Summary
Background:
In early-stage HER2-positive (HER2+) breast cancer, escalation or de-escalation of systemic therapy is a controversial topic. As an aid to treatment decisions, we present a prognostic assay that integrates multiple data types for predicting survival outcome in newly diagnosed HER2+ breast cancer.
Methods:
Clinicopathological data, stromal tumour infiltrating-lymphocytes (TILs), PAM50 subtypes and expression of 55 genes were obtained from 435 patients (34·7%) who participated in the Short-HER phase III trial, which randomised patients with newly diagnosed node-positive HER2+ breast cancer or, if node negative, with at least one risk factor (tumour size > 2·0 cm, histological grade 3, lympho-vascular invasion, Ki-67>20%, age ≤35 years, or hormone receptor negativity), to adjuvant anthracycline/taxane-based combinations with either 9 weeks or 1 year of trastuzumab. Trastuzumab was administered intravenously every 3 weeks (8 mg/kg loading dose at first cycle, and 6 mg/kg thereafter) for 18 doses or weekly (4 mg/kg loading dose at first week, and 2 mg/kg thereafter) for 9 weeks, starting concomitantly with the first taxane dose. The primary objective of this study was to derive and evaluate a combined score associated with distant metastasis-free survival (DMFS). Patient samples in the training dataset were split into a training set (n=290) and a testing set (n=145), balancing for event and treatment arm. The training set was further stratified into 100 iterations of Monte-Carlo cross validation (MCCV). Cox proportional hazard models were fit to MCCV training samples using Elastic-Net. A maximum of 92 features were evaluated. The final prognostic model was evaluated in an independent combined dataset of 267 patients with early-stage HER2+ breast cancer treated with different neoadjuvant and adjuvant anti-HER2-based combinations and disease-free survival (DFS) outcome data.
Findings:
In Short-HER, tumour stage (T1 vs. rest), nodal stage (N0 vs. rest), TILs (continuous variable), subtype (HER2-enriched and Basal-like vs. rest) and 13 genes composed the final model (HER2DX). HER2DX was significantly associated with DMFS as a continuous variable (p<0·001). Two cut-offs defined low-risk (50%), med-risk (25%) and high-risk (25%) populations. The 5-year DMFS of the low-, med- and high-risk populations were 98·1% (95% CI 96·3–99·9), 88·9% (83·2–95·0) and 73·9% (66·0–82·7), respectively (hazard ratio [HR] low- vs. high-risk=0·04, 0·0–0·1, p<0·0001). In the evaluation cohort, HER2DX was significantly associated with DFS as a continuous variable (HR=2·77, 1·4–5·6, p=0·0040) and as group categories (low- vs. high-risk HR=0·27, 0·1–0·7, p=0·010). The 5- and 8-year DFS of the HER2DX low-risk group was 93·5% (89·0–98·3%) and 91·7% (86·2–97·6%), respectively.
Interpretation:
HER2DX identifies patients with early-stage HER2+ breast cancer candidates for escalated or de-escalated systemic treatment. Future clinical validation of HER2DX seems warranted.
Funding:
Instituto Salud Carlos III, Save the Mama, Pas a Pas, AECC, SEOM, NIH, Agenzia Italiana del Farmaco, IARC and Veneto Institute of Oncology.
Introduction
HER2-positive (HER2+) breast cancer is responsible for a substantial proportion of deaths1. In early stages, (neo)adjuvant chemotherapy and anti-HER2 therapy (plus endocrine therapy in hormone receptor-positive disease) have consistently shown significant and long-term clinical benefits, in terms of disease-free survival (DFS) and overall survival1. However, substantial heterogeneity exists in HER2+ disease regarding tumour biology2–6, patient’s prognosis7 and treatment benefit7.
Strategies to either escalate or de-escalate systemic therapy in early-stage HER2+ disease have been explored8, such as decreasing the amount of chemotherapy9 and the duration of trastuzumab10 or increasing HER2 blockade with pertuzumab11, neratinib12 or switching the anti-HER2 therapy to T-DM1 in patients who did not achieve a pathological complete response (pCR) following neoadjuvant trastuzumab-based chemotherapy13. Despite all these efforts to improve survival outcomes, the crude reality is that the vast majority of patients with early-stage HER2+ disease are cured with chemotherapy and trastuzumab14.
In early-stage hormone receptor-positive/HER2-negative disease, several prognostic tools allow a better individualization of systemic treatments and are widely available. For example, gene expression-based assays such as OncotypeDX help identify low-risk patients who do not need (neo)adjuvant chemotherapy. Second generation genomic tests, such as PAM50/Prosigna, which include clinical parameters such as tumour size in the final risk assessment, might better discriminate patients who may not need chemotherapy from those who are likely to benefit.
To date, variables beyond the TNM classification have been associated with prognosis in early-stage HER2+ disease. Examples are stromal tumour-infiltrating lymphocytes (TILs)14–16 and PAM50 subtypes2,16,17. Similarly, these biomarkers and PIK3CA mutations18 have been associated with the probability to achieve a pCR18,19, which is also associated with long-term outcome20. However, decisions today about escalation or de-escalation of systemic therapies are based on nodal status, hormone receptor status and therapy response21. Therefore, a multi-parameter prognostic tool that integrates variables already known, as well as additional ones, to help guide systemic therapies in early-stage HER2+ breast cancer is urgently needed. Here, we aimed to develop a prognostic tool based on multiple variables.
Methods
The combined prognostic model was derived using retrospective clinical, pathological and genomic data from a subset of patients who participated in the Short-HER trial. The final prognostic model was evaluated retrospectively in a combined and independent cohort of patients with early-stage HER2+ breast cancer.
Study designs
Short-HER was a randomized, investigator-driven phase 3 study, aimed to assess the non-inferiority in terms of DFS of 9 weeks versus 1 year of adjuvant trastuzumab combined with chemotherapy22. Briefly, women aged 18–75 with surgically resected, HER2+ breast cancer were eligible. Women had to have node positivity, or in case of node-negativity, at least one of the following features: tumour size >2 cm, grade 3, presence of lympho-vascular invasion, Ki67 > 20%, age ≤35 years or hormone receptor negativity. A total of 1,254 patients with a performance status of 0–1 were randomised from 17th December 2007 to 6th October 2013 to arm A or arm B. Chemotherapy in arm A consisted of adriamycin 60 mg/m2 or epirubicin 90 mg/m2 plus cyclophosphamide 600 mg/m2 every 3 weeks for 4 courses followed by paclitaxel 175 mg/m2 or docetaxel 100 mg/m2 every 3 weeks for 4 courses. Trastuzumab was administered every 3 weeks for 18 doses, starting with the first taxane dose. Chemotherapy in arm B consisted of docetaxel 100 mg/m2 every 3 weeks for 3 courses followed by 5-fluorouracil 600 mg/m2, epirubicin 60 mg/m2, cyclophosphamide 600 mg/m2 every 3 weeks for 3 courses. Trastuzumab was administered weekly for 9 weeks, starting concomitantly with docetaxel. When indicated, radiation and hormonal therapy were carried out according to local standard. Median follow-up was 91·4 months (IQR 75·1–105·6). In Short-HER, DMFS was an exploratory endpoint.
CHER-LOB23 was a randomised, noncomparative, investigator-driven phase 2 study from 8th August 2006 to 25th November 2010 of preoperative taxane-anthracycline consisting of paclitaxel (80 mg/m2) for 12 weeks followed by fluorouracil, epirubicin, and cyclophosphamide for 4 courses every 3 weeks, in combination with trastuzumab, lapatinib (1,500 mg daily) or combined trastuzumab plus lapatinib (1,000 mg daily) for 26 weeks in patients with HER2+, stage II to IIIA operable breast cancer and with a performance status of 0–1. The primary aim was to estimate the pCR rate. Treatment after surgery was left to treating physician discretion. Median follow-up was 60·0 months (IQR 46·9–69·4). In CHER-LOB, DFS was an exploratory endpoint.
PAMELA was a single-group, phase 2 trial from 22nd October 2013 to 30th November 2015 aimed to the ability of the PAM50 HER2-enriched subtype to predict pCR at the time of surgery19. Patients with HER2+ disease, stage I–IIIA and a performance status of 0–1 were given lapatinib (1,000 mg per day) and trastuzumab for 18 weeks; hormone receptor-positive patients were additionally given letrozole (2·5 mg per day) or tamoxifen (20 mg per day) according to menopausal status. Treatment after surgery was left to treating physician discretion. Median follow-up was 68·1 months (IQR 57·1–72·3). In PAMELA, DFS was an exploratory endpoint.
The Hospital Clinic and Padova University cohorts are consecutive series of patients with early-stage HER2+ disease and a performance status of 0–1 treated, as per standard practice, from 28th June 2005 to 26th September 2018 (Hospital Clinic) and 23rd February 2009 to 26th May 2016 (Padova University cohort), with neoadjuvant trastuzumab-based chemotherapy for 3–6 months, followed by surgery. Adjuvant treatment was completed with trastuzumab for up to 1 year. When indicated, radiation and hormonal therapy were carried out according to local standard. Median follow-up of Hospital Clinic and Padova University cohorts were 39·3 (IQR 29·6–55·8) and 38·5 (IQR 30.1–65.7) months, respectively. In both cohorts, DFS was an exploratory endpoint.
The study was performed in accordance with Good Clinical Practice guidelines and the World Medical Association Declaration of Helsinki. Approvals for the study were obtained from independent ethics committees.
Procedures
PAM50 and single gene analyses were performed at IDIBAPS from formalin-fixed paraffin-embedded tumours. Samples analysed from Short-HER were from surgical specimens, whereas samples analysed from the neoadjuvant cohorts were from baseline samples before starting neoadjuvant therapy. A minimum of ∼125 ng of total RNA was used to measure the expression of the 50 PAM50 subtype predictor genes and 5 genes (i.e. CD8A, PDL1, PD1, CD4 and AR). Normalization and PAM50 subtyping was performed as previously described19. Regarding samples from CHER-LOB, PAM50 gene expression and subtyping was obtained from PAM50-based microarray data as previously described24. Genomic analyses were performed blinded from clinical data. Nodal and tumour stages were obtained from clinical report forms. Finally, TILs were assessed according to pre-defined criteria25.
Outcomes
The primary objective of this study was to derive and evaluate a combined prognostic score, named HER2DX, as a continuous variable. In the training dataset (i.e. Short-HER), the chosen survival endpoint was DMFS, similarly as other gene expression-based prognostic biomarkers such as the PAM50 Risk of Recurrence in hormone receptor-positive/HER2-negative breast cancer. DMFS was defined as the time between randomization and distant recurrence or death before recurrence. In the evaluation dataset, the survival endpoint was DFS due to the availability of the data, calculated as the time between treatment initiation and any of the following events, whichever first: local, regional and distant recurrence; contralateral breast cancer, other second invasive primary cancer, death before recurrence or second primary cancer. For description purposes, 5- and 8-year DMFS and DFS estimates were calculated.
The secondary objectives were: 1) to describe the clinical-pathological and genomic features of the HER2DX risk groups; 2) to explore the association of HER2DX score with DFS in the evaluation dataset according to the type of pathological response; 3) to evaluate the association of HER2DX score, and other individual variables, with pCR in the breast and axilla in the evaluation dataset. We also performed an ad-hoc analysis of the association of HER2DX with DFS in Short-HER.
Statistical analysis
The prognostic model was developed using a training dataset of 435 patients (34·7%) enrolled in the Short-HER trial (webappendix p. 1). The rule to define a patient assessable in Short-HER was availability of gene expression, clinical-pathological and TILs data. Patients were split into a training set (n=290 [67·0%] patients and 42 events [14·5%]) and a testing set (n=145 [33·0%] patients and 21 events [14·5%]), balancing for distant metastasis-free survival (DMFS) event and treatment arm. The training set was further stratified into 100 iterations of Monte-Carlo cross validation (MCCV). Cox proportional hazard models were fit to MCCV training cases using Elastic-Net (package glmnet). A maximum of 92 features were evaluated. Elastic-Net parameters (alpha and lambda) were selected to reduce partial likelihood deviance and increase Harrell’s C-index evaluated in the MCCV test sets. Selected values were then used to fit our final model against the complete training set. A total of 17 variables were selected with the following survival coefficients: nodal stage 1 (0·680), tumour stage 2–4 (0·339), MMP11 (0·200), PAM50 HER2-Enriched or Basal-like (0·156), CDC6 (0·087), CDH3 (0·076), TMEM45B (0·048), EXO1 (0·024), FGFR4 (0·021), RRM2 (0·008), TILs (−0·009), MLPH (−0·022), KRT5 (−0·024), KRT14 (−0·040), MYC (−0·050), PHGDH (−0·050) and BAG1 (−0·168).
Two cut-offs based on quartiles were defined to split patients into low- (quartile 1 and 2), medium- (quartile 3) and high-risk (quartile 4) groups. The final model was tested, as a continuous variable and using the pre-specified cut-offs, in 267 patients from the evaluation dataset (webappendix p. 2). The evaluation dataset was composed of patients from CHER-LOB (n=74 [61·2%] of 121), PAMELA (n=88 [58·3%] of 151), Padova cohort (n=37) and Hospital Clinic cohort (n=68). Missing data were not included in our analyses. This study was not pre-specified in any registry.
Cox proportional hazard regression analyses were used to investigate the association of each variable with survival outcome. Genes associated with HER2DX risk groups were identified using a multi-class Significance Analysis of Microarrays and a false discovery rate <5%. Categorical variables were expressed as number (%) and compared by χ2 test or Fisher’s exact test. Logistic regression analyses were performed to investigate the association of each variable with pCR. The significance level was set to a 2-sided alpha of 0·05. The software used was R code v3.6.2.
Role of the funding source
The study was designed by investigators from Padova University and Hospital Clinic. Funding sources had no role in the design and conduct of this study, and in the analysis and interpretation of data. All authors had full access to all data and had final responsibility for the decision to submit for publication.
Results
To build a prognostic model, clinical-pathological and molecular data were available from 435 patients of the Short-HER trial (Table 1). Briefly, mean age was 55·4 (25–78) and most tumours were ≤2·0 cm (54·0%), node-negative (60·7%), hormone receptor-positive (71·0%), histological grade 3 (71·9%) and had ≤10% TILs (72·6%). Concordant with previous studies4,26, most tumours (52·9%) were PAM50 HER2-Enriched and the proportion of HER2-Enriched disease was higher in hormone receptor-negative disease (69·8%) compared to hormone receptor-positive disease (46·0%). As expected, most Luminal A/B and Basal-like subtypes were hormone receptor-positive (99·2%) and hormone receptor-negative (70%), respectively.
Table 1.
Patient baseline characteristics of the Short-HER dataset.
| All patients | HER2DX Low | HER2DX Med/High | |||||
|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | p-value* | |
| N | 435 | - | 218 | 50·1% | 217 | 49·9% | - |
| Age (mean, SD) | 55·4 (10·2) | 55·0 (10·1) | 55·7 (10·4) | 0·48 | |||
| TILs | 0·0001 | ||||||
| TILs 0–29 | 379 | 87·1% | 176 | 80·7% | 203 | 93·5% | |
| TILs ≥30 | 56 | 12·9% | 42 | 19·3% | 14 | 6·5% | |
| pT | <0·0001 | ||||||
| T1 | 235 | 54·0% | 157 | 72·0% | 78 | 35·9% | |
| T2–4 | 200 | 46·0% | 61 | 28·0% | 139 | 64·1% | |
| pN | <0·0001 | ||||||
| N0 | 264 | 60·7% | 187 | 85·8% | 77 | 35·5% | |
| N1–3 | 171 | 39·3% | 31 | 14·2% | 140 | 64·5% | |
| PIK3CA mutations | |||||||
| WT | 339 | 77·9% | 169 | 77·5% | 170 | 78·3% | 1·000 |
| MUT | 92 | 21·1% | 46 | 21·1% | 46 | 21·2% | |
| NA | 4 | 1·0% | 3 | 1·4% | 1 | 0·5% | |
| Hormone receptor status | |||||||
| Positive | 309 | 71·0% | 163 | 74·8% | 146 | 67·3% | 0·092 |
| Negative | 126 | 29·0% | 55 | 25·2% | 71 | 32·7% | |
| Treatmet arm | |||||||
| Arm A (long) | 222 | 51·0% | 114 | 52·3% | 108 | 49·8% | 0·63 |
| Arm B (short) | 213 | 49·0% | 104 | 47·7% | 109 | 50·2% | |
| Grade | 0·25 | ||||||
| Grade 1 | 6 | 1·4% | 5 | 2·3% | 1 | 0·5% | |
| Grade 2 | 115 | 26·7% | 58 | 27·0% | 57 | 26·5% | |
| Grade 3 | 309 | 71·9% | 152 | 70·7% | 157 | 73·0% | |
| PAM50 | <0·0001 | ||||||
| Luminal A | 87 | 20·0% | 63 | 28·9% | 24 | 11·1% | |
| Luminal B | 43 | 9·9% | 24 | 11·0% | 19 | 8·8% | |
| HER2-enriched | 230 | 52·9% | 75 | 34·4% | 155 | 71·4% | |
| Basal-like | 27 | 6·2% | 17 | 7·8% | 10 | 4·6% | |
| Normal-like | 48 | 11·0% | 39 | 17·9% | 9 | 4·1% | |
TILs: tumour-infiltrating lymphocytes; MUT: mutated; WT: wild-type
p-values represent comparison between HERDX low-risk and med/high-risk groups.
Four variables had previously shown to provide independent prognostic information in Short-HER14,27: 1) Tumour size, 2) Nodal status, 3) TILs and 4) PAM50 subtype. A multivariable Cox model analysis of DMFS confirmed these findings on the 435 Short-HER patient-dataset (webappendix p. 3). Next, we evaluated the ability of 31 variables to provide additional prognostic information using cross-validated elastic net Cox models. The final score (called HER2DX) included 17 variables: tumour size (i.e. T1 vs. rest), nodal status (N0 vs. rest), TILs (as a continuous variable) and PAM50 subtype (HER2-enriched and Basal-like vs. rest), together with 13 individual genes. Among them, 7 had survival coefficients associated with poor survival outcome and were mostly tracking proliferation-related genes (i.e. CDC6, EXO1 and RRM2), HER2-enriched-related biology (i.e. TMEM45B and FGFR4) and Basal-like-related biology (i.e. CDH3). The other 6 genes had survival coefficients associated with better outcome and were mostly tracking Luminal A-related biology (i.e. BAG1), Normal-like (i.e. KRT5, KRT14, MLPH and MYC) and Basal-like-related biology (i.e. PHGDH). The predictive performance (C-index) of HER2DX in Short-HER was 0·80 (all patients), 0·83 (training set) and 0·72 (testing set).
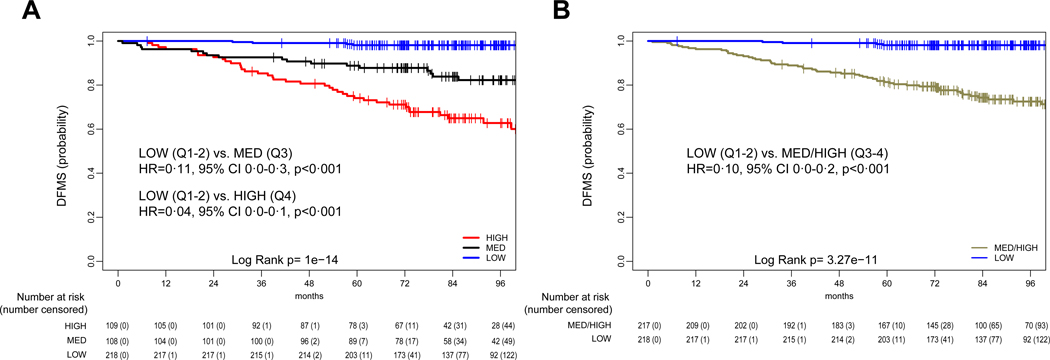
HER2DX measured as a continuous variable was found significantly (p<0·0001) associated with DMFS in the Short-HER 435 patient-dataset. According to HER2DX scoring based on quartiles (webappendix p. 4), the 5-year DMFS of quartile 1 (Q1), Q2, Q3 and Q4 were 97·1% (95% confidence interval [CI] 94·0–100·0), 99·1% (95% CI 97·3–100·0), 88·9% (95% CI 83·2–95·0) and 73·9% (95% CI 66·0–82·7), respectively. No statistically significant difference in DMFS was observed between Q2 vs. Q1 (hazard ratio [HR]=0·92, 95% CI 0·23–3·70, p=0·91). Q3 and Q4 had significant worse DMFS compared to Q1 (Q3: HR=4·57, 95% CI 1·5–13·6, p=0·010; Q4: HR=12·0, 95% CI 4·30–33·5, p<0·0001).
Based on these findings, HER2DX median score (i.e. Q1–2) was identified as the cut-off to identify low-risk patients (Fig. 1A). The 5-year DMFS of Q1–2 group was 98·1% (95% CI 96·3–99·9) (Fig. 1B). The HER2DX score that discriminates Q3 from Q4 was identified as the cut-off to distinguish medium- to high-risk patients. The low-risk group (Q1–2) had a significant better DMFS compared to the high-risk (Q4) group (HR=0·04, 95% CI 0·0–0·1, p<0·0001) and to the medium/high-risk (Q3-Q4) group (HR=0·10, 95% CI 0·1–0·2, p<0·0001). An ad-hoc analysis of HER2DX versus DFS obtained similar results (webappendix p. 4).
Figure 1. Distant metastasis-free survival (DFMS) outcomes based on HER2DX score in the Short-HER training dataset.
(A) DMFS according to low- (quartiles 1 and 2 combined), med- (quartile 3) and high-risk (quartile 4) scores; (B) DMFS according to low- (quartiles 1 and 2 combined) and med/high-risk (quartiles 3 and 4 combined) scores. Q, quartile.
Clinical-pathological and molecular features of the HER2DX low-risk patients in Short-HER were compared to med/high-risk patients (Table 1). No clinical-pathological or molecular feature was unique of HER2DX low-risk patients and features previously identified as being associated with poor survival outcome were also represented in the HER2DX low-risk group. Similarly, 7–36% of HER2DX med/high-risk patients had features previously reported to be associated with better survival outcome such as high TILs (>30%), T1 tumours or node-negative disease (Table 1).
Next, we explored the underlying biology of the HER2DX risk groups (low-, med- and high-). A total of 41 (75·0%) genes were found differentially expressed across the three risk groups (webappendix p. 5). Luminal-related genes (e.g. PGR, ESR1 and BCL2) and immune-related gene CD8A were found more expressed in HER2DX low-risk group compared to the other risk-groups. In contrast, HER2-enriched-related genes (e.g. ERBB2 and FGFR4) and proliferation-related genes (e.g. CCNE1 and UBE2T) were more expressed in the high-risk group compared to the other risk-groups. Of note, the medium-risk group had an intermediate gene expression profile, more like the high-risk group than the low-risk group.
A dataset of 267 patients with early-stage HER2+ disease obtained from a combined cohort of 4 neoadjuvant studies was used for an independent evaluation of the HER2DX score, which was determined at baseline before starting neoadjuvant therapy (Table 2). All patients received chemotherapy, 1-year of trastuzumab, 43·4% (116/267) of patients received dual HER2 blockade with lapatinib and trastuzumab for 4·5 to 6·0 months and 7·5% (20/267) received 4 cycles of neoadjuvant pertuzumab. In PAMELA, chemotherapy was administered after surgery. Despite heterogeneity in systemic therapies, no statistically significant differences in DFS were observed across the 4 cohorts (webappendix p. 6).
Table 2.
Patient baseline characteristics of the combined evaluation dataset.
| All patients | HER2DX Low | HER2DX Med/High | |||||
|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | p-value* | |
| N | 267 | - | 117 | 43·8% | 150 | 56·2% | - |
| Age (mean, range) | 54·5 (11.8) | 53·4 (11.8) | 55·4 (11.8) | 0·48 | |||
| TILs | 0·0090 | ||||||
| TILs 0–29 | 220 | 82·4% | 88 | 75·2% | 132 | 88·0% | |
| TILs ≥30 | 47 | 17·6% | 29 | 24·8% | 18 | 12·0% | |
| cT | 0·010 | ||||||
| T1 | 57 | 21·3% | 34 | 29·1% | 23 | 15·3% | |
| T2–4 | 210 | 78·7% | 83 | 70·9% | 127 | 84·7% | |
| cN | <0·0001 | ||||||
| N0 | 148 | 55·4% | 101 | 86·3% | 47 | 31·3% | |
| N1–3 | 119 | 44·6% | 16 | 13·7% | 103 | 68·7% | |
| Pathological response | 0·90 | ||||||
| pCR | 98 | 36·7% | 42 | 35·9% | 56 | 37·3% | |
| Residual disease | 169 | 63·3% | 75 | 64·1% | 94 | 62·7% | |
| Hormone receptor status | 0·0001 | ||||||
| Positive | 172 | 64·4% | 91 | 77·8% | 81 | 54·0% | |
| Negative | 95 | 35·6% | 26 | 22·2% | 69 | 46·0% | |
| Grade | 0·34 | ||||||
| Grade 1 | 15 | 5·9% | 5 | 4·6% | 10 | 6·8% | |
| Grade 2 | 71 | 28·0% | 35 | 32·4% | 36 | 24·7% | |
| Grade 3 | 168 | 66·1% | 68 | 63·0% | 100 | 68·5% | |
| PAM50 | <0·0001 | ||||||
| Luminal A | 51 | 19·1% | 38 | 32·5% | 13 | 8·7% | |
| Luminal B | 33 | 12·4% | 20 | 17·1% | 13 | 8·7% | |
| HER2-enriched | 138 | 51·7% | 35 | 29·9% | 103 | 68·7% | |
| Basal-like | 21 | 7·9% | 7 | 6·0% | 14 | 9·3% | |
| Normal-like | 24 | 9·0% | 17 | 14·5% | 7 | 4·7% | |
| Study | 0·37 | ||||||
| PAMELA | 88 | 33·0% | 33 | 28·2% | 55 | 36·7% | |
| CHER-LOB | 74 | 27·7% | 38 | 32·5% | 36 | 24·0% | |
| HOSPITAL CLINIC | 68 | 25·5% | 30 | 25·6% | 38 | 25·3% | |
| PADOVA | 37 | 13·9% | 16 | 13·7% | 21 | 14·0% | |
TILs: tumour-infiltrating lymphocytes; pCR: pathological complete response
p-values represent comparison between HERDX low-risk and med/high-risk groups.
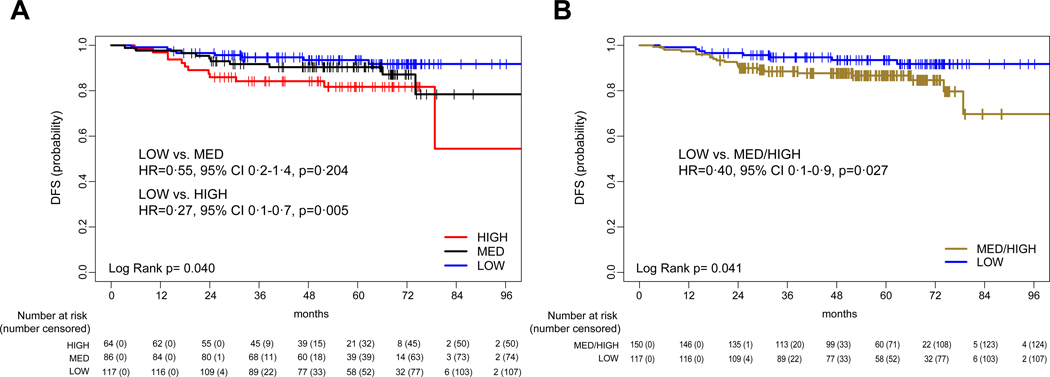
In the evaluation dataset, HER2DX score as a continuous variable was significantly associated with DFS (HR=2·77, 95% CI 1·4–5·6, p=0·0040) (webappendix p. 7). According to the pre-specified cut-offs, HER2DX low-risk group showed a better DFS compared to the high-risk groups (Fig. 2A and B). The 5-year DFS of the HER2DX low-, high- and med/high- groups were 93·5% (95% CI 89·0–98·3%), 81·1% (95% CI 71·5–92·1%) and 86·7% (95% CI 81·2–92·5%), respectively. The 8-year DFS of the HER2DX low-, high- and med/high- groups were 91·7% (95% CI 86·2–97·6%), 54·1% (95% CI 24·1–100%) and 78·7% (95% CI 62·6–98·9%), respectively.
Figure 2. Disease-free survival (DFS) outcomes based on HER2DX score in the combined evaluation dataset.
(A) DFS according to low- (quartiles 1 and 2 combined), med- (quartile 3) and high-risk (quartile 4) scores; (B) DFS according to low- (quartiles 1 and 2 combined) and med/high-risk (quartiles 3 and 4 combined) scores.
Concordant with previous studies2,14–16, TILs as a continuous variable (odds ratio [OR]=1·04, 95% CI 1·0–1·1, p<0·0001) and HER2-enriched subtype (OR=3·25, 95% CI 1·8–5·7, p<0·0001) were associated with pCR. On the contrary, HER2DX score as a continuous variable was not associated with pCR (OR=1·02, 95% CI 0·6–1·6, p=0·93). According to the pre-specified cut-offs, the pCR rates in the HER2DX low-, high- and med/high- groups were 35·8% (42/117), 38·6% (34/88) and 35·5% (22/62). Among 169 patients with residual disease, the distribution of HER2DX low-, med- and high- risk groups was 44·4%, 32·0% and 23·7%, respectively. In this setting, HER2DX low-risk group showed a better DFS compared to the high-risk group (HR=0·34, 95% CI 0·1–0·9, p=0·030) but not to med-risk group (HR=0·63, 95% CI 0·2–1·7, p=0·38) and med/high-risk group (HR=0·47, 95% CI 0·2–1·1, p=0·10) (webappendix p. 7). The 5-year DFS of the HER2DX low- and high- groups were 90·0% (95% CI 83·2–97·4%) and 78·2% (95% CI 65·6–93·2%), respectively. The 8-year DFS of the HER2DX low- and high- groups were 87·6% (95% CI 79·7–96·3%) and 39·1% (95% CI 0·1–100·0%), respectively. Among 98 patients who achieved a pCR, the distribution of HER2DX low-, med- and high- risk groups was 42·9%, 34·7% and 22·4%, respectively. In this setting, 0 and 6 events were observed in the low-risk and med/high-risk groups, respectively.
Discussion
To our knowledge, this is the first study attempting to build a combined prognostic score (called HER2DX) based on 17 clinicopathological and genomic variables in early-stage HER2+ breast cancer using tumour samples from a Phase III trial. Specifically, our results reveal that HER2DX is associated with long-term survival outcome and has the ability to identify groups of patients with different risks of relapsing following standard therapy. In addition, our study provides insights about the relationship between response to therapy in the neoadjuvant setting and long-term prognosis. From a clinical point of view, HER2DX could identify patients with early-stage HER2+ disease candidates for escalated or de-escalated systemic treatment. Future validation of HER2DX seems warranted.
Escalation or de-escalation of systemic therapies in early-stage HER2+ disease is a controversial topic. In stage 1 disease, APT trial28 demonstrated DFS rates of 93·3% following 3-months of adjuvant paclitaxel plus 1-year of trastuzumab in a single-arm trial of 410 patients. This treatment strategy is now widely adopted28, although controversy exists in patients with hormone receptor-negative disease28. Regarding de-escalation of trastuzumab, several non-inferiority studies, including Short-HER trial22, have shown a narrow reduction in recurrence risk with 12 months of therapy compared with shorter durations10,28,29. This treatment strategy, however, has not been widely adopted worldwide, despite its potential impact in low-income countries where trastuzumab is not reimbursed21.
In stage 2–3 disease, escalated systemic treatments with pertuzumab, neratinib and T-DM1 are approved by the US Food and Drug Administration and the European Medicines Agency11–13. However, the absolute benefit of pertuzumab and neratinib is very low (i.e. <3% in invasive DFS)11,12. T-DM1, contrarily, has demonstrated clinically meaningful results with an absolute increase in invasive DFS at 3-years of 11·3% in patients with HER2+ disease who do not achieve a pCR following standard anti-HER2-based chemotherapy13. However, 3 of 4 patients in the control arm of the pivotal trial13 did not present an event at 3-years. Overall, there is an urgent need to better define the populations of patients with early-stage HER2+ disease candidates for escalated or de-escalated systemic therapies.
To our knowledge, our study is the first to report a clinically valuable prognostic biomarker in HER2+ disease. Specifically, the HER2DX score can split the population of early-stage HER2+ breast cancer and identify 2 prognostically distinct groups. To accomplish this, the assay integrates multiple data types and presents a single prognostic score as a continuous variable and proposes specific cut-offs. Importantly, the HER2DX low-risk group cannot be identified by classical clinical-pathological parameters and a substantial proportion of HER2DX low-risk patients have individual features known to be associated with poor survival outcome such as a large tumour size, nodal-positivity, low TILs and residual disease after neoadjuvant therapy. Finally, an intriguing finding is that HER2DX is not associated with the probability to achieve a pCR following anti-HER2-based therapy.
Our study has several limitations. First, the evaluation dataset is a heterogeneous cohort of patients. Second, the survival endpoint from the training dataset (i.e. DMFS) is different from the evaluation dataset (i.e. DFS). The reason is that PAMELA had DFS data recorded. Third, the confidence intervals of the survival estimates at 5- and 8-years of the different risk-groups overlap. Fourth, a substantial proportion of patients in the evaluation dataset also received dual HER2 blockade with lapatinib and trastuzumab. However, the absolute impact of dual HER2 blockade with these 2 drugs in terms of survival outcomes is small (i.e. absolute increase of 2% at 4-years)30. Fifth, HER2DX was developed from primary tumour specimens and staging was based on surgical pathology reports. This is different from the neoadjuvant setting where a core biopsy is the only available tissue and staging is based on imaging. Despite this limitation, HER2DX performed well in the combined neoadjuvant dataset, arguing in favour of its ability to predict outcome at diagnosis before any treatment is initiated using core biopsies. Sixth, the Short-HER cohort was powered for another primary endpoint, which was to compare the DFS between 2 treatment arms. The analysis presented here used all available subjects from this study. Thus, we did not perform a formal power analysis, and focused on statistically significant results. Finally, the HER2DX assay is not standardized and specific cut-offs will need to be defined.
Following our results, it remains the question whether HER2DX will guide the use of systemic therapy in early-stage HER2+ disease. Our opinion is that we are not ready yet to embrace this biomarker and further validation studies should establish its clinical utility in different scenarios with a particular focus in the neoadjuvant setting, where the type of pathological response might be incorporated in the HER2DX algorithm. To accomplish this, the HER2DX assay should be standardised and applied retrospectively in tumour samples from ≥2 large and completed phase III pivotal clinical trials such as APHINITY, NeoALTTO, ExteNET, PERSEPHONE or KATHERINE. For example, patients with HER2DX low-risk disease at diagnosis and who do not achieve a pCR following anti-HER2-based neoadjuvant therapy could be spared 14 cycles of adjuvant T-DM1. Finally, HER2DX should help the design of prospective clinical trials to test novel escalation or de-escalation treatment strategies.
Supplementary Material
Research in context.
Evidence before this study
We searched PubMed between Jan 1, 2010 and May 1, 2020, for clinical trials or studies published in English assessing HER2 inhibition in early-stage breast cancer, using the search terms “HER2+”, “early-stage”, “escalation”, “de-escalation”, “biomarker”, “breast cancer” and “anti-HER2 therapy”. To date, several variables associated with survival outcome have been identified in early-stage HER2+ disease such as TNM staging before and after neoadjuvant therapy, hormone receptor status, tumour-infiltrating lymphocytes (TILs), PAM50 intrinsic subtype and PIK3CA mutations. However, validation and clinical utility of these biomarkers, either alone or in combination, remains unknown.
Implementation of clinical decision support tools to help inform decisions regarding the use of systemic therapy in early-stage HER2 breast cancer are urgently needed. International guidelines support the administration of (neo)adjuvant anti-HER2-based chemotherapy in patients with T1b-T4 or lymph-node positive disease. In the last decade, however, many studies have evaluated various strategies to either escalate or de-escalate systemic therapy in early-stage HER2+ disease, such as 1) decreasing the amount of chemotherapy, 2) decreasing the duration of trastuzumab, 3) increasing HER2 blockade with either the addition of 1-year of pertuzumab to trastuzumab or the addition of 1-year neratinib after trastuzumab and 4) switching the type of anti-HER2 therapy to T-DM1 in patients who do not achieve a pathological complete response following neoadjuvant trastuzumab-based chemotherapy. Despite the successes and limitations of these treatment strategies, the reality is that most patients with early-stage HER2+ disease are cured with chemotherapy and trastuzumab.
Added value of this study
To our knowledge, this is the first study attempting to build a combined prognostic score (called HER2DX) based on 17 clinicopathological and genomic variables in early-stage HER2+ breast cancer using tumour samples from a Phase III clinical trial. In addition, the prognostic score was evaluated in a combined neoadjuvant dataset of patients with newly diagnosed HER2+ breast cancer who received anti-HER2-based therapy, providing insights about the relationship between response to therapy in the neoadjuvant setting and long-term survival outcome.
Implications of all the available evidence
The evidence suggests that HER2DX identifies a substantial proportion of patients with early-stage HER2+ breast cancer who might not need additional therapies, such as pertuzumab, neratinib or T-DM1, due to their outstanding survival outcomes with chemotherapy and trastuzumab (plus endocrine therapy if hormonal receptor-positive). Further studies should establish the clinical utility of HER2DX in this context and explore its value to help further de-escalate systemic treatments such as the duration of trastuzumab and/or the amount of chemotherapy. Finally, multi-parameter prognostic models should be explored in other breast cancer subtypes, such as triple-negative disease, as well as other cancer-types.
Acknowledgments
This study was funded by Instituto de Salud Carlos III (to AP), Save the Mama (to AP), Pas a Pas (to AP), Fundación Científica Asociación Española Contra el Cáncer (to FBM), Fundación SEOM (to TP), Agenzia Italiana del Farmaco (FARM62MC97; to PC), Italian Association for Cancer Research (MFAG 2014–15938; to VG), the Veneto Institute of Oncology (5 × 1000 program; to MVD) and R01CA229409 (to LAC).
Declaration of interest
AP reports grants and personal fees from Roche, grants and personal fees from AstraZeneca, personal fees from Seattle Genetics, grants and personal fees from Daiichi Sankyo, personal fees from Lilly, grants and personal fees from MSD, personal fees from Pfizer, personal fees from Guardant Health, grants and personal fees from Puma, grants and personal fees from Novartis, grants and personal fees from Nanostring Technologies, personal fees from Oncolytics Biotech, personal fees from Abbvie, outside the submitted work; In addition, Dr. Prat has a patent WO2018/103834A1 licensed to Nanostring Technologies, a patent WO/2018/096191 issued, and a patent HER2DX pending. VG reports personal fees from Lilly, personal fees from Novartis, personal fees from Roche, grants from Roche, outside the submitted work; In addition, Dr. Guarneri has a patent HER2DX prognostic score pending. LP has a patent HER2DX pending. ALLC reports grants, personal fees and non-financial support from Novartis, grants, personal fees and non-financial support from Roche, grants, personal fees and non-financial support from Astra Zeneca, grants, personal fees and non-financial support from Lilly, grants and non-financial support from Eisai, grants and personal fees from Genomic Health, grants, personal fees and non-financial support from Pfizer, personal fees from MSD, grants and personal fees from GSK Tesaro, personal fees and non-financial support from Bristol, outside the submitted work. MO reports grants from GSK, during the conduct of the study; grants, personal fees and non-financial support from Roche, grants, personal fees and non-financial support from Novartis, grants and personal fees from Seattle Genetics, personal fees from GSK, grants and personal fees from AstraZeneca, grants and personal fees from PUMA Biotechnology, grants from Genentech, grants from Immunomedics, grants from Boehringer-Ingelheim, grants from Zenith Epigenetics, non-financial support from Pierre-Fabre, non-financial support from GP Pharma, non-financial support from Grünenthal, non-financial support from Eisai, outside the submitted work. AM reports grants and personal fees from Roche, personal fees from Lilly, personal fees from Macrogenics, grants and personal fees from Eisai, grants from Pfizer, personal fees from Novartis, outside the submitted work. RF reports grants and other from Lilly, grants and other from Novartis, grants and other from Pfizer, grants and other from Merck, grants and other from Roche, grants and other from BMS, other from Pharmamar, other from Bayer, other from Pierre Fabre, outside the submitted work. LM reports personal fees from Roche; AstraZeneca, Novartis, Tesaro, Pfizer, GSK, Clovis, outside the submitted work. CMP reports personal fees from Bioclassifier LLC, outside the submitted work; In addition, Dr. Perou has a patent U.S. Patent No. 12,995,459 with royalties paid to Veracyte/Nanostring. SP reports personal fees and non-financial support from Novartis, personal fees from Roche, grants and personal fees from Polyphor, outside the submitted work. CS reports personal fees from Puma biotechnology, personal fees from Pfizer, personal fees from F. Hoffmann - La Roche Ltd, personal fees from Astra Zeneca, personal fees from Celgene, personal fees from Daiichi Sankyo, personal fees from Genomic health, personal fees from Novartis, personal fees from Pierre Fabre, personal fees from Synthon biopharmaceuticals, grants from Roche-Genentech, grants from Macrogenics, grants from Pfizer, grants from Piqur therapeutics, grants from Puma biotechnology, grants from Synthon biopharmaceuticals, grants from Novartis, grants from BMS, personal fees from Merck, Sharp and Dhome España S.A., , personal fees from Odonate Therapeutics, , personal fees from Philips Healthwork, personal fees from prIME Oncology , personal fees from Sanofi Aventis. , outside the submitted work. JSP has a patent “Method of classifying a breast cancer intrinsic subtype” with royalties paid to Nanostring. JC reports grants and personal fees from Roche, personal fees from Celgene, personal fees from Cellestia, grants and personal fees from AstraZeneca, personal fees from Biothera Pharmaceutical, personal fees from Merus, personal fees from Seattle Genetics, grants and personal fees from Daiichi Sankyo, personal fees from Erytech, personal fees from Athenex, personal fees from Polyphor, personal fees from Lilly, personal fees from Servier, grants and personal fees from MSD, personal fees from GSK, personal fees from Leuko, personal fees from Bioasis, personal fees from Clovis Oncology, personal fees from Boehringer Ingelheim, grants and other from Pfizer, other from Bayer, other from Eisai, other from Guardant Health, other from Puma, other from Ariad Pharmaceuticals, other from Baxalta GMBH, outside the submitted work; In addition, Dr. Cortes has a patent WO2018/103834A1 licensed to Nanostring Technologies. MV has declared personal honoraria from Pfizer, Novartis, Roche and Daiichi Sankyo, travel, accommodations and expenses paid by Roche and Pfizer, consulting/advisory role for Roche and Novartis. EC reports personal fees from Pfizer, personal fees from Roche, personal fees from Novartis, personal fees from Lilly, during the conduct of the study. MVD reports personal fees from Lilly, personal fees from Genomic Health, personal fees from Celgene, outside the submitted work; In addition, MVD has a patent HER2DX pending. AF reports personal fees from Roche, personal fees from Novartis, personal fees from Pfizer, outside the submitted work. PV reports personal fees from Nanostring Technologies, outside the submitted work, PC reports grants from Merck KGA, grants and personal fees from Roche, personal fees from Novartis, personal fees from Lilly, outside the submitted work; In addition, PC has a patent HER2DX pending.
Footnotes
Data sharing
The data collected for the study will not be made available to others. We encourage investigators interested in data sharing and collaboration to contact the corresponding author.
Contributor Information
Aleix Prat, SOLTI Breast Cancer Research Group, Barcelona, Spain Department of Medical Oncology, Hospital Clinic of Barcelona, Barcelona, Spain; Translational Genomics and Targeted Therapeutics in Solid Tumors, August Pi i Sunyer Biomedical Research Institute (IDIBAPS), Barcelona, Spain; Department of Medicine, University of Barcelona, Barcelona, Spain.
Valentina Guarneri, Department of Surgery, Oncology and Gastroenterology, University of Padova, Padova, Italy; Medical Oncology 2, Istituto Oncologico Veneto (IOV), IRCCS, Padova, Italy.
Laia Paré, SOLTI Breast Cancer Research Group, Barcelona, Spain.
Gaia Griguolo, Department of Surgery, Oncology and Gastroenterology, University of Padova, Padova, Italy; Medical Oncology 2, Istituto Oncologico Veneto (IOV), IRCCS, Padova, Italy.
Tomás Pascual, SOLTI Breast Cancer Research Group, Barcelona, Spain; Translational Genomics and Targeted Therapeutics in Solid Tumors, August Pi i Sunyer Biomedical Research Institute (IDIBAPS), Barcelona, Spain; Lineberger Comprehensive Cancer Center, Department of Genetics, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
Maria Vittoria Dieci, Department of Surgery, Oncology and Gastroenterology, University of Padova, Padova, Italy; Medical Oncology 2, Istituto Oncologico Veneto (IOV), IRCCS, Padova, Italy.
Núria Chic, SOLTI Breast Cancer Research Group, Barcelona, Spain; Department of Medical Oncology, Hospital Clinic of Barcelona, Barcelona, Spain; Translational Genomics and Targeted Therapeutics in Solid Tumors, August Pi i Sunyer Biomedical Research Institute (IDIBAPS), Barcelona, Spain.
Blanca González-Farré, SOLTI Breast Cancer Research Group, Barcelona, Spain; Translational Genomics and Targeted Therapeutics in Solid Tumors, August Pi i Sunyer Biomedical Research Institute (IDIBAPS), Barcelona, Spain; Department of Pathology, Hospital Clinic of Barcelona, Barcelona, Spain.
Antonio Frassoldati, Clinical Oncology, Department of Morphology, Surgery and Experimental Medicine, S Anna University Hospital, Ferrara.
Esther Sanfeliu, SOLTI Breast Cancer Research Group, Barcelona, Spain; Translational Genomics and Targeted Therapeutics in Solid Tumors, August Pi i Sunyer Biomedical Research Institute (IDIBAPS), Barcelona, Spain; Department of Pathology, Hospital Clinic of Barcelona, Barcelona, Spain.
Juan Miguel Cejalvo, Department of Medical Oncology, Hospital Clínico Universitario of Valencia, Valencia, Spain.
Montserrat Muñoz, SOLTI Breast Cancer Research Group, Barcelona, Spain; Department of Medical Oncology, Hospital Clinic of Barcelona, Barcelona, Spain; Translational Genomics and Targeted Therapeutics in Solid Tumors, August Pi i Sunyer Biomedical Research Institute (IDIBAPS), Barcelona, Spain.
Giancarlo Bisagni, Pathology Unit, Azienda Unità Sanitaria Locale - IRCCS di Reggio Emilia, Reggio Emilia, Italy.
Fara Brasó-Maristany, SOLTI Breast Cancer Research Group, Barcelona, Spain; Translational Genomics and Targeted Therapeutics in Solid Tumors, August Pi i Sunyer Biomedical Research Institute (IDIBAPS), Barcelona, Spain.
Loredana Urso, Medical Oncology 2, Istituto Oncologico Veneto (IOV), IRCCS, Padova, Italy.
Maria Vidal, SOLTI Breast Cancer Research Group, Barcelona, Spain; Department of Medical Oncology, Hospital Clinic of Barcelona, Barcelona, Spain; Translational Genomics and Targeted Therapeutics in Solid Tumors, August Pi i Sunyer Biomedical Research Institute (IDIBAPS), Barcelona, Spain.
Alba Ariela Brandes, Medical Oncology, Azienda Unità Sanitaria Locale di Bologna-IRCCS Istituto delle Scienze Neurologiche, Bologna.
Barbara Adamo, SOLTI Breast Cancer Research Group, Barcelona, Spain; Department of Medical Oncology, Hospital Clinic of Barcelona, Barcelona, Spain; Translational Genomics and Targeted Therapeutics in Solid Tumors, August Pi i Sunyer Biomedical Research Institute (IDIBAPS), Barcelona, Spain.
Antonino Musolino, Department of Medicine and Surgery, University Hospital of Parma; Medical Oncology and Breast Unit, University Hospital of Parma, Piacenza.
Federica Miglietta, Department of Surgery, Oncology and Gastroenterology, University of Padova, Padova, Italy; Medical Oncology 2, Istituto Oncologico Veneto (IOV), IRCCS, Padova, Italy.
Benedetta Conte, Translational Genomics and Targeted Therapeutics in Solid Tumors, August Pi i Sunyer Biomedical Research Institute (IDIBAPS), Barcelona, Spain; Department of Medical Oncology U·O. Oncologia Medica 2, IRCCS Ospedale Policlinico San Martino, Largo R. Benzi 10, 16132, Genova, Italy.
Mafalda Oliveira, SOLTI Breast Cancer Research Group, Barcelona, Spain; Department of Medical Oncology, Vall d’Hebron University Hospital; Vall d’Hebron Institute of Oncology (VHIO), Barcelona, Spain.
Cristina Saura, SOLTI Breast Cancer Research Group, Barcelona, Spain; Department of Medical Oncology, Vall d’Hebron University Hospital; Vall d’Hebron Institute of Oncology (VHIO), Barcelona, Spain.
Sònia Pernas, SOLTI Breast Cancer Research Group, Barcelona, Spain; Department of Medical Oncology, Institut Català d’Oncologia Hospitalet, Hospitalet de Llobregat, Spain.
Jesús Alarcón, SOLTI Breast Cancer Research Group, Barcelona, Spain; Hospital Universitario Son Espases, Carretera de Valldemossa, 79, 07120, Palma de Mallorca, Spain.
Antonio Llombart-Cussac, Department of Medical Oncology, Hospital Arnau de Vilanova, Valencia, Spain.
Javier Cortés, Department of Medical Oncology, Vall d’Hebron University Hospital; Vall d’Hebron Institute of Oncology (VHIO), Barcelona, Spain; IOB Institute of Oncology, Quiron Group, Plaça d’Alfonso Comín, 5, 08023, Barcelona, Spain.
Luis Manso, SOLTI Breast Cancer Research Group, Barcelona, Spain; Department of Medical Oncology, Hospital 12 de Octubre, Madrid, Spain.
Rafael López, SOLTI Breast Cancer Research Group, Barcelona, Spain; Department of Medical Oncology, Complejo Universitario de Santiago de Compostela-CIBERONC, Spain.
Eva Ciruelos, SOLTI Breast Cancer Research Group, Barcelona, Spain; Department of Medical Oncology, Hospital 12 de Octubre, Madrid, Spain.
Francesco Schettini, SOLTI Breast Cancer Research Group, Barcelona, Spain; Department of Medical Oncology, University of Naples Federico II, Naples, Italy.; Translational Genomics and Targeted Therapeutics in Solid Tumors, August Pi i Sunyer Biomedical Research Institute (IDIBAPS), Barcelona, Spain
Patricia Villagrasa, SOLTI Breast Cancer Research Group, Barcelona, Spain.
Lisa A. Carey, Lineberger Comprehensive Cancer Center, Department of Genetics, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
Charles M. Perou, Lineberger Comprehensive Cancer Center, Department of Genetics, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
Federico Piacentini, Department of Medical and Surgical Sciences, University of Modena and Reggio Emilia, Modena, Italy.
Roberto D’Amico, Department of Medical and Surgical Sciences, University of Modena and Reggio Emilia, Modena, Italy; Center for Genome Research, University of Modena and Reggio Emilia, Modena, Italy.
Enrico Tagliafico, Department of Medical and Surgical Sciences, University of Modena and Reggio Emilia, Modena, Italy; Center for Genome Research, University of Modena and Reggio Emilia, Modena, Italy.
Joel. S. Parker, Lineberger Comprehensive Cancer Center, Department of Genetics, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
Pierfranco Conte, Department of Surgery, Oncology and Gastroenterology, University of Padova, Padova, Italy Medical Oncology 2, Istituto Oncologico Veneto (IOV), IRCCS, Padova, Italy.
References
- 1.Perez EA, Romond EH, Suman VJ, et al. Trastuzumab Plus Adjuvant Chemotherapy for Human Epidermal Growth Factor Receptor 2–Positive Breast Cancer: Planned Joint Analysis of Overall Survival From NSABP B-31 and NCCTG N9831. Journal of Clinical Oncology 2014; 32(33): 3744–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prat A, Carey LA, Adamo B, et al. Molecular Features and Survival Outcomes of the Intrinsic Subtypes Within HER2-Positive Breast Cancer. JNCI: Journal of the National Cancer Institute 2014; 106(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferrari A, Vincent-Salomon A, Pivot X, et al. A whole-genome sequence and transcriptome perspective on HER2-positive breast cancers. Nature Communications 2016; 7(1): 12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prat A, Pascual T, De Angelis C, et al. HER2-Enriched Subtype and ERBB2 Expression in HER2-Positive Breast Cancer Treated with Dual HER2 Blockade. JNCI: Journal of the National Cancer Institute 2019; 112(1): 46–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brasó-Maristany F, Griguolo G, Pascual T, et al. Phenotypic changes of HER2-positive breast cancer during and after dual HER2 blockade. Nature Communications 2020; 11(1): 385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandão M, Caparica R, Malorni L, Prat A, Carey LA, Piccart M. What Is the Real Impact of Estrogen Receptor Status on the Prognosis and Treatment of HER2-Positive Early Breast Cancer? Clinical Cancer Research 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayes DF. HER2 and Breast Cancer — A Phenomenal Success Story. New England Journal of Medicine 2019; 381(13): 1284–6. [DOI] [PubMed] [Google Scholar]
- 8.Veeraraghavan J, De Angelis C, Reis-Filho JS, et al. De-escalation of treatment in HER2-positive breast cancer: Determinants of response and mechanisms of resistance. The Breast 2017; 34: S19–S26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tolaney SM, Barry WT, Dang CT, et al. Adjuvant Paclitaxel and Trastuzumab for Node-Negative, HER2-Positive Breast Cancer. New England Journal of Medicine 2015; 372(2): 134–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pivot X, Romieu G, Debled M, et al. 6 months versus 12 months of adjuvant trastuzumab in early breast cancer (PHARE): final analysis of a multicentre, open-label, phase 3 randomised trial. The Lancet 2019; 393(10191): 2591–8. [DOI] [PubMed] [Google Scholar]
- 11.von Minckwitz G, Procter M, de Azambuja E, et al. Adjuvant Pertuzumab and Trastuzumab in Early HER2-Positive Breast Cancer. New England Journal of Medicine 2017; 377(2): 122–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin M, Holmes FA, Ejlertsen B, et al. Neratinib after trastuzumab-based adjuvant therapy in HER2-positive breast cancer (ExteNET): 5-year analysis of a randomised, double-blind, placebo-controlled, phase 3 trial. The Lancet Oncology 2017; 18(12): 1688–700. [DOI] [PubMed] [Google Scholar]
- 13.von Minckwitz G, Huang C-S, Mano MS, et al. Trastuzumab Emtansine for Residual Invasive HER2- Positive Breast Cancer. New England Journal of Medicine 2018; 380(7): 617–28. [DOI] [PubMed] [Google Scholar]
- 14.Conte PF, Griguolo G, Dieci MV, et al. PAM50 HER2-enriched subtype as an independent prognostic factor in early-stage HER2+ breast cancer following adjuvant chemotherapy plus trastuzumab in the ShortHER trial. Journal of Clinical Oncology 2019; 37(15_suppl): 544-. [Google Scholar]
- 15.Salgado R, Denkert C, Campbell C, et al. Tumor-Infiltrating Lymphocytes and Associations With Pathological Complete Response and Event-Free Survival in HER2-Positive Early-Stage Breast Cancer Treated With Lapatinib and Trastuzumab: A Secondary Analysis of the NeoALTTO Trial. JAMA Oncology 2015; 1(4): 448–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krop IE, Paulson J, Campbell C, et al. Genomic correlates of response to adjuvant trastuzumab (H) and pertuzumab (P) in HER2+ breast cancer (BC): Biomarker analysis of the APHINITY trial. Journal of Clinical Oncology 2019; 37(15_suppl): 1012-.30811295 [Google Scholar]
- 17.Schettini F, Pascual T, Conte B, et al. HER2-enriched subtype and pathological complete response in HER2-positive breast cancer: A systematic review and meta-analysis. Cancer Treatment Reviews 2020; 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loibl S, Majewski I, Guarneri V, et al. PIK3CA mutations are associated with reduced pathological complete response rates in primary HER2-positive breast cancer: pooled analysis of 967 patients from five prospective trials investigating lapatinib and trastuzumab†. Annals of Oncology 2016; 27(8): 1519–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Llombart-Cussac A, Cortés J, Paré L, et al. HER2-enriched subtype as a predictor of pathological complete response following trastuzumab and lapatinib without chemotherapy in early-stage HER2-positive breast cancer (PAMELA): an open-label, single-group, multicentre, phase 2 trial. The Lancet Oncology 2017; 18(4): 545–54. [DOI] [PubMed] [Google Scholar]
- 20.Cortazar P, Zhang L, Untch M, et al. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. The Lancet 2014; 384(9938): 164–72. [DOI] [PubMed] [Google Scholar]
- 21.Pondé N, Gelber RD, Piccart M. PERSEPHONE: are we ready to de-escalate adjuvant trastuzumab for HER2-positive breast cancer? npj Breast Cancer 2019; 5(1): 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conte P, Conte P, Bisagni G, et al. Final analysis of the phase III multicentric Italian study Short-HER: 9 weeks vs 1 year adjuvant trastuzumab for HER2+ early breast cancer. Annals of Oncology 2018; 29(12): 2328–33. [DOI] [PubMed] [Google Scholar]
- 23.Guarneri V, Frassoldati A, Bottini A, et al. Preoperative Chemotherapy Plus Trastuzumab, Lapatinib, or Both in Human Epidermal Growth Factor Receptor 2–Positive Operable Breast Cancer: Results of the Randomized Phase II CHER-LOB Study. Journal of Clinical Oncology 2012; 30(16): 1989–95. [DOI] [PubMed] [Google Scholar]
- 24.Dieci MV, Prat A, Tagliafico E, et al. Integrated evaluation of PAM50 subtypes and immune modulation of pCR in HER2-positive breast cancer patients treated with chemotherapy and HER2-targeted agents in the CherLOB trial. Annals of Oncology 2016; 27(10): 1867–73. [DOI] [PubMed] [Google Scholar]
- 25.Salgado R, Denkert C, Demaria S, et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Annals of Oncology 2015; 26(2): 259–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prat A, Pineda E, Adamo B, et al. Clinical implications of the intrinsic molecular subtypes of breast cancer. The Breast 2015; 24: S26–S35. [DOI] [PubMed] [Google Scholar]
- 27.Dieci MV, Conte P, Bisagni G, et al. Association of tumor-infiltrating lymphocytes with distant disease-free survival in the ShortHER randomized adjuvant trial for patients with early HER2+ breast cancer. Annals of Oncology 2019; 30(3): 418–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tolaney SM, Guo H, Pernas S, et al. Seven-Year Follow-Up Analysis of Adjuvant Paclitaxel and Trastuzumab Trial for Node-Negative, Human Epidermal Growth Factor Receptor 2–Positive Breast Cancer. Journal of Clinical Oncology 2019; 37(22): 1868–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joensuu H, Fraser J, Wildiers H, et al. Effect of Adjuvant Trastuzumab for a Duration of 9 Weeks vs 1 Year With Concomitant Chemotherapy for Early Human Epidermal Growth Factor Receptor 2–Positive Breast Cancer: The SOLD Randomized Clinical Trial. JAMA Oncology 2018; 4(9): 1199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piccart-Gebhart M, Holmes E, Baselga J, et al. Adjuvant Lapatinib and Trastuzumab for Early Human Epidermal Growth Factor Receptor 2–Positive Breast Cancer: Results From the Randomized Phase III Adjuvant Lapatinib and/or Trastuzumab Treatment Optimization Trial. Journal of Clinical Oncology 2016; 34(10): 1034–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.