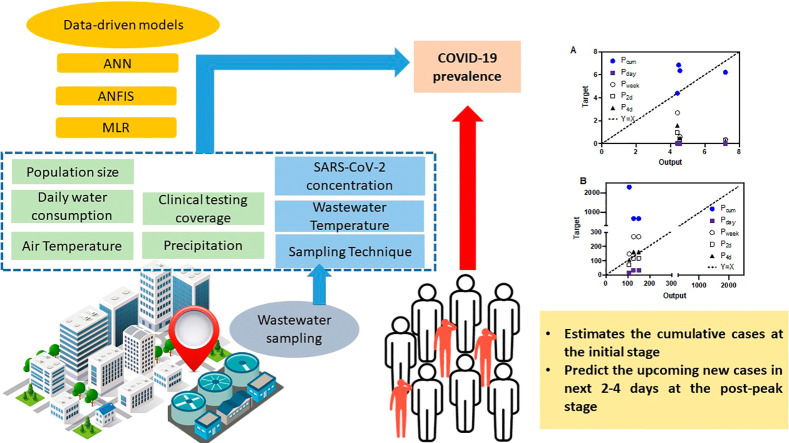
Abstract
Wastewater-based epidemiology (WBE) has been regarded as a potential tool for the prevalence estimation of coronavirus disease 2019 (COVID-19) in the community. However, the application of the conventional back-estimation approach is currently limited due to the methodological challenges and various uncertainties. This study systematically performed meta-analysis for WBE datasets and investigated the use of data-driven models for the COVID-19 community prevalence in lieu of the conventional WBE back-estimation approach. Three different data-driven models, i.e. multiple linear regression (MLR), artificial neural network (ANN), and adaptive neuro fuzzy inference system (ANFIS) were applied to the multi-national WBE dataset. To evaluate the robustness of these models, predictions for sixteen scenarios with partial inputs were compared against the actual prevalence reports from clinical testing. The performance of models was further validated using unseen data (data sets not included for establishing the model) from different stages of the COVID-19 outbreak. Generally, ANN and ANFIS models showed better accuracy and robustness over MLR models. Air and wastewater temperature played a critical role in the prevalence estimation by data-driven models, especially MLR models. With unseen datasets, ANN model reasonably estimated the prevalence of COVID-19 (cumulative cases) at the initial phase and forecasted the upcoming new cases in 2–4 days at the post-peak phase of the COVID-19 outbreak. This study provided essential information about the feasibility and accuracy of data-driven estimation of COVID-19 prevalence through the WBE approach.
Keywords: SARS-CoV-2, COVID-19, Wastewater-based epidemiology, Data-driven models, Artificial neural network
Graphical abstract
1. Introduction
The current global pandemic of coronavirus disease 2019 (COVID-19) has been circulating world-widely for more than a year. As of 15th February 2021, more than 109 million people were infected and more than 2.4 million deaths were reported around 216 countries and territories in the world (WHO, 2020). The surveillance of COVID-19 for its spreading and resurgence are crucial for the governments to reduce adverse effects of such pandemic and implement timely control measures. Currently, the surveillance heavily relies on the clinical testing of individuals, which is highly time-consuming, and might be cost-prohibitive and region-biased especially in resource-poor regions (Hart and Halden, 2020a). Moreover, long incubation periods (i.e., the period between exposure to an infection and appearance of the first symptom) and asymptomatic patients have been commonly observed, resulting in a delayed awareness of community transmission through clinical testing (Backer et al., 2020; Long et al., 2020).
During the pandemic, a significant amount of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) virus shedding has been found in stools and urines of infected patients (both symptomatic and asymptomatic) (Tang et al., 2020; Zhang et al., 2020a; Zhang et al., 2020b). This provides an alternative approach for the population-wide COVID-19 prevalence estimation through wastewater-based epidemiology (WBE) to target the affected communities. To date, although successful detections of SARS-CoV-2 virus RNA in wastewater have been reported globally (Haramoto et al., 2020; Hata et al., 2021; Medema et al., 2020; Randazzo et al., 2020a; Randazzo et al., 2020b; Westhaus et al., 2020), the usage of WBE in COVID-19 prevalence estimation is greatly limited due to the complexity and uncertainties associated with the process. Currently, COVID-19 prevalence estimation using WBE is based on two equations (two different approaches) (Eqs. (1), (2)).
| (1) |
| (2) |
PWBE: prevalence as the number of COVID-19 cases per 100,000 people; F: daily wastewater flow (Mega-liter/day); Pc: population of the community (×100,000 people); C RNA: concentration of SARS-CoV-2 RNA in wastewater (gene copies/L); R s: scaling ratio due to in-sewer decay of SARS-CoV-2 (−); E: daily excretion rate of SARS-CoV-2 by a COVID-19 patient (gene copies/day∙person); Q w: the average daily water usage (L/day∙person).
For viral shedding, variations in the magnitude, probability, and duration were commonly observed across different studies. For instance, the magnitude of viral shedding varied between 102 and 108 RNA copies per gram of feces and the shedding may last for up to 7 weeks even after the patient has been recovered from the illness (Chen et al., 2020; Joynt and Wu, 2020; Pan et al., 2020; Wölfel et al., 2020; Wu et al., 2020b). Through a qualitative analysis of 2149 SARS-CoV-2 patients, a positive proportion of the fecal samples has been found to be 51.8% (95% CI 43.8–59.7%) (van Doorn et al., 2020). After excretion, SARS-CoV-2 RNA is transported in sewer systems till the sampling location. Although the fate of SARS-CoV-2 RNA in real sewers remains unclear, its decay in bulk wastewater was facilitated by longer hydraulic retention time (HRT) and higher wastewater temperature (Ahmed et al., 2020b; Weidhaas et al., 2020). The variability of HRT, operational conditions and wastewater properties led to great uncertainties of R s. Furthermore, one recent study observed a ~10-fold increase in composite samples compared to corresponding grab samples of primary effluent, highlighting variability in the SARS-CoV-2 concentration due to the sampling technique (Gerrity et al., 2021). These uncertainties make the back-estimation through conventional WBE difficult and potentially lead to discrepancies between WBE estimated COVID-19 cases and clinically confirmed cases (Wu et al., 2020a).
In recent decades, the use of data-driven models to solve complicated problems is gaining popularity. Data-driven models have been applied successfully in various fields including wastewater and sewer processes (Dalmau et al., 2015; Jiang et al., 2016; Kenny et al., 2009; Khademi and Behfarnia, 2016; Li et al., 2019). These models ‘learn’ the patterns of the underlying processes from past data and generalize new ‘knowledge’ (or mathematical relationships between input and output data) to predict an output when being given a new set of input variables from the problem domain (Jiang et al., 2016). Hence, data-driven models could be a potential tool for the prevalence prediction with reasonable accuracy before the uncertainties associated with WBE can be thoroughly delineated. To date, data-driven models have not been applied in the prevalence prediction of COVID-19 through the WBE approach.
This study investigated the use of data-driven models to estimate the prevalence of COVID-19 cases in the community using multi-national WBE data collected through a systematic literature search. Three types of data-driven models, i.e., multiple linear regression (MLR), artificial neural network (ANN), adaptive neuro fuzzy inference system (ANFIS), were applied and their performance was evaluated for predicting the prevalence of COVID-19 cases in the area covered by a wastewater treatment plant (WWTP). The prevalence was chosen as the only output, with the average clinically testing ratio, SARS-CoV-2 RNA concentration detected in wastewater (corrected by recovery efficiency or not), wastewater temperature, air temperature, inhabitant population, average daily water consumption, sampling technique, and precipitation as inputs to the data-driven models. Furthermore, in real applications, it is normally difficult to obtain a complete input dataset, thus the robustness and accuracy of predictions using partial input parameters for each model were assessed in sixteen partial input scenarios. More importantly, the performance of data-driven models was evaluated for different stages of the outbreak using unseen datasets (data not used in model training). Our preliminary findings demonstrate the applicability and accuracy of data-driven models as a potential tool for COVID-19 prevalence surveillance and early-warning at the community level.
2. Materials and methods
2.1. Collection and meta- analysis of wastewater surveillance data
The electronic search for available literature was conducted on 25 January 2021, following PRISMA guidelines (Silverman and Boehm, 2020) to collect a comprehensive and large set of data. Databases (i.e., Web of Science core collection, Scopus, and PubMed) were searched using the term “SARS-CoV-2 AND wastewater”. A total of 487 unique papers were identified after removing duplicates using the EndNote Reference Manager software. Titles and abstracts of the retained articles were screened and assessed for eligibility following these criteria: 1) reported the detection of SARS-CoV-2 RNA in raw wastewater; 2) reported clear data regarding the population size, sampling approach, sampling date, and clinically confirmed cases in the catchment area; 3) the article is in English and is peer-reviewed. Relevant articles were further assessed by full-text reading and finally, 7 articles were included in this study for establishing the data-driven models. Details of the review process are provided in the supplementary information (SI). The methodology applied in these 7 articles, including sampling, storage and analytical methods (Table S1), was peer-reviewed and commonly applied in WBE studies for COVID-19, which can be good representatives of other WBE studies. Other WBE reports were not included in this study due to the lack of required information or inability to provide such information after communicating with their authors.
Wastewater surveillance data including SARS-CoV-2 RNA concentration in wastewater, and inhabitant population and active cases (confirmed cases minus recovered cases) in the catchment area, were summarized from seven recent papers (Ahmed et al., 2020a; Hata et al., 2021; Kitamura et al., 2020; Medema et al., 2020; Randazzo et al., 2020a; Randazzo et al., 2020b; Westhaus et al., 2020). For the studies with data at the pre-peak stage of the outbreak, cumulative cases in the catchment area of WWTP were used as active cases. Since the SARS-CoV-2 RNA concentrations were generally in 102–7 copies/L, logarithmic concentration data was included as C RNA (log10 copies/L). Active cases were converted to prevalence (P, confirmed COVID-19 cases per 100,000 people). Variations in the SARS-CoV-2 RNA concentration were observed with a wastewater sample using different concentration, extraction, and detection approaches (Wu et al., 2020a). A recent interlaboratory assessment evaluated the efficiency of 36 standard operating procedures (SOPs) including eight different concentration methods using the same wastewater sample and 80% of the recovery-corrected results based on external or internal standards fell within a band of ±1.15log10 copies/L with high reproducibility (Pecson et al., 2021). Thus, an indicator variable (F r) was included as a categorical factor to different the recovery-corrected results and non-corrected results as 1 and 0, respectively. Variability in the SARS-CoV-2 concentration due to composite and grab sampling was observed (Gerrity et al., 2021), though the impact of time-proportional or flow-proportional composite sampling and grab sampling remains unclear. Thus, the sampling technique (S T) was applied as an indicator variable to categorize grab sampling and composite sampling as 0 and 1, respectively.
During in-sewer transportation, potential decay of SARS-CoV-2 RNA would also affect the C RNA detected in wastewater samples. Recent studies found the wastewater temperature (T w) and HRT play a significant role in the RNA decay using bulk wastewater (Bivins et al., 2020). However, T w and average HRT of the catchment area were not reported in those 7 selected articles. Hart and Halden (2020b) showed that T w can be estimated based on the air temperature globally and reached a good agreement with empirical observations. Thus the estimated T w from Hart and Halden (2020b) and average air temperature (T a) of the sampling day for specific locations from Google weather data were included. HRT was found strongly correlated to the catchment size of a WWTP, ranging from several minutes to 6–10 h in small and large scale WWTPs, respectively (McCall et al., 2017). Furthermore, the clinically confirmed cases are generally classified based on Primary Health Networks (PHNs) or municipal areas, which may not be identical to the coverage of the catchment area. The population coverage of the catchment to the PHNs or municipal area was not reported in these seven articles. A larger WWTP would generally cover a higher percentage of the population in the relevant PHNs or municipal area (Ahmed et al., 2020a). In addition, it remains unclear how the distribution of patients in a catchment area impacts the prevalence estimation through WBE approach. It was reported the distribution of drug consumers became more important for the estimation of drug usage in medium and large catchments (McCall et al., 2017). Thus, the population size of a catchment area (P c, ×100,000 person) was included as a variable accounting for the catchment size. Data-driven models can also be applied to simulate the decay process of SARS-CoV-2 RNA in sewers, which can be further applied to predict the prevalence of COVID-19 in the catchment area. For such a decay model, the actual concentrations of SARS-CoV-2 RNA in wastewater before in-sewer transportation (C 0) would be required for training the model. Although some recent studies investigated the decay of SARS-CoV-2 RNA under laboratory conditions with bulk wastewater, the hydraulic pattern, operation conditions (i.e. aerobic or anaerobic), and effects of sewer biofilms etc., were not considered (Ahmed et al., 2020b; Bivins et al., 2020). Needs remain to obtain data to support a reliable model of in-sewer decay of SARS-CoV-2.
Significant dilution of SARS-CoV-2 concentration in combined sewers was observed due to the precipitation inflow (Chavarria-Miró et al., 2020). Heavy rainfall could also dilute the sewage flow in separate sewers (Jiang et al., 2013). Precipitation (P p, mm) and average daily water usage (Q w, L/person∙day) were collected from Google weather and governmental report of each country, respectively. More importantly, the clinical testing of individuals largely depends on their own motivation, contact tracing policy, and cost of each country. The testing policy, coverage, tests per confirmed case, contact tracing policy, and test cost of different countries were summarized and discussed in the supplementary information (text S2, Table S2). Overall, although the testing practice varies in each country, the testing coverage were considered adequate according to WHO guidelines (i.e., positive rates <10%) and could largely support tracking the COVID-19 prevalence in the community. In terms of the motivation of individuals for COVID-19 clinical testing, symptom-onset can be a major trigger. However, SARS-CoV-2 virus shedding has been found in stools and urines of asymptomatic patients and some patients had symptom onset several weeks after the infection (Park et al., 2020; Tang et al., 2020; Zhang et al., 2020a; Zhang et al., 2020b). A recent meta-analysis estimated that the percentage of asymptomatic infection was 15.6% (95% CI, 10.1%–23.0%), but the actual percentage varied in studies from different regions and age groups (He et al., 2020). Generally, a higher testing ratio (people being tested/total population) in an area would increase the probability of identifying asymptomatic patients among the population. The testing ratio and testing practice in the catchment area was not reported in all these seven articles, but it is publicly available for each country, which can largely reflect that of the catchment area. Thus, considering the contribution of asymptomatic patients in virus shedding, the potential impact of testing policy, and the potential detection windows (i.e. 28 days) of SARS-CoV-2 for wastewater samples (Ahmed et al., 2020a), the average testing ratio/1000 people every 30 days of each country (R T) (https://ourworldindata.org/coronavirus-testing) was included as a variable for the testing coverage. The positive ratio (confirmed cases/total testing) was not included as the clinical diagnoses can take several hours to a couple of days depending on the capacity of the testing center while the number of total testing is updated in real-time or within several hours. As all WBE analyses for wastewater were based on the same method, i.e., reverse transcription-quantitative polymerase chain reaction (RT-qPCR), the sensitivity of detection was not included as an input for the data-driven model in this study.
Since all these 7 publications conducted wastewater sampling and analysis on different days, the C RNA and P WBE measured at different sampling days with raw wastewater were collected from each publication, along with the explanatory factors (S T, F r, T w, T a, P c, P p, Q w, R T) obtained as described in paragraphs above. Finally, a total of 163 data sets were summarized from all these 7 articles. Among these 163 data points, 159 of them were applied for the distance-based redundancy analysis (db-RDA) analysis and establishing the data-driven models in the following sections. The rest data (collected at 4 different days) from (Haramoto et al., 2020) were applied to assess model performance in Section 2.3.3. These data points were chosen as a clear record of cumulative cases, daily new cases, weekly new cases, and new cases in the following 2 or 4 days was reported at the relevant sampling days (to be discussed in Section 2.3.3). The relevance of explanatory factors in explaining the distribution patterns of prevalence data from different studies was analyzed by distance-based redundancy analysis (db-RDA) using R (ver. 3.31, http://www.R-project.org/). Pearson's correlation between prevalence data and explanatory factors was calculated using R.
2.2. Data-driven models for estimating community prevalence of SARS-CoV-2
2.2.1. Multiple linear regression
The MLR is a statistical method that generates the cause-effect correlation in terms of a linear relationship between a dependent (target) and some independent variables (inputs). In MLR, the regression function between multiple input variable (X1, X2, …, Xn) and the dependent variable (Y) is defined as Eq. (3):
| (3) |
where Ŷ is the model's output, X j is the independent input variables to the model, and a 0, a 1, a 2, …, a n are the partial regression coefficients.
Considering the potential interaction between any two input parameters, the importance of interactions was determined using the F-test. Then, MLR analysis was performed on the community prevalence of SARS-CoV-2 (confirmed active COVID-19 cases per 100,000 people) with explanatory factors including R T, T w, T a, P c, Q w, S T, P p, C RNA, and F r in wastewater using R (ver. 3.6.2, http://www.R-project.org/) with the whole data sets. The coefficients for each of the input factors were determined along with the standard error.
2.2.2. Artificial neural network
ANN is a mathematical modeling approach that simulates the structure and/or functional aspects of biological neural networks to process information and produce approximate outcomes (Li et al., 2019). A typical ANN contains three layers, i.e., input, hidden and output layers, which are made of interconnected groups of artificial neurons. The general procedure of building an ANN model uses the steps of training, validation, and test as described previously (Li et al., 2019). Briefly, the training steps generate the weight of connections between neurons and an error function, i.e., the mean square error (MSE) to make the output values similar to the target values. Then validation step is applied independently to find the optimal number of hidden units or determine a stopping point for ANN (Şahin et al., 2013). Finally, the performance and accuracy of the machine learning algorithm are evaluated by the test steps. The detailed structure and function of ANN were described in the supplementary information.
In this study, ANN is used to predict the COVID-19 prevalence based on C RNA together with other variables including R T, F r, T w, T a, P c, Q w, S T, and P p as mentioned in Section 2.2.1. To determine the optimal structure of ANN, the performance of ANN with various numbers of neurons in the hidden layer was exhaustively searched using Alyuda Neuro Intelligence ver. 2.2. The final structure with the best performance was used to predict the prevalence of COVID-19 using MATLAB (R2019b). A total of 159 data points as summarized above were used for the modeling process. The proportion of observations was set as 70%, 15% and 15% in this study for the training, validation, and test steps for ANN analysis, respectively. The data included in each step was based on random choice. Based on the datasets, the sigmoidal tangent function and a linear activation function were applied for hidden nodes and the output layer, respectively. Further, the Levenberg-Marquardt (LM) algorithm was selected as the most suitable algorithms in this study.
2.2.3. Adaptive neuro fuzzy inference system
ANFIS, like ANN, is a hybrid artificial intelligence technique, which is widely accepted for solving complex problems with adequate estimations. In ANN, one of the most significant disadvantages is that the weight values between interconnected neurons are generated from the data and could not be explained. In ANFIS, the FIS function performs like a white box, where the nodes and the hidden layers of the neuron network are determined precisely by a FIS and allows the model designers to discover how the model achieved its goal (Karaboga and Kaya, 2019). The ANFIS structure is made of two parts - premise and consequence parts, which are connected by a network with fuzzy rules. Through the training process, parameters in each part are determined with an optimization algorithm, and eventually connected by IF-THEN fuzzy rules (Karaboga and Kaya, 2019). The fuzzy reasoning mechanism of ANFIS model was described in the supplementary information.
Similarly, ANFIS was applied to predict the COVID-19 prevalence using R T, F r, T w, T a, P c, Q w, S T, and P p with a total of 159 data points. The proportion of observations in this study was set as 70%, 15% and 15% for the training, validation, and test steps, respectively. The data included in each step was chosen randomly from the whole dataset. The final structure with the best performance of ANFIS was built and used to estimate the prevalence of COVID-19 using MATLAB (R2019b).
2.3. Evaluation of data-driven models for predicting community prevalence of SARS-CoV-2
2.3.1. Comparison of the performance in prevalence estimation
Three different data-driven models (i.e., MLR, ANN, and ANFIS) as stated above were constructed using the same dataset. To compare their performance in predicting SARS-CoV-2 community prevalence, coefficients of determination (R 2) were employed for all three types of models.
2.3.2. Assessment of the model robustness with partial input data
In real applications, the availability of a complete input dataset as described in Section 2.2.1, might be limited. Thus, it is essential to evaluate the robustness of these data-driven models with incomplete datasets as the input. The robustness of each model with incomplete data input can serve as an important criterion to guide the application of these data-driven models.
Considering the practical difficulty, sixteen different combination scenarios were chosen (i.e., 9 input factors, 8 input factors, 7 input factors, 6 input factors, 5 input factors, 4 input factors, 3 input factors, 2 input factors, and 1 input factor) (Table 2). These scenarios were selected based on the accessibility of input factors in real applications. Models were built under each input scenario and the accuracy of prediction with MLR, ANN, and ANFIS models was evaluated using R 2 values.
Table 2.
Coefficient of determination (R2) determined for robustness analysis using data-driven models in predicting SARS-CoV-2 prevalence with partial input parameters.
| Scenario | Coefficient of determination (R2) |
||
|---|---|---|---|
| MLR | ANN | ANFIS | |
| 1. RT, Tw, Ta, Pc, CRNA, Fr, Pp, Qw, ST | 0.58 | 0.88 | 0.79 |
| 2. RT, Tw, Ta, Pc, CRNA, Fr, Qw, ST | 0.57 | 0.87 | 0.70 |
| 3. RT, Tw, Ta, CRNA, Fr, Qw, ST | 0.56 | 0.84 | 0.45 |
| 4. RT, Tw, Ta, CRNA, Qw, ST | 0.55 | 0.85 | 0.44 |
| 5. RT, Ta, CRNA, Qw, ST | 0.43 | 0.76 | 0.60 |
| 6. RT, Tw, CRNA, Qw, ST | 0.32 | 0.83 | 0.82 |
| 7. RT, Tw, Ta, CRNA, Qw | 0.51 | 0.75 | 0.84 |
| 8. RT, Tw, Ta, CRNA, ST | 0.48 | 0.84 | 0.81 |
| 9. Tw, Ta, CRNA | 0.44 | 0.83 | 0.81 |
| 10. Ta, CRNA, Qw | 0.41 | 0.74 | 0.54 |
| 11. Tw, CRNA, Qw | 0.30 | 0.82 | 0.79 |
| 12. RT, Ta, CRNA | 0.35 | 0.70 | 0.58 |
| 13. RT, Tw, CRNA | 0.30 | 0.73 | 0.72 |
| 14. Ta, CRNA, ST | 0.32 | 0.77 | 0.48 |
| 15. Tw, CRNA | 0.30 | 0.72 | 0.54 |
| 16. CRNA | 0 | 0.33 | 0.21 |
Note: RT, average testing ratio/1000 people every 30 days, Tw, wastewater temperature (°C); Ta, air temperature (°C); Pc, community population (×100,000 person); CRNA, the virus RNA concentration (log10 copies/L) in wastewater; Fr, a categorical factor to different the recovery-corrected results and non-corrected results for CRNA; Qw, average daily water consumption (L/person∙day); ST, sampling technique (grab or composite); and Pp, precipitation (mm).
2.3.3. Assessment of the model performance at different phases of COVID-19 outbreak
As aforementioned, all these studies were carried out at the initial pre-peak stage with a rapid increase of confirmed cases, where cumulative cases can largely represent the active cases. However, with the implication of effective control/protective measures, the daily new cases gradually decrease to lower numbers after reaching the peak and the cumulative cases become relatively stable with slow growth (Maier and Brockmann, 2020). Thus, the application of data-driven models for the prevalence estimation would potentially differ at different phases of the outbreak. Recently, significant correlations were observed between C RNA in wastewater and daily (p < 0.001) or weekly (p < 0.05) new cases, for COVID-19 prevalence (Weidhaas et al., 2020). Furthermore, through 74-day monitoring, the concentration of SARS-CoV-2 in wastewater was found to foreshadow the upcoming cases by 2–4 days (Nemudryi et al., 2020). The performance of the data-driven models built in this study for the prevalence estimation, was further validated using unseen data (i.e., datasets not used for establishing the model) from the COVID-19 outbreak at the initial stage (i.e., within 1–2 detection window of SARS-CoV-2 for wastewater samples and before the number of daily new cases reached its peak) and post-peak stage (i.e. after the number of daily new cases reached its peak). Data (from four different sampling days, not used for establishing the models) from Japan at the initial stage of the outbreak (Haramoto et al., 2020), and a study (not included in the seven studies used for establishing the model) from USA (Sherchan et al., 2020) at the post-peak phase of the outbreak, were applied to assess the performance of data-driven models established in this study. Due to the data availability, the sampling technique (S T) and the SARS-CoV-2 concentration (C RNA) were summarized from the report. The prevalence estimated by data-driven models with S T, C RNA, and T a as inputs was compared against the prevalence determined by cumulative cases (P cum), daily new cases (P day), weekly new cases (P week), and new cases in the following 2 or 4 days (P 2d, P 4d).
3. Results and discussion
3.1. Meta-analysis for the correlation between the prevalence data and explanatory factors
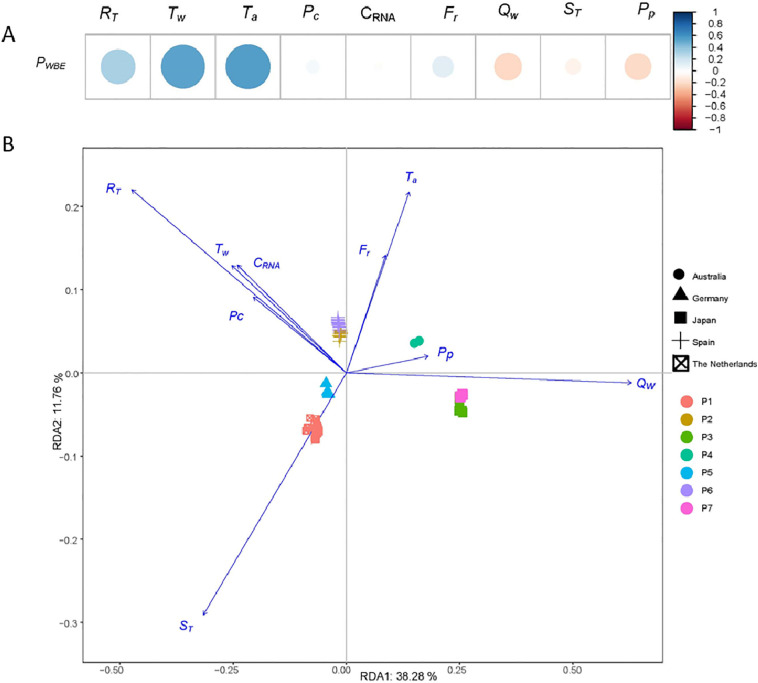
Multinational data were collected from 7 publications from 5 countries (1 each for Australia Germany, and the Netherlands, 2 each for Japan and Spain). Through Pearson's correlation analysis, R T, T w, and T a showed stronger and positive correlations with prevalence data than other factors. A higher R T was expected to be related to a higher COVID-19 prevalence. All these 7 studies were carried from March to May 2020, where the air and wastewater temperature gradually increased along with the COVID-19 development, resulting in the positive correlation between the prevalence and T w or T a. Conventionally, C RNA was considered as the most important factor for WBE studies to evaluate the COVID-19 development at the community level, where a linear relationship or strong correlation between C RNA and the prevalence data has been observed (Medema et al., 2020). However, for multi-national level and national level analysis, the correlation between C RNA and the prevalence was limited (Fig. 1A , Fig. S1). Even with the data of each publication, a strong positive correlation between P WBE and C RNA was only observed in two studies among these seven studies (Fig. S1). The strong positive correlation is likely caused by the limited data size in these two studies, (i.e. 2 data points and 18 data points, respectively). This implies that C RNA cannot be used as the sole indicator for the prevalence estimation or the comparison among different countries. This could be caused by 1) the analytical uncertainty of C RNA; 2) potential in-sewer decay of SARS-CoV-2 RNA; 3) sampling technique. Analytical uncertainty was found as one of the major uncertainties for COVID-19 prevalence estimation through WBE approach (Li et al., 2021). Variations in the recovery efficiency of the RNA concentration, extraction, and detection approaches were commonly observed, leading to the variability of the C RNA even with the same wastewater sample (Ahmed et al., 2020c). Although a recent interlaboratory assessment found that a variety of methods could produce reproducible results with the inclusion of surrogate viruses to quantify the recovery efficiency (Pecson et al., 2021), only 2 out of the 7 publications included in this study quantified the recovery efficiency and reported the recovery-corrected concentration data. Furthermore, wastewater is a complex matrix, the presence and concentrations of suspended solids concentration, dissolved oxygen, organic matters, humic-like substances likely vary due to the socioeconomic condition (i.e. age, gender, income etc.) of a country or area, which could also greatly impact the viral adsorption and decay in wastewater, resulting in the uncertainty of C RNA (Petala et al., 2020). Furthermore, the in-sewer decay of SARS-CoV-2 RNA in wastewater was greatly facilitated by higher wastewater temperature and longer HRT, leading to a lower C RNA detected in wastewater (Ahmed et al., 2020b). One recent study observed a ~ 10-fold increase in composite sampling in comparison to corresponding grab sampling of primary effluent samples (Gerrity et al., 2021), which could be another reason for the unexpected weak correlation between C RNA and P WBE. Although C RNA is indispensable for COVID-19 prevalence estimation through WBE approach, other parameters such as wastewater temperature, air temperature, catchment size etc., are also critical for the accuracy of prediction. For future COVID-19 prevalence studies through WBE approach, it is highly recommended to include these parameters in the report. In addition, negative correlations were observed with Q w and P p for the prevalence data. This revealed the inherent pattern of the collected datasets, where a catchment area with higher Q w and P p were related to a lower prevalence.
Fig. 1.
Pairwise Pearson's correlation plot between prevalence data (PWBE) and the nine explanatory factors. The color and size of the circles indicate the strength of Pearson's correlation coefficient (bigger circle = stronger link; blue = positive correlation and red = negative correlation) (A). The db-RDA diagram showing the relationship between the prevalence data and explanatory factors. Prevalence data from 7 publications were identified with different colors (P1-P7), and the countries of the 7 publications were differentiated with shapes. The % value in the RDA axes indicates the % of the total variation explained by each RDA axes (B). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
A clear national-wide difference was observed with the prevalence data through db-RDA analysis. However, the difference among data of the same country was negligible (Fig. 1). This result suggests a between-country difference in the COVID-19 development during the study period. Q W and P p had stronger correlations with the prevalence data in Australia and Japan than in other countries. T a and F r were more closely related to the prevalence data in Spain and Australia. R T, T w, C RNA and P c, showed stronger correlations with the prevalence data in Spain and Japan than in other countries. These regional-based variations suggest that a universal prevalence estimation is prone to the impact of differences from country to country.
The combination of nine explanatory factors explained 50.04% (RDA1 + RDA2) of the variations of prevalence across these 7 publications. As discussed above, various factors such as the recovery efficiency and wastewater matrix could potentially impact C RNA detection. Due to the unavailability of such kind of information for all datasets, these factors were not included in this study and may require further investigations.
3.2. Estimation of SARS-CoV-2 community prevalence using the MLR model
The interactions between input variables were found to be insignificant in the second order using F-test (Table S3), which suggests that all the input variables are independent. Thus, the MLR model is built without interaction factors. Through the MLR analysis, an equation was generated using the whole dataset to estimate the COVID-19 community prevalence (P WBE) (Eq. (4)) and the uncertainty and significance of the regression coefficients were determined as shown in Table 1 .
| (4) |
Table 1.
MLR model coefficients using the complete WBE dataset for the prediction of COVID-19 community prevalence.
| Coefficient | Estimate | Std. error | t value | P(>|t|)a | Significanceb |
|---|---|---|---|---|---|
| Intercept | 579.39 | 151.83 | 3.82 | 1.96 × 10−4 | *** |
| RT | −20.60 | 3.85 | −5.34 | 3.24 × 10−7 | *** |
| Tw | 14.36 | 2.05 | 6.99 | 8.07 × 10−11 | *** |
| Ta | 8.07 | 1.03 | 7.81 | 8.68 × 10−13 | *** |
| Pc | −1.33 | 0.58 | −2.29 | 0.02 | * |
| CRNA | −4.97 | 5.39 | −0.92 | 0.36 | |
| Fr | 10.26 | 9.79 | 1.05 | 0.30 | |
| Qw | −2.38 | 0.44 | −5.35 | 3.09 × 10−7 | *** |
| ST | −149.18 | 33.87 | −4.41 | 1.98 × 10−5 | *** |
| Pp | 0.68 | 6.44 | 0.11 | 0.92 |
P(>|t|) is the probability value using the t-test.
Significance codes represent P values of 0–0.001: ***; 0.001–0.01: **; 0.01–0.05: *.
Among all the input variables, Tw and Ta were found as the most significant factors with p values of 8.07 × 10−11 and 8.68 × 10−13, respectively (Table 1). Both T w and T a had positive coefficients, suggesting that with the same C RNA along with other factors, the actual prevalence could be higher in sewers with higher temperatures. This is consistent with the significant role of wastewater temperature in the SARS-CoV-2 RNA decay, where a higher degradation rate of SARS-CoV-2 RNA was observed in wastewater with higher wastewater temperatures (Ahmed et al., 2020b; Weidhaas et al., 2020). Since the T w was estimated using T a along with other parameters associated with soil conditions, the importance of T a in estimated prevalence is likely associated with the role of T w in the RNA decay process. The significance of T w and T a highlights the importance of in-sewer decay of SARS-CoV-2 RNA for the prevalence estimations.
RT and Q w were also found as significant parameters for the prevalence estimation with p values at around 10−7 (Table 1). As the prevalence data applied in the model was calculated based on the active cases confirmed by clinical testing, the negative coefficient of R T (−20.60) suggests a smaller difference between WBE-estimated prevalence (P WBE) and clinically confirmed prevalence with a higher testing coverage. This is likely related to the different testing policies during the study period and presence of asymptotic patients and different incubation periods for patients. For instance, among these countries, Japan had the lowest testing coverage; and clinical testing was only conducted for people who had symptoms and belong to specific groups such as key workers, hospital patients, or travelers from overseas (Table S2). In contrast, Australia had the highest R T as an open public testing policy was applied, with testing available to both symptomatic and asymptotic people. Thus, a higher testing coverage could potentially reveal more asymptotic patients as virus shedding was also observed in asymptotic patients (Tang et al., 2020; Zhang et al., 2020a; Zhang et al., 2020b). Q w had a negative coefficient value in the MLR model although a positive relationship was expected according to Eq. (2). This is likely related to the inherent pattern of the data sets collected where a higher prevalence was related to the region with lower Q w as discussed in Section 3.1.
ST was found as one of the major significant factors (p = 1.98 × 10−5) with a negative coefficient (−149.18). The negative coefficient of S T suggests that with the same C RNA along with other factors, the estimated prevalence of grab samples tends to be higher than that of composite samples. This might be related to the diurnal toilet use pattern. About 10-fold increase of SARS-CoV-2 RNA concentration in composite samples was observed in comparison to the grab samples in a recent study (Gerrity et al., 2021). However, to date, the exploration of potential impacts of sampling techniques on the SARS-CoV-2 RNA concentration in wastewater, and COVID-19 prevalence estimation through WBE approach is limited, which still needs further investigations. In addition, P c is also a significant input factor for the prevalence estimation (p = 0.02) (Table 1). Conventionally, a larger catchment area with higher P c is related to a higher HRT in the catchment area (McCall et al., 2017). The role of P c in the prevalence estimation could be related to the in-sewer decay of SARS-CoV-2 RNA, although the impact of sewer HRT remains unclear to date.
Conventionally, C RNA was regarded as the predominant parameter for COVID-19 prevalence estimation where a strong positive correlation or linear relationship was expected. However, in MLR models, the statistical significance of C RNA was limited (p = 0.36), and C RNA had a negative coefficient for the prevalence estimation. This is consistent with the correlation analysis in Section 3.1, where the correlation between C RNA and the prevalence was limited for multi-national level and national level analysis. Furthermore, C RNA also did not play a significant role in P WBE estimation in most of the articles (except one Germany study) and separate national data sets (Table S4). As discussed in Section 1, this is likely related to analytical uncertainty of C RNA, in-sewer decay of SARS-CoV-2 RNA and sampling approach. This suggests that although C RNA is critical for confirming the existence of COVID-19 patients in catchment areas, the estimation of the prevalence cannot predominantly rely on C RNA. Other factors such as T w, T a and S T, are also important for P WBE estimation.
In this study, P p was found as an insignificant input for prevalence estimation (Table 1). It is worthwhile to mention that in this study, as most of the sampling was carried out in dry weather conditions, the P p was within 0–2 mm in the dataset. Recently, significant dilution of SARS-CoV-2 concentration in combined sewers has been observed due to the storm water or precipitation inflow (Chavarria-Miró et al., 2020). Thus, the potential impact of a higher range precipitation requires future investigations. The R2 value achieved by the MLR model was 0.58, suggesting that 58% of the variability in the observed prevalence could be captured and explained by this linear model. The limited performance of MLR could be caused by the varied impact of each factor on the data from different countries or the lack of other parameters such as wastewater matrix as discussed in Section 3.1. Furthermore, the relationship between the prevalence and the input parameters here is also potentially nonlinear.
3.3. Estimation of SARS-CoV-2 prevalence using ANN and ANFIS models
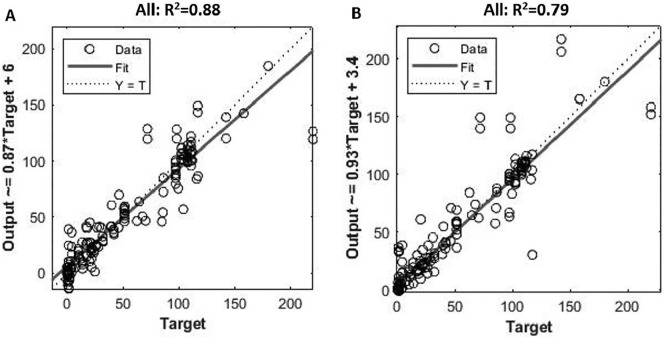
Following the regression analysis, ANN and ANFIS models were established for estimating COVID-19 prevalence using the same dataset. The final structure of the ANN model was established with 9 input, 9 hidden, and 1 output neurons after optimization. To select the best performance model, the validation and test steps were performed to prevent overfitting by measuring the error with independent datasets. During the training process, ANN achieved a satisfactory performance with an R2 value of 0.90. Consistent performances were observed in validation and test datasets with the R2 value of 0.90 and 0.80, respectively (Fig. S2). This indicates that the model established a clear relationship between the input factors and the prevalence reported. Overall, the application of ANN models greatly improved the prediction accuracy than MLR models with the overall R2 value at 0.88, although few scattered data points as likely outliers were observed (Fig. 2A ).
Fig. 2.
The outputs of the ANN model (A) and ANFIS model (B), and their correlations with the actual prevalence reported from clinical tests using all of the datasets. Target is the prevalence of active COVID-19 cases reported from the clinical testing. The output is the value obtained from the model predicting the SARS-CoV-2 prevalence using input parameters. The Y = T line is where the y-axis value equals the target value.
Following the ANN, ANFIS was built for the SARS-CoV-2 prevalence prediction. The final structure of ANFIS model has 9 inputs and 11 rules (Fig. S3). Containing ‘IF’ and ‘THEN’ parts of the fuzzy inference system, these rules are sensitive to input parameters and determine the way the input parameters change in order to minimize the measurement errors (Ausati and Amanollahi, 2016). ANFIS model achieved satisfactory performance in predicting the SARS-CoV-2 prevalence. The R2 values obtained for the whole dataset was 0.79, which was slightly lower than ANN models (Fig. 2B). Consistent performances were observed for training, validation, and test datasets, with R2 values at 0.94, 0.76 and 0.87, respectively (Fig. S4). It implies that both ANN and ANFIS models were capable of estimating the relationship adequately between COVID-19 prevalence and the chosen input factors. In comparison to ANFIS, ANN showed a slightly better performance especially for validation and test datasets. It could be due to the relatively small scale of the dataset compiled in this study and the FIS rules may become more useful when the datasets grow with more available WBE studies in the future. Since the outputs of ANN and ANFIS models were generated based on the interconnected neurons, the impact of each parameter on the prevalence estimation was not differentiated. Instead, R 2 was employed to compare the performance of these models. Overall, both ANN and ANFIS showed a highly satisfactory performance with only a few scattered data points which are likely some outliers in the compiled dataset (Fig. 2). These outliers could be due to three reasons: 1) inaccurate value of the determined parameters; 2) natural variance of the parameters; 3) impact from other variables that were not included in the current model. The inaccurate value is potentially caused by either the inaccuracy of the C RNA detected in wastewater (analytical uncertainties) or there were missed cases by clinical testing. The natural variance of parameters is likely associated with the virus shedding uncertainty, sampling uncertainty etc. The impact of other potential affecting variables is mainly related to the decay or retention of SARS-CoV-2 RNA in sewers, such as wastewater temperature variations, HRT, wastewater matrix as discussed in Section 3.1.
3.4. Analysis of model robustness subject to partial input data
In real-life applications, a complete dataset with all the input parameters is generally limited. In particular the current practice of WBE is mainly focused on detecting the concentration of SARS-CoV-2 RNA in wastewater without considering other relevant parameters. Robustness analysis is thereby applied to explore critical factors and the most suitable input scenario for the prevalence prediction (Table 2 ).
MLR models were fairly sensitive to partial inputs, especially T a and T w. Without T w or T a, R2 value of MLR models dropped from 0.55 (scenario 4) to 0.43 (scenario 5), and 0.32 (scenario 6), respectively. Although R T, P c, Q w and S T were also identified as significant inputs for MLR models with the complete dataset (Table 1), as long as T w, T a, and C RNA were included, the MLR models can still reach a reasonable prediction with R2 above 0.44 (Scenario 9). However, the overall prediction performance of MLR models is limited likely due to the linear nature.
It is evident that both ANN and ANFIS greatly improved the accuracy in estimating COVID-19 prevalence, with complete and partial input scenarios (Table 2). In particular, superior performance of ANN was observed over ANFIS with all input scenarios, especially for partial inputs. ANN and ANFIS achieved adequate performance in 9/15 and 5/15 scenarios, with R2 around or higher than 0.8 (bolded scenarios in Table 2). As long as T a and C RNA were included, ANN achieved relatively high accuracy in estimating the prevalence, with R2 value higher than 0.77. Thus, ANN is highly recommended for future WBE estimation in lieu of the conventional WBE back-estimation equation, based on the improved accuracy and high robustness to the partial input scenarios, especially for the application incorporating data from different countries.
Nevertheless, the partial input compromised the performance of all data-driven models to some extent in all the scenarios (Table 2). In particular, with C RNA as the only input (indispensable for WBE) in scenario 16, the MLR model reached the worst performance with R2 at 0. Although ANN slightly improved the performance in comparison to MLR, the accuracy of prediction was still limited (R2 = 0.33). To achieve an accurate prediction, it is thus essential to collect as many input variables as possible.
3.5. Applications of data-driven models in different stages of COVID-19 outbreak
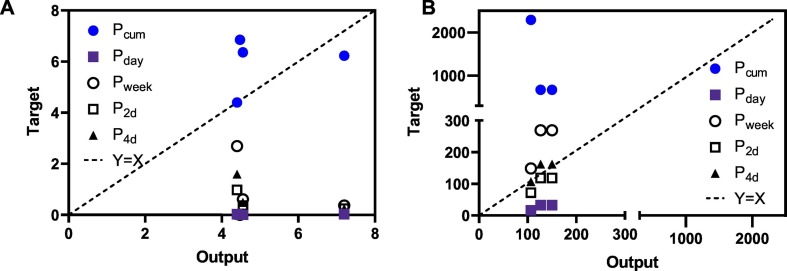
To further test the performance of data-driven models with unseen data, two studies that were not included in establishing the models were selected. Due to the limitation of other factors, the T a, S T and C RNA were collected from these studies and applied to relevant MLR, ANN and ANFIS models built in the above sections. Since ANN model showed better performance with partial inputs (Table 2), the outputs from ANN was plotted against the prevalence determined by cumulative cases (P cum), daily new cases (P day), weekly new cases (P week) and upcoming new cases in the following 2 or 4 days (P 2d, P 4d) for the outbreak at the initial pre-peak phase in Japan (Fig. 3A ) and post-peak stage in the USA (Fig. 3B).
Fig. 3.
Comparison of the output from ANN model and prevalence determined by cumulative cases (Pcum), daily new cases (Pday), weekly new cases (Pweek) and upcoming new cases in the following 2 or 4 days (P2d, P4d) for the initial (pre-peak) stage of an outbreak (A) and post-peak stage of an outbreak (B). Y = X line is where the y-axis value equals the x-axis value.
It is clear that the ANN model performance for the initial and post-peak phases of the outbreak is different. For the pre-peak phase, outputs from ANN model correlated best with P cum though some variations (within 4 people/100, 000 people) were observed (Fig. 3A). This is in line with the datasets used for the training of the ANN model, suggesting that for the initial phase of the outbreak, the ANN model can reasonably estimate the P cum with unseen data. In contrast, during the post-peak phase of the COVID-19 outbreak, outputs from the ANN model better represented P 2d and P 4d than others (Fig. 3B). This is likely related to the progression of an outbreak over time. In the initial phase, the confirmed and cumulative cases increase exponentially (Maier and Brockmann, 2020), which thereby can largely represent the number of patients excreting SARS-CoV-2. In contrast, in the post-peak phase, considering the recovery, hospitalization, and decease of the confirmed patients, the real-time prevalence of COVID-19 in the catchment area cannot be represented by P cum. Furthermore, with the implication of effective control and protective measures, the daily new cases gradually decreased to lower numbers after the peak and the cumulative cases become relatively stable with slow growth (Maier and Brockmann, 2020). This can be observed by the 1 to 2 orders of magnitude higher P cum than P day, P week, P 2d and P 4d (Fig. 3B). The better representative of model outputs for P 2d and P 4d is consistent with a recent report, where changes in C RNA foreshadowed the increase in positive tests by 2–4 days (Nemudryi et al., 2020). As currently most of the countries surpassed the initial phase, this forecasting of the COVID-19 community prevalence for the following 2–4 days provides a promising tool for the early warning of the COVID-19 resurgence (the so-called second wave) in a community. In addition, in comparison to MLR and ANFIS models, better performance was observed with ANN model (Fig. S5), which is consistent with the robustness test (Table 2).
4. Limitations and future research needs
Through the systematic literature search, seven currently available studies from five countries were included in this study. Nine explanatory factors covering the testing coverage, SARS-CoV-2 RNA concentration (recovery efficiency corrected or not), wastewater temperature, air temperature, population size, water usage pattern, precipitation and sampling technique were adapted. The meta-analysis revealed that these factors explained about 50% of the variations of the multi-national prevalence data based on redundancy analysis. Many other factors such as wastewater property, analytical uncertainties, and community socioeconomic factors such as (age, gender, income, etc.) were not included due to the unavailability of such kind of information. The impact of these factors on the prevalence estimation through WBE approach remains unknown and requires further investigations. In addition, the articles included in this study preserved samples under different temperatures (i.e. on ice, 4 °C, −20 °C or −80 °C) for several hours to a couple of days (Table S1). Recent studies indicate the SARS-CoV-2 RNA was rather stable at 4 °C for at least 14 days (Ahmed et al., 2020b; Chin et al., 2020), while a review stated that freezing and de-freezing the sample from −20 °C or −80 °C could potentially lead to degradation of the genetic material of SARS-CoV-2 (Alygizakis et al., 2021). To date, the impact of preservation strategies and time on the SARS-CoV-2 RNA concentration detected in wastewater remains unclear. Thus, the preservation conditions were not included in the models, of which the impact on the prevalence estimation requires further investigations.
Wastewater temperature and air temperature were found as significant factors for the prevalence estimation in all three types of data-driven models, which could be potentially related to the importance of the decay of SARS-CoV-2 RNA in wastewater. However, the wastewater temperature was not recorded in most WBE publications, and the data used in this study was estimated from a previous modeling approach. Thus, both wastewater temperature and air temperature were included in the models to accommodate different input scenarios of WBE studies. For future WBE studies, a comprehensive record of the analytical approach, RNA recovery efficiency, wastewater parameters (such as temperature, suspended solids, dissolved oxygen and biological oxygen demand), and socioeconomically factors are highly recommended to be included.
This is a proof-of-concept study for the application of data-driven models in multi-national or global COVID-19 prevalence estimation through WBE approach. Due to the limited size and availability of the explanatory factors of compiled data and the nature of data-driven models, ANN and ANFIS models in this study were built with the best performance under the current available dataset. In addition, the on-going vaccination progress would potentially lead to a lower or more focused testing coverage of the population, which may affect the accuracy of the established data-driven models. However, with future investigations and more detailed datasets, the ANN and ANFIS models can be improved progressively by training with accumulated WBE data, to reduce the uncertainties, and accommodate the vaccination progress for the prevalence prediction. The mortality rate due to COVID-19 is also critical for disease surveillance. Recent studies revealed that the prevalence of mortality were more closely related to the age group (> 65 years vs. < 65 years), gender, and the existing or historical disease conditions such as obesity, hypertension, diabetes, cardiovascular disease, and cancer, rather than the COVID-19 prevalence (Neil et al., 2020; Noor and Islam, 2020). Thus, the mortality rate was not considered as the output from the data-driven models established in this study using data from WBE studies. However, the data-driven approaches can be applied for future mortality rate prediction with the inclusion of relevant input parameters such as the socioeconomic parameters (i.e., age, gender, education level, etc.) and health condition (i.e., obesity, hypertension, diabetes, etc.).
5. Conclusions
This study systematically investigated the use of data-driven models as an efficient prediction tool for the COVID-19 community prevalence in lieu of the conventional WBE back-estimation approach. This leads to the following conclusions:
-
•
ANN and ANFIS are commendable candidate models for the estimation of COVID-19 community prevalence with high accuracy. In comparison, MLR is not recommended due to its limited prediction capability.
-
•
Although SARS-CoV-2 concentration in wastewater is indispensable for WBE, other relevant input parameters are also important to enhance the estimation accuracy. Especially, air temperature and wastewater temperature are critical parameters for the prevalence estimation.
-
•
ANN model showed strong robustness than the more complicated ANFIS when subject to partial data sets of input variables and unseen data sets.
-
•
The ANN model can reasonably estimate the prevalence of COVID-19 in the pre-peak phase of the outbreak and forecast the upcoming new cases in 2–4 days in the post-peak phase.
CRediT authorship contribution statement
Xuan Li: Conceptualization, Methodology, Data curation, Formal analysis, Writing – original draft. Jagadeeshkumar Kulandaivelu: Data curation, Writing – review & editing. Shuxin Zhang: Data curation. Jiahua Shi: Data curation, Writing – review & editing. Muttucumaru Sivakumar: Writing – review & editing. Jochen Mueller: Writing – review & editing. Stephen Luby: Writing – review & editing. Warish Ahmed: Writing – review & editing. Lachlan Coin: Writing – review & editing. Guangming Jiang: Conceptualization, Methodology, Writing – review & editing, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This research was supported by the Australian Research Council Discovery Project (DP190100385). Shuxin Zhang receives the support from a University of Wollongong PhD scholarship. Guangming Jiang was a recipient of the Australian Research Council DECRA Fellowship (DE170100694).
Editor: Damia Barcelo
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2021.147947.
Appendix A. Supplementary data
Supplementary material
References
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bibby K., Haramoto E., Hewitt J., Huygens F., Gyawali P., Korajkic A., Riddell S., Sherchan S.P., Simpson S.L., Sirikanchana K., Symonds E.M., Verhagen R., Vasan S.S., Kitajima M., Bivins A. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020;191 doi: 10.1016/j.envres.2020.110092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bivins A., Bibby K., Farkas K., Gathercole A., Haramoto E., Gyawali P., Korajkic A., McMinn B.R., Mueller J.F., Simpson S.L., Smith W.J.M., Symonds E.M., Thomas K.V., Verhagen R., Kitajima M. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alygizakis N., Markou A.N., Rousis N.I., Galani A., Avgeris M., Adamopoulos P.G., Scorilas A., Lianidou E.S., Paraskevis D., Tsiodras S., Tsakris A., Dimopoulos M.A., Thomaidis N.S. Analytical methodologies for the detection of SARS-CoV-2 in wastewater: protocols and future perspectives. Trends Anal. Chem. 2021;134 doi: 10.1016/j.trac.2020.116125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausati S., Amanollahi J. Assessing the accuracy of ANFIS, EEMD-GRNN, PCR, and MLR models in predicting PM2. 5. Atmos. Environ. 2016;142:465–474. [Google Scholar]
- Backer, J.A., Klinkenberg, D. and Wallinga, J. (2020) Incubation period of 2019 novel coronavirus (2019-nCoV) infections among travellers from Wuhan, China, 20–28 January 2020. Eurosurveillance 25(5), 2000062. [DOI] [PMC free article] [PubMed]
- Bivins A., Greaves J., Fischer R., Yinda K.C., Ahmed W., Kitajima M., Munster V.J., Bibby K. Persistence of SARS-CoV-2 in water and wastewater. Environ. Sci. Technol. Lett. 2020;7(12):937–942. doi: 10.1021/acs.estlett.0c00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavarria-Miró, G., Anfruns-Estrada, E., Guix, S., Paraira, M., Galofré, B., Sáanchez, G., Pintó, R. and Bosch, A. (2020) Sentinel surveillance of SARS-CoV-2 in wastewater anticipates the occurrence of COVID-19 cases. medRxiv.
- Chen C., Gao G., Xu Y., Pu L., Wang Q., Wang L., Wang W., Song Y., Chen M., Wang L., Yu F., Yang S., Tang Y., Zhao L., Wang H., Wang Y., Zeng H., Zhang F. SARS-CoV-2-positive sputum and feces after conversion of pharyngeal samples in patients with COVID-19. Ann. Intern. Med. 2020;172(12):832–834. doi: 10.7326/M20-0991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin A.W.H., Chu J.T.S., Perera M.R.A., Hui K.P.Y., Yen H.-L., Chan M.C.W., Peiris M., Poon L.L.M. Stability of SARS-CoV-2 in different environmental conditions. The Lancet Microbe. 2020;1(1) doi: 10.1016/S2666-5247(20)30003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmau M., Atanasova N., Gabarrón S., Rodriguez-Roda I., Comas J. Comparison of a deterministic and a data driven model to describe MBR fouling. Chem. Eng. J. 2015;260:300–308. [Google Scholar]
- Gerrity, D., Papp, K., Stoker, M., Sims, A. and Frehner, W. (2021) Early-pandemic wastewater surveillance of SARS-CoV-2 in Southern Nevada: methodology, occurrence, and incidence/prevalence considerations. Water Res. X 10, 100086. [DOI] [PMC free article] [PubMed]
- Haramoto E., Malla B., Thakali O., Kitajima M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 2020;737 doi: 10.1016/j.scitotenv.2020.140405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart O.E., Halden R.U. Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: feasibility, economy, opportunities and challenges. Sci. Total Environ. 2020;730 doi: 10.1016/j.scitotenv.2020.138875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart O.E., Halden R.U. Modeling wastewater temperature and attenuation of sewage-borne biomarkers globally. Water Res. 2020;172 doi: 10.1016/j.watres.2020.115473. [DOI] [PubMed] [Google Scholar]
- Hata A., Hara-Yamamura H., Meuchi Y., Imai S., Honda R. Detection of SARS-CoV-2 in wastewater in Japan during a COVID-19 outbreak. Sci. Total Environ. 2021;758 doi: 10.1016/j.scitotenv.2020.143578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Guo Y., Mao R., Zhang J. Proportion of asymptomatic coronavirus disease 2019: a systematic review and meta-analysis. J. Med. Virol. 2020;93(2):820–830. doi: 10.1002/jmv.26326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang G., Keating A., Corrie S., O’halloran K., Nguyen L., Yuan Z. Dosing free nitrous acid for sulfide control in sewers: results of field trials in Australia. Water Res. 2013;47(13):4331–4339. doi: 10.1016/j.watres.2013.05.024. [DOI] [PubMed] [Google Scholar]
- Jiang G., Keller J., Bond P.L., Yuan Z. Predicting concrete corrosion of sewers using artificial neural network. Water Res. 2016;92:52–60. doi: 10.1016/j.watres.2016.01.029. [DOI] [PubMed] [Google Scholar]
- Joynt G.M., Wu W.K.K. Understanding COVID-19: what does viral RNA load really mean? Lancet Infect. Dis. 2020;20(6):635–636. doi: 10.1016/S1473-3099(20)30237-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaboga D., Kaya E. Adaptive network based fuzzy inference system (ANFIS) training approaches: a comprehensive survey. Artif. Intell. Rev. 2019;52(4):2263–2293. [Google Scholar]
- Kenny E.D., Paredes R.S.C., de Lacerda L.A., Sica Y.C., de Souza G.P., Lázaris J. Artificial neural network corrosion modeling for metals in an equatorial climate. Corros. Sci. 2009;51(10):2266–2278. [Google Scholar]
- Khademi, F. and Behfarnia, K. (2016) Evaluation of concrete compressive strength using artificial neural network and multiple linear regression models.
- Kitamura K., Sadamasu K., Muramatsu M., Yoshida H. Efficient detection of SARS-CoV-2 RNA in the solid fraction of wastewater. Sci. Total Environ. 2020;763 doi: 10.1016/j.scitotenv.2020.144587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Khademi F., Liu Y., Akbari M., Wang C., Bond P.L., Keller J., Jiang G. Evaluation of data-driven models for predicting the service life of concrete sewer pipes subjected to corrosion. J. Environ. Manag. 2019;234:431–439. doi: 10.1016/j.jenvman.2018.12.098. [DOI] [PubMed] [Google Scholar]
- Li X., Zhang S., Shi J., Luby S.P., Jiang G. Uncertainties in estimating SARS-CoV-2 prevalence by wastewater-based epidemiology. Chem. Eng. J. 2021;415 doi: 10.1016/j.cej.2021.129039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q.-X., Tang X.-J., Shi Q.-L., Li Q., Deng H.-J., Yuan J., Hu J.-L., Xu W., Zhang Y., Lv F.-J., Su K., Zhang F., Gong J., Wu B., Liu X.-M., Li J.-J., Qiu J.-F., Chen J., Huang A.-L. Nature Medicine; 2020. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. [DOI] [PubMed] [Google Scholar]
- Maier B.F., Brockmann D. Effective containment explains subexponential growth in recent confirmed COVID-19 cases in China. Science. 2020;368(6492):742–746. doi: 10.1126/science.abb4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall A.-K., Palmitessa R., Blumensaat F., Morgenroth E., Ort C. Modeling in-sewer transformations at catchment scale–implications on drug consumption estimates in wastewater-based epidemiology. Water Res. 2017;122:655–668. doi: 10.1016/j.watres.2017.05.034. [DOI] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Environmental Science & Technology Letters; 2020. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. [DOI] [PubMed] [Google Scholar]
- Neil M., Fenton N., Osman M., McLachlan S. Bayesian network analysis of Covid-19 data reveals higher infection prevalence rates and lower fatality rates than widely reported. J. Risk Res. 2020;23(7–8):866–879. [Google Scholar]
- Nemudryi A., Nemudraia A., Wiegand T., Surya K., Buyukyoruk M., Cicha C., Vanderwood K.K., Wilkinson R., Wiedenheft B. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. Cell Rep. Med. 2020;1(6) doi: 10.1016/j.xcrm.2020.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor F.M., Islam M.M. Prevalence and associated risk factors of mortality among COVID-19 patients: a meta-analysis. J. Community Health. 2020;45(6):1270–1282. doi: 10.1007/s10900-020-00920-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020;20(4):411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.-k, Lee C.-W., Park D.-I., Woo H.-Y., Cheong H.S., Shin H.C., Ahn K., Kwon M.-J., Joo E.-J. Detection of SARS-CoV-2 in fecal samples from patients with asymptomatic and mild COVID-19 in Korea. Clin. Gastroenterol. Hepatol. 2020;S1542-3565(20) doi: 10.1016/j.cgh.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecson B.M., Darby E., Haas C., Amha Y., Bartolo M., Danielson R., Dearborn Y., Di Giovanni G., Ferguson C., Fevig S. Water Research & Technology; Environmental Science: 2021. Reproducibility and Sensitivity of 36 Methods to Quantify the SARS-CoV-2 Genetic Signal in Raw Wastewater: Findings From an Interlaboratory Methods Evaluation in the US. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petala M., Dafou D., Kostoglou M., Karapantsios T., Kanata E., Chatziefstathiou A., Sakaveli F., Kotoulas K., Arsenakis M., Roilides E., Sklaviadis T., Metallidis S., Papa A., Stylianidis E., Papadopoulos A., Papaioannou N. A physicochemical model for rationalizing SARS-CoV-2 concentration in sewage. Case study: the city of Thessaloniki in Greece. Sci. Total Environ. 2020;755(1) doi: 10.1016/j.scitotenv.2020.142855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Cuevas-Ferrando E., Sanjuán R., Domingo-Calap P., Sánchez G. Metropolitan wastewater analysis for COVID-19 epidemiological surveillance. Int. J. Hyg. Environ. Health. 2020;230 doi: 10.1016/j.ijheh.2020.113621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Şahin M., Kaya Y., Uyar M. Comparison of ANN and MLR models for estimating solar radiation in Turkey using NOAA/AVHRR data. Adv. Space Res. 2013;51(5):891–904. [Google Scholar]
- Sherchan S.P., Shahin S., Ward L.M., Tandukar S., Aw T.G., Schmitz B., Ahmed W., Kitajima M. First detection of SARS-CoV-2 RNA in wastewater in North America: a study in Louisiana, USA. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman A.I., Boehm A.B. Systematic review and meta-analysis of the persistence and disinfection of human coronaviruses and their viral surrogates in water and wastewater. Environ. Sci. Technol. Lett. 2020;7(8):544–553. doi: 10.1021/acs.estlett.0c00313. [DOI] [PubMed] [Google Scholar]
- Tang A., Tong Z.-D., Wang H.-L., Dai Y.-X., Li K.-F., Liu J.-N., Wu W.-J., Yuan C., Yu M.-L., Li P., Yan J.-B. Detection of novel coronavirus by RT-PCR in stool specimen from asymptomatic child, China. Emerg. Infect. Disease journal. 2020;26(6):1337. doi: 10.3201/eid2606.200301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorn A.S., Meijer B., Frampton C.M., Barclay M.L., de Boer N.K. Systematic review with meta-analysis: SARS-CoV-2 stool testing and the potential for faecal-oral transmission. Aliment. Pharmacol. Ther. 2020;52(8):1276–1288. doi: 10.1111/apt.16036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidhaas, J., Aanderud, Z., Roper, D., VanDerslice, J., Gaddis, E., Ostermiller, J., Hoffman, K., Jamal, R., Heck, P. and Zhang, Y. (2020) Correlation of SARS-CoV-2 RNA in wastewater with COVID-19 disease burden in sewersheds. [DOI] [PMC free article] [PubMed]
- Westhaus S., Weber F.-A., Schiwy S., Linnemann V., Brinkmann M., Widera M., Greve C., Janke A., Hollert H., Wintgens T. Detection of SARS-CoV-2 in raw and treated wastewater in Germany–suitability for COVID-19 surveillance and potential transmission risks. Sci. Total Environ. 2020;751 doi: 10.1016/j.scitotenv.2020.141750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2020) https://covid19.who.int/. WHO coronavirus disease (COVID-19) dashboard (ed).
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wu, F., Zhang, J., Xiao, A., Gu, X., Lee, W.L., Armas, F., Kauffman, K., Hanage, W., Matus, M., Ghaeli, N., Endo, N., Duvallet, C., Poyet, M., Moniz, K., Washburne, A.D., Erickson, T.B., Chai, P.R., Thompson, J. and Alm, E.J. (2020a) SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems 5(4), e00614–00620. [DOI] [PMC free article] [PubMed]
- Wu Y., Guo C., Tang L., Hong Z., Zhou J., Dong X., Yin H., Xiao Q., Tang Y., Qu X., Kuang L., Fang X., Mishra N., Lu J., Shan H., Jiang G., Huang X. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020;5(5):434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Wang S., Xue Y. Fecal specimen diagnosis 2019 novel coronavirus–infected pneumonia. J. Med. Virol. 2020;92(6):680–682. doi: 10.1002/jmv.25742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Du R.-H., Li B., Zheng X.-S., Yang X.-L., Hu B., Wang Y.-Y., Xiao G.-F., Yan B., Shi Z.-L., Zhou P. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg. Microbes Infect. 2020;9(1):386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material