Abstract
A large proportion of the complexity and redundancy of LC-MS metabolomics data comes from adduct formation. To reduce such redundancy, many tools have been developed to recognize and annotate adduct ions. These tools rely on predefined adduct lists that are generated empirically from reversed-phase LC-MS studies. In addition, hydrophilic interaction chromatography (HILIC) is gaining popularity in metabolomics studies due to its enhanced performance over other methods for polar compounds. HILIC methods typically use high concentrations of buffer salts to improve chromatographic performance. Therefore, it is necessary to analyze adduct formation in HILIC metabolomics. To this end, we developed covariant ion analysis (COVINA) to investigate metabolite adduct formation. Using this tool, we completely annotated 201 adduct and fragment ions from 10 metabolites. Many of the metabolite adduct ions were found to contain cluster ions corresponding to mobile phase additives. We further utilized COVINA to find the major ionized forms of metabolites. Our results show that for some metabolites, the adduct ion signals can be >200-fold higher than the signals from the deprotonated form, offering better sensitivity for targeted metabolomics analysis. Finally, we developed an in-source CID ramping (InCIDR) method to analyze the intensity changes of the adduct and fragment ions from metabolites. Our analysis demonstrates a promising method to distinguish the protonated and deprotonated ions of metabolites from the adduct and fragment ions.
Introduction
LC-MS-based metabolomics aims to comprehensively characterize small molecule metabolites in biological samples. Modern mass spectrometry can offer both high sensitivity and high mass resolution, making it possible to detect hundreds or thousands of metabolites in a single biological sample.1–3 As a result, raw LC-MS data are highly complex. A typical 25-minute LC-MS run on an Orbitrap instrument may produce 4000 spectra, each with >500–1000 peaks in the mass-to-charge ratio (m/z) domain. To reduce the data complexity, untargeted metabolomics data analysis starts with feature detection.4,5 Each LC-MS feature is a combination of an accurate m/z and a retention time. The features are expected to have a decent chromatographic peak shape in the time domain and may correspond to actual metabolites, isotopic peaks, dimer ions of metabolites, adduct ions, in-source fragment ions or background matrix components. To facilitate downstream metabolite identification and statistical analysis, this feature list needs to be optimized and annotated. The optimization step removes duplicate and false positive features.6 The annotation step recognizes and annotates the adduct ions and natural isotopic ions.7–9 These data processing steps dramatically reduce the complexity of metabolomics data. Mahieu et al. reported the analysis of an E. coli metabolomics dataset in which 25,000 features were reduced to fewer than 1,000 unique metabolites.10 This example highlights the complexity and redundancy of a typical metabolomics dataset.
The majority of adduct ion annotation tools fall into two categories. One involves calculating the correlations of peak intensities across samples from the feature list.6,8,11,12 The other involves reading the extracted ion chromatograms (XICs) in the raw data and calculating the correlation of chromatographic peaks.7,9 Even though each tool has demonstrated utility, the adduct ions in a metabolomics dataset are often insufficiently annotated. Glutamate, for example, was reported to produce more than 100 spectral peaks, but many of the complex adducts did not have a chemical annotation.10 Lin et al used isotope-labeled samples to facilitate the annotation of metabolites and their adduct ions and observed ions that had a large m/z but contained very few carbon atoms. These ions were considered unreasonable and were filtered out during the annotation. In fact, these ions indicated the existence of large adducts.12 It is noteworthy that many adduct annotation tools rely on a predefined list to identify the adduct ions.13,14 The incompleteness of adduct lists limits the performance of adduct annotation tools. This limitation is further amplified in hydrophilic interaction liquid chromatography (HILIC) ESI-MS metabolomics studies. In recent years, HILIC has become a popular metabolomics technique due to its resolving power for very polar cellular metabolites.15 HILIC typically involves the use of buffer salts to improve separation and peak shape.16,17 These salts have a significant impact on the ionization of metabolite molecules and adduct formation. Erngren et al. reported the formation of adduct ions that contain multiple sodium and potassium formate moieties.18 These cluster ions are not included in common adduct lists, making adduct annotation difficult. We aim to further investigate adduct ion formation under our HILIC conditions to elucidate frequently occurring m/z distances that should be included in annotation tool lists.
In this work, we set out to analyze the adduct and fragment ion formation from metabolites in HILIC ESI-MS, for which we developed an algorithm named covariant ion analysis (COVINA). Unlike existing tools, which search for coeluting peaks in an extracted feature list, COVINA is a targeted tool that analyzes one metabolite peak at a time by reading the mzXML files directly. By doing so, COVINA can avoid false positives and false negatives in the feature extraction and misgrouping in the peak alignment and grouping steps. Using this tool, we found a number of cluster adduct ions from metabolites. To help assign the chemical identity of these adduct ions, the HILIC mobile phase was prepared with 2H-labeled acetic acid. Using this method, we completely annotated 201 adduct and fragment ions resulting from 10 metabolites. We then used this new HILIC adduct list to annotate untargeted metabolomics datasets using existing tools and observed improved performance.
We also developed an in-source collision-induced dissociation (CID) ramping (InCIDR) method to help determine the neutral molecular mass of the metabolites. Adduct annotation tools implement heuristic algorithms to score hypotheses on metabolite neutral masses based on the detection of the expected adduct ions.9,19 InCIDR is a completely orthogonal approach that utilizes COVINA to analyze the intensity changes of the adduct and fragment ions with increasing in-source CID energy levels. In general, as the in-source CID energy level increases, the fragment ions increase in intensity, while the adduct ions decrease in intensity. InCIDR monitors all the covariant ions and detects such patterns to score the hypothesized neutral masses. Examples show that InCIDR is a robust and promising means of supplementing existing adduct annotation tools.
Experimental
Chemicals.
LCMS-grade methanol (A456), acetonitrile (A955), acetic acid (A35) and water (ACROS 61515) were purchased from Fisher Chemicals (Pittsburgh, PA). 2H4-Acetic acid (99.5% isotopic purity, DLM-12) was purchased from Cambridge Isotope Laboratories (Tewksbury, MA). Other chemicals, including ammonium hydroxide (09859), sodium pyruvate (P2256), sodium L-lactate (L7022), leucine (L8912), isoleucine (I5281), NAD hydrate (N7004), ATP disodium salt hydrate (A2383), D-glucose-6-phosphate dipotassium salt hydrate (G7375), D-fructose-6-phosphate disodium salt hydrate (F3726), L-malic acid (112577) and D-fructose-1,6-bisphosphate trisodium salt hydrate (F6803), were purchased from MilliporeSigma (Burlington, MA).
Cell lysate preparation.
The L3.6 human pancreatic ductal adenocarcinoma cell line was obtained from ATCC. These cells were seeded in 100 mm dishes and cultured in RPMI supplemented with 10% bovine serum (Fetal Clone III, HyClone), 100 units/ml penicillin and 100 μg/ml streptomycin. Cells were grown to ~85% confluency for metabolite extraction. Before harvesting the cells, the media was aspirated and then quickly overlaid with 1 ml 40:40:20 mixture of methanol:acetonitrile:water with 0.5% (V/V) formic acid. The plates were incubated on ice for 5 min, and then 50 μl of 15% (m/V) NH4HCO3 was added to neutralize the formic acid. Cells were scraped into microfuge tubes and centrifuged for 10 min at 15,000 x g at 4°C. The supernatant was collected for LC-MS.
Liquid chromatography.
HILIC separation was performed on a Vanquish Horizon UHPLC system (Thermo Fisher Scientific, Waltham, MA) with an XBridge BEH Amide column (150 mm × 2.1 mm, 2.5 μm particle size, Waters, Milford, MA) using a gradient of solvent A (95%:5% H2O:acetonitrile with 20 mM acetic acid, 40 mM ammonium hydroxide, pH 9.4) and solvent B (20%:80% H2O:acetonitrile with 20 mM acetic acid, 40 mM ammonium hydroxide, pH 9.4). For the 2H4-acetic acid mobile phase experiment, the acetic acid in both mobile phases A and B was replaced with 2H4-acetic acid at the same concentration. The gradient was 0 min, 100% B; 3 min, 100% B; 3.2 min, 90% B; 6.2 min, 90% B; 6.5 min, 80% B; 10.5 min, 80% B; 10.7 min, 70% B; 13.5 min, 70% B; 13.7 min, 45% B; 16 min, 45% B; 16.5 min, 100% B; and 22 min, 100% B. The flow rate was 300 μl/min. The injection volume was 5 μL, and the column temperature was set to 25°C. The autosampler temperature was set to 4°C, and the injection volume was 5 μL.
Mass spectrometry.
The mass spectrometry analysis was performed on Thermo Q Exactive PLUS instrument with a HESI source, which was set to a spray voltage of −2.7 kV in negative mode and 3.5 kV in positive mode. The sheath, auxiliary, and sweep gas flow rates were 40, 10, and 2 (arbitrary units), respectively. The capillary temperature was set to 300°C, and the aux gas heater was set to 360°C. The S-lens RF level was 45. The m/z scan range was set to 72 to 1000 m/z in either positive or negative ionization mode. The AGC target was set to 3e6, and the maximum IT was 200 ms. The in-source CID energy level was set to 0 eV unless otherwise specified. The resolution was set to 70k unless using in-source CID ramping. In the data acquisition of InCIDR, each scan cycle consisted of 8 scan events with in-source CID energy levels of 0 eV, 2 eV, 4 eV, 6 eV, 8 eV, 10 eV, 15 eV and 20 eV. InCIDR uses 17.5k resolution to achieve a scan rate of 2 Hz.
Covariant ion analysis (COVINA).
COVINA is a targeted approach to search for coeluting ion species for the metabolite of interest in the raw mzXML files. In short, COVINA builds XICs for all the ions detected together with the metabolite of interest, and returns a list of ions that are highly correlated with the metabolite of interest. The full COVINA algorithm works as the following: when the m/z of a metabolite (base m/z) is specified, COVINA first looks for the scan at the apex of the corresponding XIC (Figure 1A). If multiple peaks exist in one XIC, the scan number can be specified to focus on the metabolite of interest. This selected scan is used as the reference spectrum. For each m/z in the reference spectrum that is above a specified intensity threshold, COVINA builds an XIC within an adjustable mass tolerance. Each of these XICs is compared to the reference XIC of the metabolite of interest. Covariant ions are identified by evaluating the Pearson correlation between the base reference XIC and the query XIC (Figure 1B, Table 1). The adduct, fragment and isotopic ions usually show very good correlation (R>0.9) with the base m/z. These covariant ions plus the base m/z peak form the pseudospectrum of the metabolite (Figure 1C).
Figure 1.
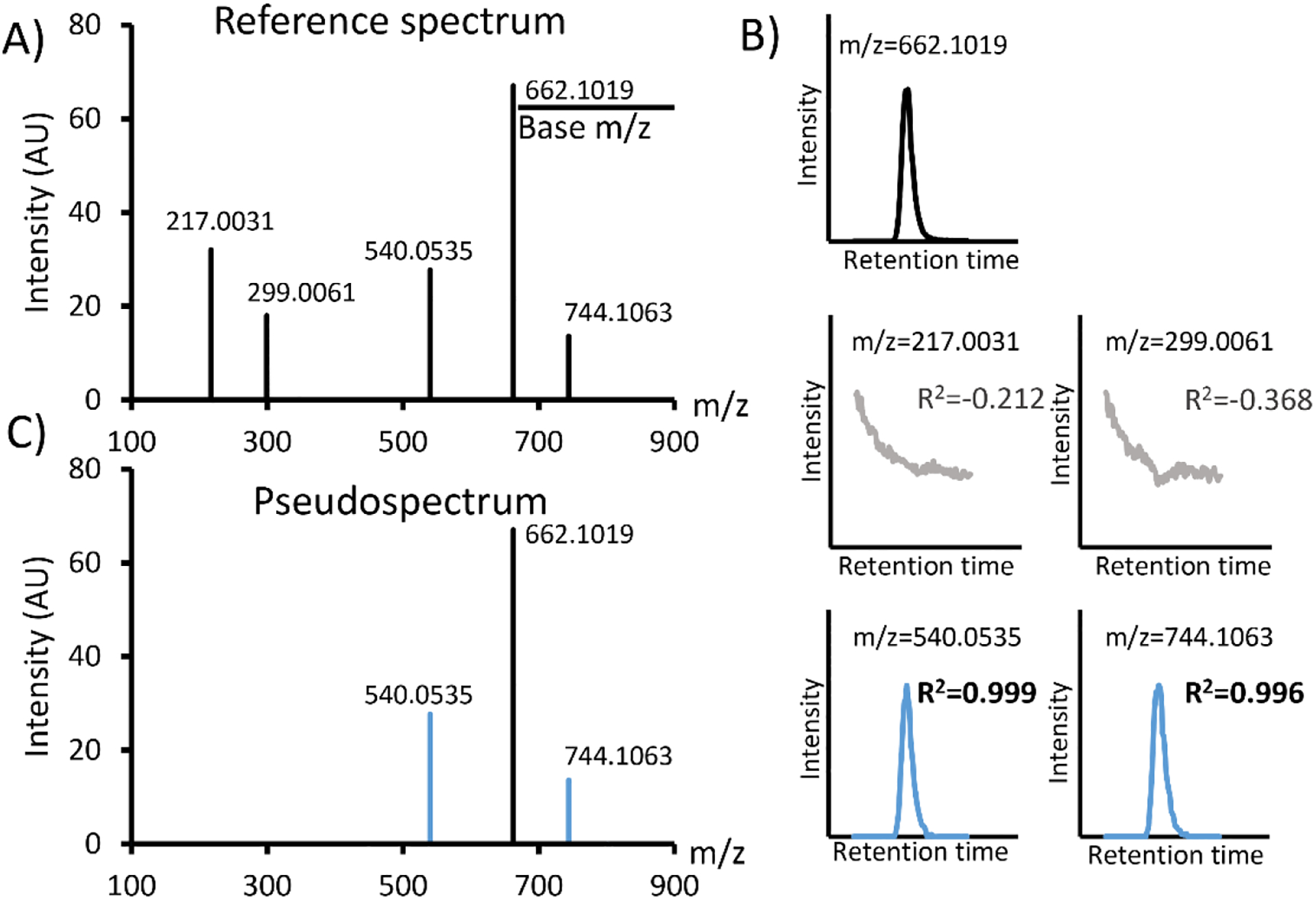
Covariant ion analysis (COVINA). (A) The reference mass spectrum at the apex of the XIC of the base m/z. (B) The XICs of the base m/z and all other m/z values in the reference spectrum. (C) COVINA generates a pseudospectrum containing the covariant ions (blue) of the base m/z.
Table 1.
Pseudocode for COVINA
| Algorithm COVINA | |
|---|---|
| 1: | procedure COVINA(input parameters: mzXML file(s), m/z; optional parameters: scan.number, |
| 2: | peak.width, mass.tolerance, intensity.threshold, correlation.threshold) |
| 3: | Use the first mzXML file to do |
| 4: | XIC.Base ← XIC of m/z within mass.tolerance |
| 5: | if scan.number is not specified then |
| 6: | scan.number ← scan with highest signal in XIC.Base |
| 7: | end if |
| 8: | scan.range ← scan.number ± peak.width |
| 9: | mz.table ← all m/z values in the scan.number-th scan that are above |
| 10: | intensity.threshold |
| 11: | Base.Chromatogram ← XIC.Base within scan.range |
| 12: | for each value in the mz.table |
| 13: | Query.Chromatogram ← corresponding XIC within mass.tolerance and |
| 14: | scan.range |
| 15: | if correlation between Base.Chromatogram and Query.Chromatogram > |
| 16: | correlation.threshold then |
| 17: | add this m/z value to the covariant.ion.list |
| 18: | end if |
| 19: | end for |
| 20: | end do |
| 21: | In each mzXML file do |
| 22: | covariant.ion.intensities ← Integrated peak area for each value in the |
| 23: | covariant.ion.list within scan.range |
| 24: | end do |
| 25: | return covariant.ion.list, covariant.ion.intensities |
| 26: | end procedure |
Covariant feature annotation.
Adduct features identified by the COVINA algorithm were annotated utilizing both mixed integer linear programming and manual identification. A list of simple ions such as Ca2+, Na+, Ac− and NO3− is provided by the user as input. For each adduct feature detected, mixed integer linear programming is used to generate a combination of simple ions that matches the observed Δm/z. If the algorithm fails to find an ion combination that is close enough to the observed Δm/z, the ion list is expanded to include more ions such as Fe2+ and H3SiO4−. The sample code, input table and output results are in our Github repository (see the link blow). All annotations were evaluated based on their mass accuracy and manual inspection.
Code and example data availability.
COVINA is implemented in R (version 3.6.1). The code as well as the sample data are available at https://github.com/XiaoyangSu/COVINA.
Results and Discussion
Investigating adduct and fragment ion formation using COVINA
A common approach to studying adduct formation is to inject chemical standards directly into the mass spectrometer via a syringe pump, either coupled to the normal LC flow or not. This method is conceptually simple, but the adduct signals are mingled with background signals, making complete adduct annotation difficult.18 Another approach used in many common adduct annotation tools is to group related m/z values in the feature lists. The feature lists used by these annotation tools are generated from a peak picking algorithm such as centWave5, which produces thousands of features from a typical dataset. Despite significant improvement and refinement of the grouping algorithms, the feature grouping is usually insufficient for placing all of the adduct and fragment ions in the same group as the parent metabolite ion. We tested the grouping performance of CAMERA7 and CliqueMS9. Both tools placed the adduct ions resulting from 10 metabolites into more than 20 different groups (Figure S1). To systematically study the patterns of metabolite adduct and fragment formation, it is necessary to optimize the detection of adducts and fragments in the data analysis process.
Instead, we utilize chromatography to analyze ion species that coelute with the metabolite of interest. The adduct ions and fragment ions should be chromatographically correlated with respect to peak shape to their parent protonated and deprotonated metabolite ions. COVINA detects such coelution correlation to filter out persistent background ions and generates a high-quality list of highly correlated covariant features. Unlike other annotation tools, COVINA does not use a feature list generated from a peak picking algorithm. Instead, COVINA is a targeted algorithm that analyzes one metabolite at a time. COVINA takes the input of the m/z value of a specific metabolite (base m/z) to build the XICs of this m/z and all other m/z values in the same spectrum and calculates the chromatographic correlation between the XIC of the base m/z and the other XICs. In the output data, COVINA produces a list of highly correlated m/z values that includes adduct and fragment ions and isotopic peaks.
Using our COVINA algorithm, we studied the adduct formation of some key metabolites in negative ionization mode. COVINA revealed multiple adducts for each of the metabolites in our chemical standard mixture. For example, Figure 2A shows the adduct occurrence of pyruvate, lactate, leucine, malate and glucose-6-phosphate (Glc6P). Interestingly, many adducts showed common m/z differences (Δm/z, m/z of an adduct – m/z of [M-H]−), suggesting that these adducts have the same chemical identity. Previous studies have shown that metabolites may form oligomer adducts ([2M-H]− or [3M-H]−) or heterodimer adducts ([M1+M2-H]−).20 These complex adducts were less frequently detected in our samples than the common adducts. The commonly observed Δm/z of 82.003 is important, as it was the only Δm/z observed for all five metabolites. Moreover, 82.003 was the common difference in some Δm/z series, such as 82.003/164.006/246.009, 97.968/179.972/261.973 and 157.989/239.992/321.995/403.999 (Figure 2A). This observation suggests that some large adducts, such as 321.995 and 403.999, may have repeating units of 82.003. We also calculated the pairwise m/z differences of all the covariant ions detected in our standard mixture samples. The histogram of these Δm/z values confirmed that 82.003 was the most common mass difference among the covariant ions (Figure 2B). 164.006, which is 82.003×2, is also a common Δm/z value, again suggesting that 82.003 represented a repeating unit in the adduct ions.
Figure 2.
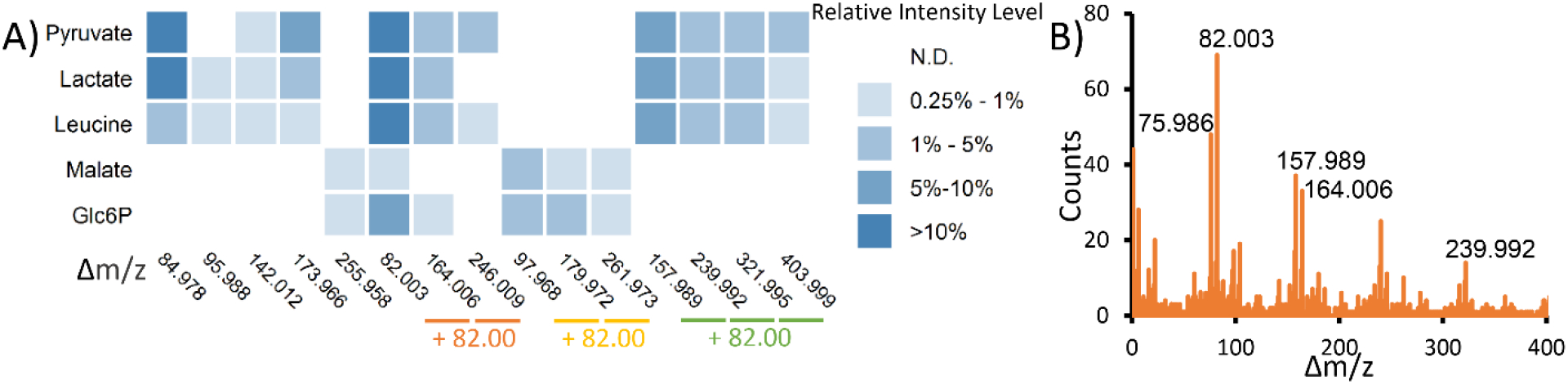
Common adducts found by COVINA. (A) Heatmap showing the intensity of the adduct ions relative to the intensity of [M-H]−. Δm/z is calculated as m/z(adduct)-m/z([M-H]−). The Δm/z values in the same arithmetic sequence are grouped together to show compounded repeating units of the same adduct. (B) Histogram of Δm/z pairwise differences of all the covariant ions within each metabolite group. The most common Δm/z values are highlighted in the plot.
Δm/z 82.003 matches the mass of sodium acetate (NaAc; calculated mass, 82.00253). While acetate is used in the mobile phase buffer, the sodium ions were not purposefully added to the mobile phase. Trace amounts of sodium may come from water, glassware or impurities in solvent additives. It is surprising to see NaAc moiety but not NH4Ac that presents in many adduct ions. To confirm that NaAc was the repeating unit in many adducts, we prepared the mobile phase using deuterium-labeled acetate ([2H3]-Ac−). In this labeled mobile phase, many adducts showed shifted m/z values (Figure 3). Pyruvate had an adduct [M-H+157.9890]− (m/z 244.9977) that was shifted to [M-H+164.027] (m/z 251.0356) in the deuterium-labeled mobile phase, showing that this adduct has 2 acetate moieties. Therefore, the chemical identity of this adduct can be determined as [M+CaAc2-H]− (Table 2). The high mass accuracy of the Orbitrap mass analyzer lends great confidence for accurate adduct identification. For example, the formula [M+CaAc2-H]− matches the detected m/z with a mass accuracy of −1.1 ppm. An alternative formula [M+KAc2]− has a mass accuracy of −37.6 ppm and should, therefore, be ruled out. Pyruvate adducts [M-H+239.9918]− and [M-H+321.9952]− are shifted by 9.056 and 12.075 Da, suggesting that they have Ac3 and Ac4, respectively. They are annotated as [M+NaCaAc3-H]− and [M+Na2CaAc4-H]−. [M-H+173.965]− did not shift in the deuterium-labeled mobile phase and, therefore, did not contain an Ac in its adduct moiety. This adduct is annotated as [M+H6Si2O7-H]−. The use of the deuterium-labeled mobile phase helps to narrow down the possible adduct formulas and leads to confident identification. The observation that many adduct ions contain cluster ions inspired us to utilized mixed integer linear programming21 to find the ion combinations for adduct annotations. The detailed procedure is described in the Method section. We also confirmed the annotations through manual inspection and curation.
Figure 3.
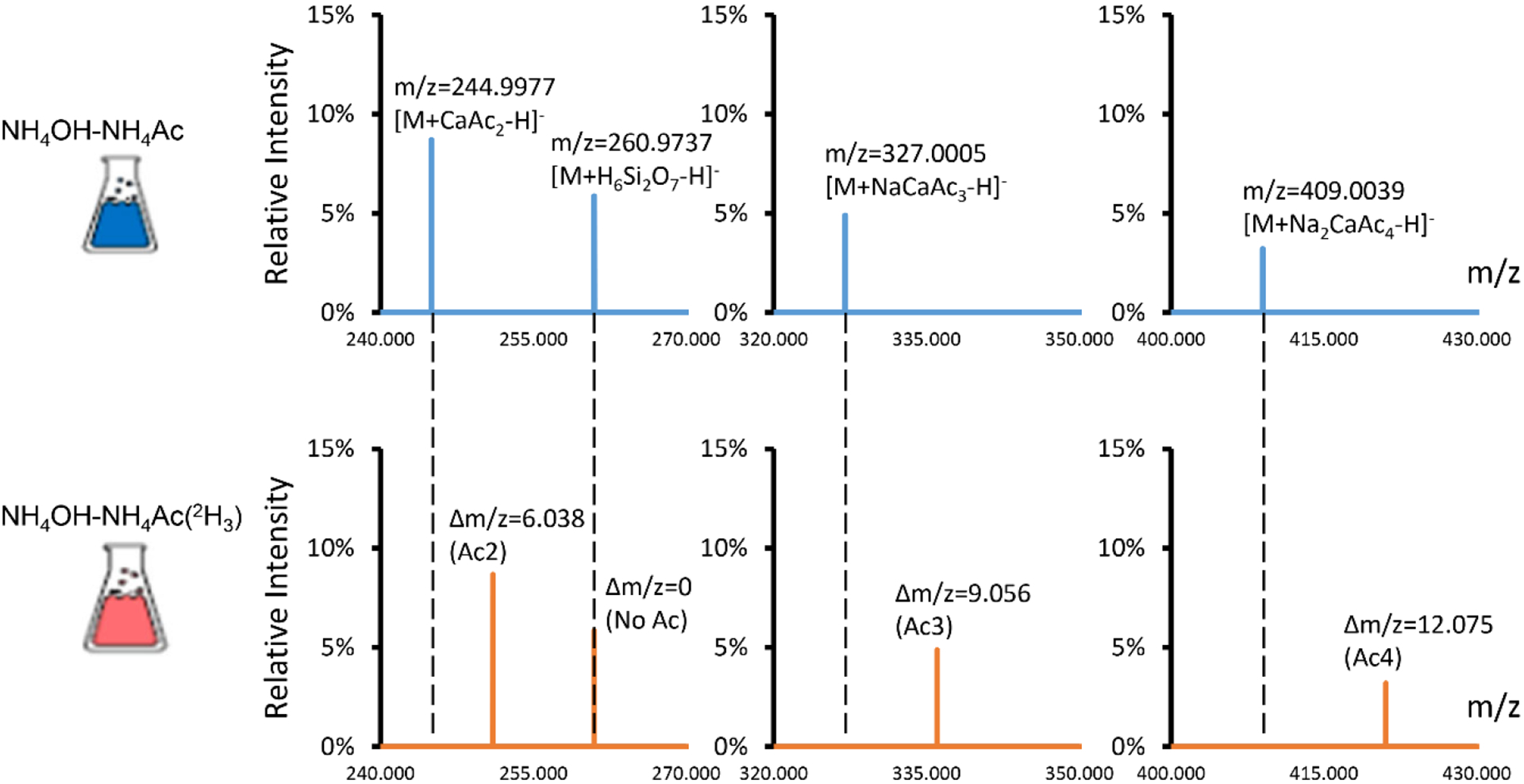
2H-acetic acid was used to determine the chemical identity of the adducts. The upper panel shows the adduct ions of pyruvate that were detected in the normal mobile phase. The lower panel shows the adduct ions of pyruvate that were detected in the mobile phase prepared with [2H4]-acetic acid. Each acetate moiety in the adduct ion should result in an m/z shift of 3.018. The adduct ions m/z 244.9977, 327.0005 and 409.0039 were shifted by 6.038, 9.056 and 12.075, respectively, in the 2H-labeled mobile phase, suggesting that 2, 3 and 4 acetate moieties are in these adduct ions, respectively. The m/z 260.9737 ion did not shift in the 2H-labeled mobile phase, suggesting that no acetate moieties are in this adduct ion.
Table 2.
Covariant ions of lactate
| Observed m/z | Relative Abundance | Δm | Chemical Annotation | Calculated m/z | Mass Accuracy (ppm) | Observed m/z in 2H3-Ac |
|---|---|---|---|---|---|---|
| 89.0244 | 100.00% | 0.0000 | [M-H]− | 89.0244 | −0.7 | 89.0245 |
| 90.0276 | 3.32% | 1.0032 | 13C1-[M-H]− | 90.0278 | −2.2 | 90.0278 |
| 91.0289 | 0.69% | 2.0045 | 18O1-[M-H]− | 91.0287 | 2.5 | 91.0287 |
| 148.9905 | 0.40% | 59.9661 | [M+SiO2-H]− | 148.9912 | −4.8 | 148.9910 |
| 171.0276 | 13.20% | 82.0032 | [M+NaAc-H]− | 171.0275 | 0.6 | 174.0462 |
| 172.0306 | 0.56% | 83.0062 | 13C1-[M+NaAc-H]− | 172.0309 | −1.7 | 175.0496 |
| 174.0020 | 36.30% | 84.9776 | [M+NaNO3-H]− | 174.0020 | 0.0 | 174.0019 |
| 175.0061 | 1.00% | 85.9817 | 13C1-[M+NaNO3-H]− | 175.0054 | 4.0 | 175.0052 |
| 177.9570 | 0.44% | 88.9327 | [M+FeO(OH)-H]− | 177.9570 | 0.1 | 177.9572 |
| 185.0120 | 0.87% | 95.9876 | [M+H4SiO4-H]− | 185.0123 | −1.7 | 185.0123 |
| 201.0387 | 6.43% | 112.0143 | [2M+Na-2H]− | 201.0381 | 3.1 | 201.0381 |
| 202.0426 | 0.43% | 113.0183 | 13C1-[2M+Na-2H]− | 202.0414 | 6.0 | 202.0415 |
| 231.0369 | 0.92% | 142.0126 | [M+MgAc2-H]− | 231.0361 | 3.7 | ND* |
| 247.0135 | 5.70% | 157.9892 | [M+CaAc2-H]− | 247.0136 | −0.4 | 253.0513 |
| 253.0312 | 1.09% | 164.0069 | [M+Na2Ac2-H]− | 253.0306 | 2.6 | 259.0681 |
| 262.9891 | 3.02% | 173.9648 | [M+H6Si2O7-H]− | 262.9896 | −1.8 | 262.9896 |
| 313.0396 | 0.51% | 224.0152 | [M+NaMgAc3-H]− | 313.0391 | 1.3 | ND* |
| 329.0170 | 2.78% | 239.9926 | [M+NaCaAc3-H]− | 329.0167 | 0.9 | 338.0732 |
| 411.0196 | 1.37% | 321.9953 | [M+Na2CaAc4-H]− | 411.0198 | −0.4 | 423.0950 |
| 493.0235 | 0.71% | 403.9991 | [M+Na3CaAc5-H]− | 493.0228 | 1.3 | 508.1173 |
ND: Not detected. The counterparts of m/z 231.0369 and 313.0396 were not detected in the 2H3-Ac mobile phase and are annotated as Mg2+-containing adduct ions. Presumably, 2H-HAc has a much lower level of Mg2+, so these adduct ions were not formed.
Similar to previous reports, complex ions such as adducts of fragments were detected.22 For Glc6P, [M-H+37.948]− was detected. This m/z matches [M+Ca-3H]− with a mass accuracy of 2.2 ppm. However, this adduct shifted to [M-H+40.965]− in the deuterated mobile phase, indicating 1 Ac group. This adduct is essentially an adduct of erythrose-4-phosphate ([E4P+CaAc-2H]−), a known fragment of Glucose6P.23 Another example of an adduct of fragment is [NAD-H+57.921]−. Δm/z 57.921 does not match any chemical formula as a simple adduct. Using the deuterated mobile phase, it was demonstrated that this ion has 2 Ac groups and is annotated as [NAD-nicotinamide+NaCaAc2-2H]−. The combined use of COVINA and the deuterated mobile phase led to the complete annotation of 201 ion covariants produced from 10 metabolites (covariant ions of lactate are shown in Table 2; see Table S1 for the complete list).
We also manually inspected and confirmed a number of adduct ions detected from L3.6 human PDAC lysate samples. Our results show that [M+NaAc-H]− was more common than [M+Na-2H]− (Figure 4A). In the annotation table, we observed a number of adducts that included salts. Due to the use of a high concentration of salt in HILIC metabolomics, some metabolites show higher ion counts in their adduct/fragment forms than in their protonated and deprotonated forms. This information is important, especially in targeted analysis of metabolomics data. Using COVINA, we investigated such cases using L3.6 cell lysates. The signal from acetyl-CoA in its divalent ion form [M-2H]2− was 8-fold higher than that in its [M-H]− form. The signal of phosphocholine was 350-fold higher in its acetate adduct form than in its [M-H]− form. S-Adenosyl-L-methionine (AdoMet) was detected in several fragment forms that had signals higher than the [M-H]− signal from AdoMet, including methylthioadenosine (MTA, 8-fold higher), the MTA acetate adduct (35-fold higher) and adenine (55-fold higher) (Figure 4B). These results enable better detection of key metabolites in the targeted metabolomics analysis workflow.
Figure 4.
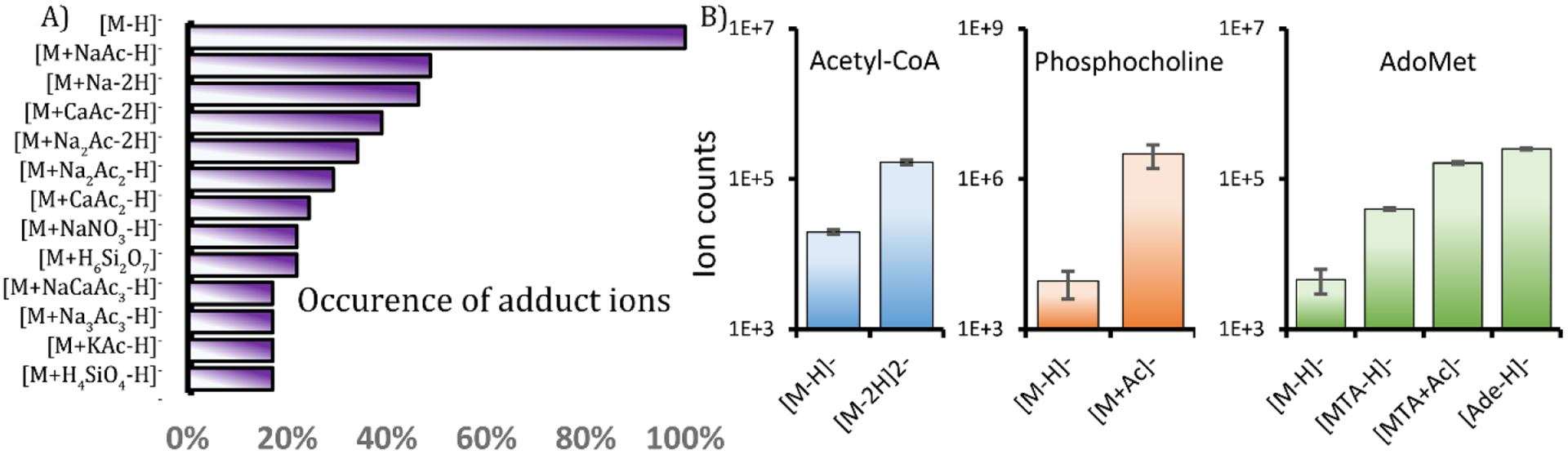
Significant adduct ions and fragment ions of key metabolites in the L3.6 human pancreatic ductal adenocarcinoma cell lysate samples. (A) Comparison of the occurrence of adduct ions. The denominator is the number of metabolites with at least one adduct ion detected. (B) Significant covariant ions that are stronger than the molecular ions. Ion counts are shown as the mean +/− standard deviation, n=3.
Adduct formation in HILIC metabolomics is largely affected by the solvent additives as well as other chemicals present in the system. NO3− adducts, for example, may have originated from the nitric acid that we used to clean the ion transfer tube of the mass spectrometer. We have developed and utilized COVINA to demonstrate the detection of many condition-specific adducts that should be added to adduct annotation lists. The systematic construction of the XIC for each feature in the reference scan, evaluation of the fidelity of the correlation between the base peak shape and the adduct peak shape, and manual verification of the feature annotation leads to confident identification of the adducts. These experimentally observed and well-credentialed adduct ions should be included in the adduct list. In addition, many adduct ions contain alkali metal ions and alkaline earth metal ions which have larger mass defects than common elements in metabolites (C, H, O, N, P and S). Because of the large mass defect of adduct ion, it is unlikely that a true metabolite ion is wrongly annotated as an adduct ion.24 Therefore, we believe the expanded adduct list will not give false positive adduct annotation, and we recommend using COVINA to generate an adduct list specific to each of the different HILIC conditions and instruments being utilized for analysis. We believe our adduct list is useful for systems operating with similar chromatographic conditions to those evaluated herein. Using such adduct lists, we can greatly improve the adduct annotation rate in untargeted metabolomics annotation tools such as CliqueMS. In L3.6 human pancreatic ductal adenocarcinoma cell lysate samples, CliqueMS annotated 23.5% and 53.5% of the features as adducts using a built-in adduct list and COVINA-generated list, respectively (Figure S2).
In essence, COVINA informs us about how metabolite molecules ionize. For instance, if the m/z and retention time of [M+H]+ are known, we can use COVINA to find all other ionized forms of M. However, not every metabolite has an easily detectable [M+H]+ form. Due to their pKa values, some metabolites, such as lactate and pyruvate, are much more predisposed to carry a negative charge than to carry a positive charge. Therefore, LC-MS metabolomics data acquisition is often performed in both positive and negative ionization modes to maximize the metabolome coverage. It is possible, however, to detect carboxylic acids such as lactate in positive ionization mode with reasonable sensitivity. We modified COVINA to analyze alternating scans in negative and positive ionization modes (Figure 5A). The XIC of [M-H]− was used as the reference, and all the XICs for negative and positive ionization modes were used to calculate the chromatographic correlation coefficients. Our results show that lactate can be detected in positive ionization mode as [M-H+2Na]+, the intensity of which is approximately 15% of the intensity of [M-H]− in negative mode (Figure 5B).
Figure 5.
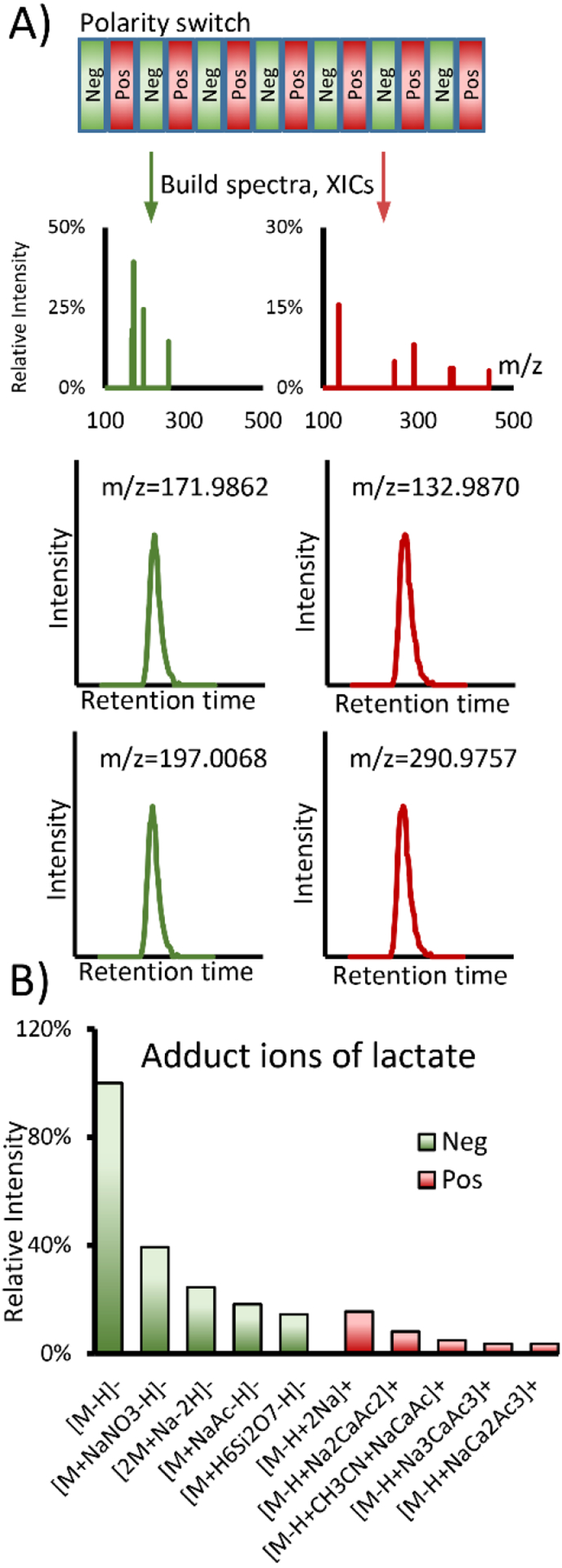
COVINA with polarity switching. (A) Scheme of COVINA with polarity switching. The XICs are built separately for each polarity, and the correlation with the reference XIC can be calculated using the standard COVINA method. (B) Example result for lactate adduct ions. Lactate is mainly detected as [M-H]− in negative ionization mode. The most abundant cation for lactate is [M-H+2Na]+, which produces a signal that is approximately 15% of the signal from [M-H]−.
Finding a metabolite neutral mass using in-source CID ramping (InCIDR)
For untargeted metabolomics data processing, the goal of adduct annotation tools is twofold: 1) to identify the adduct ions and exclude them from further analysis and 2) to find the neutral masses of the true metabolites for downstream structural elucidation and statistical analysis. To find the neutral mass of a metabolite out of many grouped coeluting ions, the annotation algorithm searches the mass differences between coeluting m/z values and matches the gaps to known adducts. The hypothesized metabolite neutral masses are scored based on the matching results and optionally the empirical frequency of the adducts.9,19 From these scores, the most plausible neutral mass is returned as the result. Our data suggest that the performance of such algorithms can be improved by having a more complete adduct list generated from COVINA (Figure S3). Nonetheless, because of the heuristic nature of these algorithms, the metabolite neutral mass may not be picked correctly. Other approaches to reinforce metabolite neutral mass prediction are needed.
Here, we propose using in-source CID ramping (InCIDR) to predict the metabolite neutral mass. Instead of using an empirical adduct list and matching the mass differences, InCIDR predicts the m/z of the protonated or deprotonated metabolite ion by analyzing the covariant ion intensity change during the ramping of the CID energy level. InCIDR is based on the assumption that when the in-source CID energy level increases, the adduct ions collapse and decrease in intensity while the fragment ions increase in intensity. Lin et al. first described such fragment ion trends and used them to filter fragment ions from a feature list.12 Based on this work, we extended this analysis to encompass the adduct ions and built a data acquisition and analysis method called in-source CID ramping (InCIDR). Unlike the original method using different CID energy levels in repeated runs, the data acquisition in InCIDR consists of a series of alternating scan events with increasing in-source CID energy levels (Figure 6A). In this way, there is no need to run the same sample repeatedly at different energy levels, reducing instrument time. Second, the single run eliminates the ion intensity variations due to inconsistent injection or chromatographic separation. Third, there is no need to group peaks in different runs by retention time. Rather, the peaks can be perfectly grouped by matching the scan numbers.
Figure 6.
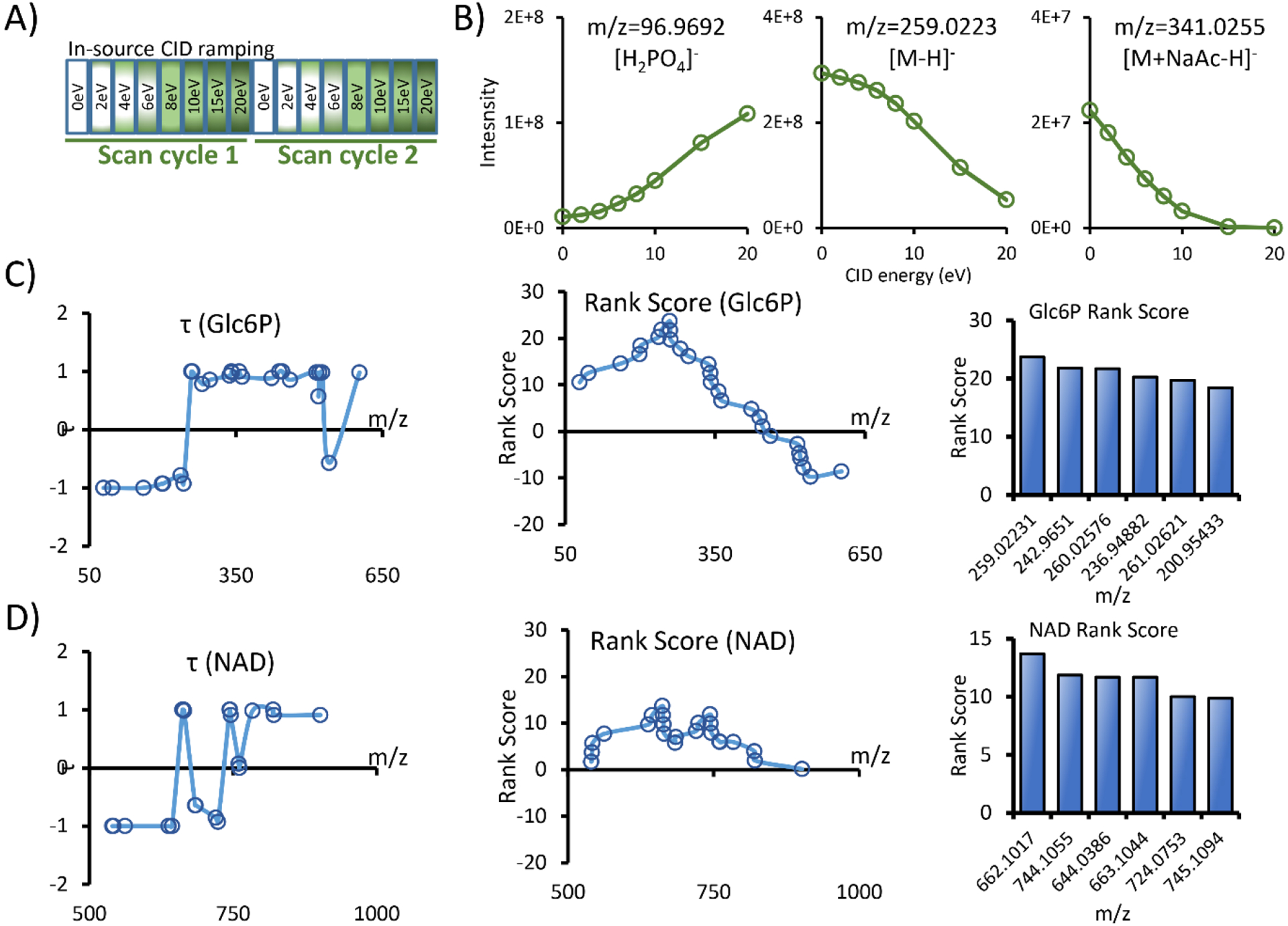
In-source CID ramping (InCIDR). (A) The data acquisition scheme of InCIDR. We used 8 scan events in each cycle to examine the intensity change of the covariant ions. (B) The trend of the intensity change as the CID energy level is ramped. The fragment ion ([H2PO4]−) and the adduct ion ([M+NaAc-H]-) of Glc6P follow the predicted pattern. (C-D) Example InCIDR results for Glc6P and NAD.
We modified COVINA to work with InCIDR data. Similar to COVINA, InCIDR starts from an m/z of interest and finds all covariant ions associated with it. This starting m/z can be any m/z in the group of covariant ions. InCIDR analyzes the intensity of the covariant ions under all in-source CID energy levels and uses the trends to predict which one of them is the protonated or deprotonated ion. To validate the basic assumption of InCIDR, we investigated the covariant ions of Glc6P. As predicted, m/z 96.9692, which is the [H2PO4]− fragment of Glc6P, increased in intensity when the CID energy level increased. In addition, m/z 341.0255, which is the [M+NaAc-H]− adduct of Glc6P, decreased in intensity when the energy level increased. Finally, m/z 259.0223, which is the deprotonated ion of Glc6P, also decreased in intensity when the energy level increased due to increased fragmentation (Figure 6B). To describe the intensity trend in quantitative terms, we calculated the rank correlation coefficient τ for each of the covariant ions.
In the above equation, n is the total number of CID energy levels, which is 8 in this study. Sgn() is the sign function that takes a value of 1, −1 or 0 if applied to a positive number, negative number or 0, respectively. Ii and Ij are the ion counts of the covariant ion at the i-th and j-th lowest CID energy level, 1≤i<j≤n. An adduct that strictly decreases in intensity as the energy level increases is assigned τ=1. A fragment ion that strictly increases in intensity as the energy level increases is assigned τ=−1. For Glc6P, all covariant ions with m/z values smaller than that of [M-H]− had negative values of τ. All ions but 1 with m/z values larger than that of [M-H]− had positive values of τ. Therefore, it is possible to use the τ pattern to find the m/z of [M-H]− among all the covariant ions. We calculated the rank score for all the covariant ions of Glc6P. For a specific m/z, the rank score is the sum of the τ values of ions above this m/z minus the sum of the τ values of ions below this m/z.
We predicted that [M-H]− would have the highest rank score out of all the covariant ions. Indeed, the [M-H]− 259.0223 had the highest rank score. In another example, we investigated the covariant ions of NAD using InCIDR. We observed abnormal adduct ions that increased in intensity when the in-source CID energy level was ramped, such as [M+Na-2H]− and [M-nicotinamide+NaCaAc2-2H]−. The increase in intensity of the [M+Na-2H]− adduct ions may have been due to the decomposition of even larger adduct ions into this sodium adduct. [M-nicotinamide+NaCaAc2-2H]− is an adduct ion of a fragment of NAD, so the higher in-source CID energy level may promote NAD fragmentation and formation of this ion. Nonetheless, m/z 662.1017 had the highest rank score out of all the covariant ions, which is indeed [M-H]− of NAD. The rank correlation coefficients and the rank scores for Glc6P and NAD are shown in Table S2. Using these two examples, we showed that the calculation of rank correlation coefficients and the rank scores ensures the robustness of InCIDR for finding the deprotonated and protonated ions and therefore the neutral mass of metabolites.
Conclusion
In this work, we investigated the ionized metabolite forms in HILIC-ESI-MS metabolomics. Using COVINA and a stable isotope-labeled mobile phase, we detected and annotated a number of adduct ions and fragment ions from metabolites. HILIC metabolomics datasets contain a large number of adduct ions, especially cluster adduct ions. We annotated and curated these adduct ions in order to improves the performance of existing annotation tools such as CliqueMS. We also investigated the intensity changes of adduct and fragment ions using InCIDR. We showed that the metabolite neutral mass can be predicted by scoring the intensity trends of the covariant ions during ramping of the in-source CID energy level.
Supplementary Material
Figure S1. The grouping performance of the adduct annotation tools
Figure S2. Statistics for the adduct annotation
Table S1. The complete annotation of covariant ions from 10 metabolites in the standard mixture
Table S2. The rank correlation coefficients and the rank scores for Glc6P and NAD calculated by InCIDR
File S1. The extended adduct list for CliqueMS (negative ionization mode)
File S2. The extended adduct list for CliqueMS (positive ionization mode)
Acknowledgment
We thank members of the Rabinowitz Laboratory at Princeton University for suggestions and constructive discussion. This research is supported by NIH grants P30 CA072720, and R01CA224550, R01CA129536, and R01CA232246 (to W.X.Z.). Chi Song was partially supported by NIH/NCATS grant UL1TR002733.
References
- (1).Fiehn O The Link between Genotypes and Phenotypes. Plant Mol. Biol 2002, 48(1–2), 155–171. [PubMed] [Google Scholar]
- (2).Wang TJ; Larson MG; Vasan RS; Cheng S; Rhee EP; McCabe E; Lewis GD; Fox CS; Jacques PF; Fernandez C; et al. Metabolite Profiles and the Risk of Developing Diabetes. Nat. Med 2011, 17 (4), 448–453. 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Jang C; Chen L; Rabinowitz JD Metabolomics and Isotope Tracing. Cell 2018, 173 (4), 822–837. 10.1016/j.cell.2018.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Smith CA; Want EJ; O’Maille G; Abagyan R; Siuzdak G XCMS: Processing Mass Spectrometry Data for Metabolite Profiling Using Nonlinear Peak Alignment, Matching, and Identification. Anal. Chem 2006, 78 (3), 779–787. 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- (5).Tautenhahn R; Bottcher C; Neumann S Highly Sensitive Feature Detection for High Resolution LC/MS. BMC Bioinformatics 2008, 9 (1), 504. 10.1186/1471-2105-9-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).DeFelice BC; Mehta SS; Samra S; Čajka T; Wancewicz B; Fahrmann JF; Fiehn O Mass Spectral Feature List Optimizer (MS-FLO): A Tool To Minimize False Positive Peak Reports in Untargeted Liquid Chromatography-Mass Spectroscopy (LC-MS) Data Processing. Anal. Chem 2017, 89 (6), 3250–3255. 10.1021/acs.analchem.6b04372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Kuhl C; Tautenhahn R; Böttcher C; Larson TR; Neumann S CAMERA: An Integrated Strategy for Compound Spectra Extraction and Annotation of Liquid Chromatography/Mass Spectrometry Data Sets. Anal. Chem 2012, 84 (1), 283–289. 10.1021/ac202450g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Uppal K; Walker DI; Jones DP XMSannotator: An R Package for Network-Based Annotation of High-Resolution Metabolomics Data. Anal. Chem 2017, 89 (2), 1063–1067. 10.1021/acs.analchem.6b01214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Senan O; Aguilar-Mogas A; Navarro M; Capellades J; Noon L; Burks D; Yanes O; Guimerà R; Sales-Pardo M CliqueMS: A Computational Tool for Annotating in-Source Metabolite Ions from LC-MS Untargeted Metabolomics Data Based on a Coelution Similarity Network. Bioinformatics 2019, 1–9. 10.1093/bioinformatics/btz207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Mahieu NG; Patti GJ Systems-Level Annotation of a Metabolomics Data Set Reduces 25 000 Features to Fewer than 1000 Unique Metabolites. Anal. Chem 2017, 89 (19), 10397–10406. 10.1021/acs.analchem.7b02380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Broeckling CD; Afsar FA; Neumann S; Ben-Hur A; Prenni JE RAMClust: A Novel Feature Clustering Method Enables Spectral-Matching-Based Annotation for Metabolomics Data. Anal. Chem 2014, 86 (14), 6812–6817. 10.1021/ac501530d. [DOI] [PubMed] [Google Scholar]
- (12).Wang L; Xing X; Chen L; Yang L; Su X; Rabitz H; Lu W; Rabinowitz JD Peak Annotation and Verification Engine for Untargeted LC-MS Metabolomics. Anal. Chem 2019, 91 (3), 1838–1846. 10.1021/acs.analchem.8b03132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Tong H; Bell D; Tabei K; Siegel MM Automated Data Massaging, Interpretation, and e-Mailing Modules for High Throughput Open Access Mass Spectrometry. J. Am. Soc. Mass Spectrom 1999, 10 (11), 1174–1187. 10.1016/S1044-0305(99)00090-2. [DOI] [Google Scholar]
- (14).Keller BO; Sui J; Young AB; Whittal RM Interferences and Contaminants Encountered in Modern Mass Spectrometry. Analytica Chimica Acta. 2008, pp 71–81. 10.1016/j.aca.2008.04.043. [DOI] [PubMed] [Google Scholar]
- (15).Wernisch S; Pennathur S Evaluation of Coverage, Retention Patterns, and Selectivity of Seven Liquid Chromatographic Methods for Metabolomics. Anal. Bioanal. Chem 2016, 408 (22), 6079–6091. 10.1007/s00216-016-9716-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Teleki A; Sánchez-Kopper A; Takors R Alkaline Conditions in Hydrophilic Interaction Liquid Chromatography for Intracellular Metabolite Quantification Using Tandem Mass Spectrometry. Anal. Biochem 2015. 10.1016/j.ab.2015.01.002. [DOI] [PubMed] [Google Scholar]
- (17).Spalding JL; Naser FJ; Mahieu NG; Johnson SL; Patti GJ Trace Phosphate Improves ZIC-PHILIC Peak Shape, Sensitivity, and Coverage for Untargeted Metabolomics. J. Proteome Res 2018, 17 (10), 3537–3546. 10.1021/acs.jproteome.8b00487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Erngren I; Haglöf J; Engskog MKR; Nestor M; Hedeland M; Arvidsson T; Pettersson C Adduct Formation in Electrospray Ionisation-Mass Spectrometry with Hydrophilic Interaction Liquid Chromatography Is Strongly Affected by the Inorganic Ion Concentration of the Samples. J. Chromatogr. A 2019. 10.1016/j.chroma.2019.04.049. [DOI] [PubMed] [Google Scholar]
- (19).Jaeger C; Méret M; Schmitt CA; Lisec J Compound Annotation in Liquid Chromatography/High-Resolution Mass Spectrometry Based Metabolomics: Robust Adduct Ion Determination as a Prerequisite to Structure Prediction in Electrospray Ionization Mass Spectra. Rapid Commun. Mass Spectrom 2017, 31 (15), 1261–1266. 10.1002/rcm.7905. [DOI] [PubMed] [Google Scholar]
- (20).Mahieu NG; Spalding JL; Gelman SJ; Patti GJ Defining and Detecting Complex Peak Relationships in Mass Spectral Data: The Mz.Unity Algorithm. Anal. Chem 2016, 88, 9037–9046. 10.1021/acs.analchem.6b01702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Baran R; Northen TR Robust Automated Mass Spectra Interpretation and Chemical Formula Calculation Using Mixed Integer Linear Programming. Anal. Chem 2013, 85 (20), 9777–9784. 10.1021/ac402180c. [DOI] [PubMed] [Google Scholar]
- (22).Mahieu NG; Spalding JL; Gelman SJ; Patti GJ Defining and Detecting Complex Peak Relationships in Mass Spectral Data: The Mz.Unity Algorithm. Anal. Chem 2016, 88 (18), 9037–9046. 10.1021/acs.analchem.6b01702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Xu YF; Lu W; Rabinowitz JD Avoiding Misannotation of In-Source Fragmentation Products as Cellular Metabolites in Liquid Chromatography-Mass Spectrometry-Based Metabolomics. Anal. Chem 2015, 87 (4), 2273–2281. 10.1021/ac504118y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).McMillan A; Renaud JB; Gloor GB; Reid G; Sumarah MW Post-Acquisition Filtering of Salt Cluster Artefacts for LC-MS Based Human Metabolomic Studies. J. Cheminform 2016, 8 (1), 44. 10.1186/s13321-016-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. The grouping performance of the adduct annotation tools
Figure S2. Statistics for the adduct annotation
Table S1. The complete annotation of covariant ions from 10 metabolites in the standard mixture
Table S2. The rank correlation coefficients and the rank scores for Glc6P and NAD calculated by InCIDR
File S1. The extended adduct list for CliqueMS (negative ionization mode)
File S2. The extended adduct list for CliqueMS (positive ionization mode)


