Abstract
Tumor infiltrating lymphocytes play an important, but incompletely understood role in chemotherapy response and prognosis. In breast cancer, there appear to be distinct immune responses by subtype, but most studies have used limited numbers of protein markers or bulk sequencing of RNA to characterize immune response, in which spatial organization cannot be assessed. To identify immune phenotypes of Basal-like vs. Luminal breast cancer we used the GeoMx® (NanoString) platform to perform digital spatial profiling of immune-related proteins in tumor whole sections and tissue microarrays (TMA). Visualization of CD45, CD68, or pan-Cytokeratin by immunofluorescence was used to select regions of interest in formalin-fixed paraffin embedded tissue sections. 44 antibodies representing stromal markers and multiple immune cell types were applied to quantify the tumor microenvironment. In whole tumor slides, immune hot spots (CD45+) had increased expression of many immune markers, suggesting a diverse and robust immune response. In epithelium-enriched areas, immune signals were also detectable and varied by subtype, with Regulatory T cell (Treg) markers (CD4, CD25, FOXP3) being higher in Basal-like vs. Luminal breast cancer. Extending these findings to TMAs with more patients (n=75), we confirmed subtype-specific immune profiles, including enrichment of Treg markers in Basal-likes. This work demonstrated that immune responses can be detected in epithelium-rich tissue, and that TMAs are a viable approach for obtaining important immunoprofiling data. In addition, we found that immune marker expression is associated with breast cancer subtype, suggesting possible prognostic or targetable differences.
INTRODUCTION
Tumor infiltrating lymphocytes (TILs), play an important, but incompletely understood role in chemotherapy response and prognosis in breast cancer1, 2. For more aggressive types of breast cancer, such as HER2+ and triple negative (TNBC) histopathologic subtypes, a higher TILs score in the primary tumor is associated with improved prognosis and response to therapy, whereas there is little or no association between TILs and outcomes for ER+ disease3, 4, 5, 6. ER+ and Luminal disease seem to be less immunogenic, although there is some heterogeneity in immune gene expression7, 8, 9.
Data from prior studies indicate that spatial heterogeneity in TILs is associated with breast cancer subtypes and outcomes10, 11, 12, 13, 14. Specifically, the location of immune cells in intratumoral stroma and/or their proximity to tumor cells15 appear to correlate with prognosis in ER− tumors13, TNBC16 and ER+ tumors14. These profiling methods using histologic images showed the importance of studying the spatial heterogeneity of TILs but were limited by the small number of immunohistochemical markers used to estimate TIL abundance.
Larger scale immunoprofiling may elucidate the relationship between specific immune subpopulations and breast tumor subtype. Recent studies have leveraged bulk RNA sequencing to identify clinically-relevant immune phenotypes and evaluate how immune cells mediate chemotherapy response and immune checkpoint blockade17, 18, 19, 20, 21, 22, 23. In a gene expression analysis of approximately 11,000 breast tumors, Ali et al. investigated 22 subsets of immune cells and found that T regulatory cells (Tregs) and M0 and M2 macrophages were most strongly associated with poor outcome, regardless of ER status23. Although these studies in bulk tissue have uncovered prognostic immune cells and signatures, they lack resolution on intratumoral heterogeneity and the spatial context of immune cells across the tumor.
Recent studies biomarkers of response to immunotherapy in melanoma incorporated high-plex immune expression data with spatial information, using the GeoMx® (NanoString) digital spatial profiling (DSP technology24, 25, 26. They demonstrated that PD-L1 expression in CD68-positive cells was a predictive marker for progression-free survival, overall survival, and response to therapy24. Conversely, PD-L1 measured in tumor cells was not prognostic. A recent paper using the same high-plex platform found that elevated expression of HLA-DR in TNBC was associated with long-term disease-free survival. Specifically, HLA-DR protein expression in the epithelial compartment was a better discriminator of outcome than stromal expression of HLA-DR27. These findings underscore that both high-plex immune marker expression and tissue context may be important variables in immune-based prognostication.
We sought to evaluate differences in immune biomarker expression, while also considering tissue context, in a population-based study of breast cancer. Specifically, we sought to compare immune marker expression in epithelium-rich and CD45+ (immune cell) infiltrated ‘hot spots’ within the tumor according to breast cancer subtype. We also compared methods for capturing immune response based on whole slides and tissue microarrays. The resulting data provide technical insights into approaches for immune profiling and point to important immune differences among breast cancer subtypes.
MATERIALS AND METHODS
Study population and samples
The Carolina Breast Cancer Study (CBCS) is a population-based study of African American and Non-African American (98% Caucasian, referred to as White) women from 44 counties of central and eastern North Carolina conducted in three phases (phase I: 1993–1996; phase II: 1996–2001; and phase III: 2008–2013); study details and sampling schema have been described previously28, 29, 30, 31, 32. Briefly, cases were women ages 20 to 74 years, diagnosed with a first primary invasive breast cancer, and identified via rapid case ascertainment. Black and younger women (age <50) were oversampled. Race was determined by self-report and categorized as white or black. Tumor characteristics for cases (e.g., tumor size, grade, hormone receptor (HR+) status, node status, and stage) were abstracted from medical records and pathology reports. For this paper, we utilized patient samples from phase III (CBCS3), which recruited women between 2008 and 2013. Patients who provided informed consent completed a baseline questionnaire regarding personal characteristics, including socioeconomics, insurance status, health behaviors, and health history, in addition to the collection of patient tumor tissue, blood, and medical records. IHC for subtype classification was performed in a central laboratory, and designations have been previously described and were defined as follows: Luminal A is ER ≥ 10% or PR ≥ 10% and Ki67 < 7%, Luminal B is ER ≥ 10% or PR ≥ 10% and Ki67 ≥ 7%, HER2+ is ER < 10% and HER2 = 3, and Basal-like is ER < 10%, PR < 10%, and any EGFR or Ki67 positive signal. For Luminal subtypes, if Ki67 was missing, grade was substituted; grade ≤ 2 for Luminal A and grade = 3 for Luminal B 33, 34. The Ki67 cutoff for the samples we used in the CBCS was developed and reporting in Allott et al.34. They identified optimal Ki67 thresholds by generating receiver operative characteristics (ROC) curve among all Luminal tumors regardless of IHC-based HER2 status and applying the Youden method35 to maximize the sum of the sensitivity and specificity for PAM50-defined Luminal tumors. They identified an optimal Ki67 threshold of 7.1%34. The study was approved by the Office of Human Research Ethics/Institutional Review Board at the University of North Carolina at Chapel Hill, conducted in accordance with U.S. Common Rule. Written informed consent and HIPAA authorization were obtained from each participant.
Three Basal-like and three Luminal A samples were chosen to perform whole slide DSP. Samples had previously been cored for tissue microarray (TMA) based on H&E analysis of slides. H&E-stained TMAs were analyzed with the Aperio (Leica Biosystems, Wetzlar, GR) GENIE algorithm to determine immune cell infiltration. Selected samples had 1% or more immune infiltrate. One un-stained, TMA-cored, whole slide per sample was used for the DSP assay. Four TMA slides from CBCS3 were chosen for analysis, encompassing cores from the 6 tumors that were analyzed on whole slides (described above). These TMAs also included cores from 69 other tumors. Each TMA had 2-4 (with the vast majority having 4) cores per patient sample and included 11 to 27 patient samples.
Digital spatial profiling (DSP)
DSP was performed using the NanoString (Seattle, WA) GeoMx® platform36. Immunofluorescence for pan-Cytokeratin (tumor), CD45 (leukocyte), CD68 (macrophage), and a DNA stain (SYTO 83) were used to visualize tissue compartments and regions of interest (ROIs). DSP analysis included 61 oligonucleotide-conjugated antibodies, including antibodies for negative control IgGs and housekeeping proteins. All immunofluorescence antibodies and oligo-tagged antibodies were from Abcam (Cambridge, UK). After hybridization of the capture probes to FFPE slide-mounted tissues, the oligo tags were released from the ROIs via targeted ultraviolet radiation exposure, and then were counted in a Nanostring nCounter assay36.
For whole slides, the Basal-like and Luminal A slides each had 12 ROIs selected from areas that were either epithelium-rich or immune hot spots based on pan-Cytokeratin and CD45 immunofluorescence signal, respectively. No regions of uniform CD68 staining were identified, but CD68 staining was often diffusely evident in CD45-positive regions. ROI selection was performed with guidance from a board-certified pathologist. Each slide had 4 small ROIs (300 μm), 4 medium ROIs (500 μm), and 4 large ROIs (650 μm). For TMAs, we used 4 TMAs, representing a total of 75 patients each with 2-4 1-mm cores. The distribution of subtypes on these TMAs included 5 HER2-positive, 31 Luminal A, 21 Luminal B, 14 Basal-like, and 4 with missing IHC subtype calls33. The sample was roughly equally divided by race, with 37 Black patients and 38 non-Black patients. We again stained with immunofluorescent markers for CD45, CD68 and pan-Cytokeratin and selected regions of interest with >70% tumor cellularity, resulting in a total of 346 ROIs selected, with 1-3 ROIs per core. ROI sizes ranged from 100-μm to 650-μm in diameter. ROIs were selected to contain 70% epithelial (pan-Cytokeratin positive) cell content in a 650 μm circular ROI. If that was not possible, a smaller circular ROI (300-100 μm) with 70% tumor cell content was selected.
Data normalization and visualization
RCC files of multiplex data for ROIs were loaded into the DSP app developed by NanoString. Within the app, sample ROIs were visualized and normalized using several normalization options, including positive control normalization (ERCC internal spike-in controls), negative control normalization (mouse and rabbit IgGs), housekeeping control normalization (Ribosomal S6 protein and Histone H3), and ROI/ultraviolet-light mask area normalization. Raw and normalized data were analyzed by unsupervised hierarchical clustering and visualized in a heatmap within the app or exported for further visualization (described below). For the final normalization, digital counts from barcodes corresponding to protein probes were normalized to internal spike-in controls (ERCC) to account for technical variation. Counts were then normalized to ultraviolet-light mask area. The same normalization method was used for TMA analysis.
All raw and normalized data were visualized using relative log expression (RLE) plots, or boxplots of log2-transformed protein expression (for all markers, including tumor proteins, immune proteins, and housekeeping proteins). Data were also analyzed by principal component analysis (PCA). Visualization of raw and normalized data was performed for all ROIs for both whole slides and TMAs. Visual assessment of normalized data for whole slides and TMAs was similar for all normalization methods, but ERCC and area normalization reduced the technical variation associated with varying ROI size. All data visualization was performed in R version 3.6.137. After normalization, protein expression values were log2-transformed and median centered using Cluster 3.038. Centroid linkage hierarchical clustering of the log2-transformed, median-centered protein expression with was also done in Cluster 3.0 and visualized using Java Tree View39. Heatmap annotations were added in Adobe Illustrator.
Statistical analyses
Supervised protein expression analyses visualized in volcano plots were performed in R. The subtype comparisons are as follows, Basal versus Luminal A, Basal versus Luminal B, Luminal A versus Luminal B. First, Wilcoxon p-values were calculated for the difference in protein expression for each protein between subtypes. The p-values had FDR-BH adjustment, with a q < 0.05 marked as significant. The p-values were then -log10-transformed. Then, the fold change in protein expression values were calculated by taking the average expression of each protein per subtype and subtracting one subtype from the other. A dot plot was made using the -log10 q-values and fold change values For the volcano plots, each data point is represented by a black dot with annotation for markers with q < 0.05 and fold change > 2. Dashed lines indicate the cut off q < 0.05 and fold change < 2.
The GeoMx® ImmunoOncology Assay does not provide single-cell colocalization of markers, but many of the immune cell types that were evaluated had more than one marker in the assay. This facilitated the development of cell type scores (using the average or median of normalized expression values of the markers of interest), similar to the approach often used in bulk sequencing or expression analysis40, 41, 42, 43. Treg scores were calculated by taking the median of the log2-transformed values for CD4, CD25, and FOXP3 for each ROI42. For whole slides, the dataset was separated into immune hot spots and epithelium-enriched ROIs, and the Treg score was calculated for each ROI. For analysis of TMAs, the average of the log2-transformed values for each protein was first calculated from all ROIs available on a given sample. Treg scores were then calculated for each sample. In an exploratory analysis, we used a ROC curve to illustrate the potential ability of the Treg score to distinguish Basal-like from Luminal A tumors. ROC curve analysis was performed in R using the Epi package44, and sensitivity was calculated. Treg score boxplots were visualized in R using ggplot245, and Wilcoxon p-values were calculated for subtype comparisons. For whole slides, immune score and p-value were stratified by dataset (immune hot spot or epithelium-enriched region).
Quality control assessment
Although immune marker expression patterns were similar in whole slides and TMAs, we formally assessed within sample agreement in whole slides versus TMAs. We compared four of the six samples used for both whole slides and TMAs, as two samples only had one epithelium-enriched ROI in the whole slides. Images of 9 epithelium-enriched ROIs measured on whole slides and on TMAs for a given sample (Sample A) are shown in Supplemental Figures 1A and B, respectively. We examined the variability in CD45 expression between each ROI and saw that expression was similar on whole slide and TMA (Supplemental Figure 1C). In Supplemental Figure 1D, we show a density plot of standard deviation across all 44 stromal and immune markers (Table 1). The standard deviation of expression for each protein was calculated as follows: for each sample, the standard deviation of single proteins was calculated for all the ROIs for the sample. Once the standard deviation was calculated for all proteins for all samples, we visualized the distribution of these standard deviations using ggplot2 density plots in R37. The distributions of standard deviation obtained from whole slides and TMAs were similar (Supplemental Figure 1D).
Table 1.
Complete list of 44 markers used in NanoString GeoMx® Digital Spatial Profiling (DSP).
| Immune, Stroma, and Tumor Markers | Housekeeping | Negative Control | ||||
|---|---|---|---|---|---|---|
| 4-1BB | CD20 | CD45 | FAPalpha | OX40L | GAPDH | MsIgG1 |
| ARG1 | CD25 | CD45RO | Fibronectin | PanCk | HistoneH3 | MsIgG2a |
| B7-H3 | CD27 | CD56 | FOXP3 | PD-1 | S6 | RbIgG |
| B2M | CD3 | CD66b | GITR | PD-L1 | ||
| CD11c | CD34 | CD68 | GZMB | PD-L2 | ||
| CD127 | CD4 | CD8 | HLA-DR | SMA | ||
| CD14 | CD40 | CD80 | ICOS | STING | ||
| CD163 | CD44 | CTLA4 | IDO1 | Tim-3 | ||
| Ki-67 | LAG3 | VISTA | ||||
Across all four samples, intraclass correlation coefficients (ICCs) were good to excellent between single sample ROIs in whole slides (0.837-0.949) and in TMAs (0.844-0.938). Concordance was also excellent between TMAs and slides (0.829-0.916). Our results suggest that efficient immunoprofiling processing of hundreds of samples using TMAs is feasible, due to similarity in expression profiles between whole tissue and TMAs when focusing on epithelium-enriched regions. For calculating the ICC within either whole slide ROIs per sample only or TMA ROIs per sample only, a one-way random effects model was chosen, with single unit measures and assaying agreement. For comparing the average protein expression for a sample in slides versus TMAs, a test-retest approach was used. The one-way random effects model was chosen, with average measures and assaying agreement.
For validation of ROI selection criteria, the expression of two B-cell lineage-specific antigens, CD19 and CD20, was compared. These markers are expressed on the surface of mature B-cells and are often used together to measure human B-cell populations46. In addition, tumor-infiltrating B-cells in breast cancer have been associated with improved clinical outcomes20, 47, and may provide an interesting avenue of future biomarker study in the CBCS. CD19 expression was analyzed with by immunofluorescence (IF) (PA0843, Leica, RTU) on the TMAs used for DSP analysis. The log2-transformed total CD19-positive cells counts were compared to log2-transformed, normalized CD20 values from DSP. The correlation between the expression of the two separate B-cell markers analyzed with different methodologies is shown in Supplemental Figure 1E (R squared = 0.3244, p < 0.0001). In addition to the CD19 vs. CD20 analysis, we confirmed correlations between the multiplexed protein quantitation and standard IHC for ER, PR, and HER2 (ER, r=0.978; PR, r=0.934; HER2, r=0.993).
RESULTS
Whole slide analysis of intratumoral immune marker heterogeneity
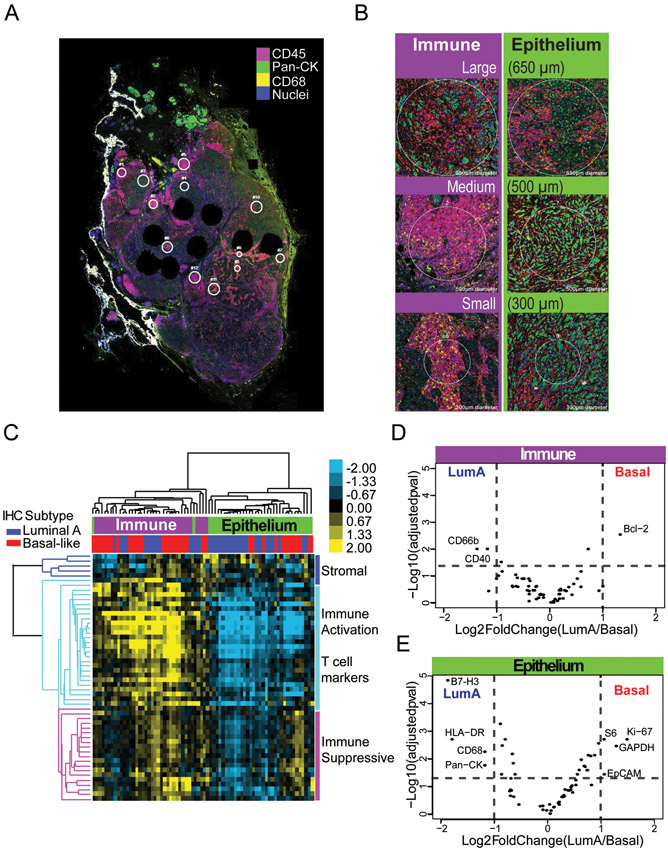
To measure differences in immune marker expression on whole slides, we selected 6 whole tumor slides with evidence of >1% infiltrating immune cells, including three invasive breast cancers of Basal-like and three of the Luminal A subtype (as defined by central IHC)33. A board-certified pathologist (B.C.C.), identified 12 regions of interest (ROIs) per slide, including areas heavily infiltrated with CD45 cells (i.e. “immune hot spots”) and areas with little evidence of CD45-positive immune infiltration (“epithelium-enriched regions”). Figure 1 shows selected ROIs with CD45 (leukocytes), CD68 (macrophages), and pan-Cytokeratin (tumor cells) immunofluorescence markers to visualize regions.
Figure 1). Subtype immune marker heterogeneity is apparent in epithelium-enriched regions versus immune hot spots.
(a) Representative whole tumor slide stained with CD45 (red), CD68 (yellow), and pan-Cytokeratin (green), in addition to 61 oligo-conjugated antibodies for immune and tumor cell markers. (b) Regions of interest (ROIs) were selected based on cellularity, large (650 μm), medium (500 μm), and small (300 μm). (c) Heatmap of protein expression for whole slide dataset with immunohistochemistry (IHC) subtype and cellularity labeled. Protein class clusters are denoted by colored bars and branches, with dark blue denoting stromal proteins, light blue denoting T cell and immune activation markers, and pink denoting immunosuppressive markers. (d-e) Volcano plots for Basal-like vs Luminal A subtypes were run separately for D) immune hot spot ROIs and E) epithelium-enriched ROIs. Data points are represented as black dots with annotation for markers with q < 0.05 and fold change > 2. Dashed lines indicate the cut off q < 0.05 and fold change < 2.
Using NanoString GeoMx® DSP, 44 targets for immune and stromal markers (Table 1), with an additional 11 tumor and proliferation markers, were measured. After normalization to control for ROI size, unsupervised hierarchical clustering was performed and heatmap visualization revealed two main clusters of protein expression, largely split by whether CD45/CD68 levels were high (indicating immune ‘hot spot’) or pan-Cytokeratin levels were high (indicating epithelium-enrichment) (Figure 1C).
To assess whether there were differences in immune profile by breast cancer subtype, we performed supervised analysis. Within immune hot spots, there were few significant differences between Basal-like and Luminal A tumors (Figure 1D), whereas in epithelium-enriched regions, there were substantial differences in immune marker expression by subtype (Figure 1E). In epithelium-enriched regions, suppressive immune markers and markers of proliferation were significantly more highly expressed in Basal-like tumors, whereas in immune hot spots, only a single anti-apoptotic marker (Bcl-2) was more highly expressed in Basal-like tumors.
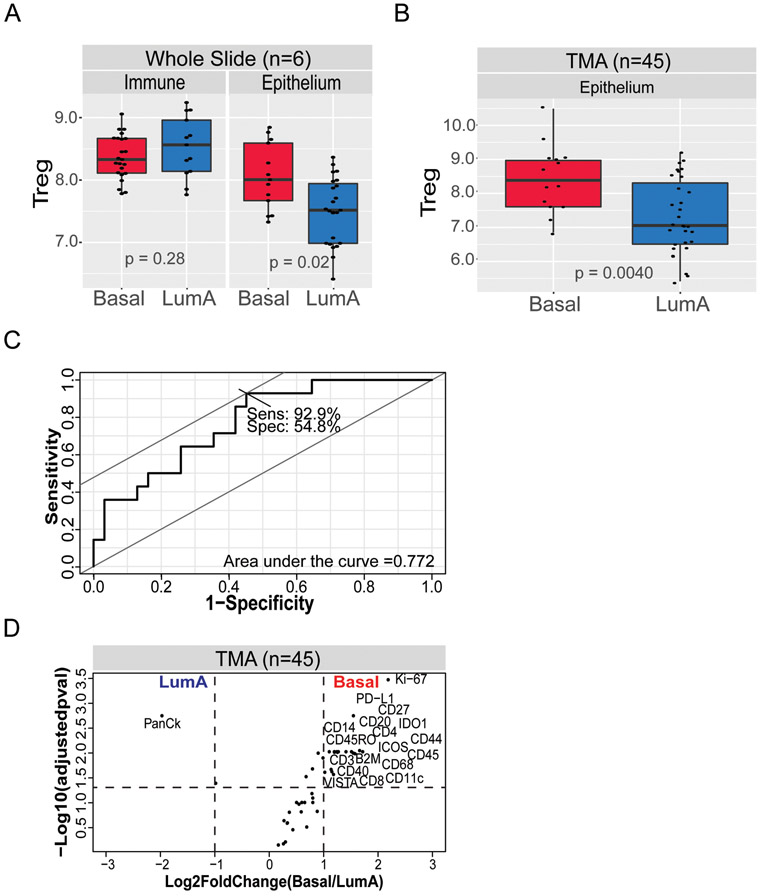
Many of the immune cell types that were evaluated had multiple markers GeoMx® ImmunoOncology Assay, facilitating the development of cell type scores to estimate the abundance of immune cell populations in the tumor microenvironment. We assessed various immune cell scores based on the median expression across multiple, related immune markers. For example, the median expression of CD8 and Granzyme B (GZMB) was used to create a CD8+ T-cell score. Within immune hot spots, there were no significant differences in the value of scores for CD8+ T-cell, B-cells, macrophages, dendritic cells, Tregs across tumor subtypes. However, within epithelium-enriched regions, Treg scores (based on CD4, CD25, and FOXP3 protein expression) were significantly higher in Basal-like samples compared to Luminal A tumors (Figure 2A).
Figure 2). Treg marker expression is higher in Basal-like tumors.
(a) T regulatory (Treg) signatures were higher in Basal-like tumors compared to Luminal A tumors only in epithelium-enriched (p = 0.02) areas but not in immune-high regions (p = 0.86). (b) Higher Treg signature expression in Basal-like tumors in tissue microarrays (TMAs) (p = 0.0078). (c) Receiver operating characteristic (ROC) analysis shows 92.9% sensitivity in Basal-like versus Luminal A classification based on Treg signature expression. (d) Significant differences in immune marker expression between Luminal A and Basal-like tumors in TMAs. Volcano plot of Basal-like versus Luminal A. Data points are represented as black dots with annotation for markers with q < 0.05 and fold change > 2. Dashed lines indicate the cut off q < 0.05 and fold change < 2.
Scaling measurement of epithelium-enriched immune expression to TMAs
To confirm these patterns within a larger dataset, we extended the DSP technology to tissue microarrays (TMAs). Comparing 14 Basal-like to 31 Luminal A cases, we identified immune profiles associated with each subtype. Similar to the findings with the whole slides, the Treg multi-marker score was significantly higher in Basal-like versus Luminal A tumors (Figure 2B). Furthermore, the Treg score discriminated between Basal-like and Luminal A cases with 92.9% sensitivity in a ROC analysis (Figure 2C). We next performed supervised analysis of immune marker expression by subtype and found significant upregulation of several other immune markers in Basal-like versus Luminal A (Figure 2D). While the emphasis of this study was on identifying markers for Basal-like immune response, we also explored qualitative differences in immune marker expression between Luminal A and Luminal B tumors and high risk of recurrence vs. low risk of recurrence (ROR-P) (Supplemental Figure 2), though sample size and power were limited. We observed a tendency towards higher immune marker expression in Luminal B and high risk of recurrence tumors.
DISCUSSION
Studying immune marker expression in both whole slides and TMAs, we found that epithelium-enriched regions show significant differences in immune marker expression by breast cancer subtype. Compared to Luminal A tumors, Basal-like breast cancers have higher expression of Treg markers, as well as a number of other immune and proliferation markers. In contrast, the ability to detect subtype-specific immune marker expression appeared to be obscured in areas with overall high immune marker expression. This suggests that in bulk tissue studies, some subtype-specific differences may be obscured due to the complex patterns of immune response in the whole section.
Our results validate a number of studies showing that Tregs are more highly infiltrated in triple-negative and Basal-like breast cancer48, 49, 50, 51, 52, 53, 54. Interestingly, we did not observe Treg differences in immune hot spots. This may provide an explanation for null findings and/or lack of focus on Tregs in previous bulk tissue studies of breast cancer, which have instead focusing on CD8+ T cells and macrophages8, 23, 55. Treg differences may have been missed in those studies if hot spots and epithelial rich sections are averaged. To contextualize our finding about the importance of Tregs in Basal-like cancers, we note that infiltrating Tregs, marked by their expression of Forkhead box protein P3 (FOXP3), have been reported to be prognostic in breast cancer56. Specifically the presence of Tregs is a poor prognostic indicator in ER+ breast cancer, but a favorable prognostic factor in HER2+/ER− disease56.
There were several strengths of this study, both technical and substantive. First, our data preserved spatial context and simultaneously evaluated multiple immune biomarkers. Interestingly, both context and multiplex may be important; the combination of Treg markers as a score showed important subtype-specific differences, even though not all the individual markers for Tregs were significantly different. These data help validate a novel method and identify sampling strategies for future immunoprofiling of breast cancers. Second, we also technically confirmed correlations between the multiplexed protein quantitation and standard IHC for ER, PR, and HER2 and evaluated intratumoral heterogeneity to inform our sampling strategy. We were able to compare whole slides and TMAs from the same breast cancers and confirm that TMAs are appropriate for studying the tumor immune microenvironment. Our results also align with at least one other study of breast cancer27, 57 in showing that epithelium-enriched regions show important immune response information. Third, we used a well-annotated data source representing a diversity of patients. Finally, despite a limited sample size, we were able to show significant differences in Basal-like vs. Luminal breast cancer, implying that future studies with larger datasets will provide even greater understanding of differences.
There were some limitations of our analysis. We recognize that this approach does not provide evidence of marker co-localization studies, which is partially offset by using multiple markers58, 59, 60, 61, 62, 63 (e.g. Treg markers CD4, CD25, and FOXP3) as a proxy for a single cell type. We also were unable to compare RNA-based molecular subtypes because data were not available for 35% of cases on our TMAs. We also lacked power to assess differences by race and age. Future expanded analyses should evaluate differences in immune response by race and in association with breast cancer progression. Previous studies in The Cancer Genome Atlas (TCGA) did not find strong differences in the breast cancer immune microenvironment by race, but this research was also limited by a small number of Black patients64. We performed ROC curve analysis to test whether an immune score, such as the Treg score, has value in predicting subtype, but we acknowledge that we have limited sample size and therefore these analyses may not be stable estimates. Finally, we acknowledge that we were unable to use ROI masking effectively. Previous studies have used masking to profile immune hot spots24, 25, however this type of masking is impractical in breast tumors, where regions of immune infiltration tend to be segregated by stromal or epithelial components.
In summary, this work demonstrated that TMAs are a viable approach for immunoprofiling, providing resolution that would be missed in single-marker studies and in bulk tissue studies. This finding is particularly impactful for large cancer epidemiological studies, which often have scarce whole slide tissue and/or only retained TMAs with epithelium-enriched cores. This work represents a first step toward developing feasible immunoprofiling approaches that could be conducted on a large scale, and ultimately combined with genomic datasets, to improve breast cancer prognostics and identify new therapeutic opportunities.
Supplementary Material
ACKNOWLEDGEMENTS AND FUNDING
Thank you to the Lineberger Comprehensive Cancer Center, a Cancer Center Support Grant (P30 CA016086), the UNC-CH Cancer Control Education Program (5T32CA057726-28), Susan G Komen for the Cure (OGUNC1202), the North Carolina University Cancer Research Fund, and the NCI-funded UNC Breast SPORE (P50 CA058223). Thank you to Liang and Erica from Nanostring. We would also like to thank the Carolina Breast Cancer Study participants and staff. We also want to acknowledge Robert C. Millikan, founder of the Carolina Breast Cancer Study Phase 3.
Footnotes
CONFLICT OF INTEREST
Dr. Calhoun is a member of the Oncology Advisory Board for Luminex Corp
REFERENCES
- 1.Denkert C, von Minckwitz G, Darb-Esfahani S, Lederer B, Heppner BI, Weber KE, et al. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: a pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol 2018;19:40–50. [DOI] [PubMed] [Google Scholar]
- 2.Salgado R, Denkert C, Demaria S, Sirtaine N, Klauschen F, Pruneri G, et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann Oncol 2014;26:259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luen SJ, Salgado R, Fox S, Savas P, Eng-Wong J, Clark E, et al. Tumour-infiltrating lymphocytes in advanced HER2-positive breast cancer treated with pertuzumab or placebo in addition to trastuzumab and docetaxel: a retrospective analysis of the CLEOPATRA study. Lancet Oncol 2017;18:52–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loi S, Sirtaine N, Piette F, Salgado R, Viale G, Van Eenoo F, et al. Prognostic and Predictive Value of Tumor-Infiltrating Lymphocytes in a Phase III Randomized Adjuvant Breast Cancer Trial in Node-Positive Breast Cancer Comparing the Addition of Docetaxel to Doxorubicin With Doxorubicin-Based Chemotherapy: BIG 02-98. J Clin Oncol 2013;31:860–867. [DOI] [PubMed] [Google Scholar]
- 5.Watanabe T, Hida AI, Inoue N, Imamura M, Fujimoto Y, Akazawa K, et al. Abundant tumor infiltrating lymphocytes after primary systemic chemotherapy predicts poor prognosis in estrogen receptor-positive/HER2-negative breast cancers. Breast Cancer Res Treat 2018;168:135–145. [DOI] [PubMed] [Google Scholar]
- 6.Hida AI, Sagara Y, Yotsumoto D, Kanemitsu S, Kawano J, Baba S, et al. Prognostic and predictive impacts of tumor-infiltrating lymphocytes differ between Triple-negative and HER2-positive breast cancers treated with standard systemic therapies. Breast Cancer Res Treat 2016;158:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luen S, Virassamy B, Savas P, Salgado R, Loi S. The genomic landscape of breast cancer and its interaction with host immunity. Breast 2016;29:241–250. [DOI] [PubMed] [Google Scholar]
- 8.Zhu B, Tse LA, Wang D, Koka H, Zhang T, Abubakar M, et al. Immune gene expression profiling reveals heterogeneity in luminal breast tumors. Breast Cancer Res 2019;21:147–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Al-Saleh K, Abd El-Aziz N, Ali A, Abozeed W, Abd El-Warith A, Ibraheem A, et al. Predictive and prognostic significance of CD8(+) tumor-infiltrating lymphocytes in patients with luminal B/HER 2 negative breast cancer treated with neoadjuvant chemotherapy. Oncol Lett 2017;14:337–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bera K, Schalper KA, Rimm DL, Velcheti V, Madabhushi A. Artificial intelligence in digital pathology - new tools for diagnosis and precision oncology. Nat Rev Clin Oncol 2019;16:703–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saltz J, Gupta R, Hou L, Kurc T, Singh P, Nguyen V, et al. Spatial Organization and Molecular Correlation of Tumor-Infiltrating Lymphocytes Using Deep Learning on Pathology Images. Cell Rep 2018;23:181–193.e187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan Y Spatial Heterogeneity in the Tumor Microenvironment. Cold Spring Harb Perspect Med 2016;6:a026583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nawaz S, Heindl A, Koelble K, Yuan Y. Beyond immune density: critical role of spatial heterogeneity in estrogen receptor-negative breast cancer. Mod Pathol 2015;28:766–777. [DOI] [PubMed] [Google Scholar]
- 14.Heindl A, Sestak I, Naidoo K, Cuzick J, Dowsett M, Yuan Y. Relevance of Spatial Heterogeneity of Immune Infiltration for Predicting Risk of Recurrence After Endocrine Therapy of ER+ Breast Cancer. J Natl Cancer Inst 2018;110: 10.1093/jnci/djx1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ali HR, Provenzano E, Dawson SJ, Blows FM, Liu B, Shah M, et al. Association between CD8+ T-cell infiltration and breast cancer survival in 12 439 patients. Ann Oncol 2014;25:1536–1543. [DOI] [PubMed] [Google Scholar]
- 16.Hida AI, Watanabe T, Sagara Y, Kashiwaba M, Sagara Y, Aogi K, et al. Diffuse distribution of tumor-infiltrating lymphocytes is a marker for better prognosis and chemotherapeutic effect in triple-negative breast cancer. Breast Cancer Res Treat 2019;178:283–294. [DOI] [PubMed] [Google Scholar]
- 17.Danaher P, Warren S, Lu R, Samayoa J, Sullivan A, Pekker I, et al. Pan-cancer adaptive immune resistance as defined by the Tumor Inflammation Signature (TIS): results from The Cancer Genome Atlas (TCGA). J Immunother Cancer 2018;6:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, et al. The Immune Landscape of Cancer. Immunity 2018;48:812–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shahamatdar S, He MX, Reyna MA, Gusev A, AlDubayan SH, Van Allen EM, et al. Germline Features Associated with Immune Infiltration in Solid Tumors. Cell Rep 2020;30:2900–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hollern DP, Xu N, Thennavan A, Glodowski C, Garcia-Recio S, Mott KR, et al. B Cells and T Follicular Helper Cells Mediate Response to Checkpoint Inhibitors in High Mutation Burden Mouse Models of Breast Cancer. Cell 2019;179:1191–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tekpli X, Lien T, Røssevold AH, Nebdal D, Borgen E, Ohnstad HO, et al. An independent poor-prognosis subtype of breast cancer defined by a distinct tumor immune microenvironment. Nat Comm 2019;10:5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anurag M, Zhu M, Huang C, Vasaikar S, Wang J, Hoog J, et al. Immune Checkpoint Profiles in Luminal B Breast Cancer (Alliance). J Natl Cancer Inst 2020;112:737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ali HR, Chlon L, Pharoah PDP, Markowetz F, Caldas C. Patterns of Immune Infiltration in Breast Cancer and Their Clinical Implications: A Gene-Expression-Based Retrospective Study. PLoS Med 2016;13:e1002194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toki MI, Merritt CR, Wong PF, Smithy JW, Kluger HM, Syrigos KN, et al. High-Plex Predictive Marker Discovery for Melanoma Immunotherapy–Treated Patients Using Digital Spatial Profiling. Clin Cancer Res 2019;25:5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blank CU, Rozeman EA, Fanchi LF, Sikorska K, van de Wiel B, Kvistborg P, et al. Neoadjuvant versus adjuvant ipilimumab plus nivolumab in macroscopic stage III melanoma. Nat Med 2018;24:1655–1661. [DOI] [PubMed] [Google Scholar]
- 26.Amaria RN, Reddy SM, Tawbi HA, Davies MA, Ross MI, Glitza IC, et al. Neoadjuvant immune checkpoint blockade in high-risk resectable melanoma. Nat Med 2018;24:1649–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stewart RL, Matynia AP, Factor RE, Varley KE. Spatially-resolved quantification of proteins in triple negative breast cancers reveals differences in the immune microenvironment associated with prognosis. Sci Rep 2020;10:6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Millikan RC, Newman B, Tse CK, Moorman PG, Conway K, Dressler LG, et al. Epidemiology of basal-like breast cancer. Breast Cancer Res Treat 2008;109:123–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, et al. Race, Breast Cancer Subtypes, and Survival in the Carolina Breast Cancer Study. JAMA 2006;295:2492–2502. [DOI] [PubMed] [Google Scholar]
- 30.Newman B, Moorman PG, Millikan R, Qaqish BF, Geradts J, Aldrich TE, et al. The Carolina Breast Cancer Study: integrating population-based epidemiology and molecular biology. Breast Cancer Res Treat 1995;35:51–60. [DOI] [PubMed] [Google Scholar]
- 31.Hair BY, Hayes S, Tse C-K, Bell MB, Olshan AF. Racial differences in physical activity among breast cancer survivors: implications for breast cancer care. Cancer 2014;120:2174–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wheeler SB, Spencer J, Pinheiro LC, Murphy CC, Earp JA, Carey L, et al. Endocrine Therapy Nonadherence and Discontinuation in Black and White Women. J Natl Cancer Inst 2019;111:498–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allott EH, Cohen SM, Geradts J, Sun X, Khoury T, Bshara W, et al. Performance of Three-Biomarker Immunohistochemistry for Intrinsic Breast Cancer Subtyping in the AMBER Consortium. Cancer Epidemiol Biomarkers Prev 2016;25:470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allott EH, Geradts J, Cohen SM, Khoury T, Zirpoli GR, Bshara W, et al. Frequency of breast cancer subtypes among African American women in the AMBER consortium. Breast Cancer Res 2018;20:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruopp MD, Perkins NJ, Whitcomb BW, Schisterman EF. Youden Index and optimal cut-point estimated from observations affected by a lower limit of detection. Biometrical journal. Biometrische Zeitschrift 2008;50:419–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merritt CR, Ong GT, Church S, Barker K, Geiss G, Hoang M, et al. High multiplex, digital spatial profiling of proteins and RNA in fixed tissue using genomic detection methods. bioRxiv 2019:559021. [DOI] [PubMed] [Google Scholar]
- 37.Team RC. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. 2013. [Google Scholar]
- 38.de Hoon MJL, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics 2004;20:1453–1454. [DOI] [PubMed] [Google Scholar]
- 39.Saldanha AJ. Java Treeview—extensible visualization of microarray data. Bioinformatics 2004;20:3246–3248. [DOI] [PubMed] [Google Scholar]
- 40.Iglesia MD, Parker JS, Hoadley KA, Serody JS, Perou CM, Vincent BG. Genomic Analysis of Immune Cell Infiltrates Across 11 Tumor Types. J Natl Cancer Inst 2016;108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf Anna C, et al. Spatiotemporal Dynamics of Intratumoral Immune Cells Reveal the Immune Landscape in Human Cancer. Immunity 2013;39:782–795. [DOI] [PubMed] [Google Scholar]
- 42.Danaher P, Warren S, Dennis L, D’Amico L, White A, Disis ML, et al. Gene expression markers of Tumor Infiltrating Leukocytes. J Immunother Cancer 2017;5:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiménez-Sánchez A, Cast O, Miller ML. Comprehensive Benchmarking and Integration of Tumor Microenvironment Cell Estimation Methods. Cancer Res 2019;79:6238–6246. [DOI] [PubMed] [Google Scholar]
- 44.Carstensen BPM, Laara E, Hills M. Epi: A Package for Statistical Analysis in Epidemiology. Vol. R package version 2.40, 2019. [Google Scholar]
- 45.H W. ggplot2:Elegant Graphics for Data Analysis. New York: Springer-Verlag, 2016. [Google Scholar]
- 46.Maecker HT, McCoy JP, Nussenblatt R. Standardizing immunophenotyping for the Human Immunology Project. Nat Rev Immunol 2012;12:191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garaud S, Buisseret L, Solinas C, Gu-Trantien C, de Wind A, Van den Eynden G, et al. Tumor infiltrating B-cells signal functional humoral immune responses in breast cancer. JCI Insight 2019;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Droeser R, Zlobec I, Kilic E, Güth U, Heberer M, Spagnoli G, et al. Differential pattern and prognostic significance of CD4+, FOXP3+ and IL-17+tumor infiltrating lymphocytes in ductal and lobular breast cancers. BMC Cancer 2012;12:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miyan M, Schmidt-Mende J, Kiessling R, Poschke I, de Boniface J. Differential tumor infiltration by T-cells characterizes intrinsic molecular subtypes in breast cancer. J Transl Med 2016;14:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu F, Lang R, Zhao J, Zhang X, Pringle GA, Fan Y, et al. CD8(+) cytotoxic T cell and FOXP3(+) regulatory T cell infiltration in relation to breast cancer survival and molecular subtypes. Breast Cancer Res Treat 2011;130:645–655. [DOI] [PubMed] [Google Scholar]
- 51.Glajcar A, Szpor J, Hodorowicz-Zaniewska D, Tyrak KE, Okoń K. The composition of T cell infiltrates varies in primary invasive breast cancer of different molecular subtypes as well as according to tumor size and nodal status. Virchows Archiv 2019;475:13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taylor NA, Vick SC, Iglesia MD, Brickey WJ, Midkiff BR, McKinnon KP, et al. Treg depletion potentiates checkpoint inhibition in claudin-low breast cancer. J Clin Invest 2017;127:3472–3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Plitas G, Konopacki C, Wu K, Bos PD, Morrow M, Putintseva EV, et al. Regulatory T Cells Exhibit Distinct Features in Human Breast Cancer. Immunity 2016;45:1122–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bohling SD, Allison KH. Immunosuppressive regulatory T cells are associated with aggressive breast cancer phenotypes: a potential therapeutic target. Mod Pathol 2008;21:1527–1532. [DOI] [PubMed] [Google Scholar]
- 55.Quigley DA, Tahiri A, Lüders T, Riis MH, Balmain A, Børresen-Dale A-L, et al. Age, estrogen, and immune response in breast adenocarcinoma and adjacent normal tissue. OncoImmunology 2017;6:e1356142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu S, Foulkes WD, Leung S, Gao D, Lau S, Kos Z, et al. Prognostic significance of FOXP3+ tumor-infiltrating lymphocytes in breast cancer depends on estrogen receptor and human epidermal growth factor receptor-2 expression status and concurrent cytotoxic T-cell infiltration. Breast Cancer Res 2014;16:432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gruosso T, Gigoux M, Manem VSK, Bertos N, Zuo D, Perlitch I, et al. Spatially distinct tumor immune microenvironments stratify triple-negative breast cancers. J Clin Invest 2019;129:1785–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Preston CC, Maurer MJ, Oberg AL, Visscher DW, Kalli KR, Hartmann LC, et al. The ratios of CD8+ T cells to CD4+CD25+ FOXP3+ and FOXP3− T cells correlate with poor clinical outcome in human serous ovarian cancer. PLoS One 2013;8:e80063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Preston CC, Goode EL, Hartmann LC, Kalli KR, Knutson KL. Immunity and immune suppression in human ovarian cancer. Immunotherapy 2011;3:539–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kryczek I, Liu R, Wang G, Wu K, Shu X, Szeliga W, et al. FOXP3 defines regulatory T cells in human tumor and autoimmune disease. Cancer Res 2009;69:3995–4000. [DOI] [PubMed] [Google Scholar]
- 61.Shevach EM. CD4+ CD25+ suppressor T cells: more questions than answers. Nat Rev Immunol 2002;2:389–400. [DOI] [PubMed] [Google Scholar]
- 62.Sakaguchi S Naturally arising Foxp3-expressing CD25+CD4+ regulatory T cells in immunological tolerance to self and non-self. Nat Immunol 2005;6:345–352. [DOI] [PubMed] [Google Scholar]
- 63.von Herrath MG, Harrison LC. Antigen-induced regulatory T cells in autoimmunity. Nat Rev Immunol 2003;3:223–232. [DOI] [PubMed] [Google Scholar]
- 64.O'Meara T, Safonov A, Casadevall D, Qing T, Silber A, Killelea B, et al. Immune microenvironment of triple-negative breast cancer in African-American and Caucasian women. Breast Cancer Res Treat 2019;175:247–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.