Abstract
PURPOSE
Patients with myelodysplastic syndrome (MDS) are at risk of relapse after allogeneic hematopoietic cell transplantation. The utility of ultra-deep genomic testing to predict and the impact of conditioning intensity to prevent MDS relapse are unknown.
METHODS
Targeted error-corrected DNA sequencing was performed on preconditioning blood samples from patients with MDS (n = 48) from the Blood and Marrow Transplant Clinical Trials Network 0901 phase III randomized clinical trial, which compared outcomes by allogeneic hematopoietic cell transplantation conditioning intensity in adult patients with < 5% marrow myeloblasts and no leukemic myeloblasts in blood on morphological analysis at the time of pretransplant assessment. Clinical end points (53-month median follow-up) included transplant-related mortality (TRM), relapse, relapse-free survival (RFS), and overall survival (OS). Of the 48 patients examined, 14 experienced TRM, 23 are relapse-free, and 11 relapsed, of which 7 died.
RESULTS
Using a previously described set of 10 gene regions, 42% of patients (n = 20) had mutations detectable before random assignment to reduced intensity conditioning (RIC) or myeloablative conditioning (MAC). Testing positive was associated with increased rates of relapse (3-year relapse, 40% v 11%; P = .022) and decreased OS (3-year OS, 55% v 79%, P = .045). In those testing positive, relapse rates were higher (3-year relapse, 75% v 17%; P = .003) and RFS was lower (3-year RFS, 13% v 49%; P = .003) in RIC versus MAC arms. Testing additional genes, including those associated with MDS, did not improve prognostication.
CONCLUSION
This study provides evidence that targeted DNA sequencing in patients with MDS before transplant can identify those with highest post-transplant relapse rates. In those testing positive, random assignment to MAC lowered but did not eliminate relapse risk.
INTRODUCTION
Myelodysplastic syndrome (MDS), one of the most common hematologic disorders, is a collection of clinically and genetically heterogeneous diseases. Allogeneic hematopoietic cell transplantation (alloHCT) is currently the only curative treatment for MDS, but its usage is limited, in part, by the risk of transplant-related mortality (TRM).1 Reduced intensity conditioning (RIC) regimens have helped to decrease toxicity although multiple retrospective studies found that the decreased TRM is counterbalanced by increased risk of relapse compared with more intense myeloablative conditioning (MAC) regimens.2,3 Two randomized phase III trials comparing outcomes following RIC and MAC conditioning regimens in patients with MDS are yet to provide a definitive answer regarding which regimen should be used when a patient is eligible for either approach.4-6
CONTEXT
Key Objective
Allogeneic hematopoetic cell transplantation (alloHCT) is the only curative therapy for Myelodysplastic syndrome (MDS) but is associated with suboptimal rates of transplant-related mortality (TRM). The ability to predict the risk of relapse and tailor alloHCT conditioning regimens to minimize TRM risk would be beneficial. Using samples from a randomized phase III clinical trial comparing alloHCT conditioning intensity in patients with MDS, this study examined the prognostic significance of pretransplant genetic markers on post-transplant outcomes.
Knowledge Generated
The presence of mutations within a previously identified set of 10 gene regions before alloHCT was associated with increased rates of relapse and decreased relapse-free survival in patients with MDS who received reduced intensity versus myeloablative conditioning. Examination of additional MDS-associated gene regions was not beneficial.
Relevance
This study provides rationale for clinical trials using pretransplant genomic testing to personalize alloHCT approaches based on the risk of relapse.
Aside from conditioning regimen, a variety of other factors, including the presence and type of genetic mutation before and after transplant, have been found to influence clinical outcomes.7-9 We recently demonstrated that patients with acute myeloid leukemia (AML) with detectable mutations before alloHCT had decreased relapse and improved overall survival (OS) when randomly assigned to MAC versus RIC.10 To determine if a similar benefit from increased conditioning intensity is seen for patients with MDS, we performed ultra-deep error-corrected DNA sequencing on blood samples collected immediately before random assignment to either MAC or RIC for alloHCT.
METHODS
Clinical Cohort
The Blood and Marrow Transplant Clinical Trials Network (BMT CTN) 0901 (ClinicalTrials.gov identifier: NCT01339910) study was a phase III randomized clinical trial comparing outcomes by conditioning intensity in adult patients with myeloid malignancy undergoing alloHCT with < 5% marrow myeloblasts and no leukemic myeloblasts in blood on morphological analysis at the time of pretransplant assessment.6 Frozen whole blood collected after enrollment before the conditioning regimen was available from 48 of the 54 patients with MDS. Extended follow-up of patients enrolled on this protocol was extracted from the Center for International Blood and Marrow Transplant Research (CIBMTR) research database. Clinical characteristics and outcomes were defined as outlined in the 0901 trial.6 Patients provided written informed consent to participate in both the BMT CTN 0901 trial and the CIBMTR research database. This post hoc study was approved by the BMT CTN and CIBMTR and conducted with the approval of the National Marrow Donor Program institutional review board.
Sequencing
DNA sequencing using a custom anchored multiplex polymerase chain reaction–based panel (ArcherDx, Boulder, CO) designed to incorporate molecular barcode or unique molecular identifiers (UMIs) and cover regions of 29 genes commonly mutated in myeloid malignancies (Data Supplement), including 10 gene regions previously shown to be prognostic in patients with AML from the same trial,10 was performed on 200 ng of genomic DNA isolated from each preconditioning blood sample. Library preparation and paired-end 150-bp sequencing were performed using unique dual-sample indices on an Hiseq 2500 (rapid run mode; Illumina, San Diego, CA) as previously described.10 An average of 43 million paired-end reads were acquired per sample (Data Supplement). Details are provided in the Data Supplement.
Bioinformatics
Consensus sequences based on UMI read families were mapped to human genome version hg19 (build GRCh37). De novo variant calls were made using a minimum allele frequency of 0.001 and were filtered using information regarding UMI read families, background error rate models, unique start sites, strand-specific priming, and homopolymer runs. Alternative approaches were used for insertional mutations in NPM1 and FLT3 internal tandem duplication. Details are provided in the Data Supplement.
Statistical Analysis
Kaplan-Meier estimation and log-rank test were used for analysis of OS and relapse-free survival (RFS) and Gray’s test for competing risks of TRM and relapse. Details are provided in the Data Supplement.
RESULTS
Patient Cohort
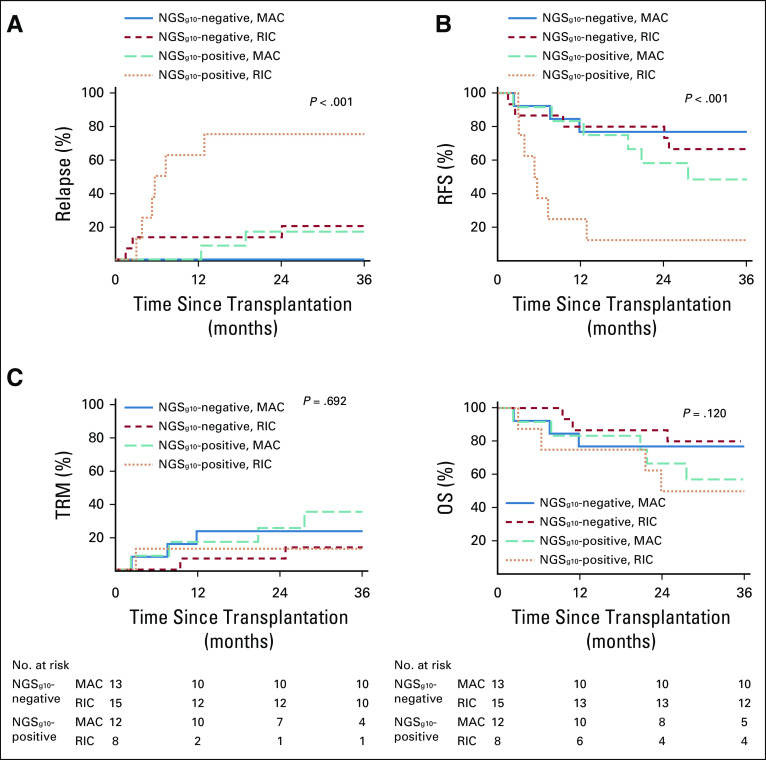
A total of 54 patients with MDS participated in the BMT-CTN 0901 clinical trial, of which 48 had blood collected after study enrollment before random assignment to MAC (n = 25) or RIC (n = 23) conditioning regimens, which was used for genomic analysis in this study. This subset of patients was well-matched for baseline characteristics (Table 1), and clinical outcomes were aligned with those previously reported (Data Supplement). The median follow-up in survivors was in excess of 53 months.
TABLE 1.
Patient Clinical Characteristics
Presence of Mutations Pretransplant Predicts Post-Transplant Clinical Outcome
Previously, we demonstrated that detection of mutations within 10 gene regions (FLT3, IDH1, IDH2, JAK2, KIT, NPM1, NRAS, RUNX1, SF3B1, and TP53) before alloHCT in patients with AML in remission was associated with higher relapse and lower survival in those randomly assigned to RIC.10 Evaluating the same gene regions in this MDS cohort, a total of 54 mutations with a median variant allele frequency (VAF) of 0.7% were detected in the blood of 42% (n = 20) of patients before conditioning treatment (Data Supplement). In samples with mutations detected, the median number of variants was 2 (range, 1-11).
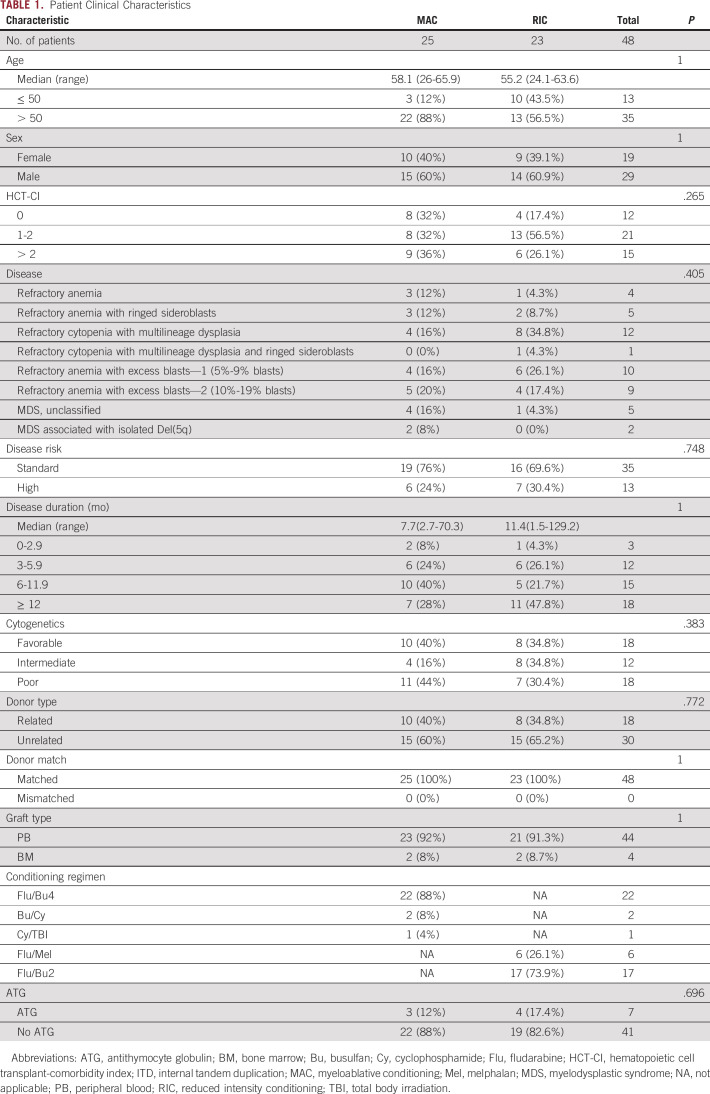
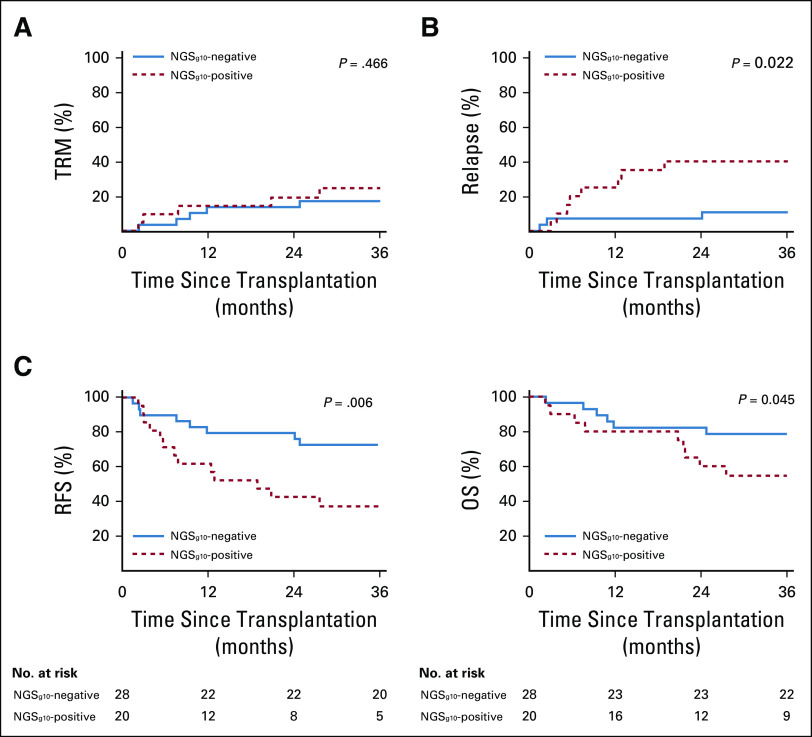
The presence of a mutation in the 10-gene panel (next-generation sequencing [NGSg10]) in the blood of patients with MDS before conditioning was found to be prognostic. NGSg10-positive patients experienced significantly higher rates of relapse (3-year relapse, 40% v 11%; P = .022) and decreased RFS (3-year RFS, 34% v 71%, P = .006) and OS (3-year OS, 55% v 79%, P = .045) compared with NGSg10-negative patients (Fig. 1 and Table 2). NGSg10 mutational status served as a strong predictor of relapse in this cohort of patients with MDS, predicting 73% of relapses at 24 months (100% for those receiving MAC), with a specificity of 78% (100% for those receiving RIC) (Data Supplement).
FIG 1.
Impact of 10-gene mutational status on clinical outcomes of patients with MDS. (A) No difference in transplant-related mortality was observed between patients with MDS based on 10-gene mutational status (P = .466). (B) Rates of relapse were significantly higher in 10-gene NGS–positive (NGSg10-positive) versus NGS-negative patients (NGSg10-negative) (3-year relapse 40% v 11%, P = .022). (C) 10-gene NGS–positive patients had significantly decreased RFS (3-year RFS, 34% v 71%, P = .006) and OS (3-year OS, 55% v 79%, P = .045) compared with NGS-negative patients. MDS, myelodysplastic syndrome; NGS, next-generation sequencing; OS, overall survival; RFS, relapse-free survival; TRM, transplant-related mortality.
TABLE 2.
Outcomes Analysis by 10-Gene NGS Status and Treatment Regimen
In contrast to NGSg10 mutational status, no significant difference in rates of relapse or OS was observed when stratifying patients by disease classification, disease risk group, or cytogenetic prognostic group (Data Supplement). Rates of relapse trended higher in patients with poor cytogenetics or categorized as high risk. Inclusion of patients with poor cytogenetics before transplant with NGSg10 mutational status improved prediction of relapse at 24 months from 73% to 91% (Data Supplement) and was associated with significantly higher rates of relapse compared with NGSg10-negative patients (Data Supplement).
Presence of Mutations Pretransplant Predicts Relapse by Conditioning Intensity
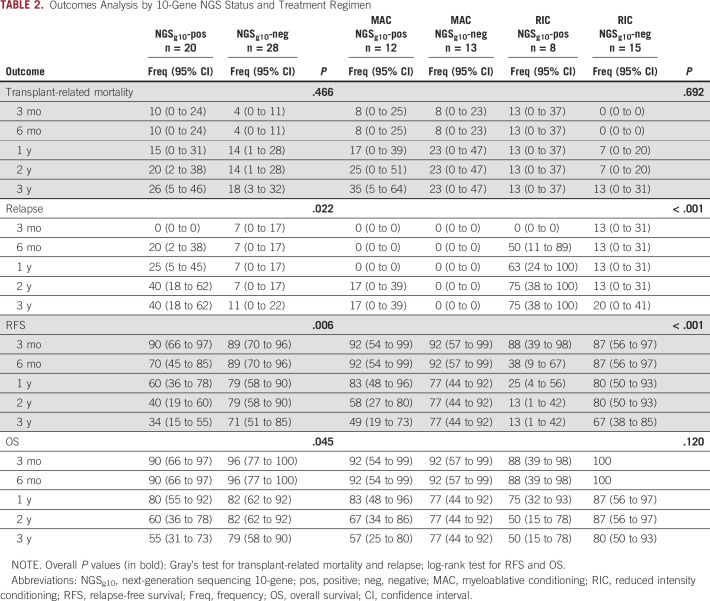
Next, the impact of conditioning intensity and NGSg10 mutational status was examined (Data Supplement). Mutations were detected in the preconditioning blood samples of 48% of MAC and 35% of RIC patients. Patients testing positive experienced higher rates of relapse (3-year relapse, 75% RIC v 17% MAC; P = .003) and lower RFS (3-year RFS, 13% RIC v 49% MAC; P = .003) when randomly assigned to RIC rather than MAC (Fig. 2A-B and Table 2). Neither of the two relapses observed after MAC occurred in the first 12 months after transplant, and the median time to relapse in those randomly assigned to RIC was 4 months (range, 2-24 months). No significant difference was observed in rates of TRM or OS when stratifying patients by both NGSg10 status and conditioning intensity (Fig. 2C and Table 2).
FIG 2.
Impact of conditioning intensity and 10-gene mutational status on clinical outcomes of patients with MDS. (A) Differences in rates of relapse were identified between subgroups defined by conditioning intensity (RIC or MAC) and mutational status (P < .001), with the highest rate occurring in 10-gene NGS–positive (NGSg10-positive) patients with MDS receiving RIC. (B) In patients with no mutations detected within the 10-gene region (NGSg10-negative), no difference in RFS was observed between conditioning intensities (3-year RFS, 67% RIC v 77% MAC, P = .634). However, in NGS-positive patients, RFS was significantly worse in those who received RIC (3-year RFS, 13% RIC v 49% MAC, P < .001). (C) No difference in transplant-related mortality (P = .692) or OS (P = .120) was observed between subgroups. MAC, myeloablative; MDS, myelodysplastic syndrome; NGS, next-generation sequencing; RIC, reduced intensity conditioning; TRM, transplant-related mortality.
For patients testing NGSg10-negative, no differences in clinical outcomes between the two conditioning intensity arms were detected, including rates of relapse (3-year relapse, 20% RIC v 0% MAC, P = .095), RFS (3-year RFS, 67% RIC v 77% MAC, P = .634), or OS (3-year OS, 80% RIC v 77% MAC, P = .845) (Fig. 2 and Table 2).
The presence of an NGSg10 mutation in the blood before conditioning predicted 67% of RIC and 100% of MAC relapses at 24 months, with specificities of 100% and 64%, respectively (Data Supplement). Inclusion of patients with poor cytogenetics before transplant with NGSg10 mutational status improved prediction of relapse at 24 months in the RIC group to 89%, with a specificity of 89%, and is associated with significantly increased rates of relapse and decreased RFS compared with patients receiving MAC (Data Supplement).
Screening for Additional Genes Does Not Improve Test Performance
The mutational spectrum of MDS is complex, with many additional genes recurrently mutated beyond the 10 AML-associated genes described above.7,11,12 Therefore, we expanded our testing to also cover regions in an additional 19 genes including those commonly found in MDS at diagnosis (ASXL1, BCOR, CBL, CUX1, DNMT3A, ETV6, EZH2, GATA2, KRAS, PHF6, PPM1D, PTPN11, SETBP1, SRSF2, STAG2, TET2, U2AF1, WT1, and ZRSR2). This resulted in the detection of an additional 82 mutations (136 in total), with a median VAF of 0.9% and a median of 2.5 variants (range, 1-12) per patient (Data Supplement). Expanding testing (total of 29 genes) resulted in the majority of patients having a mutation detectable before conditioning (80% MAC and 78% RIC).
We examined if testing for mutations in a larger number of genes would help better risk stratify patients classified as NGSg10-negative (Fig. 3, Data Supplement). The inclusion of DTA (DNMT3A, TET2, and ASXL1) genes, known to be associated with age-related clonal hematopoiesis,13,14 resulted in reclassification of 21% (n = 6) of NGSg10-negative patients to positive. Similar to what we observed in patients with AML,10 samples testing positive for only DTA mutations showed no difference in relapse rates or OS compared with those testing negative. Furthermore, although inclusion of DTA variants marginally improved the sensitivity of NGS mutational status for predicting relapse at 24 months (from 73% to 82%), it had a detrimental impact on specificity (78%-35%, Data Supplement).
FIG 3.
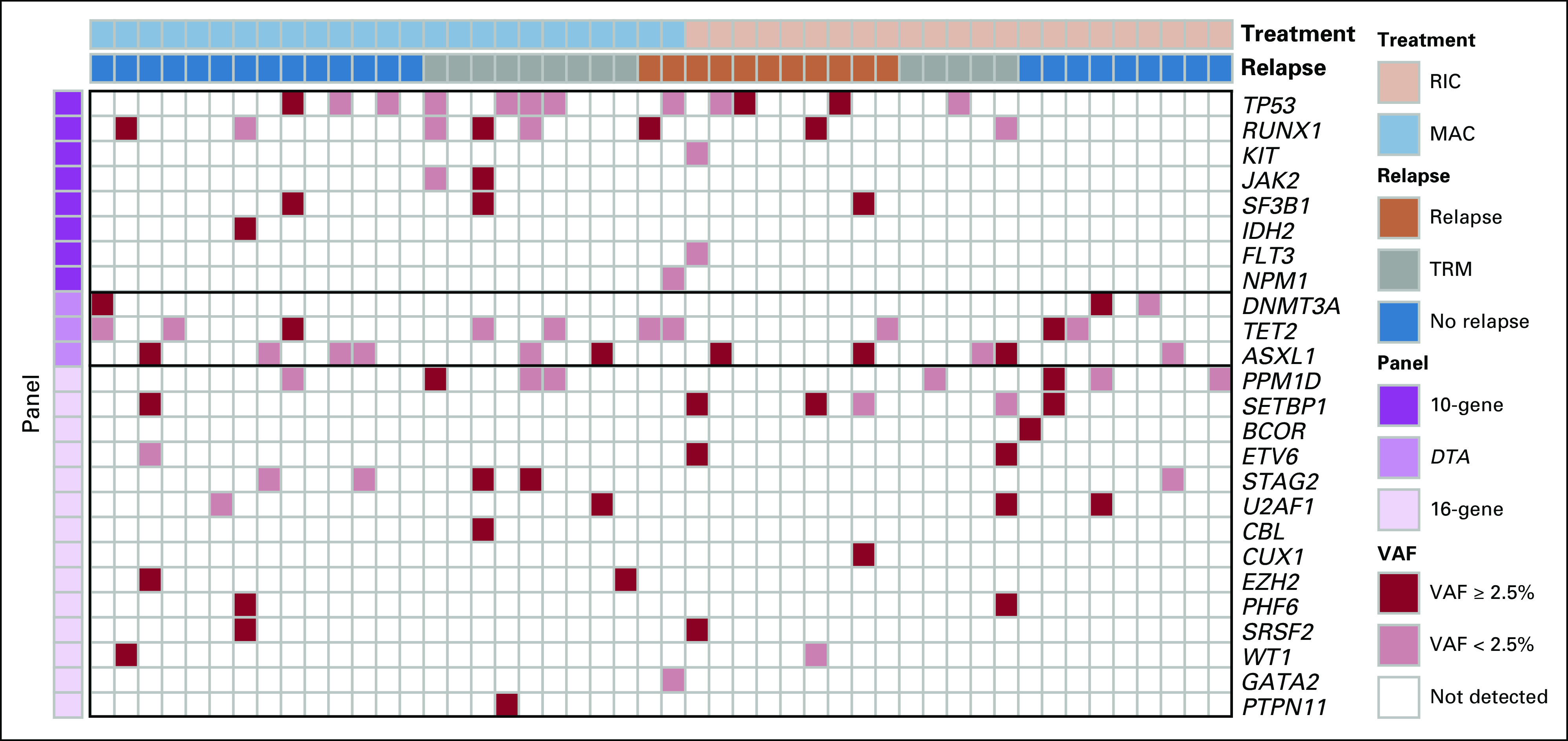
Mutational spectrum of patients with MDS before conditioning. The heatmap displays mutations detected in the preconditioning blood of patients with MDS across 29 gene regions. Patients are displayed in columns and grouped by conditioning intensity (RIC or MAC]) and clinical outcome (relapse, no relapse, or transplant-related mortality [TRM]). Genes are displayed in rows and sorted by diagnostic panel groupings, including 10-gene (prognostic in AML), DTA (associated with age-related clonal hematopoiesis), and 16-gene (commonly mutated in MDS). The presence of a mutation within a gene is denoted in the heatmap, with the color corresponding to the highest AF within each gene per patient. VAF, variant allele frequency; AML, acute myeloid leukemia; MAC, myeloablative conditioning; MDS, myelodysplastic syndrome; RIC, reduced intensity conditioning
Inclusion of 16 MDS-associated genes in the testing strategy not only resulted in reclassification of 43% (n = 12) of NGSg10-negative patients to positive but also provided no additional prognostic significance (Fig. 3, Data Supplement). No relapses were observed in patients testing positive only for these 16 genes regardless of conditioning intensity, and only one death was observed (TRM in the MAC group). Additionally, although inclusion of these additional gene regions provided no additional sensitivity for predicting relapse at 24 months, as with DTA variants, it also reduced specificity (Data Supplement).
Variant Allele Frequency and Mutation Number Do Not Provide Prognostic Significance
Since expanding testing to include all 29 gene regions resulted in the majority of patients having a mutation detected, we examined the impact of defining as positive only those patients with at least one mutation detectable at a higher level (VAF ≥ 2.5%). This resulted in reclassification of seven NGSg10-negative and five NGSg10-positive patients, resulting in 46% of patients (n = 22) being high VAF NGS-positive. However, unlike NGSg10-positive patients, no significant difference was observed for rates of relapse, RFS, or OS in high VAF NGS-positive versus NGS-negative patients (Data Supplement). Further stratification of patients by conditioning intensity revealed that 48% of MAC (n = 12) and 43% of RIC (n = 10) patients were high VAF NGS-positive. Higher rates of relapse were observed in high VAF NGS-positive patients randomly assigned to RIC versus MAC (3-year relapse, 60% v 8%; P = .010), but this effect was driven exclusively by NGSg10 mutation status, whereas no difference was observed in TRM, RFS, or OS (Data Supplement). Limiting analysis to those testing positive for an NGSg10 mutation at or above 2.5% VAF reduced the sensitivity at 24 months from 73% to 45% (Data Supplement). Similarly, we examined the impact of mutation load on patient outcomes and found no significant difference in OS based on the mutation number (Data Supplement) or number of mutated genes (Data Supplement).
Patients With MDS With Excess Blasts
Although the requirement for inclusion in the BMT-CTN 0901 study was < 5% marrow myeloblasts and no leukemic myeloblasts in blood on morphological analysis at the time of pretransplant assessment, a total of 22 of the 54 patients with MDS (41%) treated on the BMT-CTN 0901 trial had an initial diagnosis of refractory anemia with excess blasts (RAEB) I or II (ie, between 5% and 19% blasts). The median time from diagnosis to alloHCT in these patients was 6 months (range, 3-129 months). These patients accounted for 19 (40%) of the available blood samples from before conditioning (nine MAC and 10 RIC). Survival for the 19 RAEB patients with samples available was 74%. Seven of these samples tested positive for NGS10g, for which survival was 0% for RIC (n = 2, relapse) and 60% for MAC-treated patients (with one death from each relapse and transplant-related mortality). NGS10g-positive patients accounted for four of six relapses observed in the RAEB cohort, and the other two patients had high-risk cytogenetics with a monosomal karyotype that would not be detectable using this NGS assay.
DISCUSSION
Here, we present the impact of genetic mutations detected before random assignment to RIC or MAC, identified without knowledge of the mutations originally present at diagnosis, on transplant outcomes in patients with MDS. Detection of mutations using a DNA-sequencing panel covering regions of 10 genes, previously shown to have utility at the same timepoint in patients with AML,10 was associated with increased rates of relapse and decreased RFS and OS. Many of these genes (including TP53, RUNX1, JAK2, and RAS pathway genes) have shown prognostic significance in other MDS studies7-9 or are indicative of potential leukemic progression (FLT3 and NPM1). However, it should be noted that mutations in IDH1, IDH2, JAK2, and NRAS were not seen in relapsing patients in this cohort. The prognostic significance of the remaining mutations on relapse was dependent on conditioning intensity, with higher rates of relapse and lower RFS observed in patients randomly assigned to receive RIC compared with MAC.
This study provides evidence that the benefit of MAC versus RIC in reducing relapse is found in patients with MDS with detectable mutations using a 10-gene region DNA-sequencing panel before alloHCT. In 58% of patients with MDS testing negative, no difference between conditioning arms was seen for relapse, RFS, or OS. It was already well-established that detection of mutations in TP53 before alloHCT in patients with MDS was associated with increased relapse and decreased survival7,9 and that MAC was unable to overcome these risks.9 In our series, 12 of 48 subjects had a TP53 mutation, although nine (75%) of these were below the cutoff (VAF < 2.5%) used in previous studies (mean VAF 1.3%, range, 0.1%-19.74%). Four of these subjects relapsed (one of eight in MAC group and three of four in RIC group), five died of transplant-related complications (four of eight in MAC group and one of four in RIC group), and three survived without relapse (all in the MAC group). All three TP53-mutated survivors who did not relapse had a pre-alloHCT VAF of < 5% (Fig. 3). Although the size of this cohort limits extrapolation, it is possible that the prognostic implications of TP53 mutation detection in patients with MDS before alloHCT and the impact of conditioning intensity differ from those reported previously when those variants are found at a level below the limit of detection of routinely used NGS assays. In addition to VAF, other factors such as persistence since initial diagnosis, development during therapy, mutation type, functional classification, and zygosity are likely to be important for determining the relapse risk associated with detecting a TP53 mutation.15-22
Although mutation detection by targeted DNA sequencing before alloHCT was strongly associated with increased relapse and decreased RFS and these outcomes could be improved with MAC, unlike in patients10 with AML, we did not detect an OS advantage for conditioning intensification in those testing positive. This might have been due to limited sample size, predominately nonrelapse causes of death in this MDS cohort (67% of deaths, compared with 39% in the previously reported AML cohort), or other factors.
MDS is a genetically heterogenous disease.1,11,12,23 As was observed previously,8 we found that inclusion of a large number of genes resulted in the majority of patients having mutations present before transplant. Furthermore, mutations in genes associated with clonal hematopoiesis (DTA) or 16 other MDS-associated genes outside of 10-gene mutational status did not improve test performance. Reanalysis of pretransplant mutational status using only the 10 genes reported here, from another study of patients with MDS with < 5% myeloblasts before alloHCT,8 confirms our finding of significantly increased relapse rates in patients testing positive and receiving RIC versus MAC. Analysis of larger MDS cohorts using broad NGS panels will have to be performed to define the mutations with greatest prognostic significance at the pre-alloHCT timepoint.
There are several limitations to this work. The modest sample size here offers only a partial sampling of the heterogenous genetics associated with MDS, whereas differences in pretransplantation disease burden, biology, and prior treatment history further limit exact comparisons. The significance of mutation detection before alloHCT might be influenced by history of prior treatment, and previous work using the same testing in patients with AML included only those treated to cytomorphological remission before transplant. Without knowledge of genomics at initial MDS diagnosis, we cannot comment on the optimal clinical timepoint or sequencing breadth for testing. Without single-cell sequencing, we cannot determine clonal architecture to distinguish if detected mutations are present in clonal hematopoiesis rather than MDS clones.24 Nevertheless, we show here a clear association of pretransplant genomics with post-transplant relapse and a significant impact of conditioning intensity in reducing that relapse risk.
Unlike in AML, relapse does not account for the clear majority of mortality in patients with MDS undergoing alloHCT.9 Two randomized studies of conditioning intensity in MDS have failed to show a difference in survival between MAC and RIC,4,6,25 although we show here a trend toward improved survival with increased conditioning intensity for those with detectable mutations particularly in the RAEB group (ie, biology and treatment history most analogous to AML). Prior work had shown a benefit for increased conditioning intensity in patients with MDS with RAS pathway mutations (NRAS, KRAS, PTPN11, CBL, NF1, RIT1, FLT3, and KIT) undergoing alloHCT.9 As relapse is not the primary cause of death in patients with MDS undergoing alloHCT, the choice of conditioning intensity is currently dependent on the estimated ability of a patient to tolerate transplant-related toxicity.26 AlloHCT remains the only curative therapy for MDS; however, given the median age at diagnosis, many patients will not be eligible for myeloablative approaches.27,28 As it is now possible to determine the genomic basis of disease before and after transplantation, personalized approaches for alloHCT of patients with MDS are now conceivable with targeted strategies for those with detectable TP53, RAS pathway (including FLT3), IDH and JAK2 mutations, immune augmentation, or other therapies potentially able to supplement the impact of chosen conditioning intensity to minimize relapse risk.29-31
In conclusion, we show that in adult patients with MDS with < 5% marrow myeloblasts and no leukemic myeloblasts in blood before alloHCT, ultra-deep DNA sequencing for mutations in 10 gene regions previously shown to be high risk in patients with AML could identify a subset of 42% of patients who experienced the majority of post-transplant relapses. In those patients with MDS testing positive, MAC rather than RIC could dramatically lower the relapse rate, but this benefit was counterbalanced by increased transplant-related mortality. This study provides the rationale for clinical trials of personalized post-transplant maintenance for patients with MDS based on genetic assessment before transplant.
ACKNOWLEDGMENT
The authors would like to thank R. Coleman Lindsley, MD, PhD, for careful reading of this work.
EQUAL CONTRIBUTION
L.W.D. and G.G. contributed equally.
SUPPORT
Supported by the Intramural Research Program of the National Heart, Lung, and Blood Institute (NHLBI) of the National Institutes of Health (NIH) and by grants U10HL069294 and U24HL138660 to the Blood and Marrow Transplant Clinical Trials Network (BMT CTN) from the NHLBI and the National Cancer Institute (NCI). The CIBMTR registry is supported primarily by the U24-CA76518 from NHLBI, NCI, and the National Institute of Allergy and Infectious Diseases and from HHSH234200637015C (HRSA/DHHS) to the Center for International Blood and Marrow Transplant Research. This study utilized BMT CTN 0901 research materials, biospecimens, and clinical trial data, provided by the BMT CTN. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. This work utilized the NHLBI Sequencing and Genomics Core and the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov).
AUTHOR CONTRIBUTIONS
Conception and design: Laura W. Dillon, Gege Gui, Brent R. Logan, Sergio Giralt, Mehdi Hamadani, Bart L. Scott, H. Joachim Deeg, Christopher S. Hourigan
Administrative support: Mingwei Fei
Provision of study materials or patients: Mingwei Fei, Asad Bashey, Alan Howard, Richard T. Maziarz, David L. Porter, Mitchell E. Horwitz, H. Joachim Deeg
Collection and assembly of data: Laura W. Dillon, Gege Gui, Mingwei Fei, Jack Ghannam, Yuesheng Li, Asad Bashey, Hugo F. Fernandez, Sergio Giralt, Mehdi Hamadani, Alan Howard, Marcelo C. Pasquini, Bart L. Scott, Mitchell E. Horwitz, H. Joachim Deeg, Christopher S. Hourigan
Data analysis and interpretation: Laura W. Dillon, Gege Gui, Brent R. Logan, Mingwei Fei, Jack Ghannam, Abel Licon, Edwin P. Alyea, Asad Bashey, Steven M. Devine, Sergio Giralt, Mehdi Hamadani, Alan Howard, Richard T. Maziarz, David L. Porter, Erica D. Warlick, Marcelo C. Pasquini, Bart L. Scott, Christopher S. Hourigan
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO’s conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
No potential conflicts of interest were reported.
REFERENCES
- 1.Malcovati L, Hellstrom-Lindberg E, Bowen D, et al. Diagnosis and treatment of primary myelodysplastic syndromes in adults: Recommendations from the European LeukemiaNet Blood 1222943–29642013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luger SM, Ringden O, Zhang MJ, et al. Similar outcomes using myeloablative vs reduced-intensity allogeneic transplant preparative regimens for AML or MDS Bone Marrow Transpl 47203–2112012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shimoni A, Hardan I, Shem-Tov N, et al. Allogeneic hematopoietic stem-cell transplantation in AML and MDS using myeloablative versus reduced-intensity conditioning: The role of dose intensity Leukemia 20322–3282006 [DOI] [PubMed] [Google Scholar]
- 4.Kroger N, Iacobelli S, Franke GN, et al. Dose-reduced versus standard conditioning followed by allogeneic stem-cell transplantation for patients with myelodysplastic syndrome: A prospective randomized phase III study of the EBMT (RICMAC trial) J Clin Oncol 352157–21642017 [DOI] [PubMed] [Google Scholar]
- 5.Pulsipher MA.Reduced intensity for myelodysplastic syndrome: Worth the gamble? J Clin Oncol 352106–21082017 [DOI] [PubMed] [Google Scholar]
- 6.Scott BL, Pasquini MC, Logan BR, et al. Myeloablative versus reduced-intensity hematopoietic cell transplantation for acute myeloid leukemia and myelodysplastic syndromes J Clin Oncol 351154–11612017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bejar R, Stevenson KE, Caughey B, et al. Somatic mutations predict poor outcome in patients with myelodysplastic syndrome after hematopoietic stem-cell transplantation J Clin Oncol 322691–26982014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duncavage EJ, Jacoby MA, Chang GS, et al. Mutation clearance after transplantation for myelodysplastic syndrome N Engl J Med 3791028–10412018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindsley RC, Saber W, Mar BG, et al. Prognostic mutations in myelodysplastic syndrome after stem-cell transplantation N Engl J Med 376536–5472017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hourigan CS, Dillon LW, Gui G, et al. Impact of conditioning intensity of allogeneic transplantation for acute myeloid leukemia with genomic evidence of residual disease. J Clin Oncol. 2019:JCO1903011. doi: 10.1200/JCO.19.03011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haferlach T, Nagata Y, Grossmann V, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes Leukemia 28241–2472014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Papaemmanuil E, Gerstung M, Malcovati L, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes Blood 1223616–36272013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Genovese G, Kahler AK, Handsaker RE, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence N Engl J Med 3712477–24872014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaiswal S, Fontanillas P, Flannick J, et al. Age-related clonal hematopoiesis associated with adverse outcomes N Engl J Med 3712488–24982014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Montalban-Bravo G, Kanagal-Shamanna R, Benton CB, et al. Genomic context and TP53 allele frequency define clinical outcomes in TP53-mutated myelodysplastic syndromes Blood Adv 4482–4952020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sallman DA, Komrokji R, Vaupel C, et al. Impact of TP53 mutation variant allele frequency on phenotype and outcomes in myelodysplastic syndromes Leukemia 30666–6732016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coombs CC, Zehir A, Devlin SM, et al. Therapy-related clonal hematopoiesis in patients with non-hematologic cancers is common and associated with adverse clinical outcomes Cell Stem Cell 21374–382.e4, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong TN, Miller CA, Jotte MRM, et al. Cellular stressors contribute to the expansion of hematopoietic clones of varying leukemic potential. Nat Commun. 2018;9:455. doi: 10.1038/s41467-018-02858-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dutta S, Pregartner G, Rucker FG, et al. Functional classification of TP53 mutations in acute myeloid leukemia. Cancers (Basel) 2020;12:637. doi: 10.3390/cancers12030637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prochazka KT, Pregartner G, Rucker FG, et al. Clinical implications of subclonal TP53 mutations in acute myeloid leukemia Haematologica 104516–5232019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boettcher S, Miller PG, Sharma R, et al. A dominant-negative effect drives selection of TP53 missense mutations in myeloid malignancies Science 365599–6042019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giacomelli AO, Yang X, Lintner RE, et al. Mutational processes shape the landscape of TP53 mutations in human cancer Nat Genet 501381–13872018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bejar R, Stevenson K, Abdel-Wahab O, et al. Clinical effect of point mutations in myelodysplastic syndromes N Engl J Med 3642496–25062011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dillon LW, Ghannam J, Nosiri C, et al. Personalized single-cell proteogenomics to distinguish acute myeloid leukemia from non-malignant clonal hematopoiesis. medRxiv. 2020;2020 doi: 10.1158/2643-3230.BCD-21-0046. doi: 10.1101/2020.10.22.20216028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hourigan CS, McCarthy P, de Lima M.Back to the future! The evolving role of maintenance therapy after hematopoietic stem cell transplantation Biol Blood Marrow Transpl 20154–1632014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koenig K, Mims A, Levis MJ, et al. The changing landscape of treatment in acute myeloid leukemia Am Soc Clin Oncol Educ Book 401–122020 [DOI] [PubMed] [Google Scholar]
- 27.Al-Kali A, Zblewski D, Foran JM, et al. Outcome of myelodysplastic syndromes over time in the United States: A National Cancer Data Base Study From 2004-2013 Mayo Clin Proc 941467–14742019 [DOI] [PubMed] [Google Scholar]
- 28.Sorror ML, Sandmaier BM, Storer BE, et al. Long-term outcomes among older patients following nonmyeloablative conditioning and allogeneic hematopoietic cell transplantation for advanced hematologic malignancies JAMA 3061874–18832011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Appelbaum FR. Maintenance therapy after allogeneic hematopoietic cell transplantation for acute myeloid leukemia. Best Pract Res Clin Haematol. 2019;32:101109. doi: 10.1016/j.beha.2019.101109. [DOI] [PubMed] [Google Scholar]
- 30.Craddock C.Conditioning intensity in HCT for AML: The jury is still out Lancet Haematol 5e132–e1332018 [DOI] [PubMed] [Google Scholar]
- 31.Maslah N, Salomao N, Drevon L, et al. Synergistic effects of PRIMA-1Met (APR-246) and azacitidine in TP53-mutated myelodysplastic syndromes and acute myeloid leukemia Haematologica 1051539–15512020 [DOI] [PMC free article] [PubMed] [Google Scholar]