PURPOSE
Children with acute lymphoblastic leukemia (ALL) are treated according to risk-based protocols defined by the Children's Oncology Group (COG). Alignment between real-world clinical practice and protocol milestones is not widely understood. Aggregate deidentified electronic health record (EHR) data offer a useful resource to evaluate real-world clinical practice.
METHODS
A cohort of children with ALL was identified in the Cerner Health Facts deidentified aggregate EHR data. Manual review identified candidate procedural milestones. Automated methods were developed to classify likely standard-risk precursor B-cell ALL patients. Milestone procedures were adjusted relative to initiation of therapy and then aligned to the COG protocols for standard induction therapy.
RESULTS
We identified 7,728 patients with pediatric ALL with 188,187 encounters. Records for lumbar punctures (LP) and bone marrow biopsies were frequently present in the data and were appropriate targets to evaluate guideline performance. Alluvial graph analysis of 14 health systems indicated that none of the systems have data from all three COG-recommended lumbar procedures for all patients but alignment demonstrated that most systems test at the recommended times.
CONCLUSION
Source-system variation introduces inconsistency and incompleteness into aggregate EHR data. Data visualization was helpful in characterizing and interpreting the data. Health systems with patients meeting the inclusion criteria demonstrated strong alignment with the recommended milestones for LP. Large-scale aggregate EHR data are useful to evaluate alignment of recommended versus actual clinical milestones in support of treating children with ALL. This work can inform other guideline and protocol driven care.
INTRODUCTION
Variation in healthcare delivery affects patient outcomes when real-world practice deviates from widely accepted evidence-based protocols. Pediatric oncology is distinguished by the widespread use of protocols provided by the Children's Oncology Group (COG) for the management of common childhood cancers.1 The American Academy of Pediatrics recommends treatment of pediatric cancer at a tertiary center, with board-certified pediatric oncologists.2 Most tertiary pediatric academic institutions are members of the COG, with more than 90% of patients with pediatric cancer cared for at COG sites.3 In addition to treatment guidelines, COG protocols also include detailed information regarding timing of procedures, laboratory and diagnostic evaluations, treatment, and follow-up after therapy.
CONTEXT
Key Objective
How can data science methods using aggregate, deidentified, electronic health record (EHR) data inform oncologists about the alignment between protocol defined milestones and real-world clinical practice?
Knowledge Generated
Using lumbar puncture timing specified by the Children's Oncology Group (COG) as an example, we found a strong level of alignment between real-world practice and protocol recommendations for children with standard-risk acute lymphoblastic leukemia.
Working with large, deidentified data sets benefits from the application of data science methods and visualizations to account for missing data and other challenges.
Relevance
This work establishes data science methods that can be reapplied to aggregate analysis of other guideline and protocol-based events and serves as a precursor to evaluating the impact of deviation from guidelines on clinical outcomes.
Large-scale analysis of the alignment between real-world clinical practice and standardized protocols is challenging. Patients with cancer experience complex care that includes evaluation with lab and other diagnostic tests followed by treatment that often includes chemotherapy, radiation therapy, surgery, and/or transplantation. The treatment protocol assigned to a patient depends on personal risk level. For example, pediatric precursor B-cell acute lymphoblastic leukemia (ALL) cases are classified into standard or high risk categories based on National Cancer Institute (NCI) criteria.4 Patients with NCI standard-risk ALL must have an age between ≥ 1 and < 10 years and an initial WBC count of < 50 ×103/mcL. Patients are considered high risk if they are 10 years of age or older or have a WBC count ≥ 50 × 103/mcL. Children younger than 1 year of age are considered to be a distinct ALL risk category with a distinct protocol. Patients with ALL with Down syndrome have inferior survival compared to those without Down syndrome5-7 and are also a separate risk cohort.
Several database resources are used for pediatric cancer research, each with their own strengths and limitations. For example, the SEER registry is an established source of cancer data and has been used to evaluate pediatric leukemia.8 Registries such as SEER are standardized and can include details from pathology and radiology. However, registries are episodic and are often populated by manual data entry, limiting the number of patients and volume of data. SEER has limited information about comorbidities and only represents 12 states.9 Data derived from billing and claims can provide a view into patient interactions within the healthcare system. For example, the pediatric health information system database has been used extensively to characterize pediatric cancer10,11 and has demonstrated the value of combining diagnosis data with medication information.12 Key limitations of registries and billing data are the lack of temporal specificity, the absence of results, and limited ability to scale up.
Electronic health record (EHR) systems have become widely adopted following the Meaningful Use funding of the American Recovery and Reinvestment Act (ARRA).13 EHR systems include results, serve as the legally binding medical record, and are rich in date- and time-stamped details. Data from EHR systems can be applied to clinical research, including the use of EHR data to characterize the trajectories of patients treated within a single organization.14 One major EHR vendor, Cerner, has developed a large-scale aggregate data warehouse, Health Facts (HF), in which a subset of their client base has provided data rights to assemble and analyze a subset of their EHR data. The data are deidentified to Health Insurance Portability and Accountability Act (HIPAA) standards and are scrubbed of all protected health information. The HF data have been applied to research in cardiology and other disease states.15-19 A comparison of HF to the National Inpatient Survey demonstrated high correlation in the frequency of diagnoses between HF and a nationally representative survey.20 The HF data include laboratory data, inpatient medications, demographics, surgical data, billing data, and a wide variety of clinical events including vitals.
We demonstrate processes to use large-scale aggregate EHR data from healthcare organizations in the United States to compare real-world clinical practice to COG treatment protocols for managing NCI standard-risk ALL cases.
METHODS
To develop a representation of the COG ALL protocols, we reviewed the COG pre-B-cell ALL protocols for standard- and high-risk regimens1 to develop a reference framework against which to map scheduled events in the data from HF database (Cerner Corporation, Kansas City, MO). HF includes deidentified, HIPAA-compliant, EHR data from Cerner clients who agree to participate. The version of HF data in use at Children's Mercy includes more than 68 million patients, from 664 facilities associated with 100 nonaffiliated organizations, 4 billion lab results, 734 million inpatient medication orders, and other data. Significantly, the data do not include text reports as those cannot be reliably deidentified. Children's Mercy received the 2018 version of the HF data and installed the data into Microsoft Azure (Redmond, WA). Queries were performed with Microsoft SQL Server Management Studio version 17.9 and R Studio version 1.1.453 with R version 3.5.2.21,22 Queries evaluated data from 2000 to 2017.
A preliminary query to identify pediatric ALL diagnoses was performed and included the following: International Classification of Diseases (ICD)-9 diagnosis codes (204.0, 204.00, and 204.01), ICD-10-CM diagnosis codes (C91.0, C91.00, and C91.01) and patients 0 to 18 years of age. We excluded nonclinical patient encounters and ALL diagnosis codes related to relapse. To align standard-risk (SR-ALL) patient trajectories with standard COG protocols at a large scale, we needed to develop analytical methods, in the absence of risk-specific ICD codes and text notes, to exclude patients from other risk categories before inferring compliance with COG guidelines. We also sought reliable, consistent, and widely available milestone procedures in the EHR data.
HF data from seventy pediatric patients with ALL were randomly selected for manual evaluation. Oncologists from Children's Mercy reviewed each patient to assess the availability of laboratory data, diagnoses, diagnostic procedures, inpatient medications, and demographic and clinical data. This information was used to identify the milestone events likely to be well represented in the data.
The Children's Mercy Institutional Review Board has deemed work with HF to be nonhuman subjects research.
RESULTS
We developed a reference framework representing the milestone events in the care of a child treated following the COG protocols for NCI SR-ALL versus those with NCI high-risk ALL (HR-ALL) (Fig 1). This framework was used to define machine-readable inclusion and exclusion criteria and served as the reference timeline against which date- and time-stamped data found in HF would be aligned.
FIG 1.
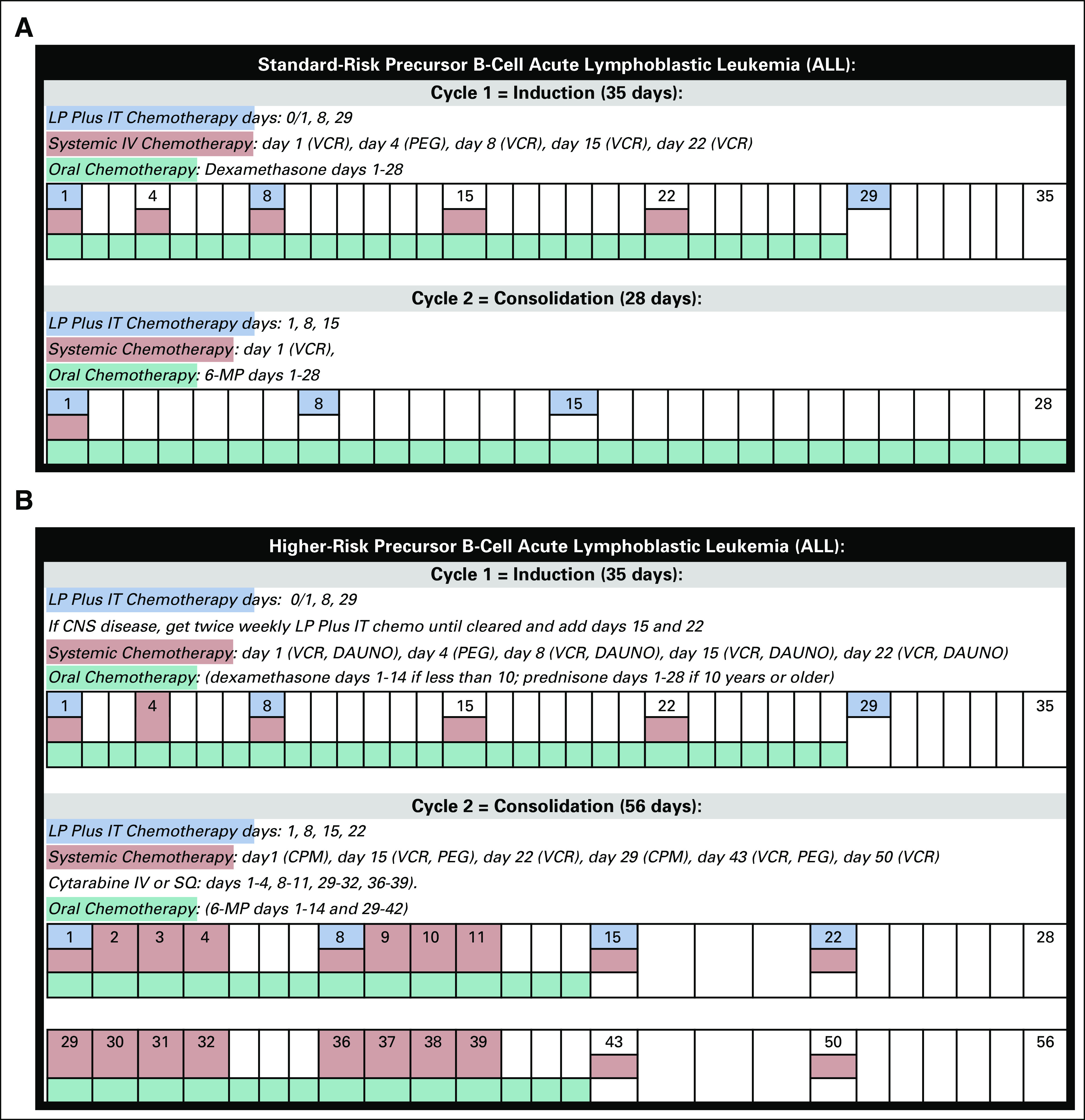
Milestone events for B-cell ALL. (A) Milestone events for standard-risk ALL, treatment cycles 1 (induction) and 2 (consolidation). (B) Milestone events for higher-risk ALL, treatment cycles 1 (induction) and 2 (consolidation). ALL, acute lymphoblastic leukemia; CPM, cyclophosphamide; DAUNO, daunorubicin; IT, intrathecal; LP, lumbar puncture; MP, mercaptopurine; PEG, pegaspargase; VCR, vincristine.
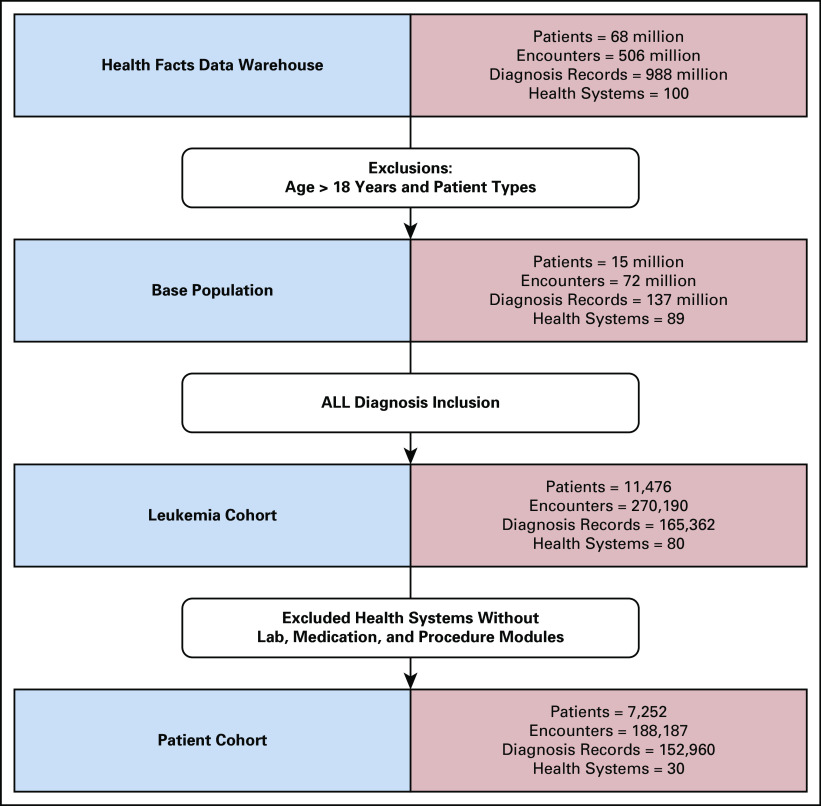
The preliminary query to identify pediatric ALL diagnoses yielded 11,476 patients with pediatric ALL in HF from 80 nonaffiliated health systems (Fig 2). These patients have 270,190 ALL-diagnosis-related encounters in the data. We then excluded health systems without adequate lab, medication, or procedure data,23 resulting in a subset of 7,252 patients with pediatric ALL from 30 health systems, with 188,187 diagnosis-related encounters.
FIG 2.
ALL cohort development. Iterative inclusions and exclusions to develop the preliminary ALL cohort. Data from organizations that do not consistently capture labs, medications, or procedures in Cerner were excluded. ALL, acute lymphoblastic leukemia.
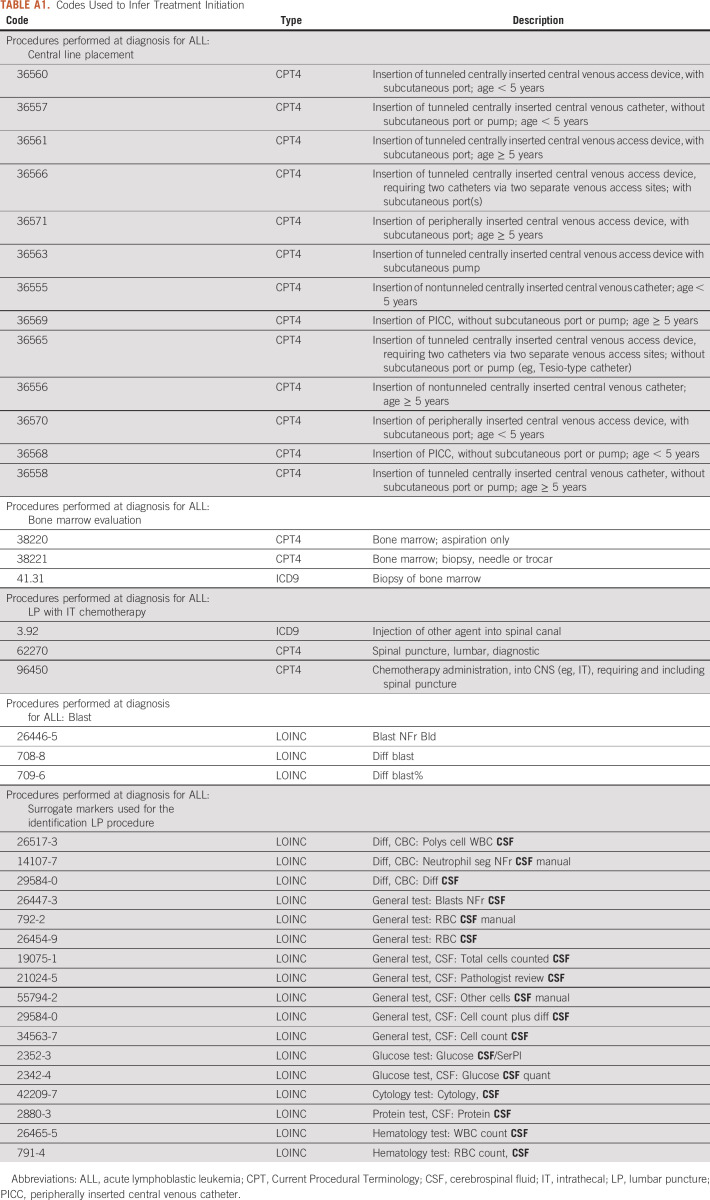
Manual review of data from 70 patients from this group was instructive in identifying data from procedures required by the COG protocol that are well represented in HF. Lumbar puncture (LP) procedures or a related CSF lab were found in 83% (58/70) of these cases. Current Procedural Terminology (CPT) and ICD procedure codes were used to directly or indirectly identify lumbar procedures (Appendix Table A1). The timing for LP recommended by COG protocols did not change during the period covered by HF, 2000-2017.
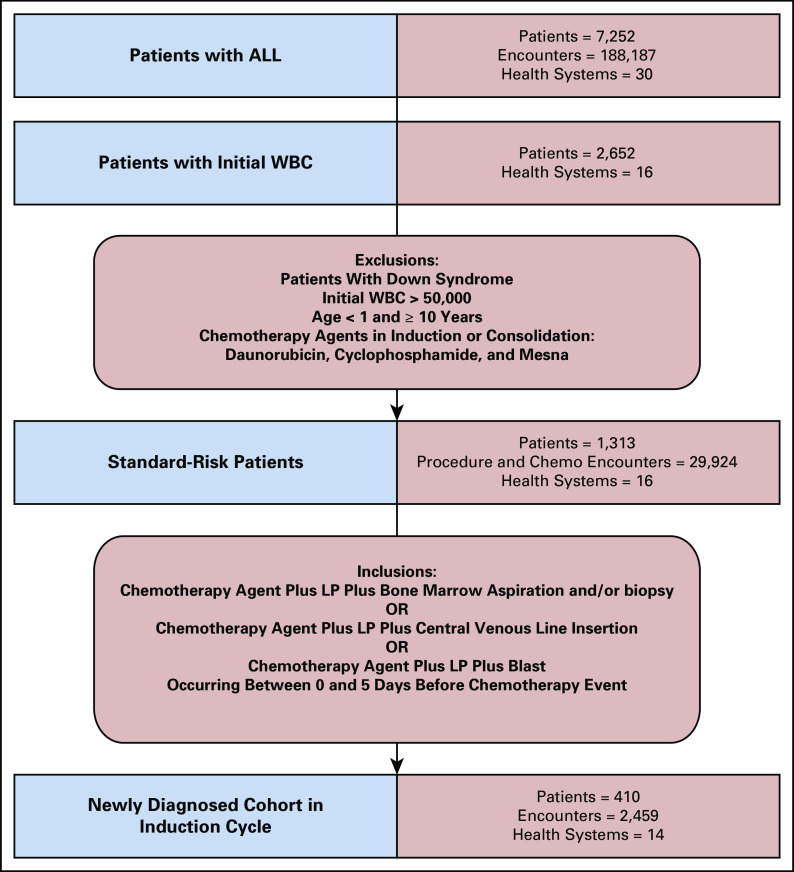
Treatment protocols are based on risk category. To include the greatest number of patients, we narrowed our protocol alignment work to likely SR-ALL and excluded patients with additional risk factors (Fig 3). Risk status is not explicitly documented, requiring us to develop analytical methods based on patient, lab, and medication factors to infer risk status. Patient-level exclusions were children younger than the age of one year, age of 10 years or older, and patients with a diagnosis code for Down syndrome (ICD-9 758.0 and ICD-10 Q90.0). Lab-based exclusion was a WBC result of > 50,000 within 30 days of initiation of therapy. Patients without a WBC available in the 30-day window of initiation of treatment were excluded. This requirement reduced our initial cohort from 7,252 to 2,652.
FIG 3.
Likely standard-risk ALL cohort development. Iterative inclusions and exclusions to develop a cohort of patients likely to have standard-risk ALL and likely to have been newly diagnosed with ALL. ALL, acute lymphoblastic leukemia; LP, lumbar puncture.
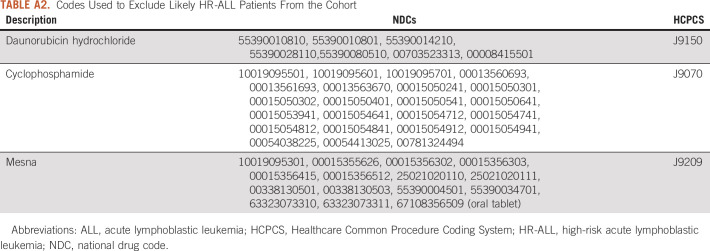
Patients with SR-ALL receive a three-drug induction chemotherapy regimen: vincristine, dexamethasone, and pegaspargase. Patients with HR-ALL receive an additional chemotherapy agent, daunorubicin, during induction chemotherapy and cyclophosphamide during consolidation. Mesna is often used to provide chemoprotection to patients with HR-ALL during consolidation. We queried the medication table, coded with national drug code values, and the procedure table, coded with CPT and Healthcare Common Procedure Coding System (HCPCS) values to exclude patients with the HR-ALL drugs (Appendix Table A2). These filters narrowed the cohort to 1,313 patients with SR-ALL from 16 nonaffiliated health systems (Fig 3). Other information potentially indicative of high risk, for example, molecular markers or cytogenetics, was not available.
Initiation of therapy is not provided as a discrete event in HF. To infer the start of induction chemotherapy (day 1) and to exclude relapsed patients, we used the earliest date of a chemotherapy and a combination of other events. Start of therapy was defined as first chemotherapy event in the same period as a lumbar procedure and at least one other identifier: bone marrow aspiration and/or biopsy, central venous line insertion, or blasts on the same day or within 5 days before the first chemotherapy event (Appendix Table A1). Of the 1,313 patients with SR-ALL, 1,005 patients had codes related to LP or lab tests involving CSF as a surrogate (126 lumbar only, 342 CSF only, and 540 both). The medication codes used to identify the earliest date of chemotherapy included cytarabine, intravenous vincristine, and injection or infusion of cancer chemotherapeutic substance and dexamethasone (Appendix Table A3).
Using the methods described above, excluding patients missing data for the events associated with day 1 of treatment, we identified 410 patients with an LP event (32% of SR-ALL cohort) (Fig 3). Based on the available data, these patients met the criteria for SR-ALL and had documentation in HF of the treatment or procedures needed to infer the date on which treatment was initiated. The remaining patients with SR-ALL were not analyzed in this study.
The 410 patients with at least one LP were analyzed using an UpSet plot24 to characterize the availability of data for LP events at predicted times (Fig 4A). For day 1, we used the date on which the standard of care induction therapy is first noted and positioned all other dates relative to day 1. For clarity, we grouped all days before treatment as diagnostic, up to and including day 1, days 7, 8, and 9, and 28, 29, 30, 31, and 32. The vertical lines connect the days and represent the day sequence relationship. The unique number of patients in the sequence relationship is shown at the top of the bar chart. If data are not available for a time categorical, a light gray circle is shown. The number of patients with an LP on a given day is in the left side of the bar chart. We noted 206 patients had data from the three COG recommended times for LP, before or on day 1, 8, and 29 of induction chemotherapy (Fig 4A). Fifty eight patients had data only for the diagnostic or day 1 milestone.
FIG 4.
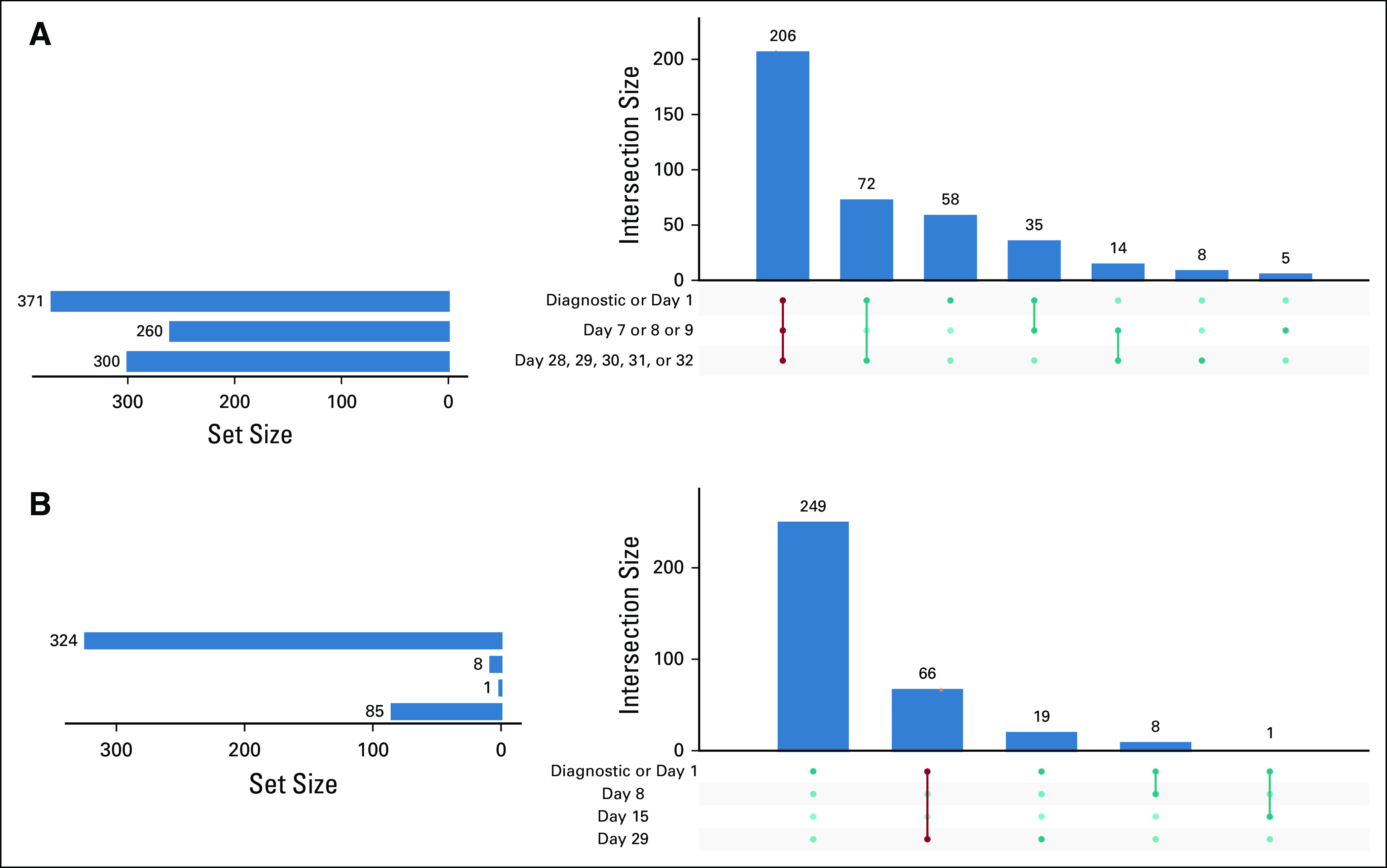
UpSet graph analysis of data availability. Horizontal bars represent the sets being compared; vertical bars represent the intersections. The values in blue represent the combination of milestone events expected from the protocol. A procedure classified as diagnostic occurred within 7 days before the inferred date of initial treatment. (A) UpSet graph of LP data availability. Dates are relative to initiation of treatment. Rows represent COG milestone date ranges. (B) Bone marrow data availability. LP, lumbar puncture.
We performed a similar UpSet graph data availability analysis for bone marrow procedural codes with the newly diagnosed cohort. Of the 410 patients, 363 had a bone marrow procedure. Most of these (324, 89%) had data available from a bone marrow encounter during the diagnostic or day 1 milestone period, whereas only 85 (23%) had a bone marrow encounter on day 29 (Fig 4B). We identified 66 patients who aligned with COG protocol for disease evaluation at days 0, 1, and 29. We also noted that 19 patients had a bone marrow procedure on day 29. The UpSet graph indicated that the availability and sequence of bone marrow procedures were not as widely available as LP procedures.
One potential explanation for the variation in data availability demonstrated in the UpSet graphs is that some contributing organizations consistently lack data from particular COG recommended dates. To examine this, we created categorical variables representing whether a patient did or did not have an LP for each of the three recommended dates. We then used an Alluvial graph25 in R to investigate the patterns of LP usage for each of the 14 organizations with the 410 patients (Fig 5). All 14 organizations had multiple pathways. For example, most patients at organization 1 follow a trajectory that includes all three recommended lumbar dates. One minor track of patients at organization 1 (uppermost track) does not have data for the first date but does for the second but not the third. Another small group has the first date, not the second but does have the third. Organization 3 has major groups following distinct trajectories.
FIG 5.
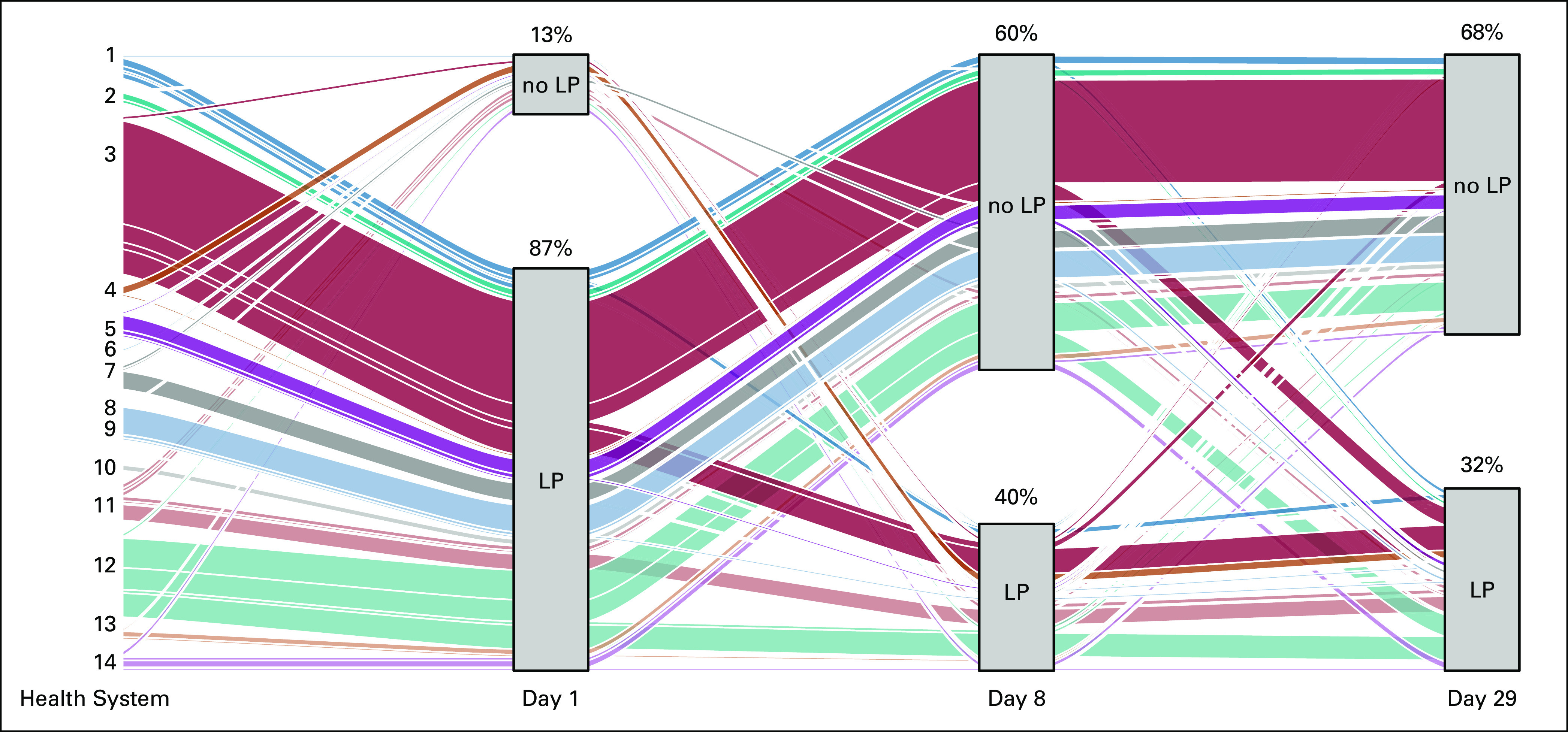
Alluvial graph of patient trajectories at 14 organizations. Data from 14 organizations representing the number of patients in LP cohort with each milestone procedure—day 0, 1, day 7, 8, 9, and day 28, 29, 30, 31, 32. The width of each band represents the proportional number of patients following each trajectory. LP, lumbar puncture.
To further examine variation between organizations, we analyzed the granular timing of LP, relative to treatment initiation, for eight HF Health Systems with at least 40 encounters including an LP (Fig 6). Here, we show alignment of the LP milestone timing in concordance with standard of care during induction therapy, highlighting those LP that would align with LP on days 1, 8, and 29. Because of expected minor modifications in timing of therapy due to scheduling or patient-related delays, these days were grouped as follows: days 0-1, days 7-9, and days 28-32.
FIG 6.
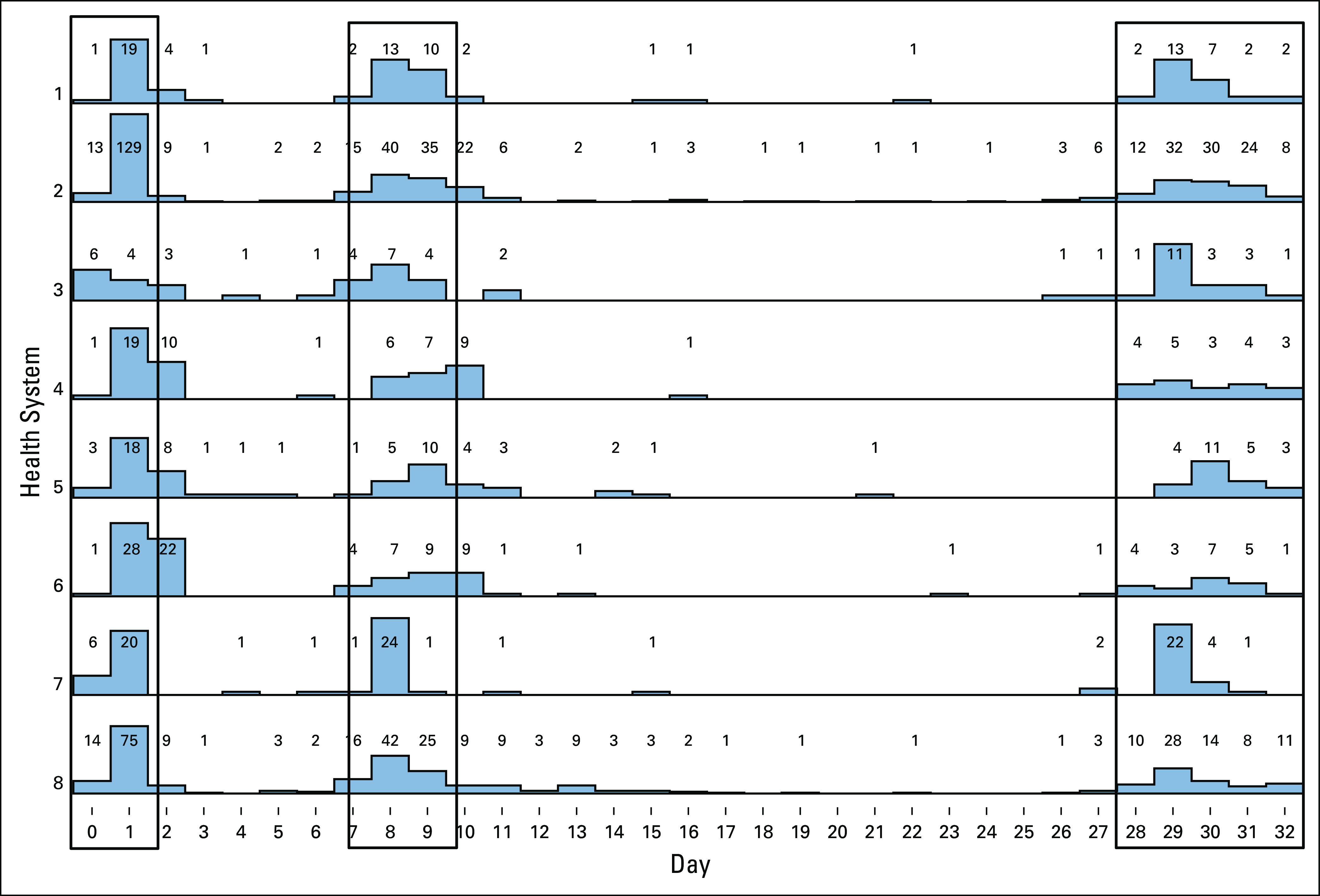
Alignment of lumbar puncture (LP) timing. Dates of lumbar punctures, relative to day 0 before treatment, were aligned for all qualifying patients from eight nonaffiliated health systems. Boxes indicate the COG recommended timing for LPs.
DISCUSSION
Evidence-based protocols guide patient care. Many organizations perform internal analyses comparing real-world practice to protocol recommendations within their institution using resources such as an enterprise data warehouse. Deeper understanding of alignment between clinical behavior and recommended practices will be enabled by cross-organization analyses. For pediatric leukemia, 90% of patients are treated at COG member institutions. We identified a large group of patients with pediatric ALL using aggregate deidentified EHR data from nonaffiliated organizations. The outcomes of pediatric ALL correlate with the risk level. Unfortunately, risk level is not currently documented as a discrete diagnosis code, and the text notes that would clarify risk level are not accessible in a deidentified data resource. Likewise, the date of initiation of treatment is not clearly available. To develop the capabilities needed to map real-world machine-readable clinical data to the events required by COG treatment protocols for SR-ALL, we developed a reference timeline visualizing representative milestones of care.
Beginning with manual review of a small subset of these patients with ALL, and then extending that to an automated data extraction, we developed informatics methods to infer risk classification and the date of treatment initiation. We applied stringent criteria to maximize the likelihood of accurately classifying patients. For example, we rejected 3,338 patients whose data did not include a WBC count within the 30 days before the initiation of treatment.
Manual data review suggested widespread availability of LP data, an event with timing specified by the COG protocol. We also found wide availability of laboratory tests but the frequency of those is not as explicitly articulated in the COG protocol. We used UpSet graphs to evaluate the availability of LP data from the three required times. Day 0, 1 procedures were the most widely available. A significant group of patients included all three COG required LP but there are also gaps in the data. This could reflect later phases of care provided at facilities not contributing to HF, variations in coding practices, and other process factors. HF does not provide the care setting (ie, infusion center) or medications that were not ordered from an in-house pharmacy, preventing us from further investigating the gaps.
A key challenge in analyses of EHR data is missing data; this issue is amplified when the data are deidentified and not traceable to the source. Our use of UpSet and Alluvial graphs to understand data availability for pediatric cancer was helpful in characterizing the complexity of this real-world data. The Alluvial graph demonstrated that the gaps in data are not because of contributing organizations consistently failing to follow COG standards for the timing of LP because some threads through each milestone were visible. For example, organization 3 had a majority of patients who missed the day 8 milestone, but significant strands traverse the day 8 milestone. Possible explanations for the data gaps could include patients receiving care at a separate organization, discrepancies in documentation practices between providers or coders, and patient mortality. Although missing data are problematic, the use of visualization to place missingness in context is helpful to the researcher; likewise, familiarity with the nuances of EHR source systems and workflows is particularly important.
Having identified methods to impute day 1 of care, we developed an alignment method to map all other events to a COG-based timeline of care. We demonstrated that LP performed at eight independent healthcare organizations align closely with the required timing of this procedure. This novel approach of aligning time-based events harvested from fully deidentified (date-shifted) data from nonaffiliated organizations against protocol recommended events can be extended to other required and ad hoc procedures. This is analogous to aligning unknown DNA sequences to a reference sequence. Although there was little deviation from the recommended timing of LP, future work can evaluate higher risk cohorts and alignment with other milestone events.
Working with a large-scale aggregate data resource derived from EHR data has a number of inherent challenges based on factors specific to each contributor and to the process of aggregating and deidentifying the data. EHR systems are generally designed and implemented to support clinical workflow and documentation, and generating high data quality for secondary analysis has generally been a limited focus. Data quality concerns are well known in EHR-derived data.26 For example, variations in the quality of diagnosis coding for brain neoplasms have been demonstrated to correlate with workflow, care setting, and personnel.27 Resolution of these challenges requires the use of emerging systems to provide monitoring of EHR data for data quality issues28,29 and the inclusion of data quality considerations during EHR system implementations.
The strict deidentification process used to generate HF removes text notes that could confirm the risk status and the date of treatment initiation. Likewise, EHR implementation variations among contributing organizations affect the data. For example, although procedure codes for LP were available from 65 organizations, a time series analysis focusing exclusively on a lab test might yield more qualifying patients because the EHR laboratory modules are widely used across the Cerner organizations contributing to HF.23 By combining several factors (first chemotherapy event, presence of blasts, and procedures), we were able to raise the likelihood that our initiation of therapy phenotype is specific to initiation of SR-ALL therapy and unlikely to represent similar sequences of events during reinduction therapy for relapsed ALL.
Despite these limitations, the power of using large-scale data to understand real-world health care is significant. The process of aligning patient experiences to a widely accepted protocol establishes the basis for future outcomes research. For example, do children whose care deviates from the protocol have different outcomes from those whose care aligns with the guideline? Likewise, the methods developed for this work have broad utility for additional data science research to evaluate the trajectories of patients with cancer using EHR data.
ACKNOWLEDGMENT
The authors would like to acknowledge the contributions of Bourke Hutchinson.
Appendix
TABLE A1.
Codes Used to Infer Treatment Initiation
TABLE A2.
Codes Used to Exclude Likely HR-ALL Patients From the Cohort
TABLE A3.
Medication Codes Used to Infer Treatment Initiation
SUPPORT
Supported by the Masonic Cancer Alliance, Partners Advisory Board Funding.
AUTHOR CONTRIBUTIONS
Conception and design: Nicole M. Wood, Karen Lewing, Janelle Noel-MacDonnell, Doina Caragea, Mark A. Hoffman
Collection and assembly of data: Sierra Davis, Karen Lewing, Earl F. Glynn, Mark A. Hoffman, Nicole M. Wood
Data analysis and interpretation: All authors
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Karen Lewing
Stock and Other Ownership Interests: St Luke's Surgicenter, Centerpointe Surgicenter, Independence
Janelle Noel-MacDonnell
Research Funding: Merck, Genzyme
Mark A. Hoffman
Stock and Other Ownership Interests: Various
No other potential conflicts of interest were reported.
REFERENCES
- 1.Hunger SP Loh ML Whitlock JA, et al. : Children's Oncology Group's 2013 blueprint for research: acute lymphoplastic leukemia. Pediatr Blood Cancer 60:957-963, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corrigan JJ, Feig SA: Guidelines for pediatric cancer centers. Pediatrics 113:1833-1835, 2004 [DOI] [PubMed] [Google Scholar]
- 3.O'Leary M Krailo M Anderson JR, et al. : Progress in childhood cancer: 50 years of research collaboration, a report from the Children's Oncology Group. Semin Oncol 35:484-493, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith M Arthur D Camitta B, et al. : Uniform approach to risk classification and treatment assignment for children with acute lymphoblastic leukemia. J Clin Oncol 14:18-24, 1996 [DOI] [PubMed] [Google Scholar]
- 5.Athale UH Puligandla M Stevenson KE, et al. : Outcome of children and adolescents with Down syndrome treated on Dana-Farber Cancer Institute Acute Lymphoblastic Leukemia Consortium Protocols 00-001 and 05-001. Pediatr Blood Cancer 65:e27256, 2018 [DOI] [PubMed] [Google Scholar]
- 6.Bassal M La MK Whitlock JA, et al. : Lymphoblast biology and outcome among children with Down syndrome and ALL treated on CCG-1952. Pediatr Blood Cancer 44:21-28, 2005 [DOI] [PubMed] [Google Scholar]
- 7.Whitlock JA Sather HN Gaynon P, et al. : Clinical characteristics and outcome of children with Down syndrome and acute lymphoblastic leukemia: A Children's Cancer Group study. Blood 106:4043-4049, 2005 [DOI] [PubMed] [Google Scholar]
- 8.Nasir SS Giri S Nunnery S, et al. : Outcome of adolescents and young adults compared with pediatric patients with acute myeloid and promyelocytic leukemia. Clin Lymphoma Myeloma Leuk 17:126-132.e1, 2017 [DOI] [PubMed] [Google Scholar]
- 9.Yu JB Gross CP Wilson LD, et al. : NCI SEER public-use data: Applications and limitations in oncology research. Oncology (Williston Park) 23:288-295, 2009 [PubMed] [Google Scholar]
- 10.Desai AV Kavcic M Huang YS, et al. : Establishing a high-risk neuroblastoma cohort using the Pediatric Health Information System Database. Pediatr Blood Cancer 61:1129-1131, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winestone LE Getz KD Miller TP, et al. : The role of acuity of illness at presentation in early mortality in black children with acute myeloid leukemia. Am J Hematol 92:141-148, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fisher BT Harris T Torp K, et al. : Establishment of an 11-year cohort of 8733 pediatric patients hospitalized at United States free-standing children's hospitals with de novo acute lymphoblastic leukemia from health care administrative data. Med Care 52:e1-e6, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adler-Milstein J, Jha AK: HITECH Act drove large gains in hospital electronic health record adoption. Health Aff (Millwood) 36:1416-1422, 2017 [DOI] [PubMed] [Google Scholar]
- 14.Pham T Tran T Phung D, et al. : Predicting healthcare trajectories from medical records: A deep learning approach. J Biomed Inform 69:218-229, 2017 [DOI] [PubMed] [Google Scholar]
- 15.Campbell R Dean B Nathanson B, et al. : Length of stay and hospital costs among high-risk patients with hospital-origin Clostridium difficile-associated diarrhea. J Med Econ 16:440-448, 2013 [DOI] [PubMed] [Google Scholar]
- 16.Campbell RS Chaudhari P Hays HD, et al. : Outcomes associated with conventional versus lipid-based formulations of amphotericin B in propensity-matched groups. Clinicoecon Outcomes Res 5:507-517, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goyal A Spertus JA Gosch K, et al. : Serum potassium levels and mortality in acute myocardial infarction. JAMA 307:157-164, 2012 [DOI] [PubMed] [Google Scholar]
- 18.Vogel TR, Kruse RL: Risk factors for readmission after lower extremity procedures for peripheral artery disease. J Vasc Surg 58:90-97.e1-4, 2013 [DOI] [PubMed] [Google Scholar]
- 19.Shafiq A Goyal A Jones PG, et al. : Serum magnesium levels and in-hospital mortality in acute myocardial infarction. J Am Coll Cardiol 69:2771-2772, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeShazo JP, Hoffman MA: A comparison of a multistate inpatient EHR database to the HCUP Nationwide Inpatient Sample. BMC Health Serv Res 15:384, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.RCoreTeam : R: A Language and Environment for Statistical Computing. Vienna, Austria, R Foundation for Statistical Computing, 2019 [Google Scholar]
- 22.RStudioTeam : RStudio: Integrated Development for R. Boston, MA, R Studio, 2015 [Google Scholar]
- 23.Glynn EF, Hoffman MA: Heterogeneity introduced by EHR system implementation in a de-identified data resource from 100 non-affiliated organizations. JAMIA Open 2:554-561, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lex A Gehlenborg N Strobelt H, et al. : UpSet: Visualization of intersecting sets. IEEE Trans Vis Comput Graph 20:1983-1992, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosvall M, Bergstrom CT: Maps of random walks on complex networks reveal community structure. Proc Natl Acad Sci U S A 105:1118-1123, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Botsis T Hartvigsen G Chen F, et al. : Secondary use of EHR: Data quality issues and informatics opportunities. Summit Transl Bioinform 2010:1-5, 2010 [PMC free article] [PubMed] [Google Scholar]
- 27.Diaz-Garelli F Strowd R Lawson VL, et al. : Workflow differences affect data accuracy in oncologic EHRs: A first step toward detangling the diagnosis data Babel. JCO Clin Cancer Inform 4:529-538, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dziadkowiec O Callahan T Ozkaynak M, et al. : Using a data quality framework to clean data extracted from the electronic health record: A case study. EGEMS (Wash DC) 4:1201, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feder SL: Data quality in electronic health records research: Quality domains and assessment methods. West J Nurs Res 40:753-766, 2018 [DOI] [PubMed] [Google Scholar]