Abstract
Purpose
The purpose of this study was to investigate the relationship between microRNA-29b-3p (miR-29b-3p) and myc-associated zinc finger protein (MAZ) expression and the effects of this interaction on the proliferation, migration, and invasion of gastric cancer cells.
Methods
qPCR and Western blots were used to detect the expression of miR-29b-3p and MAZ. The dual luciferase reporter gene system was used to explore whether MAZ is the target of miR-29b-3p. Cell function experiments and a mouse tumorigenesis model were used to determine the effects of miR-29b-3p overexpression and MAZ depletion on proliferation, migration, and invasion in gastric cancer cell lines and on tumor growth.
Results
The expression level of miR-29b-3p was low and the expression level of MAZ was high in gastric cancer cells compared with normal human gastric mucosal epithelial cells. MAZ was the target gene of miR-29b-3p. The upregulation of miR-29b-3p reduces the expression of MAZ. Overexpression of miR-29b-3p and downregulation of MAZ inhibited the proliferation and migration of cancer cells and induced apoptosis by controlling the expression of autophagy-related proteins. MiR-29b-3p mimics inhibit tumor growth in mice.
Conclusion
MiR-29b-3p inhibits the migration and invasion of gastric cancer cells by regulating the autophagy-related protein MAZ.
Keywords: gastric cancer, microRNA, autophagy, migration, invasion
Introduction
Gastric cancer is one of the most common malignant tumors of the digestive tract and seriously threatens human health.1 Although great progress has been made in clinical research and treatment of gastric cancer in the past decade, highly sensitive, and specific tumor biomarkers are still lacking in clinical practice. Biomarkers are particularly important because gastric cancer is the second deadliest cancer in the world.2 Tumor recurrence and metastasis contribute to the high mortality of gastric cancer and are closely related to abnormal gene expression, proliferation, migration, and invasion of cancer cells.3
MicroRNAs are highly conserved endogenous noncoding regulatory RNAs of 17–25 nucleotides. MicroRNAs play an important regulatory role in the development of cancer by inhibiting the translation of RNA and the expression of target genes.4,5 Each miRNA can control hundreds of gene targets and participate in multiple gene signaling pathways.6 Therefore, an miRNA can control a series of biological functions, such as cell proliferation, migration, invasion, and apoptosis. An in-depth understanding of abnormal miRNA expression may be of great significance for early diagnosis, prediction, and treatment of gastric cancer.
The microRNA-29 family has three members, including microRNA-29a, microRNA-29b, and microRNA-29c, which play important roles in inhibiting tumors. The miR-29 family exerts tumor suppressor effects by inhibiting the proliferation and migration of cancer cells via downregulation of oncogenes and/or upregulation of tumor suppressor genes. As diagnostic markers for tumors, miR-29 family members have good sensitivity and specificity, high diagnostic value, and good recognition ability.7 The expression trends of all the miRNA-29 family members are similar. We selected miRNA-29b as the research subject of this study.
Myc-associated zinc finger (SAF/MAZ) (GenBank MIM: 600,999) is a CysHis2-type transcription factor widely expressed in different tissues in humans.8 The MAZ gene is located on chromosome 16p11.2 and is transcribed as an mRNA of 2.7 kilobases (kb), which encodes for the 60-kDa MAZ protein.9 MAZ is a growth inhibitor protein in normal cells, which affects the levels of cell cycle regulatory proteins.10 MAZ protein is a transcription factor that plays a dual regulatory role in the process of transcription; MAZ initiates the transcription of certain genes and terminates transcription of target genes.11 Abnormal expression of the MAZ gene is closely related to the occurrence and development of tumors and MAZ has abnormally high expression in many malignant tumors, such as prostate cancer,12 breast cancer,13 pancreatic cancer,14 colon cancer,15 liver cancer,16 and gastric cancer.17 In highly metastatic cancer tissues, MAZ levels are elevated and the number of autophagic vesicles is significantly reduced. MAZ mainly activates the mTORC1 signaling pathway and the AKT signaling pathway negatively regulates MAZ. MAZ binds to the c-Myc promoter region to regulate the transcription of genes related to cancer development that inhibit cell autophagy and, thus, promote the migration and invasion of cancer cells.
In this study, the expression of miR-29b-3p and MAZ in human gastric mucosal epithelial cells (GES1) and gastric cancer cells was detected by qPCR. GES1 can be stably passaged in vitro for a long time after being infected with the SV40 virus for 3 weeks. In addition to the transformation characteristics, GES-1 human gastric mucosal cells retain the normal cytoskeleton structure and were non-tumorigenic in nude mice for 6 months. Thus, this cell line provides an important model system for further study of gastric epithelial cell carcinogenesis. MAZ is the target gene for miR-29b-3p, as determined by qPCR, Western blot, and dual luciferase reporter assays. The effects of miR-29b-3p and MAZ on the proliferation, migration, and invasion of gastric cancer cells were studied by cell transfection and tumor formation in nude mice. In addition, the possible mechanism of action of miR-29b-3p in gastric cancer cells was investigated.
Materials and Methods
Cell Culture
After rapid thawing, AGS and MGC803 cells (Beina Biotechnology Co., Ltd.) were centrifuged at 1000 rpm/min for 5 min. The supernatant was discarded and the cells were resuspended in 2 mL of complete medium. Then, the cells were inoculated in flasks containing 8 mL medium and cultured in 37°C and 5% CO2 incubators. The cell status was observed every other day. Cells were passaged when the cell density reached approximately 90%.
Cell Transfection
Cells in the logarithmic growth phase were inoculated into 6-well plates with 3 x 105 pores. When the cell density reached approximately 80%, cells were transfected using Lipofectamine 2000 (Corning) following the manufacturer’s instructions. The transfected cells were grown in a 37°C, 5% CO2 incubator for 24 h for further use.
RNA Extraction and Real-Time PCR
Cells were washed with precooled PBS three times. Then, total RNA and microRNA were extracted according to the instructions of the RNA and microRNA extraction kits (Invitrogen Co., Ltd.). Following the instructions of the reverse transcription kit and SYBR Green Q PCR kit (Promega Co., USA), the related genes were detected with a Veriti 96-well thermal cycler PCR. GAPDH was used as an internal control. The PCR conditions were as follows: initial denaturation at 92°C for 5 min, denaturation at 95°C for 10 s, annealing at 60°C for 15 s, and extension at 72°C for 40 s, for 40 cycles. The relative quantitative method was used to obtain the relative expression of the target gene. The experiment was repeated three times and the average value was obtained. Primers are shown in Table 1.
Table 1.
Primer Sequences of the Genes for Real-Time PCR
| Gene | Sequence | |
|---|---|---|
| GAPDH | Forward (5ʹ-3ʹ) | ACACCCACTCCTCCACCTTT |
| Reverse (5ʹ-3ʹ) | TTACTCCTTGGAGGCCATGT | |
| U6 | Forward (5ʹ-3ʹ) | CGCTTCGGCAGCACATATAC |
| Reverse (5ʹ-3ʹ) | TTCACGAATTTGCGTGTCAT | |
| MAZ | Forward (5ʹ-3ʹ) | TGCACAAGCCCTACAACTGCTC |
| Reverse (5ʹ-3ʹ) | GCACTTGTCTGACGTGACTGTTGA | |
| miR-29b-3p | Forward (5ʹ-3ʹ) | ACACTCCAGCTGGGTAGCA |
| Reverse (5ʹ-3ʹ) | TGGTGTCGTGGAGTCG | |
| LC3 | Forward (5ʹ-3ʹ) | CATGCCGTCCGAGAAGACCT |
| Reverse (5ʹ-3ʹ) | GATGAGCCGGACATCTTCCACT | |
| beclin 1 | Forward (5ʹ-3ʹ) | GAAACTGGACACGAGCTTCAAGA |
| Reverse (5ʹ-3ʹ) | ACCATCCTGGCGAGTTTCAATA | |
| P62 | Forward (5ʹ-3ʹ) | TCGCTATGGCGTCGCTC |
| Reverse (5ʹ-3ʹ) | CACCCGAAGTGTCCGTGTTT | |
MTT Assay
Cellular metabolic activity was measured using an MTT assay. Cell suspensions (100 μL) were added to each well. After incubation for 24 h at 37°C and 5% CO2, 20 μL of MTT solution was added to each well and the incubation was continued for 4 h. After 150 μL of dimethyl sulfoxide was added to each well and the plate was shaken for 10 min to fully dissolve the crystals, the absorbance of each well was measured with an enzyme-linked immunosorbent detector at OD490 nm. Replicates were performed for each experimental condition.
Transwell Assay
Cell invasion was detected using a Transwell chamber with a hydrated basement membrane coated with Matrigel and cell migration was detected using a Transwell chamber without Matrigel coating. Cell suspensions (100 μL) were added to each Transwell chamber and 600 μL RPMI culture medium containing 10% FBS was added into the lower compartment. Each experimental group was divided into triplicate wells. After 48 h of culture, the cells on the small chamber membrane were wiped off with a cotton swab, fixed with 4% polyformaldehyde, and stained with 1% crystal violet. After three washes with PBS, the cells were photographed and counted under a microscope.
Colony-Forming Assay
Using cells in the logarithmic growth phase, 100 cells/well were inoculated in 6-well plates at 37°C and cultured in a 5% CO2 incubator for 6 h. The cells were divided into three groups; one group was a blank control group without any treatment. The other two groups were transfected with mimics-NC (negative control) or miR-29b-3p mimics. The miR-29b-3p mimics and control were purchased from Shanghai Jima Pharmaceutical Technology Ltd. The plates were incubated at 37°C and 5% CO2 for 2–3 weeks. The culture was terminated when colonies were visible to the naked eye. The cells were washed twice with PBS and allowed to dry naturally. The cells were fixed with 4% methanol at room temperature for 15 min and were allowed to dry naturally after the methanol was discarded. The cells were stained with Giemsa dye solution (Sigma) for 10 min, washed slowly with running water, and air-dried.
Dual Luciferase Reporter Assay
The 293T cells (Beina Biotechnology Co., Ltd.) were cultured in 12-well plates and divided into three groups with three wells per group. The 293T cells can be transfected with high expression levels, which can exclude differences in transfection efficiency, and they rarely express endogenous receptors required by extracellular ligands, so they are commonly used to express exogenous genes. TargetScan was used to predict the binding site of miR-29b-3p in wild-type MAZ and the complementary binding site was mutated. The MAZ 3ʹ-UTR sequence or the 3ʹ-UTR mutated sequence was inserted into the pGL3 plasmid. The recombinant and control plasmids (Mimics-NC and mimics-29b-3p) were transfected into 293T cells using Lipofectamine 2000. After 24 h, the plasmids were detected according to the instructions of the dual luciferase reporter gene detection kit (Promega Ltd, USA).
Western Blot Analysis
Cells were washed once with precooled PBS. Then, the cells were lysed with RIPA cell lysis buffer on ice, scraped with a cell scraper, collected in a microcentrifuge tube, and boiled at 100°C for 10 min to extract the total protein. For protein assays, the protein content of each group of cells was determined using a BCA kit (Biyuntian Biotechnology Research Institute of Shanghai). The protein lysates (40 μg) were subjected to Western blot analysis. Blots were imaged with LabWorks gel imaging and analysis system. Using GAPDH as an internal reference, ImageJ software was used to perform grayscale analysis and analyze the relative expression of each protein.
Animal Experiment
Five-week-old female BALB/c-nu nude mice (Beijing Weitonglihua Laboratory Animal Technology Ltd.) were divided into two groups for tumorigenesis experiments. Five mice in each group were injected subcutaneously at the left axilla with an AGS cell suspension transfected with the miR-29b-3p mimics or mimics-NC. Starting with the day the tumor was visible to the naked eye, the weight of the mouse and the tumor long axis diameter a and the short diameter b were determined every 3 days. The tumor volume was calculated using the formula: Volume (mm3) = a * b2/2. After five weeks, the mice were sacrificed and the weight and volume of the tumors were measured. All procedures were performed in accordance with national (D.L.n.26, March 4th, 2014) and international laws and policies (directive 2010/63/EU).
Statistical Analysis
The experimental data were analyzed using GraphPad Prism 5 software. Experimental data are expressed as means ± standard deviations. Statistical differences between the groups were compared using t-tests. Statistical differences were denoted as * for P < 0.05, ** for P < 0.01, or *** for P < 0.001.
Results
Expression of miR-29b-3p and MAZ by qPCR and Identification of Target Genes
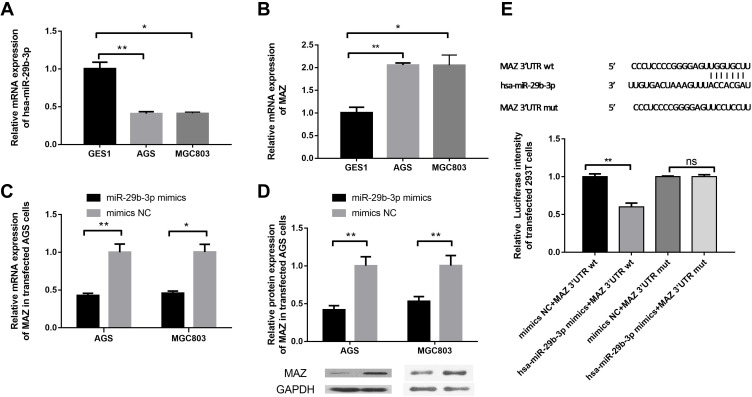
We evaluated the expression profile of miR-29b-3p in AGS and MGC803 cells using qRT-PCR. MiR-29b-3p is aberrantly and significantly downregulated by about 60% in AGS and MGC803 cells compared with GES1 cells (Figure 1A). The expression of MAZ was about twice as high in AGS and MGC803 cells compared with GES1 cells (Figure 1B). The mRNA expression level of MAZ in cells transfected with the miR-29b-3p mimics was 60% lower than that of the blank control (Figure 1C). The expression levels of MAZ protein in AGS and MGC cells transfected with the miR-29b-3p mimics were 0.42 and 0.53 of the blank control group, respectively, and the results were statistically significant (Figure 1D). In cells transfected with the wild-type MAZ gene 3ʹUTR plasmid, the activity of the miR-29b-3p mimics significantly decreased by 40% and 54% compared with the blank group (Figure 1E). When the mutant MAZ gene 3ʹUTR plasmid was transfected, there were no significant differences between the activity of the miR-29b-3p mimics and that of the blank group. These results indicate that miR-29b-3p binds to the 3ʹUTR of the MAZ gene and regulates the expression of MAZ. MiR-29b-3p did not affect expression when binding to the MAZ mutant, that is to say the regulatory relationship between miR-29b-3p and MAZ was no longer observed. These results indicate that the predicted binding sites are correct and MAZ is a target gene of mir-29b-3p.
Figure 1.
Detection of the expression of miR-29b-3p and MAZ and identification of target genes. (A) The mRNA expression of miR-29b-3p in cancer cell lines and AGS cells. (B) The mRNA expression of MAZ in cancer cell lines and AGS cells. (C) The mRNA expression of MAZ in cells transfected with the miR-29b-3p mimics. (D) The protein expression of MAZ in cells transfected with the miR-29b-3p mimics. (E) Dual luciferase reporter verification of the target gene. *P < 0.05, **P < 0.01.
Abbreviation: ns, no significant difference.
miR-29b-3p Overexpression Inhibits Cell Migration and Invasion
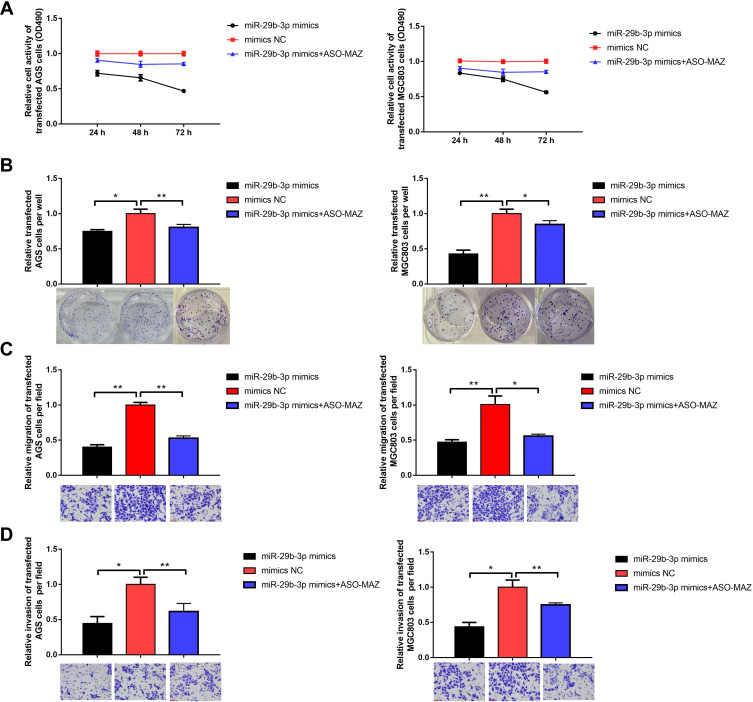
AGS and MGC cells were transfected with the miR-29b-3p mimics to overexpress miR-29b-3p. Then, MTT, colony-forming, and Transwell assays were conducted to study the effects of miR-29b-3p on metabolic activity, proliferation, migration, and invasion. The results of the MTT assay showed that 72 hours after the transfection of AGS and MGC cells with the miR-29b-3p mimics, cell survival rate decreased significantly to 0.47 and 0.56, respectively, compared with the blank control group. And up-regulation of MAZ after over-expression of miR-29b-3p in the two cell lines increased cell survival compared to miR-29b-3p overexpression, suggesting that the effect of miR-29b-3p overexpression is offset by up-regulation of the direct target gene MAZ. (Figure 2A). The colony formation assay showed that transfection of AGS cells and MGC cells with the miR-29b-3p mimics reduced the number of colonies to 0.74 and 0.43 of the blank control group, respectively (Figure 2B). The Transwell assay showed that miR-29b-3p overexpression reduced the migration (Figure 2C) and invasion (Figure 2D) of transfected cells to almost 45% of the blank control groups per field. And these effects were attenuated by up-regulation of MAZ after overexpression of miR-29b-3p. Overall, these results were statistically significant; high expression of miR-29b-3p reduced the proliferative ability, colony-forming capability, and migration and invasive potential of gastric cancer cells by targeting MAZ.
Figure 2.
Effects of overexpression of miR-29b-3p on cellular function. (A) MTT assay detected cell viability. (B) Colony forming assay. (C) Transwell assay detected cell migration. (D) Transwell assay detected cell invasion. *P < 0.05, **P < 0.01.
MAZ Interference Inhibits Cell Migration and Invasion
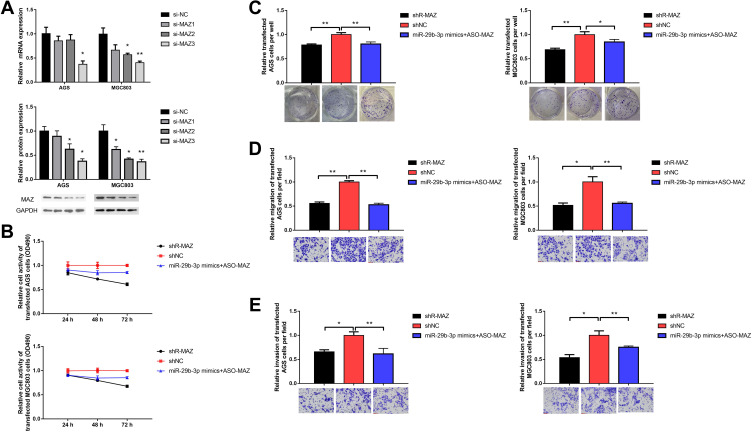
We designed three short hairpin RNAs (shRNA) to construct plasmid vectors; the interference sequences are shown in Table 2. One shRNA had a MAZ knockout efficiency below 50% steadily (Figure 3A). AGS and MGC cells were transfected with the MAZ shRNA to block MAZ expression. The MTT assay showed that cell survival rate decreased to 0.61 and 0.67 compared with the blank control group after AGS and MGC cells were transfected with MAZ shRNA (Figure 3B). And up-regulation of MAZ after over-expression of miR-29b-3p in the two cell lines increased cell survival compared to MAZ interference. The inhibitory effects of MAZ depletion on the proliferation of transfected cells were further verified with a colony formation assay; MAZ depletion reduced colonies in AGS and MGC cells to 0.79 and 0.69 of the blank control groups (Figure 3C). MAZ depletion reduced the migration (Figure 3D) and invasion (Figure 3E) of transfected cells to 45% and 40% less than blank control groups. Overall, these results indicated that MAZ depletion reduced the proliferative ability, colony-forming capability, and migration and invasive potential of AGS and MGC cells.
Table 2.
The Interference Sequences of the shRNAs for MAZ Knocking Out
| Gene | Sequence | |
|---|---|---|
| Si-NC | Forward (5ʹ-3ʹ) | UUCUCCGAACGUGUCACGUTT |
| Reverse (5ʹ-3ʹ) | ACGUGACACGUUCGGAGAATT | |
| Si-MAZ1 | Forward (5ʹ-3ʹ) | GAACAUCAUUAGACUCUAAGA |
| Reverse (5ʹ-3ʹ) | UUAGAGUCUAAUGAUGUUCAU | |
| Si-MAZ2 | Forward (5ʹ-3ʹ) | CAACAGUCACGUCAGACAAGU |
| Reverse (5ʹ-3ʹ) | UUGUCUGACGUGACUGUUGAG | |
| Si-MAZ3 | Forward (5ʹ-3ʹ) | GAGAAGAGAUGGAGUCUUAGG |
| Reverse (5ʹ-3ʹ) | UAAGACUCCAUCUCUUCUCUG | |
Figure 3.
Effects of blocking MAZ expression on cellular function. (A) MAZ knockout efficiency of siRNA. (B) MTT assay detected cell viability. (C) Colony forming assay. (D) Transwell assay detected cell migration. (E) Transwell assay detected cell invasion. *P < 0.05, **P < 0.01.
miR-29b-3p Overexpression and MAZ Depletion Can Regulate Autophagy
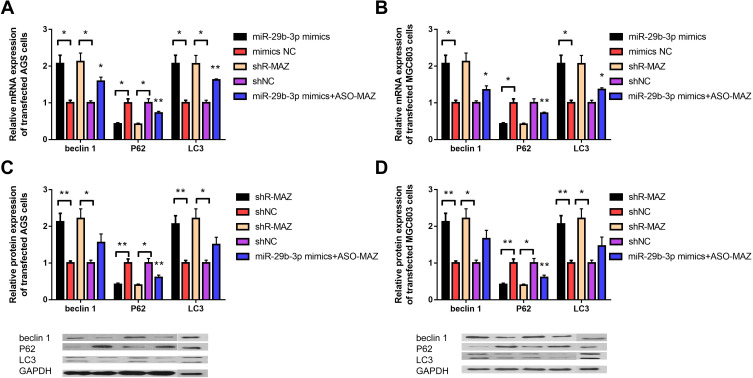
To further explore the molecular basis of miR-29b-3p and MAZ effects on cell proliferation and invasion, we detected the regulatory effects of miR-29b-3p overexpression, MAZ depletion and up-regulation of MAZ after over-expression of miR-29b-3p on autophagy-related genes. The mRNA and protein expressions of the autophagy-related genes, beclin 1, P62, and LC3, were measured in gastric cancer cells transfected with the miR-29b-3p mimics or with MAZ shRNA. The results showed that the mRNA expression levels of beclin 1 and LC3 in both AGS and MGC cells were about twice as high as those of the blank control group, and the expression level of P62 was about 68% lower than the blank control group (Figure 4A and B). The protein expression levels of these autophagy-related genes were similar to the mRNA levels; beclin 1 and LC3 protein levels were about two times that of the control groups. The expression level of P62 was 36% of the control group (Figure 4C and D). MiR-29b-3p overexpression and MAZ interference can both regulate autophagy, but the regulatory relationship is no longer obvious when up-regulation of MAZ after overexpression of MiR-29b-3p in the two cell lines, proving that miR-29b-3p regulate autophagy by MAZ. The results were statistically significant, indicating that both high expression of miR-29b-3p and depletion of MAZ contribute to the regulation of cell autophagy in AGS and MGC cells by regulating the expression of the autophagy-related genes, beclin 1, P62, and LC3, thereby inhibiting cancer cell migration and invasion.
Figure 4.
Expression levels of autophagy-related genes beclin 1, P62, and LC3 in transfected cells. (A) The mRNA expression level of autophagy-related genes in AGS cells transfected with the miR-29b-3p mimics and MAZ interference plasmid. (B) The mRNA expression level of autophagy-related genes in MGC cells transfected with the miR-29b-3p mimics and MAZ interference plasmid. (C) The protein expression level of autophagy-related genes in transfected AGS cells. (D) The protein expression level of autophagy-related genes in transfected MGC cells. *P < 0.05, **P < 0.01.
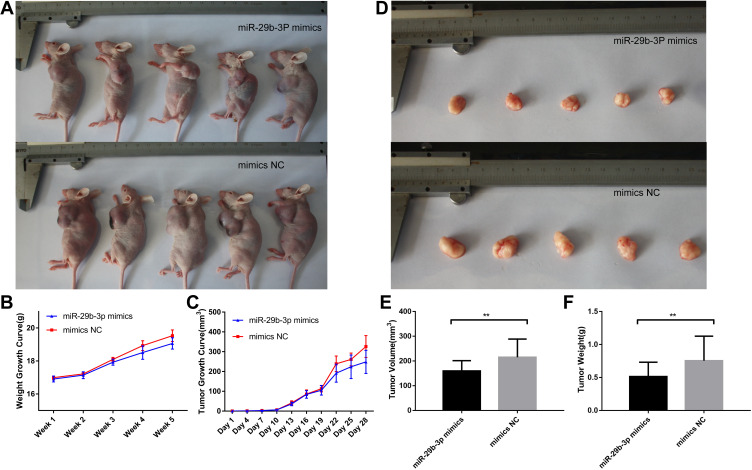
Injection of miR-29b-3p Mimics Inhibits Tumor Growth in Mice
The antitumor effects of miR-29b-3p in vivo were investigated in nude mice (5 per group). The number, size, and weight of tumors in nude mice injected with AGS cells transfected with the miR-29b-3p mimics and in nude mice injected with AGS cells transfected with mimics-NC were compared. The results showed that mice injected with AGS cells transfected with the miR-29b-3p mimics were slightly smaller in size (Figure 5A) and weighed 0.5 g less (Figure 5B) than the control group (mimics-NC) in the fifth week. Analysis of the growth rate of AGS tumors over the 28-day experimental period indicated that tumors of mice treated with AGS cells transfected with the miR-29b-3p mimics grew much slower than the control group (Figure 5C). After sacrifice 5 weeks later, the tumors in the experimental group were significantly smaller than tumors in the control group (Figure 5D). Tumor volume was also significantly lower (55 mm3 less) in the mice injected with AGS cells transfected with the miR-29b-3p mimics than tumor volume in the control group (Figure 5E). Similar observations were observed in tumor weights, which were 0.24 g lighter in the experimental group (Figure 5F). Thus, the growth and metastasis of tumors were inhibited by the miR-29b-3p mimics, confirming the significant inhibitory effect of miR-29b-3p on tumor growth in vivo.
Figure 5.
Body characteristics and tumour growth of nude mice injected with transfected AGS cells. (A) Nude mice body shape comparison, (B) Nude mice body weight growth curve, (C) Nude mice tumour growth curve, (D) Nude mice tumour comparison, (E) Nude mice tumour volume after removal. (F) Nude mice tumour weight after removal. **P < 0.01.
Discussion
Gastric cancer is one of the most common malignant tumors in the world, with the highest incidence in East Asia.18 Patients with early gastric cancer can be treated successfully to some extent.19 While some patients are cured by surgery and adjuvant therapy,20 once patients advance to the middle and late stages, gastric cancer becomes almost incurable. Since there are no obvious symptoms in the early stages of gastric cancer, many patients are not diagnosed until the advanced stage.21 Cancer recurrence and metastasis are the main obstacles to improving the survival of patients with gastric cancer. Therefore, finding new treatment strategies and effective biomarkers are particularly important. To achieve successful medical treatment for gastric cancer patients, targeted gene therapy is a viable and potentially important option.
In recent years, the role of microRNAs in tumors has attracted considerable attention. One of the important steps in understanding the regulatory role of microRNAs is the identification of their targeted mRNAs.22 In many human tumors, microRNA-29b can alter cell proliferation, migration, and invasiveness by targeting multiple genes.23 Overexpression of microRNA-29b inhibits the proliferation of cancer cells and induces apoptosis.24 MAZ plays an important role in regulating the transcriptional initiation of some microRNA-29b target genes, and MAZ expression is abnormally increased in malignant tumors.25 MAZ can inhibit autophagy and promote metastasis of cancer cells by activating the mTORC1 pathway, which is also regulated by AKT signaling. In addition, MAZ downregulates the expression of c-Myc to enhance autophagic flow and increase the number of autophagic vesicles. Thus, c-myc/MAZ interactions inhibit the physiological function of cancer cells. Considering the importance of microRNAs and MAZ in protein regulation and tumorigenesis, we investigated the effects of microRNA-29b and its target genes on the proliferation, migration, and invasion of gastric cancer cells using cell function and animal model experiments to identify regulatory mechanisms in cancer cells.
First, the transcriptional expression levels of hsa-miR-29b-3p and MAZ in GES1 cells and gastric cancer cells were investigated. The qRT-PCR results show that the expression of hsa-miR-29b-3p was significantly higher in GES1 cells compared with gastric cancer cells. Conversely, the expression of MAZ was lower in GES1 cells than in gastric cancer cells. Subsequently, the expression of MAZ in AGS and MGC cells transfected with miR-29b-3p mimics was detected by qRT-PCR, Western blot, and fluorescence reporter assay. The results showed that the mRNA and protein expression levels of MAZ in AGS and MGC cells transfected with the miR-29b-3p mimics were lower than those in the blank control. The dual luciferase reporter assay confirmed that MAZ is the target gene of hsa-miR-29b-3p and MAZ is regulated by miR-29b-3p. To verify the effects of miR-29b-3p and MAZ on invasion and migration of gastric cancer cells, we overexpressed miR-29b-3p or depleted MAZ with shRNA in AGS and MGC cells. Colony-forming, Transwell (72 h), and MTT assays were conducted to measure cell migration, invasion, and cell viability. The upregulation of miR-29b-3p or silencing of MAZ weakened the viability of AGS and MGC cells. The proliferation, migration, and invasion of AGS and MGC cells transfected with miR-29b-3p mimics or MAZ shRNA were significantly lower than those of the control groups. These results indicate that overexpression of miR-29b-3p and depletion of MAZ inhibit the growth and survival of cancer cells via inhibition of proliferation, migration, and invasion.
Autophagy is a process in which cells remove harmful substances to avoid or repair damage. This process involves a variety of autophagy genes, such as beclin 1, microtubule-associated protein light chain 3 (LC3), and P62. Beclin 1, the most important positive regulator of autophagy, regulates the maturation and formation of autophagy, induces the initiation of autophagy, and enhances autophagy activity.26 LC3 localizes to the products of autophagy-related genes on the surface of autophagic membranes, promotes the maturation of autophagosomes, and has specificity. During autophagy, the LC3 enzyme cleaves the polypeptide into LC3-I and is, subsequently, conjugated to PE to form LC3-II. P62, also known as sequestosome 1 (SQSTM1), plays an important role in regulating autophagy and apoptosis as a selective autophagy adaptor protein.27 P62 binds to intracellular ubiquitinated proteins during autophagy and forms a complex with LC3-II, which is finally degraded in the autophagosomes.28 The expression level of LC3-II negatively correlates with autophagy levels.29 In this study, qPCR and Western blotting were used to detect the expression of beclin 1, P62, and LC3 in transfected cells. The mRNA and protein expression levels of beclin 1 and LC3 in cells transfected with miR-29b-3p mimics or with MAZ shRNA were higher than those in the blank control group. Inversely, the expression level of P62 was lower in transfected cells. These consistent findings support the idea that the upregulation of miR-29b-3p in AGS and MGC cells functional inhibit tumor progression. The results of tumor formation in nude mice demonstrate that the upregulation of miR-29b-3p inhibits tumor growth. Thus, overexpression of miR-29b-3p downregulates MAZ to reduce tumor growth by controlling autophagy, resulting in a reduction in proliferation, migration, and invasion of cancer cells. Overall, those experimental results showed that high expression of miR-29b-3p and depletion of MAZ regulates autophagy in cancer cells, which is of great significance for inhibiting tumors.
Conclusions
This study demonstrates that hsa-miR-29b-3p is expressed at low levels and MAZ is expressed at high levels in gastric cancer cells. The target gene of miR-29b-3p is MAZ, and the upregulation of miR-29b-3p can reduce the expression of MAZ. Overexpression of miR-29b-3p and inhibition of MAZ expression significantly inhibits cell viability, proliferation, migration, and invasion of AGS and MGC cells by regulating the expression of autophagy-related proteins beclin 1, P62, and LC3, and up-regulation of MAZ after overexpression of miR-29b-3p offset the effects, proving that miR-29b-3p regulate autophagy by MAZ. The regulation of autophagy has a significant inhibitory effect on cancer. These results provide a new strategy for targeted therapy in patients with gastric cancer.
Funding Statement
This work was financially supported by National Key Research and Development Project of China (2020YFA0907900).
Data Sharing Statement
All data generated or analyzed during this study are included in this published article.
Ethics Approval and Consent to Participate
The animal experiment was approved by the ethics committee of Tianjin Medical University (approval Number: TMUaMEC2019012). All procedures were performed in accordance with national (D.L.n.26, March 4th, 2014) and international laws and policies (directive 2010/63/EU).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi: 10.3322/caac.21166 [DOI] [PubMed] [Google Scholar]
- 3.Okugawa Y, Mohri Y, Tanaka K, et al. Metastasis-associated protein is a predictive biomarker for metastasis and recurrence in gastric cancer. Oncol Rep. 2016;36(4):1893–1900. doi: 10.3892/or.2016.5054 [DOI] [PubMed] [Google Scholar]
- 4.Tuysuz EC, Gulluoglu S, Yaltirik CK, et al. Distinctive role of dysregulated miRNAs in chordoma cancer stem-like cell maintenance. Exp Cell Res. 2019;380(1):9–19. doi: 10.1016/j.yexcr.2019.03.039 [DOI] [PubMed] [Google Scholar]
- 5.Sun -J-J, Chen G-Y, Xie Z-T. MicroRNA-361-5p inhibits cancer cell growth by targeting CXCR6 in hepatocellular carcinoma. Cell Physiol Biochem. 2016;38(2):777–785. doi: 10.1159/000443033 [DOI] [PubMed] [Google Scholar]
- 6.Esquela-Kerscher A, Slack FJ. Oncomirs — microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–269. doi: 10.1038/nrc1840 [DOI] [PubMed] [Google Scholar]
- 7.Rostas JW 3rd, Pruitt HC, Metge BJ, et al. microRNA-29 negatively regulates EMT regulator N-myc interactor in breast cancer. Mol Cancer. 2014;13(1):200. doi: 10.1186/1476-4598-13-200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ren J, Guo D, Wang X, Zhang C, Wang B, Gao Z. Revealing the alternative promoter usage of SAF/MAZ gene by bichromatic fluorescent reporter construct. Biosci Rep. 2019;39(1). doi: 10.1042/BSR20171668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song J, Murakami H, Tsutsui H, et al. Genomic organization and expression of a human gene for Myc-associated zinc finger protein (MAZ). J Biol Chem. 1998;273(32):20603–20614. doi: 10.1074/jbc.273.32.20603 [DOI] [PubMed] [Google Scholar]
- 10.Stubbs MC, Min IS, Izzo MW, Rallapalli R, Derfoul A, Hall DJ. The ZF87/MAZ transcription factor functions as a growth suppressor in fibroblasts. Biochem Cell Biol. 2000;78(4):477–485. doi: 10.1139/o00-053 [DOI] [PubMed] [Google Scholar]
- 11.Maity G, Haque I, Ghosh A, et al. The MAZ transcription factor is a downstream target of the oncoprotein Cyr61/CCN1 and promotes pancreatic cancer cell invasion via CRAF–ERK signaling. J Biol Chem. 2018;293(12):4334–4349. doi: 10.1074/jbc.RA117.000333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiao L, Li Y, Shen D, et al. The prostate cancer-up-regulated Myc-associated zinc-finger protein (MAZ) modulates proliferation and metastasis through reciprocal regulation of androgen receptor. Med Oncol. 2013;30:570. [DOI] [PubMed] [Google Scholar]
- 13.Ray A, Ray BK. Induction of Ras by SAF-1/MAZ through a feed-forward loop promotes angiogenesis in breast cancer. Cancer Med. 2015;4:224–234. doi: 10.1002/cam4.362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maity G, Sarkar S, Dhar K, et al. Transcription factor MAZ promotes cell growth and aggressive behavior of human pancreatic cancer cells. Cancer Res. 2014;74. [Google Scholar]
- 15.Triner D, Castillo C, Hakim JB, et al. Myc-associated zinc finger protein regulates the proinflammatory response in colitis and colon cancer via STAT3 signaling. Mol Cell Biol. 2018;38(22). doi: 10.1128/MCB.00386-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo W, Zhu X, Liu W, et al. MYC associated zinc finger protein promotes the invasion and metastasis of hepatocellular carcinoma by inducing epithelial mesenchymal transition. Oncotarget. 2016;7(52):86420–86432. doi: 10.18632/oncotarget.13416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu N, Liu X, Zhou N, Wu Q, Zhou L, Li Q. Gene expression profiling and bioinformatics analysis of gastric carcinoma. Exp Mol Pathol. 2014;96(3):361–366. doi: 10.1016/j.yexmp.2014.02.007 [DOI] [PubMed] [Google Scholar]
- 18.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- 19.Ichikawa H, Nagahashi M, Shimada Y, et al. Actionable gene-based classification toward precision medicine in gastric cancer. Genome Med. 2017;9(1):93. doi: 10.1186/s13073-017-0484-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mizrak Kaya D, Harada K, Shimodaira Y, Amlashi FG, Lin Q, Ajani JA. Advanced gastric adenocarcinoma: optimizing therapy options. Expert Rev Clin Pharmacol. 2017;10(3):263–271. doi: 10.1080/17512433.2017.1279969 [DOI] [PubMed] [Google Scholar]
- 21.Kim HJ, Oh SC. Novel systemic therapies for advanced gastric cancer. J Gastric Cancer. 2018;18:1–19. doi: 10.5230/jgc.2018.18.e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naifang S, Minping Q, Minghua D. Integrative approaches for microRNA target prediction: combining sequence information and the paired mRNA and miRNA expression profiles. Curr Bioinform. 2013;8:37–45. doi: 10.2174/1574893611308010008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langsch S, Baumgartner U, Haemmig S, et al. miR-29b mediates NF-kappaB signaling in KRAS-induced non-small cell lung cancers. Cancer Res. 2016;76:4160–4169. doi: 10.1158/0008-5472.CAN-15-2580 [DOI] [PubMed] [Google Scholar]
- 24.Lv M, Zhong Z, Huang M, Tian Q, Jiang R, Chen J. lncRNA H19 regulates epithelial-mesenchymal transition and metastasis of bladder cancer by miR-29b-3p as competing endogenous RNA. Biochim Biophys Acta Mol Cell Res. 2017;1864:1887–1899. doi: 10.1016/j.bbamcr.2017.08.001 [DOI] [PubMed] [Google Scholar]
- 25.Yao Y, Ma J, Xue Y, et al. MiR-449a exerts tumor-suppressive functions in human glioblastoma by targeting Myc-associated zinc-finger protein. Mol Oncol. 2015;9(3):640–656. doi: 10.1016/j.molonc.2014.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang M, Yang XM, Yin DH, et al. Beclin1 enhances cisplatin-induced apoptosis via Bcl-2-modulated autophagy in laryngeal carcinoma cells Hep-2. Neoplasma. 2018;65:42–48. doi: 10.4149/neo_2018_161102N528 [DOI] [PubMed] [Google Scholar]
- 27.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140(3):313–326. doi: 10.1016/j.cell.2010.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim J, Lachenmayer ML, Wu S, et al. Proteotoxic stress induces phosphorylation of p62/SQSTM1 by ULK1 to regulate selective autophagic clearance of protein aggregates. PLoS Genet. 2015;11(2):e1004987. doi: 10.1371/journal.pgen.1004987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamark T, Svenning S, Johansen T, Lane JD, Korolchuk VI, Murray JT. Regulation of selective autophagy: the p62/SQSTM1 paradigm. Essays Biochem. 2017;61(6):609–624. doi: 10.1042/EBC20170035 [DOI] [PubMed] [Google Scholar]