Abstract
Background
Identification of infected healthcare workers (HCWs) is an important step in severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) transmission control. Rapid antigen tests (RATs) are considered an important addition to molecular tests in diagnosing coronavirus disease 2019 (COVID-19), mainly because of their fast turnaround time, easier analytical procedure and lower price. However, real-life studies on the usefulness of such testing for screening of HCWs are limited.
Methods
Physicians, nurses and hospital attendants currently working at the University Clinic of Respiratory and Allergic Diseases Golnik were invited to participate in the pilot study. Nasopharyngeal swabs were obtained three times per week for two consecutive weeks and tested with a point-of-care RAT and reverse transcription polymerase chain reaction (RT-PCR). Serum samples were obtained at the beginning of the study and 2 weeks after the last swab was collected to evaluate the serological status.
Results
A total of 191 nasopharyngeal swabs from 36 HCWs were obtained. None of the samples tested was positive for the presence of SARS-CoV-2 antigen, whereas two HCWs tested positive on RT-PCR. Of these, one HCW had a newly identified SARS-CoV-2 infection, whereas RT-PCR probably detected a previous but recent infection in the other HCW.
Conclusion
Based on the results of this pilot study, it is unlikely that RAT will reliably detect novel SARS-CoV-2 infections among asymptomatic HCWs despite serial sampling. Although RT-PCR-based screening of HCWs may not be feasible due to high sample volume, molecular methods may identify SARS-CoV-2-infected HCWs already during the presymptomatic stage.
Trial registration number NCT04716088, 19.1.2021, retrospectively registered.
Keywords: SARS-CoV-2, Healthcare workers, Rapid antigen test, RT-PCR, Screening
To the editor,
Healthcare workers (HCW) represent a potential source of infection for the patients at non-coronavirus disease 2019 (COVID-19) hospital wards, so many hospitals use regular testing for their employees. A testing method used for SARS-CoV-2 surveillance regimen needs to return results quickly and should be sufficiently inexpensive and easy to perform to allow frequent testing (e.g. multiple times per week) [1]. Besides, a test should detect SARS-CoV-2 infection as early as possible, preferably in the presymptomatic stage of infection and in asymptomatic carriers. Rapid antigen tests (RATs) have lower sensitivity compared to reverse transcription real-time polymerase chain reaction (RT-PCR) tests but have rapid test turnaround time [2]. In contrast, because seroconversion in SARS-CoV-2-infected patients occurs between 7 and 14 days after symptom onset, antibody testing is not useful for screening or diagnosis of current SARS-CoV-2 infection but may aid in clarifying possible past infection [3]. Data on the performance of antigen tests for detecting SARS-CoV-2 in healthcare settings are limited. To the best of our knowledge, this is the first study to assess the usefulness of RATs for detection of SARS-CoV-2 among HCWs in Slovenia.
Physicians, nurses and hospital attendants currently working at the University Clinic of Respiratory and Allergic Diseases Golnik were invited to enrol in the pilot study. HCWs directly involved in COVID-19 patient care and those who had a previous laboratory-confirmed SARS-CoV-2 infection were not eligible for inclusion in the study. For each participating HCW, nasopharyngeal swabs were obtained. Swabs were collected on Mondays, Wednesdays and Fridays for two consecutive weeks with a maximum of six samplings per person scheduled. Recruitment began on 18 and 20 November 2020, and the last scheduled swab was obtained on 30 November and 2 December 2020, respectively.
Swabs were inserted into universal transport medium (UTM) and tested with SARS-CoV-2 rapid antigen test (Roche Diagnostics GmbH, Mannheim, Germany), following the manufacturer’s instructions. Briefly, samples were thoroughly vortexed and 350 μL of UTM was mixed with the extraction buffer included in the testing kit. Three drops of the mixture were applied to the specimen well of the lateral flow test device, and results were visually read after a 15-min incubation at room temperature.
Nasopharyngeal swab samples were subsequently tested for the presence of SARS-CoV-2 RNA without intermitting freeze–thaw cycle. Viral RNA was isolated from the specimens using the QIAamp Viral RNA Mini Kit (QIAGEN GmbH, Hilden, Germany). PCR amplification was set up on CFX96 Touch Real-Time PCR Detection System (Bio-Rad Laboratories—Dubai Branch, Dubai, United Arab Emirates), and a commercially available PCR Kit Allplex™2019-nCoV (Seegene, Seoul, South Korea) was used. The allplex™2019-nCoV assay enables simultaneous detection and identification of three target genes, specific for SARS-CoV-2 (e.g. the E, RdRp and N gene). Samples were considered positive for SARS-CoV-2 only if all three targeted genes were amplified.
Besides, serum samples were collected at the beginning of the study and 14 days after the last nasopharyngeal swab was obtained. The presence of antibodies against SARS-CoV-2 was evaluated using IDK anti-SARS-CoV-2 IgM and IgG ELISA Kit (Immundiagnostik AG, Bensheim, Germany), following the manufacturer’s instructions.
The study was conducted in accordance with the Code of Ethics of the World Medical Association (Declaration of Helsinki) and was approved by the National Medical Ethics Committee at the Slovenian Ministry of Health of the Republic of Slovenia (consent number: 0120-542/2020/4).
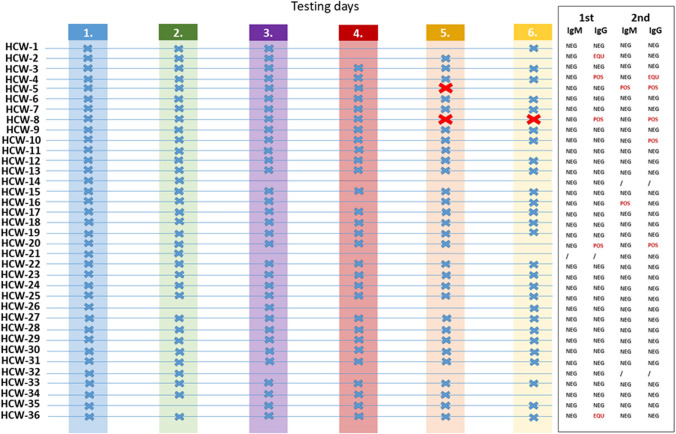
A total of 191 nasopharyngeal swabs from 36 HCWs were obtained. More than half of participating HCWs (24/36; 66.7%) provided nasopharyngeal swab samples on all 6 testing days, and only three HCWs (8.3%) provided less than half of the predicted swab samples (Fig. 1).
Fig. 1.
Nasopharyngeal swab and serology testing. Days on which swabs were obtained for an individual healthcare worker (HCW) are marked with a blue X. All samples tested using rapid antigen test (RAT) were negative. Samples that were RT-PCR positive are marked with a red X. Serology was performed on the day of the first swab and two weeks after the last swab: POS = positive, NEG = negative, EQU = equivocal
All antigen tests performed had a visible control line and thus yielded valid results. None of the samples tested was positive for the presence of SARS-CoV-2 antigen, whereas three samples tested positive for the presence of SARS-CoV-2 RNA (Fig. 1). At the beginning of the study, serum samples were obtained for 35/36 (97.2%) of HCWs, whereas follow-up serology was available for 34/36 (94.4%) of participating HCWs (Fig. 1). At the beginning of the study, none of the HCWs had detectable anti-SARS-CoV-2 IgM antibodies, whereas one HCW (2.6%) had equivocal and three HCWs (8.6%) positive results for anti-SARS-CoV-2 IgG antibodies, suggesting possible previous asymptomatic infection with SARS-CoV-2.
HCW-5 was RT-PCR positive on the 5th sampling day while being asymptomatic. She developed symptoms consistent with SARS-CoV-2 infection the following day, and infection was confirmed with an RT-PCR-positive result on that day. Because the results of the RT-PCR test were not available until the end of that day work shift, the HCW was not removed from the work process during the presymptomatic phase of infection. Seroconversion for both anti-SARS-CoV-2 IgM and IgG was observed on follow-up serology testing. A negative RAT, in this case, can be explained by the fact that the RT-PCR Ct values were > 30, which is consistent with previous reports suggesting antigen tests are less sensitive than RT-PCR and may reliably detect infections only when the viral load is high (e.g. RT-PCR Ct values < 25) [4]. Accordingly, the performance of Roche SARS-CoV-2 RAT progressively declines as RT-PCR Ct values increase, with sensitivity rates in samples with Ct values between 30 and < 37 dropping to only 12–18% [5].
On the other hand, while initially negative on four consecutive testing days by both RAT and RT-PCR, HCW-8 tested positive for the presence of SARS-CoV-2 RNA on the 5th and 6th testing day. HCW-8 had detectable anti-SARS-CoV-2 IgG at the beginning of the study. The Ct values for the SARS-CoV-2 E, RdRp and N genes were > 35, so it is highly likely that a so-called RNA-positive tail was detected and that this HCW was identified after the infectious period has passed [1]. RT-PCR-based assays may detect SARS-CoV-2 RNA for weeks to even months after acute infection [6–8]. Moreover, a large study on 29,686 samples has shown that several outcomes are possible in case of repeating molecular testing, including prolonged viral RNA shedding, false-negative results and alternating positive and negative results [8].
Interestingly, HCW-10 fell ill with COVID-19 shortly after completion of the pilot study; however, second serology testing only showed the presence of IgG, whereas IgM antibodies were non-reactive. HCW-16 was asymptomatic during the study period and had several negative RT-PCR tests after completion of the study on 30 November 2020, but tested positive on 10 January 2021, indicating that the IgM antibodies detected on second serology testing might be false positive (Fig. 1). Although serology testing is considered highly specific, as confirmed by a recent meta-analysis [9], IgM tends to exhibit somewhat lower specificity compared to IgG in COVID-19 patients (0.98 [95% CI, 0.96–0.99] versus 0.95 [95% CI, 0.91–0.98]).
Our study had several limitations, including small sample size and short duration of the observation period.
In conclusion, careful interpretation of both RAT, as well as confirmatory RT-PCR test results, is needed for accurate identification of SARS-CoV-2-infected HCWs during screening. Even serial antigen testing might miss infected persons in the early stages of infection. On the other hand, RT-PCR tests are time-consuming and results may not be available early enough to allow for possible removal of HCW from the work process during the presymptomatic phase of infection. Moreover, regular testing (e.g. several times per week) with nasopharyngeal swabs may be too bothersome for HCWs. The usefulness of alternative and less invasive sampling methods, such as saliva, mouth rinse, gargle lavage or anterior nasal swab, should be further evaluated for this purpose [10–13].
Acknowledgements
We would like to thank Gregor Ostanek for his help with the coordination of the study. Thanks to Maruša Ravnik, BSc, Dane Lužnik, PhD, and Vesna Špendal for performing RT-PCR tests.
Author contributions
AS implemented the study design, performed nasopharyngeal swabs and RAT and wrote the manuscript. VT was responsible for PCR testing and contributed to the interpretation of the data. UBS was responsible for serologic testing and contributed to the interpretation of the data. KV enrolled HCWs and coordinated the testing facility. AR supported the study design setup and the interpretation of the data. MZ designed the study and wrote the manuscript. All authors reviewed the manuscript.
Funding
This study was funded by University Clinic of Respiratory and Allergic Diseases Golnik, Golnik, Slovenia, internal funds.
Data availability
All data are available upon a request to the corresponding author.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Consent to participate and to publish
Informed consent was obtained from all individual participants included in the study.
Ethics approval
The study was conducted in accordance with the Code of Ethics of the World Medical Association (Declaration of Helsinki) and was approved by the National Medical Ethics Committee at the Slovenian Ministry of Health of the Republic of Slovenia (consent number: 0120-542/2020/4).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mina MJ, Parker R, Larremore DB. Rethinking Covid-19 test sensitivity: a strategy for containment. N Engl J Med. 2020;383:e120. doi: 10.1056/NEJMp2025631. [DOI] [PubMed] [Google Scholar]
- 2.European Centre for Disease Prevention and Control. Options for the use of rapid antigen tests for COVID-19 in the EU/EEA and the UK. ECDC: Stockholm; 2020. https://www.ecdc.europa.eu/en/publications-data/options-use-rapid-antigen-tests-covid-19-eueea-and-uk#no-link. Accessed 24 Dec 2020.
- 3.Vabret N, Britton GJ, Gruber C, et al. Immunology of COVID-19: current state of the science. Immunity. 2020;52:910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.European Centre for Disease Prevention and Control. Options for the use of rapid antigen tests for COVID-19 in the EU/EEA and the UK. 19 November 2020. ECDC: Stockholm; 2020.
- 5.Salvagno GL, Gianfilippi G, Bragantini D, et al. Clinical assessment of the Roche SARS-CoV-2 rapid antigen test. Diagnosis. 2021 doi: 10.1515/dx-2020-0154. [DOI] [PubMed] [Google Scholar]
- 6.Zhou B, She J, Wang Y, Ma X. Duration of viral shedding of discharged patients with severe COVID-19. Clin Infect Dis. 2020;71:2240–2242. doi: 10.1093/cid/ciaa451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noh JY, Yoon JG, Seong H, et al. Asymptomatic infection and atypical manifestations of COVID-19: Comparison of viral shedding duration. J Infect. 2020;81:816–846. doi: 10.1016/j.jinf.2020.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gniazdowski V, Morris CP, Wohl S, et al. Repeat COVID-19 molecular testing: correlation of SARS-CoV-2 culture with molecular assays and cycle thresholds. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang H, Ai J, Loeffelholz MJ, et al. Meta-analysis of diagnostic performance of serology tests for COVID-19: impact of assay design and post-symptom-onset intervals. Emerg Microbes Infect. 2020;9:2200–2211. doi: 10.1080/22221751.2020.1826362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Azzi L, Carcano G, Gianfagna F, Grossi P, Gasperina DD, Genoni A, Fasano M, Sessa F, Tettamanti L, Carinci F, Maurino V, Rossi A, Tagliabue A, Baj A. Saliva is a reliable tool to detect SARS-CoV-2. J Infect. 2020;81(1):e45–e50. doi: 10.1016/j.jinf.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maricic T, Nickel O, Aximu-Petri A, Essel E, Gansauge M, Kanis P, Macak D, Richter J, Riesenberg S, Bokelmann L, Zeberg H, Meyer M, Borte S, Pääbo S. A direct RT-qPCR approach to test large numbers of individuals for SARS-CoV-2. PLoS One. 2020;15(12):e0244824. doi: 10.1371/journal.pone.0244824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malecki M, Lüsebrink J, Teves S, Wendel AF. Pharynx gargle samples are suitable for SARS-CoV-2 diagnostic use and save personal protective equipment and swabs. Infect Control Hosp Epidemiol. 2020 doi: 10.1017/ice.2020.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindner AK, Nikolai O, Kausch F, et al. Head-to-head comparison of SARS-CoV-2 antigen-detecting rapid test with self-collected anterior nasal swab versus professional-collected nasopharyngeal swab. Eur Respir J. 2020 doi: 10.1183/13993003.03961-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are available upon a request to the corresponding author.