Abstract
How cells respond to DNA damage is key to maintaining genome integrity or facilitating genetic change. In fungi, DNA damage responses have been extensively characterized in the model budding yeast Saccharomyces cerevisiae, which is generally not pathogenic. However, it is not clear how closely these responses resemble those in fungal pathogens, in which genetic change plays an important role in the evolutionary arms race between pathogen and host and the evolution of antifungal drug resistance. A close relative of S. cerevisiae, Candida glabrata, is an opportunistic pathogen that displays high variability in chromosome structure among clinical isolates and rapidly evolves antifungal drug resistance. The mechanisms facilitating such genomic flexibility and evolvability in this organism are unknown. Recently we characterized the DNA damage response of C. glabrata and identified several features that distinguish it from the well characterized DNA damage response of S. cerevisiae. First, we discovered that, in contrast to the established paradigm, C. glabrata effector kinase Rad53 is not hyperphosphorylated upon DNA damage. We also uncovered evidence of an attenuated DNA damage checkpoint response, wherein in the presence of DNA damage C. glabrata cells did not accumulate in S-phase and proceeded with cell division, leading to aberrant mitoses and cell death. Finally, we identified evidence of transcriptional rewiring of the DNA damage response of C. glabrata relative to S. cerevisiae, including an upregulation of genes involved in mating and meiosis – processes that have not been reported in C. glabrata. Together, these results open new possibilities and raise tantalizing questions of how this major fungal pathogen facilitates genetic change.
Keywords: Candida glabrata, genome stability, DNA damage response, DNA damage checkpoint, Rad53, HO endonuclease, mating, meiosis
Candida glabrata – a major fungal pathogen with high chromosomal variability and rapid evolution of drug resistance
An ability to regulate the capacity for genetic change, or evolvability, is especially important for pathogenic microbes that exist in the frequently changing environment of the host. Fungal pathogens are well known for their capacity to rapidly adapt to environmental pressures, such as cytotoxic antifungal drugs, by inducing genetic variants (e.g. aneuploidies and segmental duplications) with increased drug tolerance (Coste, et al. 2007, Selmecki, et al. 2006, Selmecki, et al. 2009). Genetic variation is likewise induced by passaging fungi through a mammalian host, likely in response to the host’s immune defense mechanisms (Ene, et al. 2018, Forche, et al. 2018). These phenomena have been largely described in diploid and polyploid fungi, such as Candida albicans and Cryptococcus neoformans (Bennett, et al. 2014). However, haploid fungi also show high capacity for generating genetic variation. Candida (Nakaseomyces) glabrata is a haploid commensal and opportunistic pathogen fungus closely related to baker’s yeast Saccharomyces cerevisiae (Shen, et al. 2018). C. glabrata is currently the second most prevalent cause of invasive candidiasis in North America and Europe, and its incidence is rising due to its reduced intrinsic susceptibility to azole class antifungals, which have been in wide clinical use for the last 40 years, and its ability to rapidly evolve drug resistance (Barber, et al. 2019, Bizerra, et al. 2014, Bordallo-Cardona, et al. 2017, Perlin, et al. 2017). Interestingly, C. glabrata clinical isolates display highly variable chromosome structures (karyotypes) due to translocations and copy number variation (Carrete, et al. 2018, Healey, et al. 2016, Lopez-Fuentes, et al. 2018, Muller, et al. 2009, Polakova, et al. 2009, Shin, et al. 2007). It is thought that the propensity of C. glabrata to rapidly evolve drug resistance is related to its ability to generate and tolerate extensive genetic variation; however, the mechanisms enabling this variation are so far unknown.
Attenuated DNA damage checkpoint signaling in C. glabrata
How cells respond to DNA damage is at the crux of maintenance of genome integrity. Eukaryotic cells have evolved intricate DNA damage checkpoint programs, which, among other things, slow down S-phase and prevent cell division in the presence of damaged DNA (Branzei and Foiani 2010, Friedel, et al. 2009). Defective checkpoint responses, for example in cancer cells, are associated with increased genome instability (Lobrich and Jeggo 2007), and in S. cerevisiae mutations in checkpoint genes greatly increase chromosome rearrangements, producing the variable karyotypes reminiscent of C. glabrata clinical isolates (Myung, et al. 2001, Serero, et al. 2014).
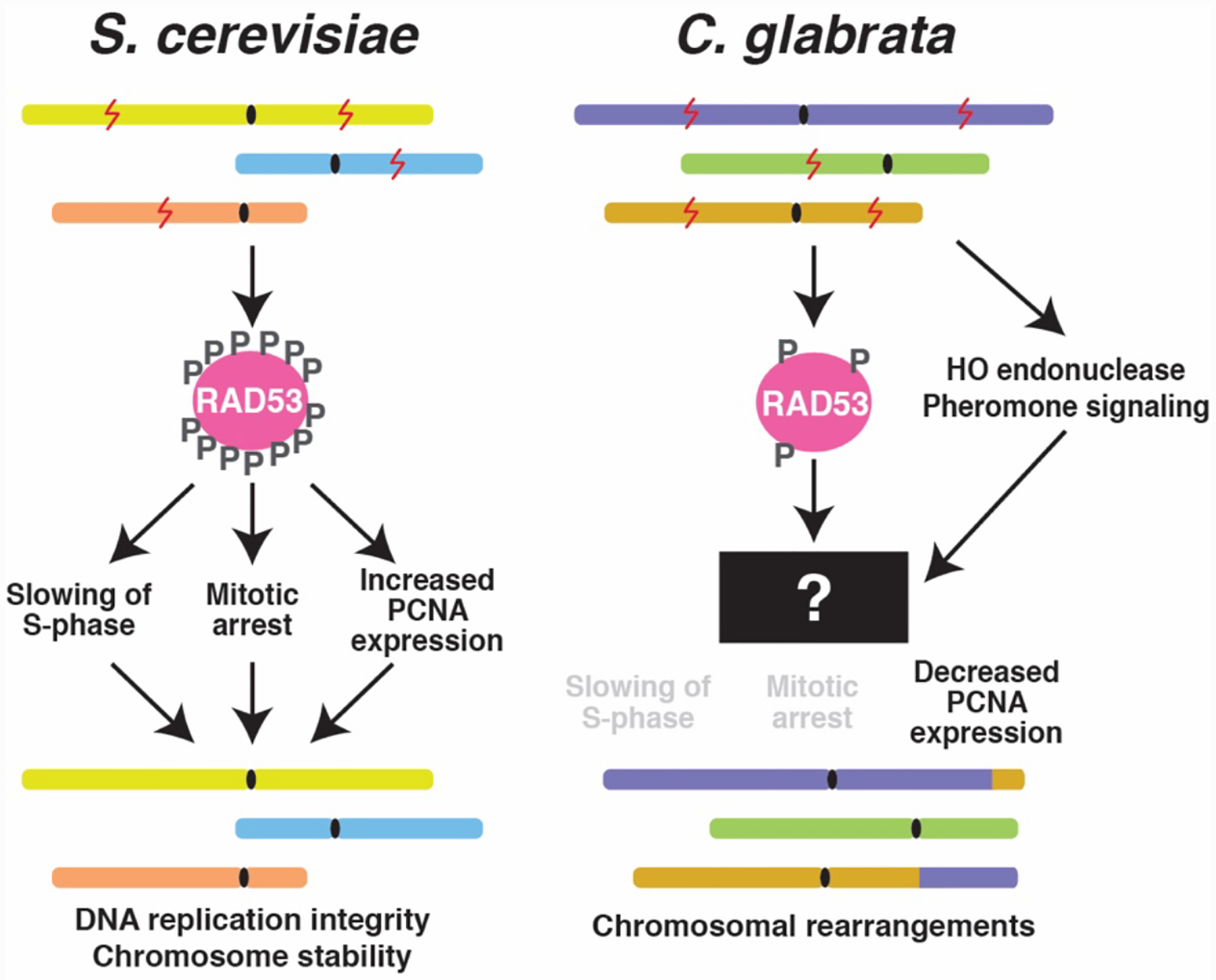
To investigate how C. glabrata responds to DNA damage, we examined the phosphorylation state of Rad53 (CHK2 in higher eukaryotes), a conserved effector kinase that is extensively phosphorylated upon DNA damage in S. cerevisiae and other organisms (Chen, et al. 2014, Jung, et al. 2019, Lee, et al. 2003, Pellicioli and Foiani 2005, Sanchez, et al. 1996, Sweeney, et al. 2005, Wang, et al. 2012). This hyper-phosphorylation, carried out by upstream DNA damage sensing kinases as well as by autophosphorylation, is a key signal transduction event necessary for multiple aspects of a functional DNA damage response (Lee, et al. 2003). Rad53 is also hyperphosphorylated upon genotoxic stress in C. albicans (Sun, et al. 2011, Wang, et al. 2012), where it is required for the DNA damage-induced switch from the yeast to the filamentous form (Shi, et al. 2007). In S. cerevisiae, dozens of Rad53 serines and threonines are phosphorylated upon DNA damage (Chen, et al. 2014), such that its mobility in acrylamide gels is significantly slowed to produce a characteristic band shift on a Western blot, which has become one of the hallmarks of a functional DNA damage response. Interestingly, unlike S. cerevisiae Rad53, C. glabrata Rad53 (CgRad53) did not show a size shift in the presence of heavy DNA damage – either 0.1% MMS (alkylating damage) or 10 mM H2O2 (oxidative damage) (Shor, et al. 2020). Importantly, both of these treatments produced robust phosphorylation of Ser129 of histone H2A (a.k.a. γH2A.X), indicating that cells were experiencing DNA damage but that CgRad53 was nevertheless not hyperphosphorylated (Shor, et al. 2020). We also confirmed the lack of DNA damage-induced CgRad53 hyperphosphorylation by mass spectrometry of immunoprecipitated endogenous CgRad53 (Shor, et al. 2020). These results were the first to indicate that the DNA damage checkpoint response in C. glabrata deviates from the paradigmatic response elucidated by studies of S. cerevisiae.
A key function of the DNA damage checkpoint is to slow down DNA replication, stabilize replication forks, and prevent cell division in the presence of DNA damage; failure to do this, for example in checkpoint mutants, results in chromosomal instability and cell death (Myung, et al. 2001, Segurado and Diffley 2008). Thus, we investigated these aspects of the DNA damage response in C. glabrata. We found that, unlike S. cerevisiae cells, C. glabrata cells did not accumulate in S phase in the presence of MMS (Shor, et al. 2020). Furthermore, time-lapse microscopy showed that some C. glabrata cells proceed with cell division even in the presence of MMS, and marking the nucleoplasm with nuclear-localized RFP allowed us to detect aberrant mitoses (i.e. those marked by unequal nuclear division), which were followed by cell death (Shor, et al. 2020). In accordance with these results, we also observed that C. glabrata cells exhibited higher lethality in the presence of moderate and high doses of MMS than S. cerevisiae (Shor, et al. 2020). These results are consistent with a lack of DNA damage-induced CgRad53 phosphorylation in C. glabrata and indicate that its DNA damage checkpoint response is attenuated relative to that of S. cerevisiae, suggesting that C. glabrata is more “permissive” of DNA replication and cell division in the presence of damaged DNA. This “permissiveness” may be one mechanism contributing to the high karyotype diversity of C. glabrata clinical isolates. Thus, it is possible that the obligate commensal and opportunistic pathogenic lifestyle of C. glabrata has been accompanied by relaxed DNA damage checkpoint activity and increased genetic flexibility, allowing chromosomal rearrangements at the cost of higher DNA damage-induced lethality.
Transcriptional response to DNA damage response in C. glabrata
In S. cerevisiae, DNA damage-activated Rad53 in turn phosphorylates multiple transcription factors that activate or repress hundreds of genes, resulting in the stereotypical transcriptional reprogramming in response to DNA damage (Edenberg, et al. 2014, Gasch, et al. 2001, Jaehnig, et al. 2013, Smolka, et al. 2007). We compared the transcriptional response to 0.1% MMS in S. cerevisiae and C. glabrata and found that, with a few intriguing differences highlighted below, the global transcriptional responses to DNA damage are quite similar in the two organisms (Shor, et al. 2020). Indeed, most of the genes whose orthologs’ up- or downregulation is dependent on Rad53 in S. cerevisiae were still up- or downregulated by DNA damage in C. glabrata within the timeframe (1 hour) where no CgRad53 hyper-phosphorylation was occurring (Shor, et al. 2020). This result suggests that some transcriptional rewiring has occurred in C. glabrata, wherein genes that are Rad53-dependent in S. cerevisiae are regulated by Rad53-independent means in C. glabrata. Alternatively, it is possible that CgRad53 is mildly phosphorylated upon DNA damage (which our results do not exclude), and that this is sufficient for downstream transcriptional activation.
Interestingly, several genes involved in the maintenance of genome stability (DNA repair, replication integrity, chromosome segregation) were differentially regulated by DNA damage in S. cerevisiae and C. glabrata, most notably POL30, which encodes the proliferating cell nuclear antigen (PCNA). Consistent with previous studies, ScPOL30 was strongly induced by DNA damage whereas, in contrast, CgPOL30 was repressed by DNA damage both at RNA and protein levels (Shor, et al. 2020). The reasons for these differences are not yet clear, yet they are likely to have profound implications for how the two organisms maintain chromosome integrity in the presence of DNA damage because of the multiple roles of PCNA in preserving the integrity of the replisome and mediating its interactions with DNA repair complexes (Amin and Holm 1996, Ayyagari, et al. 1995, Brothers and Rine 2019, Chen, et al. 1999).
DNA damage induces orthologs of mating and sporulation genes in C. glabrata
One of the more mysterious aspects of the transcriptional response of C. glabrata to MMS was revealed by gene ontology (GO) analysis of downregulated and upregulated genes. Consistent with previous studies, the major functional classes downregulated by DNA damage in both S. cerevisiae and C. glabrata were involved in growth and protein synthesis, whereas genes upregulated by DNA damage were involved in stress and damage responses (Shor, et al. 2020). However, an additional functional category was found for MMS-induced genes only in C. glabrata: genes whose orthologs in S. cerevisiae functioned in meiosis. This was surprising and intriguing because, as mentioned above, sexual reproduction has not been observed in C. glabrata in the lab, and only genome analyses of clinical isolates have provided indirect evidence of rare sex and recombination in this species (Carrete, et al. 2018, Lott, et al. 2010). Nevertheless, C. glabrata possesses the full complement of genes whose orthologs in S. cerevisiae function in mating, meiosis, and sporulation, suggesting either that sexual reproduction does occur in C. glabrata under as yet unknown conditions, or that these genes have perhaps been coopted for another function in this fungus (Muller, et al. 2008, Wong, et al. 2003). To begin to understand the connection between DNA damage and mating and meiosis, we more closely scrutinized the transcriptional changes in mating and meiosis-related genes in C. glabrata.
The process of sexual reproduction in S. cerevisiae can be broken down into several distinct stages. First, two cells of the opposite mating type signal their presence to each other by secreting pheromones. Mating type identity is determined by the MAT locus, which contains either MATa- or MATα-specific transcription factors that regulate pheromone production and other mating factors (Heitman, et al. 2013). Yeast cells can switch their mating type by expressing HO endonuclease, which creates a DNA double strand break at the MAT locus, triggering a gene conversion event from a transcriptionally silenced locus carrying MAT genes of the opposite mating type (HMRa or HMLα) (Haber 2012). Most lab strains of S. cerevisiae contain point mutations in HO and are therefore incapable of mating type switching. Interestingly, our transcriptome analysis showed that the HO gene was downregulated by DNA damage in S. cerevisiae but significantly upregulated in C. glabrata (Figure 1, “Mating type determination, pheromone production”), suggesting that DNA damage may promote mating type switching in C. glabrata or perhaps other reconfiguring of the MAT locus. Whether this is indeed true remains to be experimentally determined.
Figure 1. DNA damage-induced transcriptional changes of C. glabrata orthologs of genes involved in sexual reproduction.
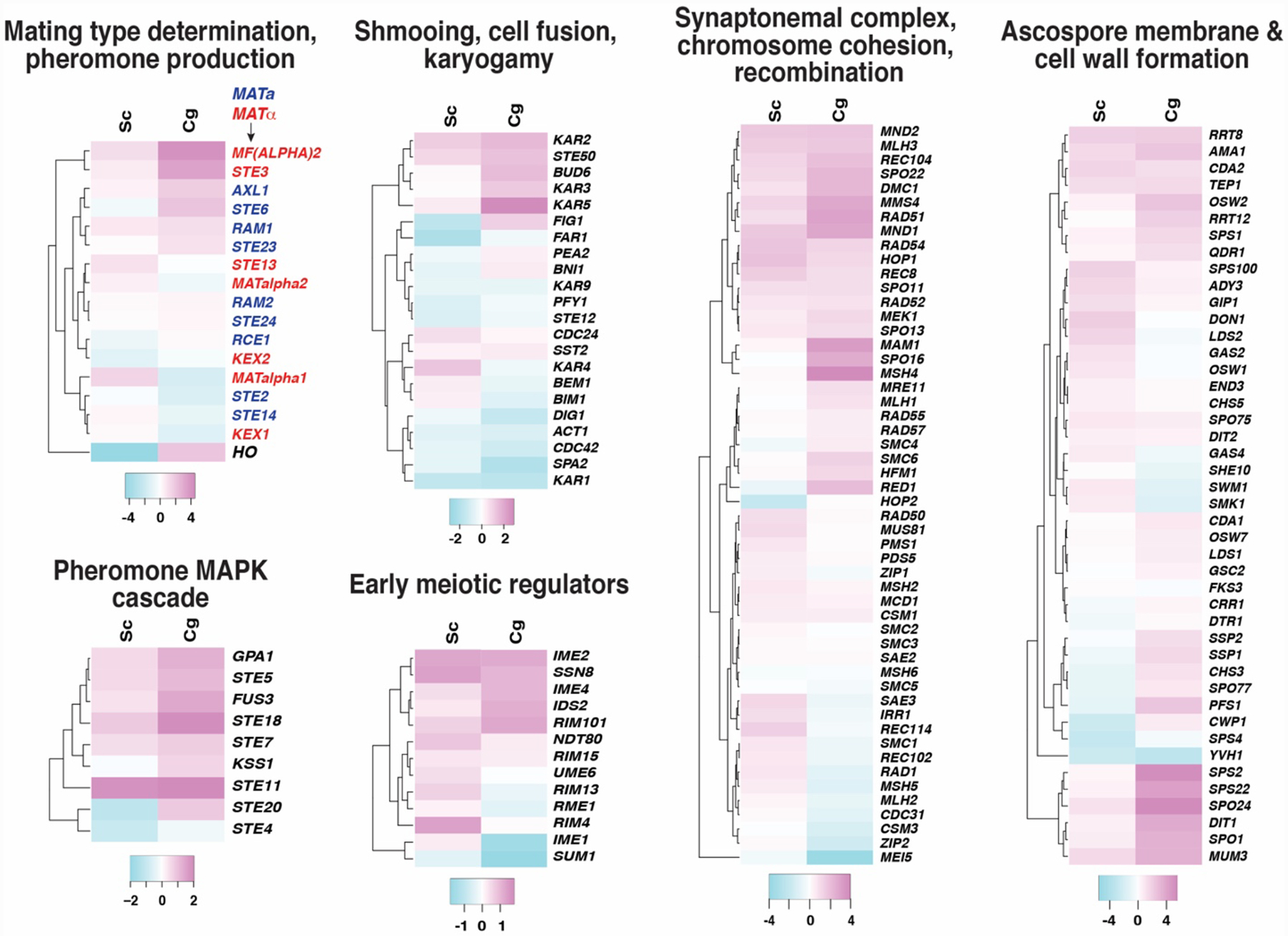
The color scheme represents transcript abundance log2 ratios (MMS/no MMS). The RNAseq data are available at the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE155701) and have been previously described (Shor, et al. 2020). The heatmaps were generated using the R studio gplots package.
Because the focus of our study had been on the DNA damage checkpoint, with no anticipation of the effects of DNA damage on mating-related processes, the S. cerevisiae and C. glabrata strains exposed to MMS and used for RNAseq happened to be of different mating types: the C. glabrata strain was MATα whereas the S. cerevisiae strains was MATa. Interestingly, MF(alpha)2 gene, which encodes the MATα-specific pheromone α-factor, and STE3, which encodes the receptor for the MATa pheromone a-factor, were both strongly induced by MMS in C. glabrata, as if in preparation for mating (Figure 1, “Mating type determination, pheromone production”). Furthermore, all genes involved in the pheromone-signaling mitogen-activated protein kinase (MAPK) cascade (Wang and Dohlman 2004) were also upregulated by MMS in C. glabrata relative to S. cerevisiae, with the exception of STE4, which was downregulated in S. cerevisiae but unchanged in C. glabrata (Figure 1, “Pheromone MAPK cascade”). However, genes involved in subsequent steps of mating (cell shape change known as “shmooing”, cell fusion, karyogamy) were not, as a whole, more likely to be induced in C. glabrata than in S. cerevisiae (Figure 1, “Shmooing, cell fusion, karyogamy”). Together, these data suggest several possibilities. First, it is possible that DNA damage, or perhaps another kind of damage caused by MMS, truly triggers mating type switching and mating in C. glabrata. Consistent with this hypothesis, the early genes involved in pheromone signaling were induced within the timeframe of our experiment (1 hour MMS exposure), whereas the later genes involved in cell fusion were not. An alternative explanation is also possible, however: that the orthologs of mating genes in S. cerevisiae have been coopted for another process in C. glabrata, one that is induced by MMS. There is precedent for non-mating functions of the mating gene cascade: for example, in C. albicans pheromone signaling also promotes cell adhesion and biofilm formation (Alby and Bennett 2011).
The second stage of sexual reproduction is meiosis, upon which a diploid cell forms four haploid spores. In S. cerevisiae mating and meiosis are two distinct processes occurring in haploid or diploid cells, respectively, and triggered by different stimuli: presence of the opposite mating type and nutrient limitation, respectively. Likewise, mating and meiosis are associated with distinct transcriptional programs. In contrast, in the haploid pathogenic yeast, C. lusitaniae, mating and meiosis occur together in quick succession, and the two gene sets are coordinately upregulated by the same set of stimuli and are controlled of the same transcription factor (Ste12) (Sherwood, et al. 2014). We examined the transcription changes in orthologs of meiosis-related genes in C. glabrata, including the early regulators of meiosis, genes involved in meiosis-specific chromosomal transactions, and genes involved in spore development. None of the orthologs of well-defined early meiotic regulators (e.g. IME1, IME2, NDT80) were induced more strongly in C. glabrata than in S. cerevisiae (Figure 1, “Early meiotic regulators”). Furthermore, although a few genes involved in regulating meiosis-specific chromosomal transactions (MAM1, SPO16, MSH4, RED1) were strongly induced by MMS in C. glabrata relative to S. cerevisiae, most such genes were as likely to be repressed as induced in C. glabrata (Figure 1, “Synaptonemal complex, chromosome cohesion, recombination”). Finally, genes involved in spore development (mostly those promoting the formation of the spore membrane or cell wall) did tend to be upregulated in C. glabrata relative to S. cerevisiae, with several showing very strong induction by MMS (log2ratio > 4, Figure 1, “Ascospore membrane & cell wall formation”). Given the importance of the cell wall in C. glabrata (Lopez-Fuentes, et al. 2018), it is possible that the upregulation of these genes reflects not their meiotic functions but perhaps the fact that in C. glabrata they serve to maintain cell wall integrity in the face of stress, such as MMS exposure. In sum, these data do not provide strong evidence of induction of the meiotic program within the timeframe of the experiment, and further experiments, with longer DNA damage exposures, are required to clarify this issue.
Summary and future directions
Our study of the DNA damage response in C. glabrata revealed several unique features, some easier to interpret than others, which likely impact genetic flexibility and karyotype diversity in this organism. The absence of Rad53 hyper-phosphorylation, the attenuated checkpoint response, and downregulation of PCNA expression upon DNA damage are all expected to affect chromosome stability and may contribute to the variable karyotypes that characterize C. glabrata clinical isolates (Figure 2). The observation that HO endonuclease and pheromone signaling genes are strongly induced by MMS in C. glabrata but not S. cerevisiae suggests that mating in C. glabrata – which has never been observed as yet – may be connected to its DNA damage response or another stress response triggered by MMS. If true, this would fit with the paradigm of generating genetic diversity – via sexual reproduction – in the presence of stress (Ram and Hadany 2016). Alternatively, DNA damage may induce other functions mediated by the orthologs of mating and sporulation genes, e.g. cell adhesion, biofilm formation, or cell wall integrity. If this is the case, it would support the conclusion that C. glabrata is largely asexual, and perhaps its relaxed DNA damage checkpoint function helps generate genetic diversity in the absence of sex. In any case, what these data show is that C. glabrata responds to DNA damage differently than its close non-pathogenic relative S. cerevisiae, and understanding these differences will help explain how C. glabrata regulates genetic change and evolvability, and may identify routes towards inhibiting rapid emergence of antifungal drug resistance.
Figure 2. Schematic representation of the differences between the responses to DNA damage ( ) in C. glabrata and S. cerevisiae.
) in C. glabrata and S. cerevisiae.
Funding.
This work was supported by NIH 5R01AI109025 to D.S.P.
Footnotes
Conflicts of interest/Competing interests. D.S.P. has received funding from the U.S. National Institutes of Health and contracts with The Centers for Disease Control and Prevention, Amplyx, Astellas, Cidara, and Scynexis. He serves on advisory boards for Amplyx, Astellas, Cidara, Matinas, N8 Medical, and Scynexis.
Availability of data and material. The RNAseq data are available at the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE155701) and have been previously described (Shor, et al. 2020).
Code availability. The heatmaps were generated using the R studio gplots package, and the code is available upon request.
REFERENCES
- Alby K, Bennett RJ (2011) Interspecies pheromone signaling promotes biofilm formation and same-sex mating in Candida albicans. Proc Natl Acad Sci U S A 108: 2510–2515 doi: 10.1073/pnas.1017234108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin NS, Holm C (1996) In vivo analysis reveals that the interdomain region of the yeast proliferating cell nuclear antigen is important for DNA replication and DNA repair. Genetics 144: 479–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyagari R, Impellizzeri KJ, Yoder BL, Gary SL, Burgers PM (1995) A mutational analysis of the yeast proliferating cell nuclear antigen indicates distinct roles in DNA replication and DNA repair. Mol Cell Biol 15: 4420–4429 doi: 10.1128/mcb.15.8.4420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber AE, Weber M, Kaerger K, Linde J, Golz H, Duerschmied D, Markert A, Guthke R, Walther G, Kurzai O (2019) Comparative Genomics of Serial Candida glabrata Isolates and the Rapid Acquisition of Echinocandin Resistance during Therapy. Antimicrob Agents Chemother 63 10.1128/AAC.01628-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RJ, Forche A, Berman J (2014) Rapid mechanisms for generating genome diversity: whole ploidy shifts, aneuploidy, and loss of heterozygosity. Cold Spring Harb Perspect Med 4 10.1101/cshperspect.a019604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizerra FC, Jimenez-Ortigosa C, Souza AC, Breda GL, Queiroz-Telles F, Perlin DS, Colombo AL (2014) Breakthrough candidemia due to multidrug-resistant Candida glabrata during prophylaxis with a low dose of micafungin. Antimicrob Agents Chemother 58: 2438–2440 doi: 10.1128/AAC.02189-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordallo-Cardona MA, Escribano P, de la Pedrosa EG, Marcos-Zambrano LJ, Canton R, Bouza E, Guinea J (2017) In Vitro Exposure to Increasing Micafungin Concentrations Easily Promotes Echinocandin Resistance in Candida glabrata Isolates. Antimicrob Agents Chemother 61 10.1128/AAC.01542-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Foiani M (2010) Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol 11: 208–219 doi: 10.1038/nrm2852 [DOI] [PubMed] [Google Scholar]
- Brothers M, Rine J (2019) Mutations in the PCNA DNA Polymerase Clamp of Saccharomyces cerevisiae Reveal Complexities of the Cell Cycle and Ploidy on Heterochromatin Assembly. Genetics 213: 449–463 doi: 10.1534/genetics.119.302452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrete L, Ksiezopolska E, Pegueroles C, Gomez-Molero E, Saus E, Iraola-Guzman S, Loska D, Bader O, Fairhead C, Gabaldon T (2018) Patterns of Genomic Variation in the Opportunistic Pathogen Candida glabrata Suggest the Existence of Mating and a Secondary Association with Humans. Curr Biol 28: 15–27 e17 doi: 10.1016/j.cub.2017.11.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Merrill BJ, Lau PJ, Holm C, Kolodner RD (1999) Saccharomyces cerevisiae pol30 (proliferating cell nuclear antigen) mutations impair replication fidelity and mismatch repair. Mol Cell Biol 19: 7801–7815 doi: 10.1128/mcb.19.11.7801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ES, Hoch NC, Wang SC, Pellicioli A, Heierhorst J, Tsai MD (2014) Use of quantitative mass spectrometric analysis to elucidate the mechanisms of phospho-priming and auto-activation of the checkpoint kinase Rad53 in vivo. Mol Cell Proteomics 13: 551–565 doi: 10.1074/mcp.M113.034058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coste A, Selmecki A, Forche A, Diogo D, Bougnoux ME, d’Enfert C, Berman J, Sanglard D (2007) Genotypic evolution of azole resistance mechanisms in sequential Candida albicans isolates. Eukaryot Cell 6: 1889–1904 doi: 10.1128/EC.00151-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg ER, Vashisht A, Benanti JA, Wohlschlegel J, Toczyski DP (2014) Rad53 downregulates mitotic gene transcription by inhibiting the transcriptional activator Ndd1. Mol Cell Biol 34: 725–738 doi: 10.1128/MCB.01056-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ene IV, Farrer RA, Hirakawa MP, Agwamba K, Cuomo CA, Bennett RJ (2018) Global analysis of mutations driving microevolution of a heterozygous diploid fungal pathogen. Proc Natl Acad Sci U S A 115: E8688–E8697 doi: 10.1073/pnas.1806002115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forche A, Cromie G, Gerstein AC, Solis NV, Pisithkul T, Srifa W, Jeffery E, Abbey D, Filler SG, Dudley AM, Berman J (2018) Rapid Phenotypic and Genotypic Diversification After Exposure to the Oral Host Niche in. Genetics 209: 725–741 doi: 10.1534/genetics.118.301019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedel AM, Pike BL, Gasser SM (2009) ATR/Mec1: coordinating fork stability and repair. Curr Opin Cell Biol 21: 237–244 doi: 10.1016/j.ceb.2009.01.017 [DOI] [PubMed] [Google Scholar]
- Gasch AP, Huang M, Metzner S, Botstein D, Elledge SJ, Brown PO (2001) Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast ATR homolog Mec1p. Mol Biol Cell 12: 2987–3003 doi: 10.1091/mbc.12.10.2987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber JE (2012) Mating-type genes and MAT switching in Saccharomyces cerevisiae. Genetics 191: 33–64 doi: 10.1534/genetics.111.134577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healey KR, Jimenez Ortigosa C, Shor E, Perlin DS (2016) Genetic Drivers of Multidrug Resistance in Candida glabrata. Front Microbiol 7: 1995 doi: 10.3389/fmicb.2016.01995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J, Sun S, James TY (2013) Evolution of fungal sexual reproduction. Mycologia 105: 1–27 doi: 10.3852/12-253 [DOI] [PubMed] [Google Scholar]
- Jaehnig EJ, Kuo D, Hombauer H, Ideker TG, Kolodner RD (2013) Checkpoint kinases regulate a global network of transcription factors in response to DNA damage. Cell Rep 4: 174–188 doi: 10.1016/j.celrep.2013.05.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung KW, Lee Y, Huh EY, Lee SC, Lim S, Bahn YS (2019) Rad53- and Chk1-Dependent DNA Damage Response Pathways Cooperatively Promote Fungal Pathogenesis and Modulate Antifungal Drug Susceptibility. mBio 10 10.1128/mBio.01726-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Schwartz MF, Duong JK, Stern DF (2003) Rad53 phosphorylation site clusters are important for Rad53 regulation and signaling. Mol Cell Biol 23: 6300–6314 doi: 10.1128/mcb.23.17.6300-6314.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobrich M, Jeggo PA (2007) The impact of a negligent G2/M checkpoint on genomic instability and cancer induction. Nat Rev Cancer 7: 861–869 doi: 10.1038/nrc2248 [DOI] [PubMed] [Google Scholar]
- Lopez-Fuentes E, Gutierrez-Escobedo G, Timmermans B, Van Dijck P, De Las Penas A, Castano I (2018) Candida glabrata’s Genome Plasticity Confers a Unique Pattern of Expressed Cell Wall Proteins. J Fungi (Basel) 4 10.3390/jof4020067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lott TJ, Frade JP, Lockhart SR (2010) Multilocus sequence type analysis reveals both clonality and recombination in populations of Candida glabrata bloodstream isolates from U.S. surveillance studies. Eukaryot Cell 9: 619–625 doi: 10.1128/EC.00002-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H, Hennequin C, Gallaud J, Dujon B, Fairhead C (2008) The asexual yeast Candida glabrata maintains distinct a and alpha haploid mating types. Eukaryot Cell 7: 848–858 doi: 10.1128/EC.00456-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H, Thierry A, Coppee JY, Gouyette C, Hennequin C, Sismeiro O, Talla E, Dujon B, Fairhead C (2009) Genomic polymorphism in the population of Candida glabrata: gene copy-number variation and chromosomal translocations. Fungal Genet Biol 46: 264–276 doi: 10.1016/j.fgb.2008.11.006 [DOI] [PubMed] [Google Scholar]
- Myung K, Chen C, Kolodner RD (2001) Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature 411: 1073–1076 doi: 10.1038/35082608 [DOI] [PubMed] [Google Scholar]
- Pellicioli A, Foiani M (2005) Signal transduction: how rad53 kinase is activated. Curr Biol 15: R769–771 doi: 10.1016/j.cub.2005.08.057 [DOI] [PubMed] [Google Scholar]
- Perlin DS, Rautemaa-Richardson R, Alastruey-Izquierdo A (2017) The global problem of antifungal resistance: prevalence, mechanisms, and management. Lancet Infect Dis 17: e383–e392 doi: 10.1016/S1473-3099(17)30316-X [DOI] [PubMed] [Google Scholar]
- Polakova S, Blume C, Zarate JA, Mentel M, Jorck-Ramberg D, Stenderup J, Piskur J (2009) Formation of new chromosomes as a virulence mechanism in yeast Candida glabrata. Proc Natl Acad Sci U S A 106: 2688–2693 doi: 10.1073/pnas.0809793106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram Y, Hadany L (2016) Condition-dependent sex: who does it, when and why? Philos Trans R Soc Lond B Biol Sci 371 10.1098/rstb.2015.0539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Y, Desany BA, Jones WJ, Liu Q, Wang B, Elledge SJ (1996) Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271: 357–360 doi: 10.1126/science.271.5247.357 [DOI] [PubMed] [Google Scholar]
- Segurado M, Diffley JF (2008) Separate roles for the DNA damage checkpoint protein kinases in stabilizing DNA replication forks. Genes Dev 22: 1816–1827 doi: 10.1101/gad.477208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmecki A, Forche A, Berman J (2006) Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science 313: 367–370 doi: 10.1126/science.1128242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmecki AM, Dulmage K, Cowen LE, Anderson JB, Berman J (2009) Acquisition of aneuploidy provides increased fitness during the evolution of antifungal drug resistance. PLoS Genet 5: e1000705 doi: 10.1371/journal.pgen.1000705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serero A, Jubin C, Loeillet S, Legoix-Ne P, Nicolas AG (2014) Mutational landscape of yeast mutator strains. Proc Natl Acad Sci U S A 111: 1897–1902 doi: 10.1073/pnas.1314423111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen XX, Opulente DA, Kominek J, Zhou X, Steenwyk JL, Buh KV, Haase MAB, Wisecaver JH, Wang M, Doering DT, Boudouris JT, Schneider RM, Langdon QK, Ohkuma M, Endoh R, Takashima M, Manabe RI, Čadež N, Libkind D, Rosa CA, DeVirgilio J, Hulfachor AB, Groenewald M, Kurtzman CP, Hittinger CT, Rokas A (2018) Tempo and Mode of Genome Evolution in the Budding Yeast Subphylum. Cell 175: 1533–1545.e1520 doi: 10.1016/j.cell.2018.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood RK, Scaduto CM, Torres SE, Bennett RJ (2014) Convergent evolution of a fused sexual cycle promotes the haploid lifestyle. Nature 506: 387–390 doi: 10.1038/nature12891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi QM, Wang YM, Zheng XD, Lee RT, Wang Y (2007) Critical role of DNA checkpoints in mediating genotoxic-stress-induced filamentous growth in Candida albicans. Mol Biol Cell 18: 815–826 doi: 10.1091/mbc.e06-05-0442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JH, Chae MJ, Song JW, Jung SI, Cho D, Kee SJ, Kim SH, Shin MG, Suh SP, Ryang DW (2007) Changes in karyotype and azole susceptibility of sequential bloodstream isolates from patients with Candida glabrata candidemia. J Clin Microbiol 45: 2385–2391 doi: 10.1128/JCM.00381-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shor E, Garcia-Rubio R, DeGregorio L, Perlin DS (2020) A Noncanonical DNA Damage Checkpoint Response in a Major Fungal Pathogen. mBio 11 10.1128/mBio.03044-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolka MB, Albuquerque CP, Chen SH, Zhou H (2007) Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc Natl Acad Sci U S A 104: 10364–10369 doi: 10.1073/pnas.0701622104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun LL, Li WJ, Wang HT, Chen J, Deng P, Wang Y, Sang JL (2011) Protein phosphatase Pph3 and its regulatory subunit Psy2 regulate Rad53 dephosphorylation and cell morphogenesis during recovery from DNA damage in Candida albicans. Eukaryot Cell 10: 1565–1573 doi: 10.1128/EC.05042-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney FD, Yang F, Chi A, Shabanowitz J, Hunt DF, Durocher D (2005) Saccharomyces cerevisiae Rad9 acts as a Mec1 adaptor to allow Rad53 activation. Curr Biol 15: 1364–1375 doi: 10.1016/j.cub.2005.06.063 [DOI] [PubMed] [Google Scholar]
- Wang H, Gao J, Li W, Wong AH, Hu K, Chen K, Wang Y, Sang J (2012) Pph3 dephosphorylation of Rad53 is required for cell recovery from MMS-induced DNA damage in Candida albicans. PLoS One 7: e37246 doi: 10.1371/journal.pone.0037246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Dohlman HG (2004) Pheromone signaling mechanisms in yeast: a prototypical sex machine. Science 306: 1508–1509 doi: 10.1126/science.1104568 [DOI] [PubMed] [Google Scholar]
- Wong S, Fares MA, Zimmermann W, Butler G, Wolfe KH (2003) Evidence from comparative genomics for a complete sexual cycle in the ‘asexual’ pathogenic yeast Candida glabrata. Genome Biol 4: R10 doi: 10.1186/gb-2003-4-2-r10 [DOI] [PMC free article] [PubMed] [Google Scholar]


