetic in an in vitro whole blood model following an endotoxin challenge. Lymphok Cytok Res. 1993;12:115–20. [PubMed] [Google Scholar]
- 44.Haga S, Yamamoto N, Nakai-Murakami C, Osawa Y, Tokunaga K, Sata T, et al. Modulation of TNF-α-converting enzyme by the spike protein of SARS-CoV and ACE2 induces TNF-production and facilitates viral entry. Proc Natl Acad Sci USA. 2008;105:7809–14. doi: 10.1073/pnas.0711241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang M, Guo Y, Luo Q, Huang Z, Zhao R, Liu SY, et al. T-cell subset counts in peripheral blood can be used as discriminatory for diagnosis and severity prediction of coronavirus disease 2019. J Infect Dis. 2020;222:198–202. doi: 10.1093/infdis/jiaa252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim DB, Pretorius E. Serum ferritin is an important inflammatory disease marker, as it is mainly a leakage product from damaged cells. Metallomics. 2014;6:748–73. doi: 10.1039/c3mt00347g. [DOI] [PubMed] [Google Scholar]
- 47.Lin Z, Long Fei, Yang Y, Chen X, Xu L, Yang M. Serum ferritin as an independent risk factor for severity in COVID-19 patients. Letters to the Editor. J Infect. 2020;81:647–79. doi: 10.1016/j.jinf.2020.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang W, Knovich MA, Coffman LG, Torti FM, Torti SV. Serum ferritin: Past, present and future. Biochim Biophys Acta. 2010;1800:760–9. doi: 10.1016/j.bbagen.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Müller B, Becker KL, Schächinger H, Rickenbacher PR, Huber PR, Zimmerli W, et al. Calcitonin precursors are reliable markers of sepsis in a medical intensive care unit. Crit Care Med. 2000;28:977–83. doi: 10.1097/00003246-200004000-00011. [DOI] [PubMed] [Google Scholar]
- 50.Wang R, Pan M, Zhang X, Han M, Fan X, Zhao F, et al. Epidemiological and clinical features of 125 Hospitalized Patients with COVID-19 in Fuyang, Anhui, China. Int J Infect Dis. 2020;95:421–8. doi: 10.1016/j.ijid.2020.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]