Abstract
Purpose
Coronavirus disease 2019 (COVID-19) caused by severe acute respiratory coronavirus 2 (SARS-CoV-2) has spread around the world. Differentiation between pure viral COVID-19 pneumonia and secondary infection can be challenging. In patients with elevated C-reactive protein (CRP) on admission physicians often decide to prescribe antibiotic therapy. However, overuse of anti-infective therapy in the pandemic should be avoided to prevent increasing antimicrobial resistance. Procalcitonin (PCT) and CRP have proven useful in other lower respiratory tract infections and might help to differentiate between pure viral or secondary infection.
Methods
We performed a retrospective study of patients admitted with COVID-19 between 6th March and 30th October 2020. Patient background, clinical course, laboratory findings with focus on PCT and CRP levels and microbiology results were evaluated. Patients with and without secondary bacterial infection in relation to PCT and CRP were compared. Using receiver operating characteristic (ROC) analysis, the best discriminating cut-off value of PCT and CRP with the corresponding sensitivity and specificity was calculated.
Results
Out of 99 inpatients (52 ICU, 47 Non-ICU) with COVID-19, 32 (32%) presented with secondary bacterial infection during hospitalization. Patients with secondary bacterial infection had higher PCT (0.4 versus 0.1 ng/mL; p = 0.016) and CRP (131 versus 73 mg/L; p = 0.001) levels at admission and during the hospital stay (2.9 versus 0.1 ng/mL; p < 0.001 resp. 293 versus 94 mg/L; p < 0.001). The majority of patients on general ward had no secondary bacterial infection (93%). More than half of patients admitted to the ICU developed secondary bacterial infection (56%). ROC analysis of highest PCT resp. CRP and secondary infection yielded AUCs of 0.88 (p < 0.001) resp. 0.86 (p < 0.001) for the entire cohort. With a PCT cut-off value at 0.55 ng/mL, the sensitivity was 91% with a specificity of 81%; a CRP cut-off value at 172 mg/L yielded a sensitivity of 81% with a specificity of 76%.
Conclusion
PCT and CRP measurement on admission and during the course of the disease in patients with COVID-19 may be helpful in identifying secondary bacterial infections and guiding the use of antibiotic therapy.
Keywords: COVID-19, C-reactive protein, Procalcitonin, Antimicrobial stewardship, Secondary bacterial infections
Introduction
The novel beta-coronavirus SARS-CoV-2 caused a major outbreak of respiratory illness starting in Wuhan, China at the end of 2019. In February 2020, World Health Organization (WHO) named the novel coronavirus disease as COVID-19 [1]. Until 15th December 2020, more than 76 million cases and over 1.6 million deaths due to COVID-19 have been confirmed worldwide. Although the vast majority of people suffered from mild or uncomplicated illness, severe disease requiring hospitalization is also seen in a subset of patients [2].
The knowledge about symptoms, clinical course, and risk factors of COVID-19 disease is increasing [3–5], but data on co-infections and the use of antibiotic therapy are limited. Most studies did not address secondary infections [6]. In patients with mild COVID-19, bacterial co-infections are rare, but in severe disease, co-infections have been reported in up to 50% of the affected patients [6–9]. The prevalence of confirmed co-infections in patients with COVID-19 on general ward was 3.5% in a series from the US (n = 1705) [10], 4% in a systematic review and meta-analysis from the UK (3834 patients) [7] and 8% from another review from the UK (n = 806) [6]. Rates of co-infections in ICU patients varied from 14 to 50% [7, 9]. Despite the overall low rates of confirmed secondary bacterial infections, the vast majority (57–86%) of COVID-19 patients received empirical antibiotic therapy [6, 8, 10].
Serum procalcitonin (PCT) may be useful in identifying secondary infections in patients with COVID-19. In isolated COVID-19, as in other viral infections, PCT levels usually remain normal. The lack of a PCT rise in viral infections may be due to virus-stimulated production of interferon-γ by macrophages, which inhibits TNF-α in the immune response [11]. PCT has emerged as a promising tool to facilitate decisions about antibiotic therapy in lower respiratory tract infections [12–14].
An association of PCT levels with bacterial co-infections in hospitalized patients with COVID-19 has been demonstrated in a few studies [3]. Van Berkel et al. found that a PCT level of < 0.25 µg/L had a negative predictive value of 81% and a PCT > 1 µg/L had a positive predictive value of 93% for bacterial co-infection [9]. The authors concluded that antibiotic therapy can be safely withheld in patients with COVID-19 and low PCT levels on the ICU.
CRP is usually increased on presentation in patients with COVID-19 [9]. As CRP is consistently elevated, this biomarker might not have predictive value for bacterial infections in the initial phase of COVID-19. Serial CRP with an in- or decrease over time may help to identify or rule out nosocomial bacterial infections and prompt appropriate use of antibiotic therapy [9]. Patients with severe disease had also higher CRP levels than those with non-severe disease [2].
Our institution implemented a COVID-19 standard operation procedure (SOP) in March 2020 for diagnostic measures and antimicrobial treatment. The collection of two sets of blood cultures and urinary antigen test for Legionella pneumophila and Streptococcus pneumoniae in each patient with confirmed COVID-19 disease was part of the routine diagnostic workup. In case of suspicion of secondary bacterial or fungal infection a bronchoscopy with broncho alveolar lavage (BAL) or tracheal secret and urine sampling were supplemented. In non-ICU patients with PCT levels < 0.5 ng/mL, antibiotic therapy was not recommended. For ICU patients no standard recommendations regarding antibiotic therapy were made.
In the present study, we investigated the diagnostic value of PCT and CRP to detect secondary bacterial infections in patients with COVID-19.
Materials and methods
Study design and participants
This was a retrospective single centre study which has been approved by the internal review board (No. 9491_BO_K_2020). All hospitalized patients aged 18 years and older with confirmed Covid-19 pneumonia, admitted between 6th March and 30th October 2020 to a large university hospital, located in Hannover (Hannover Medical School, MHH), Germany, were included in the study (Fig. 1). The final date of follow up was 15th December 2020. Patient characteristics including baseline demographics and chronic comorbidities were obtained. PCT and CRP levels were obtained on admission and on every second day on general ward and daily on the ICU. Data were recorded according to good clinical practice (GCP) guidelines including patient demographics, comorbidities, clinical parameters, laboratory findings, microbiology, outcome, antibiotic, antifungal and antiviral regime. Laboratory confirmation of SARS-CoV-2 was achieved by reverse transcription polymerase chain reaction (RT-PCR). All patients underwent radiological chest examination to confirm pneumonia. Patients without PCT or CRP results were excluded from the study as patients without evidence for pneumonia on their chest radiograph. Urine samples were used for lateral flow antigen test detection of L. pneumophila serogroup 1 and S. pneumoniae (AlereBinaxNOW, Abbott, Wiesbaden, Germany). For microbiological workup, blood was inoculated into aerobic and anaerobic Blood culture (BC) media via an automated BC system (Bactec, Becton Dickinson GmbH, Heidelberg, Germany). Culture bottles were incubated for 5–7 days according to the manufacturer’s recommendations. Considering the difficulty in determining the clinical significance of coagulase-negative staphylococci (CoNS) in BC, these isolates were reviewed separately based on the number of positive culture sets, the presence of intravascular devices, and patient’s characteristics. Isolates were considered clinically significant (true bacteremia) if two or more bottles yielded the same CoNS. Respiratory samples (tracheal secret or BAL) for microbiological tests were obtained and cultures were performed on Columbia sheep blood agar, chocolate agar, and sabouraud dextrose agar (Becton Dickinson GmbH, Heidelberg, Germany). In the case of a clinically suspected infection by Legionella, a buffered charcoal yeast extract (BCYE) agar was used in addition (Becton Dickinson GmbH, Heidelberg, Germany).
Fig. 1.
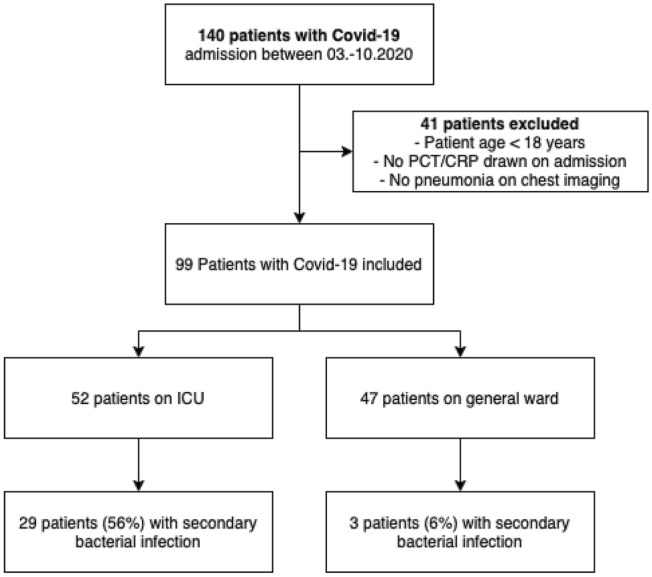
Flow diagram of study cohort
The primary outcome of this study was secondary bacterial infections (yes or no) and associated PCT and CRP levels. Secondary bacterial infection was defined as “any infectious episode” evidenced by the presence of a bacterial pathogen in positive blood cultures, respiratory samples or urine. A specialist in infective medicine not involved in the patient’s care classified the clinical relevance of the detected pathogen. In case of multiple secondary bacterial infections, only the first infectious episode was analysed. The patients were divided into two groups according to the presence or absence of secondary bacterial infection. Disease severity was classified based on the place of treatment (ICU/Non-ICU) and the WHO Ordinal Scale for Clinical Improvement [15].
Statistical analysis
IBM SPSS Statistics (version 27.0, IBM Corp., Armonk, New York) and Stata 13.0 (State Corp LP, College Station, Texas, USA) statistical software programs were used for statistical analysis. Categorical data were presented as numbers and percentages, and for group comparisons, Chi-square, or Fisher’s exact test were used as appropriate. Continuous variables were presented as medians with the first and third quartile (Q1 and Q3), and for group comparisons two-sided t test or Mann–Whitney U test were used as appropriate. Receiver operating characteristic (ROC) analysis was performed to assess the predictive performance of PCT resp. CRP and secondary bacterial infection and to calculate the AUC. Using ROC curve analysis with calculation of the area under curve (AUC), the best cut-off value of PCT and CRP with the corresponding sensitivity and specificity were calculated. All tests were two-sided; a p value of ≤ 0.05 was considered statistically significant [16].
Results
Demographic, clinical information and laboratory findings
During the study period, 140 confirmed COVID-19 patients were evaluated for the study. After exclusion of patients < 18 years, no PCT/CRP on admission or no pneumonic infiltrates on chest imaging, 99 patients were eligible for this analysis (Fig. 1), of whom 52 were treated on ICU and 47 on general ward. COVID-19 was diagnosed by RT-PCR in all patients. The median age of the cohort was 57 years (range 18–91), and 72 (73%) patients were male. Comorbidities comprised of obesity (BMI ≥ 30), any history of cardiac disease, pulmonary disease, hypertension, diabetes mellitus, renal disease, liver disease, any kind of immunosuppression, solid organ transplantation, and malignancies. Arterial hypertension, diabetes, and coronary heart disease were the most frequent comorbidities. Only a minority of 16% had no comorbidities.
The median interval between symptom onset and hospital admission was 7 days. PCT and CRP levels on admission and during the hospital stay were higher in ICU patients. During hospitalization, 19 patients died. Detailed characteristics of the subjects and laboratory findings are shown in Table 1.
Table 1.
Patients characteristics and laboratory findings of 99 patients with COVID-19 pneumonia
| Total cohort (n = 100) | ICU COVID-19 patients (n = 52) | Non-ICU COVID-19 patients (n = 47) | |
|---|---|---|---|
| Median age (range) | 57 (18–91) | 55 (18–82) | 58 (18–91) |
| Male sex | 72 (73%) | 45 (87%) | 27 (57%) |
| Median WHO scale | 5 (3–8) | 7 (5–8) | 3 (3–8) |
| Comorbidities | |||
| No comorbidities as listed below | 16 (16.2%) | 5 (9.6%) | 11 (23.4%) |
| Obesity | 10 (10.1%) | 8 (15.4%) | 2 (4.3%) |
| Arterial Hypertension | 39 (29.4%) | 23 (44.2%) | 16 (34%) |
| Diabetes | 19 (19.2%) | 15 (28.8%) | 4 (8.5%) |
| Coronary heart disease | 17 (17.2%) | 7 (13.5%) | 10 (21.3%) |
| Congestive heart failure | 3 (3%) | 2 (3.8%) | 1 (2.1%) |
| COPD | 8 (8.1%) | 1 (2.1%) | 8 (8.1%) |
| Asthma | 2 (2%) | 1 (1.9%) | 1 (2.1%) |
| Chronic kidney disease | 3 (3%) | 1 (2.1%) | 2 (3.8%) |
| Cancer | 8 (8.1%) | 5 (9.6%) | 3 (6.4%) |
| Immunodeficiency | 4 (4%) | 1 (1.9%) | 3 (6.4%) |
| Organ transplantation | 5 (5.1%) | 2 (3.8%) | 3 (6.4%) |
| Chronic liver disease | 4 (4%) | 2 (3.8%) | 2 (4.3%) |
| Antifungal therapy | 12 (12%) | 12 (23.1%) | 0 (0%) |
| Antiviral therapy | 32 (32%) | 24 (46.2%) | 8 (17%) |
| Antibiotic therapy | 68 (68.7%) | 49 (94%) | 19 (40.4%) |
| Median interval between symptoms and hospital admission (days) | 7 (6–9) | 7.5 (7–13.3) | 7 (5–7) |
| Secondary bacterial infection | 32 (32.3%) | 29 (55.8%) | 3 (6.4%) |
| Nosocomial/community acquired infection | 84.3%/ 15.7% | 82.8%/ 17.2% | 100%/0% |
| Laboratory findings | |||
| PCT (ng/mL) on admission | 0.2 (0.1–0.4) | 0.35 (0.2–0.9) | 0.1 (0.1–0.1) |
| Highest PCT (ng/mL) | 0.3 (0.1–2.4) | 1.75 (0.1–80) | 0.1 (0.1–0.2) |
| Day of highest PCT after dmission | 1 (1–3) | 3 (1–7.5) | 1 (1–2) |
| CRP (mg/L) on admission | 84.4 (42.7–147.2) | 122 (77.5–185,1) | 52.4 (16.8–93.5) |
| Highest CRP (mg/L) | 135 (73.8–220) | 193.5 (133.3–332.8) | 73.8 (26–126) |
| Day of highest CRP after admission | 2 (1–5) | 3 (2–9) | 1 (1–3) |
| Deceased during hospital stay | 19 (19.2%) | 15 (28.8%) | 4 (8.5%) |
Data are presented as absolute numbers and relative frequencies (n(%)) or as median (IQR)
COVID-19 coronavirus disease 2019; ICU intensive care unit; WHO World health organisation; COPD chronic obstructive pulmonary disease; CRP C-reactive protein; PCT procalcitonin
Most patients received antibiotic therapy within 24 h of admission (n = 68; 69%). More patients on ICU were treated with antibiotics than on general ward (94% versus 40%). The antibiotic regimens most commonly used in the ICU were Piperacillin/Tazobactam in 26 patients, followed by Meropenem in 22 patients and Ampicillin/Sulbactam in 14 patients. On general ward, the most common antibiotic therapy was Piperacillin/Tazobactam in eight patients, followed by Ampicillin/Sulbactam in five patients and Azithromycin in four patients. The majority of patients (n = 55; 81%) received more than one antibiotic regimen during their hospital stay. Thirty-two patients (32%) were treated with remdesivir as antiviral therapy.
Microbiologic results
Most patients received a complete microbiological workup during their hospital stay according to local SOP. Blood cultures (BC) were collected from 87 patients (88%) with a positive rate of 14% (12 patients, after exclusion of presumed contaminations). The most common bacterial pathogen in BC was coagulase-negative staphylococci (n = 5), after excluding possible contaminations. All of these were catheter associated blood stream infections. Respiratory samples (tracheal secret or BAL) for microbiological tests were obtained in 44 patients (44%). BAL samples were obtained from 79% of the patients admitted to the ICU with a positive result in 29%. The most common pathogens in respiratory samples were Escherichia coli (n = 5), Klebsiella pneumoniae (n = 4), Haemophilus influenzae (n = 4) and Staphylococcus aureus (n = 3). Four patients had clinically relevant pathogens in urine (Pseudomonas aeruginosa (2), Klebsiella pneumoniae (1) and Escherichia coli (1)). Urinary antigen tests for L. pneumophila and S. pneumoniae were performed for 74 patients (75%), with negative results in all patients. Table 2 summarizes the results of the microbiological testing from the study.
Table 2.
Results of microbiologic diagnostics
| Total cohort | ICU COVID-19 patients (n = 52) | Non-ICU COVID-19 patients (n = 47) | |
|---|---|---|---|
| Blood cultures collected | 87 (87.9%) | 51 (98.1%) | 36 (76.6%) |
| Blood cultures positive | 19 (19.2%) | 17 (32.7%) | 2 (4.3%) |
| Contamination only | 3 (3%) | 2 (3.8%) | 1 (2.1%) |
| Bronchoscopy performed | 44 (44.4%) | 41 (78.8%) | 3 (6.4%) |
| Positive result BAL or TS | 15 (15.2%) | 15 (28.8%) | 0 |
| L. pneumophila + S. pneumoniae urinary test performed; positive rate | 74 (74.7%); 0 | 45 (86.5%); 0 | 29 (61.7%); 0 |
| Clinically relevant bacterial pathogens in BAL/TS | Haemophilus influenzae (n = 4), Pseudomonas aeruginosa (n = 2), Klebsiella pneumoniae (n = 4), Acinetobacter baumannii (n = 1), Staphylococcus aureus (n = 3), Escherichia coli (n = 5), Enterobacter cloacae (n = 1), Serratia marcescens (n = 1), Aspergillus fumigatus (n = 1) | ||
| Clinically relevant bacterial pathogens in BC | Pseudomonas aeruginosa (n = 1), Klebsiella pneumoniae (n = 1), Escherichia coli (n = 3), Staphylococcus epidermidis (n = 5), Staph. hominis (n = 1), Enterococcus faecium (n = 1), Candida albicans (n = 3), Candida glabrata (n = 1) | ||
| Clinically relevant bacterial pathogens in urine | Pseudomonas aeruginosa (n = 2), Klebsiella pneumoniae (n = 1), Escherichia coli (n = 1) | ||
Data are presented as absolute numbers and relative frequencies (n(%))
BAL broncho alveolar lavage; TS tracheal secret; BC blood culture
Analysis of secondary bacterial infections
Around one-third of patients (32%) developed secondary bacterial infection during hospitalization. Patients with secondary bacterial infection had higher PCT and CRP levels on admission and during their hospital stay. Details are shown in Table 3. The time of highest PCT and CRP levels correspond with the diagnosis of secondary bacterial infection.
Table 3.
Procalcitonin and CRP levels of COVID-19 patients who did and did not develop a secondary bacterial infection
| Secondary infection | No secondary infection | p value | |
|---|---|---|---|
| All patients | 32 (32.3%) | 67 (67.7%) | |
| PCT (ng/mL) on admission | 0.4 (0.1–1.1) | 0.1 (0.1–0.2) | 0.016 |
| PCT (ng/mL) highest value | 2.9 (0.9–15.8) | 0.1 (0.1–0.4) | < 0.001 |
| Day of highest PCT | 4.5 (1–10.8) | 1 (1–2) | < 0.001 |
| Rise/Fall of PCT (ng/mL) per day | 0.2 (0–1.1) | 0 (0–0.03) | 0.011 |
| CRP (mg/L) on admission | 130.6 (68.8–186.65) | 73.4 (31.2–119.5) | 0.001 |
| CRP (mg/L) highest value | 292.5 (183.5–341.8) | 93.9 (50–171) | < 0.001 |
| Day of highest CRP | 6 (2.3–11.8) | 2 (1–3) | < 0.001 |
| Rise/Fall of CRP (mg/L) per day | 15.2 (4.1–28.6) | 2.8 (0–10) | 0.002 |
| Deceased during hospital stay | 11 (34.4%) | 8 (11.9%) | 0.008 |
Data are presented as absolute numbers and relative frequencies (n(%)) or as median (IQR)
COVID-19 coronavirus disease 2019; CRP C-reactive protein; PCT procalcitonin
Only three patients (6.3%) with COVID-19 admitted to general wards had secondary bacterial infection. All other non-ICU patients had no secondary bacterial infections (n = 45; 93.8%). In 41 of these patients, PCT levels on admission were < 0.55 ng/mL while CRP was elevated (70.7 mg/L, IQR 2.4–96.2). Of note, 40% of these patients received empirical antibiotic therapy.
Half of ICU patients had secondary bacterial infection (55.8%). In all of these patients, PCT levels on highest value were > 0.55 ng/mL. Regarding ICU patients without secondary bacterial infection median PCT levels were 0.5 (IQR 0.3–2.4) while CRP was considerably elevated (155.8, IQR 78.8–204.6).
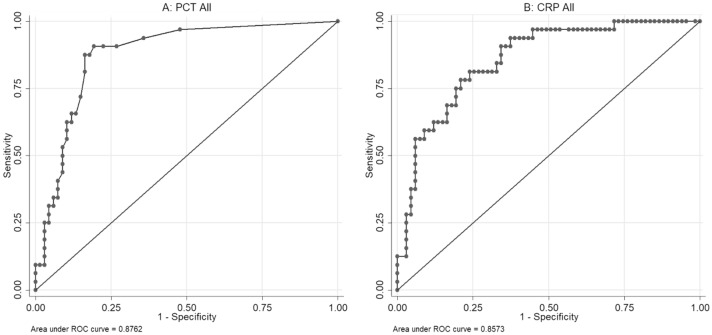
ROC analysis of PCT yielded AUCs of 0.88 (p < 0.001) for all patients (Fig. 2a). At a cut-off value of 0.55 ng/mL, PCT had a sensitivity of 91% and a specificity of 81% for the detection of secondary bacterial infection. In patients with PCT < 0.55 ng/mL, the negative predictive value was 94%, whereas PCT levels of > 0.55 ng/mL had a positive predictive value of 69%.
Fig. 2.
ROC curve analysis for highest PCT level (a) and CRP level (b) as a marker for secondary infection in inpatients with COVID-19 pneumonia: Analysis revealed an area under the curve of 0.88 (p < 0.001) for PCT and 0.86 (p < 0.001) for CRP
ROC analysis of CRP yielded AUCs of 0.86 (p < 0.001) for all patients (Fig. 2b). At a cut-off of 172 mg/L (AUC of 86% (95% CI 78–93%), CRP had a sensitivity of 81% and a specificity of 76% for the detection of secondary bacterial infection. In patients with CRP < 172 mg/L, the negative predictive value was 90%, whereas CRP levels of > 172 mg/L had a positive predictive value of 62%.
Discussion
The present study showed that PCT levels were normal in most patients with COVID-19 unless secondary bacterial infection was present. PCT levels < 0.55 ng/mL ruled out secondary bacterial infections with a negative predictive value of 94%. For CRP levels, the cut-off value was considerably elevated with 172 mg/L and predictive values slightly less robust. Inferring from this, PCT and CRP measurement may help identifying patients with secondary bacterial infections and allow a targeted use of antimicrobials thus promoting antibiotic stewardship.
Bacterial co-infections are uncommon in patients with mild COVID-19. However, secondary bacterial infections occur in an appreciable number of critically ill, hospitalized patients, since risk factors for nosocomial infections such as prolonged mechanical ventilation are prominent features of severe disease [17, 18]. Bacterial pneumonia, especially ventilator-associated pneumonia, is the most common secondary bacterial infection, but patients with severe COVID-19 are also susceptible to urinary tract and bloodstream infections [19].
On the other hand, many patients with COVID-19 have no secondary bacterial infection and don’t require antibiotic therapy. In a report from 552 hospitals in 30 Chinese provinces, 58% of patients were treated with antibiotics despite a low number of bacterial infections [2]. Antibiotics were administered to 80–100% of critically ill COVID-19 patients in Chinese ICUs [17, 20–22]. Based on past experience, clinicians often decide to take more antibiotic treatment for patients with severe illness. Empirical use of antibiotics is common but in the absence of bacterial co-infection, antibacterial therapy has no known benefit in patients with COVID-19 [10].
The presence of lower PCT levels has been shown to have a 94% negative predictive value for bacterial co-infection in intensive care unit patients with confirmed influenza A(H1N1)pdm09 [23]. Some biomarker studies have been conducted in hospitalized patients with COVID-19. Initial reports from China have shown that most patients with COVID-19 did not have elevated PCT (> 0.5 μg/L) [2, 17]. However, elevated levels were found more frequently in severe cases and in patients who died [2, 17, 24]. Our results showed, not surprisingly, that patients with secondary bacterial infections were more likely to have negative clinical outcomes than those with no evidence of bacterial infection. Even if CRP and PCT are also elevated in systemic response to COVID-19, the cut-off values in our study are much higher than described to discriminate between mild and severe disease [25]. In line with this observation, a recent review paper suggested that about 10% of deaths associated with viral disease are attributable to secondary bacterial infections [26].
In our study, a significant increase in both PCT and CRP levels was observed in patients with secondary bacterial infection. The receiver operated curve analysis of highest PCT and CRP yielded AUCs of 0.88 and 0.86, respectively. In patients with PCT < 0.55 ng/mL, the negative predictive value was 93%. With regard to antimicrobial stewardship, initiation of empirical antibacterial therapy in patients with low PCT can probably be avoided. CRP levels were also significantly higher in patients with bacterial infection but the cut-off value between both groups was with 172 mg/L considerably elevated (< 5 mg/L) and the predictive values slightly less robust. These results are in line with a recent investigation of Van Berkel et al. [9].
The available evidence together with our findings suggest that high proportion of COVID-19 patients received unnecessary antibiotic treatment [26, 27]. This increase in antibiotic administration can cause pressure on bacterial pathogens resulting antibiotic resistance [28, 29]. Hence, a potential consequence of the COVID-19 pandemic may be an accelerated propagation of antimicrobial resistance [30]. Clinicians, hospitals, microbiology labs, and public health organizations must be vigilant in monitoring the potential impact of increased antimicrobial consumption on emergence of resistance in individual patients and at institutional and regional levels.
Our study has several limitations, including the relatively small sample size, the monocentric setting, the retrospective design, the missing validation cohort, and the exclusion of patients with COVID-19 who were not hospitalized. In addition, the diagnosis of secondary bacterial infections was based not only on microbiology results but also on clinician’s decision about the relevance of the detected pathogen; e.g., bias and misclassifications cannot be excluded. It should also be noted that not all blood cultures, respiratory and urine samples were performed prior antibiotic treatment, which might influence the detection of bacterial infection.
In conclusion, PCT measurement on admission and during the course of the stay in patients with COVID-19 may help identifying secondary bacterial infections and allow for more targeted use of antimicrobial therapy. The use of CRP to detect secondary bacterial infection is also feasible but the cut-off value is considerably elevated. As the COVID-19 pandemic proceeds, prospective studies are needed to systematically collect clinical, microbiologic, and antimicrobial resistance data on superinfections. Results from carefully designed studies can be used to inform rational antimicrobial treatment and stewardship strategies, and to develop diagnostic criteria for secondary bacterial infections.
Abbreviations
- AUC
Area under curve
- BI
Betalactamase inhibitor
- BC
Blood culture
- BAL
Broncho alveolar lavage
- CDI
Clostridioides difficile Infection
- CoNS
Coagulase-negative staphylococci
- CAP
Community-acquired pneumonia
- COVID-19
Coronavirus disease 2019
- CRP
C-reactive protein
- GCP
Good clinical practice
- ICU
Intensive care unit
- LRTI
Lower respiratory tract infections
- MHH
Medical School Hannover
- MERS-CoV
Middle East respiratory syndrome coronavirus
- POCT
Point-of-care testing
- RT-PCR
Reverse transcription polymerase chain reaction
- PCT
Procalcitonin
- ROC
Receiver operating characteristic
- SARS-CoV-2
Severe acute respiratory coronavirus 2
- SARS-CoV-1
Severe acute respiratory coronavirus 1
- SOP
Standard operation procedure
- TS
Tracheal secret
- TNF-α
Tumor necrosis factor-α
- WBC
White blood count
- WHO
World Health Organization
Author contributions
IP and JR conceived of the presented idea. IP, DR, and JR collected the data, JF was responsible for the statistical evaluation. RV, TW, and MMH contributed to the interpretation of the results. All authors discussed the results and contributed to the final manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL. Not applicable.
Availability of data and materials
Availability upon reasonable request.
Code availability
Not applicable.
Declarations
Conflict of interest
IP received grants and/or advisory/lecture/clinical trial fees from Chiesi, outside the submitted work. MMH received grants and/or advisory/lecture/clinical trial fees from Acceleron, Actelion, Bayer, GSK, Janssen, MSD and Pfizer, outside the submitted work. TW received grants and/or advisory/lecture/clinical trial fees from DFG (German Research Council), BMBF (German Ministry of Research and Education), BMG (German Ministry of Health), EU, WHO (research grants), AstraZeneca, Basilea, Biotest, Bayer, Boehringer, Berlin Chemie, GSK, Infectopharm, MSD, Novartis, Pfizer, Roche (fees for lectures), AstraZeneca, Basilea, Biotest, Bayer, Boehringer, Gilead, GSK, Janssens, Novartis, Pfizer, Roche (advisory boards), outside the submitted work. JR received grants and/or advisory/lecture/clinical trial fees and/or non-financial support from AstraZeneca, Bayer, Brahms, Chiesi, Esanum, Grifols and Novartis, outside the submitted work. DR, JF, and RV declare that they have no conflict of interest.
Ethical approval
Ethical approval was waived by the local Ethics Committee of University A in view of the retrospective nature of the study and all the procedures being performed were part of the routine care.
Consent to participate
All patients or their next of kin gave prior written consent.
Footnotes
Isabell Pink and David Raupach contributed equally.
Contributor Information
Isabell Pink, Email: Pink.Isabell@mh-hannover.de.
David Raupach, Email: Raupach.david@stud.mh-hannover.de.
Jan Fuge, Email: Fuge.Jan@mh-hannover.de.
Ralf-Peter Vonberg, Email: Vonberg.Ralf@mh-hannover.de.
Marius M. Hoeper, Email: Hoeper.marius@mh-hannover.de
Tobias Welte, Email: Welte.Tobias@mh-hannover.de.
Jessica Rademacher, Email: Rademacher.Jessica@mh-hannover.de.
References
- 1.WHO . Coronvirus disease 2019 (COVID-19) situation report. Geneva: WHO; 2020. [Google Scholar]
- 2.Guan W-J, Ni Z-Y, Hu Y, Liang W-H, Ou C-Q, He J-X, et al. Clinical characteristics of Coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Izcovich A, Ragusa MA, Tortosa F, Lavena Marzio MA, Agnoletti C, Bengolea A, et al. Prognostic factors for severity and mortality in patients infected with COVID-19: a systematic review. PLoS ONE. 2020;15:e0241955. doi: 10.1371/journal.pone.0241955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang JJY, Lee KS, Ang LW, Leo YS, Young BE. Risk factors for severe disease and efficacy of treatment in patients infected with COVID-19: a systematic review, meta-analysis, and meta-regression analysis. Clin Infect Dis Off Publ Infect Dis Soc Am. 2020;71:2199–2206. doi: 10.1093/cid/ciaa576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet Lond Engl. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rawson TM, Moore LSP, Zhu N, Ranganathan N, Skolimowska K, Gilchrist M, et al. Bacterial and fungal co-infection in individuals with coronavirus: a rapid review to support COVID-19 antimicrobial prescribing. Clin Infect Dis Off Publ Infect Dis Soc Am. 2020 doi: 10.1093/cid/ciaa530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lansbury L, Lim B, Baskaran V, Lim WS. Co-infections in people with COVID-19: a systematic review and meta-analysis. J Infect. 2020;81(2):266–275. doi: 10.1016/j.jinf.2020.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rothe K, Feihl S, Schneider J, Wallnöfer F, Wurst M, Lukas M, et al. Rates of bacterial co-infections and antimicrobial use in COVID-19 patients: a retrospective cohort study in light of antibiotic stewardship. Eur J Clin Microbiol Infect Dis Off Publ Eur Soc Clin Microbiol. 2020 doi: 10.1007/s10096-020-04063-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Berkel M, Kox M, Frenzel T, Pickkers P, Schouten J. Biomarkers for antimicrobial stewardship: a reappraisal in COVID-19 times? Crit Care Lond Engl. 2020;24:600. doi: 10.1186/s13054-020-03291-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vaughn VM, Gandhi T, Petty LA, Patel PK, Prescott HC, Malani AN, et al. Empiric antibacterial therapy and community-onset bacterial co-infection in patients hospitalized with COVID-19: a multi-hospital cohort study. Clin Infect Dis Off Publ Infect Dis Soc Am. 2020 doi: 10.1093/cid/ciaa1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Müller B, Becker KL, Schächinger H, Rickenbacher PR, Huber PR, Zimmerli W, et al. Calcitonin precursors are reliable markers of sepsis in a medical intensive care unit. Crit Care Med. 2000;28:977–983. doi: 10.1097/00003246-200004000-00011. [DOI] [PubMed] [Google Scholar]
- 12.Schuetz P, Wirz Y, Sager R, Christ-Crain M, Stolz D, Tamm M, et al. Procalcitonin to initiate or discontinue antibiotics in acute respiratory tract infections. Cochrane Database Syst Rev. 2017;10:CD007498. doi: 10.1002/14651858.CD007498.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Briel M, Schuetz P, Mueller B, Young J, Schild U, Nusbaumer C, et al. Procalcitonin-guided antibiotic use vs a standard approach for acute respiratory tract infections in primary care. Arch Intern Med. 2008;168:2000–2007. doi: 10.1001/archinte.168.18.2000. [DOI] [PubMed] [Google Scholar]
- 14.Burkhardt O, Ewig S, Haagen U, Giersdorf S, Hartmann O, Wegscheider K, et al. Procalcitonin guidance and reduction of antibiotic use in acute respiratory tract infection. Eur Respir J. 2010;36:601–607. doi: 10.1183/09031936.00163309. [DOI] [PubMed] [Google Scholar]
- 15.COVID-19_Treatment_Trial_Design_Master_Protocol_synopsis_Final_18022020.pdf. 2020. https://www.who.int/blueprint/priority-diseases/key-action/COVID-19_Treatment_Trial_Design_Master_Protocol_synopsis_Final_18022020.pdf. Accessed 16 Dec 2020.
- 16.Mehta HB, Mehta V, Girman CJ, Adhikari D, Johnson ML. Regression coefficient-based scoring system should be used to assign weights to the risk index. J Clin Epidemiol. 2016;79:22–28. doi: 10.1016/j.jclinepi.2016.03.031. [DOI] [PubMed] [Google Scholar]
- 17.Chen T, Wu D, Chen H, Yan W, Yang D, Chen G, et al. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ. 2020;368:m1091. doi: 10.1136/bmj.m1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel Coronavirus-infected pneumonia in Wuhan. China JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng K, He M, Shu Q, Wu M, Chen C, Xue Y. Analysis of the risk factors for nosocomial bacterial infection in patients with COVID-19 in a tertiary hospital. Risk Manag Healthc Policy. 2020;13:2593–2599. doi: 10.2147/RMHP.S277963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao J, Tu W-J, Cheng W, Yu L, Liu Y-K, Hu X, et al. Clinical Features and Short-term Outcomes of 102 Patients with Coronavirus Disease 2019 in Wuhan, China. Clin Infect Dis Off Publ Infect Dis Soc Am. 2020;71:748–755. doi: 10.1093/cid/ciaa243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong X, Cao Y-Y, Lu X-X, Zhang J-J, Du H, Yan Y-Q, et al. Eleven faces of coronavirus disease 2019. Allergy. 2020;75:1699–1709. doi: 10.1111/all.14289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan. China Lancet Lond Engl. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodríguez AH, Avilés-Jurado FX, Díaz E, Schuetz P, Trefler SI, Solé-Violán J, et al. Procalcitonin (PCT) levels for ruling-out bacterial coinfection in ICU patients with influenza: a CHAID decision-tree analysis. J Infect. 2016;72(2):143–151. doi: 10.1016/j.jinf.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 24.Chen G, Wu D, Guo W, Cao Y, Huang D, Wang H, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu F, Li L, Xu M, Wu J, Luo D, Zhu Y, et al. Prognostic value of interleukin-6, C-reactive protein, and procalcitonin in patients with COVID-19. J Clin Virol. 2020;127:104370. doi: 10.1016/j.jcv.2020.104370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manohar P, Loh B, Nachimuthu R, Hua X, Welburn SC, Leptihn S. Secondary Bacterial Infections in Patients With Viral Pneumonia. Front Med. 2020;7:420. doi: 10.3389/fmed.2020.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Antimicrobial resistance in the age of COVID-19. Nat Microbiol. 2020 Jun;5(6):779. 10.1038/s41564-020-0739-4. PMID: 32433531 [DOI] [PubMed]
- 28.Usman M, Farooq M, Hanna K. Environmental side effects of the injudicious use of antimicrobials in the era of COVID-19. Sci Total Environ. 2020;745:141053. doi: 10.1016/j.scitotenv.2020.141053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mirzaei R, Goodarzi P, Asadi M, Soltani A, Aljanabi HAA, Jeda AS, et al. Bacterial co-infections with SARS-CoV-2. IUBMB Life. 2020;72:2097–2111. doi: 10.1002/iub.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rawson TM, Ming D, Ahmad R, Moore LSP, Holmes AH. Antimicrobial use, drug-resistant infections and COVID-19. Nat Rev Microbiol. 2020;18:409–410. doi: 10.1038/s41579-020-0395-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Availability upon reasonable request.
Not applicable.