PURPOSE
External control (EC) arms derived from electronic health records (EHRs) can provide appropriate comparison groups when randomized control arms are not feasible, but have not been explored for metastatic colorectal cancer (mCRC) trials. We constructed EC arms from two patient-level EHR-derived databases and evaluated them against the control arm from a phase III, randomized controlled mCRC trial.
METHODS
IMblaze370 evaluated atezolizumab with or without cobimetinib versus regorafenib in patients with mCRC. EC arms were constructed from the Flatiron Health (FH) EHR-derived de-identified database and the combined FH/Foundation Medicine Clinico-Genomic Database (CGDB). IMblaze370 eligibility criteria were applied to the EC cohorts. Propensity scores and standardized mortality ratio weighting were used to balance baseline characteristics between the IMblaze370 and EC arms; balance was assessed using standardized mean differences. Kaplan-Meier method estimated median overall survival (OS). Cox proportional hazards models estimated hazard ratios with bootstrapped 95% CIs to compare differences in OS between study arms.
RESULTS
The FH EC included 184 patients; the CGDB EC included 108 patients. Most characteristics were well-balanced (standardized mean difference < 0.1) between each EC arm and the IMblaze370 population. Median OS was similar between the IMblaze370 control arm (8.5 months [95% CI, 6.41 to 10.71]) and both EC arms: FH (8.5 months [6.93 to 9.92]) and CGDB (8.8 months [7.85 to 9.92]). OS comparisons between the IMblaze370 experimental arm and the FH EC (hazard ratio, 0.85 [0.64 to 1.14]) and CGDB EC (0.86 [0.65 to 1.18]) yielded similar results as the comparison with the IMblaze370 control arm (1.01 [0.75 to 1.37]).
CONCLUSION
EC arms constructed from the FH database and the CGDB closely replicated the control arm from IMblaze370. EHR-derived EC arms can provide meaningful comparators in mCRC trials when recruiting a randomized control arm is not feasible.
INTRODUCTION
Clinical trials for cancer treatments face special challenges related to the ethics and feasibility of including control arms, particularly for trials with highly restrictive eligibility criteria, when an active comparator is nonexistent or considered an inadequate treatment option, and for trials in rare cancers.1,2 Although randomized controlled trials remain the gold standard for clinical research, single-arm trials may be acceptable for regulatory consideration when, for example, an unprecedented treatment effect is observed for a patient population with a high unmet medical need. However, controlled confirmatory trials may also be needed.3-6 When evaluating a therapeutic landscape, it can be difficult or impossible to contextualize data from single-arm studies because of differences in the study designs, participants, and treatment comparators used over time.2 However, the urgency of cancer care often necessitates interpreting single-arm study findings in the context of available therapeutic options before the results of a confirmatory trial become available.
CONTEXT
Key Objective
How can real-world data be used to generate external control (EC) arms for colorectal cancer clinical trials? We constructed EC arms from real-world databases in the first application of this method to metastatic colorectal cancer (mCRC) trials.
Knowledge Generated
This proof-of-concept study showed that EC arms generated from the Flatiron Health database and the Flatiron Health/Foundation Medicine Clinico-Genomic Database successfully replicated the randomized control arm of the IMblaze370 trial. To our knowledge, this is the first study to demonstrate the feasibility of using EC arms built from electronic health record–derived databases in clinical trials in patients with mCRC receiving third-line or later treatment.
Relevance
Our methodological approach may be replicated or adapted to other real-world data sources to generate EC arms for other trials in mCRC.
External control (EC) arms can provide useful information in the absence of randomized control arms and when the scientific literature trails behind emerging research. EC arms using detailed patient information from real-world data (RWD) sources, such as electronic health records (EHRs), can be used to construct statistically and clinically appropriate comparison cohorts for patients receiving active interventions in single-arm trials.7-9 Data from patient registries and control groups from completed trials have been used as EC arms to support accelerated approvals, such as of blinatumomab for acute lymphoblastic leukemia and avelumab for Merkel cell carcinoma.10-12 However, analytic challenges have been raised to these approaches related to the relevance of treatment comparators and other period effects.13
The sophistication of RWD sources and statistical methods has made individual patient-level data derived from EHRs a viable option for EC arms in cancer research.14,15 EC arms built from the Flatiron Health (FH) de-identified EHR-derived database were recently shown to approximate the results of several randomized trials in non–small-cell lung cancer, serving as a proof-of-concept for future work.16 We aimed to extend this work to metastatic colorectal cancer (mCRC) research using patient-level data from the FH database and the combined FH/Foundation Medicine (FMI) Clinico-Genomic Database (CGDB). The CGDB data set includes EHR information and genomic data, which can be particularly relevant in mCRC for clinical development programs targeting biomarker-specific patients. We analyzed the comparability of results from the RWD EC arms with the randomized comparator arm (regorafenib) of the IMblaze370 trial of atezolizumab with or without cobimetinib as a third- or later-line (3L+) therapy in patients with mCRC.17
METHODS
Trial Data Source
We used patient-level data from the IMblaze370 clinical trial (ClinicalTrials.gov identifier: NCT02788279) whose methods and primary findings have been previously reported.17 IMblaze370 was a phase III, open-label, randomized trial investigating atezolizumab with or without cobimetinib versus regorafenib as 3L+ therapy for adults with unresectable locally advanced or metastatic CRC. Patients were randomly assigned (2:1:1) to atezolizumab plus cobimetinib, atezolizumab monotherapy, or regorafenib. The primary end point was overall survival (OS). The IMblaze370 study protocol was approved by the institutional review boards or independent ethics committees of each study site, and all patients gave written informed consent.
External Data Sources
Two EC arms were constructed from RWD sources, one from the FH EHR-derived de-identified database (Flatiron Health, New York, NY)18 and the other from the combined FH/FMI CGDB (Foundation Medicine, Cambridge, MA).19
The nationwide FH database is a longitudinal database, including patient-level information from structured data (eg, laboratory values and prescribed treatments) and unstructured data (eg, biomarker reports) collected via technology-enabled chart abstraction from physicians’ notes and other documents. During the study period, the de-identified data originated from approximately 280 cancer clinics (approximately 800 sites of care, primarily community-based cancer centers), representing > 2.4 million patients with cancer in the United States. The data were de-identified with provisions in place to prevent re-identification. We used data collected between January 2013 and June 2019.
The CGDB includes patients from the FH database who underwent comprehensive genomic profiling by FMI. In addition to the information in the FH database, the CGDB provides de-identified patient-level genomic data, including specimen features (eg, tumor mutation burden and pathologic purity), alteration-level details (eg, genomic position, reference, and alternate alleles), and targeted therapeutic options reported to the clinician at the time of testing. We used CGDB data collected between January 2011 and December 2019.
Both databases comprise patients from the same underlying population. However, each database is subject to different selection criteria, and the FH database also applies a sampling fraction. Therefore, these are separate but overlapping populations. The overlap varies with each update of the databases. Approximately 10%-15% of the patients with mCRC in the FH database are present in the CGDB, and 55%-65% of patients with mCRC in the CGDB are present in the FH database. Both databases consist of retrospective observational de-identified anonymized patient-level data; as such, this study was exempt from informed consent and institutional review board requirements.
Construction of the EC Arms
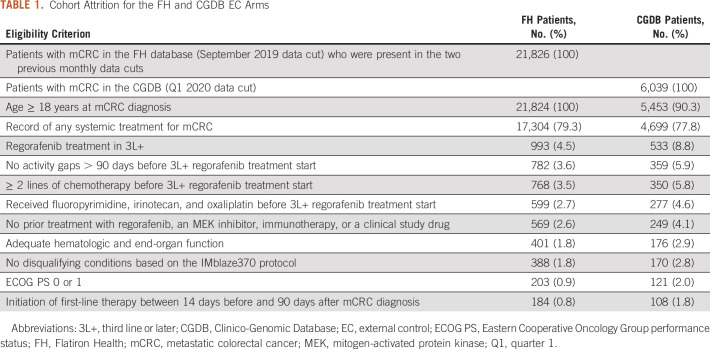
An EC arm was built from each database to mimic the randomized control arm of IMblaze370. Initially, IMblaze370 eligibility criteria were applied (in addition to a requirement of receiving 3L+ regorafenib) to create a cohort of RWD patients comparable to the trial patients (Table 1). Details on the IMblaze370 eligibility criteria have been described elsewhere.17 In some instances, applying the IMblaze370 eligibility criteria to the RWD patients required adaptation because of differences in the nature of assessments between these research settings. Time windows (around the 3L+ regorafenib treatment initiation) for assessing Eastern Cooperative Oncology Group (ECOG) performance status (−60 to +7 days) and laboratory values (−30 to +7 days) were defined. Patients with abnormal laboratory measurements or an ECOG status ≥ 2 within these timeframes were excluded. Patients in the EC arms also had to have started their first-line therapy between 14 days before and 90 days after their mCRC diagnosis, to exclude patients who possibly received treatment outside the FH network and may have had incomplete treatment information. Similarly, patients with activity gaps in their data of more than 90 days between diagnosis and 3L+ regorafenib treatment initiation were excluded.
TABLE 1.
Cohort Attrition for the FH and CGDB EC Arms
Propensity Scoring
Once the trial-like RWD patients were determined, propensity scores (PSs) were calculated for each patient using multivariate logistic regression. These reflect each patient’s probability of being assigned to the trial interventional arm or EC arm given a set of relevant baseline covariates (those predictive of OS). An example of pseudocode is provided elsewhere.20 PSs were used to assess imbalances in terms of these covariates between the IMblaze370 experimental arm and EC arms. Covariates included age (at initiation of 3L+ regorafenib treatment), sex, race, ECOG performance status, time from diagnosis to 3L+ regorafenib treatment initiation, and number of prior lines of therapy. Standardized mortality ratio weighting (SMRW) was then applied to achieve balance in these covariates between the IMblaze370 experimental arm (atezolizumab with cobimetinib) and the EC arms.21 That is, a weight of PS/(1 − PS) was applied to each EC patient, whereas the patients in the trial experimental arm were fixed to a weight of 1. The SMRW method was chosen to reflect that in a prospective single-arm trial, the aim would likely be to leave the trial arm unadjusted and to use the EC arm weighting to address balance issues. Weighting both arms would estimate the average treatment effect for the overall population and not for the population in the experimental arm.
After weighting, standardized mean differences (SMDs) were calculated and used to quantify balance between the arms, with an SMD ≤ 0.1 for each covariate indicating that adequate balance was achieved.22 This weighted pseudopopulation was then used for the primary analysis.
Statistical Analyses
OS was the outcome of interest, defined as time from random assignment (trial patients) or initiation of 3L+ regorafenib treatment (EC patients) to death from any cause. Trial patients had to initiate treatment within 3 days of random assignment. EC patients were censored at the earlier of either their last contact date (date of last activity recorded in the database) or the maximum follow-up time of the trial experimental arm. OS estimates were determined using the Kaplan-Meier method, and differences between arms were assessed using the log-rank test. Median OS and corresponding 95% CIs were summarized for each arm along with Kaplan-Meier curves. Cox proportional hazards models estimated hazard ratios (HRs) for OS, with 95% CIs calculated using bootstrapping. The bootstrap method was used because weighting induces within-subject correlations in the pseudopopulation, and the lack of independence between subjects can cause a naïve model–based variance estimator to be biased. The bootstrap method has been shown to be one of the most unbiased methods for variance estimation.23
HRs were assessed for the following comparisons:
EC arm (regorafenib) versus IMblaze370 control arm (regorafenib)
IMblaze370 experimental arm (atezolizumab-cobimetinib) versus EC arm (regorafenib)
IMblaze370 experimental arm (atezolizumab-cobimetinib) versus IMblaze370 control arm (regorafenib)
Sensitivity Analyses
The following sensitivity analyses were conducted to examine how different methodological approaches to building the EC arms and analyzing the data affected the results. A sensitivity analysis was performed using trimming in addition to weighting, to remove EC patients with PS outside of the PS distribution of the trial population. Another sensitivity analysis used stabilized inverse probability of treatment weighting instead of SMRW, where all patients were assigned a weight equal to the inverse of the probability of receiving the treatment they actually received.21 This analysis was performed both with and without trimming of trial and EC patients from nonoverlapping PS regions. A third sensitivity analysis was performed with a doubly robust estimator using a weighted Cox regression adjusted for the same baseline covariates included in the PS model and additional selected variables not included in the primary analysis because of high proportions of missingness in one or both external data sources.24 These variables were disease stage at initial diagnosis, RAS mutational status, and location of the primary tumor (side of the colon).
All analyses were conducted using R Studio version 1.3.0 and R version 4.0.0.
RESULTS
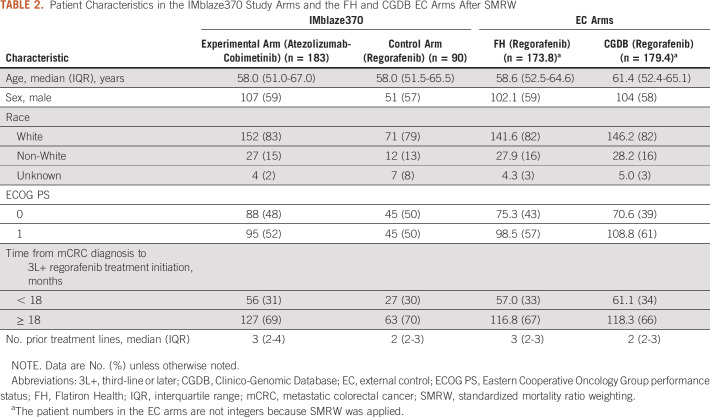
A total of 184 patients from the FH database and 108 from the CGDB met the eligibility criteria and were included in the EC arms (Table 1). After applying SMRW, the baseline covariates were well-balanced between the FH EC arm and the IMblaze370 population (Table 2, Fig 1A). With the exception of ECOG performance status, the baseline covariates were also well-balanced between the IMblaze370 study arms and the CGDB EC arm (Table 2, Fig 1B).
TABLE 2.
Patient Characteristics in the IMblaze370 Study Arms and the FH and CGDB EC Arms After SMRW
FIG 1.
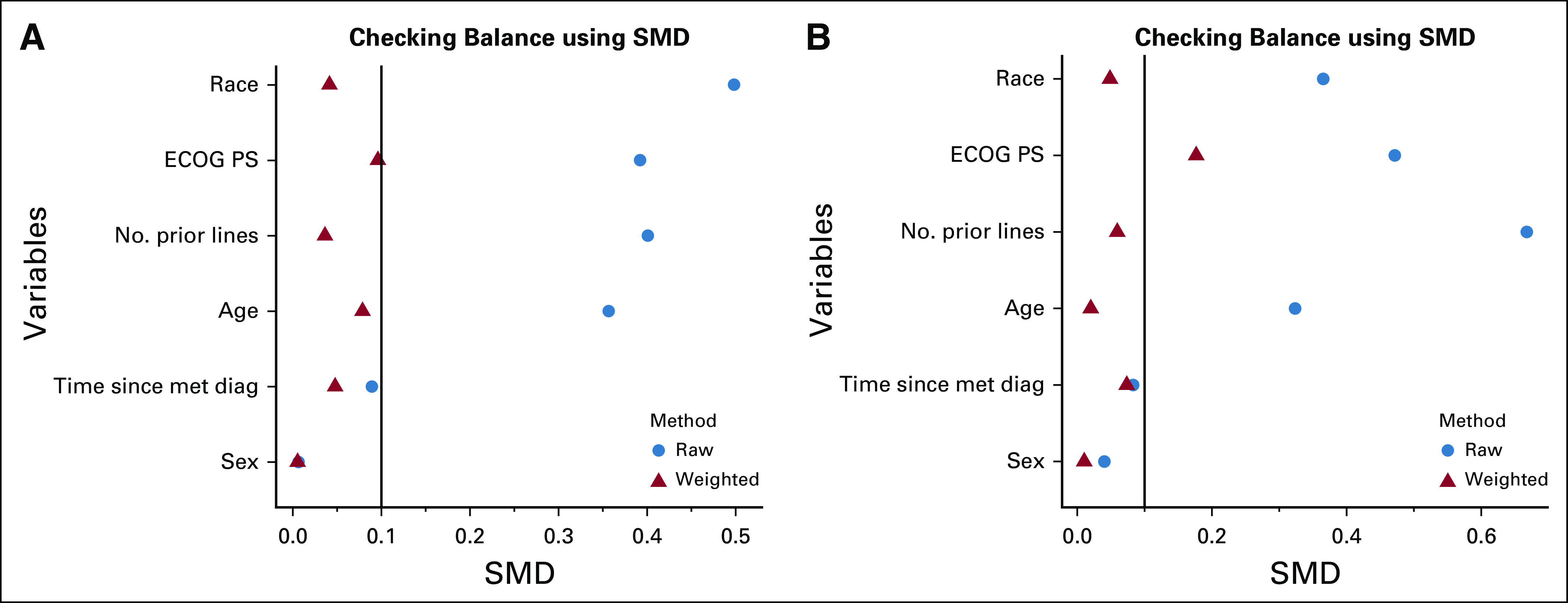
Covariate balance assessment between the IMblaze370 experimental arm and the EC arms using SMDs. (A) FH EC arm. (B) CGDB EC arm. CGDB, Clinico-Genomic Database; diag, diagnosis; EC, external control; ECOG PS, Eastern Cooperative Oncology Group performance status; FH, Flatiron Health; met, metastatic; SMD, standardized mean difference.
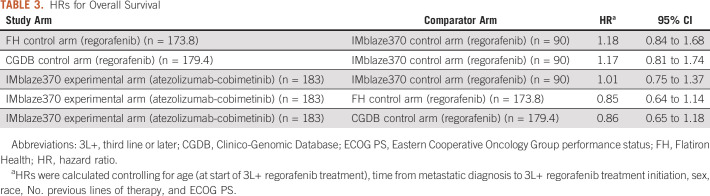
OS was similar between the FH EC arm (median, 8.48 months [95% CI, 6.93 to 9.92]) and the IMblaze370 control arm (median: 8.51 months [6.41 to 10.71]), with an almost perfect congruence up to 10 months of follow-up (Fig 2A, Table 3). OS was also similar between the EC arm built from the CGDB (median, 8.77 months [95% CI, 7.85 to 9.92]) and the IMblaze370 control arm (median, 8.51 months [6.41 to 10.71]; Fig 2B, Table 3). Regarding the HRs for OS (adjusted for age, time from metastatic diagnosis to 3L+ regorafenib treatment start, sex, race, number of previous lines of therapy, and ECOG status), and as previously reported,17 there was no statistically significant difference in OS between the atezolizumab plus cobimetinib experimental arm and the regorafenib control arm. Comparison of the IMblaze370 experimental arm with the EC arms built either from the FH database or from the CGDB led to consistent HRs, and CIs as were observed in the IMblaze370 regorafenib arm (Table 3).
FIG 2.
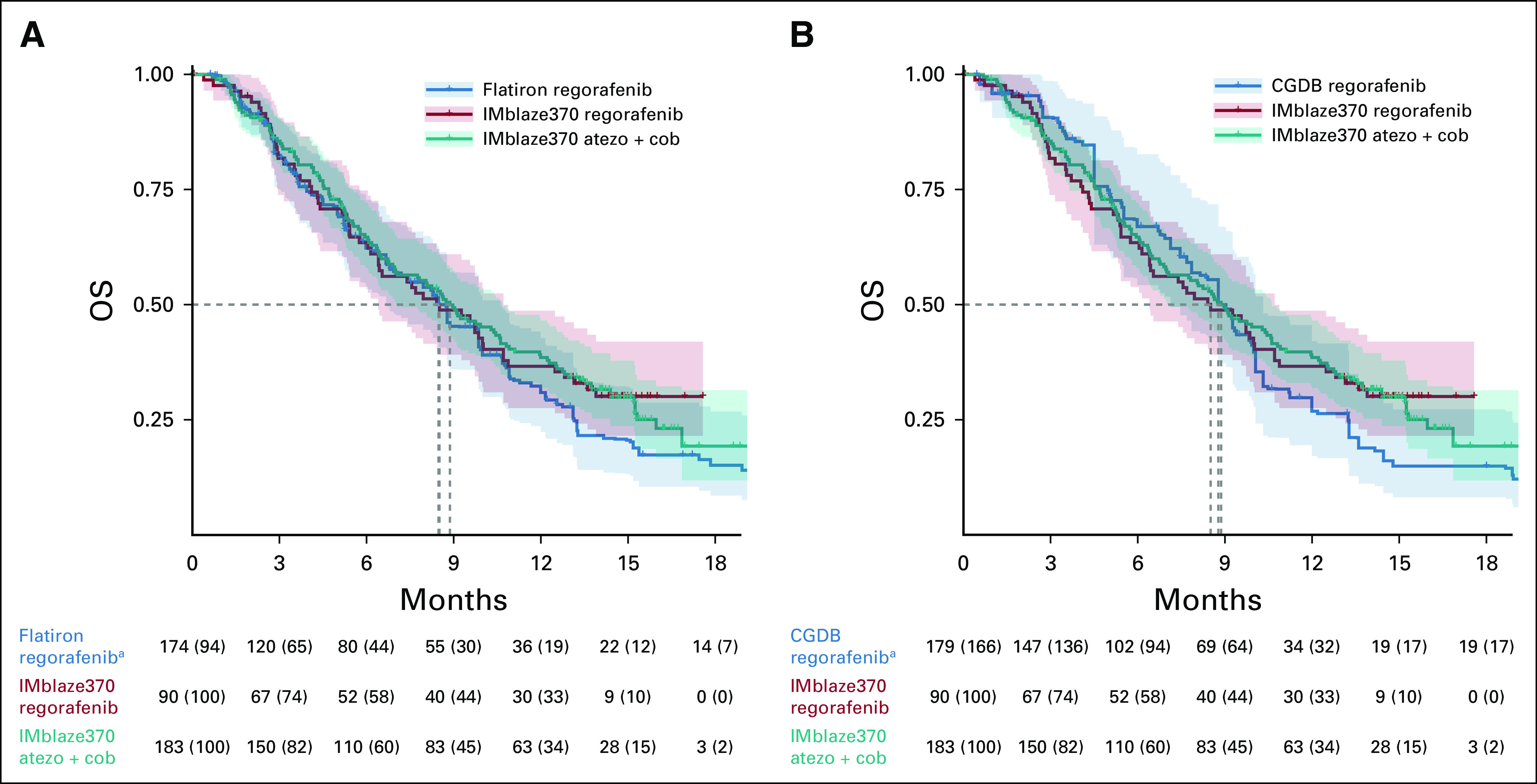
OS in the EC arm and the IMblaze370 arms. (A) EC arm built from the FH database. (B) EC arm built from the CGDB. aPercentages for the EC arm refer to the study population before SMRW was applied. Atezo, atezolizumab; CGDB, Clinico-Genomic Database; Cob, cobimetinib; EC, external control; FH, Flatiron Health; OS, overall survival; SMRW, standardized mortality ratio weighting.
TABLE 3.
HRs for Overall Survival
All sensitivity analyses were consistent with the primary findings. HRs for OS in both the FH and the CGDB EC arms versus the IMblaze370 experimental arm yielded consistently lower but similar estimates as the original IMblaze370 analysis (Figs 3A and 3B).
FIG 3.
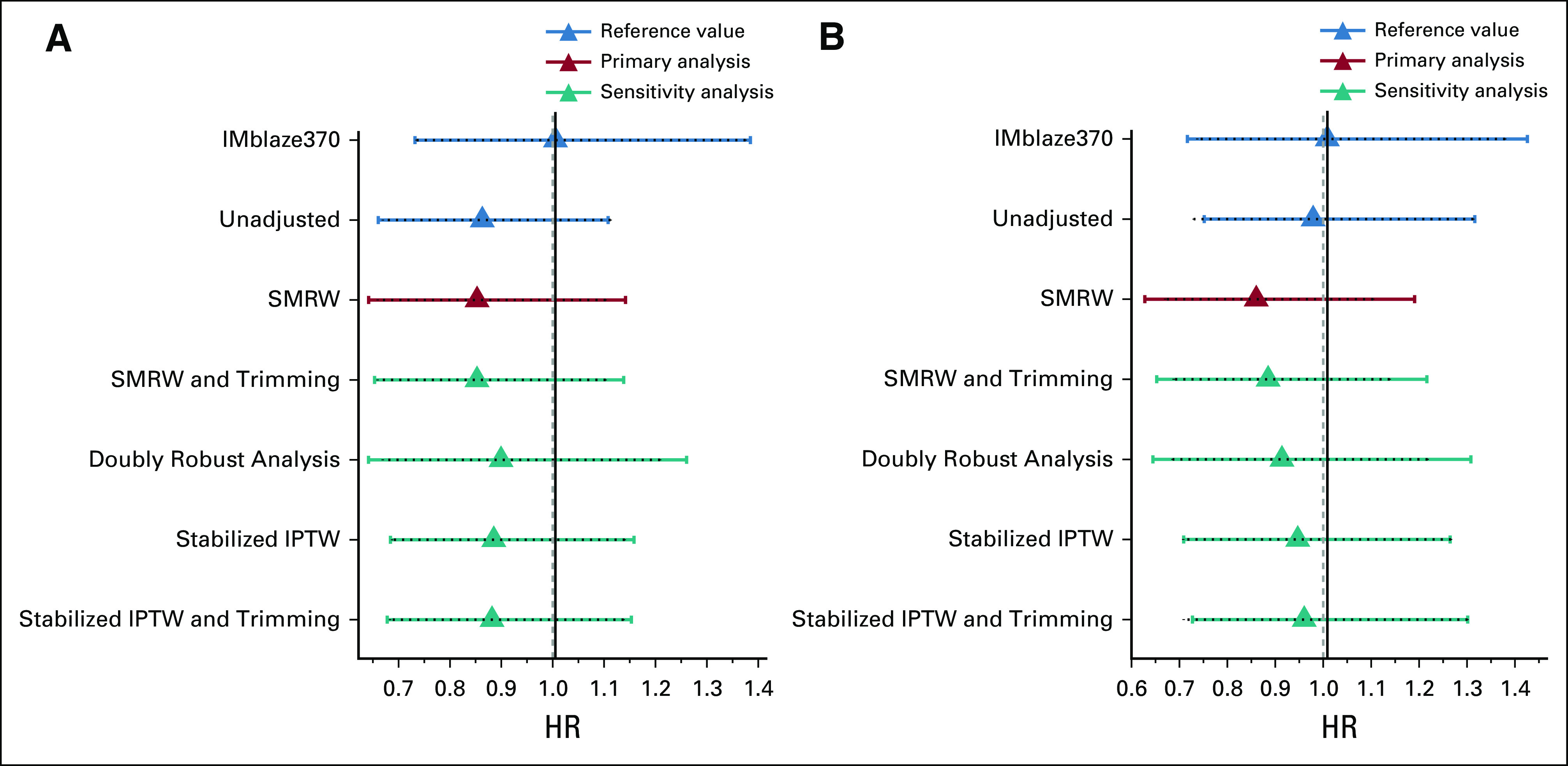
HRs (with 95% CIs) for OS between the EC arms and the IMblaze370 experimental arm (primary and sensitivity analyses). (A) EC arm built from the FH database. (B) EC arm built from the CGDB. CGDB, Clinico-Genomic Database; EC, external control; FH, Flatiron Health; HR, hazard ratio; IPTW, inverse probability of treatment weighting; OS, overall survival; RCT, randomized clinical trial; SMRW, standardized mortality ratio weighting.
DISCUSSION
This study demonstrated the feasibility of constructing EHR-derived EC arms to replicate a randomized control arm from a clinical trial in patients with mCRC. Median OS was almost identical between the EC arms and the control arm from the IMblaze370 trial. HRs for OS comparing the EC arms with the IMblaze370 experimental arm were consistent with the original comparison between the randomized IMblaze370 study arms and lead to the same conclusions. Analyses from the FH and CGDB EC arms showed consistent results, supported by robust sensitivity analyses.
This study extends the work of others who have used EC arms derived from historical controls, patient registries, and patient-level EHR-derived data.10-12 Carrigan recently used the FH database to construct and compare EHR-derived EC arms with standard-of-care treatment arms from several immunotherapy and targeted treatment trials in patients with advanced non–small-cell lung cancer.16 The EC arms were similar to the trials’ active control arms with the exception of one comparison for a small trial of a mesenchymal-to-epithelial transition (MET) inhibitor in a highly MET-positive patient population, but MET expression assessments were not part of the EHR database.16,25 This insight underscores the importance of available information to appropriately match patients from clinical trials and EHR databases on confounding or prognostic factors. For our study, the information included in the FH database appeared adequate to build EC arms from patients with similar characteristics as those in IMblaze370. The ability to examine these comparisons with substantial detail may lend confidence to future work in this area where the evaluation of EC arms can employ granular patient-level data-driven assessments.
Our findings support the EHR-derived database approach and should be interpreted in the context of certain strengths and limitations. The generalizability of these findings to other patient subgroups and therapeutic modalities, and to other cancer types, remains uncertain and warrants further analysis. The FH database and the CGDB were used for the breadth and depth of EHR-derived information for patients with mCRC. The additional information available in the CGDB has the potential to integrate genomic biomarkers in future studies in the field of personalized medicine (eg, in tumor-agnostic and biomarker-specific indications), which suggests a strong rationale for the use of the CGDB for the design and evaluation of EC arms. An advantage of both databases is their data recency, which makes it possible to mitigate confounding period effects. The FH database also captures mortality data with high accuracy, which is critical when assessing OS.26 Since the adjusted HRs from the two data sources were similar, the most important confounding factors appeared to have been accounted for in our models.
Not all inclusion and exclusion criteria from IMblaze370 could be applied to the EHR-derived patients. In some cases, the relevant information recorded in the trial was not collected in the EHR. In other cases, such as with medical history and comorbidities, because the EHR data were specific to oncology care, additional information beyond what was captured for oncology care might have been unreliably ascertained, such as on acute or chronic conditions. Although FH aims to increase data completeness by abstracting unstructured information, such as from physician notes, some details not related to oncology care may still be missing since this is dependent on information recorded by different physicians. The eligibility criteria based on baseline assessments in IMblaze370, such as ECOG performance status and laboratory values, have been adapted to include a short window of time to account for variability in assessment and recording practices inherent in EHR data. Future EC studies should consider aligning the time period when EC patients are selected with the trial recruitment period. IMblaze370 patients were recruited between July 2016 and January 2017. Because of the limited number of eligible EC patients, we did not apply any time restriction, using all available data from the databases. This likely had a limited impact for mCRC as there were no substantial changes to the standard of care during the study period; however, this may be different in other settings.
Sensitivity analyses used SMRW and inverse probability of treatment weighting to balance covariates between the IMblaze370 experimental arm and the EC arms. Both are common weighting methods that aim to balance covariates between two populations in a relevant manner.21 Individual patient weights are adjusted to create a reweighted pseudopopulation where the treatment assignment is independent of the observed covariates. These two weighting methods are used to address different targets of inferences (estimands), with the former estimating the average treatment effect in the treated and the latter estimating the average treatment effect in the whole population. In our study, both methods led to similar findings. Important considerations for interpreting HRs because of deviations from proportional hazards from 10 months onward should be noted (reflected in the differences between the EC and IMblaze370 survival curves after approximately 10 months), as this affects the trial results where hazards deviate from proportionality around the same time.
Unlike arms in randomized controlled trials, EHR-derived EC arms cannot control for unmeasured or unknown confounders; they are restricted to known confounders that are available in both the trial data and the EHR database. EHRs also reflect real-world patterns of care and vary in data quality and completeness. As with any EHR database where data are collected for clinical care, misclassification and incomplete or delayed data entry might have been inherent in the used databases. FH data are obtained primarily (> 80%) from participating community-based cancer centers; consequently, selection bias may exist. Broader applications of these findings should exercise caution just as with clinical trials, which usually include highly selected patient populations.
The FH database and the CGDB are drawn from the same patient pool. However, because each database is subject to different selection criteria and the FH database also applies a sampling fraction, these were considered separate but overlapping populations. Since our eligibility criteria were unrelated to the selection criteria for the databases, we did not expect the overlap between the cohorts to differ significantly from the underlying patient population.
This proof-of-concept study showed that EC arms generated from EHR-derived databases successfully replicated the randomized control arm of the IMblaze370 trial in patients with mCRC. EC arms can serve as meaningful comparators for clinical trials where recruiting a control arm is not feasible. Future research in other patients with CRC and treatment settings, and other tumor types, could help strengthen trust in EC arms and provide further insights relevant to study design and statistical methods.
ACKNOWLEDGMENT
The authors would like to thank Matt Secrest of Genentech, Inc, for his significant contributions to ensuring the quality of the analyses. We would also like to thank Louise Roberts of Genentech, Inc, for reviewing the manuscript.
Carsten Schröder
Employment: Roche
Stock and Other Ownership Interests: Roche, Pfizer, Bayer, Sanofi
Marcus Lawrance
Employment: Roche/Genentech
Stock and Other Ownership Interests: Roche/Genentech, GlaxoSmithKline
Chen Li
Employment: Roche
Christelle Lenain
Employment: Roche
Stock and Other Ownership Interests: Roche
Travel, Accommodations, Expenses: Roche
Shivani K. Mhatre
Employment: Genentech/Roche
Stock and Other Ownership Interests: Genentech/Roche
Marwan Fakih
Honoraria: Amgen
Consulting or Advisory Role: Amgen, Taiho Pharmaceutical, Bayer, Pfizer, Array BioPharma
Speakers' Bureau: Amgen, Guardant Health
Research Funding: Novartis, Amgen, AstraZeneca
Irmarie Reyes-Rivera
Employment: Roche Pharma AG
Stock and Other Ownership Interests: Roche/Genentech
Research Funding: Roche Pharma AG
Travel, Accommodations, Expenses: Roche Pharma AG
Michael T. Bretscher
Employment: F. Hoffmann-La Roche
Stock and Other Ownership Interests: F. Hoffmann-La Roche
Travel, Accommodations, Expenses: F. Hoffmann-La Roche
No other potential conflicts of interest were reported.
SUPPORT
Support for third-party writing assistance for this manuscript, furnished by Jeff Frimpter, MPH, of Health Interactions, was provided by F. Hoffmann-La Roche Ltd.
C.S. and M.L. are co-first authors.
AUTHOR CONTRIBUTIONS
Conception and design: Carsten Schröder, Marcus Lawrance, Christelle Lenain, Shivani K. Mhatre, Irmarie Reyes-Rivera, Michael T. Bretscher
Collection and assembly of data: Carsten Schröder, Marcus Lawrance, Irmarie Reyes-Rivera, Michael T. Bretscher
Data analysis and interpretation: Carsten Schröder, Marcus Lawrance, Chen Li, Christelle Lenain, Marwan Fakih, Irmarie Reyes-Rivera, Michael T. Bretscher
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO’s conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Carsten Schröder
Employment: Roche
Stock and Other Ownership Interests: Roche, Pfizer, Bayer, Sanofi
Marcus Lawrance
Employment: Roche/Genentech
Stock and Other Ownership Interests: Roche/Genentech, GlaxoSmithKline
Chen Li
Employment: Roche
Christelle Lenain
Employment: Roche
Stock and Other Ownership Interests: Roche
Travel, Accommodations, Expenses: Roche
Shivani K. Mhatre
Employment: Genentech/Roche
Stock and Other Ownership Interests: Genentech/Roche
Marwan Fakih
Honoraria: Amgen
Consulting or Advisory Role: Amgen, Taiho Pharmaceutical, Bayer, Pfizer, Array BioPharma
Speakers' Bureau: Amgen, Guardant Health
Research Funding: Novartis, Amgen, AstraZeneca
Irmarie Reyes-Rivera
Employment: Roche Pharma AG
Stock and Other Ownership Interests: Roche/Genentech
Research Funding: Roche Pharma AG
Travel, Accommodations, Expenses: Roche Pharma AG
Michael T. Bretscher
Employment: F. Hoffmann-La Roche
Stock and Other Ownership Interests: F. Hoffmann-La Roche
Travel, Accommodations, Expenses: F. Hoffmann-La Roche
No other potential conflicts of interest were reported.
REFERENCES
- 1.Beaver JA Howie LJ Pelosof L, et al. : A 25-year experience of US Food and Drug Administration accelerated approval of malignant hematology and oncology drugs and biologics: A review. JAMA Oncol 4:849-856, 2018 [DOI] [PubMed] [Google Scholar]
- 2.DeLoughery EP, Prasad V: The US Food and Drug Administration’s use of regular approval for cancer drugs based on single-arm studies: Implications for subsequent evidence generation. Ann Oncol 29:527-529, 2018 [DOI] [PubMed] [Google Scholar]
- 3.European Medicines Agency (EMA) : Guideline on Procedures for the Granting of a Marketing Authorisation under Exceptional Circumstances, Amsterdam, European Medicines Agency, 2005 [Google Scholar]
- 4.US Food and Drug Administration (FDA) : Guidance for Industry: Expedited Programs for Serious Conditions – Drugs and Biologics, Silver Spring, MD, US Food and Drug Administration, 2014 [Google Scholar]
- 5.International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use : ICH Harmonised Tripartite Guideline: Choice of Control Group and Related Issues in Clinical Trials E10, 2000. https://database.ich.org/sites/default/files/E10_Guideline.pdf [Google Scholar]
- 6.Simon R Blumenthal GM Rothenberg ML, et al. : The role of nonrandomized trial in the evaluation of oncology drugs. Clin Pharmacol Ther 97:502-507, 2015 [DOI] [PubMed] [Google Scholar]
- 7.Lim J Walley R Yuan J, et al. : Minimizing patient burden through the use of historical subject-level data in innovative confirmatory clinical trials: Review of methods and opportunities. Ther Innov Regul Sci 52:546-559, 2018 [DOI] [PubMed] [Google Scholar]
- 8.Neuenschwander B Capkun-Niggli G Branson M, et al. : Summarizing historical information on controls in clinical trials. Clin Trials 7:5-18, 2010 [DOI] [PubMed] [Google Scholar]
- 9.van Rosmalen J Dejardin D van Norden Y, et al. : Including historical data in the analysis of clinical trials: Is it worth the effort? 27:3167-3182, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gökbuget N Kelsh M Chia V, et al. : Blinatumomab vs historical standard therapy of adult relapsed/refractory acute lymphoblastic leukemia. Blood Cancer J 6:e473, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barlev A Lin VW Katz A, et al. : Estimating long-term survival of adults with philadelphia chromosome-negative relapsed/refractory b-precursor acute lymphoblastic leukemia treated with blinatumomab using historical data. Adv Ther 34:148-155, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cowey CL Mahnke L Espirito J, et al. : Real-world treatment outcomes in patients with metastatic Merkel cell carcinoma treated with chemotherapy in the USA. Future Oncol 13:1699-1710, 2017 [DOI] [PubMed] [Google Scholar]
- 13.Berger ML Curtis MD Smith G, et al. : Opportunities and challenges in leveraging electronic health record data in oncology. Future Oncol 12:1261-1274, 2016 [DOI] [PubMed] [Google Scholar]
- 14.US Food and Drug Administration : Framework for FDA's Real-World Evidence Program. https://www.fda.gov/media/120060/download [Google Scholar]
- 15.Sharpless NE, Doroshow JH: Modernizing clinical trials for patients with cancer. JAMA 321:447-448, 2019 [DOI] [PubMed] [Google Scholar]
- 16.Carrigan G Whipple S Capra WB, et al. : Using electronic health records to derive control arms for early phase single-arm lung cancer trials: Proof-of-concept in randomized controlled trials. Clin Pharmacol Ther 107:369-377, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eng C Kim TW Bendell J, et al. : Atezolizumab with or without cobimetinib versus regorafenib in previously treated metastatic colorectal cancer (IMblaze370): A multicentre, open-label, phase 3, randomized, controlled trial. Lancet Oncol 20:849-861, 2019 [DOI] [PubMed] [Google Scholar]
- 18.Flatiron Health, New York, NY, https://flatiron.com [Google Scholar]
- 19.Foundation Medicine, Cambridge, MA, https://www.foundationmedicine.com [Google Scholar]
- 20.Olmos A, Govindasamy P: A practical guide for using propensity score weighting in R. Pract Assess Res Eval 20. http://pareonline.net/getvn.asp?v=20&n=13, 2015 [Google Scholar]
- 21.Brookhart MA Wyss R Layton JB, et al. : Propensity score methods for confounding control in nonexperimental research. Circ Cardiovasc Qual Outcomes 6:604-611, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stuart EA, Lee BK, Leacy FP: Prognostic score-based balance measures for propensity score methods in comparative effectiveness research. J Clin Epidemiol 66:S84-S90, 2013. (8 suppl) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Austin PC: Variance estimation when using inverse probability of treatment weighting (IPTW) with survival analysis. Stat Med 35:5642-5655, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Funk MJ Westreich D Wiesen C, et al. : Doubly robust estimation of causal effects. Am J Epidemiol 173:761-767, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eichler H-G Bloechi-Daum B Bauer P, et al. : “Threshold-crossing”: A useful way to establish the counterfactual in clinical trials? Clin Pharmacol Ther 100:699-712, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Curtis MD Griffith SD Tucker M, et al. : Development and validation of a high-quality composite real-world mortality endpoint. Health Serv Res 53:4460-4476, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]