Abstract
Background
Asthma is usually associated with airway inflammation and airway remodeling. Epithelial mesenchymal transition (EMT) often occurs in airway remodeling. The purpose of this study is to identify the effect of miR-21-5p and Smad7 signaling pathway in macrophage-derived exosomes on EMT of airway epithelial cells.
Methods
HE staining and Masson staining were used to verify the successful establishment of the asthma model. The levels of epithelial cell adhesion factor and stromal cell markers were detected by Western blot. The levels of miR-21-5p were detected by qRT-PCR. The expression of miR-21-5p in lung tissue was further verified by fluorescence in situ hybridization (FISH). Exosome morphology was observed by transmission electron microscopy (TEM) and nanoparticle tracking analysis (NTA). Luciferase reporter assay was applied to analyze the interaction of miR-21-5p with Smad7.
Results
The expression of miR-21-5p was upregulated in macrophages of rats in vivo with OVA-induced asthma. In vitro cultured alveolar macrophages stimulated by LPS could secrete exosomes with high levels of miR-21-5p. The exosome-derived miR-21-5p promotes EMT in rat tracheal epithelial cells through TGFβ1/Smad signaling pathway by downregulating Smad7. This process can be blocked by miR-21-5p inhibitor.
Conclusion
Rat alveolar macrophages produced high levels of miR-21-5p-containing exosomes, which transported miR-21-5p to tracheal epithelial cells, thus promoting EMT through TGF-β1/Smad signaling pathway by targeting Smad7.
Keywords: asthma, exosomes, miRNA, miR-21-5p, Smad7, epithelial mesenchymal transition
Background
Asthma is one of the most common chronic diseases in the world.1 According to the World Health Organization, there are more than 300 million people suffering from asthma in the world,2 which places a heavy burden on global health and economy.
Asthma is a heterogeneous disease characterized by chronic airway inflammation.3 It involves in a variety of cell types, such as eosinophils, mast cells, T lymphocytes, neutrophils and airway epithelial cells.4 From the perspective of pathophysiology, asthma usually associated with airway inflammation and airway remodeling.5 The main changes of airway remodeling are the increase of airway smooth muscle, airway wall thickening, inflammatory cell infiltration, angiogenesis and airway mucus gland hyperplasia.6 It is also worth noting that epithelial mesenchymal transition (EMT) often occurs in airway remodeling.7 EMT is characterized by loss of epithelial cell polarity, decreased production of adhesion factors (such as E-cadherin), and increased production of stromal cell markers (such as α-SMA and vimentin) and the phenotypic characteristics of stromal cells.8 These processes lead to structural and functional changes of airway. Currently, little about the mechanisms that lead to airway remodeling in asthma are known. The origin of the mesenchymal cells that contribute to fibrosis of the airway is poorly understood. However, emerging evidence suggested that environmental challenge of the airway epithelium could induce downregulation of epithelial cell–cell adhesions and promote mesenchymal gene expression both in vitro and in vivo, contribute to airway remodeling through the process of epithelial–mesenchymal transition in asthma.9
There are few effective treatments for asthma, and microRNA is an important potential therapeutic target. MicroRNA is a small non-coding single stranded RNA, containing 18–25 nucleotides, which is widely distributed in various organisms from viruses to humans.10 To date, more than 1000 human miRNAs have been found. They act as regulators of gene expression, regulating the expression of most genes by inhibiting protein translation, and play a key role in cell signal transduction, tissue and organ development and other biological processes.11 Accumulating evidence indicates the key role of microRNA in normal lung development and respiratory diseases, such as asthma, chronic obstructive pulmonary disease, cystic fibrosis, idiopathic pulmonary fibrosis and lung cancer.12,13 It has been reported that there are significant differences in microRNA expression spectrum between normal lung tissues and asthmatic tissues, indicating that microRNA plays a regulatory role in the pathogenesis of asthma.14
Exosomes as extracellular vesicles can transmit signals from donor cells to recipient cells, thus acting as a bridge for intercellular communication.15 It has been known that exosome-derived cargos play a crucial role in many biological processes, such as apoptosis, coagulation, inflammation and intercellular signal transduction.16–19 Many reports highlight the important roles of exosomes in lung disease, including asthma.20 Exosomes can be released from asthma-related cells, such as mast cells, eosinophils, dendritic cells, T cells and bronchial epithelial cells, thereby enhance the allergic response during asthma.21 However, there are few reports about the effect of exosomes produced by macrophages on the process of asthma at present.
In the present study, we report that alveolar macrophages of rats stimulated by LPS could secrete high levels of miR-21-5p-containing exosomes. These exosomes could transport miRNA to tracheal epithelial cells, and finally promote the epithelial mesenchymal transition (EMT) through TGFβ1/Smad signal pathway by targeting Smad7.
Methods
Animal Study
All animal experiments were conducted in accordance with the requirements of “guidelines for the care and use of laboratory animals”. Six- to seven-week-old male Sprague-Dawley rats were purchased from Charles River. The animals were acclimatized prior to experiment at room temperature to a 12 hour light and 12 hour dark cycle with free access to water and food ad libitum. The animals were randomly divided into two groups (n = 6); Group I served as normal control (sensitized with saline), Group II served as asthma control (challenged with ovalbumin, OVA). The specific operation is as follows: on days 1, 8 and 15, rats were injected with 1 mg OVA in the peritoneal cavity, dissolved in 0.8 mL sterile saline containing 0.3 mL Aluminium Hydroxide Gel. From day 23, intranasal exposure of 100 μL 1% OVA (in normal saline) was performed once a day for 7 days. The study was carried out in compliance with the ARRIVE guidelines, and all research protocols and guidelines for the care and use of laboratory animals had been approved by the Experimental Animal Welfare and Ethics Committee of China Medical University (IACUC No.CMU2018081). The animal experiments involved in this paper were carried out in accordance with the NIH Guide for the Care and Use of Laboratory Animals.
Hematoxylin and Eosin (HE) Staining
The paraffin slices were heated in an oven at 65°C for 30 min to melt the paraffin completely. The tissue was dewaxed in xylene and then hydrated in gradient ethanol. After washing with water, the tissue was stained with hematoxylin for 2 min, then washed, soaked in 1% hydrochloric acid ethanol for 3 s, and then stained with eosin for 1 min, and then dehydrated. The tissue was immersed in xylene for transparency. A piece of neutral gum was used for sealing and the sample was observed under a microscope.
Masson Staining
The paraffin sections were dewaxed and washed with distilled water. The nuclei were stained with Weigert sappan semen for 5–10 min. After washing the slices with water, Masson Ponceau acid red solution was used to stain for 5–10 min. The sections were soaked in 2% glacial acetic acid solution for a while, and then differentiated with 1% molybdophosphoric acid solution for 3–5 min. The slices were dyed with aniline blue for 5 min without washing, and then soaked in 0.2% glacial acetic acid solution for a minute. Finally, anhydrous alcohol was used for dehydration. Xylene was used for transparency and neutral gum was used for sealing.
Quantitative Real-Time PCR (qRT-PCR)
The differentially expressed genes were examined by real-time quantitative RT-PCR. Total RNA was extracted by Trizol reagent. Nanodrop ultra-micro spectrophotometer was used to measure the concentration and purity of RNA samples. The cDNA was synthesized by reverse transcription according to the instructions of TaqMan micro RNA reverse transcription kit (ThermoFisher, #4366596). Finally, real-time quantitative PCR was carried out on real-time PCR system (Agilent). The following primer sequences were used: miR-21-5p, forward 5′-TCGCTCGAGATTTTTTTTTATCAAGAGGG-3′, reverse 5′-TCGGCGGCCGCGACAAGAATGAGACTTTAATC-3′; U6, forward 5′-GCTTCGGCAGCACATATACTAAAAT-3′, reverse 5′-CGCTTCACGAATTTGCGTGTCAT-3′. The U6 small nuclear RNA was used as an internal control. The data were analyzed by 2 −ΔΔCt relative quantitative method. Samples were analyzed in triplicates.
Fluorescence in situ Hybridization (FISH)
Formalin-fixed paraffin-embedded tissues were used for miRNA detection by in situ hybridization using digoxigenin (DIG) double-labeled, LNA-enhanced probe for miR-21-5p (miRCURY LNA miRNA detection probe, Qiagen). The probe was denatured at 83°C for 5 min. The tissue sections were heated in 65°C oven for 2 h, denatured and dehydrated in sodium citrate normal saline (SCC). The denatured probe (40 nM) was mixed with the tissue slice and incubated overnight at 37°C. Slides were extensively washed and then incubated for 1 hour at 37°C with anti-DIG fluorescein isothiocyanate (FITC) conjugated antibody, washed again, drained and counterstained with 4′-6′Diamidino-2-phenylindole (DAPI). LNA-FISH signals were visualized on a fluorescence microscope.
Luciferase Reporter Assays
The plasmids containing the WT 3ʹ-UTR of Smad7 or the 3ʹ-UTR mutated in the predicted binding site of miR-21-5p were co-transfected into rat tracheal epithelial cells with miRNA mimics or mimic-NC. After 36 hours, the medium was discarded and the cells were washed with PBS. According to the instruction manual of double luciferase detection kit, double luciferase analysis was carried out. To ensure the authenticity and reliability of the data, all experiments were repeated at least three times.
Exosomes Isolation and Labeling
Cells were cultured with medium supplemented with exosome-depleted FBS for exosome preparation. Exosomes were isolated by ultracentrifuge according to the published protocol22 and then labeled with PKH67 lipophilic membrane dyes (Sigma-Aldrich) according to the manufacturer’s guide. Briefly, the exosome pellet was suspended using the Diluent C (sigma-Aldrich) for general membrane labeling. PKH67 dye working solution was added into the exosome. The mixture was mixed by vortexing, and then incubated for 10 min. The exosomes were extracted using sucrose solution and centrifugation to remove the excess dyes. PBS was used to resuspend the precipitate containing the dyed exosomes.
Inhibition of Exosome Secretion
GW4869 (sigma-Aldrich, D1692) was used to inhibit exosome secretion. To test the effect of GW4869 on cell proliferation, the cells were cultured with 5% DMSO and 5 μg/mL GW4869 for 24 h, and the proliferation of cells was examined by MTT assay kit (Beyotime, C0009S). For GW4869 treatment, the cells were seeded in the culture dish. The next day, cells were refed with fresh medium with or without 5 μg/mL GW4869. Exosomes were collected and the protein concentration was determined to determine the inhibitory effect of GW4869 on exosome secretion.
Cell Culture and LPS Treatment
NR8383 (rat alveolar macrophages) was purchased from ATCC. The cells were cultured in humidified incubator with 5% CO2 at 37°C. DMEM supplemented with 10% exosome-depleted FBS (ThermoFisher) were used for cell culture. For macrophage stimulation with LPS, the cells were treated with 1 µg/mL LPS for 12 h.
Transfection
When the cell confluence reached 70–90%, miR-21-5p mimic (UAGCUUAUCAGAC UGAUGUUGA) or inhibitor (UAGCUUAUCAGACUGAUGUUGA) was transfected into rat tracheal epithelial cells to a final concentration of 50 nM with Lipofectamine 3000 (Invitrogen) according to the manufacturer's guide.
For siRNA knockdown, final concentration of 40 nM was used for transfection. Target siRNAs included in this study are: rat siTGFβ1 (Sigma-Aldrich, 59086), rat siSmad7 (ThermoFisher, 155242). Cells were analyzed two to three days after transfection.
Western Blotting
After SDS-PAGE electrophoresis, the proteins were transferred to a nitrocellulose membrane. Then the membrane was blocked in 5% skimmed milk for 1.5 h at room temperature. After washing three times with TBST, the membranes were incubated with primary antibodies against E-cadherin (Santa Cruz, sc-8426, 1:1000), α-SMA (Santa Cruz, sc-53142, 1:1000), vimentin (Santa Cruz, sc-6260, 1:1000), Smad7 (Santa Cruz, sc-365846, 1:500), TGFβ1 (Santa Cruz, sc-130348, 1:500), TGFβRII (Santa Cruz, sc-17799, 1:500), p-Smad2 (Abcam, ab188334, 1:1000), p-Smad3 (Abcam, ab63403) and GAPDH (Santa Cruz, sc-365062, 1:2000) at 4°C overnight. The membrane was incubated with horseradish peroxidase (HRP)-bound secondary antibody (1:5000) for 1.5 h at room temperature, and visualized with BeyoECL Moon chemiluminescence kit (Beyotime, P0018FS).
Statistical Analysis
In order to ensure the accuracy of the experimental results, all experiments were repeated three times or more, and the data were expressed as mean ± SD. SPSS 19.0 was used to analyze the data. Differences among multiple groups were evaluated by one-way analysis of variance (ANOVA) followed by Tuckey post hoc tests. P < 0.05 means the difference is statistically significant.
Results
OVA Treatment Increased Levels of miR-21-5p in Lung Tissue of Rats
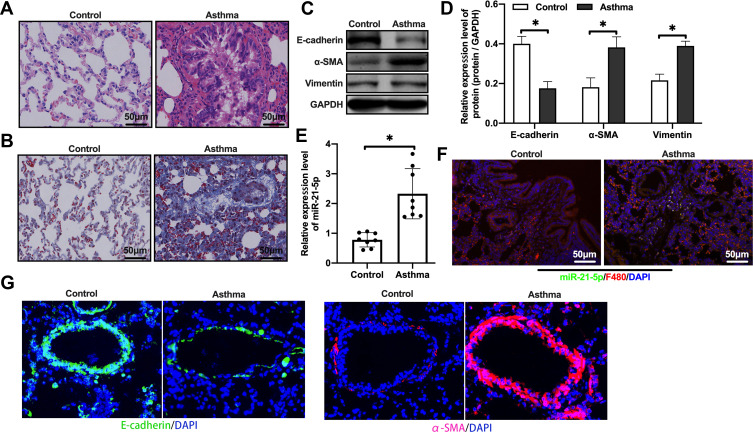
We first evaluated the expression level of miR-21-5p in rats with OVA-induced asthma. In order to verify whether our rat model was successful, H&E staining and Masson staining were performed. These results showed inflammatory cell infiltration, airway thickening and collagen deposition (Figure 1A–B). In view of the important role of epithelial mesenchymal transition (EMT) in airway remodeling of asthma, we analyzed the levels of epithelial cell adhesion factor and stromal cell markers by Western blot. The results showed that, compared with the control group, the protein levels of E-cadherin in the asthma group were significantly reduced, and the levels of α-SMA and Vimentin were increased (Figure 1C–D). qRT-PCR analysis showed that the expression level of miR-21-5p in the asthma group was significantly increased (Figure 1E). The expression of miR-21-5p in lung tissue was further verified by FISH. We found miR-21-5p was upregulated in rats with OVA-induced asthma, and it was mainly expressed in macrophages (Figure 1F). Immunofluorescent staining showed that the expression of α-SMA was increased in the epithelial cells and E-cadherin was decreased, further confirmed the enhanced EMT in the asthma group (Figure 1G). Overall, these results indicated that OVA treatment induced asthma and airway remodeling, together with increased expression of miR-21-5p in the macrophages.
Figure 1.
The expression level of miR-21-5p was increased in rats with OVA-induced asthma. Histological changes were analyzed by H&E staining assay (A) (Scale bar = 50 μm) and Masson staining assay (B) (Scale bar = 50 μm). The levels of epithelial cell adhesion factor and stromal cell markers were analyzed by Western blot (C and D, n=6). The expression level of miR-21-5p were analyzed by qRT-PCR analysis (E, n=6) and FISH (F). (G) Representative immunofluorescent staining of lung tissues from control and asthma group showing the expression of α-SMA and E-cadherin. Data were expressed as mean ± SD. *Indicates P < 0.05.
LPS Stimulation Caused Release of miR-21-5p-Containing Exosomes from Alveolar Macrophages
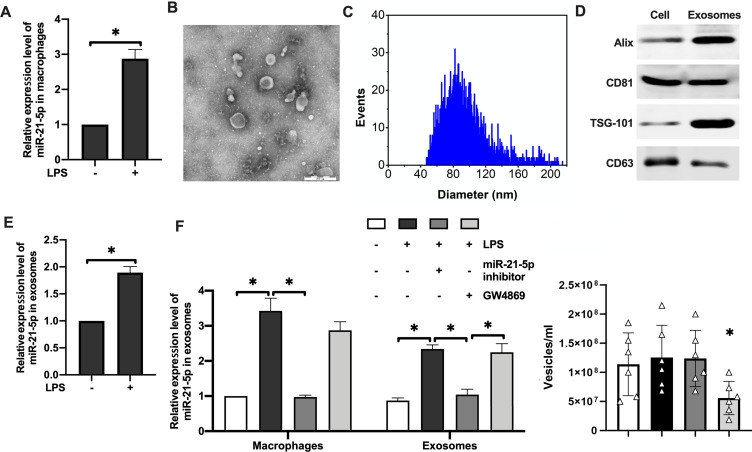
LPS was used to stimulate rat alveolar macrophages to establish inflammatory macrophage microenvironment in vitro. qRT-PCR analysis showed that miR-21-5p content in macrophages of the experimental group was significantly increased (Figure 2A). Exosomes were isolated from the cell culture supernatant of macrophages by high-speed centrifugation, and their morphology was verified by transmission electron microscopy (TEM) and nanoparticle tracking analysis (NTA) (Figure 2B–C). The purity of exosomes was further verified by detecting the levels of exosome surface markers using Western blotting (Figure 2D), and enrichment of marker proteins including Alix, CD81 and TSG-101 were detected in the purified exosomes. Similarly, miR-21-5p was also significantly upregulated in the exosomes derived from rat alveolar macrophages stimulated by LPS (Figure 2E). In contrast, miR-21-5p inhibitor significantly inhibited the expression of miR-21-5p in exosomes (Figure 2F, left). GW4869 (exosome release inhibitor) did not change the relative levels of miR-21-5p in the exosomes, but it caused about 2 fold reduction in the number of vesicles isolated from the same amount of medium (Figure 2F, right). These results demonstrated that alveolar macrophages stimulated by LPS could secrete exosomes with high levels of miR-21-5p.
Figure 2.
Alveolar macrophages stimulated by LPS could secreted exosomes with high levels of miR-21-5p. The expression level of miR-21-5p in macrophages were analyzed by qRT-PCR analysis (A, n=6). Exosomes were isolated from the cell culture supernatant of macrophages by high-speed centrifugation, and their morphology was verified by transmission electron microscopy (B) and nanoparticle tracking analysis of the particle size distribution (C). The expression of exosomal surface markers were analyzed by Western blot (D, n=5). The expression level of miR-21-5p in the exosomes were analyzed by qRT-PCR analysis (E and F). The concentration of exosomes (vesicles/mL) was measured by nanoparticle tracking analysis (n=6). Data were expressed as mean ± SD. *Indicates P < 0.05.
MiR-21-5p Was Transported to Tracheal Epithelial Cells Through Exosomes and Promoted EMT
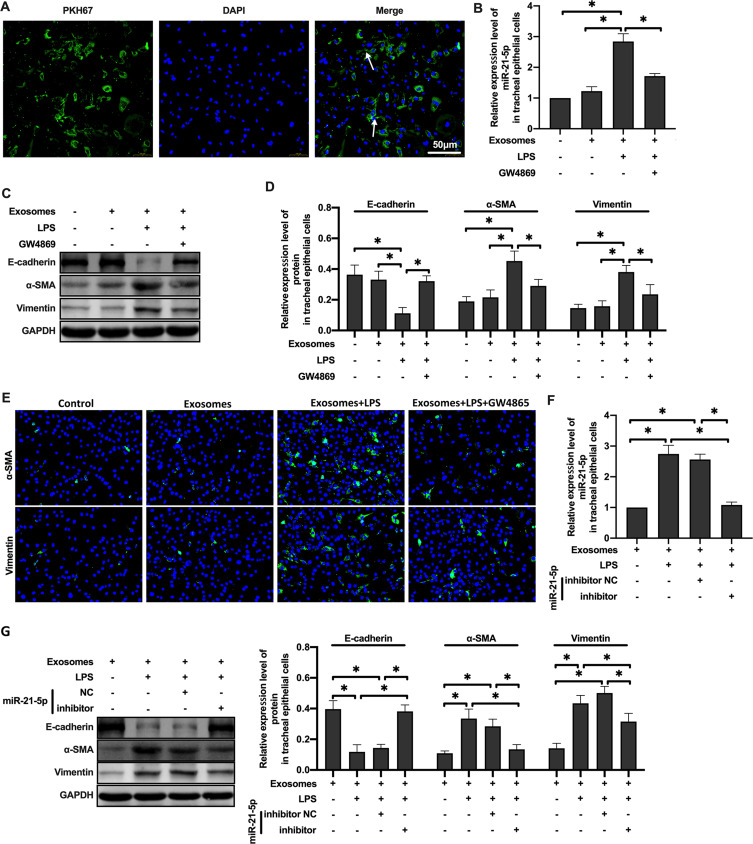
Exosomes mediate intercellular communication through the transfer of their cargos and participate in lung dynamic balance. Therefore, we hypothesized that miR-21-5p could be transferred from LPS-treated rat alveolar macrophages to rat tracheal epithelial cells via exosomes. To determine whether the exosomes from alveolar macrophages can be absorbed by epithelial cells, the exosomes were labeled with fluorescent dye PKH67 and then coincubated with epithelial cells. The results suggested that epithelial cells can absorb exosomes (Figure 3A). And, qRT-PCR analysis showed that the level of miR-21-5p in rat tracheal epithelial cells co-cultured with exosomes was higher, which was inhibited by GW4869 (Figure 3B). Overall, these results demonstrated that macrophages can secrete miRNA-containing exosomes and transport them to epithelial cells.
Figure 3.
MiR-21-5p was transported to tracheal epithelial cells through exosomes and promoted EMT. The exosomes were labeled with fluorescent dye PKH67 and observed under a fluorescence microscope (A). The level of miR-21-5p in rat tracheal epithelial cells cocultured with exosomes isolated from the same amount of culture medium were analyzed by qRT-PCR analysis (B and F) (n=3–6). The levels of epithelial cell adhesion factor and stromal cell markers were analyzed by Western blot (C, D and G) (n=5). The levels of α-SMA and vimentin were analyzed by Immunofluorescence assay (E). Data were expressed as mean ± SD. *Indicates P < 0.05.
In order to further detect whether the phenotype of rat tracheal epithelial cells changed, Western blot was used to analyze the expression of marker proteins in epithelial cells. The results showed that the expression of E-cadherin protein decreased and the expression of α-SMA and vimentin protein increased in rat tracheal epithelial cells co-cultured with the exosomes of rat alveolar macrophages treated with LPS. GW4869 inhibited this change (Figure 3C–D). miR-21-5p inhibitors significantly inhibited the level of miR-21-5p in exosomes, and resulted in low levels of miR-21-5p in rat tracheal epithelial cells (Figure 3E). The expression of EMT marker proteins was well correlated with the levels of exosome-derived miR-21-5p (Figure 3F–G). Taken together, these results demonstrate that after treated with LPS, exosomes from rat alveolar macrophages can promote EMT of rat tracheal epithelial cells through up-regulating miR-21-5p.
MiR-21-5p in Exosomes Derived from LPS-Treated Alveolar Macrophages Targets Smad7 in Rat Tracheal Epithelial Cells
In order to explore how miR-21-5p exerted its function in EMT of rat tracheal epithelial cells, we predicted the downstream targets of miR-21-5p through three independent online databases, including Targetscan (targetscan.org), miRTarBase (miRtarbase.mbc.nctu.edu.tw) and miRanda (microrna.org). We found that Smad7 was the potential targets of miR-21-5p.
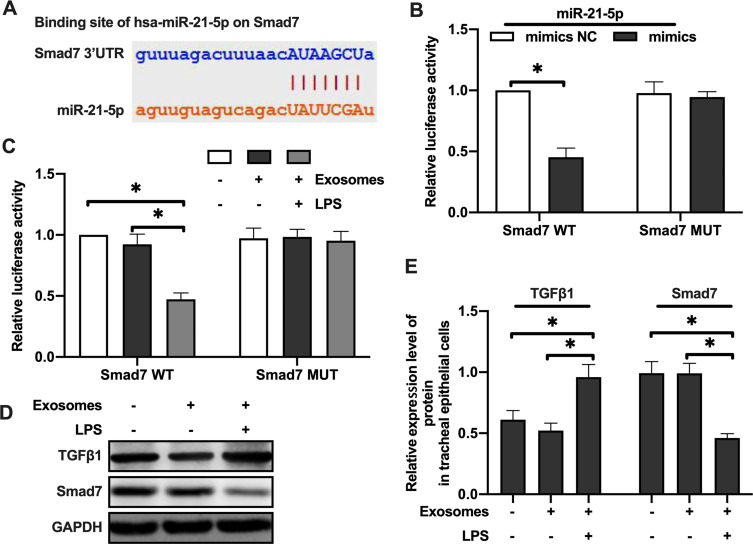
Luciferase assay was performed using plasmids containing wild-type 3ʹUTR of Smad7 containing the predicted binding site of miR-21-5p, or the mutant 3ʹUTR without binding site (Figure 4A). Luciferase activity was decreased after co-transfecting rat tracheal epithelial cells with plasmid containing wild-type Smad7 3ʹUTR and miR-21-5p mimics. There was no significant change in luciferase activity when the miR-21-5p mimic was co-transfected with Smad7 3ʹUTR mutants (Figure 4B). LPS-treated rat alveolar macrophage exosomes reduced the luciferase activity of Smad7 3ʹUTR construct, but did not change the luciferase activity of the mutants’ 3ʹUTR Smad7 construct (Figure 4C). The rat bronchial epithelial cells were treated with exosomes (50 μg/mL) derived from normal or LPS-treated rat alveolar macrophages for 24 hours, and Western blotting results showed that the Smad7 protein level was decreased in tracheal epithelial cells exposed to the LPS-treated rats exosomes of alveolar macrophages while TGF-β1 protein levels was increased (Figure 4D–E). These results suggested that the miR-21-5p targeted Smad7 in rat tracheal epithelial cells.
Figure 4.
MiR-21-5p in exosomes derived from LPS-treated alveolar macrophages targeted Smad7 in rat tracheal epithelial cells. Luciferase was determined using plasmids containing wild-type or mutant 3ʹUTR Smad7 proteins and the binding site of miR-21- 5p (A–C) (n=3–6). The expression of Smad7 and TGFβ1 protein were analyzed by Western blot (D and E) (n=3). Data were expressed as mean ± SD. *Indicates P < 0.05.
MiR-21-5p Promoted EMT of Rat Airway Epithelial Cells by Inhibiting the Expression of Smad7
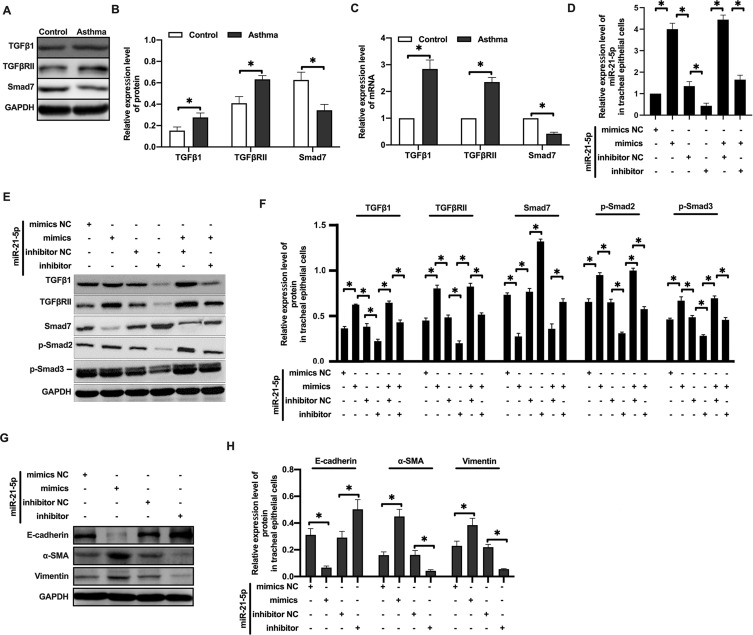
Since Smad7 has been well established to be a key negative regulator of TGF-β signaling, we analyzed the expression of proteins in the TGF-β/Smad signaling pathway subsequently. Western blot analysis showed that the expression of TGFβ1 and TGFβRII protein were increased, while Smad7 protein expression was decreased in rats with OVA-induced asthma (Figure 5A–B). qRT-PCR analysis showed similar results (Figure 5C).
Figure 5.
MiR-21-5p promoted EMT of rat airway epithelial cells by inhibiting the expression of Smad7. The expression of TGFβ1, TGFβRII and Smad7 protein were analyzed by Western blot (A and B) (n=3–4). The expression of TGFβ1, TGFβRII and Smad7 mRNA were analyzed by qRT-PCR analysis (C) (n=3–4). The level of miR-21-5p in rat tracheal epithelial cells were analyzed by qRT-PCR analysis (D) (n=3–5). The expression of Smad7, TGFβ1, TGFβRII, p-Smad2 and p-Smad3 protein were analyzed by Western blot (E and F) (n=3). The levels of epithelial cell adhesion factor and stromal cell markers were analyzed by Western blot (G and H) (n=3). Data were expressed as mean ± SD. *Indicates P < 0.05.
When rat tracheal epithelial cells were transformed with miR-21-5p mimics (Figure 5D), Smad7 protein level was decreased, and TGFβ1, TGFβRII, p-Smad2, p-Smad3 proteins were increased. After transfection of miR-21-5p inhibitor (Figure 5D), Smad7, TGFβ1, TGFβRII, p-Smad2 and p-Smad3 showed opposite trend (Figure 5E–F). In miR-21-5p mimic transfected airway epithelial cells, the expression of E-cadherin protein was decreased, and the expression of α-SMA and vimentin proteins was increased. These effects were reversed by transfection with miR-21-5p inhibitor (Figure 5G–H). Overall, our data suggested that miR-21-5p targeted and downregulated Smad7, leading to enhanced activity of TGFβ1/Smad signaling pathway, and thus promoted EMT.
MiR-21-5p in Exosomes Produced by LPS-Treated Macrophages Promote EMT of Rat Tracheal Epithelial Cells Through TGF-β1/Smad7 Signal Pathway
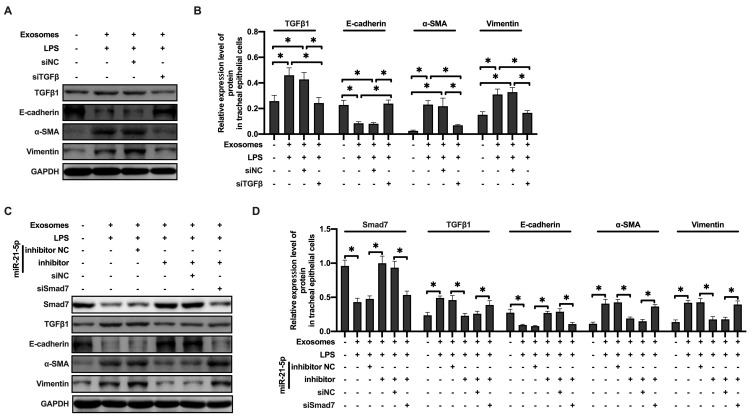
To further explore the specific mechanism of exosome-derived miR-21-5p in promoting EMT, the expression of TGFβ1 in rat tracheal epithelial cells was downregulated by siRNA. We found that decrease in TGFβ1 expression prevented the increased protein levels of α-SMA and vimentin induced by exosome-derived miR-21-5p (Figure 6A–B). Moreover, rat tracheal epithelial cells co-transfected with Smad7 siRNA and miR-21-5p inhibitor were exposed to exosomes. Western blot results showed that the down regulation of miR-21-5p inhibited the expression level of TGFβ 1, α-SMA and vimentin, while increased the levels of Smad7 and E-cadherin. Smad7 knockdown attenuated the effect of miR-21-5p inhibitor (Figure 6C–D). These observations indicated that LPS-stimulated alveolar macrophages secreted exosomes containing miR-21-5p, which promoted EMT in rat tracheal epithelial cells through downregulating Smad7 and upregulating TGFβ1/Smad signaling pathway.
Figure 6.
MiR-21-5p in exosomes produced by LPS-treated macrophages promoted EMT of rat tracheal epithelial cells through TGFβ1/Smad signal pathway. The levels of epithelial cell adhesion factor and stromal cell markers and TGFβ1 were analyzed by Western blot (A and B) (n=3–4). The levels of epithelial cell adhesion factor and stromal cell markers, Smad7 and TGFβ1 were analyzed by Western blot (C and D) (n=3–4). Data were expressed as mean ± SD. *Indicates P < 0.05.
Discussion
OVA is frequently used to build asthma model, and OVA-sensitized rat exhibit different asthma characteristics including excessive mucus production, airway hyperresponsiveness, and eosinophilic airway inflammation.23 Similarly, LPS is the well-known activator of macrophages, and the in vitro model of LPS-stimulated macrophages has been widely used for studying the inflammatory responses and molecular mechanisms of allergic asthma.24 Macrophages are key regulators of the development and progression of asthma, facilitating deleterious airway remodeling in affected patients.25 Immune cell function is tightly regulated by microRNAs (miRNAs), but how these miRNAs impact macrophage-mediated airway remodeling in the context of asthma remains to be determined.
It has been reported that the expression profile of miRNAs in serum of asthmatic patients has changed significantly.26 Prior studies have demonstrated that miR-19a is upregulated, which is related to cell proliferation, and miR-181-5p is downregulated in bronchial epithelium of asthmatic patients, regulating the expression of IL13, IL-1 β and CCL11.27,28 Milger et al identified five combinations of miRNAs (miR-21-5p/miR-15a-5p; miR-27a-3p/miR-15a-5p; miR-29c-3p/miR-15a-5p; miR-223-3p/miR-425-5p; miR-15a-5p/miR-342-3p) as biomarkers for asthma treatment.29 Recent evidence suggests that miR-21-5p has a significant correlation with asymptomatic bronchiectasis. It is regarded as a biomarker of airway reversibility.30
In the present study, OVA was used to create an asthma model, H&E staining and Masson staining were used to verify the success of the model. We found that the expression of miR-21-5p was upregulated in asthma rats induced by OVA, and it was mainly expressed in alveolar macrophages. Similarly, in vitro LPS treatment of alveolar macrophages increased the intracellular and exosome-derived miR-21-5p. Since we observed enhanced EMT of tracheal epithelial cells in the asthma rat, we hypothesized that miR-21-5p produced by macrophages could be transported to tracheal epithelial cells and promote the EMT, which was tested using in vitro cell culture models.
Using fluorescently labeled exosomes, together with miR-21-5p mimics/inhibitor and GW4869 (exosome release inhibitor), we found that alveolar macrophages could transfer miR-21-5p to rat tracheal epithelial cells through exosomes and promote EMT. Furthermore, Western blot and luciferase reporter assay indicated that the exosome-derived miR-21-5p targeted Smad7 in rat tracheal epithelial cells and affected the TGFβ1/Smad signaling pathway. The role of TGFβ1 in EMT in asthma has been intensively investigated.31 It is a potent profibrogenic factor whose expression is increased in the asthmatic airways and is a prime candidate for the initiation and persistence of airway remodeling in asthma. TGFβ1 initiates signals by binding to TGF-βRII.32 Smads are important intracellular effectors of TGF-β1 signaling superfamily.33 Phosphorylated Smad2 and Smad3 are translocate to the nucleus, where they regulate transcription of target genes.33 In contrast, the inhibitory Smad7 inhibit activation of the receptor-regulated Smads.32
miRNA inhibitors are possible therapies for asthma.34,35 They are a class of antisense oligonucleotides that complement with specific miRNA targets. They bind to miRNAs and miRNAs binding sites of mRNA targets. Therefore, the inhibitor miRNAs compete with the target mRNA, resulting in functional inhibition of miRNAs and decompression of direct targets.36 The application in animal models has been studied so far. In mice, anti-miR-221 reduced eosinophils in bronchoalveolar lavage fluid.37 Anti-miR-145 and anti-miR-126 can reduce eosinophil infiltration, Th2 cytokine production and airway hyper-responsiveness.38,39 In contrast, anti-miR-155-5p did not achieve such results. Although miR-155-5p was highly upregulated in asthma mice, intranasal administration of antagonists had no effect on airway hyper-responsiveness or Th2 cytokine expression.
In summary, we found that rat alveolar macrophages produced high levels of miR-21-5p in the exosomes under stress condition, which transported miR-21-5p to tracheal epithelial cells, thus promoting EMT through TGFβ1/Smad signaling pathway by targeting Smad7 in rat tracheal epithelial cells. This process can be blocked by miR-21-5p inhibitor. Our study suggested that exosomes play an important role in chronic asthma, and may provide new insights into the clinical treatment of asthma.
Conclusions
MiR-21-5p promotes epithelial mesenchymal transition (EMT) of rat airway epithelial cells through TGF-β1/Smad signal pathway by targeting Smad7.
Funding Statement
This work was supported by National Natural Science Foundation of China (81800029), China Medical University (3110118042) and 345 Talent Project of Shengjing Hospital of China Medical University (M0421).
Abbreviations
EMT, epithelial mesenchymal transition; FISH, fluorescence in situ hybridization; TEM, transmission electron microscopy; NTA, nanoparticle tracking analysis; OVA, ovalbumin; ASMCs, airway smooth muscle cells.
Data Sharing Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
This research has been approved by Experimental Animal Welfare and Ethics Committee of China Medical University (IACUC No.CMU2018081), and carried out in compliance with the ARRIVE guidelines.
Disclosure
The authors declare that they have no conflicts of interest for this work.
References
- 1.Pinnock H, Shah R. Asthma. BMJ. 2007;334(7598):847–850. doi: 10.1136/bmj.39140.634896.BE [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ebmeier S, Thayabaran D, Braithwaite I, Benamara C, Weatherall M, Beasley R. Trends in international asthma mortality: analysis of data from the WHO mortality database from 46 countries (1993-2012). Lancet. 2017;390(10098):935–945. doi: 10.1016/S0140-6736(17)31448-4 [DOI] [PubMed] [Google Scholar]
- 3.Mims JW. Asthma: definitions and pathophysiology. Int Forum Allergy Rhinol. 2015;5(Suppl 1):S2–6. doi: 10.1002/alr.21609 [DOI] [PubMed] [Google Scholar]
- 4.Umetsu DT, Dekruyff RH. Natural killer T cells are important in the pathogenesis of asthma: the many pathways to asthma. J Allergy Clin Immunol. 2010;125(5):975–979. doi: 10.1016/j.jaci.2010.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elliot JG, Noble PB, Mauad T, et al. Inflammation-dependent and independent airway remodelling in asthma. Respirology. 2018;23(12):1138–1145. doi: 10.1111/resp.13360 [DOI] [PubMed] [Google Scholar]
- 6.Fehrenbach H, Wagner C, Wegmann M. Airway remodeling in asthma: what really matters. Cell Tissue Res. 2017;367(3):551–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pu Y, Liu Y, Liao S, Miao S, Zhou L, Wan L. Azithromycin ameliorates OVA-induced airway remodeling in Balb/c mice via suppression of epithelial-to-mesenchymal transition. Int Immunopharmacol. 2018;58:87–93. doi: 10.1016/j.intimp.2018.03.016 [DOI] [PubMed] [Google Scholar]
- 8.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hackett TL. Epithelial-mesenchymal transition in the pathophysiology of airway remodelling in asthma. Curr Opin Allergy Clin Immunol. 2012;12(1):53–59. doi: 10.1097/ACI.0b013e32834ec6eb [DOI] [PubMed] [Google Scholar]
- 10.Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141(4):1202–1207. doi: 10.1016/j.jaci.2017.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11(9):597–610. doi: 10.1038/nrg2843 [DOI] [PubMed] [Google Scholar]
- 12.Rupani H, Sanchez-Elsner T, Howarth P. MicroRNAs and respiratory diseases. Eur Respir J. 2013;41(3):695–705. doi: 10.1183/09031936.00212011 [DOI] [PubMed] [Google Scholar]
- 13.Cushing L, Jiang Z, Kuang P, Lu J. The roles of microRNAs and protein components of the microRNA pathway in lung development and diseases. Am J Respir Cell Mol Biol. 2015;52(4):397–408. doi: 10.1165/rcmb.2014-0232RT [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams AE, Larner-Svensson H, Perry MM, et al. MicroRNA expression profiling in mild asthmatic human airways and effect of corticosteroid therapy. PLoS One. 2009;4(6):e5889. doi: 10.1371/journal.pone.0005889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pegtel DM, Gould SJ. Exosomes. Annu Rev Biochem. 2019;88(1):487–514. doi: 10.1146/annurev-biochem-013118-111902 [DOI] [PubMed] [Google Scholar]
- 16.Meldolesi J. Exosomes and ectosomes in intercellular communication. Curr Biol. 2018;28(8):R435–R444. doi: 10.1016/j.cub.2018.01.059 [DOI] [PubMed] [Google Scholar]
- 17.Kapustin AN, Schoppet M, Schurgers LJ, et al. Prothrombin loading of vascular smooth muscle cell-derived exosomes regulates coagulation and calcification. Arterioscler Thromb Vasc Biol. 2017;37(3):e22–e32. doi: 10.1161/ATVBAHA.116.308886 [DOI] [PubMed] [Google Scholar]
- 18.Cheng X, Zhang G, Zhang L, et al. Mesenchymal stem cells deliver exogenous miR-21 via exosomes to inhibit nucleus pulposus cell apoptosis and reduce intervertebral disc degeneration. J Cell Mol Med. 2018;22(1):261–276. doi: 10.1111/jcmm.13316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan BD, Wong WY, Lee MM, et al. Exosomes in inflammation and inflammatory disease. Proteomics. 2019;19(8):e1800149. doi: 10.1002/pmic.201800149 [DOI] [PubMed] [Google Scholar]
- 20.Kubo H. Extracellular vesicles in lung disease. Chest. 2018;153(1):210–216. doi: 10.1016/j.chest.2017.06.026 [DOI] [PubMed] [Google Scholar]
- 21.van den Berge M, Tasena H. Role of microRNAs and exosomes in asthma. Curr Opin Pulm Med. 2019;25(1):87–93. doi: 10.1097/MCP.0000000000000532 [DOI] [PubMed] [Google Scholar]
- 22.Coughlan C, Bruce KD, Burgy O, et al. Exosome isolation by ultracentrifugation and precipitation and techniques for downstream analyses. Curr Protoc Cell Biol. 2020;88(1):e110. doi: 10.1002/cpcb.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Casaro M, Souza VR, Oliveira FA, Ferreira CM. OVA-induced allergic airway inflammation mouse model. Methods Mol Biol. 2019;1916:297–301. [DOI] [PubMed] [Google Scholar]
- 24.Maruthamuthu V, Henry LJK, Ramar MK, Kandasamy R. Myxopyrum serratulum ameliorates airway inflammation in LPS-stimulated RAW 264.7 macrophages and OVA-induced murine model of allergic asthma. J Ethnopharmacol. 2020;255:112369. doi: 10.1016/j.jep.2019.112369 [DOI] [PubMed] [Google Scholar]
- 25.Saradna A, Do DC, Kumar S, Fu QL, Gao P. Macrophage polarization and allergic asthma. Transl Res. 2018;191:1–14. doi: 10.1016/j.trsl.2017.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kho AT, Sharma S, Davis JS, et al. Circulating microRNAs: association with lung function in asthma. PLoS One. 2016;11(6):e0157998. doi: 10.1371/journal.pone.0157998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haj-Salem I, Fakhfakh R, Berube JC, et al. MicroRNA-19a enhances proliferation of bronchial epithelial cells by targeting TGFbetaR2 gene in severe asthma. Allergy. 2015;70(2):212–219. doi: 10.1111/all.12551 [DOI] [PubMed] [Google Scholar]
- 28.Huo X, Zhang K, Yi L, et al. Decreased epithelial and plasma miR-181b-5p expression associates with airway eosinophilic inflammation in asthma. Clin Exp Allergy. 2016;46(10):1281–1290. doi: 10.1111/cea.12754 [DOI] [PubMed] [Google Scholar]
- 29.Milger K, Gotschke J, Krause L, et al. Identification of a plasma miRNA biomarker signature for allergic asthma: a translational approach. Allergy. 2017;72(12):1962–1971. doi: 10.1111/all.13205 [DOI] [PubMed] [Google Scholar]
- 30.Mathieu M, Martin-Jaular L, Lavieu G, Thery C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol. 2019;21(1):9–17. doi: 10.1038/s41556-018-0250-9 [DOI] [PubMed] [Google Scholar]
- 31.Boxall C, Holgate ST, Davies DE. The contribution of transforming growth factor-beta and epidermal growth factor signalling to airway remodelling in chronic asthma. Eur Respir J. 2006;27(1):208–229. doi: 10.1183/09031936.06.00130004 [DOI] [PubMed] [Google Scholar]
- 32.Zimowska M. [Signaling pathways of transforming growth factor beta family members]. Postepy Biochem. 2006;52(4):360–366. [Indonesian] [PubMed] [Google Scholar]
- 33.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425(6958):577–584. doi: 10.1038/nature02006 [DOI] [PubMed] [Google Scholar]
- 34.Qin HB, Xu B, Mei JJ, et al. Inhibition of miRNA-221 suppresses the airway inflammation in asthma. Inflammation. 2012;35(4):1595–1599. doi: 10.1007/s10753-012-9474-1 [DOI] [PubMed] [Google Scholar]
- 35.Mei D, Tan WSD, Wong WSF. Pharmacological strategies to regain steroid sensitivity in severe asthma and COPD. Curr Opin Pharmacol. 2019;46:73–81. doi: 10.1016/j.coph.2019.04.010 [DOI] [PubMed] [Google Scholar]
- 36.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79(1):351–379. doi: 10.1146/annurev-biochem-060308-103103 [DOI] [PubMed] [Google Scholar]
- 37.Zhang K, Liang Y, Feng Y, et al. Decreased epithelial and sputum miR-221-3p associates with airway eosinophilic inflammation and CXCL17 expression in asthma. Am J Physiol Lung Cell Mol Physiol. 2018;315(2):L253–L264. doi: 10.1152/ajplung.00567.2017 [DOI] [PubMed] [Google Scholar]
- 38.Zhang XH, Zhang YN, Li HB, et al. Overexpression of miR-125b, a novel regulator of innate immunity, in eosinophilic chronic rhinosinusitis with nasal polyps. Am J Respir Crit Care Med. 2012;185(2):140–151. doi: 10.1164/rccm.201103-0456OC [DOI] [PubMed] [Google Scholar]
- 39.Yang Y, Yin X, Yi J, Peng X. MiR-146a overexpression effectively improves experimental allergic conjunctivitis through regulating CD4(+)CD25(-)T cells. Biomed Pharmacother. 2017;94:937–943. doi: 10.1016/j.biopha.2017.07.157 [DOI] [PubMed] [Google Scholar]