PURPOSE
This study tested whether a composite mortality score could overcome gaps and potential biases in individual real-world mortality data sources. Complete and accurate mortality data are necessary to calculate important outcomes in oncology, including overall survival. However, in the United States, there is not a single complete and broadly applicable mortality data source. It is further likely that available data sources are biased in their coverage of sex, race, age, and socioeconomic status (SES).
METHODS
Six individual real-world data sources were combined to develop a high-quality composite mortality score. The composite score was benchmarked against the gold standard for mortality data, the National Death Index. Subgroup analyses were then conducted to evaluate the completeness and accuracy by sex, race, age, and SES.
RESULTS
The composite mortality score achieved a sensitivity of 94.9% and specificity of 92.8% compared with the National Death Index, with concordance within 1 day of 98.6%. Although some individual data sources show significant coverage gaps related to sex, race, age, and SES, the composite score maintains high sensitivity (84.6%-96.1%) and specificity (77.9%-99.2%) across subgroups.
CONCLUSION
A composite score leveraging multiple scalable sources for mortality in the real-world setting maintained strong sensitivity, specificity, and concordance, including across sex, race, age, and SES subgroups.
INTRODUCTION
Real-world data (RWD) collected from routine patient care are valuable for expediting and enhancing outcomes research in oncology. It provides an opportunity to characterize cancer care and outcomes among a broader set of patients, including groups often underrepresented in traditional prospective clinical trials and population studies.1-3 RWD applications are expanding, particularly in oncology, with regard to research, clinical care, regulatory, and commercial applications. However, capturing accurate and complete RWD necessary to power meaningful research is challenging given the fragmented nature of healthcare delivery and associated data collection in the United States (US).
CONTEXT
Key Objective
What are the gaps in completeness and accuracy among real-world sources for mortality data, and can a composite mortality score overcome these gaps?
Knowledge Generated
Compared with the gold-standard source for vital status and dates of death, real-world data sources were generally accurate but had significant gaps in data coverage. Development of a mortality composite score that combines data across sources addressed these gaps and achieved a sensitivity of 94.9% compared with the gold standard.
Relevance
Mortality data from electronic health records and registries at community health systems tend to be complete but have notable gaps and biases. Other real-world data sources, including digitized obituaries, do not capture all deaths and may underrepresent patients on the basis of age, race, sex, and socioeconomic factors. However, combining across sources to develop a composite mortality score addresses biases associated with individual sources and provides more complete capture of patient vital status and dates of death.
Complete and accurate mortality data are necessary to calculate important outcomes in oncology, including overall survival. Incomplete mortality data can cause inaccurate estimation of survival and result in erroneous conclusions in comparative studies.4 In the United States, there is no single mortality data source that is both complete and broadly applicable.
Available data sources for mortality each have limitations. The Centers for Disease Control and Prevention’s National Death Index (NDI) captures all US death certificates and is considered the gold standard, but access to and use of NDI data is limited. The National Cancer Institute’s SEER database also captures death certificates but has limited geographic coverage.5 Online obituary aggregators and the Social Security Death Index and/or Death Master File (SSDI) capture broad but incomplete data. Documentation in electronic health records (EHRs) is also often incomplete, particularly for deaths occurring outside of healthcare facilities. Finally, many cancer registries often rely on SSDI, EHRs, or manual curation (eg, by searching online obituaries) and do not include all patients.6
Mortality data sources in the real world likely have biases that influence which patients are represented. It is well-documented that obituaries underrepresent women.7,8 Additional studies have shown differences in obituary descriptions by race for those individuals who receive obituaries.9 Beyond these known biases, disparities in coverage of mortality data across race and/or ethnicity, age, or socioeconomic status (SES) have not been well-documented.
The hypothesis for this work was that a composite mortality score could overcome gaps and biases in real-world mortality data sources. Potential areas of bias by age, sex, race, and SES were evaluated. Previous research has examined approaches to improving the accuracy of real-world mortality data by combining data from multiple sources;10,11 however, to our knowledge, this is the first study to evaluate whether a composite score overcomes biases pertaining to age, sex, race, and SES associated with individual sources.
METHODS
Data Sources and Study Population
A sample of patients diagnosed with cancer between 2011 and 2017 was selected from the Syapse Learning Health Network (LHN), a proprietary database of patients from US health systems data sources including EHRs, enterprise data warehouses, laboratories, tumor registries, digitized obituaries, and other clinical sources. International Classification of Disease diagnosis codes and tumor-specific data tables were used to identify patients with cancer.
Vital status was based on the presence of death data obtained from six sources: (1) hospital EHR data feeds, (2) hospital tumor registries, (3) digitized obituaries, (4) SSDI, (5) SEER, and (6) manual curation. EHR data feeds and hospital tumor registry sources were obtained via direct integrations with health systems. Digitized obituaries and SSDI were sourced through publicly available US mortality data sources and linked using probabilistic patient matching.12 Patient-level identifiable data from SEER was obtained via direct bidirectional data sharing with health systems. Manual curation was conducted by Certified Tumor Registrars with access to enterprise-wide EHRs in partner health systems.
If the patient was listed as deceased from any source, that patient was marked as deceased in the composite score. Date of death was determined as the first death date found when searching these sources in a waterfall method. The waterfall method examined data sources in ascending order to resolve discrepancies: hospital tumor registries, SEER, EHRs, SSDI, digitized obituaries, and manual curation. Data across sources were linked at the patient level (Fig 1).
FIG 1.
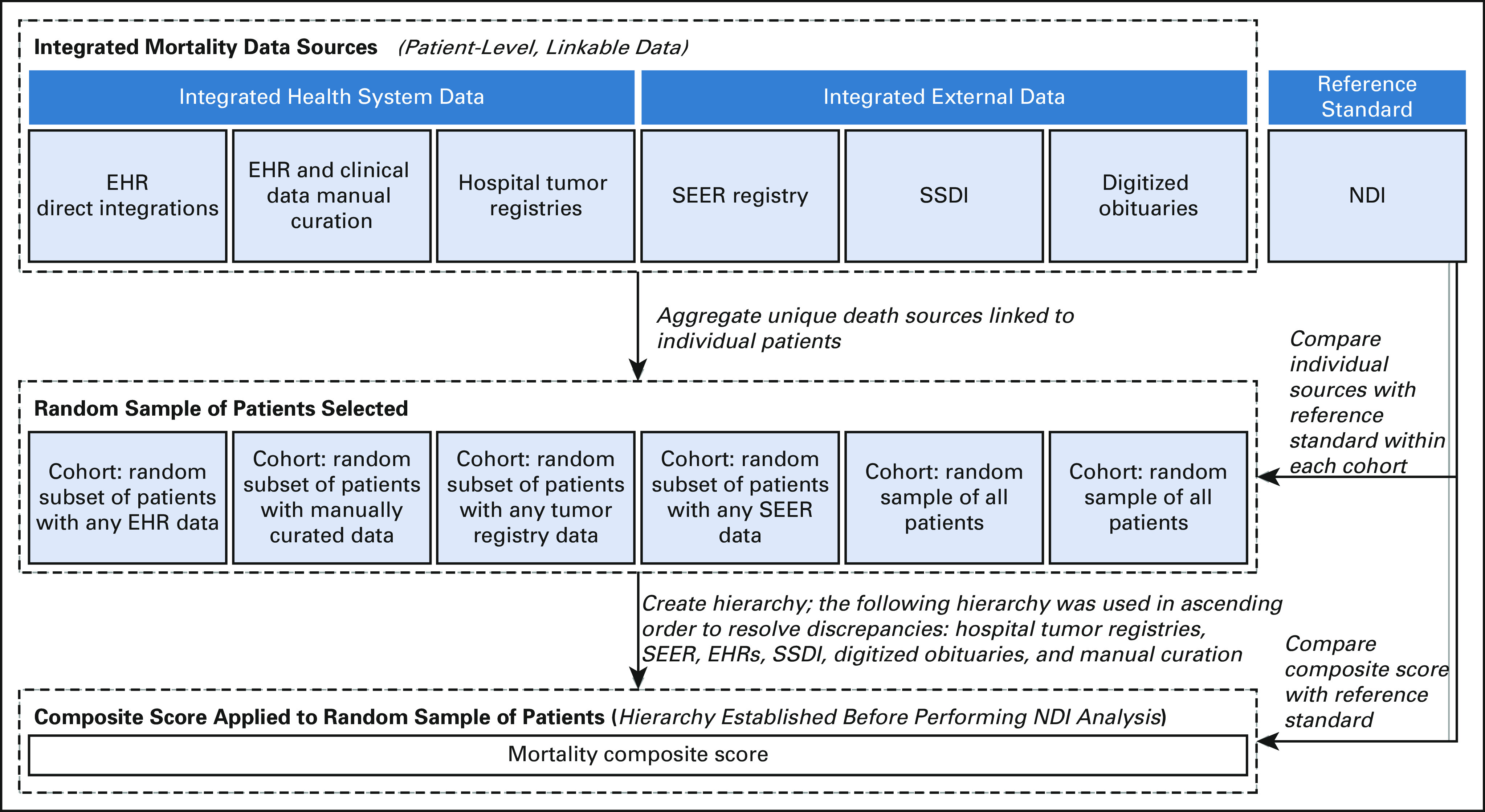
Composite score development flowchart. EHR, electronic health record; NDI, National Death Index; SSDI, Social Security Death Index.
At the time of this study, NDI reported dates of death occurring until the end of 2017. To ensure an appropriate comparison, other mortality data sources were limited to the same time window (eg, 2011-2017).
NDI was selected as the comparator given its status as the gold standard for mortality in the United States.13-16 Patient identifiers (first and last name, sex, race, dates of birth and death, age at death, and location of death by state) for these patients were submitted for matching with NDI. On the basis of NDI User's Guide,6 matches with a score above 27 were considered positive matches, with an estimated accuracy of > 99%. Patients with NDI matching scores above this threshold were therefore marked as deceased according to the NDI.
Statistical Analysis
Extraction of patients and division of patients into subsets for analysis was performed using RStudio v. 3.6.1, along with the Open Database Connection (obdc) v. 1.2.2, and dplyr v. 1.0.2 libraries for query creation, eeptools v. 1.2.0 for age calculation, and data.table v. 1.13.2 for representing data structures.
Calculation of sensitivity and specificity was performed with Python 3.8, using pandas v 1.1.4 and numpy v 1.17.4 for dataframe representation and transformation and datetime.datetime for representing dates. CIs were calculated using Python 3.8 using the math and scipy v. 1.4.1 libraries. Chi-square tests were conducted with the scipy.stats chi2_contingency library.
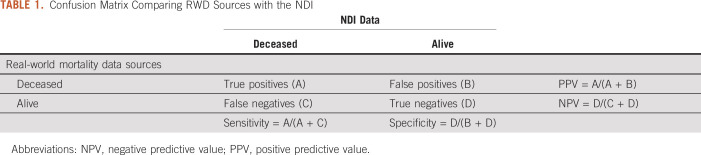
Sensitivity, specificity, positive predictive value, negative predictive value, and date concordance were evaluated against the NDI (Table 1). Assessments were conducted for each data source individually versus NDI and for the composite mortality score versus NDI. This composite mortality score was also assessed with and without data from manual curation.
TABLE 1.
Confusion Matrix Comparing RWD Sources with the NDI
Patients were considered deceased according to NDI if they matched an NDI record with a matching score above 27; otherwise, the patients were considered alive according to NDI. For individual data sources, analyses were performed using the subset of patients for whom each individual source was relevant (eg, only patients with tumor registry data available) or all patients (eg, SSDI and digitized obituaries), as appropriate.
Sensitivity and specificity were calculated for subgroups defined by extent of cancer spread (metastatic and nonmetastatic), race (White, Black, Asian or Pacific islander, American Indian or Alaskan Native, and Unknown), age (younger than 30, 30-59 years, 60 and older), sex, and SES on the basis of residential zip code (household income < $30,000 in US dollars [USD], $30,000-$59,999 USD, $60,000-$99,999 USD, ≥ $100,000 USD). If multiple zip codes were recorded for a patient, then the last location on file was used to establish the median income category.
CIs for sensitivity, specificity, and concordance were computed using Wilson's method, chosen to give accurate CIs for mismatched and small sample sizes. Chi-squared statistics with P ≤ .05 indicate statistical significance.
Additional details on the code used to generate the analysis are available in the Appendix.
Ethical Considerations
The research conducted by Syapse is exempt from Institutional Review Board review because this is a retrospective research project using de-identified patient data. Syapse received this exemption from Advarra, an external and independent Institutional Review Board.
RESULTS
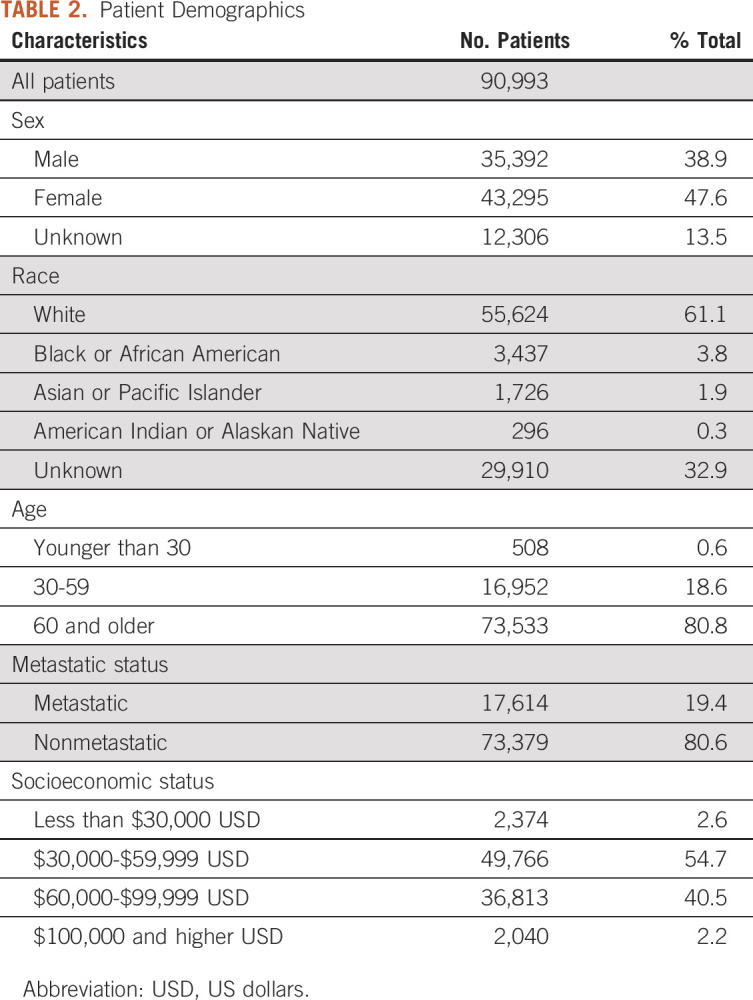
The final study population included 90,993 unique patients after removing duplicates, of whom 17,614 patients had metastatic cancer (Table 2). Approximately 47.6% of the population was female (13.5% unknown sex), 80.8% were 60 years and older, and 61.1% were White. Patients were concentrated in the low to middle household income range ($30,000-$59,999 USD: 54.7%; $60,000-$99,999 USD: 40.5%).
TABLE 2.
Patient Demographics
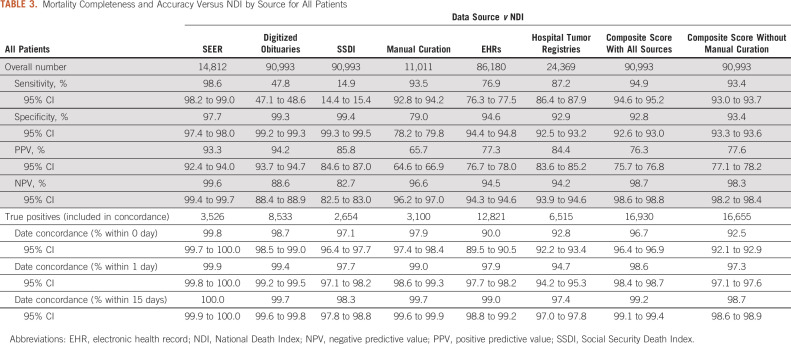
Overall, the sensitivity for vital status across individual data sources ranged from 14.9% (SSDI) to 98.6% (SEER) (Table 3). Specificity ranged from 79.0% (manual curation) to 99.4% (SSDI). For true positive dates of death, concordance (date agreement) within 1 day of NDI date of death ranged from 94.7% (hospital tumor registries) to 99.9% (SEER) across sources.
TABLE 3.
Mortality Completeness and Accuracy Versus NDI by Source for All Patients
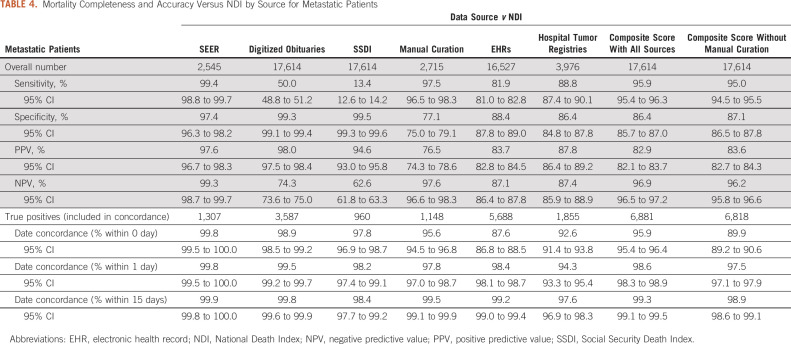
The composite score achieved a sensitivity of 94.9% with a specificity of 92.8% and concordance within 1 day of 98.6% (Table 3). Excluding dates of death obtained via manual curation, the composite score achieved sensitivity of 93.4%, specificity of 93.4%, and concordance within 1 day of 97.3%. The composite score achieved similarly high sensitivity (95.9%) for patients with metastatic cancer (Table 4), with 86.4% specificity and 98.6% concordance within 1 day.
TABLE 4.
Mortality Completeness and Accuracy Versus NDI by Source for Metastatic Patients
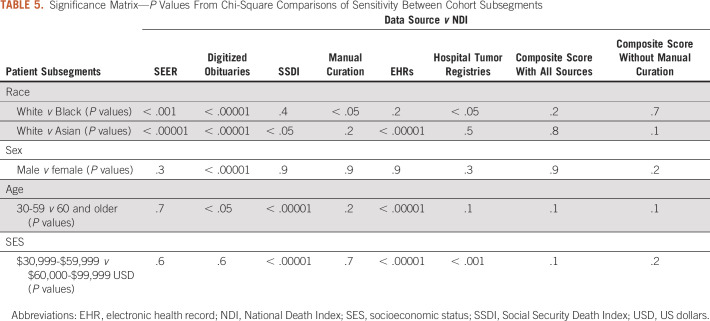
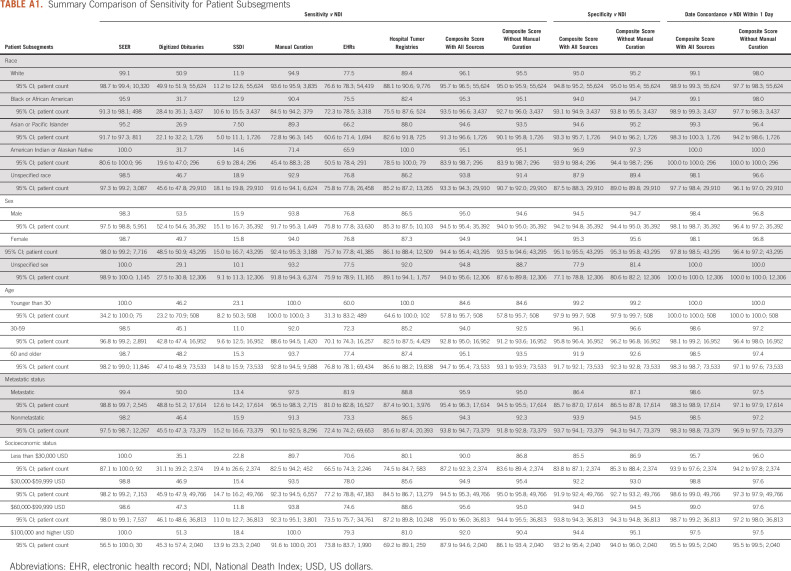
There were a number of statistically significant differences in sensitivity for subgroup analyses from individual data sources (Table 5 and Appendix Table A1). Mortality data in obituaries, SSDI, and EHRs were more complete for patients age 60 years and older compared with their younger counterparts (all P < .05). Black patients and Asian patients were also less likely to have data captured in obituaries and SEER compared with White patients (all P < .05). Manual curation and registries were less likely to capture Black patients compared with White patients, whereas SSDI and EHRs were less likely to capture Asian patients compared with White patients (all P < .05). SSDI was more likely to capture patients residing in areas with median household income of $30,000-$59,000 USD compared with patients in the $60,000-$99,999 USD category (P < .05). By contrast, EHRs and tumor registries were less likely to capture patients in the $30,000-$59,000 USD category compared with patients in the $60,000-$99,999 USD category (P < .05). Differences in sensitivity were not observed in the composite mortality score for any subgroup, either including or not including manual curation (all P > .05). By combining data across sources, the composite score was able to overcome these biases and maintain high sensitivity (84.6%-96.1%), specificity (77.9%-99.2%), and date concordance within one day (95.7%-100.0%) across subgroups.
TABLE 5.
Significance Matrix—P Values From Chi-Square Comparisons of Sensitivity Between Cohort Subsegments
DISCUSSION
Individual data sources for death have intrinsic biases in specific populations that limit their utility in determining mortality for a group of unselected patients. A composite mortality score developed from these sources overcomes these biases, in particular regarding race and sex.
This study demonstrated that a composite mortality score was capable of achieving sensitivity of 94.9% and specificity of 92.8% compared with the NDI, with concordance within 1 day of 98.6%.
Excluding manual curation from the mortality composite score, sensitivity and concordance decreased by 1.6 percentage points or less across these measures (sensitivity 93.4%, specificity 93.4%, and concordance within 1 day 97.3%), indicating that a high-quality mortality composite score can be built from scalable sources alone.
The concordance for all individual data sources versus NDI showed high degrees of accuracy (94.7%-99.9% concordance within 1 day). Thus, if a date of death was present in any source, it could be reasonably trusted to be accurate.
Sensitivity varied by data source, ranging from 14.9% to 98.6%. Additionally, not all sources are relevant to all patient populations, further influencing data completeness by source. Hospital tumor registries are specific to each health system, and include only patients newly diagnosed within that health system, leaving gaps in coverage. SEER mortality data had the highest sensitivity (98.6%), specificity (97.7%), and concordance (99.9% within 1 day) of any source; however, SEER is only relevant for patients treated within its coverage areas (currently ∼35% of the US population),5 and only 16.3% of the randomly selected patient population matched SEER data available at the time of the study. No single source achieved high levels of sensitivity with relevance across the entire real-world patient population. When combined, however, the composite score achieves trustworthy levels of completeness (sensitivity of 94.9%) and accuracy (concordance within 1 day of 98.6%).
When evaluating individual data sources, there were a number of significant differences in sensitivity between different patient subgroups. These potential biases are important because disparities in data completeness and accuracy can cause researchers to draw incorrect conclusions in real-world studies.4 Differential accuracy of mortality data by demographic or other patient characteristics, as observed for individual data sources in the current study, can invalidate comparative analyses.
First, men were more likely than women to have their deaths captured in digitized obituaries. This is consistent with prior analyses of digitized obituaries that showed that women were awarded significantly fewer obituaries compared with men, potentially because of survivor bias and/or the value placed on women’s life achievements. 8 Obituaries were the only mortality source for which a sex bias was statistically significant, and biases were not observed for the composite score.
Second, every individual source exhibited statistically significant differences in sensitivity of vital status by race. Black patients were less likely to have dates of death captured than White patients by SEER, obituaries, manual curation, and tumor registries, whereas Asian patients were less likely than White patients to have dates of death captured by SEER, obituaries, SSDI, and EHRs. The greatest racial bias within these sources was observed in obituary data, which achieved 50.90% sensitivity for White patients but only 31.66% for Black patients and 26.87% for Asian patients. This suggests that Black and Asian patients are either less likely than White patients to have their deaths memorialized in an obituary or that their obituaries are less likely to be captured by digital obituary scrapers. To our knowledge, this appears to be the first documentation of racial bias in digitized obituary data within the United States. These racial disparities in mortality documentation can potentially lead to healthcare disparities within RWD and real-world evidence. First, observational research studies relying on the completeness of these data may misrepresent outcomes in these populations. Second, searchable online obituaries are sometimes used as a resource when manually abstracting registries; thus, biases within digitized obituaries may be propagated through other, seemingly unrelated sources. The composite mortality score was able to overcome these data source biases.
Comparing across age, patients 60 years and older were significantly more likely to have death data captured in digitized obituaries, SSDI, and EHRs than patients between 30 and 59 years. The population age 30-59 years also had more missing identifiers than the population age 60 years and older (data not shown), which may have affected matching rates with NDI in some cases. It is possible that the availability of Medicare among the older population improves the availability of patient identifiers. As with sex and race, significant biases by age were not observed in the mortality composite score.
Finally, some intriguing trends were observed in segmenting patients by the median income level within their zip code that warrant further investigation. Given the small size of the lowest and highest income populations, trend variability across sources, and the use of an income proxy rather than income, it is difficult to draw clear conclusions.
This study has a number of limitations. Most importantly, NDI has limitations in both patient linkage and recency. Patients are matched to NDI on the basis of available patient identifiers including name, date of birth, sex, race, state of birth, and state of residence. If a match to NDI is found, the corresponding date of death and the confidence level in the match are returned. If a match is not found, the patient is assumed to be alive. If there are errors matching patients, these patients would be erroneously labeled as alive according to NDI. This could result in errant false positives (deceased in source but alive in NDI) and reduced specificity. When limiting analysis to only patients with complete patient identifiers, however, the resulting composite score showed only slightly improved specificity (from 92.8% to 94.9%), sensitivity (from 94.9% to 95.8%), and concordance within 1 day (from 98.5% to 98.9%), indicating that matching errors, while a potential factor, likely did not have a significant impact on the results.
Additionally, NDI mortality data are delayed relative to typical real-world data sets. Data in the LHN real-world database are updated regularly, with many automated real-time or daily feeds. At the time of the analysis, data available from NDI covered deaths occurring only until the end of 2017. Thus, this study was unable to determine the sensitivity, specificity, and concordance of patient deaths occurring between January 2018 and November 2019, when the study was initiated.
The mortality composite score tested in this study was developed a priori, prior to measuring each source in comparison with NDI. If a patient had dates of death from multiple sources, the mortality composite score used the following hierarchy, in ascending order, to resolve discrepancies: hospital tumor registries, SEER, EHRs, SSDI, digitized obituaries, and manual curation. It is likely that a reordering of this composite score ranking methodology could further improve the concordance, given that hospital tumor registries and EHRs were ranked first and third but had among the lowest NDI concordance.
Results are also dependent on availability of multiple high-quality data sources. Patients within the database are treated within large, integrated community health systems with enterprise-wide EHR capture across the system (eg, outpatient oncology clinic, inpatient hospital, and hospice), allowing for greater capture of mortality data. Indeed, the level of sensitivity for EHR data alone within this study (76.9%) exceeds a comparable published report of outpatient oncology EHR data only (65.97%).10 It further exceeds coverage by the Department of Veterans Affairs Medical SAS Inpatient Datasets (sensitivity of 12.0% compared with NDI).17 In addition, this study relies on data from both hospital tumor registries and SEER. Because of reporting requirements, these sources are typically maintained by and available to hospitals or health systems, but less so to independent community practices, giving health systems an advantage in creating a strong composite mortality score.
In summary, this study shows that although there were several significant sensitivity biases within individual mortality sources, a composite score was able to successfully overcome these biases. This indicates that merging multiple high-quality but incomplete sources together is able to overcome biases in individual sources and can generate a trustworthy mortality score for an entire real-world patient population.
ACKNOWLEDGMENT
The authors thank Sally Wadedaniel, Chenan Zhang, Yanina Natanzon, Joshua Loving, Andrew Schrag, Jay Ronquillo, Jonathan Hirsch, Alakananda Iyengar, Raghu Warrier, Sheetal Walters, Lydie Prevost, Arnon Moscona, and Antoinette Cummins.
APPENDIX Code Used to Generate Analyses
Libraries
Libraries and versions used:
True positives, false positives, true negatives, and false negatives were derived by comparing death status from Syapse patients and the corresponding death status from NDI.
Python 3.8 was used for coding.
Libraries used:
#######
For general dataframe loading
#######
pandas - 1.1.4
numpy - 1.17.4
datetime.datetime - standard library
######
For confidence interval
######
math.sqrt - standard library (no version)
scipy.special.ndtri - 1.4.1
Confusion Matrix
import pandas as pd
import numpy as np
# For regex
# Input dataframe contains columns with comparison values of our various sources vs NDI.
# These can take on four values: TP, FP, TN, FN for True and False Positives and Negatives
# There are also columns with the difference in days of dates of death - these can be from 0 and up - columns are ints.
inputDataFrame = pd.read_csv("∼/patientFileForDoDWhitePatients.csv", low_memory=False)
standardDataFrame = pd.read_csv("∼/patientFileForDoDAllPatients.csv")
# There are also dataframes containing only a subset of patients
# These include dfs with patients split up by:
# - Tumor type (Any Lung Cancer, NSCLC, SCLC, AML, All CRC, Advanced CRC, All Breast cancer, Metastatic Breast Cancer, Unknown)
# - Race (White, Black or African American, Asian / Pacific Islander, American Indian / Alaskan Native, Unknown)
# - Age (0-30, 30-60, 60 and up, Unknown)
# - Sex (Male, Female, Unknown)
# - SES (as determined by patient zip code: 0-30K, 30-60K, 60-100K, 100-185K, >185K, Unknown)
# - Metastatic Status (Metastatic, Non-Metastatic)
# We want to go column by column comparing the positive/negative status and the date agreement for our various sources.
positivesValues = ['1. SEER vs. NDI', '2. Datavant vs. NDI', '2a. Obit vs. NDI', '2b. SSA vs. NDI', '4. Manual Abstraction vs. NDI', '5. EMR vs. NDI', '6. Hospital Registries vs. NDI', '7. Rolled up Syapse View vs. NDI', 'Rolled Up Syapse View Without MA Vs NDI']
datesValues = ['1. SEER DoD Agreement', '2. Datavant DoD Agreement', '2a. Obit DoD Agreement', '2b. SSA DoD Agreement', '4. Manual Abstraction DoD Agreement', '5. EMR DoD Agreement', '6. Hospital Registries DoD Agreement', '7. Rolled up Syapse View DoD Agreement', 'Rolled Up Syapse View Without MA Date Diff']
for x in range(0, len(positivesValues)):
# Total number is the overall # of patients
# Class number is the # of patients in this 'class', AKA the group under review
totalNumber = len(standardDataFrame.index)
classNumber = len(inputDataFrame.index)
# Our input data frame will be the one we get values from
columnPos = positivesValues[x]
columnDates = datesValues[x]
print(columnPos)
TP = len(inputDataFrame[inputDataFrame[columnPos] == 'TP'])
TN = len(inputDataFrame[inputDataFrame[columnPos] == 'TN'])
FP = len(inputDataFrame[inputDataFrame[columnPos] == 'FP'])
FN = len(inputDataFrame[inputDataFrame[columnPos] == 'FN'])
print('overall # -', (TP + TN + FP + FN))
if totalNumber > 0 and classNumber > 0:
print('percent coverage - all patients', ((TP + TN + FP + FN) / totalNumber))
print('percent coverage - this subgroup', ((TP + TN + FP + FN) / classNumber))
print('tp - ', str(TP))
print('fp - ', str(FP))
print('tn - ', str(TN))
print('fn - ', str(FN))
# This is to handle situations with very low patient numbers:
# We'll to avoid dividing by 0, we'll set 0 numbers to just be very small - this will still show up as 0s
if TP == 0 and FP == 0:
FP = 0.00000000000000000000000000001
TP = 0.00000000000000000000000000001
if TN == 0 and FN == 0:
FN = 0.00000000000000000000000000001
TN = 0.00000000000000000000000000001
if TP == 0 and FN == 0:
TP = 0.00000000000000000000000000001
FN = 0.00000000000000000000000000001
if TP == 0:
TP = 0.00000000000000000000000000001
print('Sensitivity - ' + str(TP / (TP + FN)))
print('Specificity - ' + str(TN / (FP + TN)))
print('PPV - ' + str(TP / (TP + FP)))
print('NPV - ' + str(TN / (TN + FN)))
# Find the number off by zero days - we're also only interested in ones where the patient is in fact deceased
# FP will mean NDI doesn't have a DoD
# FN will mean Syapse doesn't have a DoD
# TN will mean neither has a DoD
numWithExact = len(inputDataFrame[(inputDataFrame[columnDates] == 0) & (inputDataFrame[columnPos] == 'TP')]) + len(
inputDataFrame[(inputDataFrame[columnDates].isna()) & (inputDataFrame[columnPos] == 'TP')])
print('% within 1 day - ' + str((numWithExact + len(inputDataFrame[(inputDataFrame[columnDates] == 1) & (inputDataFrame[columnPos] == 'TP')])) / TP))
print('# within 0 day - ' + str(numWithExact))
print('# within 1 day - ' + str(len(inputDataFrame[(inputDataFrame[columnDates] == 1) & (inputDataFrame[columnPos] == 'TP')])))
print('% within 1 day - ' + str(len(inputDataFrame[inputDataFrame[columnDates] == 1]) / len(inputDataFrame[inputDataFrame[columnDates].notna()])))
print('# within 7 days - ' + str(len(inputDataFrame[(inputDataFrame[columnDates] <= 7) & (inputDataFrame[columnDates] > 1) & (inputDataFrame[columnPos] == 'TP')])))
print('% within 7 days - ' + str(len(inputDataFrame[(inputDataFrame[columnDates] <= 7) & (inputDataFrame[columnDates] > 1)]) / len(inputDataFrame[inputDataFrame[columnDates].notna()])))
print('# within 15 days - ' + str(len(inputDataFrame[(inputDataFrame[columnDates] <= 15) & (inputDataFrame[columnDates] > 7) & (inputDataFrame[columnPos] == 'TP')])))
print('% within 15 days - ' + str(len(inputDataFrame[(inputDataFrame[columnDates] <= 15) & (inputDataFrame[columnDates] > 7)]) / len(inputDataFrame[inputDataFrame[columnDates].notna()])))
print('# within 30 days - ' + str(len(inputDataFrame[(inputDataFrame[columnDates] <= 30) & (inputDataFrame[columnDates] > 15) & (inputDataFrame[columnPos] == 'TP')])))
print('% within 30 days - ' + str(len(inputDataFrame[(inputDataFrame[columnDates] <= 30) & (inputDataFrame[columnDates] > 15)]) / len(inputDataFrame[inputDataFrame[columnDates].notna()])))
print('# past 30 days - ' + str(len(inputDataFrame[(inputDataFrame[columnDates] > 30) & (inputDataFrame[columnPos] == 'TP')])))
print('% past 30 days - ' + str(len(inputDataFrame[inputDataFrame[columnDates] > 30]) / len(inputDataFrame[inputDataFrame[columnDates].notna()])))
# input()
print('')
print('')
print('###############################')
print('PPV - ' + str(TP/(TP+FP)))
print('NPV - ' + str(TN/(TN+FN)))
Confidence Intervals
from __future__ import print_function, division
from math import sqrt
from scipy.special import ndtri
# Find the confidence interval given a proportion
# This uses the notation described in
# "Proportions and their differences 2nd Ed."
# by, D. G. Altman et al.
def confidence_interval_prop(a, b, c):
X = 2a + c^2
Y = z * sqrt(c^2 + 4a * (1 - (a/b)))
Z = 2 * (b + c^2)
return (str(round(((X - Y) / Z) * 100,2)) + '%', str(round(((X + Y) / Z)*100,2)) + '%')
# Sensitivity and specificity confidence intervals derived using Wilson's method
def sens_spef_conf_interval(TP, FP, FN, TN, alpha=0.95):
z = -ndtri((1.0 - alpha) / 2)
# Compute sensitivity using wilson's method
sens_point_est = TP / (TP + FN)
sens_conf_int = confidence_interval_prop(TP, TP + FN, z)
# Compute specificity using same method
spec_point_est = TN / (TN + FP)
spec_conf_int = confidence_interval_prop(TN, TN + FP, z)
# Compute PPV and NPV
PPV_point_estimate = TP / (TP + FP)
NPV_point_estimate = TN / (TN + FN)
PPV_confidence_interval = confidence_interval_prop(TP, TP + FP, z)
NPV_confidence_interval = confidence_interval_prop(TN, TN + FN, z)
return sens_point_est, spec_point_est, sens_conf_int, spec_conf_int, PPV_point_estimate, PPV_confidence_interval, NPV_point_estimate, NPV_confidence_interval
# Takes the #s of patients who have dates of death off by:
# 0 for same
# 1 for 'next'
# More than 1, less than or equal to 7 for 'week'
# More than 7, less than or equal to 14 for 'twoweek'
# More than 14, less than or equal to 30 for 'month'
#
def days_matching_with_confidence_interval(same, next, week, twoweek, month, total):
zero_day_point = same / total
one_day_point = (same + next) / total
week_point = (same + next + week) / total
two_week_point = (same + next + week + twoweek) / total
month_point = (same + next + week + twoweek + month) / total
zero_day_conf = sqrt(((zero_day_point * (1-zero_day_point)) / total)) * 1.96
one_day_conf = sqrt(((one_day_point * (1-one_day_point)) / total)) * 1.96
week_conf = sqrt(((week_point * (1-week_point)) / total)) * 1.96
two_week_conf = sqrt(((two_week_point * (1-two_week_point)) / total)) * 1.96
month_conf = sqrt(((month_point * (1-month_point)) / total)) * 1.96
zero_day_conf = (str(round((zero_day_point - zero_day_conf) * 100, 2)) + '%', str(round((zero_day_point + zero_day_conf) * 100, 2)) + '%')
one_day_conf = (str(round((one_day_point - one_day_conf) * 100, 2)) + '%', str(round((one_day_point + one_day_conf) * 100, 2)) + '%')
week_conf = (str(round((week_point - week_conf) * 100, 2)) + '%', str(round((week_point + week_conf) * 100, 2)) + '%')
two_week_conf = (str(round((two_week_point - two_week_conf) * 100, 2)) + '%', str(round((two_week_point + two_week_conf) * 100, 2)) + '%')
month_conf = (str(round((month_point - month_conf) * 100, 2)) + '%', str(round((month_point + month_conf) * 100, 2)) + '%')
zero_day_point = str(round(zero_day_point * 100, 2)) + '%'
one_day_point = str(round(one_day_point * 100, 2)) + '%'
week_point = str(round(week_point * 100, 2)) + '%'
two_week_point = str(round(two_week_point * 100, 2)) + '%'
month_point = str(round(month_point * 100, 2)) + '%'
return zero_day_point, zero_day_conf, one_day_point, one_day_conf, week_point, week_conf, two_week_point, two_week_conf, month_point, month_conf
# These counts are derived From the TP/FP/TN/FN of a particular group, as found in the DoD_Stats.py program
counts = [236, 88, 1691, 25]
# These days are the # that have
# [Exact match, Off by 1, More than 1 <= 7 off, more than 7 <= 14 off, more than 14 <= 30 off, > 30 off]
days = [214, 16, 3, 0, 0, 3]
TP = counts[0]
FP = counts[1]
TN = counts[2]
FN = counts[3]
zeroDay = days[0]
oneDay = days[1]
sevenDay = days[2]
fifteenday = days[3]
thirtyDay = days[4]
past = days[5]
a = 0.95
sens_point_est, spec_point_est, sens_conf_int, spec_conf_int, PPV, PPV_confidence, NPV, NPV_confidence \
= sens_spef_conf_interval(TP, FP, FN, TN, alpha=a)
print("Sensitivity: %f, Specificity: %f" % (sens_point_est*100, spec_point_est*100))
print("sensitivity:", '(' + ', '.join(sens_conf_int) + ')')
print("specificity:", '(' + ', '.join(spec_conf_int) + ')')
print("PPV: %f, NPV: %f" % (PPV*100, NPV*100))
print("PPV:", '(' + ', '.join(PPV_confidence) + ')')
print("NPV:", '(' + ', '.join(NPV_confidence) + ')')
print("")
zdp, zdc, odp, odc, wp, wc, twp, twc, mp, mc = days_matching_with_confidence_interval(zeroDay, oneDay, sevenDay, fifteenday, thirtyDay, TP)
print('zero day')
print(zdp)
print('(' + ', '.join(zdc) + ')')
print('one day')
print(odp)
print('(' + ', '.join(odc) + ')')
print('week')
print(wp)
print('(' + ', '.join(wc) + ')')
print('two week')
print(twp)
print('(' + ', '.join(twc) + ')')
print('month')
print(mp)
print('(' + ', '.join(mc) + ')')
# Chi-Squared calculation
from scipy.stats import chi2_contingency
print("Chi squared with Yates' Correction is - ", chi2_contingency(ar)[0], "\nWith a p-value of - ", chi2_contingency(ar)[1])
print("Chi squared without Yates Correction is - ", chi2_contingency(ar, correction=False)[0], "\nWith a p-value of - ", chi2_contingency(ar, correction=False)[1])
TABLE A1.
Summary Comparison of Sensitivity for Patient Subsegments
Michelle H. Lerman
Employment: Syapse
Stock and Other Ownership Interests: Syapse
Benjamin Holmes
Employment: Syapse
Stock and Other Ownership Interests: Syapse
Travel, Accommodations, Expenses: Syapse
Daniel St Hilaire
Employment: Syapse
Stock and Other Ownership Interests: Syapse
Mary Tran
Employment: Syapse
Stock and Other Ownership Interests: Syapse
Matthew Rioth
Employment: Syapse
Stock and Other Ownership Interests: Syapse
Vinod Subramanian
Employment: Syapse
Leadership: Syapse
Stock and Other Ownership Interests: Syapse
Travel, Accommodations, Expenses: Syapse
Alissa M. Winzeler
Employment: Syapse
Leadership: Syapse
Stock and Other Ownership Interests: Syapse
Travel, Accommodations, Expenses: Syapse
Thomas Brown
Employment: GenomiCare Biotechnology, Syapse
Leadership: Syapse
Stock and Other Ownership Interests: GenomiCare Biotechnology, Syapse
Honoraria: Novartis
Consulting or Advisory Role: Jiahui Health, GenomiCare Biotechnology, Lug Healthcare Technology, Syapse
Speakers' Bureau: Syapse, Novartis
Travel, Accommodations, Expenses: Syapse, Jiahui Health, GenomiCare, Lug Healthcare Technology, Novartis
No other potential conflicts of interest were reported.
DISCLAIMER
Syapse is a privately held company. All research was performed in-house by Syapse employees using company resources. There were no sources of external funding used to support this research, including grants, contracts, or philanthropy.
M.H.L., B.H., and D.S.H. contributed equally to this work.
DATA SHARING STATEMENT
Syapse obtains the legal rights to use the de-identified patient data in its possession to conduct research from its business partners. These rights restrict Syapse's ability to publicly share or provide access to the de-identified data.
AUTHOR CONTRIBUTIONS
Conception and design: Michelle H. Lerman, Daniel St Hilaire, Mary Tran, Vinod Subramanian, Alissa M. Winzeler, Thomas Brown
Administrative support: Thomas Brown
Collection and assembly of data: Benjamin Holmes, Daniel St Hilaire, Thomas Brown
Data analysis and interpretation: All authors
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO’s conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Michelle H. Lerman
Employment: Syapse
Stock and Other Ownership Interests: Syapse
Benjamin Holmes
Employment: Syapse
Stock and Other Ownership Interests: Syapse
Travel, Accommodations, Expenses: Syapse
Daniel St Hilaire
Employment: Syapse
Stock and Other Ownership Interests: Syapse
Mary Tran
Employment: Syapse
Stock and Other Ownership Interests: Syapse
Matthew Rioth
Employment: Syapse
Stock and Other Ownership Interests: Syapse
Vinod Subramanian
Employment: Syapse
Leadership: Syapse
Stock and Other Ownership Interests: Syapse
Travel, Accommodations, Expenses: Syapse
Alissa M. Winzeler
Employment: Syapse
Leadership: Syapse
Stock and Other Ownership Interests: Syapse
Travel, Accommodations, Expenses: Syapse
Thomas Brown
Employment: GenomiCare Biotechnology, Syapse
Leadership: Syapse
Stock and Other Ownership Interests: GenomiCare Biotechnology, Syapse
Honoraria: Novartis
Consulting or Advisory Role: Jiahui Health, GenomiCare Biotechnology, Lug Healthcare Technology, Syapse
Speakers' Bureau: Syapse, Novartis
Travel, Accommodations, Expenses: Syapse, Jiahui Health, GenomiCare, Lug Healthcare Technology, Novartis
No other potential conflicts of interest were reported.
REFERENCES
- 1.Loree JM Anand S Dasari A, et al. : Disparity of race reporting and representation in clinical trials leading to cancer drug approvals from 2008 to 2018. JAMA Oncol 5:e191870, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nazha B Mishra M Pentz R, et al. : Enrollment of racial minorities in clinical trials: Old problem assumes new urgency in the age of immunotherapy. Am Soc Clin Oncol Educ Book 39:3-10, 2019 [DOI] [PubMed] [Google Scholar]
- 3.Sharrocks K Spicer J Camidge DR, et al. : The impact of socioeconomic status on access to cancer clinical trials. Br J Cancer 111:1684-1687, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bretscher MT, Sanglier T: Quantifying the Impact of Mortality Underreporting on Analyses of Overall Survival. Presented at the 34th International Conference for Pharcoepidemiology and Risk Management, Prague, Czech Republic, August 24, 2018
- 5.National Cancer Institute : About the SEER Registries. Bethesda, MD, NCI [Google Scholar]
- 6.National Center for Health Statistics : National Death Index User’s Guide. Hyattsville, MD, National Center for Health Statistics, 2013 [Google Scholar]
- 7.Maybury KK: Invisible lives: Women, men and obituaries. Omega 32:27-37, 1996 [Google Scholar]
- 8.Ogletree SM, Pena D, Figueroa P: A double standard in death? Gender differences in obituaries. Omega 51:337-342, 2005 [Google Scholar]
- 9.Marks A, Piggee T: Obituary analysis and describing a life lived: The impact of race, gender, age, and economic status. Omega 31:37-57, 1999 [Google Scholar]
- 10.Curtis MD Griffith SD Tucker M, et al. : Development and validation of a high-quality composite real-world mortality endpoint. Health Serv Res 53:4460-4476, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turchin A, Shubina M, Murphy SN: I am not dead yet: Identification of false-positive matches to death master file. AMIA Annu Symp Proc 2010:807-811, 2010 [PMC free article] [PubMed] [Google Scholar]
- 12.Datavant : Further Evidence That COVID-19 Disproportionately Impacts African-American, Hispanic, and Low-Income Populations. San Francisco, CA, Datavant [Google Scholar]
- 13.Cowper DC Kubal JD Maynard C, et al. : A primer and comparative review of major US mortality databases. Ann Epidemiol 12:462-468, 2002 [DOI] [PubMed] [Google Scholar]
- 14.Fillenbaum GG, Burchett BM, Blazer DG: Identifying a national death index match. Am J Epidemiol 170:515-518, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rich-Edwards JW, Corsano KA, Stampfer MJ: Test of the National Death Index and Equifax Nationwide Death Search. Am J Epidemiol 140:1016-1019, 1994 [DOI] [PubMed] [Google Scholar]
- 16.Wentworth DN, Neaton JD, Rasmussen WL: An evaluation of the social security administration master beneficiary record file and the National Death Index in the ascertainment of vital status. Am J Public Health 73:1270-1274, 1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sohn MW Arnold N Maynard C, et al. : Accuracy and completeness of mortality data in the department of veterans affairs. Popul Health Metr 4:2, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Syapse obtains the legal rights to use the de-identified patient data in its possession to conduct research from its business partners. These rights restrict Syapse's ability to publicly share or provide access to the de-identified data.