PURPOSE
Many institutions throughout the world have launched precision medicine initiatives in oncology, and a large amount of clinical and genomic data is being produced. Although there have been attempts at data sharing with the community, initiatives are still limited. In this context, a French task force composed of Integrated Cancer Research Sites (SIRICs), comprehensive cancer centers from the Unicancer network (one of Europe's largest cancer research organization), and university hospitals launched an initiative to improve and accelerate retrospective and prospective clinical and genomic data sharing in oncology.
MATERIALS AND METHODS
For 5 years, the OSIRIS group has worked on structuring data and identifying technical solutions for collecting and sharing them. The group used a multidisciplinary approach that included weekly scientific and technical meetings over several months to foster a national consensus on a minimal data set.
RESULTS
The resulting OSIRIS set and event-based data model, which is able to capture the disease course, was built with 67 clinical and 65 omics items. The group made it compatible with the HL7 Fast Healthcare Interoperability Resources (FHIR) format to maximize interoperability. The OSIRIS set was reviewed, approved by a National Plan Strategic Committee, and freely released to the community. A proof-of-concept study was carried out to put the OSIRIS set and Common Data Model into practice using a cohort of 300 patients.
CONCLUSION
Using a national and bottom-up approach, the OSIRIS group has defined a model including a minimal set of clinical and genomic data that can be used to accelerate data sharing produced in oncology. The model relies on clear and formally defined terminologies and, as such, may also benefit the larger international community.
INTRODUCTION
Most national and international funding agencies have acknowledged the importance of making clinical and genomic data publicly available to obtain enough information1 for correlative studies on biomarkers associated with response to specific targeted immunotherapies.
CONTEXT
Key Objective
How can we improve standardization of clinical and genomic data to improve data sharing and interoperability by capturing the disease course in the context of precision medicine in Oncology?
Knowledge Generated
We propose an event-based data model of a minimal set of clinical and genomic items using international standards and terminologies enabling a strong interoperability. OSIRIS common data model is modular and extensible to other types of data.
Relevance
OSIRIS was developed in the context of large precision medicine clinical trials to incorporate the longitudinal changes associated with disease progression and resistance to therapeutic interventions. It could also provide an effective real-world data ecosystem by developing a data standard, which, if used, could improve the compatibility, quality, and consistency of electronic health record. In both cases, OSIRIS may facilitate the application of artificial intelligence and enhance supervised machine learning and data science in the context of clinical care and clinical research.
A large amount of cancer genomics data has been provided to the scientific community through significant national and international collaborations (ie, the CIT research program,2 the Cancer Genome Atlas [TCGA],3 and the International Cancer Genome Consortium [ICGC]4). Although these data sets are freely available, researchers and clinicians face major obstacles using them because of (1) a lack of standards regarding genomic data characterization and quality5 and (2) limited clinical data in particular related to disease outcomes.
Despite these obstacles, the number of data sharing initiatives continues to grow. Several international data-sharing networks have recently emerged to help researchers get access to clinical and genomic data in genomic cancer medicine. For examples, international networks such as PCORnet,6 OHDSI Research Network,7 GENIE,8 BRCA Exchange from GA4GH7,9 CancerLinQ,10 ORIEN,11 and ICGC ARGO,12 and European networks such as Europe lung cancer data collection13 and the German Cancer Consortium (DKTK)14 are some important initiatives in this field. Although such initiatives are interesting, access to full data is restricted to consortium members or lacks relevant clinical information to link genomic alterations with clinical benefit under specific drugs.
Between 2011 and 2013, precision medicine was becoming pervasive and Integrated Cancer Research (SIRICs) sites in France conducted molecular profiling clinical trials (eg, SHIVA, MOSCATO, and ProfiLER). These clinical trials meant a radical paradigm shift by providing innovative therapies guided by genomic alterations identified in a tumor. The idea in 2013 was to link multiple SIRIC-program clinical trials to reach a critical number of patients, which would enable a meaningful analysis. To empower the joint analysis of these trials, a common minimal data set along with a Common Data Model (CDM) was needed to reach our goal. We wanted a CDM to be modular, limited in size and agile, extensible beyond clinical and genomic data, and able to capture the longitudinal changes associated with disease progression and resistance to therapeutic interventions.
In this context, the OSIRIS (Interoperability and data sharing of clinical and biological data in oncology) initiative15 was launched in 2015 to address data heterogeneity with four major commitments: (1) using a bottom-up approach by keeping the OSIRIS set as minimal as possible, (2) reaching a national consensus from all stakeholders involved in cancer research, (3) using internationally established terminologies as much as possible, and finally (4) defining implementation rules to guarantee data consistency of the OSIRIS set across institutions.
A French national task force composed of Comprehensive Cancer Centers from the Unicancer network,16 University Hospitals, and the eight SIRICs was formed under the auspices of the French Institut National du Cancer (INCa).17 Herein, we propose a minimum set of clinical and genomic data relevant to the field of precision medicine in oncology, a data model that allows us to capture the disease course, including therapy response and toxicity. OSIRIS was compared with other similar initiatives such as mCODE18 and OMOP19 and was tested in a set of 300 patients included in six different multicentric studies as a proof-of-concept.
MATERIALS AND METHODS
CDM: Identification of the Data Concepts
The goal of the CDM was to share a set of standardized, extensible data concepts (eg, patient, tumor events, treatment, response evaluation, and adverse event) that enabled consistency of both data and their meaning across applications. The group focused its efforts on (1) providing a comprehensive resource of cancer event–based data and their temporal relationships, that is, to help clinicians discover longitudinal changes associated with disease progression and resistance to therapeutic interventions; (2) creating a modular and extensible data model, that is, to integrate omics data from different experiments on the same samples (vertical integration) or across studies on the same variables (horizontal integration); and (3) creating the technical conditions for interoperability.
Minimum Data Set: Standardization and Interoperability of the Data Elements
A data set was defined as a collection of variables, such as the gender and age of a patient, with a list of possible values for each of them. Each variable is called a Data Element (DE), and the list of possible values a Code List (CL). To define a minimum set of valuable DEs at the national level, our approach was based on the following steps: (1) identifying the most relevant items based on a 2015 Institut Curie data model, the experience of the Unicancer network of comprehensive cancer centers with Consore,20,21 the case report forms of precision medicine clinical trials promoted by participating institutions, and the national database Conticabase22-27; (2) extracting the DEs of those clinical trials; (3) identifying and comparing the most relevant DEs; and finally, (4) standardizing the CLs of the Common Data Elements (CDEs) using appropriate international terminologies when possible (Fig 1). To ensure the accuracy and quality of data collected through the different studies, the OSIRIS group used an adapted data collection strategy that focused on a limited number of mandatory CDEs to facilitate structured data extraction and limit missing data.
FIG 1.
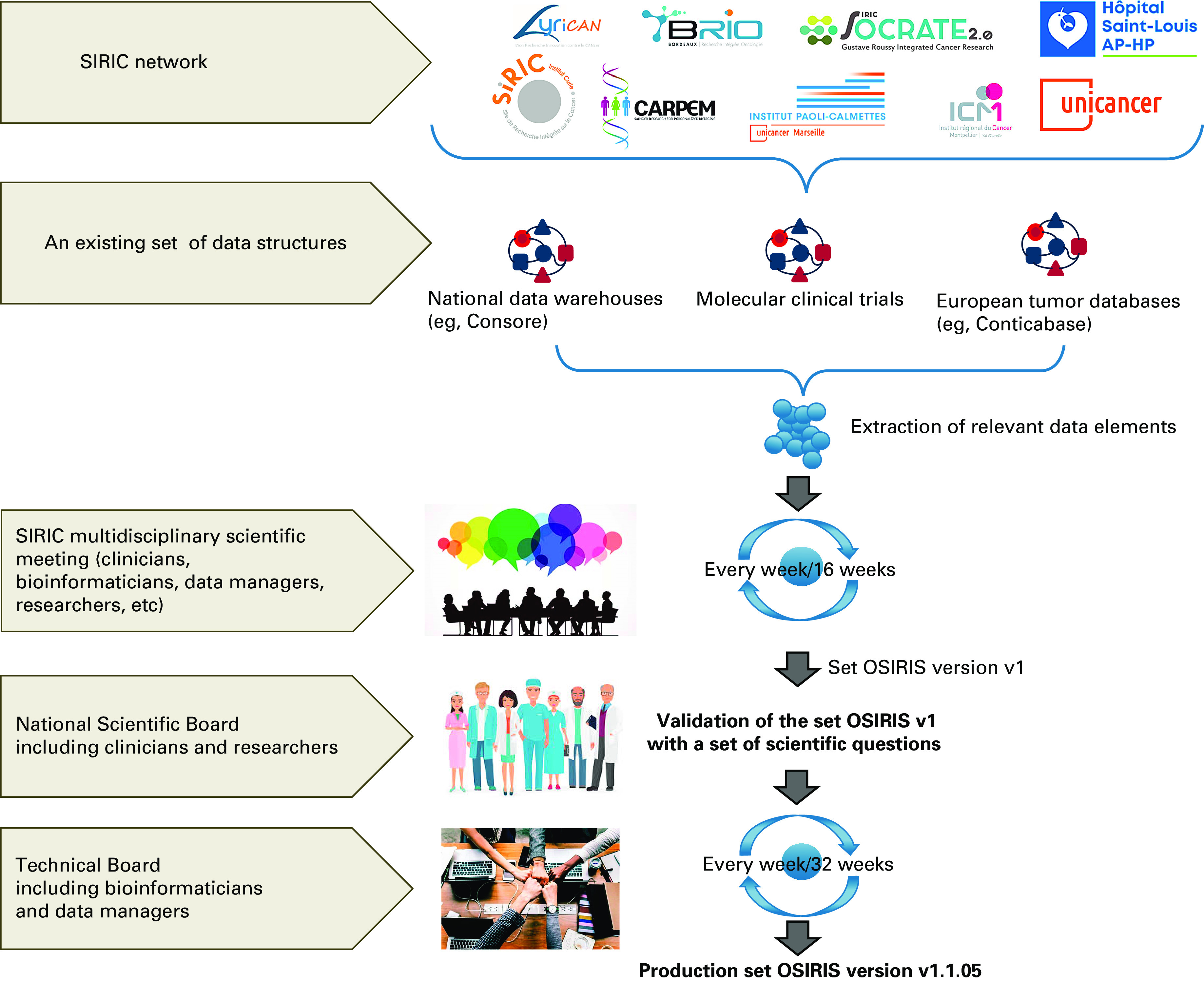
The overall methodology used to deliver the first release of the OSIRIS set. During several months, weekly meetings of several national groups (SIRIC multidisciplinary group and scientific and technical boards) were held to release the first version of the OSIRIS set. SIRIC, Integrated Cancer Research Sites.
Identification of French Genomic-Driven Clinical Trials and National Databases and Extraction of the DEs
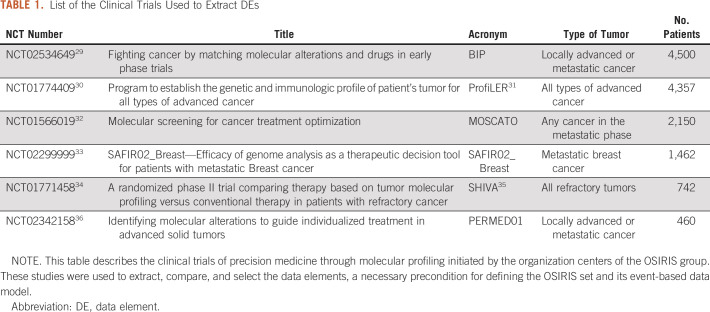
Major clinical trials were conducted in France28 using targeted sequencing to identify actionable alterations and assign therapy to patients with refractory cancers. Data from these clinical trials are of high quality but sorely lack the standards to enable data sharing across institutions. Six broad clinical trials (Table 1) were selected as a case study to extract the DEs.
TABLE 1.
List of the Clinical Trials Used to Extract DEs
Identification and Comparison of the Most Relevant DEs
To define a set of CDEs representing a minimal data set to answer scientific and medical questions, the OSIRIS group used a multidisciplinary approach including oncologists, medical informatics specialists, bioinformaticians, epidemiologists, biostatisticians, and regulatory specialists in iteration planning meetings. This method provided different perspectives and prompted extensive discussions to select the most relevant DEs.37 After a year of weekly iteration meetings, the OSIRIS group reached a consensus.
Validation of the CDE
A national board composed of clinicians and translational researchers in oncology and data protection officers first examined whether the CDEs were compliant with the General Data Protection Regulation38 and the National Commission for Data Protection and Liberties (CNIL-France).39 They oversaw the centralization of common scientific questions from different medical specialties and cancer pathologies (eg, identification of actionable genomic alterations, identification of rare recurrent genetic alterations, and understanding genetic trajectories). Common scientific questions that emerged can be classified into three broad categories: (1) clinical and biological cohorts, (2) descriptive and functional translational studies, and (3) interventional clinical trials. Then, national board proceeded to ensure that the OSIRIS CDEs could answer those questions.
Standardizing the CLs of the CDE
CL standardization was a prerequisite step to ensure greater interoperability of the OSIRIS set. A technical board composed of bioinformaticians and data managers was responsible for identifying the most relevant international and national terminologies (eg, CCAM code of the medical act). Most of the CDEs were mapped to these terminologies (Table 2). For some CDEs, the group created its own terminologies for terms that did not have yet a common standard definition (eg, biomarker code, sample origin, and panel name). Genomic CDEs were based on Fast Healthcare Interoperability Resources (FHIR) v3 genomic items when available (eg, AminoAcidChange, DNARegionName, and GenomicSourceClass).
TABLE 2.
Main International and National Terminologies Used in the OSIRIS Set
Validation of the OSIRIS Set
To validate this approach, the group federated retrospective data to assess a first cohort of 300 patients included in six clinical trials. Minor functional changes, such as modifying a CDE or supplementing a CL, were needed to strengthen the model.
RESULTS
OSIRIS CDM
A first version of the CDM (v1) was released in 2018,40 which formed the basis of discussion for the SIRIC multidisciplinary scientific meetings and, later, for the technical board meetings. Several institutions tested the model (ie, completeness level and inconsistency rate), and the group proposed and implemented substantial revisions through functional analysis and data integration feedback. This helped establish a reliable, robust, and comprehensive model to describe cancer. Figure 2 shows the clinical data model with the CDEs and the relationships between them. Given that health data change over time, the OSIRIS group focused its efforts on modeling an event-based temporal data model. Each DE of the OSIRIS CDM is linked to a particular TumorPathologyEvent (TPE) (ie, Primary, Recurrent, and Metastatic tumor) concept. Each TPE occurs not only over a period of time but also relative to the time of another TPE to follow the course of the disease precisely. Patient follow-up represented on a timeline (ie, Analysis and Treatment concepts) is another key element of the model, and derived concepts such as AdverseEvent, ResponseEvaluation, and Biomarker are also included. Rules were defined when the time of a TPE is incomplete or uncertain by looking at the timeline of the follow-up care plan. This provides a high-quality OSIRIS-formatted data set required for training machine learning models to address questions like relapse prediction and covers the necessary topics for patient similarity identification.41 To date, the model covers the description of solid tumors and minor changes will be needed to handle hematological tumors.
FIG 2.
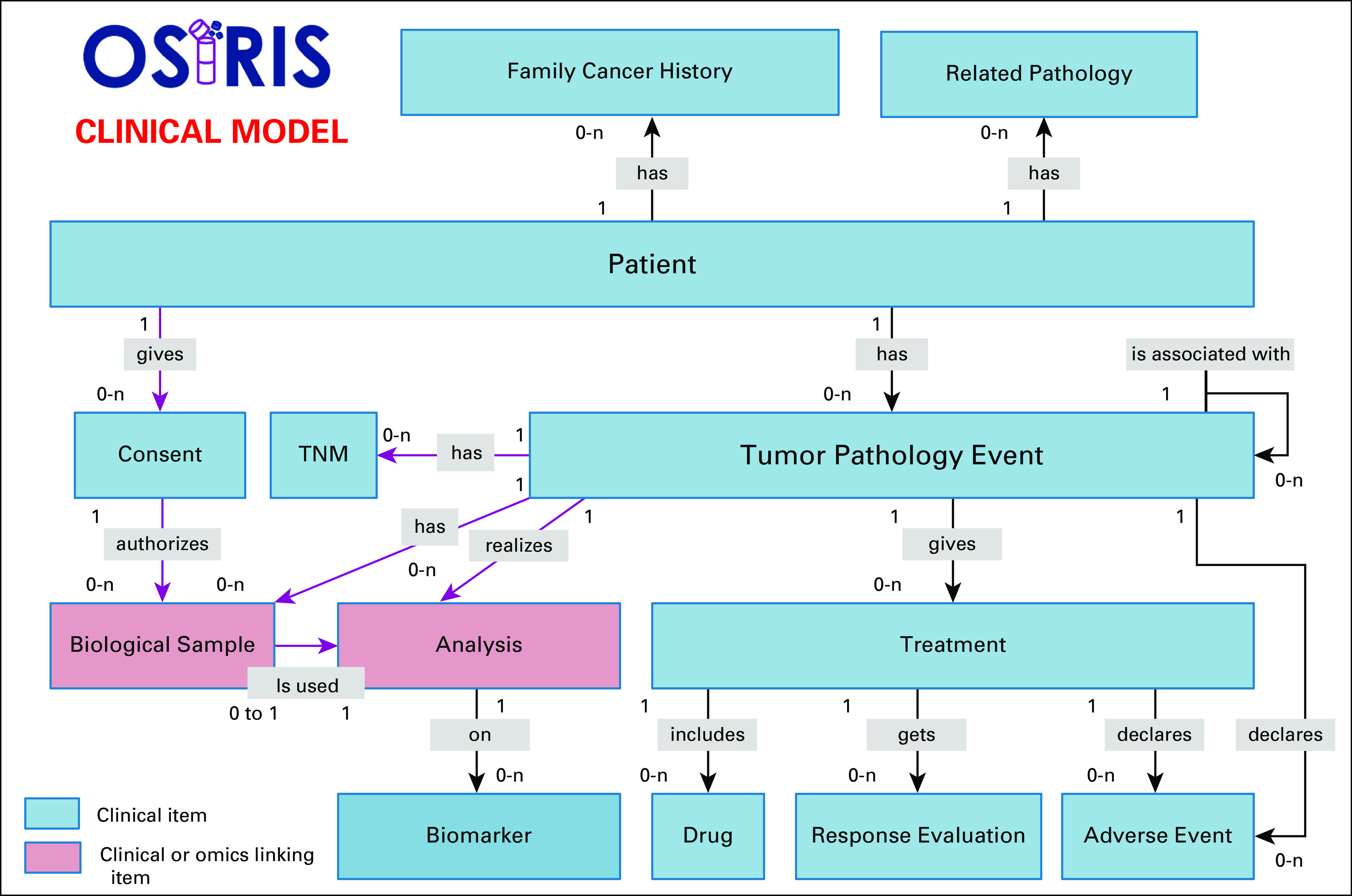
OSIRIS clinical data model. This figure shows the OSIRIS event–based clinical data model to follow the disease course longitudinally. For each event type (primary tumor and local and metastatic relapse), the response and adverse events of a treatment are associated. Moreover, any analysis carried out on a sample (imaging, omics, biology, pathologic examination) is also linked to a specific event.
Genomics is an integral component and is considered as a separate and distinct type of analysis in the data model as shown in Figure 3. All analysis concepts are linked to the clinical model using the TumorPathologyEvent concept as a key component. Some concepts define the analysis itself to ensure reproducibility, tracking provenance, and context of the results (ie, sequencing technology, target panel, and bioinformatics pipeline including analysis parameters), whereas others give a confidence level of the predictions (ie, validation) or variant annotation for cancer diagnosis (ie, annotation). The AlterationOnSample class is an umbrella concept of shared attributes common to the different types of genomic alterations. Entities related to this generic concept enable the storage of specific alteration data type. To date, the data model covers copy number alterations, fusions, gene expression data, and somatic mutations. The flexibility of the model is a major asset to easily add new omics data (eg, epigenetic and proteomic data) that will allow multiomics layer integration.
FIG 3.
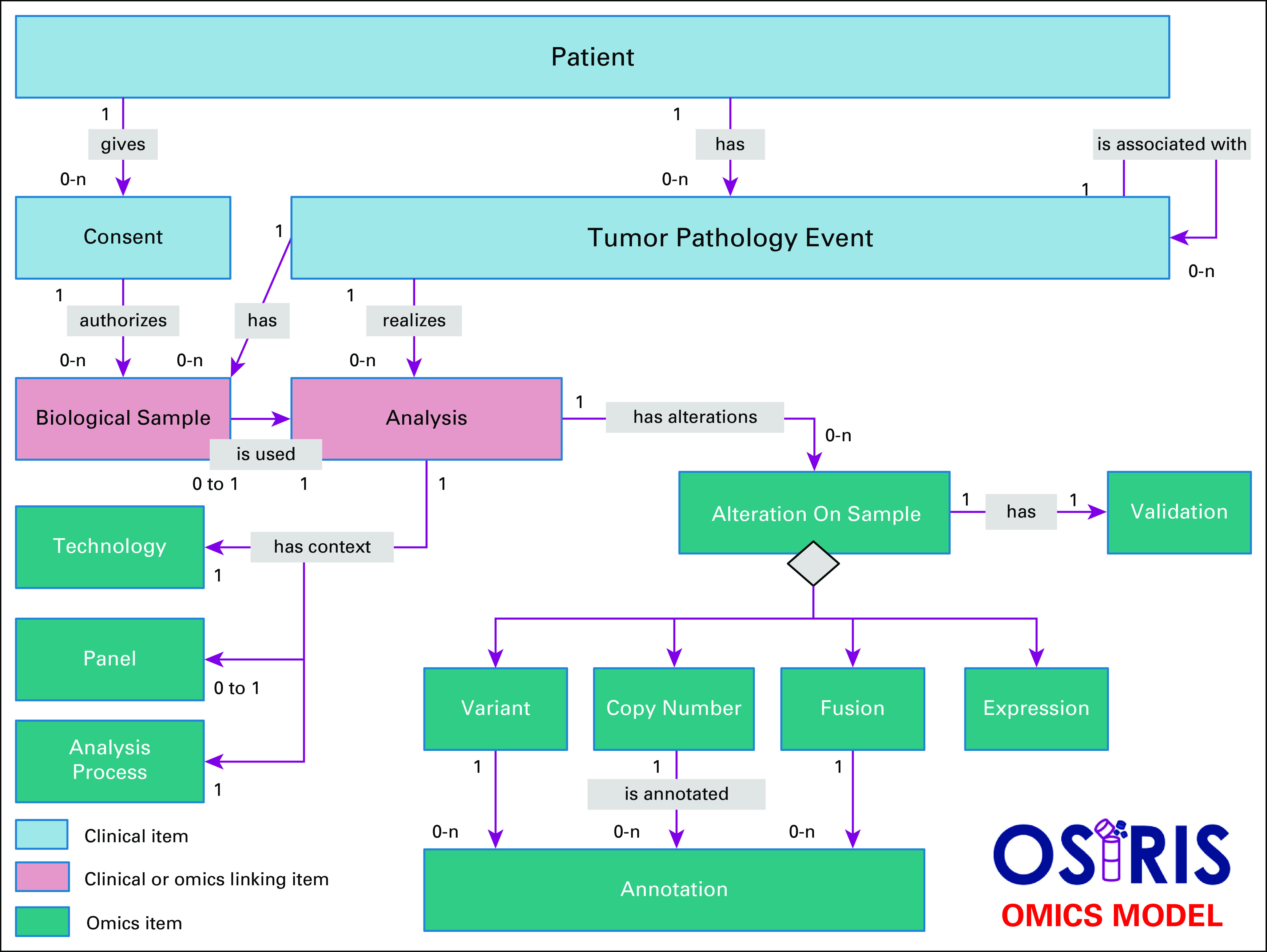
OSIRIS omics data model. Thanks to an object-oriented model, the omics concepts were designed to be scalable and modular. The model uses inheritance to store common (ie, AlterationOnSample concept) and specific attributes of various kinds of genomic alterations. Each genomic alteration is annotated for cancer diagnosis (ie, Annotation concept) along with the confidence level of the prediction (ie, validation concept).
The OSIRIS Set
Once the OSIRIS set reached a steady state in terms of its semantic content, a first version (1.1.05) was released in February 2019. This current release, built on an event-based data model, describes a minimum data set that allows capture of the disease course longitudinally. This release, freely available to the community on GitHub,42 consists of a minimal data set of 67 clinical and 65 omics items (described in the Data Supplement) required and validated by the national scientific board. CLs are based on international and national terminologies, complemented by their own terminologies for terms that do not yet have a common standard definition. The Data Supplement presents precisely the list of possible values for each DE.
As a proof-of-concept, clinical and genomic data from 300 patients included in six different clinical trials were used to allow basic descriptive statistics. As a result of these statistics, there is much heterogeneity of the clinical CDEs being used within each clinical trial. Some clinical concepts are well represented (ie, Patient, BiologicalSample, TumorPathologyEvent, Treatment, AdverseEvent, and Drug), whereas others are hardly used (ie, FamilyCancerHistory and RelatedPathology). By comparing the CDE terminologies, substantial clinical differences were found. For instance, some French pathologists rely on a national multiaxial terminology (Association pour le Développement de l'Informatique en Cytologie et Anatomie Pathologique, ADICAP), whereas others use the international standard ICD-O-3 (International Classification of Diseases for Oncology) to standardize pathology reports. Translating ADICAP codes to ICD-O-3 was necessary, and a quality control program was applied to assess data quality, correcting errors and adding missing data. All the clinical trials used targeted next-generation sequencing (NGS) as a tool for precision medicine, which ensured the consistent use of the genomic concepts. Variability in genomic data reporting was observed, rooted in the wide variety of NGS technologies and analysis tools with no emphasis on interoperability. Genetic reports provided by the different stakeholders often result in a partial or heterogeneous description of genomic alterations (eg, the reference sequence of single nucleotide variants is not always properly reported). We noted as well a variability of the metadata that add further clinical interpretation of variants (ie, cross-references to external databases and precomputed in silico algorithm-based predictions). To ensure the sustainability, expandability, and interoperability of the OSIRIS set with other systems, the first release was made compatible with the growing health information exchange standard HL7 FHIR in June 2020. For instance, the genomic CDM was mapped to FHIR v4 genomic resources. The new resource, MolecularSequence, was used for annotating data with external repositories and the existing resources such as Observation. In the Data Supplement, OSIRIS was compared with other similar initiatives such as mCODE and OMOP.
List of Structured Flat Files to Share Data
To share the OSIRIS set, each participating institution generated a list of structured flat files in comma separated format (“.csv”). The layout, content, and coding of eighteen flat files were standardized43 to structure data in each center. The flat files follow the relational OSIRIS CDM to be directly interoperable with any third-party software and readable by the clinicians. By providing a single entry point, any data source can be processed and represented in our data model (described in Appendix Fig A1). For instance, we used these files to store real-world data (RWD) either to complete the OSIRIS set for clinical data (ie, FamilyCancerHistory and RelatedPathology concepts) or to integrate genomics data not used to guide targeted therapy. To ensure the correct use of the CDEs, OSIRIS users must follow the DE specification guideline and carry out the necessary checks to guarantee compliance therewith.44
DISCUSSION
Within the context of the SIRIC efforts and supported by INCa, a need to enhance data sharing in genomic-driven clinical trials to clinicians and translational researchers was identified. The OSIRIS project emerged in 2015 to find creative, technological, and regulatory solutions for improving standardization and data sharing at the national level.
We identified common challenges to promote data sharing in precision medicine,45 which led to the constitution of three working groups (ie, regulatory and data policy, data standards, and technical data sharing solutions). Governance required transparent decision making, and this transparency offers a constructive basis for engagement with new institutions.
The group emphasized the value of data collected within the context of clinical trials and beyond. For example, artificial intelligence (AI) projects have emerged over the past few years and rely heavily on standardized data sets to train and validate statistical models, which could be provided by the OSIRIS data set. Moreover, OSIRIS provides an effective RWD ecosystem by developing a data standard, which, if used, could improve the compatibility, quality, and consistency of Electronic Health Record.
The OSIRIS set is a useful standardization tool for clinical cancer research data within the SIRIC research network. Since it relies on already established terminologies, it may also be useful within the context of a larger international community across different networks. In addition, the OSIRIS set greatly facilitates data model harmonization and data integration with other repositories. At an international level, data interoperability in health care is a major challenge and some standards have emerged such as FHIR (Fast Healthcare Interoperability Resources).45 FHIR is the latest HL7 (Health Level 7's) healthcare data representation standard for data exchange by (1) using established code sets (eg, LOINC, SNOMED CT, and ICD-9/-10) or (2) FHIR-specific value sets to maximize standardization. Moreover, FHIR offers the ability for individual users and organizations to build extensions to capture data that are not explicitly defined by HL7.46 Accordingly, the group adopted its data model to bring it in line with this international standard. We profiled the overlap and gaps between the OSIRIS schema (version 1.1.05) and the corresponding FHIR resources (version 4.0.1).
The main values of the OSIRIS set and its event-based data model are the following: (1) a minimum data set organized as a temporal model of cancer events enabling the data longitudinal search; (2) an original blend of clinical and omics concepts; (3) a data model designed to be scalable and modular to integrate other specific terminology aspects according to localization, treatment (eg, radiation therapy), and other types of biological and omics analysis (eg, proteomic); and (4) the OSIRIS set that relies mostly on international terminologies. Some limitations remain and must be addressed by the OSIRIS group. For instance, OSIRIS does not provide data quality checks of these files that could be run against an OSIRIS CDM instance. In 2019, OMOP developed a Data quality Dashboard47 based on the Kahn Framework, which uses a system of categories and contexts that represent strategies for assessing data quality. This framework could be used to provide quality metrics as (1) conformity of the OSIRIS CDM specifications and (2) level of completeness of the OSIRIS set. Another limitation of OSIRIS is the lack of procedure to follow the different versions of the terminologies used. This task is necessary to reproduce different analysis done on different versions of the OSIRIS set. Versioning the data for reproducible analysis is not an easy task in a fast-moving field, which requires ongoing updating and comparison of the different versions. To ensure the dissemination of the OSIRIS set, INCa promotes its use in clinical trials and data sharing projects (eg, React-4kids network) that they fund. The OSIRIS set may also be useful in projects involving healthcare data storage and retrieval (eg, Health Data Hub48 and in the French Genomic Medicine Plan 202549 as part of the Data Collector and Analyzer).
To date, the OSIRIS consortium is working on (1) the set and CDM's evolution, (2) improved interoperability with international standards (eg, OMOP), and finally (3) training support and advice to all stakeholders interested in using the OSIRIS set.
ACKNOWLEDGMENT
The authors thank the patients included in the clinical trials used in the proof of concept. OSIRIS acknowledges the eight SIRICs (Integrated Cancer Research Site) funded by INCa, DGOS, and Inserm.
Appendix
FIG A1.
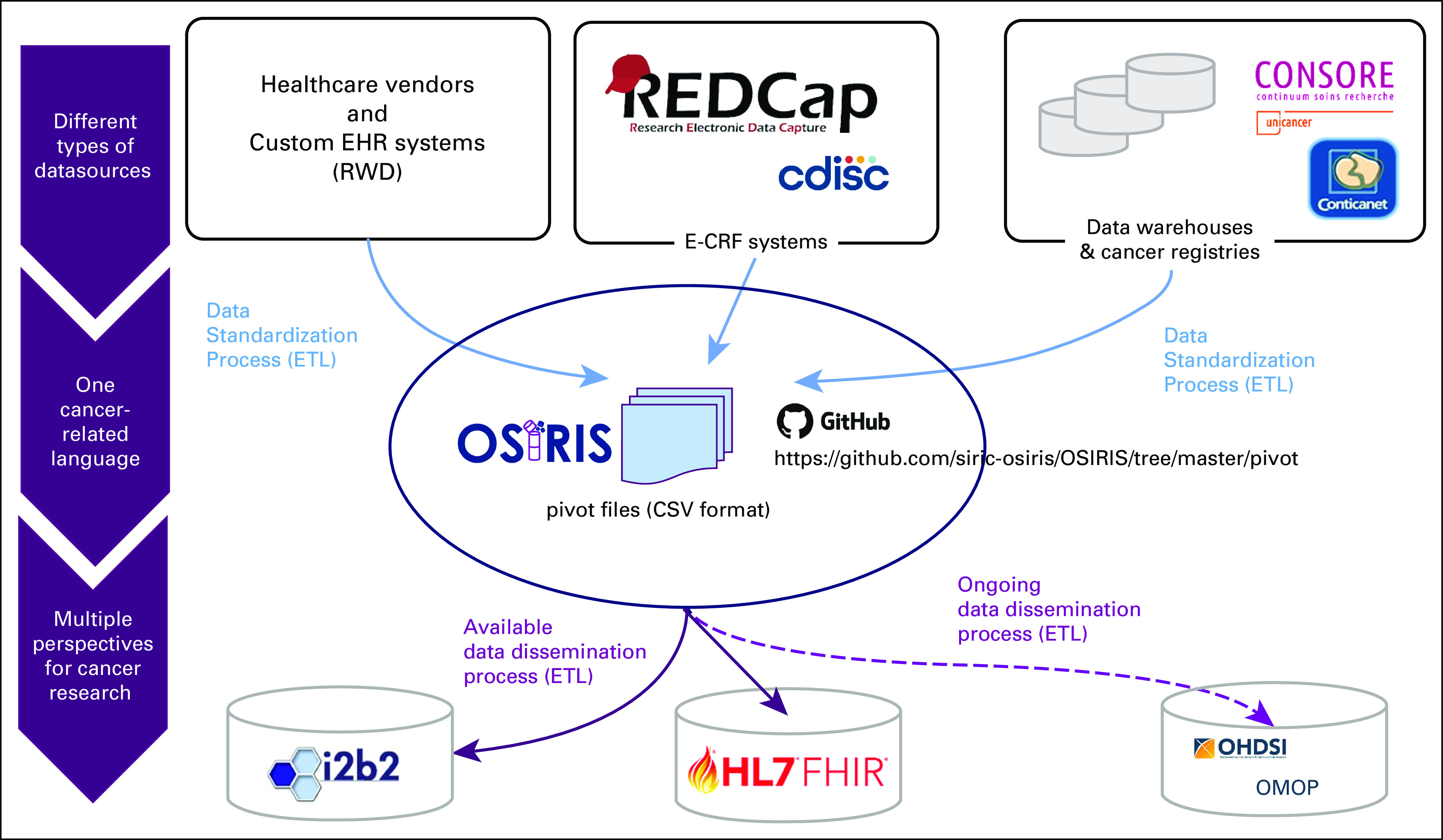
Description of the use of the OSIRIS structured flat files. We use the OSIRIS flat files as an entry point to standardize data from different data sources (ie, EHRs, eCRFs, data warehouses, and cancer registries). These pivot files are then used to facilitate interoperability with other standards. For instance, we used them to construct ETLs with I2B2 CDM instances and the FHIR API. API, application programming interface; CDM, Common Data Model; EHR, Electronic Health Record; ETL, extract, transform, and load; FHIR, Fast Healthcare Interoperability Resources.
SUPPORT
OSIRIS (GrOupe inter-SIRIC sur le paRtage et l'Intégration des donnéeS clinico-biologiques en cancérologie) was supported by the Institut National du Cancer (INCa_12600).
J.G., Y.L., and V.L.T. contributed equally to this work.
AUTHOR CONTRIBUTIONS
Conception and design: Julien Guérin, Yec'han Laizet, Vincent Le Texier, Laetitia Chanas, François Lion, Sophie Gourgou, Anne-Laure Martin, Manuel Tejeda, Elisabeth Hess, Marina Rousseau-Tsangaris, Vianney Jouhet, Pierre Saintigny
Provision of study materials or patients: Yec'han Laizet, François Lion, Sophie Gourgou, Anne-Laure Martin, Pierre Saintigny, Julien Guérin, Vincent Le Texier
Collection and assembly of data: Julien Guérin, Yec'han Laizet, Vincent Le Texier, Laetitia Chanas, Bastien Rance, Florence Koeppel, Sophie Gourgou, Manuel Tejeda, Maud Toulmonde, Vianney Jouhet
Data analysis and interpretation: Yec'han Laizet, Vincent Le Texier, Sophie Gourgou, Manuel Tejeda, Maud Toulmonde, Stéphanie Cox, Pierre Saintigny, Julien Guérin
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Vincent Le Texier
Travel, Accommodations, Expenses: Roche/Genentech
Sophie Gourgou
Consulting or Advisory Role: Celgene, Roche
Elisabeth Hess
Stock and Other Ownership Interests: Sanofi, GENFIT, Innate Pharma
Pierre Saintigny
Honoraria: HTG Molecular Diagnostics, Inivata, Archer, Bristol-Myers Squibb, Roche Molecular Diagnostics
Research Funding: Roche, AstraZeneca, Novartis, Bristol-Myers Squibb
Travel, Accommodations, Expenses: Illumina, Bristol-Myers Squibb, AstraZeneca, Roche
No other potential conflicts of interest were reported.
REFERENCES
- 1.Lawrence MS Stojanov P Mermel CH, et al. : Discovery and saturation analysis of cancer genes across 21 tumor types. Nature 505:495–501, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.CIT Program. Carte d'Identité des Tumeurs—Accueil. https://cit.ligue-cancer.net/ [Google Scholar]
- 3.The Cancer Genome Atlas Program. National Cancer Institute. https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga, 2018 [Google Scholar]
- 4.International Cancer Genome Consortium. https://icgc.org/, 2019 [Google Scholar]
- 5.Learned K Durbin A Currie R, et al. : Barriers to accessing public cancer genomic data. Sci Data 6:98, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.PCORnet® : The National Patient-Centered Clinical Research Network. https://www.pcori.org/research-results/pcornet%C2%AE-national-patient-centered-clinical-research-network, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.OHDSI : Observational Health Data Sciences and Informatics. https://www.ohdsi.org/ [Google Scholar]
- 8.Micheel CM Sweeney SM LeNoue-Newton ML, et al. : American Association for Cancer Research Project genomics evidence neoplasia information exchange: From inception to first data release and beyond-lessons learned and member institutions' perspectives. JCO Clin Cancer Inform 2:1–14, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.BRCA Exchange. https://brcaexchange.org/ [Google Scholar]
- 10.Rubinstein SM, Warner JL: CancerLinQ: Origins, implementation, and future directions. JCO Clin Cancer Inform 2:1–7, 2018 [DOI] [PubMed] [Google Scholar]
- 11.ORIEN : Oncology Research Information Exchange Network. http://oriencancer.org/, 2019 [Google Scholar]
- 12.ICGC ARGO : Home. ICGC ARGO. https://www.icgc-argo.org/ [Google Scholar]
- 13.Rich A, Beckett P, Baldwin D: Status of lung cancer data collection in Europe. JCO Clin Cancer Inform 2:1–12, 2018 [DOI] [PubMed] [Google Scholar]
- 14.Lablans M, Schmidt EE, Ückert F: An architecture for translational cancer research as exemplified by the German Cancer Consortium. JCO Clin Cancer Inform 2:1–8, 2018 [DOI] [PubMed] [Google Scholar]
- 15.OSIRIS : A National Data Sharing Project. https://en.e-cancer.fr/OSIRIS-a-national-data-sharing-project, 2020 [Google Scholar]
- 16.UNICANCER. http://www.unicancer.fr/en, 2020 [Google Scholar]
- 17.The French National Cancer Institute : https://en.e-cancer.fr/, 2020
- 18.mCODE : Minimal Common Oncology Data Elements. https://mcodeinitiative.org/ [Google Scholar]
- 19.Overhage JM Ryan PB Reich CG, et al. : Validation of a common data model for active safety surveillance research. J Am Med Inform Assoc 19:54–60, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heudel P Livartowski A Arveux P, et al. : The ConSoRe project supports the implementation of big data in oncology. Bull Cancer 103:949–950, 2016 [DOI] [PubMed] [Google Scholar]
- 21.UNICANCER : Big Data Analysis of Clinical Records for Cancer Care. http://www.unicancer.fr/sites/default/files/big-data-analysis-clinical-records-paper-ConSoRe-white-paper.pdf [Google Scholar]
- 22.Boughzala-Bennadji R Stoeckle E Le Péchoux C, et al. : Localized myxofibrosarcomas: Roles of surgical margins and adjuvant radiation therapy. Int J Radiat Oncol Biol Phys 102:399–406, 2018 [DOI] [PubMed] [Google Scholar]
- 23.Penel N Coindre JM Giraud A, et al. : Presentation and outcome of frequent and rare sarcoma histologic subtypes: A study of 10,262 patients with localized visceral/soft tissue sarcoma managed in reference centers. Cancer 124:1179–1187, 2018 [DOI] [PubMed] [Google Scholar]
- 24.Le Guellec S Chibon F Ouali M, et al. : Are peripheral purely undifferentiated pleomorphic sarcomas with MDM2 amplification dedifferentiated liposarcomas? Am J Surg Pathol 38:293–304, 2014 [DOI] [PubMed] [Google Scholar]
- 25.Le Guellec S Decouvelaere A-V Filleron T, et al. : Malignant peripheral nerve sheath tumor is a challenging diagnosis: A systematic pathology review, immunohistochemistry, and molecular analysis in 160 patients from the French Sarcoma Group Database. Am J Surg Pathol 40:896–908, 2016 [DOI] [PubMed] [Google Scholar]
- 26.Szablewski V Neuville A Terrier P, et al. : Adult sinonasal soft tissue sarcoma: Analysis of 48 cases from the French Sarcoma Group database. Laryngoscope 125:615–623, 2015 [DOI] [PubMed] [Google Scholar]
- 27.Cassier PA Kantor G Bonvalot S, et al. : Adjuvant radiotherapy for extremity and trunk wall atypical lipomatous tumor/well-differentiated LPS (ALT/WD-LPS): A French Sarcoma Group (GSF-GETO) study. Ann Oncol 25:1854–1860, 2014 [DOI] [PubMed] [Google Scholar]
- 28.Nowak F, Soria JC, Calvo F: Tumour molecular profiling for deciding therapy—The French initiative. Nat Rev Clin Oncol 9:479–486, 2012 [DOI] [PubMed] [Google Scholar]
- 29.Institut Bergonié : Bergonie Institut Profiling: Fighting Cancer by Matching Molecular Alterations and Drugs in Early Phase Trials. https://clinicaltrials.gov/ct2/show/NCT02534649, 2020 [Google Scholar]
- 30.Smart Patients : Program to Establish the Genetic and Immunologic Profile of Patient's Tumor for All Types of Advanced Cancer. ClinicalTrials.gov. https://clinicaltrials.gov/ct2/show/NCT01774409, 2020 [Google Scholar]
- 31.Trédan O Wang Q Pissaloux D, et al. : Molecular screening program to select molecular-based recommended therapies for metastatic cancer patients: Analysis from the ProfiLER trial. Ann Oncol 30:757–765, 2019 [DOI] [PubMed] [Google Scholar]
- 32.Institut Gustave Roussy : Molecular Screening for Cancer Treatment Optimization. ClinicalTrials.gov. https://clinicaltrials.gov/ct2/show/NCT01566019, 2019 [Google Scholar]
- 33.UNICANCER : SAFIR02_Breast: Efficacy of Genome Analysis as a Therapeutic Decision Tool for Patients With Metastatic Breast Cancer. ClinicalTrials.gov. https://clinicaltrials.gov/ct2/show/NCT02299999, 2019 [Google Scholar]
- 34.Institut Curie : A Randomized Phase II Trial Comparing Therapy Based on Tumor Molecular Profiling Versus Conventional Therapy in Patients With Refractory Cancer. ClinicalTrials.gov. https://clinicaltrials.gov/ct2/show/NCT01771458, 2019 [DOI] [PubMed] [Google Scholar]
- 35.Le Tourneau C Delord J-P Gonçalves A, et al. : Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): A multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol 16:1324–1334, 2015 [DOI] [PubMed] [Google Scholar]
- 36.Institut Paoli-Calmettes : Identifying Molecular Alterations to Guide Individualized Treatment in Advanced Solid Tumors. ClinicalTrials.gov. https://clinicaltrials.gov/ct2/show/NCT02342158, 2019 [Google Scholar]
- 37.Choi BCK, Pak AWP: Multidisciplinarity, interdisciplinarity and transdisciplinarity in health research, services, education and policy: 1. Definitions, objectives, and evidence of effectiveness. Clin Invest Med 29:14, 2006 [PubMed] [Google Scholar]
- 38.General Data Protection Regulation (GDPR) : Official Legal Text. General Data Protection Regulation (GDPR). https://gdpr-info.eu/, 2020 [Google Scholar]
- 39.Homepage. CNIL. https://www.cnil.fr/en/home, 2020 [Google Scholar]
- 40.siric-osiris/OSIRIS . GitHub. https://github.com/siric-osiris/OSIRIS/blob/master/documentation/MPD_OSIRIS_model_v1.1.05.png, 2020 [Google Scholar]
- 41.Seligson ND Warner JL Dalton WS, et al. : Recommendations for patient similarity classes: Results of the AMIA 2019 workshop on defining patient similarity. J Am Med Inform Assoc 27:1808-1812, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.siric-osiris : siric-osiris/OSIRIS. https://github.com/siric-osiris/OSIRIS, 2020 [Google Scholar]
- 43.siric-osiris/OSIRIS : GitHub. https://github.com/siric-osiris/OSIRIS/tree/master/pivot, 2020 [Google Scholar]
- 44.siric-osiris : siric-osiris/OSIRIS. https://github.com/siric-osiris/OSIRIS/blob/master/documentation/OSIRIS_Conventions_alimentation_v1.0_EN.pdf, 2020 [Google Scholar]
- 45.Texier VL Henda N Cox S, et al. : Data sharing in the era of precision medicine: A scientometric analysis. Precision Cancer Med 2, 2019 [Google Scholar]
- 46.HL7 : Index—FHIR v4.0.1. https://www.hl7.org/fhir/, 2019 [Google Scholar]
- 47.GitHub : OHDSI/DataQualityDashboard. Observational Health Data Sciences and Informatics. https://github.com/OHDSI/DataQualityDashboard, 2020 [Google Scholar]
- 48.Health Data Hub, Plateforme Des Données De Santé, France : Healthdatahub. https://www.health-data-hub.fr?lang=en, 2020 [Google Scholar]
- 49.Lévy Y: Genomic medicine 2025: France in the race for precision medicine. Lancet 388:2872, 2016 [DOI] [PubMed] [Google Scholar]