Abstract
One major challenge in synthetic biology is the deleterious impacts of cellular stress caused by expression of heterologous pathways, sensors and circuits. Feedback control and dynamic regulation are broadly proposed strategies to mitigate this cellular stress by optimizing gene expression levels temporally and in response to biological cues. While a variety of approaches for feedback implementation exist, they are often complex and cannot be easily manipulated. Here, we report a strategy that uses RNA transcriptional regulators to integrate additional layers of control over the output of natural and engineered feedback responsive circuits. Called riboregulated switchable feedback promoters (rSFPs), these gene expression cassettes can be modularly activated using multiple mechanisms, from manual induction to autonomous quorum sensing, allowing control over the timing, magnitude and autonomy of expression. We develop rSFPs in Escherichia coli to regulate multiple feedback networks and apply them to control the output of two metabolic pathways. We envision that rSFPs will become a valuable tool for flexible and dynamic control of gene expression in metabolic engineering, biological therapeutic production, and many other applications.
Keywords: Gene Regulation, RNA, Feedback, Quorum-Sensing, Metabolic Engineering, Isoprenoids
Graphical Abstract
Introduction
The fine tuning of gene expression to improve system performance is a long standing goal of synthetic biology for applications ranging from chemical synthesis via metabolic engineering1,2 to advanced therapeutics3 and diagnostics4. A nearly universal challenge within synthetic biology is the burden and toxicity engineered genetic systems place on host cells due to high levels of heterologous gene expression and possible accumulation of toxic biochemical intermediates5,6. This burden creates a selective pressure for mutations that can break the introduced genetic system and lead to loss of productivity, effectiveness, or entire function7, creating a continuous need for strategies that alleviate or avoid these pitfalls. This is a non-trivial challenge, because each application presents unique sources of stresses that can change over time, making it difficult to find generalizable solutions.
Current approaches to solve expression-related challenges range from static tuning of gene expression to utilizing dynamic gene expression control systems8-10. For static control, promoter strength, ribosomal binding site (RBS) strength, plasmid copy number, or the location or number of genetic integrations are varied and screened to find an optimal solution11-16. Dynamic control systems can be implemented in multiple ways. For example, endogenous feedback networks can be harnessed by utilizing regulatory elements, such as stress-response promoters, that integrate signals from natural genetic networks to modulate mRNA synthesis in response to cellular cues like membrane, oxidative, pH extremes and nutrient deprivation stresses17. These have been incorporated to regulate heterologous genetic systems, leading to notable improvements to productivity and yield for protein expression18 and engineered metabolic pathways for the production of the artemisinin precursor amorphadiene19 and n-butanol20 as examples. Alternatively, synthetic circuits can be built using ligand-inducible transcription factors21-23 or ribozymes24 that sense and respond to metabolic pathway intermediates so that expression can adapt dynamically to maintain optimal enzyme concentration over time9,10,25,26. Synthetic feedback circuits have also been constructed to enable additional useful features, such as engineered stabilized promoters that maintain constant gene expression regardless of changes or fluctuation in DNA copy number27.
While each of the above strategies has moved the field of synthetic biology forward, there are still significant limitations. For example, hard-coded static solutions cannot adapt to stresses that vary in time, and may no longer be optimal upon inclusion of additional genetic components or within a new environment8. Natural dynamic feedback-responsive circuits such as stress-response promoters could resolve this but have not been widely adopted, as their unknown architecture and interconnectedness to native regulatory systems makes it difficult to fine-tune their behavior for specific applications. Synthetic feedback circuits that sense pathway intermediates are useful in specific contexts, but often do not respond to general aspects of the cellular environment such as growth phase, fermentation conditions and cellular stresses that are important sources of variation that affect system performance across many applications. A unifying limitation for both natural and synthetic feedback systems is the difficulty in integrating additional external points of control that can tune either the timing or overall magnitude of their transcriptional outputs – two key parameters for optimizing system performance28.
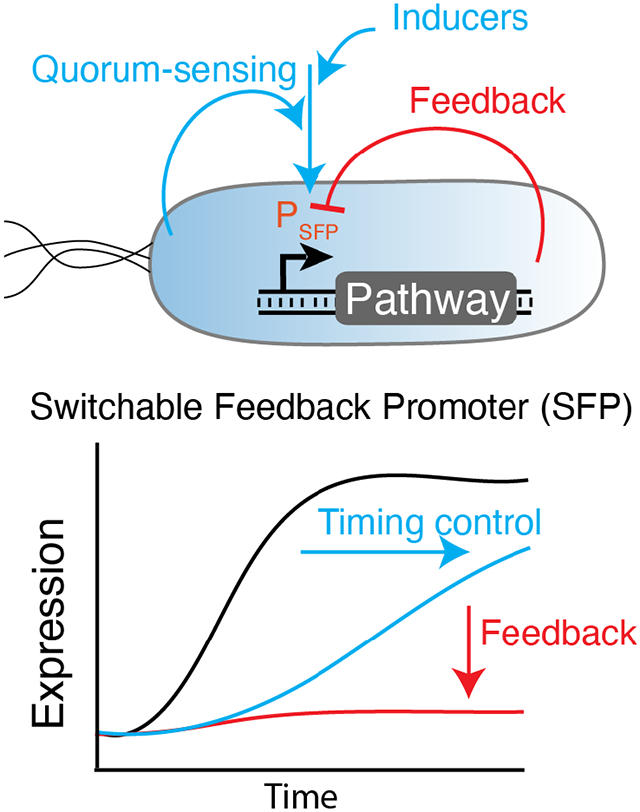
To address this limitation, we created a new regulatory motif called a switchable feedback promoter (SFP) that combines the properties of natural and synthetic feedback-responsive promoter systems, with integrated regulators that offer additional control of the timing and overall magnitude of transcriptional outputs (Fig. 1A-D). The SFP concept is general, relying on a trans-acting synthetic regulator to gate the transcription of the feedback promoter system. Here, we focus on utilizing small transcription activating RNAs (STARs)29 to make riboregulated SFPs (rSFPs) in Escherichia coli, as their well-defined composition rules enables them to be inserted into a gene expression construct without modification or disruption of the desired promoter sequence. This enables the rSFP output to be controlled with any strategy that can regulate the expression of the trans-acting RNA.
Figure 1.
Synthetic regulation of feedback responsive promoters enables combined control of gene expression timing and tuning with feedback regulation. (a) Commonly used constitutive promoters can be manually tuned to alter gene expression levels but are not designed to respond to sources of toxicity, stress, and other biological signals. (b) Natural stress-response promoter and engineered feedback promoter systems use regulatory networks to enable responsiveness to sources of toxicity, stress, and other biological signals, but gene expression timing and tuning is difficult to manipulate. (c) A switchable feedback promoter (SFP) integrates external control with feedback-responsive promoters, allowing induction timing and tuning with inducible regulators and autonomous quorum sensing systems. (d) A riboregulated SFP (rSFP) is composed of a feedback-responsive promoter and an RNA transcriptional switch. (e) Schematic of the small transcription activating RNA (STAR) mechanism. A target sequence (switch symbol) is placed downstream of a feedback-responsive promoter. The transcribed target RNA is designed to fold into an intrinsic transcription terminator hairpin that causes RNA polymerase to terminate transcription upstream of the gene to be regulated (gene OFF). A separately transcribed STAR RNA (colored red) can bind to the target RNA, preventing hairpin formation and allowing transcription elongation (gene ON).
Results
We report the creation and characterization of STAR-mediated feedback responsive promoters in E. coli using both natural stress-responsive promoters as well as engineered stabilized promoters27. First, we created a set of 18 stress-responsive rSFPs by interfacing STARs with natural E. coli stress-response promoters and placing trans-acting STAR production under control of an inducible promoter. We then characterized select rSFPs for their response to sources of cellular stress, including membrane protein expression and toxic metabolite accumulation. Second, we create stabilized rSFPs and show them to maintain constant gene expression over different plasmid copy numbers while simultaneously introducing inducible control. To demonstrate the applicability of rSFPs, we next apply them to regulate two important metabolic pathways, one for amorphadiene, a precursor to the antimalarial artemsinin, and the other for an oxygenated taxane precursor to the anticancer drug Taxol. Finally, to demonstrate the use of other control points for rSFPs, we engineer quorum sensing rSFPs that offer autonomous pathway expression regulation with titers similar to manual induction but without costly external inducers. Overall, rSFPs represent a novel and general strategy to add additional points of control to feedback-responsive gene regulation systems to enhance their use and optimizations for broad synthetic biology applications. The rSFP methodology works in multiple contexts and should be readily applied to many other engineered bacterial organisms.
rSFPs enable inducible control of feedback responsive promoters in E. coli
We used STARs to construct rSFPs because they exhibit low leak and high dynamic range comparable to exemplary protein-based regulators and can be computationally designed to not interfere with other RNA elements required for downstream gene expression30. STARs activate transcription by disrupting the folding pathway of a terminator hairpin sequence, called a target, that is placed upstream of the gene to be regulated (Fig. 1E). In the absence of a STAR, the target region folds into an intrinsic terminator hairpin which stops transcription before reaching the downstream gene. When present, a STAR RNA can bind to the 5’ portion of the terminator hairpin, preventing its formation, and allowing transcription. rSFPs are then created by inserting a target sequence downstream of a candidate feedback-responsive promoter. In this way, the introduction of the STAR/target adds an additional layer of control, gating its transcriptional output through the regulation of STAR RNA expression, which can be controlled using a variety of mechanisms.
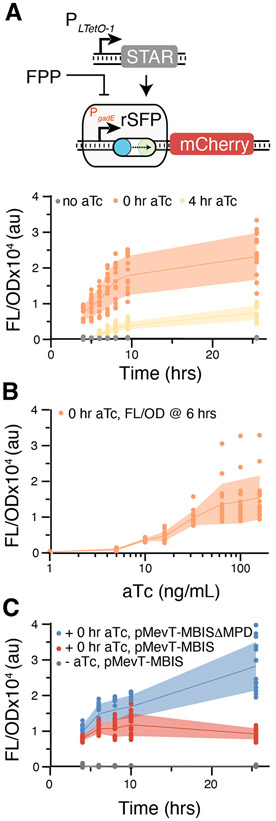
We began rSFP development with the previously characterized PgadE acid stress-response promoter that has been shown to improve amorphadiene pathway production by responding to accumulation of the toxic metabolite farnesyl pyrophosphate (FPP)19. Our initial rSFP design utilized a previously developed STAR30 under the well-characterized inducible system TetR/PLTetO-131 promoter to control its expression. This STAR was interfaced with the PgadE promoter by cloning a target sequence immediately after the promoter and 5’ UTR, and directly before the start codon of the natural gene regulated by the stress-response promoter in E. coli. This sequence was followed by an mRNA region containing an RBS and mCherry. We found that induction of PLTetO-1-STAR resulted in activation (~40x) from the PgadE stress-response promoter (Fig. 2A). Additionally, we found that timing control of PgadE expression could be achieved by delaying induction, albeit with lower endpoint expression levels (Fig. 2A). We characterized the transfer curve of the PgadE rSFP by titrating inducer levels and found that it exhibited a monotonically increasing induction profile (Fig. 2B), reflecting the properties of the PLTetO-1 promoter system and providing evidence that other transfer curve profiles might be achieved by selecting different inducible promoter systems for STAR expression.
Figure 2.
Creation of the PgadE rSFP. (a) Time-course characterization of the PgadE rSFP. Fluorescence characterization was performed on E. coli DH1 transformed with plasmids encoding the PgadE rSFP controlling mCherry expression in the absence (grey) and presence of 100 ng/mL aTc added at 0 hrs (orange) or 4 hrs (yellow). (b) The PgadE rSFP under different aTc induction levels measured after 6 hrs of growth. (c) Responsiveness of the PgadE rSFP to FPP accumulation. Fluorescence characterization was performed on E. coli DH1 transformed with a PLTetO-1 inducible STAR plasmid and a plasmid encoding the PgadE rSFP controlling mCherry expression in the absence (grey) and presence (colored) of 100 ng/mL aTc added at inoculation. Plasmids were co-expressed with pMevT-MBIS (FPP production, red) or pMevT-MBIS DMPD (no FPP production, blue) induced with IPTG at 2.5 hrs. Reduced endpoint expression was observed for P gadE rSFP with pMevT-MBIS. Lines in a-c represent mean values in units of arbitrary fluorescence/optical density (FL/OD) and shaded areas represent mean values +/− s.d. of at least n = 16 biological replicates. Colored points represent individual data points for each condition. FPP, farnesyl pyrophosphate. MPD, mevalonate pyrophosphate decarboxylase.
Next, we characterized downregulation of the PgadE rSFP by FPP accumulation. In addition to the PgadE rSFP and PLTetO-1-STAR plasmid, we co-expressed either pMevT-MBIS that results in accumulation of FPP or pMevT-MBIS ΔMPD that is defective in pyrophosphate decarboxylase activity involved in conversion of mevalonate to FPP. We found the PgadE rSFP expression was repressed over time in the presence of pMevT-MBIS in comparison with pMevT-MBIS ΔMPD (Fig. 2C), while similar repression was not observed with a constitutive promoter replacing PgadE (Fig. S1).
We expanded the rSFP designs to include a library of 17 putative membrane stress-responsive promoters20, chosen as several had been previously identified to regulate a biofuel transporter protein in E. coli20 and could therefore be valuable for dynamic regulation of membrane proteins in metabolic pathways. We found that induction of PLTetO-1-STAR resulted in activation from all members of the stress-response promoter library (Fig. 3A-B), exemplifying the modularity of the rSFP concept. Eight library members were activated by >25x fold upon induction, with a maximum activation of nearly 150x fold (Fig. S2). We characterized a subset of high-performing rSFPs for stress-responsiveness to a model stress from the oligosaccharyltransferase membrane protein PglB from Campylobacter jejuni32 and for other features of their expression. The expression of each was affected by PglB, with PgntK and PompF showing the largest repression (Fig. 4A-B). We examined the transfer curves of select rSFPs (Fig. S3A,B) and found that they were monotonically increasing. Characterization of the expression profile over time showed that all were activated at the earliest measured time point (4 hrs) and achieved maximal activation by ~10 hrs (Fig. S3C). Finally, comparison of select rSFPs with corresponding unregulated stress-response promoters revealed profiles with lower overall endpoint expression levels for rSFPs (Fig. S3D), due to the incorporation of the STAR target sequence that likely exhibits an inherent level of termination even upon STAR expression. Previous work30 suggests that the overall rSFP expression could be further tuned by changing plasmid copy number or RBS strength as needed.
Figure 3.
Creation of an rSFP library with unique envelope stress-responsive promoters. (a) Characterization of rSFP variants containing unique envelope stress-response promoters. PLTetO-1 inducible STAR expression is used to activate rSFPs containing a natural stress-response promoter upstream of a STAR target sequence, a ribosome binding site, and a red fluorescent protein (mCherry) coding sequence. (b) Fluorescence characterization was performed on E. coli Tax1 transformed with plasmids encoding each rSFP controlling mCherry expression in the absence and presence of 100 ng/mL aTc added at inoculation. Data in b represent mean values in units of arbitrary fluorescence/optical density (FL/OD) and error bars represent s.d. of n = 9 biological replicates. * indicate a statistically significant difference in FL/OD by a Welch’s t-test (two-tailed, unequal variances) (* = P < 0.05, ** = P < 0.005) between no aTc and 100 ng/mL aTc conditions. P values for each condition are reported in Table S19. Grey points represent individual data points for each condition.
Figure 4.
Analysis of feedback-responsiveness of selected stress-response promoters to C. jejuni PglB-induced stress. (a) Schematic of plasmids used for fluorescence characterization of rSFP stress response. PJ23119-STAR8 was used to constitutively activate expression from select rSFP plasmids. PBAD-PglB was used to induce PglB stress. Monitoring rSFP controlled expression of mCherry then allows the response to PglB stress to be characterized. (b) Fluorescence characterization of cells containing select PJ23119-STAR activated rSFPs controlling mCherry expression, and PBAD-PglB induced with either 0 μg/mL or 200 μg/mL Larabinose. Pconstitutive = PJ23115. Overall expression levels differed significantly from Fig. 3 in media conditions used for PglB expression (see methods). Bars represent mean values in units of arbitrary fluorescence/optical density (FL/OD) and error bars represent s.d. of n = 16 biological replicates. Individual colored data points represent FL/OD data.
To demonstrate that rSFPs can be configured to control other feedback architectures, including engineered feedback promoter systems, we created rSFPs utilizing the recently developed stabilized promoter system that buffers gene expression from changes and fluctuations in DNA copy number using an incoherent feedforward loop (iFFL)27. Stabilized promoters work by configuring promoter expression to be responsive to a co-expressed transcription-activator-like effector (TALE) repressor. In this way, increased DNA copy number results in increased repressor expression, which interacts with the stabilized promoter to counter changes in gene expression. Stabilized promoters are of interest because they enable more precise control of gene expression by buffering against changes in DNA copy number that occur over time and between cells34, in different host strains35, and in different growth conditions such as medium36,37, temperature38, and growth rate36. Furthermore, stabilized promoter systems are useful to buffer genetic constructs from changes in copy number that are caused throughout the engineering/optimization process, such as adding new pathway enzymes39-41, accumulating mutations that influence plasmid maintenance during a bioprocess27, or integrating genes into the host genome27.
Much like natural stress-response promoters, stabilized promoters lack the ability to control gene expression timing, which is critical for creating a separation of growth and production phase in biomanufacturing processes9. Therefore, we applied the rSFP promoter gating concept to create stabilized promoter rSFPs. Similar to stress-response promoter rSFPs, a STAR target sequence was cloned immediately downstream of a stabilized promoter to regulate expression of an sfGFP reporter with the cognate PLTetO-1-STAR construct (Fig. 5A, blue shaded region). For comparison, a STAR-regulated constitutive promoter lacking the TALE repressor protein was also cloned to regulate expression of an sfGFP reporter with the same PLTetO-1-STAR construct (Fig. 5A, red shaded region). We found that induction of PLTetO-1-STAR resulted in activation of the stabilized promoter of ~33x-fold (Fig. 5B, S4A, blue bars) and the STAR-regulated constitutive promoter lacking the TALE repressor protein of ~95x-fold (Fig. 5B, S4A, red bars). It is notable that the stabilized rSFP has lower overall expression levels than the constitutive STAR-regulated promoter system. This is an inherent feature of the previously developed iFFL27 utilized here and could likely be compensated through tuning of RBS strength16, promoter strength27, or use of a STAR/TARGET pair with different expression level features30.
Figure 5.
Creation of stabilized rSFPs. (a) Architecture of a STAR-regulated constitutive promoter (shaded red) and the stabilized rSFP (shaded blue). Expression activation is mediated by the inducible system PLTetO-1-STAR vector in both cases. The stabilized rSFP uses a previously developed co-expressed TALE repressor27 that counters changes in gene expression from higher plasmid DNA copy number resulting in outputs that are uniform across different plasmids. Constructs were cloned into SC101 plasmid variants with a range of copy numbers. (b) Fluorescence characterization of the stabilized rSFP system. Experiments were performed on E. coli transformed with plasmids encoding the stabilized rSFP controlling sfGFP expression in the absence (grey) and presence of 100 ng/mL aTc for the STAR-regulated constitutive promoter (red) and stabilized rSFP (blue) for each SC101 plasmid. Bars in b represent mean values in units of molecules of equivalent fluorescein (MEFL) determined by flow cytometry and error bars represent s.d. of n = 11 biological replicates. Color filled points represent individual data points of MEFL.
To demonstrate the effect of plasmid copy number on both a STAR-regulated constitutive promoter and the stabilized rSFP, we cloned mutants of the commonly used pSC101 plasmid backbone that exhibit a range of different copy numbers, between ~2 to ~3027,33. We observed that the STAR-regulated constitutive promoter system increased sfGFP expression as the pSC101 plasmid backbone increased in copy number, as expected. Importantly, there was negligible change in sfGFP expression when the stabilized rSFP was expressed from the various pSC101 mutant backbones (Fig. 5B). Interestingly, analysis of cell-to-cell variability observed across distributions of individual flow cytometry samples revealed lower variability for stabilized rSFPs compared with STAR-regulated constitutive promoters at high copy numbers (Fig. S4B), an anticipated feature of iFFLs27.
Overall, these results demonstrate our ability to leverage STARs to generate novel switchable feedback promoter circuits with different underlying feedback mechanisms. Our library provides tunable control of gene expression level by selecting different stress-response promoters or engineered promoter systems and manipulating inducer concentration. In addition, these rSFPs exhibit responsiveness to various sources of feedback (FPP and PglB stress19,20, transcription factor repression27), suggesting that the additional layer of regulation does not interfere with the feedback-responsiveness of these promoters.
rSFPs enable switchable control of metabolic pathway genes with stress-response promoters
We next tested the ability of stress-response promoter rSFPs to regulate expression of metabolic pathway genes. First, we sought to regulate a pathway for amorphadiene biosynthesis that involves the toxic intermediate metabolite farnesyl pyrophosphate (FPP). In this pathway, FPP production is encoded by the previously engineered MevT-MBIS operon with final conversion by an amorphadiene synthase from Artemisia annua (ADS)42 (Fig. 6A). Previous work showed that the PgadE stress-response promoter is downregulated by FPP stress and that amorphadiene production is improved when PgadE is configured to control expression of the MevT-MBIS pathway for FPP synthesis19. We constructed a variant of the MevT-MBIS pathway under PgadE rSFP control and performed small-scale amorphadiene fermentations to compare these variants with MevT-MBIS under an unregulated PgadE promoter. Upon analysis, we found that the PgadE rSFP produced 238+/−136 mg/L of amorphadiene, which was comparable with the amorphadiene titer of the unregulated PgadE variant (260+/−178 mg/L) (Fig. 6B), but with the additional ability to regulate induction that can be essential in industrial scale-up7. In comparison, cultivations with MevT-MBIS under control of a STAR-regulated constitutive promoter showed more heterogeneity in production between biological replicates, but with greater average amorphadiene titers (Fig. S5A,B). While this system would require further optimization for eventual application, these results confirm the ability of rSFPs to enable inducible control of multi-gene metabolic pathway operons expressed from a stress-response promoter. We also found that the stabilized promoter rSFP can control the amorphadiene pathway with similar fermentation experiments (Fig. S5C,D), however delivers relatively weak induction.
Figure 6.
Switchable control of an amorphadiene pathway with the PgadE rSFP. (a) Amorphadiene biosynthesis schematic depicting control of the MevT-MBIS pathway with the PgadE rSFP. The MevT module produces mevalonate from Acetyl-CoA and the MBIS module produces FPP from mevalonate. Amorphadiene synthase (ADS) converts FPP to amorphadiene. (b) Fermentation titers with E. coli DH1 containing pTrc-ADS and MevT-MBIS under control of the unregulated PgadE promoter or PgadE rSFP in the absence or presence of 100 ng/mL aTc added at inoculation. Fermentations were performed in supplemented M9 minimal media with 0.5 mM IPTG added at inoculation to induce ADS expression. Bars in b represent mean values of amorphadiene titers measured with GCMS after 72 hrs of fermentation and error bars represent s.d. of n = 6 biological replicates. Colored points represent individual data points of amorphadiene.
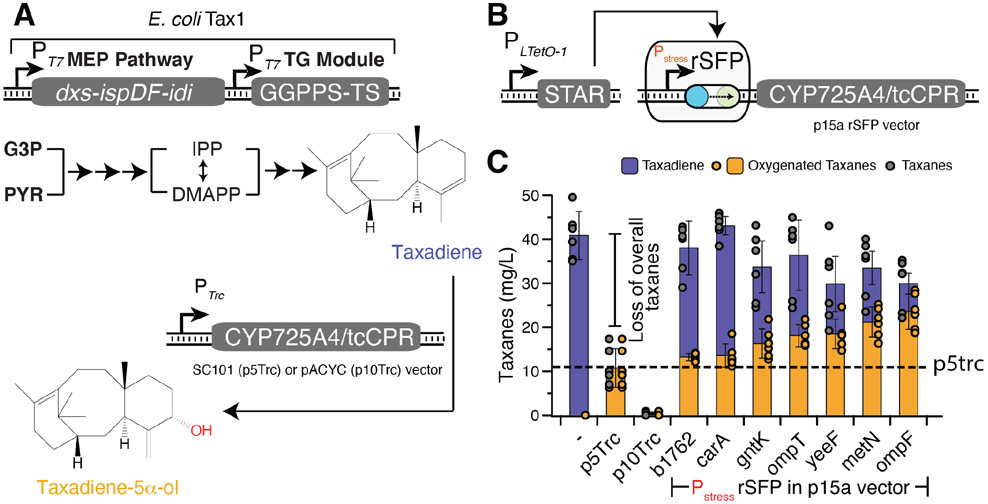
To demonstrate the modularity of rSFPs and their ability to improve pathway expression over a previous gold-standard, we next used them to regulate a portion of the anticancer drug paclitaxel’s biosynthesis pathway that has been previously reconstituted in E. coli43. We focused on the first P450-mediated step where taxadiene is oxygenated by the membrane anchored cytochrome P450 CYP725A4 (Fig. 7A) and can be converted to Taxol through additional enzymatic or synthetic routes44. Previous work has shown that expression level of CYP725A4 and its reductase partner is critical to achieving high titers of oxygenated taxanes in E. coli43. A previously optimized low-copy expression vector (p5Trc-CYP725A4/tcCPR) (Fig. 7A) transformed into the E. coli Tax1 strain containing genomic modifications to maximize the synthesis of the taxadiene precursor, produces ~11 mg/L of oxygenated taxanes in our experiments (Fig. 7C). However, as found before, increasing expression of the enzyme using a medium copy expression vector (p10Trc) does not increase titer, but causes a complete loss of pathway productivity (Fig. 7C), presumably due to the enzyme’s membrane stress crossing a critical threshold and triggering a global response.
Figure 7.
Control of a taxadiene oxygenation pathway with rSFPs. (a) Taxol biosynthesis schematic depicting an abbreviated overview of the Taxol precursor pathway involving the toxic cytochrome P450 (CYP) 725A4 enzyme. In the E. coli strain Tax1, the methylerythritol phosphate (MEP) pathway and taxadiene synthase/geryanlgeranyl diphosphate (GGPP) synthase (TS) module convert glyceraldehyde-3-phosphate (G3P) and pyruvate (PYR) into the 20-carbon backbone taxa-4 (5),11 (12)-diene (taxadiene). Taxadiene is oxygenated by the CYP725A4/tcCPR fusion enzyme to form taxadiene-5α-ol. CYP725A4/tcCPR was initially expressed from a standard IPTG-inducible PTrc promoter in either a low copy (p5Trc) or medium copy (p10Trc) plasmid. IPP = isopentenyl diphosphate, DMAPP = dimethylallyl diphosphate. (b) rSFPs are applied to control CYP725A4/tcCPR utilizing the PLTetO-1-STAR vector. (c) Fermentation titers with E. coli Tax1 containing an empty vector (−), p5Trc, p10Trc, or an rSFP plasmid for expression of CYP725A4/tcCPR with 100 ng/mL aTc added at inoculation. Dashed line represents production of oxygenated taxanes from p5Trc. Bars in c represent mean values of taxane titers measured with GCMS after 96 hrs of fermentation and error bars represent s.d. of at least n = 4 biological replicates. Grey filled points represent individual data points of overall taxanes, and orange filled points represent individual data points of oxygenated taxanes.
We hypothesized we could achieve greater pathway productivity over the p5Trc benchmark strain by identifying relevant rSFPs for control of CYP725A4/tcCPR. To test this, the CYP725A4/tcCPR coding sequence was introduced into each of the 17 rSFP constructs (Fig. 7B). E. coli Tax1 was transformed with each rSFP construct and the PLTetO-1-STAR plasmid and tested in the context of taxadiene oxygenation cultivations, with the STAR induced from inoculation. Using this approach, we found that several performed well against the p5Trc benchmark strain (Fig. S6). In particular, 7 of the rSFPs had greater titers of oxygenated taxanes than the p5Trc strain (Fig. 7C), with all also improving overall taxane production. Furthermore, the PompF rSFP resulted in ~2.2x fold greater oxygenated taxanes (~23.5+/−4.0 mg/L) and ~2.8x fold greater overall taxanes (29.9+/−5.8 mg/L) than the p5Trc strain.
To confirm that rSFPs can indeed be feedback regulated by CYP725A4/tcCPR stress, we performed fluorescence analysis of E. coli cells containing plasmids for rSFP expression of an mCherry reporter and the p10Trc plasmid separately expressing CYP725A4/tcCPR, in order to monitor changes in rSFP expression caused by membrane stress (Fig. S7A). We observed reduced expression from PompF when p10Trc was present in place of an empty vector (Fig. S7B), suggesting that it is indeed responsive to CYP725A4/tcCPR induced stress. A constitutive promoter control had no response as expected (Fig. S7B). Interestingly, the PmetN rSFP did not exhibit responsiveness to CYP725A4/tcCPR expression, despite our earlier observation that it did respond to PglB expression (Fig. 4). This finding indicates that not all rSFPs respond to stresses in the same way and that CYP725A4/tcCPR presents a unique stress compared to PglB, highlighting the need to pair different stresses to appropriate stress-response promoters.
Controls with varied strength constitutive promoters regulated by STARs were also run and one combination was found to achieve similar titers to the PompF rSFP (Fig. S8A-E). This suggests that in this pathway the introduction of a STAR to control promoter output may help contribute to improved pathway performance but requires the highest strength promoter (PapFAB45). To further explore the impact of introducing STAR regulation, we performed fermentations with unregulated PmetN and PompF promoters replacing the corresponding rSFPs (Fig. S8F). We observed that rSFPs outperformed unregulated stress-response promoters in both cases with regards to total taxane production and, in the case of the PmetN rSFP, oxygenated taxane production (Fig. S8G).
We next explored how the external control offered by rSFPs can be used to further optimize induction level and timing of stress-response promoter activity. To test this, we selected the two best rSFP systems and performed a matrix of aTc induction at four levels (0, 16, 32, and 100 ng/mL aTc), which were added at six different induction times (0, 3, 6, 12, 24, 48 hrs) post inoculation (Fig. 8A). We found that oxygenated taxane production with both rSFPs was sensitive to induction level and timing (Fig. 8B,C, S9A,B) and that optimizing induction of PmetN and PompF rSFPs could improve final titers of oxygenated taxanes further to 25.4+/−0.9 and 25.1+/−1.3 mg/L, respectively, and overall taxanes to 39.0+/−4.8 and 31.0+/−2.9 mg/L (Fig. 8D,E), representing an overall 2.4x and 2.3x fold improvement over the previous gold standard benchmark in terms of oxygenated taxanes, and 3.6x and 2.9x fold improvements in terms of overall taxanes, demonstrating potential performance advantages of inducible control in rSFPs.
Figure 8.
Induction optimization of the best performing taxadiene oxygenation rSFP strains. (a) Conditions of aTc induction level and timing used for rSFP CYP725A4/tcCPR expression optimization. (b, c) Induction level and timing optimization of fermentations with E. coli Tax1 containing the (b) PmetN or (c) PompF rSFP controlling CYP725A4/tcCPR. Heatmaps show mean values of oxygenated taxane titers measured with GCMS for different combinations of aTc concentration and induction time after 96 hrs of fermentation. (d, e) Fermentation titers with E. coli Tax1 containing the (d) PmetN or (e) PompF rSFP without aTc induction, with aTc induction before optimization (100 ng/mL aTc at 0 hrs), and with aTc induction after optimization. Dashed line represents production of oxygenated taxanes from p5Trc in Fig. 7C. Bars in d, e represent mean values of taxane titers measured with GCMS after 96 hrs of fermentation and error bars represent s.d. of at least n = 4 biological replicates. Grey filled points represent individual data points of overall taxanes, and orange filled points represent individual data points of oxygenated taxanes. P-values indicate a statistically significant difference in oxygenated taxane production by a Welch’s t-test (two-tailed, unequal variances) between the unoptimized and optimized conditions.
Overall, we demonstrate that the rSFP regulation concept is modular, effectively enabling inducible control and optimization of metabolic pathway production using different stress-response promoters and different metabolic pathways. Importantly, rSFPs enable tuning of expression timing and overall magnitude of stress-response promoter output to further enhance fermentation titers.
Quorum-sensing activated rSFPs allow autonomous regulation of pathway expression
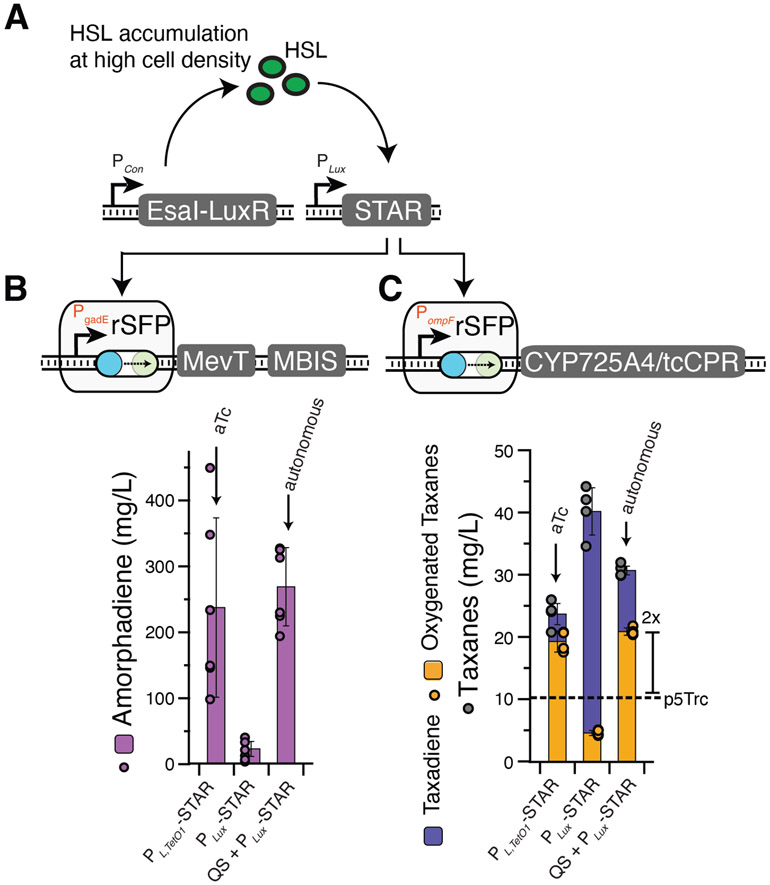
Though inducible systems offer flexibility for screening of optimal induction timing, the cost of inducers can be prohibitive at an industrial scale45,46, and several efforts have been carried out to design autonomous means of induction. Quorum-sensing (QS) systems that are activated in a cell-density dependent manner offer one such route to this behavior47. QS systems have been used with great utility in metabolic engineering to create a separation of cell growth and pathway production phases without the need for a chemical inducer and provide a natural means for balancing carbon utilization with biomass production48-50. We therefore utilized this strategy within our model pathways by leveraging the modularity of rSFPs to be easily configured to utilize different input systems. Specifically, we chose the PLux promoter that is activated by the LuxR transcriptional activator upon sufficient production of the C6-homoserine lactone (HSL) signaling molecule51, since we had previously used PLux/LuxR to control STAR production30. In addition, we chose the EsaI HSL synthase52 because it had previously been used successfully in metabolic engineering applications49. We cloned a STAR under control of PLux and integrated an operon with the EsaI and LuxR into the genome of the E. coli DH1 or Tax1 to create DH1-QS and Tax1-QS strains (Fig. 9A). When plasmids encoding the expression of PLux-STAR and the PgadE rSFP controlling mCherry expression were transformed into E. coli DH1 or DH1-QS, we found that activation only occurred in the engineered DH1-QS strain containing EsaI and LuxR (Fig. 9B). Similarly, transformation of a PLux-STAR vector and the PompF rSFP into E. coli Tax1 or Tax1-QS resulted in mCherry fluorescence activation only in Tax1-QS (Fig. 9C). These QS-activated rSFPs produced mCherry autonomously over time with comparable fold activation to manual induction with PLTetO-1.
Figure 9.
Characterization of autonomous quorum-sensing rSFPs. (a) Schematic showing the PLux rSFP activation system in the presence of EsaI and LuxR to allow autonomous control of pathway expression. LuxR is activated by C6-HSL produced by the EsaI HSL synthase upon sufficient accumulation due to an increase in cell density. LuxR activation results in STAR production from the PLux promoter, thereby activating rSFP expression. (b, c) Fluorescence experiments showing autonomous activation of mCherry expression over time with (b) the PgadE rSFP transformed in E. coli DH1 or E. coli DH1-QS and (c) the PompF rSFP transformed in E. coli Tax1 or E. coli Tax1-QS. Lines in b, c represent mean values in units of arbitrary fluorescence/optical density (FL/OD) and shaded areas represent mean values +/− s.d. of at least n = 9 biological replicates. Colored points represent individual data points for each condition.
To demonstrate that QS-activated rSFPs could be used to autonomously control the expression of metabolic pathway enzymes, we applied the PgadE QS-activated rSFP to control expression of MevT-MBIS within the amorphadiene pathway and the PompF QS-activated rSFP to control CYP725A4/tcCPR expression within the taxadiene oxygenation pathway. Cultivations were performed by inoculating cell cultures into media without addition of exogenous inducer. Upon cultivation and analysis, we found that QS-based activation in both systems resulted in comparable titers of amorphadiene or oxygenated taxanes to those obtained from manual induction (Fig. 10). Importantly, this was achieved with a completely autonomous genetic feedback network without the need for costly inducers and, for the oxygenated taxane pathway, this represented a 2x-fold improvement over the previous gold standard. These results demonstrate the composability of rSFPs, showing that QS systems can be configured to autonomously activate expression of rSFPs to regulate metabolic pathways with favorable performance when compared to manually induced rSFP configurations.
Figure 10.
Autonomous control of metabolic pathways with quorum-sensing rSFPs. (a) Schematic showing the PLux rSFP activation system in the presence of EsaI and LuxR to allow autonomous control of pathway expression. (b) Amorphadiene fermentation titers with the PgadE rSFP controlling MevT-MBIS expression and pTrc-ADS. Left: E. coli DH1 (without QS insert) containing a PLTetO-1-STAR plasmid induced by aTc; middle: E. coli DH1 containing a PLux-STAR plasmid; right: E. coli DH1-QS containing a PLux-STAR plasmid. (c) Oxygenated taxadiene fermentation titers with the PompF rSFP controlling CYP725A4/tcCPR expression. Left, middle, and right bars contain same conditions as a, but for oxygenated taxadiene fermentations in E. coli Tax1. Dashed line represents production of oxygenated taxanes from p5Trc in Fig. 7C. Bars in a represent mean values amorphadiene titers measured with GCMS after 72 hrs of fermentation and error bars represent s.d. of n = 6 biological replicates. Color filled points represent individual data points. Bars in b represent mean values taxane titers measured with GCMS after 96 hrs of fermentation and error bars represent s.d. of n = 4 biological replicates. Grey filled points represent individual data points of overall taxanes, and orange filled points represent individual data points of oxygenated taxanes.
Discussion
Here we report the development, characterization and application of switchable feedback promoters that enable an additional synthetic layer of control over natural stress-response promoters and engineered feedback promoter systems. Stress-response promoters are a promising route to achieving dynamic control of heterologous metabolic pathways by acting as sensor-actuators to stresses caused by pathway expression, intermediate metabolites and other fermentation conditions19,20. While stress-response promoters have previously been shown to improve production of desired chemicals by regulating expression in response to toxic pathway intermediates and enzymes, their use is constrained by their complexity in terms of their specific signaling pathways and regulatory architecture, which may not be fully understood. This has led to a lack of control over the timing and overall magnitude of their transcriptional output, which is essential to achieving a separation of growth phase and production phase in large-scale fermentations7,53. This same limitation is also true of many engineered promoter systems, including stabilized promoters that buffer gene expression from changes in copy number27.
By design, the rSFP concept enables switchable control by introducing an additional regulatory layer within the natural or engineered feedback pathway by gating stress-response promoter outputs with trans-acting RNA regulators. The use of an exogenous small molecule-inducible system to control RNA regulator synthesis allows modification of the timing and overall magnitude of the feedback promoter outputs. Furthermore, the use of QS systems allows the autonomous activation of rSFPs in a cell-density dependent manner. In this way, rSFPs are a composable element and have modularity at the level of their activation inputs, gene expression outputs, and the types of stresses they can respond to through changing of the regulated feedback promoter. By utilizing transcriptional RNA regulators, rSFPs offer the flexible implementation of controllable feedback networks in a single compact locus that is convenient for expression of operons. However, translational RNA regulators such as toehold switches54 or antisense RNAs55, configured to appropriately regulate the individual genes within an operon, could be used with similar results. Although transcriptional activation is expected to be optimal in many applications, an additional benefit of the rSFP system is the flexible ability to swap transcriptional activation of STARs with alternative modalities for transcriptional repression56,57. Alternative technologies, such as CRISPR interference58, would be difficult to implement to control stress-response promoter outputs in metabolic pathways due to the need for expressing additional burdensome components (e.g. dCas9) and the reliance on repression, rather than activation afforded by STARs. Along these lines, technologies such as the burden-driven feedback controller that leverages stress-response promoters to dynamically regulate CRISPR gRNAs18 could be enhanced by our rSFP approach by enabling inducible control of gRNA expression while maintaining burden-driven feedback.
We demonstrate that rSFPs are both modular and tunable – the rSFP concept can be applied to many unique stress-response promoters as well as the engineered stabilized promoter system in a plug-and-play fashion, activator inputs can be easily interchanged, and activated output levels can be modulated by titrating inducer concentrations. To demonstrate their utility in the context of optimizing metabolic pathway production, we applied rSFPs to regulate expression of a multi-gene operon in amorphadiene biosynthesis and a toxic cytochrome P450 enzyme in a synthetic Taxol precursor pathway in E. coli43,59, enabling inducible control of pathway expression and improvements in production of the desired oxygenated taxane. We also showed that optimizing rSFP induction timing and magnitude in the oxygenated taxane fermentation enabled additional improvements, highlighting an advantage of the rSFP system to enable the control of pathway expression timing. We next showed that rSFPs can be controlled by QS systems that do not require addition of an exogenous inducer, enabling fully autonomous control of pathway expression. We developed rSFPs in E. coli and the system is likely adaptable to bacterial metabolic engineering hosts such as Pseudomonas putida, Bacillus subtilis, or Acinetobacter baylyi. However, future development of RNA transcriptional regulators or implementing rSFPs with other types of genetic control could be used to adapt the concept for yeast or other organisms.
Notably, we found that all 18 of the stress-response promoters, and the stabilized promoter, were activated significantly, strongly suggesting that the concept can be used with new feedback promoters as they are discovered in nature or engineered. In the case where an individual promoter does not perform well in rSFPs, which can be caused by extra 5’ UTR sequence downstream of the stress-response promoter, alternative STAR/target pairs30 may be screened or the extra 5’ UTR may be trimmed for improved fold activation. Characterization of selected promoters under stress caused by buildup of a toxic intermediate metabolite or expression of toxic proteins showed that rSFPs remain responsive to stress as expected based on the behavior of unregulated stress-response promoters. These features allow rapid screening of rSFP libraries within combinatorial strain engineering procedures60 that could be used by industry to identify effective implementations of dynamic control by capturing the unique temporal profiles and feedback responsiveness of different stress-response promoters. In addition, the ability of rSFPs to naturally adapt to an optimal expression level may allow for rapid prototyping of potentially toxic enzymes and pathways without the requisite need to first balance expression levels with constitutive static regulators – speeding the pace of pathway construction for new chemical products, especially if rSFPs become well characterized for use with specific types of stress.
Dynamic pathway regulation is a promising approach in the construction of genetic systems but can be difficult to implement. The rSFP strategy enables modular and tunable control of natural and engineered feedback-responsive promoters that have sophisticated transcriptional responses to a range of cellular stresses and cues. Due to their simplicity, we envision that the rSFP concept will enable streamlined implementation of dynamic regulation into metabolic pathways. Furthermore, given their modularity, we imagine rSFPs will be useful for dynamic control in other applications, such as high-level expression of difficult or toxic proteins, living therapeutics3, and cellular diagnostics4 where endogenous and engineered promoters could be used as sensor-actuators for numerous environments.
Methods
Plasmid assembly.
All plasmids used in this study can be found in Table S13 with key sequences provided in Tables S13 and S14. Gibson assembly and inverse PCR (iPCR) was used for construction of all plasmids. All assembled plasmids were verified using DNA sequencing. rSFPs for the 17 envelope stress-response promoters and the stabilized promoter all used STAR/target variant 8 and the PgadE rSFP used STAR/target variant 3. The downstream end of each stress-response promoter was defined as the 5’ adjacent nucleotide to the start codon of its endogenously regulated gene. Cognate STARs were cloned in a second PLTetO-1 or PLux plasmid.
Integration of QS operon into the E. coli genome.
Strains containing genomic insertions of the EsaI-LuxR operon were created using the clonetegration61 platform for creation of E. coli Tax1-QS or λ Red recombineering62 for E. coli DH1-QS as summarized in Table S15. For clonetegration, the HK022 plasmid was used to integrate constructs into the attB site of the E. coli genome. Successful integrations were identified by antibiotic selection and colony PCR according to the published protocol. For recombineering, double-stranded DNA fragments flanked upstream and downstream by 40 bp of homology to the attB site were generated for both the cat-sacB cassette and the EsaI-LuxR operon. Homology to the attB site was included in oligos and appended to each insert via PCR. The cat-sacB cassette was amplified from a purified E. coli TUC01 genome. E. coli DH1 carrying the pSIM6 plasmid was subjected to two rounds of recombineering according to the published protocol62. The first round inserted the cat-sacB cassette at the attB locus, and the second round replaced the cat-sacB cassette with the EsaI-LuxR operon. Successful integrations were identified by resistance to chloramphenicol (first round) or growth on sucrose and colony PCR (second round). Insertion of the complete EsaI-LuxR operon was confirmed by Sanger sequencing.
Strains, growth media, in vivo bulk fluorescence measurements.
Fluorescence characterization experiments for all envelope stress-response promoters (Fig. 3B, 4B, 9C) were performed in E. coli strain Tax143 containing the synthetic pathway for taxadiene biosynthesis or modified Tax1-QS containing the QS operon. Experiments involving the PgadE promoter (Fig. 2A-C, 9B) were performed in E. coli strain DH1 (F– λ– endA1 recA1 relA1 gyrA96 thi-1 glnV44 hsdR17(rK–mK–)) or modified DH1-QS containing the QS operon. Experiments were performed for at least 7 biological replicates collected over two separate days. For each day of fluorescence measurements, plasmid combinations were transformed into chemically competent E. coli cells and plated on LB+Agar (Difco) plates containing combinations of 100 μg/mL carbenicillin, 34 μg/mL chloramphenicol and/or 50 μg/mL spectinomycin depending on plasmids used (see Table 13 for plasmids used in each experiment), and incubated approximately 17 hours (h) overnight at 37 °C. Plates were taken out of the incubator and left at room temperature for approximately 7 h. Three colonies were used to inoculate three cultures of 300 μL of LB containing antibiotics at the concentrations described above in a 2 mL 96-well block (Costar), and grown for approximately 17 h overnight at 37 °C at 1,000 rpm in a Vor-Temp 56 (Labnet) bench top shaker. Fig. 2B, 3B: 4 μL of each overnight culture were added to 196 μL (1:50 dilution) of supplemented M9 minimal media (1 × M9 minimal salts, 1 mM thiamine hydrochloride, 0.4 % glycerol, 0.2 % casamino acids, 2 mM MgSO4, 0.1 mM CaCl2) containing the selective antibiotics and grown for 6 h at the same conditions as the overnight culture. Appropriate concentrations of anhydrotetracycline (Sigma) were added to culture media as indicated. Fig. 2A, 2C, 9B, 9C: 20 μL of each overnight culture were added to 980 μL of M9 minimal media containing selective antibiotics and grown for 24 h at 37C. Periodic samples of 10-200 μL of culture were collected for characterization by bulk fluorescence measurements. Fig. 4B: 4 μL of each overnight culture were added to 196 μL of LB media containing selective antibiotics and grown for 2.5 h at 37C. 200 μg/mL L-arabinose was added to appropriate conditions at 2.5 h. After another 4 hrs of growth at 37C, 100 μL were sampled for characterization by bulk fluorescence measurements. For all bulk fluorescence measurements: 10-200 μL of sampled culture were transferred to a 96-well plate (Costar) containing 0-190 μL of phosphate buffered saline (PBS). Fluorescence (FL) and optical density (OD) at 600 nm were then measured using a Synergy H1 plate reader (Biotek). The following settings were used: mCherry fluorescence (560 nm excitation, 630 nm emission).
Bulk fluorescence data analysis.
On each 96-well block there were two sets of controls; a media blank and E. coli Tax1 cells transformed with combination of control plasmids JBL002 and JBL644 (blank cells) and thus not expressing mCherry (Table S12). The block contained three replicates of each control. OD and FL values for each colony were first corrected by subtracting the corresponding mean values of the media blank. The ratio of FL to OD (FL/OD) was then calculated for each well (grown from a single colony) and the mean FL/OD of blank cells was subtracted from each colony’s FL/OD value. Three biological replicates were collected from independent transformations, with three colonies characterized per transformation (9 colonies total). Occasional wells were discarded due to poor growth (OD < 0.1 at measurement), however, all samples contained at least 7 replicates over the three experiments. Means of FL/OD were calculated over replicates and error bars represent standard deviations (s.d). Fold activation was calculated as FL/OD for each colony grown with 100 ng/mL aTc over the same colony with 0 ng/mL aTc. Means of fold activation were calculated over replicates and error bars represent standard deviations (s.d).
Flow cytometry data collection and analysis for sfGFP fluorescence analysis of stabilized rSFP variants.
All flow cytometry experiments were performed in E. coli strain TG1 (F'traD36 lacIq Delta(lacZ) M15 pro A+B+/supE Delta(hsdM-mcrB)5 (rk- mk- McrB-) thi Delta(lac-proAB)). Plasmid combinations were transformed into chemically competent E. coli cells, plated on Difco LB+Agar plates containing 100 μg/mL carbenicillin and 34 μg/mL chloramphenicol (see Table S12 for plasmids used in each experiment), and grown overnight at 37 °C. Following overnight incubation, plates were left at room temperature for approximately 7 h. Individual colonies were grown overnight in LB, then diluted 1:50 into M9 minimal media. After 6 h, cells were diluted 1:100 in 1x Phosphate Buffered Saline (PBS) containing 2 mg/mL kanamycin. A BD Accuri C6 Plus flow cytometer fitted with a high-throughput sampler was then used to measure sfGFP fluorescence. Measurements were taken for 11 biological replicates collected over two separate experiments on different days.
Flow cytometry data analysis was performed using FlowJo (v10.4.1). Cells were gated by FSC-A and SSC-A, and the same gate was used for all samples prior to calculating the geometric mean fluorescence for each sample. All fluorescence measurements were converted to Molecules of Equivalent Fluorescein (MEFL) using CS&T RUO Beads (BD cat#661414). The average fluorescence (MEFL) over replicates of cells expressing empty plasmids (pJBL001 and pJBL002) was then subtracted from each measured fluorescence value. Robust CV was calculated for each measurement using FlowJo (v10.7.1).
Amorphadiene fermentation.
Small-scale batch fermentations were used to evaluate amorphadiene production. Experiments were performed with at least 5 biological replicates over two independent experiments. E. coli DH1 cells were transformed with PTrc-ADS (subcloned into the pCDF vector), the appropriate MevT-MBIS plasmid, and the PLTetO-1-STAR or PLux-STAR plasmid as appropriate. An inadvertent T303N mutation was present in the MK gene of all MevT-MBIS plasmid variants used in this study. Individual colonies were inoculated into culture tubes containing LB and appropriate antibiotics and incubated at 37°C for roughly 16 hrs overnight to achieve an approximate OD600 of 3. 125 uL of overnight cells were inoculated into tubes of 4.875 mL supplemented M9 minimal media (1 × M9 minimal salts, 1 mM thiamine hydrochloride, 0.2 % casamino acids, 2 mM MgSO4, 0.1 mM CaCl2) with 1% glucose and a 10% dodecane overlay to capture amorphadiene. Cultures were induced with 0.5 mM IPTG and 100 ng/mL aTc as appropriate. Tubes were incubated at 37°C and 250 rpm for 72 hrs. After the fermentations were complete, the cultures were centrifuged to collect the dodecane overlay. These overlays were subsequently diluted into hexane for analytical procedures described below.
Small-scale “Hungate” fermentation.
Small-scale fermentation assays were used to quantify oxygenated taxanes and taxadiene production in E. coli Tax1 or Tax1-QS. Experiments were performed with six biological replicates collected over three independent experiments (Fig. 7C) or four biological replicates collected over two independent experiments (Fig. 8B, 8C, 8D, 8E, 10C). For each experiment, plasmid combinations (Table S12) were transformed into chemically competent E. coli cells and plated on LB+Agar (Difco) plates containing appropriate antibiotics (100 μg/mL carbenicillin, 34 μg/mL chloramphenicol and/or 50 μg/mL spectinomycin). Plates were incubated approximately 17 hrs overnight at 30 °C. Individual colonies were inoculated into culture tubes containing LB and appropriate antibiotics and incubated at 30°C for roughly 16 hrs overnight to achieve an approximate OD600 of 3. For 2 mL batch fermentations, 50 μL of overnight cells were added to 1.95 mL of complete R-media (Tables S16-S18) and appropriate antibiotics in glass hungate tubes (ChemGlass). 0.1 mM IPTG was added for induction of the upstream pathway enzymes and p5Trc/p10Trc expression. 16-100 ng/mL aTc was added, as indicated, to induce PLTetO-1-STAR activated rSFPs. A 10% v/v dodecane layer (200 μL) was added in all fermentations. Hungate tubes were sealed with a rubber septum and plastic screwcap (ChemGlass). PrecisionGlide 18G hypodermic needles (BD) were inserted into the rubber septa to allow for gas exchange. Hungate tubes were incubated at 22°C and 250 rpm for 96 hrs. After the fermentations were completed, the culture was centrifuged to collect the dodecane overlay. This overlay was subsequently diluted into hexane for analytical procedures described below.
GC-MS analysis.
Dodecane samples collected from batch fermentations were diluted at a ratio of 1:20 (for taxadiene fermentations) or 1:200 (for amorphadiene fermentations) in n-hexane containing 5 mg/L ß-caryophyllene. The 5 mg/L ß-caryophyllene was utilized as a standard to calculate titer of taxadiene and oxygenated taxanes. GC-MS analysis was performed with an Agilent 7890 GC and Agilent HP-5ms-UI column (Ultra Inert, 30 m, 0.25 mm, 025 μm, 7 in cage). Helium was utilized as a carrier gas at a flow rate of 1 mL/min and the sample injection volume was 1 μL. The splitless method begins at 50 °C hold for 1 minute followed by a 10°C/min ramp to 200 °C and a final 5°C/min ramp to 270 °C (final ramp excluded for amorphadiene analysis). Mass spectroscopy data was collected for 22.5 minutes with an 11-minute solvent delay. m/z values ranging from 40-500 were scanned with a scan time of 528ms. MassHunter Workstation Qualitative Analysis software (vB.06.00) was utilized to integrate peaks on the chromatograms and determine their respective mass spectrums (Fig. S10). The ratio of peak area of taxadiene (m/z 272) and amorphadiene (m/z 204) to the standard ß-caryophyllene (m/z 204) was used to calculate titer of taxadiene and amorphadiene, while the ratio of the sum of all peaks of oxygenated taxanes (m/z 288) to ß-caryophyllene was used to calculate titer of the oxygenated taxanes. Overall taxanes were calculated by summing taxadiene and oxygenated taxane titers for each sample. Means of titers were calculated over replicates and error bars represent s.d.
Data and materials availability
All data presented in this manuscript are available as supporting data files. The E. coli Tax1 strain and P450/tcCPR fusion were obtained under an MTA with Manus Bio and cannot be distributed by the authors. Requests for those materials must be made to Manus Bio directly. All other biological materials will be made available upon request or through Addgene at publication and may require a material transfer agreement (Addgene Link: https://www.addgene.org/browse/article/28207639/).
Supplementary Material
ACKNOWLEDGMENTS
The authors gratefully acknowledge Dr. Ryan Philippe for careful reading of the manuscript, the gift of E. coli Tax1 and plasmids p5Trc and p10Trc from Manus Bio, and Taylor Nichols for helpful discussions. The pOSIP plasmid kit used for clonetegration was a gift from Drew Endy and Keith Shearwin (Addgene kit # 1000000035). E. coli DH1, pPgadE-MevT-MBIS and pTrc-ADS were gifts from Jay Keasling. pTHSSe_59 was a gift from Christopher Voigt (Addgene plasmid # 109253; http://n2t.net/addgene:109253; RRID:Addgene 109253).
Funding Sources
This work was supported by an NSF CAREER award (1452441 to J.B L.), an NSF CBET award (1803747 to J.B.L., K.E.J.T. and D.T.-E.), an NSF Graduate Research Fellowship (DGE-1144153 to C.J.G.), an NIH Biotechnology Training Grant (T32-GM008449-23 to B.W.B.) and an NSF Synthetic Biology REU (DBI-1757973 to A.V.).
ABBREVIATIONS
- rSFP
riboregulated switchable feedback promoter
- RBS
ribosomal binding site
- STAR
small transcription activating RNA
- FPP
farnesyl pyrophosphate
- aTc
anhydrotetracycline
- HSL
homoserine lactone
- IPP
isopentenyl diphosphate, DMAPP, dimethylallyl diphosphate
- MEP
methylerythritol phosphate
- GGPP
geryanlgeranyl diphosphate G3P, glyceraldehyde-3-phosphate
- PYR
pyruvate
Footnotes
Conflicts of Interest
A non-provisional patent application has been filed by C.J.G., B.W.B., D.T.-E., K.E.J.T., and J.B.L. for the invention and application of riboregulated switchable feedback promoters (PCT/US2019/051133).
Supporting Information
Supplemental Figures and Tables: Glasscock_ACS_SynBiol_SI.pdf.
Source Data: Glasscock_ACS_SynBiol_Source_Data.xlsx
REFERENCES
- 1.Keasling JD (2010) Manufacturing molecules through metabolic engineering. Science 330, 1355–1358. doi: 10.1126/science.1193990. [DOI] [PubMed] [Google Scholar]
- 2.Nielsen J and Keasling JD (2016) Engineering Cellular Metabolism. Cell 164, 1185–1197. doi: 10.1016/j.cell.2016.02.0043. [DOI] [PubMed] [Google Scholar]
- 3.Isabella VM, Ha BN, Castillo MJ, Lubkowicz DJ, Rowe SE, Millet YA, Anderson CL, Li N, Fisher AB, West KA, Reeder PJ, Momin MM, Bergeron CG, Guilmain SE, Miller PF, Kurtz CB, and Falb D (2018) Development of a synthetic live bacterial therapeutic for the human metabolic disease phenylketonuria. Nat. Biotechnol. 36, 857–864. doi: 10.1038/nbt.4222. [DOI] [PubMed] [Google Scholar]
- 4.Watstein DM, McNerney MP and Styczynski MP (2015) Precise metabolic engineering of carotenoid biosynthesis in Escherichia coli towards a low-cost biosensor. Metab. Eng. 31, 171–180. doi: 10.1016/j.ymben.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biggs BW, De Paepe B, Santos CNS, De Mey M and Ajikumar PK (2014) Multivariate modular metabolic engineering for pathway and strain optimization. Curr. Opin. Biotechnol. 29, 156–162. doi: 10.1016/j.copbio.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 6.Sun J, Jeffryes JG, Henry CS, Bruner SD and Hanson AD (2017) Metabolite damage and repair in metabolic engineering design. Metab. Eng. 44, 150–159. doi: 10.1016/j.ymben.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 7.Rugbjerg P and Sommer MOA (2019) Overcoming genetic heterogeneity in industrial fermentations. Nat. Biotechnol. 37, 869–876. doi: 10.1038/s41587-019-0171-6. [DOI] [PubMed] [Google Scholar]
- 8.Holtz WJ and Keasling JD (2010) Engineering Static and Dynamic Control of Synthetic Pathways. Cell 140, 19–23. doi: 10.1016/j.cell.2009.12.029. [DOI] [PubMed] [Google Scholar]
- 9.Brockman IM and Prather KLJ (2015) Dynamic metabolic engineering: New strategies for developing responsive cell factories. Biotechnol. J. 10, 1360–1369. doi: 10.1002/biot.201400422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan SZ and Prather KL (2017) Dynamic pathway regulation: recent advances and methods of construction. Curr. Opin. Chem. Biol. 41, 28–35. doi: 10.1016/j.cbpa.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 11.Alper H, Fischer C, Nevoigt E and Stephanopoulos G (2005) Tuning genetic control through promoter engineering. Proc. Natl. Acad. Sci. 102, 12678–12683. doi: 10.1073/pnas.0504604102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jensen PR and Hammer K (1998) The sequence of spacers between the consensus sequences modulates the strength of prokaryotic promoters. Appl. Environ. Microbiol. 64, 82–87. doi: 10.1128/AEM.64.1.82-87.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pfleger BF, Pitera DJ, Smolke CD and Keasling JD (2006) Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes. Nat. Biotechnol. 24, 1021–2032. doi: 10.1038/nbt1226. [DOI] [PubMed] [Google Scholar]
- 14.Barrick D, Villanueba K, Childs J, Kalil R, Schneider TD, Lawrence CE, Gold L, and Stormo GD (1994) Quantitative analysis of ribosome binding sites in E. coli. Nucleic Acids Res. 22, 1287–1295. doi: 10.1093/nar/22.7.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mutalik VK, Guimaraes JC, Cambray G, Lam C, Christoffersen MJ, Mai QA, Tran AB, Paul M, Keasling JD, Arkin AP, and Endy D (2013) Precise and reliable gene expression via standard transcription and translation initiation elements. Nat. Methods. doi: 10.1038/nmeth.2404. [DOI] [PubMed] [Google Scholar]
- 16.Salis HM, Mirsky EA and Voigt CA (2009) Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 27, 946–950. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gottesman S (2019) Trouble is coming: Signaling pathways that regulate general stress responses in bacteria. J. Biol. Chem. 294, 11685–11700. doi: 10.1074/jbc.REV119.005593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ceroni F Boo A, Furini S, Gorochowski TE, Borkowski O, Ladak YN, Awan AR, Gilbert C, Stan GB, and Ellis T (2018) Burden-driven feedback control of gene expression. Nat. Methods 15, 387–393. doi: 10.1038/nmeth.463519. [DOI] [PubMed] [Google Scholar]
- 19.Dahl RH Zhang F, Alonso-Gutierrez J, Baidoo E, Batth TS, Redding-Johanson AM, Petzold CJ, Mukhopadhyay A, Lee TS, Adams PD, and Keasling JD (2013) Engineering dynamic pathway regulation using stress-response promoters. Nat. Biotechnol. 31, 1039–1046. doi: 10.1038/nbt.2689. [DOI] [PubMed] [Google Scholar]
- 20.Boyarskiy S, Davis López S, Kong N and Tullman-Ercek D (2016) Transcriptional feedback regulation of efflux protein expression for increased tolerance to and production of n-butanol. Metab. Eng. 33, 130–137. doi: 10.1016/j.ymben.2015.11.00521. [DOI] [PubMed] [Google Scholar]
- 21.Zhang F, Carothers JM and Keasling JD (2012) Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nat. Biotechnol. 30, 354–359. doi: 10.1038/nbt.2149. [DOI] [PubMed] [Google Scholar]
- 22.Xu P, Li L, Zhang F, Stephanopoulos G and Koffas M (2014) Improving fatty acids production by engineering dynamic pathway regulation and metabolic control. Proc. Natl. Acad. Sci. U.S.A. 111, 11299–11304. doi: 10.1073/pnas.1406401111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moser F Borujeni AE, Ghodasara AN, Cameron E, Park Y, and Voigt CA (2018) Dynamic control of endogenous metabolism with combinatorial logic circuits. Mol. Syst. Biol. 14, e8605. doi: 10.15252/msb.20188605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carothers JM, Goler JA, Juminaga D and Keasling JD (2011) Model-driven engineering of RNA devices to quantitatively program gene expression. Science 334, 1716–1719. doi: 10.1126/science.1212209. [DOI] [PubMed] [Google Scholar]
- 25.Farmer WR and Liao JC (2000) Improving lycopene production in Escherichia coli by engineering metabolic control. Nat. Biotechnol. 18, 533–537. doi: 10.1038/75398. [DOI] [PubMed] [Google Scholar]
- 26.Venayak N, Anesiadis N, Cluett WR and Mahadevan R (2015) Engineering metabolism through dynamic control. Curr. Opin. Biotechnol. 34, 142–152. doi: 10.1016/j.copbio.2014.12.022 [DOI] [PubMed] [Google Scholar]
- 27.Segall-Shapiro TH, Sontag ED and Voigt CA (2018) Engineered promoters enable constant gene expression at any copy number in bacteria. Nat. Biotechnol. 36, 352–358. doi: 10.1038/nbt.4111. [DOI] [PubMed] [Google Scholar]
- 28.Jones JA, Vernacchio VR, Lachance DM, Leboish M, Fu Li., Shirke AN, Schultz VL, Cress B, Linhardt RJ, and Koffas MAG (2015) EPathOptimize: A combinatorial approach for transcriptional balancing of metabolic pathways. Sci. Rep. 5, 11301. doi: 10.1038/srep11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chappell J, Takahashi MK and Lucks JB (2015) Creating small transcription activating RNAs. Nat. Chem. Biol. 11, 214–220. doi: 10.1038/nchembio.1737. [DOI] [PubMed] [Google Scholar]
- 30.Chappell J, Westbrook A, Verosloff M and Lucks JB (2017) Computational design of small transcription activating RNAs for versatile and dynamic gene regulation. Nat. Commun. 8, 1051. doi: 10.1038/s41467-017-01082-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lutz R and Bujard H (1997) Independent and tight regulation of transcriptional units in escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2regulatory elements. Nucleic Acids Res. 25, 1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feldman MF, Wacker M, Hernandez M, Hitchen PG, Marolda CL, Kowarik M, Morris HR, Dell A, Valvano MA, and Aebi M (2005) Engineering N-linked protein glycosylation with diverse O antigen lipopolysaccharide structures in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 102, 3016–3021. doi: 10.1073/pnas.0500044102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong Ng J, Chatenay D, Robert J and Poirier MG (2010) Plasmid copy number noise in monoclonal populations of bacteria. Phys. Rev. E. Stat. Nonlin. Soft. Matter Phys. 81, e011909. doi: 10.1103/PhysRevE.81.011909. [DOI] [PubMed] [Google Scholar]
- 34.Lopilato J, Bortner S and Beckwith J (1986) Mutations in a new chromosomal gene of Escherichia coli K-12, pcnB, reduce plasmid copy number of pBR322 and its derivatives. Mol. Genet. Genomics 205, 285–290. doi: 10.1007/BF00430440. [DOI] [PubMed] [Google Scholar]
- 35.Lin-Chao S & Bremer H Effect of the bacterial growth rate on replication control of plasmid pBR322 in Escherichia coli. Mol. Genet. Genomics 203, 143–149 (1986). doi: 10.1007/BF00330395 [DOI] [PubMed] [Google Scholar]
- 36.Wegrzyn G (1999) Replication of plasmids during bacterial response to amino acid starvation. Plasmid 41, 1–16. doi: 10.1006/plas.1998.1377. [DOI] [PubMed] [Google Scholar]
- 37.Lin-Chao S, Chen W-T and Wong T-T (1992) High copy number of the pUC plasmid results from a Rom/Rop-suppressible point mutation in RNA II. Mol. Microbiol. 6, 3385–3393. doi: 10.1111/j.1365-2958.1992.tb02206.x. [DOI] [PubMed] [Google Scholar]
- 38.Cheah UE, Weigand WA and Stark BC (1987) Effects of recombinant plasmid size on cellular processes in Escherichia coli. Plasmid 18, 127–134. doi: 10.1016/0147-619X(87)90040-0. [DOI] [PubMed] [Google Scholar]
- 39.Corchero JL and Villaverde A (1998) Plasmid maintenance in Escherichia coli recombinant cultures is dramatically, steadily, and specifically influenced by features of the encoded proteins. Biotechnol. Bioeng. 58, 625–632. doi:. [DOI] [PubMed] [Google Scholar]
- 40.Stueber D and Bujard H (1982) Transcription from efficient promoters can interfere with plasmid replication and diminish expression of plasmid specified genes. EMBO J. 1, 1399–1404. doi: 10.1002/j.1460-2075.1982.tb01329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peterson J and Phillips GJ (2008) New pSC101-derivative cloning vectors with elevated copy numbers. Plasmid 59, 193–201. doi: 10.1016/j.plasmid.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin VJJ, Pitera DJ, Withers ST, Newman JD and Keasling JD (2003) Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat. Biotechnol. 21, 796–802. doi: 10.1038/nbt833. [DOI] [PubMed] [Google Scholar]
- 43.Biggs BW, Lim CG, Sagliani K, Shankar S, Stephanopoulos G, De Mey M, and Ajikumar PK (2016) Overcoming heterologous protein interdependency to optimize P450-mediated Taxol precursor synthesis in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 113, 3209–3214. doi: 10.1073/pnas.1515826113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanda Y, Nakamura H, Umemiya S, Puthukanoori RK, Appala VRM, Gaddamaniugu GK, Paraselli BR, and Baran PS (2020) Two-Phase Synthesis of Taxol. J. Am. Chem. Soc. 142, 10526–10533. doi: 10.1021/jacs.0c03592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van Dien S (2013) From the first drop to the first truckload: Commercialization of microbial processes for renewable chemicals. Curr. Opin. Biotechnol. 24, 1061–1068. doi: 10.1016/j.copbio.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 46.Weber W and Fussenegger M (2007) Inducible product gene expression technology tailored to bioprocess engineering. Curr. Opin. Biotechnol 18, 399–410. doi: 10.1016/j.copbio.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 47.Papenfort K and Bassler BL (2016) Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 14, 576–588. doi: 10.1038/nrmicro.2016.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsao CY, Hooshangi S, Wu HC, Valdes JJ and Bentley WE (2010) Autonomous induction of recombinant proteins by minimally rewiring native quorum sensing regulon of E. coli. Metab. Eng. 12, 291–297. doi: 10.1016/j.ymben.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 49.Gupta A, Reizman IMB, Reisch CR and Prather KLJ (2017) Dynamic regulation of metabolic flux in engineered bacteria using a pathway-independent quorum-sensing circuit. Nat. Biotechnol 35, 273–279. doi: 10.1038/nbt.3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim EM Woo HM, Tian T, Yilmaz S, Javidpour P, Keasling JD, and Lee TS (2017) Autonomous control of metabolic state by a quorum sensing (QS)-mediated regulator for bisabolene production in engineered E. coli. Metab. Eng 44, 325–336. doi: 10.1016/j.ymben.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 51.Engebrecht J and Silverman M (1984) Identification of genes and gene products necessary for bacterial bioluminescence. Proc. Natl. Acad. Sci. U.S.A. 81, 4154–4158. doi: 10.1073/pnas.81.13.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Minogue TD, Wehland-Von Trebra M, Bernhard F and Von Bodman SB (2002) The autoregulatory role of EsaR, a quorum-sensing regulator in Pantoea stewartii ssp. stewartii: Evidence for a repressor function. Mol. Microbiol. 44, 1625–1635. doi: 10.1046/j.1365-2958.2002.02987.x. [DOI] [PubMed] [Google Scholar]
- 53.Malinowski JJ (2001) Two-phase partitioning bioreactors in fermentation technology. Biotechnol. Adv. 19, 525–538. doi: 10.1016/S0734-9750(01)00080-5. [DOI] [PubMed] [Google Scholar]
- 54.Green AA, Silver PA, Collins JJ and Yin P (2014) Toehold switches: De-novo-designed regulators of gene expression. Cell 159, 925–939. doi: 10.1016/j.cell.2014.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Na D, Yoo SM, Chung H, Park H, Park JH, and Lee SY (2013) Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs. Nat. Biotechnol 31, 170–174. doi: 10.1038/nbt.2461 [DOI] [PubMed] [Google Scholar]
- 56.Takahashi MK and Lucks JB (2013) A modular strategy for engineering orthogonal chimeric RNA transcription regulators. Nucleic Acids Res. 41, 7577–7588. doi: 10.1093/nar/gkt452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Westbrook AM and Lucks JB (2017) Achieving large dynamic range control of gene expression with a compact RNA transcription-translation regulator. Nucleic Acids Res. 45, 5614–5624. doi: 10.1093/nar/gkx215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, and Lim WA (2013) Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ajikumar PK, Xiao WH, Tyo KEJ, Wang Y, Simeon F, Leonard E Much O, Phon TH, Pfeifer B, and Stephanopoulos G (2010) Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science 330, 70–74. doi: 10.1126/science.1191652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smanski MJ, Bhatia S, Zhao D, Park YJ, Woodruff LBA, Giannoukos G, Ciulla D, Busby M, Calderon J, Nicol R, Gordon DB, Densmore D, and Voigt CA (2014) Functional optimization of gene clusters by combinatorial design and assembly. Nat. Biotechnol. 32, 1241–1249. doi: 10.1038/nbt.3063. [DOI] [PubMed] [Google Scholar]
- 61.St-Pierre F, Cui L Priest DG, Endy D, Dodd IB, and Shearwin KE (2013) One-step cloning and chromosomal integration of DNA. ACS Synth. Biol 2, 537–541. doi: 10.1021/sb400021j. [DOI] [PubMed] [Google Scholar]
- 62.Thomason LC, Sawitzke JA, Li X, Costantino N and Court DL (2014) Recombineering: Genetic engineering in bacteria using homologous recombination. Curr. Protoc. Mol. Biol. 106, 1–39. doi: 10.1002/0471142727.mb0116s106. [DOI] [PubMed] [Google Scholar]
- 63.Meier I, Wray LV and Hillen W (1988) Differential regulation of the Tn10-encoded tetracycline resistance genes tetA and tetR by the tandem tet operators O1 and O2. EMBO J. 7, 567–572. doi: 10.1002/j.1460-2075.1988.tb02846.x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.