Abstract
Background
The genus Rhododendron L. has been widely cultivated for hundreds of years around the world. Members of this genus are known for great ornamental and medicinal value. Owing to advances in sequencing technology, genomes and transcriptomes of members of the Rhododendron genus have been sequenced and published by various laboratories. With increasing amounts of omics data available, a centralized platform is necessary for effective storage, analysis, and integration of these large-scale datasets to ensure consistency, independence, and maintainability.
Results
Here, we report our development of the Rhododendron Plant Genome Database (RPGD; http://bioinfor.kib.ac.cn/RPGD/), which represents the first comprehensive database of Rhododendron genomics information. It includes large amounts of omics data, including genome sequence assemblies for R. delavayi, R. williamsianum, and R. simsii, gene expression profiles derived from public RNA-Seq data, functional annotations, gene families, transcription factor identification, gene homology, simple sequence repeats, and chloroplast genome. Additionally, many useful tools, including BLAST, JBrowse, Orthologous Groups, Genome Synteny Browser, Flanking Sequence Finder, Expression Heatmap, and Batch Download were integrated into the platform.
Conclusions
RPGD is designed to be a comprehensive and helpful platform for all Rhododendron researchers. Believe that RPGD will be an indispensable hub for Rhododendron studies.
Keywords: Rhododendron, Horticulture plant, Database, Functional genomics
Background
Rhododendron L. is the largest genus in the Ericaceae, which is the largest genus of woody angiosperms in China [1]. The genus is widely distributed throughout the Northern Hemisphere from tropical Southeast Asia to northeastern Australia [2]. There are more than 1000 species of Rhododendron worldwide, approximately 600 of which encompassing nine subgenera are found in China [3, 4]. Southwestern China and the eastern Himalayas are considered as centers of Rhododendron diversification and differentiation [5]. Rhododendrons are considered to have great ornamental and medicinal value [6, 7].
Horticultural interest in Rhododendron can be traced back at least several centuries, owing in part to their bright coloring and elegant posture [8, 9]. In China, its introduction and cultivation was first documented in poetry from the Tang dynasty, and rhododendrons have long been developed as one of the ten national-traditional ornamental flowers [8]. The breeding history began with gardening enthusiasts in Western countries in the late eighteenth century [9]. Currently, there are over 28,000 cultivars of Rhododendron [10], which are widely cultivated in many regions such as Asia, America, and Europe [6]. Most wild rhododendrons are found in regions with temperate climates, high rainfall, humid atmosphere, and organic acid soils with low nutrient composition [11]. Furthermore, most varieties are derived through crossbreeding by gardening enthusiasts according to their preference for ornamental traits. In general, breeding goals have previously been focused mostly on ornamental characteristics rather than adaptability and resistance, resulting in a disconnect between existing varieties and market demands. Therefore, a challenge for Rhododendron breeding is the development of varieties capable of adapting to environments with cold winters, hot summers, lower rainfall and humidity, and less optimal soils [12].
Additionally, the genus Rhododendron has a long history in traditional medicine [7]. Phytochemists have demonstrated interest in Rhododendron species due to their abundance of secondary metabolites [13]. Currently, approximately 200 compounds, mostly flavonoids and diterpenoids, have been isolated from Rhododendron. Some of the isolates have demonstrated intriguing bioactivity [14, 15]. For example, diterpenoids isolated from the flowers, roots, and fruits of R. molle exhibit significant anticancer, antiviral, antinociceptive, immunomodulatory, and sodium channel antagonistic activities.
With the rapid development of sequencing and genomic editing technology, molecular design breeding has become a more efficient and accurate plant breeding method [16]. Elucidation of the genetic mechanisms associated with ornamental traits (flower color, flower shape, etc.), adaptability, resistance, secondary metabolism, etc. will be a helpful and necessary foundation for more practical Rhododendron breeding. A great deal of omics data concerning Rhododendron have been accumulated to date and several rhododendron genomes have been sequenced. The R. delavayi genome sequence was released in 2017 [17], R. williamsianum in 2019 [18], and R. simsii in 2020 [19]. In addition, relevant transcriptomic data have also been published in recent years [20–24]. Progress in the development of high-throughput sequencing technology has greatly accelerated studies on Rhododendron [17–24]. These large genomic data sets provide a new perspective for understanding biological traits such as ornamentation, adaptability, resistance, and secondary metabolism for breeders and phytochemists alike.
Rhododendron omics data sets are currently distributed in public databases that are easily accessible [25, 26]. However, processing these data is a considerable challenge for research groups with limited bioinformatics experience. To address this problem, we have constructed a comprehensive database for data storage, categorization, online analysis, and visualization of Rhododendron omics data sets.
Here, we present the Rhododendron Plant Genome Database (RPGD; http://bioinfor.kib.ac.cn/RPGD/), a data center for Rhododendron functional genomics researchers. The database integrates the three released genome sequences, expression profiles, functional annotations, gene family ontologies, simple sequence repeats, chloroplast genome assemblies, and gene homology information. We have also incorporated bioinformatics tools such as BLAST, JBrowse, Flanking Sequence Finder, Genome Synteny Browser, Ortholog Gene Finder, Expression Heatmap, and Batch Download into the user interface. The interface is designed to be simple and user-friendly. We suggest that RPGD will be of great convenience as a “one-stop shop” to a wide range of Rhododendron researchers.
Construction and content
Genomic data
Currently, three reference genome sequences of Rhododendron - R. delavayi, R. williamsianum and R. simsii - are hosted in RPGD (Table 1). The genome sizes are 695 Mb, 532 Mb and 529 Mb, respectively; and the scaffold N50 are 637.83 kb, 218.8 kb and 36.3 Mb, respectively [17–19]. The genome of R. simsii was sequenced by PacBio long-read sequencing technology [19], while R. delavayi and R. williamsianum were based on next-generation sequencing [17, 18]. We downloaded the genome assembly, general feature format (GFF3), coding sequence (CDS), and protein sequence (PEP) of R. delavayi (http://gigadb.org/dataset/100331) from the GigaScience database [17, 26], and for R. williamsianum (https://www.ncbi.nlm.nih.gov/assembly/GCA_009746105.1) and R. simsii (https://www.ncbi.nlm.nih.gov/assembly/GCA_014282245.1) from NCBI [18, 19, 25].
Table 1.
Data statistics in RPGD database
| Data type | Number |
|---|---|
| Gene | |
| Genes for R. delavayi | 32,938 |
| Genes for R. williamsianum | 23,559 |
| Genes for R. simsii | 32,999 |
| Genome | |
| Scaffolds for R. delavayi | 193,091 |
| Chromosomes for R. williamsianum | 13 |
| Chromosomes for R. simsii | 13 |
| Gene ontology (GO) | |
| R. delavayi | |
| Genes | 21,361 |
| Annotations | 805,276 |
| R. williamsianum | |
| Gene | 17,658 |
| Annotations | 687,600 |
| R. simsii | |
| Genes | 22,235 |
| Annotations | 785,704 |
| Gene Family | |
| Gene families for R. delavayi | 4168 |
| Gene families for R. williamsianum | 3546 |
| Gene families for R. simsii | 3742 |
| Transcription factor (TF) and Transcriptional regulators (TRs) | |
| TFs and TRs for R. delavayi | 2104 |
| TFs and TRs for R. williamsianum | 1622 |
| TFs and TRs for R. simsii | 2156 |
| Simple sequence repeat (SSR) | |
| SSRs for R. delavayi | 361,268 |
| SSRs for R. williamsianum | 230,013 |
| SSRs for R. simsii | 358,705 |
| Chloroplast genome assemblies | |
| R. delavayi chloroplast genome assembly | 2 |
| R. pulchrum chloroplast genome assembly | 1 |
| InterPro | |
| Annotated to InterPro for R. delavayi | 77,221 |
| Annotated to InterPro for R. williamsianum | 60,834 |
| Annotated to InterPro for R. simsii | 81,654 |
| Gene expression | |
| RNA-Seq for R. delavayi | 2 |
| Genomic synteny | 2913 |
| OrthoFinder orthologous/paralogs group | 18,048 |
Transcriptomic data
All publicly available RNA-Seq datasets in the NCBI Sequence Read Archive (SRA) database, including data from two projects and 19 samples, were obtained. One transcriptomics project was related to drought stress (4 samples) while the other was related to the flower bud in different dormancy statuses (15 samples) [23] (Table 1). Both projects focused on R. delavayi.
We processed and analyzed the RNA-Seq datasets by a standard pipeline method. First, we used the SRA Toolkit [27] to convert the data format to FASTQ and low-quality reads were removed from raw reads by Trimmomatic [28]. We then employed Tophat2 [29] to map all clean reads onto the reference genome (R. delavayi) with default parameters, which were assembled using Cufflinks (version 2.2.1) using the reference genome as a guide [30]. Combined transcriptome assemblies were generated using Cuffmerge. Based on the alignments, the read counts of each gene were calculated and normalized to fragments per kilobase of transcript per million mapped fragments (FPKM) values in Cuffdiff. Mean and standard errors of the FPKM values were derived for the biological replicates.
Gene model and function annotation
A total of 89,496 protein-coding genes were collected from the downloaded data mentioned in the genomic data, including 32,938 from R. delavayi, 23,559 from R. williamsianum, and 32,999 from R. simsii. The protocol for annotating protein-coding genes is described as follows. Firstly, protein-coding genes were annotated using two software packages, eggNOG-mapper [31, 32] and InterProScan with default parameters [33]. Then, the results from the two different tools were combined and redundant annotations were removed to obtain complete and precise GO annotations using homemade scripts. The protein sequences were aligned against the NCBI non-redundant (nr), UniProt (Swiss-Prot and TrEMBL), and Arabidopsis protein (TAIR) databases using the BLASTP command of DIAMOND with an E-value cutoff of 1e− 5 [34]. The BLASTP results against the UniProt and TAIR databases were then fed to the AHRD program (https://github.com/groupschoof/AHRD) to obtain concise, precise, and informative gene function descriptions. All BLASTP results are shown on the detailed gene page. All of these protein sequences were further compared against the InterPro database using InterProScan to identify functional domains [33].
As a result, the genes from R. delavayi were functionally annotated to 805,276 on GO database and 77,221 on InterPro. The R. williamsianum gene were functionally annotated to 687,600 on GO and 60,834 on InterPro. The R. simsii genes were functionally annotated to 785,704 on GO and 81,654 on InterPro (Table 1).
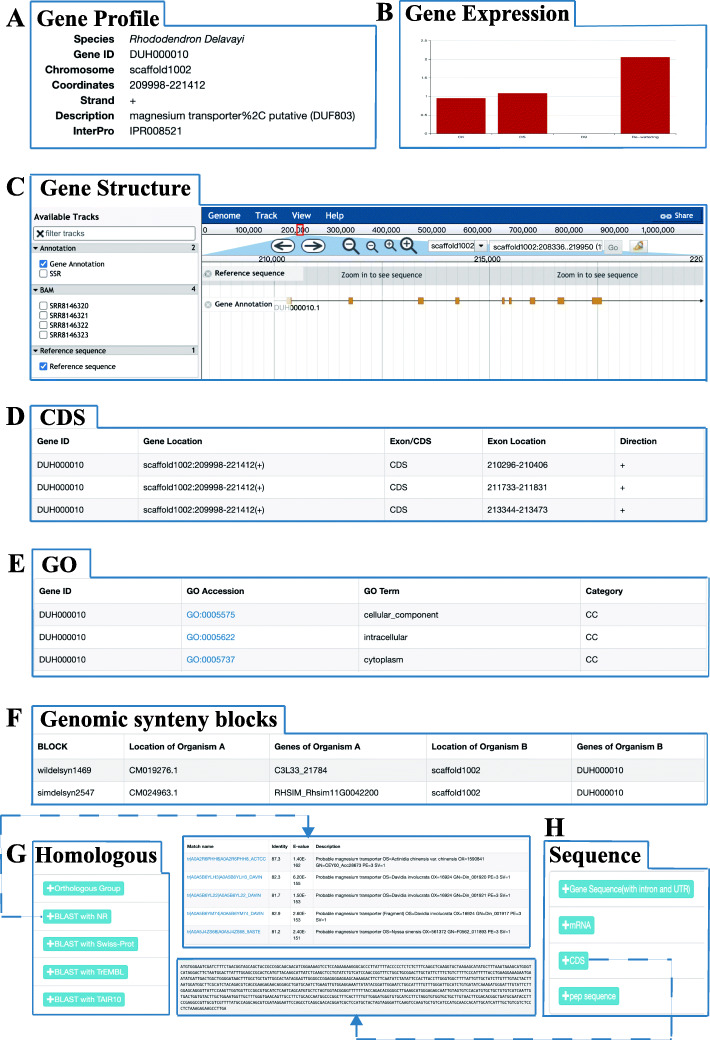
These genes were used as a “data hub” to link all data types (Fig. 1), including gene summary information (species, gene ID, location, description, InterPro and gene family) (Fig. 1a), expression profiles (Fig. 1b), JBrowse gene visualization (Fig. 1c), gene exon/CDS information (Fig. 1d), GO annotation (Fig. 1e), genomic synteny blocks (Fig. 1f), homologous genes and BLASTP results against the nr-NCBI, UniProt and TAIR databases (Fig. 1g), gene/mRNA/CDS/protein sequences (Fig. 1h). All information mentioned here is shown on an integrated interface to allow users to browse conveniently.
Fig. 1.
Gene feature page in RPGD. a Overview of gene profile information including species, gene ID, location, description, InterPro and gene family. b Expression profiles. c JBrowse gene visualization. d Exon/CDS information of gene. e GO annotation. f Genomic synteny blocks. g Homologous genes information in 6 organisms and BLASTP results against the nr-NCBI, UniProt and TAIR databases. h Gene/mRNA/CDS/protein sequences
Transcription factors and transcriptional regulators
The iTAK package was used to identify transcription factors (TFs) and transcriptional regulators (TRs) in the three Rhododendron genomes and all candidates were classified into different gene families using the default parameters [35]. Thus, R. delavayi contains 1662 TFs and 442 TRs, R. williamsianum contains 1261 TFs and 361 TRs, and R. simsii contains 1740 TFs and 416 TRs (Table 1).
Orthologous/paralogs group
OrthoFinder [36, 37] was employed to identify orthologous and paralogous genes by using default parameters among R. delavayi, R. williamsianum, R. simsii, Actinidia chinensis [38], Camellia sinensis [39] and Arabidopsis thaliana [40]. In total, 18,048 orthologous groups were identified. To ensure that the inference of orthologous genes was sufficiently accurate, we extracted 985 groups of single-copy orthologs to construct the “Orthologous Groups” module (Table 1). We also used OrthoFinder to search for pairwise homologous genes between the three Rhododendron genomes and A. thaliana respectively [36, 37]. We considered the genes of each orthologous group as belonging to one gene family and mapped gene family information from A. thaliana to R. delavayi (4168 gene families), R. williamsianum (3546 gene families), and R. simsii (3742 gene families).
Simple sequence repeats
Simple sequence repeats (SSRs) were identified in R. delavayi, R. williamsianum and R. simsii by MISA with default parameters; the total number were 361,268, 230,013, and 358,705, respectively [41] (Table 1). We also used Primer3 with default parameters to design primers for SSRs and the primers can be displayed on the SSR detail page [42].
Chloroplast genomes
We also collected full-length chloroplast genomes of R. delavayi and R. pulchrum from the NCBI database [43–45]. RPGD hosts two complete chloroplast genome assemblies of R. delavayi. One of them is 193,798 bp in length, and 123 genes were annotated, including 80 protein-coding genes, 35 tRNA genes, and 8 rRNA genes [43]. The other is 202,169 bp in length, a total of 137 genes were found, including 88 protein-coding genes, 41 tRNAs, and 8 rRNAs [44]. The chloroplast genome of R. pulchrum is 136,249 bp in length, and it contains 73 genes, comprising 42 protein-coding genes, 29 tRNA genes, and 2 rRNA genes [45] (Table 1).
Syntenic relationships among R. delavayi, R. williamsianum and R. simsii
We identified syntenic blocks and homologous gene pairs in the three Rhododendron genomes. Protein sequences were first aligned against each other (pairwise comparisons) using BLASTP with an E-value cutoff of 1e− 5 [46]. Based on the BLASTP results and gene positions, syntenic blocks were determined using MCScanX with default parameters [47]. A total of 2913 syntenic blocks and 55,590 homologous genes were identified (Table 1) with detail presented in the “Tools/Genome Synteny” module. Users should note that the current assembly of draft genomes and annotations might affect the results of syntenic relationships, and we will update the data when new versions become available.
Implementation
RPGD was constructed using the LAMP framework, including Apache2 (a free and open-source cross-platform web server software; https://www.apache.org/), MariaDB (a relational database management system; https://mariadb.org/), and PHP (a popular general-purpose scripting language; https://www.php.net/). All data were stored on a Linux platform with the MariaDB database to facilitate efficient management, search, and display. The web pages were built using HTML5, CSS3, JavaScript, and Bootstrap3 (a free and open-source CSS framework directed at responsive, mobile-first front-end web development; https://getbootstrap.com/docs/3.3/). The Bootstrap-table (an extended Bootstrap table with radio, checkbox, sort, pagination, extensions, and other added features; https://bootstrap-table.com/) and jQuery (a JavaScript library designed to simplify HTML DOM tree traversal and manipulation; http://jquery.com, version 3.4.1) were used to display the query results dynamically. Presentation of the diagram was made by Echart (a free, powerful charting and visualization library offering a way of easily adding intuitive, interactive, and highly customizable charts; https://echarts.apache.org/zh/index.html).
Utility and discussion
Browsing RPGD
Users can browse all data in RPGD easily on the “Browse” page, including genome statistics, gene models, gene function annotations, SSRs, genome syntenic blocks, gene expression profiles, gene families and transcription factor information from R. delavayi, R. williamsianum and R. simsii, respectively. The information described above is presented in tabular form on the web page using a Bootstrap-table plug. Additionally, a detailed information page for a specific gene can be accessed by clicking the gene ID hyperlink. Information about each gene is displayed on a detailed page, including the gene summary, exons, gene structure (in JBrowse), GO, family, expression, homology, and sequence information.
Searching RPGD
A series of search tools are presented on the navigation menu “Search”, such as “Gene”, “Genome”, “Gene Ontology”, “Gene Family”, “Gene Expression”, “Transcription Factor”, “Chloroplast Genome” and “SSR” to help users more easily find data of interest to them. (i). “Search Gene”: RPGD provides four different ways to search genes including gene ID, AHRD descriptions, InterPro, GO accession, and GO term. The response is a dynamic table that contains all genes associated with the entered search terms, and the list of those genes can be downloaded as a TXT file for further analysis. Additionally, the details of the genes can be viewed by clicking the gene ID hyperlink. (ii). “Search Genome”: users can use scaffold/chromosome ID to search the scaffold/chromosome information. The results are divided into a list, a table, and a chromosome viewer. The list shows basic information about the chromosome, including the species, chromosome ID, and the length of the chromosome. The table displays information about all genes on the chromosome. The chromosome viewer is embedded in JBrowse to display the chromosome profile. (iii). “Search GO”: users can use gene ID, GO accession, and GO term to query GO information of a gene. The responses are a set of genes annotated with the queried functions. Similarly, users can download the list of genes and click the gene ID hyperlink to review gene details. (iv). “Search Family”: users can find genes with gene family names specified by the user. A list of genes related to this gene family are generated as the response. Users can also download the list of genes and click the gene ID hyperlink to view gene details. (v). “Search Gene Expression”: users can input gene ID of interest to search their expression patterns based on currently provided transcriptomics results. The output is a line chart that shows graphically the expression level and can be downloaded locally for further analysis. (vi). “Search Transcription Factor”: users can search for transcription factor genes by clicking transcription factor names. The responses are a list of genes annotated as transcription factors. Users can also download the list of genes and click the gene ID hyperlink to view gene details. (vii). “Search Chloroplast Genome”: users can use the gene or product name to find the information from chloroplast genes. The response is a list of detailed information about the entered keywords. In addition, the list returned contains a number of hyperlinks which allow user to view the details about that chloroplast gene at NCBI. (viii). “Search SSR”: RPGD provides SSR location, SSR type (monomer to hexamer) and SSR motif to query the SSR detailed information, including SSR ID, type, motif, size, and location. Users can click the SSR ID hyperlink to view SSR primer information. Examples are displayed below each search field that can be clicked to autofill the search keywords on every search page.
BLAST
BLAST is a sequence similarity searching program frequently used for bioinformatics queries [46]. ViroBLAST [48], a useful and user-friendly tool for online data analysis, was integrated into RPGD (Fig. 2a). Users can input their sequence of interest or upload their sequence files to perform BLASTN, BLASTP, BLASTX, tBLASTN, and tBLASTX against a whole genome, CDS, or peptide library.
Fig. 2.
Screenshots of online tools page. a Online BLAST. b JBrowse for visualizing genome and other tracks. c Expression Heatmap showing expression patterns. d Enrichment Analysis
JBrowse
A key mission of RPGD is to help users browse genomic data in detail. Therefore, JBrowse [49], a fast, scalable, and widely used genome browser built completely with JavaScript and HTML5, was embedded in RPGD to visualize genomic information (Fig. 2b). In RPGD, JBrowse hosts different tracks, including genome sequence, gene models, SSRs, and transcriptome-aligned BAM files of R. delavayi, R. williamsianum, and R. simsii, respectively. In addition, we will integrate other data styles, such as single-nucleotide polymorphisms (SNPs), as they become available.
Flanking sequence finder
The flanking sequences of genes often contain a wealth of information including regulatory elements and promoters. To aid in research of flanking sequences, we utilized gene annotations and genome data to develop a useful tool - “Flanking Sequence Finder”. Researchers can find and download flanking sequences by inputting gene ID and specifying the length of the desired flanking sequences.
Genome syntenic browser
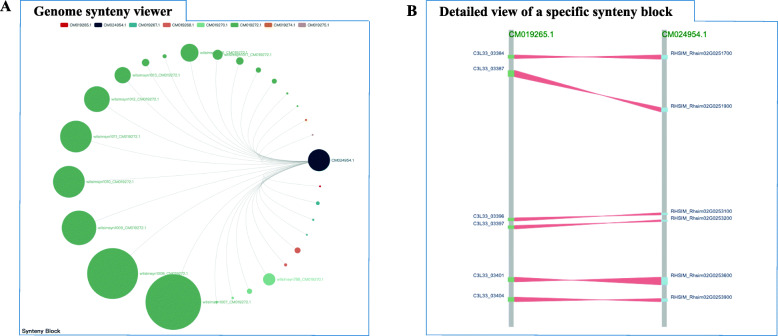
To view genome syntenic blocks and homologous gene pairs between the three Rhododendron genomes, we constructed the “Genome Syntenic Browser” module using AJAX, JavaScript and Echart. Users can browse the genome syntenic blocks or search for a specific block they want to query. Users can retrieve syntenic blocks by selecting a chromosome and subject genome together. This module returns an image to displaying all syntenic blocks for every paired query and subject genome (Fig. 3a) and a full list of the syntenic blocks. For each syntenic block, users can jump to a new page by clicking on the block ID hyperlink which contains an image to display the homologous gene pairs (Fig. 3b). The full list of genes is also provided with links to the “data hub” interface to detail the gene information for each gene (Fig. 1).
Fig. 3.
Genome synteny viewer. a Syntenic blocks displayed in a circus plot. The darkslategray circle represents the query chromosome. Besides, the same color represents the same chromosome, and different circles of the same color represent different syntenic blocks located on the same chromosome. Additionally, the lines between darkslategray and other colors represent syntenic blocks identified between the two genomes. By the way, all blocks of this chromosome can be made to disappear from the image by clicking on the color that represents that chromosome. b Detailed view of a specific synteny block. The two gray lines represent the chromosomes of different species, and the red areas represent homologous gene pairs
Orthologous groups
A common task in routine bioinformatics analysis is the identification of homologous genes. Users can input gene IDs to find orthologous groups in R. delavayi, R. williamsianum, R. simsii, as well as A. chinensis, C. sinensis, and A. thaliana. The details of the homologous genes are be presented in a table, which also provides links to “data hub” page for each gene (Fig. 1).
Expression heatmap
RPGD not only stores gene expression profiles derived from RNA-Seq datasets but also provides an “Expression Heatmap” module (Fig. 2c). “Expression Heatmap” can be used to retrieve the gene expression patterns of a group of genes from different samples. The output is a heatmap that graphically shows expression levels and can be downloaded locally for further analysis.
GO and KEGG enrichment analysis
Functional enrichment analysis is a powerful method for mining gene data, providing further insight into what biological processes these genes may be involved in. To help users to capture biological information of genes, we construct the GO and KEGG enrichment analysis tools base on the functional annotation mentioned above and clusterProfiler R package [50]. Users can input a list of interested genes to perform the enrichment analysis (Fig. 2d). The results returned the significantly enriched functional categories.
Download and batch download
All the data in RPGD were available for users to download, including genome assembly (FASTA), gene prediction (GFF3), gene function annotation (TXT), complete chloroplast genome (FASTA), gene family data (CSV), orthologous groups data (CSV), simple sequence repeat data (TXT), gene expression data (CSV), and other related data can also be downloaded in this module. “Batch Download” is provided for users to export custom datasets or bulk download datasets from RPGD. Users can download multiple types of sequences (gene, CDS, PEP, flanking sequence and gene expression profile) by inputting a list of genes.
Conclusions
RPGD is dedicated to providing a comprehensive database of Rhododendron omics data. The current implementation of RPGD integrates important data including genome sequence assemblies, gene expression profiles, functional annotations, gene families, transcription factors, homologous genes, simple sequence repeats, and chloroplast genome assemblies. It also provides a series of tools for online data analysis and visualization. The integration of these data and tools makes RPGD a valuable database. We intend to continue updating the datasets when new data are released. For instance, our team will release a novel Rhododendron genome (R. irroratum) and its phenotypic datasets, including breeds, genotypes, and phenotypes in the near future. Additionally, we will continue to develop and integrate tools for functional, evolutionary, and network analysis. We hope that researchers will take advantage of these resources and also provide comments and suggestions for improving RPGD. Believe that RPGD will be in indispensable hub for Rhododendron studies.
Acknowledgments
We would like to thank Editage (www.editage.cn) for English language editing.
Abbreviations
- RPGD
Rhododendron Plant Genome Database
- GFF3
General feature format
- CDS
Coding sequence
- PEP
Protein sequence
- GO
Gene ontology
- TF
Transcription factor
- TR
Transcriptional regulator
- SSR
Simple sequence repeat
- SRA
Sequence Read Archive
- FPKM
Fragments per kilobase of transcript per million mapped fragments
- SNP
Single-nucleotide polymorphisms
Authors’ contributions
CZ and JW designed and managed the project; NL constructed the database; LZ and YZ collected and analyzed the data; YM and DG participated in discussions; NL and MT designed the layout of the web pages; ZW will maintain the website. NL and MT wrote and revised the manuscript. All authors read, commented on, and approved the manuscript.
Funding
This study was supported by grants from the National Natural Science Foundation of China (31760231), Construction of International Flower Technology Innovation Center and Industrialization of achievements (2019ZG006), Program of Science and Technology Talents Training in Yunnan province (2016HA005), Youth Program of National Natural Science Foundation of China (32000180), Yunnan Young & Elite Talents Project.
Availability of data and materials
RPGD is freely available at http://bioinfor.kib.ac.cn/RPGD/.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jihua Wang, Email: wjh0505@gmail.com.
Chengjun Zhang, Email: zhangchengjun@mail.kib.ac.cn.
References
- 1.Yan LJ, Liu J, Möller M, Zhang L, Zhang XM, Li DZ, Gao LM. DNA barcoding of Rhododendron (Ericaceae), the largest Chinese plant genus in biodiversity hotspots of the Himalaya-Hengduan Mountains. Mol Ecol Resour. 2015;15(4):932–944. doi: 10.1111/1755-0998.12353. [DOI] [PubMed] [Google Scholar]
- 2.Chamberlain D, Hyam R, Argent G, Fairweather G, Walter KS. The genus Rhododendron: its classification and synonymy. Edinburgh: Royal Botanic Garden Edinburgh; 1996. [Google Scholar]
- 3.Tian XL, Chang YH, Neilsen J, Wang SH, Ma YP. A new species of Rhododendron (Ericaceae) from northeastern Yunnan. China Phytotaxa. 2019;395(2):66–70. doi: 10.11646/phytotaxa.395.2.2. [DOI] [Google Scholar]
- 4.Fang MY, Fang RZ, He MY, Hu LZ, Yang HB, Qin HN, Min TL, Chamberlain D, Stevens P, Wallace G, Anderberg A. Flora of China. Volume 14: Apiaceae through Ericaceae. Beijing: Science Press; 2005. [Google Scholar]
- 5.Ma YP, Wu ZK, Xue RJ, Tian XL, Gao LM, Sun WB. A new species of Rhododendron (Ericaceae) from the Gaoligong Mountains, Yunnan, China, supported by morphological and DNA barcoding data. Phytotaxa. 2013;114(1):42–50. doi: 10.11646/phytotaxa.114.1.4. [DOI] [Google Scholar]
- 6.De RJ, De KE, Calsyn E, Eeckhaut T, Van HJ, Kobayashi N. Azalea. In: Van HJ, editor. Ornamental Crops. Cham: Springer; 2018. pp. 237–271. [Google Scholar]
- 7.Popescu R, Kopp B. The genus Rhododendron: an ethnopharmacological and toxicological review. J Ethnopharmacol. 2013;147(1):42–62. doi: 10.1016/j.jep.2013.02.022. [DOI] [PubMed] [Google Scholar]
- 8.Yonghui Z, Weibing J, Mangling W. Meanings of Rhododendron and ways used in gardens. Chin Agric Sci Bull. 2007;09:376–380. [Google Scholar]
- 9.Kron KA, Gawen LM, Chase MW, et al. Evidence for introgression in azaleas (Rhododendron; Ericaceae): Chloroplast DNA and morphological variation in a hybrid swarm on Stone Mountain, Georgia. Am J Bot. 1993;80(9):1095–9. 10.1002/j.1537-2197.1993.tb15335.x.
- 10.Leslie A. The international Rhododendron register and checklist. 2. London: Royal Horticultural Society; 2004. [Google Scholar]
- 11.Cox PA. The larger species of rhododendron. 1. London: Batsford Ltd; 1979. [Google Scholar]
- 12.Perkins S, et al. More weighings: exploring the ploidy of hybrid elepidote. rhododendrons. Azalean. 2015;37:28–42. [Google Scholar]
- 13.Qiang Y, Zhou B, Gao K. Chemical constituents of plants from the genus Rhododendron. Chem Biodivers. 2011;8(5):792–815. doi: 10.1002/cbdv.201000046. [DOI] [PubMed] [Google Scholar]
- 14.Zhu YX, Zhang ZX, Yan HM, Lu D, Zhang HP, Li L, Liu YB, Li Y. Antinociceptive diterpenoids from the leaves and twigs of Rhododendron decorum. J Nat Prod. 2018;81(5):1183–1192. doi: 10.1021/acs.jnatprod.7b00941. [DOI] [PubMed] [Google Scholar]
- 15.Zhou J, Liu T, Zhang H, Zheng G, Qiu Y, Deng M, Zhang C, Yao G. Anti-inflammatory grayanane diterpenoids from the leaves of Rhododendron molle. J Nat Prod. 2018;81(1):151–161. doi: 10.1021/acs.jnatprod.7b00799. [DOI] [PubMed] [Google Scholar]
- 16.Zhu H, Li C, Gao C. Applications of CRISPR–Cas in agriculture and plant biotechnology. Nat Rev Mol Cell Biol. 2020;21(11):661–677. doi: 10.1038/s41580-020-00288-9. [DOI] [PubMed] [Google Scholar]
- 17.Zhang L, Xu PW, Cai YF, Ma LL, Li SF, Li SF, Xie WJ, Song J, Peng LC, Yan HJ, et al. The draft genome assembly of Rhododendron delavayi Franch. var. delavayi. GigaScience. 2017;6(10):11. doi: 10.1093/gigascience/gix076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soza VL, Lindsley D, Waalkes A, Ramage E, Patwardhan RP, Burton JN, Adey A, Kumar A, Qiu RL, Shendure J, Hall B. The Rhododendron genome and chromosomal organization provide insight into shared whole-genome duplications across the heath family (Ericaceae) Genome Biol Evol. 2019;11(12):3353–3371. doi: 10.1093/gbe/evz245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang FS, Nie S, Liu H, Shi TL, Tian XC, Zhou SS, Bao YT, Jia KH, Guo JF, Zhao W, et al. Chromosome-level genome assembly of a parent species of widely cultivated azaleas. Nat Commun. 2020;11(1):5269. doi: 10.1038/s41467-020-18771-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choudhary S, Thakur S, Jaitak V, Bhardwaj P. Gene and metabolite profiling reveals flowering and survival strategies in Himalayan Rhododendron arboreum. Gene. 2019;690:1–10. doi: 10.1016/j.gene.2018.12.035. [DOI] [PubMed] [Google Scholar]
- 21.Xing W, Liao J, Cai M, Xia Q, Liu Y, Zeng W, Jin X. De novo assembly of transcriptome from Rhododendron latoucheae Franch. using Illumina sequencing and development of new EST-SSR markers for genetic diversity analysis in Rhododendron. Tree Genet Genomes. 2017;13(3):53. doi: 10.1007/s11295-017-1135-y. [DOI] [Google Scholar]
- 22.Choudhary S, Thakur S, Najar RA, Majeed A, Singh A, Bhardwaj P. Transcriptome characterization and screening of molecular markers in ecologically important Himalayan species (Rhododendron arboreum) Genome. 2018;61(6):417–428. doi: 10.1139/gen-2017-0143. [DOI] [PubMed] [Google Scholar]
- 23.Cai YF, Wang JH, Zhang L, Song J, Peng LC, Zhang SB. Physiological and transcriptomic analysis highlight key metabolic pathways in relation to drought tolerance in Rhododendron delavayi. Physiol Mol Biol Plants. 2019;25(4):991–1008. doi: 10.1007/s12298-019-00685-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jia X, Tang L, Mei X, Liu H, Luo H, Deng Y, Su J. Single-molecule long-read sequencing of the full-length transcriptome of Rhododendron lapponicum L. Sci Rep. 2020;10(1):6755. doi: 10.1038/s41598-020-63814-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sayers EW, Beck J, Brister JR, Bolton EE, Canese K, Comeau DC, Funk K, Ketter A, Kim S, Kimchi A, Kitts PA, Kuznetsov A, Lathrop S, Lu Z, McGarvey K, Madden TL, Murphy TD, O’Leary N, Phan L, Schneider VA, Thibaud-Nissen F, Trawick BW, Pruitt KD, Ostell J. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2020;48(D1):D9–D16. doi: 10.1093/nar/gkz899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sneddon TP, Li P, Edmunds SC. GigaDB: announcing the GigaScience database. GigaScience. 2012. 10.1186/2047-217X-1-11. [DOI] [PMC free article] [PubMed]
- 27.Leinonen R, Sugawara H, Shumway M, on behalf of the International Nucleotide Sequence Database Collaboration The sequence read archive. Nucleic Acids Res. 2011;39(D1):D19–D21. doi: 10.1093/nar/gkq1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):13. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huerta-Cepas J, Forslund K, Coelho LP, Szklarczyk D, Jensen LJ, von Mering C, Bork P. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol Biol Evol. 2017;34(8):2115–2122. doi: 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huerta-Cepas J, Szklarczyk D, Heller D, Hernandez-Plaza A, Forslund SK, Cook H, Mende DR, Letunic I, Rattei T, Jensen LJ, et al. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019;47(D1):D309–D314. doi: 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitchell AL, Attwood TK, Babbitt PC, Blum M, Bork P, Bridge A, Brown SD, Chang HY, El-Gebali S, Fraser MI, et al. InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 2019;47(D1):D351–D360. doi: 10.1093/nar/gky1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12(1):59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 35.Zheng Y, Jiao C, Sun HH, Rosli Hernan G, Pombo Marina A, Zhang P, Banf M, Dai XB, Martin Gregory B, Giovannoni James J, et al. iTAK: a program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol Plant. 2016;9(12):1667–1670. doi: 10.1016/j.molp.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 36.Emms DM, Kelly S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015;16(1):157. doi: 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emms DM, Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20(1):238. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang S, Ding J, Deng D, Tang W, Sun H, Liu D, Zhang L, Niu X, Zhang X, Meng M, Yu J, Liu J, Han Y, Shi W, Zhang D, Cao S, Wei Z, Cui Y, Xia Y, Zeng H, Bao K, Lin L, Min Y, Zhang H, Miao M, Tang X, Zhu Y, Sui Y, Li G, Sun H, Yue J, Sun J, Liu F, Zhou L, Lei L, Zheng X, Liu M, Huang L, Song J, Xu C, Li J, Ye K, Zhong S, Lu BR, He G, Xiao F, Wang HL, Zheng H, Fei Z, Liu Y. Draft genome of the kiwifruit Actinidia chinensis. Nat Commun. 2013;4(1):2640. doi: 10.1038/ncomms3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xia EH, Li FD, Tong W, Li PH, Wu Q, Zhao HJ, Ge RH, Li RP, Li YY, Zhang ZZ, Wei CL, Wan XC. Tea plant information archive: a comprehensive genomics and bioinformatics platform for tea plant. Plant Biotechnol J. 2019;17(10):1938–1953. doi: 10.1111/pbi.13111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lamesch P, Berardini TZ, Li DH, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, Karthikeyan AS, Lee CH, Nelson WD, Ploetz L, Singh S, Wensel A, Huala E. The Arabidopsis information resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012;40(D1):D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beier S, Thiel T, Munch T, Scholz U, Mascher M. MISA-web: a web server for microsatellite prediction. Bioinformatics. 2017;33(16):2583–2585. doi: 10.1093/bioinformatics/btx198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012;40(15):e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu J, Chen T, Zhang YB, Li YK, Gong JY, Yi Y. The complete chloroplast genome of Rhododendron delavayi (Ericaceae) Mitochondrial DNA Part B-Resour. 2020;5(1):37–38. doi: 10.1080/23802359.2019.1689860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li HE, Guo QQ, Li Q, Yang L. Long-reads reveal that Rhododendron delavayi plastid genome contains extensive repeat sequences, and recombination exists among plastid genomes of photosynthetic Ericaceae. Peerj. 2020. 10.7717/peerj.9048. [DOI] [PMC free article] [PubMed]
- 45.Shen JS, Li XQ, Zhu XT, Huang XL, Jin SH. Complete chloroplast genome of Rhododendron pulchrum, an ornamental medicinal and food tree. Mitochondrial DNA Part B-Resour. 2019;4(2):3527–3528. doi: 10.1080/23802359.2019.1676181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 47.Wang YP, Tang HB, DeBarry JD, Tan X, Li JP, Wang XY, Lee TH, Jin HZ, Marler B, Guo H, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40(7):e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deng W, Nickle DC, Learn GH, Maust B, Mullins JI. ViroBLAST: a stand-alone BLAST web server for flexible queries of multiple databases and user's datasets. Bioinformatics. 2007;23(17):2334–2336. doi: 10.1093/bioinformatics/btm331. [DOI] [PubMed] [Google Scholar]
- 49.Buels R, Yao E, Diesh CM, Hayes RD, Munoz-Torres M, Helt G, Goodstein DM, Elsik CG, Lewis SE, Stein L, Holmes IH. JBrowse: a dynamic web platform for genome visualization and analysis. Genome Biol. 2016;17(1):66. doi: 10.1186/s13059-016-0924-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
RPGD is freely available at http://bioinfor.kib.ac.cn/RPGD/.